Spinal Muscular Atrophy: Why do low levels of SMN make motor neurons sick? (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 1.
Published in final edited form as: Nat Rev Neurosci. 2009 Jul 8;10(8):597–609. doi: 10.1038/nrn2670
Abstract
Many neurogenetic disorders are caused by the mutation of ubiquitously expressed genes. Spinal muscular atrophy is one such disorder and is caused by loss or mutation of the survival motor neuron 1 gene (SMN1), leading to reduced SMN protein levels and a selective dysfunction of motor neurons. SMN, in collaboration with partner proteins, functions in the assembly of small nuclear ribonucleoproteins (snRNPs), which are important for pre-mRNA splicing. It has also been suggested that SMN might function in the assembly of other RNP complexes. Two hypotheses have been proposed to explain the molecular dysfunction that gives rise to SMA and its specificity to a particular group of neurons. The first hypothesis states that the loss of SMN’s well-known function in snRNP assembly causes an alteration in the splicing of a specific gene (or genes). A second hypothesis proposes that SMN is critical for the transport of mRNA in neurons and disruption of this function results in SMA.
Introduction
Spinal muscular atrophies (SMA) are characterized by the loss of lower motor neurons and atrophy of muscle.1 Proximal SMA (which we refer to as SMA) is a common genetic cause of infant death2 and the most frequent SMA.3–6. SMA is one of a number of neurological disorders associated with genes that play important roles in RNA metabolism (Supplementary Information S1 box). Although SMA is caused by reduced levels of a ubiquitously expressed protein, Survival Motor Neuron (SMN), it affects the lower motor neurons. A number of other neurogenetic disorders are caused by mutations in ubiquitously expressed genes, including amyotrophic lateral sclerosis (caused by mutations in Super Oxide Dismutase 1)7, Huntington’s disease (caused by an expanded CAG repeat in Huntingtin)8 and several others. 9–12 Thus, understanding how SMN deficiency causes SMA could give us insight into the more general question of how a mutation in a ubiquitously expressed gene causes a neurological disease.
Two hypotheses have arisen to explain SMA. Reduction of SMN is proposed to affect pre-mRNA splicing of certain genes giving rise to SMA, or to disrupt its function in axons, resulting in reduced levels of certain transcripts in the distal axon. Papers have recently appeared providing evidence for both hypotheses. Here we discuss the function of SMN, SMA genetics and insights gained from animal models of the disease. We consider the evidence for each theory, how the proposed changes might result in SMA, and how strong evidence for the mechanism of SMA might be found. We aim in this article to provide a summary of the current debate in the field, as well as provoking critical discussion of each of the proposed disease mechanisms, so that experiments may be designed to resolve how deficiency of SMN causes SMA.
SMN Function
SMN is a 38kD protein found in the cytoplasm and nucleus of all cells.13–17 An isoform of SMN comprising only the amino terminus (exons 1–3 and a part of intron 3) and found only in axons has also been reported.18 However, this isoform is unlikely to play a role in SMA as some SMA patients carry missense mutations in exon 6 that would not alter its coding sequence.19
SMN has been reported to bind to or associate with many proteins.20–23 However, it is difficult to determine which of these interactors form functional complexes with SMN. Four criteria are useful in discerning the most likely functional SMN complexes. First, reciprocal immunoprecipitation of the endogenous forms of the interacting proteins should be shown. Second, isolation of a complex containing SMN as well as the protein in question from cells or tissues should be possible. Third, co-localization of SMN and the protein in question should be demonstrated. This co-localization may be only partial, that is, each protein may be found in separate locations (in other words only some of the immunofluorescent ‘spots’ may show overlap)however, where it occurs the overlap should be complete (that is, the spots for each protein on top of each other, rather than partially overlapping) to be considered as evidence for interaction.24 Finally, the complex containing the SMN interacting protein should show a functional deficit on SMN reduction. At the present time only the SMN complex involved in snRNP assembly has met these criteria.
SMN in snRNP assembly
The best-characterized SMN complex is involved in snRNP assembly (Figure1).20,25–31 Certain snRNPs are critical for the recognition of splice sites and the catalytic removal of introns from pre-mRNA. SnRNPs are composed of a U snRNA (U1, U2, U4, U5, U11 or U12) and for those snRNPs involved in splicing a heptameric ring of Sm proteins.25,32 Although Sm proteins can self-associate onto snRNA in vitro,32 the SMN complex is required for this process in vivo.27,28 The SMN complex that performs this function in vertebrates consists of SMN, Gemins2–8, unr interacting protein (unrip) and ATP as the assembly reaction is ATP dependent (Figure 1).25–27,33–35 Evidence to date indicates that SMN in this complex exists as an oligomer. The data include SMN’s ability to self-oligomerize in vitro,36,37 the interaction of the products of tagged SMN constructs in transfected cells38,39 and the results of genetic complementation studies in vivo40 (see below). Proteomic studies of a functional ATP dependent SMN complex that determine the precise stoichiometry of the components should be performed to further confirm the oligomeric nature of SMN as well as the number of SMN monomers in the active SMN complex.
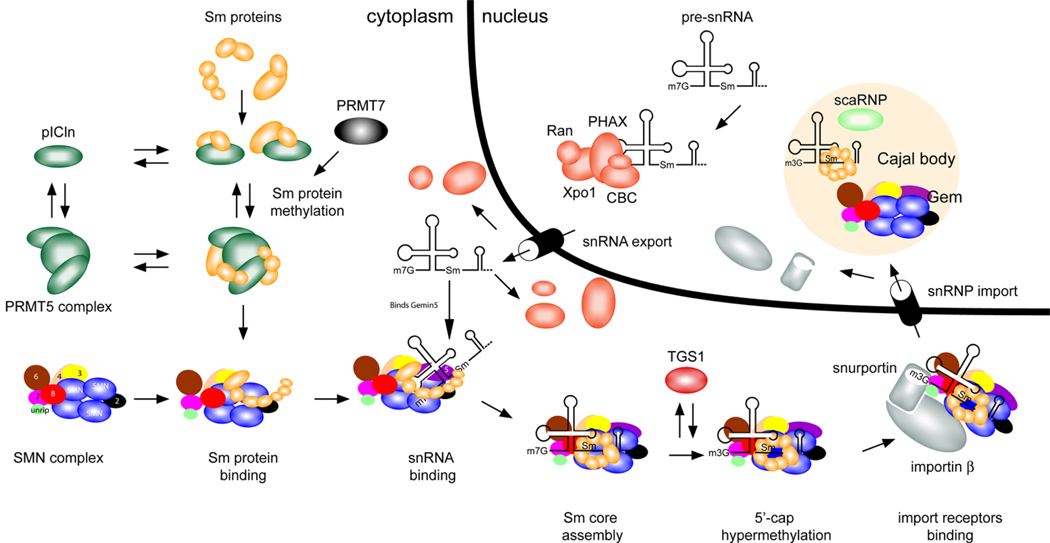
Figure 1. Function of SMN in snRNP assembly.
Small nuclear ribonucleoproteins (snRNPs) are active in recognizing and removing introns from pre-mRNA in the nucleus. Each snRNP particle is composed of small nuclear RNA (snRNA) of approximately 150 nucleotides, several Sm proteins and a number of specific proteins that are unique for each snRNP. Survival motor neuron (SMN) functions in the cytoplasm to assemble Sm proteins onto the snRNAs to produce an active snRNP particle.20, 25, 26, 32.
A) In the cytoplasm the 7 Sm proteins bind to the chloride conductance regulatory protein (pICln).42, 43 In vitro studies reveal that pICIn first binds the Sm proteins as two separate complexes: SmB, SmD3, and SmD1, SmD2.44 The latter subsequently binds SmE, SmF and SmG44 The protein arginine methyltrasferase (PRMT5 complex) and PRMT7 methylate the Sm proteins SmB, SmD1 and SmD3.42,43,156, 157 Sm proteins are released from pICln-PRMT5 complex and bind the SMN complex.
B1) The SMN complex is composed of SMN, Gemins2-8 and unrip. SMN is shown in the figure as an oligomer as it has been shown to self-associate and it has been suggested that oligomerization is critical for SMN function (see text). The exact numbers of SMN monomers in a SMN complex is unknown (it has been suggested to be an octomer).158 The Gemins are shown as single units for simplicity as the exact stoichiometry of the SMN complex has not been determined.
B2) snRNA is transcribed in the nucleus and then binds the export proteins phosphorylated adaptor for RNA export (PHAX), Cap-binding complex (CBC), exportin (Xpo1) and ras-related nuclear protein GTP (Ran), which transport it to the cytoplasm.25 In vertebrates, the snRNA is brought into the Sm protein-bound SMN complex by binding to Gemin5.46
C) The SMN complex places the Sm proteins onto the snRNA.25, 44 The m7G cap of the snRNA is hypermethylated by trimethylguanosine sythetase 1 (TGS)25, allowing the SMN complex with the snRNA to bind snurportin and importin, which mediates transport of the SMN complex with an assembled snRNP into the nucleus.25
D) In the nucleus the SMN complex and snRNPs localize to the Cajal body and snRNPs undergo further maturation.78 Depending on the cell type and developmental stage, SMN can localize as a separate body adjacent to the Cajal body.13–15
The figure is adapted from Pellizzoni.25
In snRNP assembly, Sm proteins first form a complex with the chloride conductance regulatory protein (pICIn) which is thought to prevent inappropriate binding of Sm proteins to RNA.22,25–30,41 pICln exists in a complex known as the PRMT5 complex (PRMT=protein arginine methyltransferase) or methylsome which can methylate Sm proteins and is capable of transferring Sm proteins to the SMN complex.41–44 (Figure1) The SMN complex then facilitates assembly of the Sm proteins onto the snRNA (Figure 1).22,25–30,41,45–47 Although the basic mechanisms of SnRNP assembly are well characterized, there is a number of outstanding issues on which there is not full agreement (Supplementary Information S2 box). The Tudor domain of SMN has been shown to bind arginine and glycine rich domains (RG and RGG domains) of various proteins such as the SmB, D1 and D3.48,49 Once the Sm proteins are assembled onto the snRNA the m7G-cap of the snRNA is hypermethylated to a m3G-cap and the whole SMN complex with snRNA is transported into the nucleus.50 In the nucleus snRNPs are further matured for pre-mRNA splicing but SMN’s role in these steps is poorly defined.
Does SMN assemble other RNPs?
In addition to SMN’s role in assembly of snRNPs, SMN has been called the master RNP assembler.51 However, for which RNPs SMN is important remains poorly defined. Many RNA-binding protein targets of SMN have RG and RGG domains. Of particular relevance are the Sm-like proteins (Lsm proteins),48,49,52–55 which share many features with the Sm proteins. Both Lsm and Sm proteins form heptameric ring structures and certain Lsm and Sm proteins contain RG domains which can bind the Tudor domain of SMN.48,49,52–55
Lsm10 and Lsm11 interact with Sm proteins to form a heptameric ring that binds the U7snRNAs.55 SMN is important for assembly of this ring on U7 RNA in vitro.55 Lsm2–8 proteins form a ring structure on the U6snRNA, a snRNA that is localized to the nucleus.52–54 U6 snRNA levels do not appear to be altered in tissue from mice with SMA.40,56 However specific assembly reactions to test SMN’s importance in Lsm assembly on U6snRNPs have not been reported.
The Lsm1–7 protein ring is found in the cytoplasm.52–54 The Lsm1–7 proteins have roles in mRNA decay57 and stabilization.58 Recently the Lsm1 and Lsm4 proteins have been shown to be present in dendrites and certain axons including spinal cord axons.59 This axonal/dentritic RNP complex was associated with RNAs transported in axons and contained SMN.59 Although SMN has yet to be shown to be important for Lsm ring assembly it is clear that both pICln60 and SMN can bind Lsm448,49 making it possible that the SMN complex is important for assembly of these rings on mRNA.
The Scd6 family is made up of conserved Lsm proteins that are associated with RNA granules. The C. elegans and Drosophila orthologues of Scd6 – CAR1 and Trailer Hitch respectively – also contain Sm like domains, form a 7-member ring that can bind RNA61 and have a role in RNA localization.61,62 The vertebrate orthologue of trailer hitch, Lsm14, assembles onto mRNAs and associates with decapping enzyme 1 (DCP1) and Me13B in distinct protein complexes that do not contain Lsm proteins 1–8 in non-neuronal cells .63,64,65 Lsm1 and 4 have been reported in an RNP complex containing SMN in neuronal processes,59 but whether this complex or other vertebrate axonal RNP complexs contain Lsm14 is not known. No assay to examine the effect of reduced SMN on assembly of these Lsm RNPs has been reported. However as Lsm proteins are important in RNA localization and contain domains that can bind SMN we suggest that SMN reduction might affect the assembly of RNP complexes that are critical for mRNA transport. Alternatively SMN has been hypothesized66–68 to function in a unique axonal complex with hnRNPQ/R66,67 and ZBPs69 to affect transport of β-actin message or other mRNAs yet to be identified.
Several other proteins have been reported to be able to interact with either SMN or the SMN complex.20,21,70,71 For example the proteins FMRP,72 hnRNPQ/R66,73 and profilin74 can bind SMN and thus could influence the function of SMN as an RNP assembler, although this has not yet been demonstrated.
SMN has also been suggested to be important for the assembly of snoRNPs 75,76, and knockdown of SMN alters the levels of U3snoRNA.77 SnoRNPs are small nuclear RNA protein complexes were the snoRNA acts as a guide for the modification (methylation or pseudouridylation) of ribosomal RNA, transfer RNA and snRNA.78 It is therefore necessary to determine which RNP assembly reactions are dependent on SMN and which are critical to the development of SMA.
Genetics of SMA
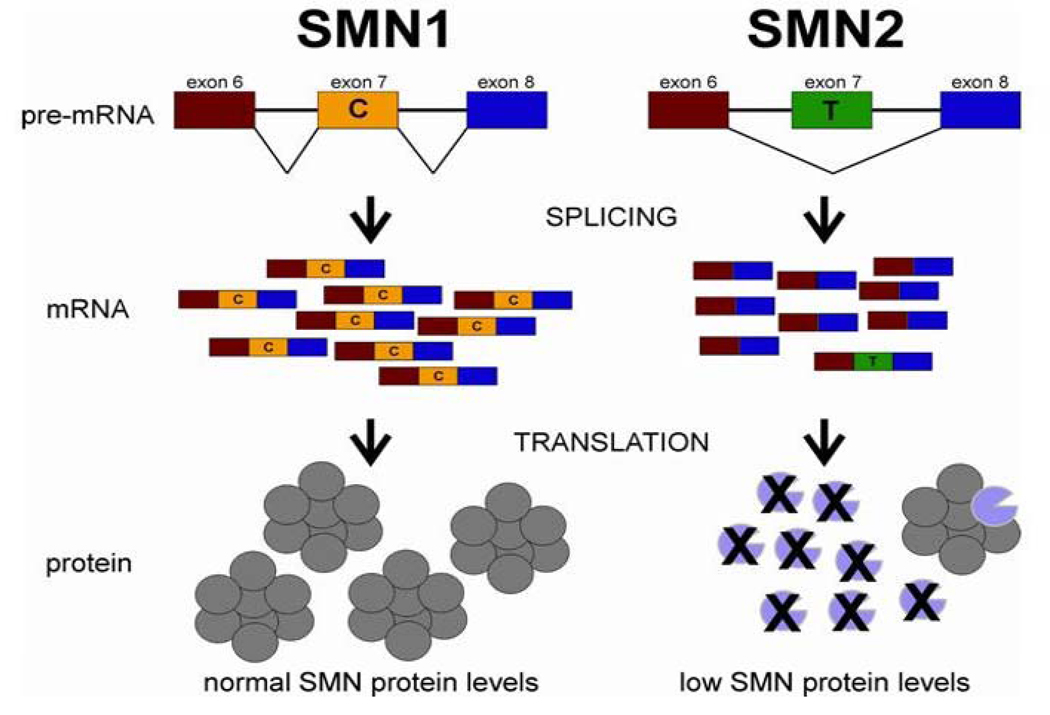
In humans, two genes, SMN1 and SMN2, encode SMN. SMA is an autosomal recessive disorder caused by loss or mutation of the SMN1 gene and retention of SMN26,79 (Figure 2). Chromosomes that lack both SMN1 and SMN2 can occur in SMA carriers but homozygous loss of both genes has not been reported, presumably as a result of lethality.79 Indeed, mice have a single Smn gene, the loss of which results in very early embryonic lethality.80 Loss of just SMN2 but not SMN1 does occur in the human population and has no consequence.79 The severity of SMA patients can be divided into five groups (0-IV) based on clinical presentation.81 The copy number of SMN2 is a major determinant of severity.5,6,79,82,83 SMN1 and SMN2 are 99.9% identical,84 have equivalent promoters,85,86 and express RNA6 and protein ubiquitously.87,88 The functional difference between the two genes is a C-T change in exon 7, which alters the amount of exon 7 that is incorporated in the final SMN transcript; most transcripts from SMN2 lack exon 7.6,84,89–92 The absence of exon 7 disrupts SMN’s ability to oligomerize, leading to an unstable protein that is rapidly degraded.36,37,93,94 In essence, a single SMN2 gene produces considerably less SMN protein than a SMN1 gene87,88 (Figure 2). Thus SMA is caused by low SMN protein levels rather than the complete loss of SMN.87,88
Figure 2A. SMN1 and SMN2 genes: structure and splicing.
The SMN1 and SMN2 have identical gene structure and are 99.9% identical at the sequence level.84 The essential difference between the two genes is a single nucleotide change in exon 7 (C or T as indicated). This single nucleotide change affects the splicing of the gene. Thus the majority of SMN transcripts from SMN2 lack exon 7 whereas those from SMN1 contain exon 7.6, 84, 89 –92, 121 However, because SMN2 does produce some full-length SMN it can be viewed as a gene with reduced function but not loss of function. The loss of amino acids that are encoded by exon 7 results in the production of SMN protein with severely decreased oligomerization efficiency and stability.36, 37, 93, 94 The SMN monomers are rapidly degraded.94 Thus, loss of SMN1 results in reduction of SMN levels in most tissues. The SMN oligomer is represented as an octomer based on gel filtration of SMN complexes formed in vitro158 Figure is taken from Butchbach and Burghes.159
In 95% of SMA cases the SMN1 gene is absent.6,79 However, five percent of SMA cases have small mutations, such as small deletions, splice mutants and missense mutations, in SMN1.95 These missense mutations can give us critical insight into the function of SMN that is disrupted in SMA. It is important to consider the context in which these missense mutations occur. SMN2 is always present and thus interactions between the mutant protein and wild-type SMN protein produced by SMN2 could occur. Table 1 summarizes the SMA missense mutations found in patients along with the activity of the resulting SMN molecules in various assays.
Table 1.
Properties of missense mutations in SMA patients.
| Mutation | Position of mutation | SMAtype | _SMN2_copynumber | Ability toRescuemouse orfish SMAmodels | Ability tobind SMN | Ability tobind Smproteins | Ability toAssemblesnRNP whenlow amounts ofnon mutantSMN arepresent | References | |
|---|---|---|---|---|---|---|---|---|---|
| Exon | Domain | ||||||||
| A2G | 1 | ND | III | 1 | High | (++) | (++) | (++) | 98, 103, 104 |
| D30N | 2a | Gemin2 | II | 2 | ND | (+++) | (+++) | (+++) | 47, 96 |
| D44V | 2a | Gemin2 | III | 1 | ND | (+++) | (+++) | (++) | 96,99 |
| W92S | 3 | Tudor | I | 3 | ND | ND | (+) | ND | 100 |
| V94G | 3 | Tudor | II | 3 | ND | ND | ND | ND | 101 |
| G95R | 3 | Tudor | III | 1 | ND | (+++) | (+++) | ND | 96 |
| A111G | 3 | Tudor | Ib | 2 | High | (+++) | (+++) | (+++)40, 47 | 40, 47 96,123 |
| I116F | 3 | Tudor | Ib | 2 | ND | ND | ND | (+) | 47, 102 |
| Y130C | 3 | Tudor | III | 2 | ND | ND | ND | ND | 163 |
| E134K | 3 | Tudor | I | 2 | ND | (+++) | (+)# | (+) | 47, 96, 97 |
| Q136E | 3 | Tudor | I | 1 | ND | ND | ND | (+) | 47, 102 |
| A188S | 4 | ND | I | ND | ND | ND | ND | ND | 164 |
| P245L | 6 | oligomer | III | ND | ND | ND | ND | ND | 165 |
| L260S | 6 | oligomer | II | 2 | ND | ND | ND | ND | 101 |
| S262G | 6 | oligomer | III | 1 | ND | ND | ND | ND | 96 |
| S262I | 6 | oligomer | III | 1 | ND | (++) | (++) | ND | 36, 166 |
| M263R | 6 | oligomer | I | 2 | ND | ND | ND | ND | 101 |
| M263T | 6 | oligomer | II | 1 | ND | ND | ND | ND | 95 |
| S266P | 6 | oligomer | II | 2 | ND | ND | ND | ND | 163 |
| Y272C* | 6 | oligomer | II | 2 | No | (+) | (+) | (+) | 36, 47, 123,166 |
| H273R | 6 | oligomer | II | ND | ND | ND | ND | ND | 163 |
| T274I | 6 | oligomer | III | 1 | ND | (++) | (++) | (+++) | 36, 47, 166 |
| G275S | 6 | oligomer | III | ND | ND | ND | ND | ND | 101 |
| G279C | 7 | oligomer | I | ND | ND | ND | ND | ND | 167 |
| G279V | 7 | oligomer | I | ND | No | (+) | (+) | (+) | 36, 47, 123,162 |
SMN can oligomerize with itself in vitro.36,37 Furthermore, when SMN tagged with different epitopes is transfected into cells, subsequent immunopreciptation reveals that it self-interacts.38,39 All mild mutations analyzed (those that occur in type II or III SMA when SMN2 copy number is limited, i.e. 1 or 2 copies) produce proteins that are able to associate with wild-type SMN to form oligomers.36,37 Other mutations, such as the loss of exon 7 and certain missense mutations in exon 6 (figure 2, table 1) severely disrupt SMN’s ability to oligomerize in vitro.36,37 All missense mutations that cause a major disruption of SMN’s ability to form oligomers can be classified as severe alleles as when they occur in patients with limited SMN2 copy number (1 or 2) they result in a severe phenotype.36,96 When SMN fails to oligomerize it is rapidly degraded and thus results in low SMN levels.94 Indeed it has been reported that SMN levels from a type I SMA patient with a mutation that disrupts oligomerization (Y272C), were similar to those of typical type I patients with complete loss of SMN187. A second group of severe SMA missense mutations disrupt the binding of the Sm proteins to SMN but are able to oligomerize (Figure2, table1).96–103 A number of reported mutations have been assayed for their ability to perform snRNP assembly in the presence of limiting amounts of wild-type SMN. All severe mutations assayed did not perform snRNP assembly whereas mild missense mutations did. 40,47,104 Thus, mild SMN missense mutations retain the ability to oligomerize with wild-type SMN and bind Sm proteins. Our understanding of how these mutated proteins function has been further clarified by genetic complementation studies (Box 1), demonstrating that SMN functions as an oligomer and mild missense mutation are not functional by themselves. Thus in vitro assays for SMN function of the missense alleles should be performed in the presence of limiting amounts of wild-type SMN.
Models of SMA and the requirement for SMN
SMN deficiency has been modeled in several organisms (Box 2).80,105–116 All species used to model SMA possess one Smn gene that is equivalent to SMN1. In all organisms, loss of SMN leads to lethality, however the exact time point of lethality is determined by the levels of maternal SMN (SMN derived from the mother’s SMN gene as opposed to the fetus). In mice lacking Smn there is minimal maternal SMN and death occurs early.80 However, in egg laying animals such as Zebrafish (unpublished observation) and Drosophila, death occurs later when maternal SMN levels drop below a critical point.111 As would be expected, loss of SMN from a specific tissue in conditional Smn mutants results in the death of that tissue.114,117,118
In transgenic mice the genetic situation that occurs in SMA has modeled by the introduction of SMN2 into mice lacking mouse Smn. When the SMN2 copy number is low (2) SMA mice develop whereas high SMN2 copy (8) number results in normal mice.107,108 High neuronal expression of SMN driven by the Prion promoter rescues motor neuron loss of SMA mice and extends survival, even though high SMN levels are not restored in all tissues.119 This suggests that two copies of SMN2 produce sufficient SMN for the normal function of most tissues, whereas motor neurons require higher levels of SMN, at least in mice.119 Further experiments to determine the precise temporal and spatial requirement for high SMN levels are required. In conditional mouse mutants, or in species in which maternal SMN prevents embryonic lethality, it is likely that SMN levels could mimic levels that occur in SMA and might in turn model some aspects of the disease. However it is difficult to distinguish these features from those that are due to SMN levels that fall below what is required for most cells. It is therefore advisable to confirm observations made in these systems in animals where reduced levels of wild-type SMN are found everywhere.
There often appears to be inconsistencies between results obtained in different model systems, but we urge caution in this interpretation. For example, in Drosophila substantial rescue of SMN deficient animals can be obtained by expressing SMN in muscle.111,120 In SMA mice, however, expression of SMN in muscle alone has little impact.119 An examination of the drivers and mutants used in each case reveals two differences. In Drosophila the muscle driver used is expressed early in dividing mesodermal tissue, whereas in mice the actin promoter used drives SMN expression in postmitotic myofibers, but not in satellite cells or other premitotic mesodermal tissues.111,119,120 Furthermore, in Drosophila functional SMN becomes depleted over time, whereas in SMA mice the human SMN2 transgene will continue to produce SMN. Thus, in SMN deficient Drosophila, the expression of SMN in a wide range of premitotic mesoderm has a major impact, but the effect of SMN expression in postmitotic muscle fibers is unknown. In SMA mice, expression of SMN in postmitotic muscle fibers has no phenotypic effect and the impact of expression in mesoderm is unknown.
Invertebrates and non-mammalian vertebrates can be used to carry out experiments that cannot be done efficiently in mammals. We would advocate exploiting the strengths of each species. Genetic screens in C. elegans and Drosophila can be used to identify suppressors and genetic interactors120, which can then be tested in both zebrafish and SMA mice. To date, the genes identified by suppressor and synthetic lethal screens in Drosophila120 (discussed below) have not been tested in vertebrate models. In zebrafish it is easy to test a large number of DNA constructs transiently, a select number of which can be followed up in transgenic mice or zebrafish. It is also advisable to confirm results obtained in vitro in cultured cells with in vivo systems.
In mice, a range of phenotypes can be obtained by titering the level of SMN or by introducing additional transgenes.39,107,108,114,117–121 The introduction of 8 copies of SMN2 completely rescues SMA mice, 2 copies of SMN2 results in SMA mice that die at 5 days,107,108 and one copy of SMN2 is embryonic lethal.109 Furthermore, introduction of a transgene that expresses SMN lacking exon 7 (SMNΔ7) in SMA mice gives mice that survive 14 days39 with specific motor defects122 and introduction of SMNA2G results in a mild SMA phenotype.98 It is clear that very small changes in SMN levels can have a major impact on phenotype.119
Understanding the cause of SMA
Two hypotheses have been put forward to explain the mechanism of SMA (Figure 3). The first suggests that the disrupted formation snRNPs affects the splicing of a select group of genes which are important for the motor neuron circuritry.20,25,104 The second suggests that SMN has a function in axons that is disrupted in SMA.20,25,67,104,113,123,124 It is also possible that the two hypothesis are linked, that is, reduced snRNP assembly could influence the splicing of a gene important for axons. Evidence for both hypotheses has been gathered, but there is still no clear indication of which is correct or whether both apply.
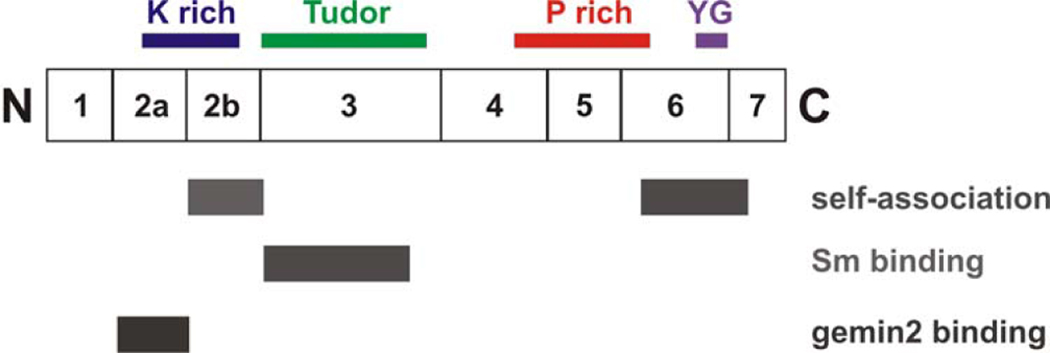
Figure 2B. Domains of SMN.
A diagram of SMN showing the exons and domains. Exon 2B encodes a domain important in binding Gemin2 as well as self-association.160 Both exon 2A and 2B are conserved. The K domain is rich in lysine, the Tudor domain is in exon 3 and has homology to other Tudor domains. The Tudor domain binds Sm proteins.97 Exon 5 and part of exon 6 contain a proline rich domain that may influence profilin binding.161 The C-terminal domain of exon 6 contains the conserved YG box and is important for self-association.160, 162
SMN in SnRNP assembly
According to the first hypothesis, SMN reduction causes an alteration in snRNP assembly, which in turn alters the amount or profile of snRNPs and thus causes an alteration in splicing. What are these alterations and how do they give rise to SMA? Assays to measure SMN’s ability to assemble Sm proteins onto snRNA in both cells from SMA patients and tissues from SMA mice have been developed.27,33,47,56,104,125–127 In mice, snRNP assembly levels change over development.126 In addition, spinal cord extracts from postnatal day 3 severe SMA mice have low snRNP assembly activity. Mild SMA mice have reduced snRNP assembly activity, when compared to normal or carrier mice, but considerably more activity than severe SMA mice.104 There is currently a complete correlation between the ability of an SMN protein complex to perform snRNP assembly and its ability to rescue SMA animals.40,104 However, the introduction of constructs that correct assembly function could affect both the assembly of Sm proteins onto snRNA and the assembly of other RNP complexes. It is therefore important to assay the formation of various RNP complexes to determine which are dependent on SMN. Moreover, snRNP assembly has not been assayed directly in motor neurons of SMA mice. Regardless, the ability of SMN to perform assembly of RNP complexes appears to be critical in SMA.
When SMN is decreased in zebrafish, motor axon defects occur.113 A study has shown that reintroduction of assembled snRNPs can rescue these defects,128 suggesting that snRNP assembly is the critical pathway affecting motor axons when SMN levels are reduced. However, as indicated by the authors, 50% of the zebrafish scored in the snRNP rescue assay were morphological defective (the study authors reported slightly delayed development and shortened and/or kinked body axis) and snRNPs also rescued these morphological defects (see supplemental table in reference 128). Thus, it is possible that the morphological defects give rise to abnormal motor axons due to the altered body muscles upon which these axons extend; thus, bringing into question the link between snRNPs and motor axon defects.113,129 Knockdown of Gemin2 and pICln were also reported to cause motor axon defects in zebrafish, again supporting a role snRNP assembly in the effects of SMN knock down.128 However, studies that knocked down both Smn and Gemin2 in individual motor neurons, thereby excluding morphological defects, demonstrated that knockdown of Gemin_2_ did not cause motor axon defects whereas knockdown of Smn did.113 To resolve if snRNPs are critical for motor axon defects it will be necessary to ask whether addition of snRNPs suppresses motor axon abnormalities when Smn is only knocked down in motor neurons as opposed to the whole organism. Then studies should be extended to genetic models of SMA in zebrafish and mice to ensure that rescue of other SMA phenotypes, including lethality, occurs.
The motor axonal phenotype observed in SMN-knockdown zebrafish can be corrected by reintroducing full-length SMN, but not mutant constructs or SMN lacking exon 7.123 It was reported that the SMN mutant SMNA111G, which can perform snRNP assembly40,47, was not capable of rescuing zebrafish axons, whereas a SMN which lacked exon 7 but had the additional amino acids VDQNQKE, and was predicted to be unable to perform snRNP assembly123, was able to do so.123 This was taken as evidence against this pathway being critical for axon function. However, we have subsequently obtained evidence that SMNA111G can correct SMA in mice and axon defects in zebrafish when expressed at high levels.40 Furthermore it has not been possible to assay snRNP assembly in fish extracts and it is not clear whether the VDQNQKE allele does or does not perform snRNP assembly in vivo.
An important question for the snRNP hypothesis in SMA is what is the consequence of reduced snRNP assembly? One would predict a uniform reduction in the levels of all of the SMN-assembled snRNPs. However, there appears to be uneven changes in the snRNP profiles of tissues in SMA mice.56,104 Most notable was a preferential reduction of minor snRNPs such as those containing U11 snRNA in the spinal cord of these mice.40,56,104 This would suggest that some of the approximately 700 genes that use the minor U12/U11-dependent splicing pathway would be disrupted.104,130,131 However, a clear demonstration of a SMN dependent downstream target that has a functional alteration in splicing has not been reported. Studies using both expression arrays and exon arrays have been undertaken to determine the effects of SMN reduction on downstream genes. Yet clear changes that can be convincingly related to SMA or even SMN levels directly as opposed to non-specific secondary changes due to the disease state of the mouse have not been obtained to date.56,132–134 For example, a recent study used exon arrays to define changes in SMA mice (SMN2+/+; Smn−/−, SMNΔ7+/+). These mice have an average life expectancy of 13 days and where examined at 11 days when the animals are rapidly declining in health. Many hundreds of genes were altered in 11 day old SMA mice in various tissues including spinal cord.56 However it is difficult to know which of these changes are due to low SMN levels as opposed to secondary changes due to the state of the mouse. Indeed, kidney cytochrome p450 (Cy27b1), a proteins known to respond to stress stimuli, had the largest fold change in expression in the SMA sample135 Furthermore, it is difficult to mechanistically relate these changes to an alteration in the minor splicing pathway. Although the list contains genes with minor introns, it also features numerous genes that lack them, in addition SMN itself is not listed in all tissues. It is thus hard to distinguish a splice or expression change due to stress, from changes due to SMN depletion. Given that there are a number of factors secondary to SMN reduction that might explain these splicing changes it is not possible to conclude that SMA is a pan-splicing disease rather than a disorder that affects splicing of specific targets important for motor neurons.
Overall, expression changes in SMA samples remain ill defined and the targets of reduced snRNPs have not been convincingly identified. Studies using arrays with exon junction probes136, and the use of younger animals as well as suitable controls will aid this approach. Furthermore, analysis of gene expression in motor neurons will be useful134 as well as the implementation of expression comparison using the new deep sequencing techniques. 137 Therefore we conclude that snRNP assembly is altered in SMA, correlates with phenotype severity, and leads to reduced levels of certain snRNPs. However what target genes are specifically affected remains unknown.
A role for SMN in axons
SMN has been found in neuronal axons and growth cones of cells in culture, where it is found in low abundance in a complex that does not contain Sm proteins.38,66,68,74,124 Although it is relatively straight forward to detect SMN in the axons of cultured cells, it has not yet been convincingly detected in either axons or synapses in vivo by light microscopy in vertebrates.138 In part this is due to the difficulty in distinguishing whether granules or spots lie in an axon as opposed to an adjacent cell. This might be resolved by the use of specific neural preparations and electron microscopy. To confirm the localization of SMN in axons, the loss or reduction of punctuate SMN staining in axons and growth cones should be observed when SMN is knocked down or in SMA animals.
A role for SMN in axons was strengthened by the finding that knockdown of SMN in zebrafish leads to specific motor neuron axonal defects, including truncated and/or, branched axons.113. However, as described above, it is unclear whether these defects result from the loss of snRNP assembly or some other function of SMN. Currently no mutant form of SMN that clearly lacks snRNP assembly activity can correct SMA in animals or humans. There is a complete correlation between reduced ability to perform assembly of Sm proteins onto snRNA and the severity of SMA.40,104 However, the reduced assembly of other RNP complexes, for example Lsm onto mRNA, could also correlate with severity of SMA and this has not been examined. It is therefore critical to both define the RNP complexes that SMN assembles and to determine whether they are disrupted in SMA.
Motor neurons cultured from SMA mice show a similar phenotype to zebrafish with reduced SMN, including short axons with small growth cones and reduced levels of β-actin mRNA transport to the growth cones.67 This results in an altered distribution of the Cav2.2 channel and corresponding electrophysiological alterations.139 An alteration of Ca channel distribution at the neuromuscular junction (NMJ) might influence neurotransmitter release and the development of active zones, as seen in mice lacking laminin β2.140,141 Interestingly SMA mice do show developmental abnormalities of the NMJ with reduced arborization.39,142–144 However, reduced folds at the NMJ do not necessarily indicate a severely impaired NMJ and do not account for the severe phenotype seen in SMA. As both Cav2.1 and Cav2.2 channels are important in the CNS synapses, it is currently not clear why motor neuron synapses and not CNS synapses are affected if alteration in Ca channels is critical to SMA. Patients with deficiency of laminin β2 do not demonstrate the classic fiber type grouping seen in type I SMA or the uniform atrophy seen in type 0 SMA and severe SMA mice.145,146 Furthermore in patients lacking laminin β2 there is clear dysfunction of the CNS.146 Thus it remains unclear whether SMA can be explained by an alteration of Ca channel distribution at the distal axon. Recently, it has been reported that in SMA mice with a relatively severe phenotype, there is reduced release of neurotransmiter at the NMJ, which could relate to altered Ca channel function at the NMJ or altered input to the motor neuron.144 Further studies in SMA mouse models and humans147 are required to resolve the specifics of what is altered at the NMJ.
In support of a critical role for β-actin in SMA, plastin3, a protein that stabilizes filamentous actin 148,149, has recently been reported to be a modifier of SMA.150 However, there were some inconsistencies. In one discordant family T-plastin was expressed at similar levels in both the affected and unaffected sibs with the male sib being more severely affected. Second, in male patients with 2 copies of SMN2 SMA was not modified by high plastin3 expression.5,151,152 The authors suggest that this discrepancy occurs because plastin3 is a sex-linked modifier with incomplete penetrance. However, it is difficult to eliminate the possibility that the expression of plastin3 is associated with, as opposed to the cause of, the modification. The possibility that a transcription factor activates or repress both plastin3 and a modifying gene, such as SMN2, in spinal cord cannot be ruled out.
SMN and plastin3 are reported to form a complex,150 however it is unclear whether SMN is associated with filamentous actin as the required assays have not been performed.148,149 How could plastin3 modify the SMA phenotype? The axonal defects that occur in cultured motor neurons from SMA mice or zerbrafish with knockdown of SMN, can be rescued by over-expression of plastin3.150 However, whether this is because plastin3 encourages axon growth or whether its effect is specific to SMA is unknown. Actin filaments are also important in the nucleus,153 and it will be important to consider whether snRNP assembly or splicing could be altered by plastin3 expression. It will be important to express plastin3 in mouse models of SMA and determine whether the lethality due to SMA is corrected.
In severe SMA mice, motor neurons grow at a normal rate and do not display axonal branching or misdirection.109 In severe SMA mice (_SMN2+/+, Smn_−/−) there are NMJs that are unoccupied by a motor neuron109 indicating denervation. In intermediate SMA mice (SMN2+/+; SMNΔ7+/+; Smn−/− that live for 14 days) both loss of occupation and partial occupation of synapses occurs.39,142,154 In addition, the motor neuron synapses that mature more rapidly during development are more vulnerable than those synapses that mature more slowly. 154 In addition the NMJ of SMA mice are generally immature with abnormal development and poor terminal arborization.142–144 In mild SMA mice (_SMN2+/+; Smn_−/−; SMNA2G) electrophysiology reveals functional deficits at the NMJ, including intermittent neurotransmission failures.142 In intermediate SMA mice (SMN2+/+; Smn−/−; SMNΔ7+/+) electrophysiological studies show a reduced quantal content release of synaptic vesicles.144 In contrast to mild SMA mice the intermediate SMA mice show facilitation during repetitive stimulation indicating a reduced probability of synaptic vesicle release144 which could correlate with the alteration of Ca channels reported in motors from SMA mice in culture. In the future it will be important to study Ca Channel disruption in vivo in the NMJ of SMA mice as well as consider alteration of inputs to the motor neuron as responsible for defective output of the motor neuron in SMA. In all studies examining NMJs, the abnormal accumulation of neurofilament in motor neurons was observed resulting in varicosities in motor neurons and in certain cases neurofilament accumulation within the NMJ.109,142,154 Interestingly, similar motor neuronal varicosities have been reported in SMA patients.155 In Drosophila, reduction of SMN results in lethality and a decreased number of boutons. The latter phenotype can be modulated by changing the BMP signaling pathway.120
In conclusion, there is evidence for a role for SMN in the axon, but conclusive evidence that it is the axonal SMN complex that is critically disrupted in SMA is still lacking. Certain critical elements are not known. What is the function of SMN that is directly affected in SMA axons and can it be assayed? Or does disruption of snRNP assembly alter the splicing of a gene that is critical in these axon/NMJ functions? In the case of an axonal function does reduction of SMN directly affect the assembly of complexes onto mRNA leading to decreased mRNA transport to the distal axon? If so, which mRNAs besides β-actin are affected and can this be detected in vivo? Lastly, how might this disruption of RNA transport disrupt the lower motor neuron so specifically when SMN is found in the axons of all neurons? In this regards it is important to assay both in vitro and in vivo the affect of reduced SMN on proposed pathways.
Conclusion
It is currently unclear how reduced SMN levels cause SMA. This issue may be resolved by combining biochemical and genetic approaches. In vitro assembly assays of SMN missense mutations, both in homomeric and heteromeric complexes, combined with allelic complementation (Box 1) can be used to discern which SMN function is critically affected in SMA. Suppression or modification of the SMA phenotype can give invaluable insight into which process is critical in SMA. A directed suppression approach in which a specific RNP assembly reaction, such as snRNP assembly, is restored in SMA mice resulting in correction would give strong evidence for this pathway. However, it is difficult to identify such a suppressor that can be studied in a relatively straight forward manner in mice. In Drosophila and C. elegans non-directed suppressor and synthetic lethal screens can be performed. These screens ask what can suppress or enhance a specific phenotype without prior assumptions about a particular pathway. Such a screen in Drosophila has indicated the bone morphogenic protein BMP signaling pathway as important in correction of the button phenotype but it is unclear whether this is a informational suppressor or relatively specific to suppression of SMA phenotypes.120 Additional screens, in particular using knockdown as opposed to more severe mutants, should provide further insight into SMN dependent pathways. Desirable features of the suppressor include strong suppression, suppression of more than one SMN linked phenotype, elimination of informational suppressors, and more than one suppressor in a pathway. Furthermore, the ability to translate suppression into a vertebrate model will be important. However useful information can be obtained without fulfilling all these criteria. The identification of plastin3 as a potential modifier of SMA is of great interest and experiments to demonstrate that plastin3 does suppress the SMA phenotype in mice are critical. If plastin3 suppresses SMA in mice then it will be important to determine whether suppression is acting in the distal axon or via the nucleus.
SnRNP assembly is clearly altered in SMA, but are other SMN dependent assembly reactions affected? Importantly, what dictates the motor neuron specificity of this disease or how motor neuron specific is the disease? Detailed expression studies using arrays or sequencing technologies are clearly required, but it will be critical to distinguish a direct target from an indirect target. Presumably in the case of snRNPs, there would be different degrees of splicing dysfunction in mice of different severity. It is tempting to speculate that reduced SMN affects either the splicing or axonal transport of a limited number of target genes that are specific to motor neurons. Indeed altered splicing of a gene could in turn give rise to altered transport of mRNA. Restoration of expression of this defective gene(s) would be predicted to suppress the SMA phenotype, thus confirming a target gene. Answers to these questions may also provide insights into other disorders, such as ALS. We believe a careful use of genetics and biochemistry can dissect the immediate molecular targets of SMN deficiency and thus give information on the critical pathway in SMA.
TEXT BOX 1 Allelic complementation of SMN missense mutations.
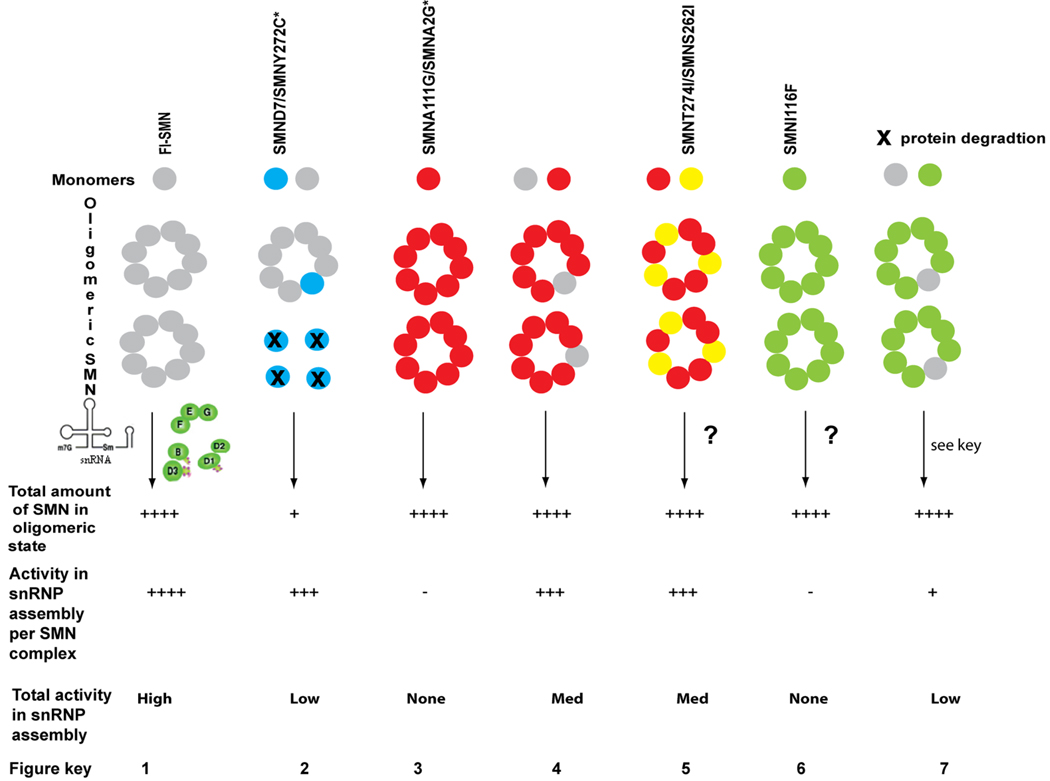
Allelic complementation occurs when the same protein, encoded by two different alleles, oligomerizes to form a functional unit with greater activity than homomers of proteins from either allele. 168,169 This is directly relevant to the analysis of SMA mild missense mutations.
Transgenic mice lacking Smn, but expressing 2 copies of human SMN2 (SMA mice) 107,108 exhibit very low snRNP assembly levels in spinal cord tissue and have severe SMA, dying at postnatal day 5 (figure, part 2).104,108 Expression of SMNA2G98 or SMNA111G40 missense alleles in this background (figure, part 4) increases their survival and increases snRNP assembly activity.40,98,104 However, neither allele expressed alone (without SMN2) can rescue Smn−/− mice (figure, part 3).40,98
The expression of one copy of SMN2 alone cannot rescue embryonic lethality in Smn−/− mice109; however the additional; expression of one SMNA111G allele can. Thus, the wild-type SMN produced by SMN2 interacts with SMNA111G to give greater total function than the addition of a SMN2 gene (SMA mice contain two copies of SMN2 and die at 5days). This indicates that two SMN proteins form a heteromer with substantial function.
Although unable to function in vivo by themselves, in the presence of limiting amounts of SMN, all mild SMA missense alleles assayed to date (figure, parts 2, 4 and 5) have substantial snRNP activity. This activity suggests that when small amounts of wild-type SMN from SMN2 are added to mild mutant SMN, the resulting heteromers are functional.40 High levels of expression of SMNA111G increases snRNP assembly in Smn−/− mice containing one copy of SMN2 to a much greater extent than an additional SMN2 copy, suggesting that the SMN complex is a heteromer of multiple subunits (an oligomer).
Severe alleles such as SMNI116F can interact with wild-type SMN but, when assayed in culture, SMNI116F does not complement low levels of wild-type SMN to result in increased snRNP assembly (figure, part 7).47 Indeed, as they do not have a dominant negative affect in carrier patients these alleles would be predicted to interact with SMN from SMN2 resulting in no net increase or decrease in total activity.
Could two missense mutations interact to produce a functional SMN oligomer? Although this situation would not arise in patients, this experiment can provide insights into the contribution of different protein domains to SMN function. Clearly, SMNA2G and SMNA111G do not complement each other.40 However, it would be interesting to determine whether these N-terminal mutations can complement mild C-terminal mutations, which reduce, but do not eliminate, the ability to bind full-length SMN (this possibility is illustrated in part 5).36,96
The ability to assay SMN’s snRNP assembly activity in vitro44 will allow analysis of both homomer and heteromer activity of SMN missense mutations. This activity can then be correlated with complementation analysis in vivo to understand both the in vitro and in vivo properties affected by SMN missense mutations. In all cases knowledge of the stability and level of mutant proteins in patient derived cell lines is useful in directing future studies and interpretation of results. The figure below shows the outcome of experiments to date on allelic complementation of SMN missense alleles. The amount of SMN or activity in assembly is defined by none −, low +, moderate +++ and high ++++. A question mark (?) indicates allele has not been experimentally tested.
Supplementary Material
Supplement 1
Supplement 2
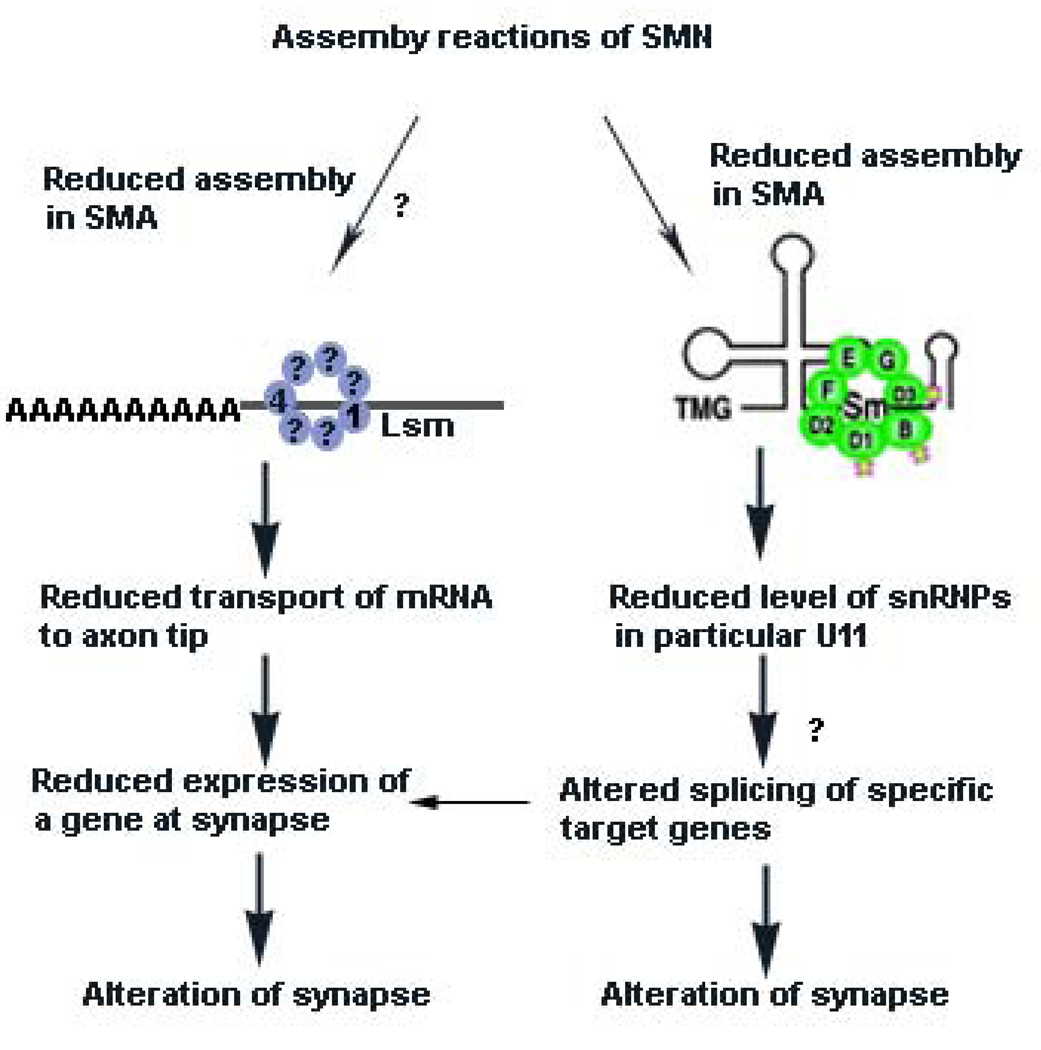
Figure 3. Mechanisms proposed to explain how reduced SMN levels cause SMA.
According to one hypothesis, reduced SMN levels result in reduced assembly of Sm proteins onto snRNA. This unevenly alters the levels of specific endogenous snRNPs, such as those used to splice minor introns (particularly U11) from pre-mRNA.40, 56, 104 It remains to be determined what the downstream target genes of the affected snRNPs are and how this specifically affects motor neuron function (indicated by a question mark (?)). One possibility is that the critical target gene is specific to motor neuron system. Alternatively, a function of critical importance to motor neurons could be disrupted.
In addition, it has been suggested that reduced levels of β-actin mRNA or other mRNA occur at the axon tip or synapse due to SMN having a function in axon RNA transport66, 67, 68 at the growth cones of motor neurons cultured from SMA mice. It has been proposed that hRNPQ/R66, 67 and ZBP69 participate with SMN in this complex and that the reduced β-actin transport leads to alteration of calcium channel distribution at the axon terminal139 which in turn could affect neurotransmitter release (see text). Lsm proteins 1 and 4 have been found in axons in an RNP complex.59 We suggest that it is possible that reduced SMN levels affect the assembly of Lsm proteins required for axonal transport of mRNA, leading to reduced expression of specific genes at the synapse. However, a functional biochemical assay linking reduced SMN levels to an alteration in the formation of the required complex for transport of mRNA is lacking (indicated by ?). Whether other Lsm proteins, such as Lsm14, associate with this complex in neurons is not known.
We have not indicated other potential or known SMN dependent assembly pathways, such as assembly of U7 snRNA, as it is not clear how alteration of this pathway would give rise to SMA. However, we cannot eliminate the possibility that other RNP assembly reactions are affected by reduced SMN levels. Lastly, it is possible to unite the two hypotheses where reduced snRNP assembly causes reduced splicing of a target gene that is critical for transport of mRNA to the motor neuron synapse.
Figure for Text box1.
Table 2.
SMN mutant organisms
| Organism | Mutation | Phenotype | References |
|---|---|---|---|
| S. pombe | Loss of smn by genedisruption | Lethal | 105, 106, 115 |
| C.elegans | RNAi knockdown andknockout of smn | Embryonic lethality anddevelopmental defects,movement defects(with later knockdownof SMN) | 110, 170 |
| Drosophila | Smn73Ao and SmnB(G202S and S201Frespectively).Mutations disrupt SMN self-association and canbe consideredequivalent to nullalleles | Larval lethal orembryonic lethal ifmaternal SMNremoved | 111 |
| SmnE33 Loss of SMNin adult thorax | Lose ability to fly andjump | 171 | |
| siRNA knockdown ofSMN | Lethality at differentstages depending onknockdown. | 120 | |
| Zebrafish | Antisense morpholinoknockdown of _Smn_resulting in reducedSmn early butrestoration of Smnlevels later | Abnormal axonpatterning and death. | 113 |
| Mouse | Loss of Smn by genedisruption of mouse_Smn_ gene | Embryonic lethal | 80 |
| Conditional (Cre-driven) loss of Smn inspecific tissues atspecific times | Death of cells in thetissue where Smn isremoved | 114, 117,118 | |
| Reduced SMN levelsthrough loss of mouse_Smn_ and introductionof SMN2 transgene. | Depends on _SMN2_copy number; 8 _SMN2_copies normal, 2copies SMN2 death at5 days. Phenotypevaries dependent onthe deletionbackground or thepresence of other SMNtransgenes | 107, 108 | |
| Human | Loss of SMN1 andretention of SMN2 | Phenotype isdependent on the copynumber of SMN2 and,more specifically, theamount of functionalSMN complex | 6 |
Acknowledgements
We would like to thank all members of the SMA community for many helpful discussions. Due to limited space we have not quoted all literature in the field and apologize to those whose articles are not referenced. We would like to thank Dr. Livio Pellizzoni for many helpful comments on the manuscript and Dr Matthew Butchbach for figure 2. We would like to thank all members of our laboratories for many useful discussions in particular Dr. Vicki McGovern who has read and edited many portions of this article in various formats. The work in our laboratories has been supported by NINDS, Families of SMA, Fight SMA, SMA Foundation, the Madison, Matthew, Preston and Cade & Katelyn funds and the SMA Angels (Savannah).
Glossary Terms
Splicing
The removal of introns from pre-mRNA to obtain mRNA
Autosomal recessive disorder
The mutation to cause the disorder lies within a gene that is on an autosomal (not sex) chromosome and both copies of the gene need to be mutated for the disorder to occur
Missense mutations
A mutation that results in the substitution of an amino acid in a protein
UsnRNA
Small nuclear RNAs which are rich in uridine. The U1, U2, U4, U5 and U6 RNA in a complex with certain proteins form the major splicesome complex important for removal of major introns. The U11, U12, U4atac, U5, and U6atac RNA in a complex with certain proteins form the minor splicesome complex important for removal of minor introns
Sm/Lsm proteins
Sm proteins were first found as antigens in a patient with systemic lupus erythematosus and named after the patient. Lsm refers to like Sm proteins. Sm/Lsm are a group of RNA binding proteins that have a common three dimensional stucture and form a heptameric or hexameric ring strucutre
Complementation
Refers to a relationship between two different mutations that in the homozygous recessive state produce the same phenotype. The ability of two mutant alleles to interact with each other to produce a normal phenotype is complementation. In general this occurs when the mutations that give rise to the phenotype occur in two different genes. However allelic complementation can arise when different mutations in the same gene can interact with each other to produce a functional complex. This often occurs in oligomeric proteins
Tudor domain
A conserved 50 amino acid stretch originally found in the Tudor protein of Drosophila and often found in proteins with known roles in RNA metabolism
RNA granules
Cytoplasmic deposits that contain RNA. These can also be found in axons and dendrites. RNA in these granules may be transported to the distal axon for localized protein translation in the growth cone
P bodies
P-bodies are cytoplasmic domains that contain proteins involved in diverse post-transcriptional processes such as mRNA degradation, nonsense-mediated mRNA decay, translational repression, and RNA-mediated gene silencing
Active zones
A portion of the presynaptic membrane that faces the postsynaptic density across the synaptic cleft. It constitutes the site of synaptic vesicle clustering, docking and neurotransmitter release
Informational suppressor
A suppressor that will modify in a non-gene specific manner. In the case discussed here over expression of plastin3 could encourage axon growth thus rescuing the SMN deficient growth phenotype. If the growth defect was caused by a mutation in another gene in a different pathway over expression of plastin3 could also rescue that phenotype
References
- 1.Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 2.Roberts DF, Chavez J, Court SD. The genetic component in child mortality. Arch Dis Child. 1970;45:33–38. doi: 10.1136/adc.45.239.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearn J. Incidence, prevalence, and gene frequency studies of chronic childhood spinal muscular atrophy. J Med Genet. 1978;15:409–413. doi: 10.1136/jmg.15.6.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melki J. Spinal muscular atrophy. Curr Opin Neurol. 1997;10:381–385. doi: 10.1097/00019052-199710000-00005. [DOI] [PubMed] [Google Scholar]
- 5.McAndrew PE, et al. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465.First copy analysis of SMN1 and SMN2 and clear indication that copy number of SMN2 modifies SMA phenotype
- 6.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3.Identification of survival motor neuron gene as the Spinal muscular atrophy gene and the similarity of SMN1 and SMN2
- 7.Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 8.Walling HW, Baldassare JJ, Westfall TC. Molecular aspects of Huntington’s disease. J Neurosci Res. 1998;54:301–308. doi: 10.1002/(SICI)1097-4547(19981101)54:3<301::AID-JNR1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 9.Park SG, Schimmel P, Kim S. Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci U S A. 2008;105:11043–11049. doi: 10.1073/pnas.0802862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salinas S, Carazo-Salas RE, Proukakis C, Schiavo G, Warner TT. Spastin and microtubules: Functions in health and disease. J Neurosci Res. 2007;85:2778–2782. doi: 10.1002/jnr.21238. [DOI] [PubMed] [Google Scholar]
- 11.Fink JK, Rainier S. Hereditary spastic paraplegia: spastin phenotype and function. Arch Neurol. 2004;61:830–833. doi: 10.1001/archneur.61.6.830. [DOI] [PubMed] [Google Scholar]
- 12.Thomas B, Beal MF. Parkinson's disease. Hum Mol Genet. 2007;16:R183–R194. doi: 10.1093/hmg/ddm159. Spec No. 2. [DOI] [PubMed] [Google Scholar]
- 13.Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. Embo J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 14.Young PJ, et al. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res. 2001;265:252–261. doi: 10.1006/excr.2001.5186. [DOI] [PubMed] [Google Scholar]
- 15.Young PJ, Le TT, thi Man N, Burghes AH, Morris GE. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- 16.Carvalho T, et al. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol. 1999;147:715–728. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matera AG, Frey MR. Coiled bodies and gems: Janus or gemini? Am J Hum Genet. 1998;63:317–321. doi: 10.1086/301992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Setola V, et al. Axonal-SMN (a-SMN), a protein isoform of the survival motor neuron gene, is specifically involved in axonogenesis. Proc Natl Acad Sci U S A. 2007;104:1959–1964. doi: 10.1073/pnas.0610660104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burghes HM. Other forms of survival motor neuron protein and spinal muscular atrophy: an opinion. Neuromuscul Disord. 2008;18:82–83. doi: 10.1016/j.nmd.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Eggert C, Chari A, Laggerbauer B, Fischer U. Spinal muscular atrophy: the RNP connection. Trends Mol Med. 2006;12:113–121. doi: 10.1016/j.molmed.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 21.Simic G. Pathogenesis of proximal autosomal recessive spinal muscular atrophy. Acta Neuropathol. 2008;116:223–234. doi: 10.1007/s00401-008-0411-1. [DOI] [PubMed] [Google Scholar]
- 22.Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol. 2002;12:472–478. doi: 10.1016/s0962-8924(02)02371-1. [DOI] [PubMed] [Google Scholar]
- 23.Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol. 2002;14:305–312. doi: 10.1016/s0955-0674(02)00332-0. [DOI] [PubMed] [Google Scholar]
- 24.Hao le T, et al. Absence of gemin5 from SMN complexes in nuclear Cajal bodies. BMC Cell Biol. 2007;8:28. doi: 10.1186/1471-2121-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gubitz AK, Feng W, Dreyfuss G. The SMN complex. Exp Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 27.Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962.Clear demonstration of the function of SMN as an assembler of Sm proteins onto snRNA
- 28.Meister G, Buhler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945.Clear demonstration of the function of SMN acting as an assembler of Sm proteins on to snRNA
- 29.Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2.Disruption of SMN by antibodies alters snRNP biogensis
- 30.Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell. 1997;90:1013–1021. doi: 10.1016/s0092-8674(00)80367-0.Identification of SMN interacting with Sm proteins indicating a role for SMN in snRNP assembly. (along with reference 29)
- 31.Ogawa C, et al. Role of survival motor neuron complex components in small nuclear ribonucleoprotein assembly. J Biol Chem. 2009:19321448. doi: 10.1074/jbc.M809031200. PMID. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raker VA, Hartmuth K, Kastner B, Luhrmann R. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol Cell Biol. 1999;19:6554–6565. doi: 10.1128/mcb.19.10.6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meister G, et al. Characterization of a nuclear 20S complex containing the survival of motor neurons (SMN) protein and a specific subset of spliceosomal Sm proteins. Hum Mol Genet. 2000;9:1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- 34.Otter S, et al. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J Biol Chem. 2007;282:5825–5833. doi: 10.1074/jbc.M608528200. [DOI] [PubMed] [Google Scholar]
- 35.Carissimi C, Saieva L, Gabanella F, Pellizzoni L. Gemin8 is required for the architecture and function of the survival motor neuron complex. J Biol Chem. 2006;281:37009–37016. doi: 10.1074/jbc.M607505200. [DOI] [PubMed] [Google Scholar]
- 36.Lorson CL, et al. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19:63–66. doi: 10.1038/ng0598-63.Demonstration that missense mutations in SMN affect ability of SMN to oligomerize with itself
- 37.Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci U S A. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167.Demonstration that SMN needs to be oligomeric to efficiently bind Sm proteins
- 38.Zhang HL, et al. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le TT, et al. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 40.Workman E, et al. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice Hum Mol Genet 200818PMID19329542.Clear indication of mild SMN missense mutations alleles complementing SMN2
- 41.Pu WT, Krapivinsky GB, Krapivinsky L, Clapham DE. pICln inhibits snRNP biogenesis by binding core spliceosomal proteins. Mol Cell Biol. 1999;19:4113–4120. doi: 10.1128/mcb.19.6.4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meister G, et al. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 43.Friesen WJ, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chari A, et al. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 45.Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. Embo J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Battle DJ, et al. The Gemin5 protein of the SMN complex identifies snRNAs. Mol Cell. 2006;23:273–279. doi: 10.1016/j.molcel.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 47.Shpargel KB, Matera AG. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102.First assay of the ability of a panel of SMN missense alleles to perform snRNP assembly
- 48.Friesen WJ, Dreyfuss G. Specific sequences of the Sm and Sm-like (Lsm) proteins mediate their interaction with the spinal muscular atrophy disease gene product (SMN) J Biol Chem. 2000;275:26370–26375. doi: 10.1074/jbc.M003299200. [DOI] [PubMed] [Google Scholar]
- 49.Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. Rna. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum Mol Genet. 2002;11:1785–1795. doi: 10.1093/hmg/11.15.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Terns MP, Terns RM. Macromolecular complexes: SMN--the master assembler. Curr Biol. 2001;11:R862–R864. doi: 10.1016/s0960-9822(01)00517-6. [DOI] [PubMed] [Google Scholar]
- 52.Khusial P, Plaag R, Zieve GW. LSm proteins form heptameric rings that bind to RNA via repeating motifs. Trends Biochem Sci. 2005;30:522–528. doi: 10.1016/j.tibs.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 53.He W, Parker R. Functions of Lsm proteins in mRNA degradation and splicing. Curr Opin Cell Biol. 2000;12:346–350. doi: 10.1016/s0955-0674(00)00098-3. [DOI] [PubMed] [Google Scholar]
- 54.Kiss T. Biogenesis of small nuclear RNPs. J Cell Sci. 2004;117:5949–5951. doi: 10.1242/jcs.01487. [DOI] [PubMed] [Google Scholar]
- 55.Pillai RS, et al. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003;17:2321–2333. doi: 10.1101/gad.274403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031.Along with reference 104 demonstrate that reduced SMN that occurs in SMA leads to reduced assembly of snRNPs which in turn reduces the steady state level of certain snRNPs which could selectively alter splicing
- 57.Tharun S, Muhlrad D, Chowdhury A, Parker R. Mutations in the Saccharomyces cerevisiae LSM1 gene that affect mRNA decapping and 3' end protection. Genetics. 2005;170:33–46. doi: 10.1534/genetics.104.034322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bergman N, et al. Lsm proteins bind and stabilize RNAs containing 5' poly(A) tracts. Nat Struct Mol Biol. 2007;14:824–831. doi: 10.1038/nsmb1287. [DOI] [PubMed] [Google Scholar]
- 59.di Penta A, et al. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J Cell Biol. 2009;184:423–435. doi: 10.1083/jcb.200807033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gandini R, et al. LSm4 associates with the plasma membrane and acts as a co-factor in cell volume regulation. Cell Physiol Biochem. 2008;22:579–590. doi: 10.1159/000185542. [DOI] [PubMed] [Google Scholar]
- 61.Decker CJ, Parker R. CAR-1 and trailer hitch: driving mRNP granule function at the ER? J Cell Biol. 2006;173:159–163. doi: 10.1083/jcb.200601153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brody T. The Interactive Fly: gene networks, development and the Internet. Trends Genet. 1999;15:333–334. doi: 10.1016/s0168-9525(99)01775-8. [DOI] [PubMed] [Google Scholar]
- 63.Tanaka KJ, et al. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem. 2006;281:40096–40106. doi: 10.1074/jbc.M609059200. [DOI] [PubMed] [Google Scholar]
- 64.Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. Rna. 2006;12:547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tritschler F, et al. Similar modes of interaction enable Trailer Hitch and EDC3 to associate with DCP1 and Me31B in distinct protein complexes. Mol Cell Biol. 2008;28:6695–6708. doi: 10.1128/MCB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rossoll W, et al. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor axons? Hum Mol Genet. 2002;11:93–105. doi: 10.1093/hmg/11.1.93.Demonstration of SMN in axons along with reference 124
- 67.Rossoll W, et al. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128.Cultured motor neurons from SMA mice show alteration in axon length and transport of beta-actin mRNA to axon tips
- 68.Zhang H, et al. Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J Neurosci. 2006;26:8622–8632. doi: 10.1523/JNEUROSCI.3967-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu W, Pan F, Zhang H, Bassell GJ, Singer RH. A predominantly nuclear protein affecting cytoplasmic localization of beta-actin mRNA in fibroblasts and neurons. J Cell Biol. 2002;156:41–51. doi: 10.1083/jcb.200105133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 71.Claus P, Bruns AF, Grothe C. Fibroblast growth factor-2(23) binds directly to the survival of motoneuron protein and is associated with small nuclear RNAs. Biochem J. 2004;384:559–565. doi: 10.1042/BJ20040801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piazzon N, et al. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J Biol Chem. 2008;283:5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 73.Mourelatos Z, Abel L, Yong J, Kataoka N, Dreyfuss G. SMN interacts with a novel family of hnRNP and spliceosomal proteins. Embo J. 2001;20:5443–5452. doi: 10.1093/emboj/20.19.5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma A, et al. A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp Cell Res. 2005;309:185–197. doi: 10.1016/j.yexcr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 75.Pellizzoni L, Charroux B, Rappsilber J, Mann M, Dreyfuss G. A functional interaction between the survival motor neuron complex and RNA polymerase II. J Cell Biol. 2001;152:75–85. doi: 10.1083/jcb.152.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones KW, et al. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J Biol Chem. 2001;276:38645–38651. doi: 10.1074/jbc.M106161200. [DOI] [PubMed] [Google Scholar]
- 77.Watkins NJ, et al. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–798. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 78.Matera AG, Terns RM, Terns MP. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 79.Burghes AH. When is a deletion not a deletion? When it is converted. Am J Hum Genet. 1997;61:9–15. doi: 10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schrank B, et al. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920.First demonstration that loss of SMN in mammals is an embryonic lethal phenotype showing Smn is an essential gene
- 81.Zerres K, Rudnik-Schoneborn S. Natural history in proximal spinal muscular atrophy. Clinical analysis of 445 patients and suggestions for a modification of existing classifications. Arch Neurol. 1995;52:518–523. doi: 10.1001/archneur.1995.00540290108025. [DOI] [PubMed] [Google Scholar]
- 82.Feldkotter M, Schwarzer V, Wirth R, Wienker TF, Wirth B. Quantitative analyses of SMN1 and SMN2 based on real-time lightCycler PCR: fast and highly reliable carrier testing and prediction of severity of spinal muscular atrophy. Am J Hum Genet. 2002;70:358–368. doi: 10.1086/338627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mailman MD, et al. Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet Med. 2002;4:20–26. doi: 10.1097/00125817-200201000-00004. [DOI] [PubMed] [Google Scholar]
- 84.Monani UR, et al. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177.Together with reference 89 show that the essential difference between SMN1 and SMN2 is the single nucleotide change in exon7 that alters splicing
- 85.Monani UR, McPherson JD, Burghes AH. Promoter analysis of the human centromeric and telomeric survival motor neuron genes (SMNC and SMNT) Biochim Biophys Acta. 1999;1445:330–336. doi: 10.1016/s0167-4781(99)00060-3. [DOI] [PubMed] [Google Scholar]
- 86.Echaniz-Laguna A, Miniou P, Bartholdi D, Melki J. The promoters of the survival motor neuron gene (SMN) and its copy (SMNc) share common regulatory elements. Am J Hum Genet. 1999;64:1365–1370. doi: 10.1086/302372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lefebvre S, et al. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265.Demonstration of the correlation of SMN level with severity of SMA as well as identification of the SMNY272C mutation reducing SMN levels in SMA
- 88.Coovert DD, et al. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205.As in reference 87 demonstration of the correlation of SMN levels with severity of SMA
- 89.Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307.Together with reference 84 show that the essential difference between SMN1 and SMN2 is the single nucleotide change in exon7 that alters splicing
- 90.Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 91.Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 92.Gennarelli M, et al. Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem Biophys Res Commun. 1995;213:342–348. doi: 10.1006/bbrc.1995.2135. [DOI] [PubMed] [Google Scholar]
- 93.Lorson CL, Androphy EJ. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 94.Burnett BG, et al. Regulation of SMN protein stability. Mol Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Alias L, et al. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum Genet. 2009;125:29–39. doi: 10.1007/s00439-008-0598-1. [DOI] [PubMed] [Google Scholar]
- 96.Sun Y, et al. Molecular and functional analysis of intragenic SMN1 mutations in patients with spinal muscular atrophy. Hum Mutat. 2005;25:64–71. doi: 10.1002/humu.20111. [DOI] [PubMed] [Google Scholar]
- 97.Buhler D, Raker V, Luhrmann R, Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum Mol Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 98.Monani UR, et al. A transgene carrying an A2G missense mutation in the SMN gene modulates phenotypic severity in mice with severe (type I) spinal muscular atrophy. J Cell Biol. 2003;160:41–52. doi: 10.1083/jcb.200208079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ogawa C, et al. Gemin2 plays an important role in stabilizing the survival of motor neuron complex. J Biol Chem. 2007;282:11122–11134. doi: 10.1074/jbc.M609297200. [DOI] [PubMed] [Google Scholar]
- 100.Kotani T, et al. A novel mutation at the N-terminal of SMN Tudor domain inhibits its interaction with target proteins. J Neurol. 2007;254:624–630. doi: 10.1007/s00415-006-0410-x. [DOI] [PubMed] [Google Scholar]
- 101.Clermont O, et al. Molecular analysis of SMA patients without homozygous SMN1 deletions using a new strategy for identification of SMN1 subtle mutations. Hum Mutat. 2004;24:417–427. doi: 10.1002/humu.20092. [DOI] [PubMed] [Google Scholar]
- 102.Cusco I, Barcelo MJ, del Rio E, Baiget M, Tizzano EF. Detection of novel mutations in the SMN Tudor domain in type I SMA patients. Neurology. 2004;63:146–149. doi: 10.1212/01.wnl.0000132634.48815.13. [DOI] [PubMed] [Google Scholar]
- 103.Parsons DW, et al. Intragenic telSMN mutations: frequency, distribution, evidence of a founder effect, and modification of the spinal muscular atrophy phenotype by cenSMN copy number. Am J Hum Genet. 1998;63:1712–1723. doi: 10.1086/302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gabanella F, et al. Ribonucleoprotein Assembly Defects Correlate with Spinal Muscular Atrophy Severity and Preferentially Affect a Subset of Spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921.Along with reference 56 demonstrate that reduced SMN that occurs in SMA leads to reduced assembly of snRNPs which in turn reduces the steady state level of certain snRNPs which could selectively alter splicing
- 105.Owen N, Doe CL, Mellor J, Davies KE. Characterization of the Schizosaccharomyces pombe orthologue of the human survival motor neuron (SMN) protein. Hum Mol Genet. 2000;9:675–684. doi: 10.1093/hmg/9.5.675. [DOI] [PubMed] [Google Scholar]
- 106.Paushkin S, et al. The survival motor neuron protein of Schizosacharomyces pombe. Conservation of survival motor neuron interaction domains in divergent organisms. J Biol Chem. 2000;275:23841–23846. doi: 10.1074/jbc.M001441200. [DOI] [PubMed] [Google Scholar]
- 107.Hsieh-Li HM, et al. A mouse model for spinal muscular atrophy. Nat Genet. 2000;24:66–70. doi: 10.1038/71709.Along with reference 108 produce the first animal model of SMA that recapitulate the situation that occurs in SMA patients. Loss of SMN1 and retention of SMN2
- 108.Monani UR, et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333.Along with reference 107 produce the first animal model of SMA that recapitulate the situation that occurs in SMA patients. Loss of SMN1 and retention of SMN2
- 109.McGovern VL, Gavrilina TO, Beattie CE, Burghes AH. Embryonic motor axon development in the severe SMA mouse. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miguel-Aliaga I, et al. The Caenorhabditis elegans orthologue of the human gene responsible for spinal muscular atrophy is a maternal product critical for germline maturation and embryonic viability. Hum Mol Genet. 1999;8:2133–2143. doi: 10.1093/hmg/8.12.2133. [DOI] [PubMed] [Google Scholar]
- 111.Chan YB, et al. Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum Mol Genet. 2003;12:1367–1376. doi: 10.1093/hmg/ddg157. [DOI] [PubMed] [Google Scholar]
- 112.Miguel-Aliaga I, Chan YB, Davies KE, van den Heuvel M. Disruption of SMN function by ectopic expression of the human SMN gene in Drosophila. FEBS Lett. 2000;486:99–102. doi: 10.1016/s0014-5793(00)02243-2. [DOI] [PubMed] [Google Scholar]
- 113.McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–932. doi: 10.1083/jcb.200303168.Demonstration that knockdown of SMN in zebrafish gives a cell autonomous defect in motor axons that can be corrected with SMN expression allowing assay of critical pathways in SMA
- 114.Frugier T, et al. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum Mol Genet. 2000;9:849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- 115.Hannus S, Buhler D, Romano M, Seraphin B, Fischer U. The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum Mol Genet. 2000;9:663–674. doi: 10.1093/hmg/9.5.663. [DOI] [PubMed] [Google Scholar]
- 116.Briese M, et al. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum Mol Genet. 2009;18:97–104. doi: 10.1093/hmg/ddn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Cifuentes-Diaz C, et al. Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J Cell Biol. 2001;152:1107–1114. doi: 10.1083/jcb.152.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vitte JM, et al. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am J Pathol. 2004;165:1731–1741. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gavrilina TO, et al. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum Mol Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379.Two copies of SMN2 produces sufficient SMN for some tissues
- 120.Chang HC, et al. Modeling spinal muscular atrophy in Drosophila. PLoS ONE. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.DiDonato CJ, et al. Regulation of murine survival motor neuron (Smn) protein levels by modifying Smn exon 7 splicing. Hum Mol Genet. 2001;10:2727–2736. doi: 10.1093/hmg/10.23.2727. [DOI] [PubMed] [Google Scholar]
- 122.Butchbach ME, Edwards JD, Burghes AH. Abnormal motor phenotype in the SMNDelta7 mouse model of spinal muscular atrophy. Neurobiol Dis. 2007;27:207–219. doi: 10.1016/j.nbd.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Carrel TL, et al. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fan L, Simard LR. Survival motor neuron (SMN) protein: role in neurite outgrowth and neuromuscular maturation during neuronal differentiation and development. Hum Mol Genet. 2002;11:1605–1614. doi: 10.1093/hmg/11.14.1605.Demonstration of SMN in axons along with reference 66
- 125.Wan L, et al. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol Cell Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gabanella F, Carissimi C, Usiello A, Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum Mol Genet. 2005;14:3629–3642. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- 127.Grimmler M, et al. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep. 2005;6:70–76. doi: 10.1038/sj.embor.7400301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Winkler C, et al. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.McWhorter ML, Boon KL, Horan ES, Burghes AH, Beattie CE. The SMN binding protein Gemin2 is not involved in motor axon outgrowth. Dev Neurobiol. 2008;68:182–194. doi: 10.1002/dneu.20582. [DOI] [PubMed] [Google Scholar]
- 130.Sheth N, et al. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. and Alioto (2006) U12DB: a database of orthologous U12-type spliceosomal introns NAR 35: database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alioto TS. U12DB: a database of orthologous U12-type spliceosomal introns. Nucleic Acids Res. 2007;35:D110–D115. doi: 10.1093/nar/gkl796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee S, et al. Genome-wide expression analysis of a spinal muscular atrophy model: towards discovery of new drug targets. PLoS ONE. 2008;3:e1404. doi: 10.1371/journal.pone.0001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Olaso R, et al. Activation of RNA metabolism-related genes in mouse but not human tissues deficient in SMN. Physiol Genomics. 2006;24:97–104. doi: 10.1152/physiolgenomics.00134.2005. [DOI] [PubMed] [Google Scholar]
- 134.Corti S, et al. Neural stem cell transplantation can ameliorate the phenotype of a mouse model of spinal muscular atrophy. J Clin Invest. 2008;118:3316–3330. doi: 10.1172/JCI35432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schauber J, et al. Injury enhances TLR2 function and antimicrobial peptide expression through a vitamin D-dependent mechanism. J Clin Invest. 2007;117:803–811. doi: 10.1172/JCI30142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ule J, et al. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 137.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Briese M, Richter DU, Sattelle DB, Ulfig N. SMN, the product of the spinal muscular atrophy-determining gene, is expressed widely but selectively in the developing human forebrain. J Comp Neurol. 2006;497:808–816. doi: 10.1002/cne.21010. [DOI] [PubMed] [Google Scholar]
- 139.Jablonka S, Beck M, Lechner BD, Mayer C, Sendtner M. Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy. J Cell Biol. 2007;179:139–149. doi: 10.1083/jcb.200703187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Nishimune H, Sanes JR, Carlson SS. A synaptic laminin-calcium channel interaction organizes active zones in motor nerve terminals. Nature. 2004;432:580–587. doi: 10.1038/nature03112. [DOI] [PubMed] [Google Scholar]
- 141.Noakes PG, Gautam M, Mudd J, Sanes JR, Merlie JP. Aberrant differentiation of neuromuscular junctions in mice lacking s-laminin/laminin beta 2. Nature. 1995;374:258–262. doi: 10.1038/374258a0. [DOI] [PubMed] [Google Scholar]
- 142.Kariya S, et al. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Narver HL, et al. Sustained improvement of spinal muscular atrophy mice treated with trichostatin A plus nutrition. Ann Neurol. 2008;64:465–470. doi: 10.1002/ana.21449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kong L, et al. Impaired synaptic vesicle release and immaturity of neuromuscular junctions in spinal muscular atrophy mice. J Neurosci. 2009;29:842–851. doi: 10.1523/JNEUROSCI.4434-08.2009.Physiological characterization of SMA mice
- 145.Dubowitz V, Sewry CA. Muscle biopsy : a practical approach. xiii. Philadelphia?: Saunders/Elsevier; 2007. p. 611. [Google Scholar]
- 146.Wuhl E, et al. Neurodevelopmental deficits in Pierson (microcoria-congenital nephrosis) syndrome. Am J Med Genet A. 2007;143:311–319. doi: 10.1002/ajmg.a.31564. [DOI] [PubMed] [Google Scholar]
- 147.Swoboda KJ, et al. Natural history of denervation in SMA: relation to age, SMN2 copy number, and function. Ann Neurol. 2005;57:704–712. doi: 10.1002/ana.20473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Glenney JR, Jr, Kaulfus P, Matsudaira P, Weber K. F-actin binding and bundling properties of fimbrin, a major cytoskeletal protein of microvillus core filaments. J Biol Chem. 1981;256:9283–9288. [PubMed] [Google Scholar]
- 149.Bretscher A. Fimbrin is a cytoskeletal protein that crosslinks F-actin in vitro. Proc Natl Acad Sci U S A. 1981;78:6849–6853. doi: 10.1073/pnas.78.11.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Oprea GE, et al. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cobben JM, et al. Deletions of the survival motor neuron gene in unaffected siblings of patients with spinal muscular atrophy. Am J Hum Genet. 1995;57:805–808. [PMC free article] [PubMed] [Google Scholar]
- 152.Jedrzejowska M, et al. Unaffected patients with a homozygous absence of the SMN1 gene. Eur J Hum Genet. 2008;16:930–934. doi: 10.1038/ejhg.2008.41. [DOI] [PubMed] [Google Scholar]
- 153.Faure C, et al. Mechanical signals modulated vascular endothelial growth factor-A (VEGF-A) alternative splicing in osteoblastic cells through actin polymerisation. Bone. 2008;42:1092–1101. doi: 10.1016/j.bone.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 154.Murray LM, et al. Selective vulnerability of motor neurons and dissociation of pre- and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008;17:949–962. doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 155.Coërs C, Woolf AL. The innervation of muscle; a biopsy study. xv. Springfield, Ill: Thomas; 1959. p. 149. [Google Scholar]
- 156.Gonsalvez GB, Praveen K, Hicks AJ, Tian L, Matera AG. Sm protein methylation is dispensable for snRNP assembly in Drosophila melanogaster. Rna. 2008;14:878–887. doi: 10.1261/rna.940708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Gonsalvez GB, et al. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol. 2007;178:733–740. doi: 10.1083/jcb.200702147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Nguyen thi M, et al. A two-site ELISA can quantify upregulation of SMN protein by drugs for spinal muscular atrophy. Neurology. 2008;71:1757–1763. doi: 10.1212/01.wnl.0000313038.34337.b1. [DOI] [PubMed] [Google Scholar]
- 159.Butchbach MER, Burghes AHM. Perspectives on models of spinal muscular atrophy for drug discovery. Drug Discovery Today Disease Models. 2004;1:151–156. [Google Scholar]
- 160.Young PJ, et al. The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding. Hum Mol Genet. 2000;9:2869–2877. doi: 10.1093/hmg/9.19.2869. [DOI] [PubMed] [Google Scholar]
- 161.Giesemann T, et al. A role for polyproline motifs in the spinal muscular atrophy protein SMN. Profilins bind to and colocalize with smn in nuclear gems. J Biol Chem. 1999;274:37908–37914. doi: 10.1074/jbc.274.53.37908. [DOI] [PubMed] [Google Scholar]
- 162.Talbot K, et al. Missense mutation clustering in the survival motor neuron gene: a role for a conserved tyrosine and glycine rich region of the protein in RNA metabolism? Hum Mol Genet. 1997;6:497–500. doi: 10.1093/hmg/6.3.497. [DOI] [PubMed] [Google Scholar]
- 163.Prior TW. Spinal muscular atrophy diagnostics. J Child Neurol. 2007;22:952–956. doi: 10.1177/0883073807305668. [DOI] [PubMed] [Google Scholar]
- 164.Zapletalova E, et al. Analysis of point mutations in the SMN1 gene in SMA patients bearing a single SMN1 copy. Neuromuscul Disord. 2007;17:476–481. doi: 10.1016/j.nmd.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 165.Rochette CF, et al. Molecular diagnosis of non-deletion SMA patients using quantitative PCR of SMN exon 7. Neurogenetics. 1997;1:141–147. doi: 10.1007/s100480050021. [DOI] [PubMed] [Google Scholar]
- 166.Hahnen E, et al. Missense mutations in exon 6 of the survival motor neuron gene in patients with spinal muscular atrophy (SMA) Hum Mol Genet. 1997;6:821–825. doi: 10.1093/hmg/6.5.821. [DOI] [PubMed] [Google Scholar]
- 167.Wang CH, Papendick BD, Bruinsma P, Day JK. Identification of a novel missense mutation of the SMN(T) gene in two siblings with spinal muscular atrophy. Neurogenetics. 1998;1:273–276. doi: 10.1007/s100480050040. [DOI] [PubMed] [Google Scholar]
- 168.Yu B, Howell PL. Intragenic complementation and the structure and function of argininosuccinate lyase. Cell Mol Life Sci. 2000;57:1637–1651. doi: 10.1007/PL00000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Rodriguez-Pombo P, et al. Towards a model to explain the intragenic complementation in the heteromultimeric protein propionyl-CoA carboxylase. Biochim Biophys Acta. 2005;1740:489–498. doi: 10.1016/j.bbadis.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 170.Burt EC, Towers PR, Sattelle DB. Caenorhabditis elegans in the study of SMN-interacting proteins: a role for SMI-1, an orthologue of human Gemin2 and the identification of novel components of the SMN complex. Invert Neurosci. 2006;6:145–159. doi: 10.1007/s10158-006-0027-x. [DOI] [PubMed] [Google Scholar]
- 171.Rajendra TK, et al. A Drosophila melanogaster model of spinal muscular atrophy reveals a function for SMN in striated muscle. J Cell Biol. 2007;176:831–841. doi: 10.1083/jcb.200610053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement 1
Supplement 2