Differential regulation of microRNA stability (original) (raw)
Abstract
MicroRNAs (miRNAs) are endogenous single-stranded RNA molecules of about 21 nucleotides in length that are fundamental post-transcriptional regulators of gene expression. Although the transcriptional and processing events involved in the generation of miRNAs have been extensively studied, very little is known pertaining to components that regulate the stability of individual miRNAs. All RNAs have distinct inherent half-lives that dictate their level of accumulation and miRNAs would be expected to follow a similar principle. Here we demonstrate that although most miRNA appear to be stable, like mRNAs, miRNAs possess differential stability in human cells. In particular, we found that miR-382, a miRNA that contributes to HIV-1 provirus latency, is unstable in cells. To determine the region of miR-382 responsible for its rapid decay, we developed a cell-free system that recapitulated the observed cell-based-regulated miR-382 turnover. The system utilizes in vitro-processed mature miRNA derived from pre-miRNA and follows the decay of the processed miRNA. Using this system, we demonstrate that instability of miR-382 is driven by sequences outside its seed region and required the 3′ terminal seven nucleotides where mutations in this region increased the stability of the RNA. Moreover, the exosome 3′–5′ exoribonuclease complex was identified as the primary nuclease involved in miR-382 decay with a more modest contribution by the Xrn1 and no detectable contribution by Xrn2. These studies provide evidence for an miRNA element essential for rapid miRNA decay and implicate the exosome in this process. The development of a biochemically amendable system to analyze the mechanism of differential miRNA stability provides an important step in efforts to regulate gene expression by modulating miRNA stability.
Keywords: miRNA stability, miRNA decay, miR-382, instability element
INTRODUCTION
MicroRNAs (miRNAs) are endogenous short noncoding single-stranded RNA molecules that post-transcriptionally regulate eukaryotic gene expression. While over 600 miRNAs have been identified in humans (Griffiths-Jones et al. 2008), as many as 1000 miRNAs have been predicted to be encoded by the human genome, representing ∼3% of the total number of genes (Bentwich 2005).
Two cellular processing steps consecutively occur in the nucleus and the cytoplasm to generate mature miRNAs. First, primary miRNA (pri-miRNA) is cleaved into an approximate 70 nt-long hairpin precursor miRNA (pre-miRNA) by Drosha (Lee et al. 2003). Drosha, in conjunction with a regulatory subunit, DGCR8/Pasha, produces a pre-miRNA containing a 5′ phosphate and a 3′ hydroxyl with a 2-nt overhang at its 3′ end that serves as a recognition site for the second processing step (Denli et al. 2004; Gregory et al. 2004; Han et al. 2004; Landthaler et al. 2004). The pre-miRNA is exported to the cytoplasm by Exportin-5 (Zeng et al. 2003; Bohnsack et al. 2004; Lund et al. 2004), where a second processing step catalyzed by Dicer cleaves the pre-miRNA ∼20-nt upstream of the 2-nt 3′ overhang recognition site to produce a miRNA duplex intermediate. One of the two strands, generally the strand containing the least extent of hydrogen bonding at the 5′ end of the duplex, becomes the mature miRNA, termed guide-strand, whereas the other strand is lost (Khvorova et al. 2003; Schwarz et al. 2003). Once selected, the mature miRNA is eventually incorporated into an RNA-induced silencing complex (RISC), and serves as the guide for RNA silencing (Murchison and Hannon 2004).
While expression of miRNA can conceivably be regulated at any step during their biogenesis, very little is known regarding the decay of miRNAs. By analogy, the regulation of mRNA stability plays a crucial part in the control of gene expression, where both the 5′ m7GpppN cap structure and the 3′ poly(A) tail contribute to the stability and translational competence of an mRNA (Furuichi and Shatkin 2000; Mangus et al. 2003). In contrast to their precursor pri-miRNAs, miRNAs as well as pre-miRNAs appear to have 5′ and 3′ unprotected ends that may render them accessible to exoribonucleases. However, in plants, miRNAs are protected from 3′–5′ exonucleolytic activity by a 2′-O-methyl group or an oligo uridine tail added to their 3′ end (Li et al. 2005; Yu et al. 2005). Methylation or uridylation of the miRNA deters the activity of the recently identified 3′–5′ exoribonuclease SDN1 that specifically degrades single-stranded small RNAs in a sequence-independent manner (Ramachandran and Chen 2008). The SDN gene family is universally present in eukaryotes, but whether the human homolog of SDN1 exerts similar functions is currently unknown. In Caenorhabditis elegans, degradation of a subset of miRNAs was recently shown to be mediated by the nuclear 5′–3′ exoribonuclease, Xrn2 (Chatterjee and Grosshans 2009). Factors that can promote miRNA stability have also been reported recently. Translin, a DNA/RNA-binding protein, was shown to bind and increase the stability of miR-122 in vivo (Yu and Hecht 2008). Furthermore, post-transcriptional regulation of an mRNA is also dependent on both _cis_-elements and _trans_-acting factors. Although these _cis_-elements could occur anywhere on the mRNA, the majority of them are localized within the 3′ UTR (Decker and Parker 1993; Jackson 1993). Interestingly, a report by Hwang et al. (2007) showed that a hexanucleotide motif at the 3′ end of miR-29b was important for its regulation and was essential for its nuclear import.
We carried out a global analysis of miRNA levels following transcriptional shutoff in Human Embryonic Kidney (HEK293) cells and demonstrate that a majority of miRNAs does not appreciably decrease during the 8-h duration tested, with several exceptions. In particular, we identified miR-382, a miRNA involved in the inhibition of HIV-1 protein contributing to viral latency (Huang et al. 2007), as an unstable miRNA in cells. To further characterize the decay of miR-382, we developed an in vitro processing-coupled decay assay and identified a sequence within miR-382 that was essential for its reduced stability.
RESULTS
Differential stability of miRNAs
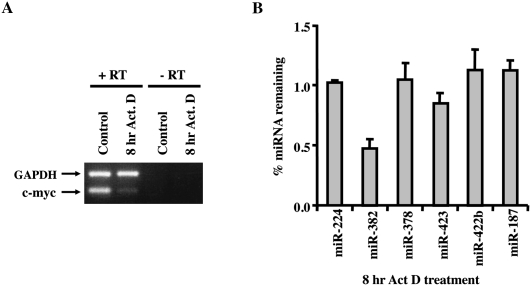
In order to analyze the extent to which miRNAs vary in their inherent stability, we used a microarray approach to specifically detect individual miRNA expression in HEK293 cells following a block in transcription. The miRNA microarray analysis was carried out with RNA extracted from cells treated with actinomycin D for 8 h to inhibit transcription. The efficiency of transcription inhibition was confirmed by RT-PCR of the unstable c-myc proto-oncogene. As expected, a significant reduction in c-myc mRNA levels were detected following 8 h of transcriptional shutoff, while the stable GAPDH mRNA was still abundantly present (Fig. 1A). Of the 310 human miRNA antisense probes contained on the NCode chip (Invitrogen), ∼30% were expressed at appreciable levels in HEK293 cells and could be detected. Interestingly, 95% of the miRNAs remained stable and did not significantly change throughout the 8-h time course of the assay, reflecting half-lives greater than 8 h (Supplemental Fig. S1; Supplemental Table S1). Five miRNAs (miR-154, miR-224, miR-382, miR-204, and miR-211) were of significantly lower abundance at the 8-h actinomycin D time-point and appeared to have half-lives less than 8 h. However, it should be noted that since actinomycin D is expected to only affect the synthesis of pri-miRNA and not the subsequent processing events, microarray results represent the sum total of pri-miRNA stability and processing, pre-miRNA stability and processing, and stability of the mature miRNA. These data indicate that the steady-state levels of only a subset of miRNAs decrease in HEK293 cells following transcriptional silencing for 8 h, while the levels of a majority of miRNAs do not change, as determined by microarray analysis.
FIGURE 1.
Differential stability of miRNAs in actinomycin D-treated HEK293 cells. (A) Total RNA from 8-h actinomycin D treated or untreated HEK293 cells were RT-PCR amplified with primers to detect the unstable c-myc proto-oncogene mRNA or stable GAPDH mRNA to confirm transcriptional inhibition. (B) Microarray results for a subset of miRNAs were verified by quantitative RT-PCR using TaqMan miRNA assay kit (Applied Biosystems). Data represent the ratio of miRNA at 8 h actinomycin D treatment relative to untreated cells and represent the mean of at least three independent experiments ± standard errors.
In an effort to validate the microarray data, an independent real-time PCR approach was used to monitor levels of a subset of miRNAs before and after 8 h of actinomycin D-directed transcriptional shutoff. Six miRNAs were further tested and included two potentially unstable miRNAs (miR-224 and miR-382), two with apparent intermediate stability (miR-378 and miR-423), and two that appeared to be stable (miR-422b and miR-187) (Supplemental Fig. S1). Quantitative TaqMan real-time PCR that specifically detects and discriminates miRNA from their precursors was used. Consistent with the microarray results, miR-382 levels were reduced more than 50% after 8 h of actinomycin D treatment, while miR-423 was reduced by 20%, and the two apparently stable miRNAs, miR-422b and miR-187, remained unchanged (Fig. 1B). In contrast to the microarray results, the levels of miR-224 and miR-378 remained unchanged. The greater sensitivity of the real-time PCR approach suggests that these results more accurately represent the cellular miRNA levels and indicate that miR-224 and miR-378 are stable in HEK293 cells, while miR-382 appears to be selectively and reproducibly unstable in HEK293 cells.
miR-382 is unstable in cells
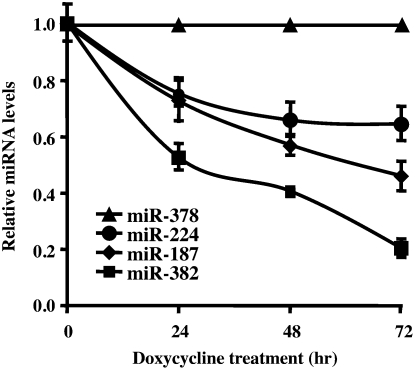
To more directly follow the stability of miRNAs, a Dicer knockdown cell line was used to curtail pre-miRNA processing and enable a more focused analysis of miRNA stability minimally affected by the extent of pre-miRNA processing. A HEK293 cell line containing a tetracycline-regulated short hairpin RNA (shRNA) directed against Dicer, which can efficiently reduce Dicer levels following 48 h of induction, was used (Schmitter et al. 2006). The fate of four miRNAs, miR-382, miR-187, mi-224, and miR-378, was followed over 3 d with quantitative real-time PCR following induction of Dicer-directed shRNA. The levels of each miRNA tested are presented relative to the stable miR-378. As shown in Figure 2, the relative levels of each miRNA are distinctly affected, demonstrating differential stability of miRNAs. Consistent with the above data, miR-382 levels were the most prominently reduced, with miR-187 and miR-224 displaying intermediate profiles compared with miR-378. Collectively, our data demonstrated that miR-382 was consistently unstable relative to miR-187, miR-224, and miR-378 in HEK293 cells.
FIGURE 2.
Differential stability of miRNAs in Dicer knockdown HEK293 cells. The level of miRNA remaining at the indicated times following Dicer-specific shRNA induction in HEK293 2b2 cells were determined by quantitative RT-PCR using the TaqMan miRNA assay kit (Applied Biosystems) and presented relative to the stable miR-378, which was set to 1. Data represent the mean of at least three independent experiments ± standard errors.
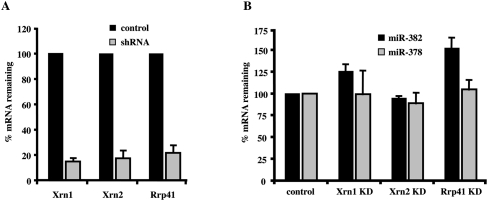
To identify the exoribonucleases that affect miR-382 levels, a lentiviral shRNA delivery system was implemented to individually knockdown the exoribonucleases, Xrn1, Xrn2, or the exosome complex. The efficiency of the knockdowns is shown in Figure 3A, where an 85% and 83% reduction in the mRNAs encoding the Xrn1 and Xrn2 5′–3′ exoribonucleases was obtained, respectively. A 78% knockdown of the mRNA encoding a core component of the 3′–5′ exosome complex, Rrp41, was attained. Interestingly, a knockdown of Rrp41 generated the most significant and specific stabilization of miR-382, with a 52% increase in miR-382 levels (Fig. 3B), while a knockdown of Xrn1 led to a more modest 26% increase in miR-382 levels. A reduction in Xrn2 levels did not lead to a measurable difference in miR-382 levels. Importantly, significant differences in the levels of miR-378 were not detected by a reduction in any of the three nucleases (Fig. 3B). Collectively, these data support a role for both the exosome and Xrn1 in the selective decay of miR-382, with the exosome appearing to fulfill a more primary role.
FIGURE 3.
miR-382 levels are primarily responsive to the exosome complex. (A) HEK293T cells were transduced with lentiviruses expressing shRNAs directed either to Xrn1, Xrn2, or Rrp41. Levels of the indicated mRNAs in control and shRNA-expressing cells as determined by quantitative real time–PCR are graphed. Xrn1, Xrn2, and Rrp41 mRNA levels were reduced by 85%, 83%, and 78% in the respective knockdown cells. (B) miR-382 and miR-378 levels were determined using the TaqMan miRNA assay kit (Applied Biosystems) in each indicated knockdown cell line and presented relative to RNU43 snoRNA. Data represent the mean of at least three independent experiments ± standard errors.
Development of an in vitro miRNA decay system
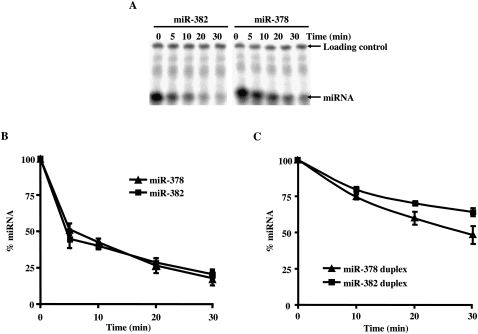
To enable a mechanistic understanding of components that govern miRNA stability, we initiated studies to analyze miRNA stability in vitro. We initially adapted an assay that we have successfully used to study mRNA decay (Wang et al. 1999) and followed the decay of the stable miR-378 and unstable miR-382. However, when using either single-stranded or double-stranded miRNA to follow their decay in extract, differential stability between miR-382 and miR-378 was not detected (Fig. 4). These data suggest that the distinct stabilities between the two miRNAs observed in cells might be due to specific ribonucleoprotein complexes that normally form on the miRNA during pre-miRNA processing, which were not present on the naked RNA added to the reaction.
FIGURE 4.
Decay of mature miRNA in vitro. Incubation of mature miRNA in an uncoupled decay reaction does not recapitulate the differential miRNA stability of miR-382 and miR-378 observed in cells. Assays were carried out with 50 μg of HEK293T cytoplasmic extract at 15°C with the indicated 32P-labeled single-stranded miRNA. (A) Reactions were terminated with stop buffer spiked with 5′-end 32P-labeled loading control to enable normalization between the lanes. The miRNA and loading controls are indicated on the right. Quantitations of the decay reactions derived from mature single-stranded (B) or double-stranded (C) miR-382 and miR-378 are shown. Data represent the mean of at least three independent experiments ± standard errors.
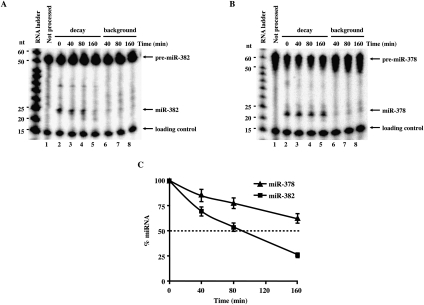
To overcome this limitation, we rationalized that miRNAs that were generated in vitro from pre-miRNA would more closely mimic endogenous conditions. Pre-miRNA processing might promote assembly of endogenous ribonucleoprotein complexes onto the miRNA. A coupled in vitro pre-miRNA processing and miRNA decay assay was developed to directly access the stability of miR-378 and miR-382. 32P-labeled pre-miRNAs were incubated with cytoplasmic extract derived from cells overexpressing Dicer protein to facilitate more efficient pre-miRNA processing (Zhang et al. 2002). The decay was initiated by the addition of cell extract lacking exogenous Dicer and a shift of the reaction to 18°C to minimize pre-miRNA processing. Consistent with the data observed in cells, miR-378 was reproducibly more stable relative to miR-382 with half-lives of >160 and 80 min, respectively (Fig. 5A,B; summarized in Fig. 5C). These data indicate that differential stability of miRNAs observed in cells can be mimicked within a cell-free assay system that utilizes miRNA derived from pre-miRNA processing.
FIGURE 5.
Coupled pre-miRNA processing and miRNA decay in vitro. Coupled processing and decay assays were carried out with 5′-end 32P-labeled pre-miR-382 (A) and pre-miR-378 (B). 32P-labeled-pre-miRNAs were incubated with 10 μg of Dicer-overexpressing HEK293 cytoplasmic S15 extract at 37°C for 30 min to process the pre-miRNA into mature miRNAs. Decay of the mature miRNA was then followed by the addition of 50 μg of HEK293T cytoplasmic S15 at 18°C (lanes 2–5). Lanes 6–8 show the extent of background processing that occurs during the decay assay. The 32P-labeled pre-miRNA in lanes 6–8 were incubated directly at 18°C with a mixture of 10 μg of cytoplasmic S15 extract derived from Dicer-overexpressing cells and 50 μg of cytoplasmic S15 extract from HEK293 cells. Reactions were terminated as in the legend to Figure 4. The pre-miRNA, mature miRNA, and loading controls are indicated on the right and the size markers on the left. (C) Graphic representation of miR-382 and miR-378 decay relative to the level of miRNA present in the “0” time point are shown. 32P-signals were quantified using ImageQuant software. Data represent the mean of at least three independent experiments ± standard errors.
The 3′ terminus of miR-382 is necessary for its instability
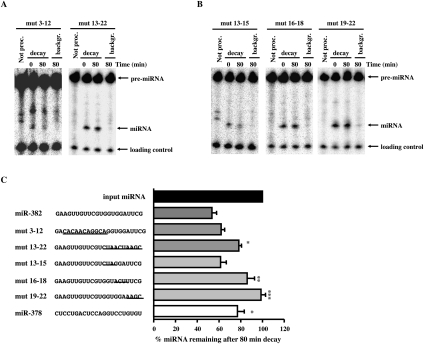
In an effort to decipher whether a specific _cis_-element contributed to the instability of miR-382, mutational analyses were carried out. Two sets of mutations were initially tested that either alter the first half or second half of miR-382. The mutations were designed such that the overall secondary structures of the pre-miRNAs were conserved to enable pre-miRNA processing. This required flipping the top and bottom half arms of the pre-miRNAs (Supplemental Fig. S2). Sequence substitutions in the first half of the miRNA did not significantly alter the miR-382 half-life (miR-382mut3-12), while substitution of sequences in the second half (miR-382mut13-22) dramatically increased miRNA stability (Fig. 6A,C), indicating that sequences within the second half of miR-382 were necessary for its rapid decay.
FIGURE 6.
Requirement of miR-382 3′ terminus for its rapid decay. (A) Processing-coupled in vitro decay assays were carried out with pre-miR-382 containing substitutions within the first or second half of the miRNA or (B) at positions 13–15, 16–18, and 19–22 in the second half of the miRNA. Reaction conditions and labeling are as described in the legend to Figure 4. (C) The sequence of wild-type and mutants miR-382 are shown at the left, where lines demarcate the substituted sequences. Bars represent the ratio of wild-type and mutant miR-382 levels expressed as a ratio of miRNA remaining after an 80-min decay reaction relative to the initial level of miRNA prior to the decay. Data represent the mean of at least three independent experiments ± standard errors. Significance was determined by ANOVA using Graph Pad InStat software, *P < 0.05, **P < 0.01, ***P < 0.001.
The essential region within the second half of miR-382 was determined with three additional mutations to further refine the minimal sequence necessary for instability (Fig. 6B,C). Substitutions at positions 13–15 (miR-382mut13-15) had no significant effect on the stability of the miRNA, indicating that these nucleotides do not contribute to the decay of miR-382. Interestingly, two sets of mutations that disrupted sequences within the 3′ terminal seven nucleotides (miR-382mut16-18 and miR-382mut19-22) both increased stability of the miRNA. These data demonstrate that the 3′ terminal seven nucleotides of miR-382 (positions 16–22) are necessary for the observed rapid decay of this miRNA.
DISCUSSION
Despite an extensive analysis of the multiple processes involved in the generation of miRNAs, relatively little is known regarding the decay of miRNAs and the contribution of miRNA stability to their accumulation in a cell. In this study, we demonstrate that miRNAs can possess intrinsic distinct stabilities. Using a global analysis approach to identify miRNAs with reduced levels following a transcriptional shutoff, we identified a subset of miRNAs whose steady-state levels decreased significantly. In particular, miR-382 was found to be unstable in HEK293 cells, and the reduced stability was dependent on the 3′ terminal seven nucleotides of the miRNA. Our data demonstrates that a specific sequence within a miRNA could dictate its stability, and suggests that other miRNAs may also have intrinsic elements that modulate their stability.
Our global analysis of miRNA levels following transcriptional shutoff indicated that the miRNA levels for a majority of miRNAs detected did not appreciably change during the 8-h transcriptional arrest (Supplemental Fig. S1). However, it should be noted that although transcription was terminated, processing of pri- and pre-miRNAs into miRNAs is expected to continue under these conditions. If the processing of a particular miRNA precursor is regulated and the processing occurs relatively slowly, miRNAs could be replenished and appear to be more stable than they are. In addition, whether actinomycin D, which has been shown to aberrantly stabilize some mRNAs (Shyu et al. 1989), could also impact miRNA stability is currently unknown. Therefore, it is possible that the observed ∼95% of the miRNAs that did not decrease in overall abundance could be an overestimation.
Dicer knockdown conditions, which minimize pre-miRNA processing and accentuate miRNA turnover, further substantiated our hypothesis that miRNAs possess distinct stabilities. Under these conditions, all four miRNAs tested displayed distinct relative decay kinetics, with miR-382 as the most unstable, miR-187 and miR-224 displaying intermediate decay rates relative to the stable miR-378. Although in our analyses only a subset of miRNAs appear to be unstable, surprisingly, a recent report detected the levels of five brain-enriched miRNAs following α-amanitin-directed transcriptional shutoff, and found the level of all five miRNAs decreased rapidly with <50% of the detected miRNAs remaining by 3 h (Sethi and Lukiw 2009). Whether the rapid decrease in these five miRNAs is a general property of neuronal cells or serendipitous, is currently unknown.
Further support for selective post-transcriptional regulation of miRNAs was recently reported. The testis-brain RNA-binding protein, Translin, was shown to stabilize miR-122 when overexpressed in HUH-7 cells and selectively bind this miRNA in vitro (Yu and Hecht 2008). Most miRNAs are tightly associated with the RISC and only a small fraction are thought to be independent of RISC in cells (Tang et al. 2008). However, it is likely that the 3′ terminus of the miRNA could be accessible to factors, regardless of their association with RISC (see below). In addition, components of the RISC may also influence the stability of miRNAs. A central component of RISC, the Ago2 protein (Filipowicz 2005), was shown to have an RNase-independent function of post-transcriptionally enhancing expression of mature miRNAs at either the processing or stability levels (Diederichs and Haber 2007). Indeed, loss of Ago2 in mouse embryonic fibroblast cells derived from Ago2 knockout mice leads to a decrease in miRNA levels that can be restored by expression of the protein.
The mechanism of miRNA turnover may be analogous to that of mRNA decay and utilize the same decay machinery. A correlation between the percent of AU or UA dinucleotides in several miRNAs and their instability has been proposed to possibly be analogous to the mRNA Adenine-Uracil-rich element that can confer mRNA instability (Sethi and Lukiw 2009). Conversely, the 3′–5′ exoribonuclease SDN1 preferentially degrade single-stranded small RNAs in a sequence-independent manner in plants (Ramachandran and Chen 2008). This activity was deterred by HEN1-mediated methylation of the last ribose residue in all miRNAs or siRNAs tested by a sequence-independent manner (Yu et al. 2005). Here, we showed that knockdowns of Rrp41 and Xrn1 resulted in an increase of miR-382 levels, suggesting that both the 5′–3′ and 3′–5′ decay pathways characteristic of mRNA degradation (Coller and Parker 2004) are potentially involved in the degradation of miR-382. Importantly, the selective decay of miR-382 appears to predominantly be dependent on the 3′–5′ decay pathway and is consistent with the location of the miR-382 destabilization element at the 3′ end. However, whether the exosome directly or indirectly targets miR-382 remains to be determined. A recent study (Chatterjee and Grosshans 2009) showed that another exoribonuclease, Xrn2, a predominantly nuclear protein (Amberg et al. 1992), is involved in the degradation of let-7 and other miRNAs in Caenorhabditis elegans. Interestingly, we did not detect a significance of Xrn2 in either miR-382 or miR-378 levels in HEK293 cells. Therefore, it is likely that different miRNAs are targeted by distinct nucleases for decay.
The development of an in vitro–coupled pre-miRNA processing and miRNA decay assay that recapitulates the differential stabilities of miR-378 and miR-382 observed in vivo provides a biochemical system to enable mechanistic analysis of miRNA stability and potentially other noncoding RNA stabilities. Using this system, we found that the last seven nucleotides of miR-382 are essential for conferring instability onto the miRNA. Similarly, a hexanucleotide motif contained at the 3′ end of miR-29b has also been implicated in miRNA regulation by directing enrichment of this miRNA into the nucleus (Hwang et al. 2007). The positioning of the required region for stability and transport within miR-382 and miR-29b, respectively, is particularly intriguing. The 5′ region of a given miRNA, referred to as the seed region, is important for directing a miRNA to its mRNA target. Structural analysis of a bacterial argonaute protein in complex with a guide DNA and target mRNA revealed that the 3′ end of a DNA-guide strand (residues 12–19) displayed fewer contacts with the protein than its extensively anchored 5′ seed counterpart (Wang et al. 2008), indicating its potential accessibility to proteins. Positioning of the miRNA regulatory region to the 3′ end and likely outside the mRNA and RISC interaction regions could render the miRNA more accessible to _trans_-factors that can exert an influence onto the miRNA, perhaps analogous to the function of the 3′-untranslated region of an mRNA. Future studies to identify _trans_-factors that can function through the miR-382 3′-end element will begin to address these possibilities.
Although currently underappreciated, regulation of miRNA stability is likely to emerge as an important step in the control of miRNA levels and, consequently, a broader range of regulation that can impact gene expression. Modulation of miR-382 can have significant biological relevance, as is illustrated by its contribution to differential HIV-1 protein production in resting and activated CD4+ T cells, as well as modulation of HIV-1 latency (Huang et al. 2007). The resistant nature of latent HIV-1 to therapeutic intervention (Lassen et al. 2004; Peterson et al. 2007) indicates that efforts aimed at preventing latency would significantly benefit therapeutic efficacy and one avenue could involve controlling stability of miR-382.
Having developed a coupled pre-miRNA processing and miRNA stability assay can now provide an appropriate platform to identify factors conferring miRNA stability and to mechanistically decipher their molecular function. The demonstration that miRNAs could possess distinct intrinsic stabilities will facilitate new areas of investigation into miRNA regulation. Moreover, an understanding of how gene expression can be controlled by miRNA stability may provide novel therapeutic applications to manipulate the expression of a broad spectrum of genes associated with human disorders.
MATERIALS AND METHODS
Plasmid generation
The Dicer coding region was excised from the pcDNA3-T7-Dicer plasmid (kindly provided by J. Liu, Sloan-Kettering Institute) with HindIII and XhoI and inserted into the same sites in pcDNA3-FLAG plasmid to generate pcDNA3-Flag-Dicer, which expresses FLAG-tagged Dicer protein.
Cell culture, transfections, and extract preparation
HEK293 and HEK293T cells were maintained in Dulbecco's Modified Eagle Media (DMEM, Invitrogen) supplemented with 10% fetal bovine serum and a mixture of antibiotic–antimycotic. HEK293T cells were transfected at 80% confluency with pcDNA3-FLAG-Dicer using Lipofectamine 2000 (Invitrogen) as described by the manufacturer. Twenty-four hours post-transfection, cells were harvested and cytoplasmic S15 extract isolated as previously described (Song and Kiledjian 2007). HEK293 cells at 80% confluency were treated with actinomycin D (5 μg/mL, Sigma) for 8 h to inhibit transcription. Knockdown of Dicer in HEK293 conditional Dicer knockdown 2b2 cells was carried out as previously described (Schmitter et al. 2006). Knockdowns of Xrn1, Xrn2, and Rrp41 were obtained in HEK293T by lentiviral RNAi-directed gene silencing and selection with puromycin (3 μg/mL) as described by the manufacturer (Sigma).
RNA isolation and synthesis
Total RNA isolations were carried out with TRIzol (Invitrogen). miRNAs were enriched from total RNAs using two Microcon columns following the protocol described by the manufacturer (Millipore) to enrich for low-molecular weight RNAs (LMW). The efficiency of actinomycin D-mediated transcriptional shutoff was verified by RT-PCR of the proto-oncogene c-myc mRNA, which has a half-life less than 1 h in mammalian cells. Briefly, total RNAs were treated with Turbo DNase (Ambion) to degrade DNA prior to oligo(dT) (IDT)-directed reverse transcription with M-MLV-RT (Promega) according to the manufacturer's instructions. Amplification of the cDNA was carried out by PCR at 94°C for 30 sec, 60°C for 20 sec, and 72°C for 30 sec, for 22 cycles using Taq DNA polymerase.
DNA templates to generate pre-miRNAs were obtained by PCR of 60 base oligonucleotide pre-miRNA templates using 5′ SP6 and 3′ pre-miRNA specific primers (Supplemental Table S2) by Advantage Taq DNA polymerase (Clontech) according to the manufacturer. Pre-miRNAs were transcribed with SP6 RNA polymerase (Promega) according to the manufacturer. The 5′ end of the resulting RNA was dephoshorylated with Calf Intestine Phosphatase (New England BioLabs) and 5′ 32P-labeled with [γ-32P]ATP (Perkin Elmer) using T4 polynucleotide kinase (New England BioLabs).
Genome-wide analysis of miRNA stability
The LMW RNAs (500 ng) were subsequently tagged using the 3DNA Array 900 miRNA direct protocol from Genisphere and used to probe a miRNA microarray chip as follows. Briefly, a poly(A) tail was first added to the 3′ hydroxyl end of the LMW RNAs. The LMW RNAs were subsequently ligated to a capture sequence tag via a bridging oligonucleotide. The capture sequence is a 31-nt-long oligonucleotide partially complementary to the bridging oligo and attached to a 3DNA dendrimer labeled with either Cy3 or Cy5 fluorochromes. The 19-nt-long bridging oligonucleotide consists of 9 nt that are complementary to the capture sequence tag and 10 nt complementary to the added poly(A) tail. Sequence-tagged LMW RNAs were hybridized to the NCode miRNA microarrays (Invitrogen) containing antisense miRNA probes according to Goff et al. (2005). A set of 10 synthetic control miRNAs (NCode Control Set, Invitrogen) are spiked into the microarray labeling reaction to minimize and monitor experimental variability between time points and across microarrays. These probes do not cross-react with any probes for endogenous miRNAs on the microarray.
Arrays were scanned using an Axon GenePix 4000B scanner (Molecular Devices) and median spot intensities collected using Axon GenePix 4.0 (Molecular Devices). Data analysis and manipulation were conducted in GeneSpring 7.0 (Agilent) using quantile analysis (with the R package limma; http://www.bioconductor.org). Results were normalized to the spike-in NCode control probe signals to correct for global changes in miRNA populations.
Real time PCR confirmation was carried out with Turbo DNase (Ambion) treated total RNA and RT-PCR using the appropriate TaqMan microRNA assay (Applied Biosystems) as described by the manufacturer.
Coupled pre-miRNA processing and miRNA decay
Coupled pre-miRNA processing and miRNA decay assays were carried out with 5′-end labeled in vitro synthesized pre-miR-382 and its derivatives or purchased pre-miR-378 (Dharmacon) (Supplemental Table S2). Pre-miRNA hairpins were formed by incubation at 95°C for 2 min and slow cooled to room temperature in annealing buffer (20 mM KOAc, 6 mM HEPES at pH 7.4, 0.4 mM MgOAc). Pre-miRNA processing reactions were carried out with 1000 cpm of 32P-labeled pre-miRNA incubated at 37°C for 30 min in IVD-1 buffer (20 mM Tris at pH 7.5, 200 mM KOAc, 4 mM MgOAc, 4 mM DTT, 20 mM creatine phosphate, 2 mM ATP, 0.8 mM GTP, 0.2 mM spermine) containing 10 μg of cytoplasmic extract from Dicer-overexpressing HEK293T cells pretreated for 10 min with 10 ng proteinase K (Sigma) to activate Dicer activity (Zhang et al. 2002). The decay reactions were subsequently initiated by the addition of 50 μg of HEK293T cytoplasmic extract and followed at 18°C for the indicated times. Control reactions to determine the extent of background processing during the decay reactions were determined by incubation of pre-miRNA with both proteinase K-pretreated Dicer-overexpressing and HEK293T cytoplasmic extracts at 18°C for the same time intervals. Reactions were stopped by addition of 150 μL of ULB (10 mM Tris at pH 7.5, 0.35 M NaCl, 2% SDS, 10 mM EDTA, 7 M Urea) spiked with 100 cpm 5′-end 32P-labeled loading control, phenol-chloroform extracted, ethanol precipitated, and resolved on a 10% polyacrylamide-7M Urea gel. Gels were dried and exposed to a PhosphorImager, scanned using a Storm Scanner and quantitated using ImageQuant software. Uncoupled in vitro decay reactions were carried out with 32P-labeled mature single-stranded or double-stranded miRNAs (IDT) (Supplemental Table S2) and 50 μg of HEK293T cytoplasmic extract at 15°C. Reactions were stopped with ULB spiked with 1000 cpm 5′-end 32P-labeled 60-nt-long loading control.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank W. Filipowicz (Friedrich Miescher Institute) for providing the HEK293 conditional knockdown cells and J. Liu (Sloan-Kettering Institute) for providing pcDN3-T7-Dicer plasmid. S.B. was supported in part by an American Heart Postdoctoral Fellowship and R.P.H. was supported by NIH R21NS054028, The New Jersey Commission on Spinal Cord Injury, the New Jersey Commission on Science & Technology, and Invitrogen, Inc. This work was supported by NIH grant GM67005 to M.K.
Footnotes
REFERENCES
- Amberg DC, Goldstein AL, Cole CN 1992. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev 6: 1173–1189 [DOI] [PubMed] [Google Scholar]
- Bentwich I 2005. Prediction and validation of microRNAs and their targets. FEBS Lett 579: 5904–5910 [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Czaplinski K, Gorlich D 2004. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10: 185–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Grosshans H 2009. Active turnover modulates mature microRNA activity in Caenorhabditis elegans. Nature 461: 546–549 [DOI] [PubMed] [Google Scholar]
- Coller J, Parker R 2004. Eukaryotic mRNA Decapping. Annu Rev Biochem 73: 861–890 [DOI] [PubMed] [Google Scholar]
- Decker CJ, Parker R 1993. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev 7: 1632–1643 [DOI] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432: 231–235 [DOI] [PubMed] [Google Scholar]
- Diederichs S, Haber DA 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Filipowicz W 2005. RNAi: The nuts and bolts of the RISC machine. Cell 122: 17–20 [DOI] [PubMed] [Google Scholar]
- Furuichi Y, Shatkin AJ 2000. Viral and cellular mRNA capping: Past and prospects. Adv Virus Res 55: 135–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff LA, Yang M, Bowers J, Getts RC, Padgett RW, Hart RP 2005. Rational probe optimization and enhanced detection strategy for microRNAs using microarrays. RNA Biol 2: 93–100 [DOI] [PubMed] [Google Scholar]
- Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R 2004. The Microprocessor complex mediates the genesis of microRNAs. Nature 432: 235–240 [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ 2008. miRBase: Tools for microRNA genomics. Nucleic Acids Res 36: D154–D158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN 2004. The Drosha–DGCR8 complex in primary microRNA processing. Genes & Dev 18: 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H 2007. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nat Med 13: 1241–1247 [DOI] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT 2007. A hexanucleotide element directs microRNA nuclear import. Science 315: 97–100 [DOI] [PubMed] [Google Scholar]
- Jackson RJ 1993. Cytoplasmic regulation of mRNA function: The importance of the 3′ untranslated region. Cell 74: 9–14 [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD 2003. Functional siRNAs and miRNAs exhibit strand bias. Cell 115: 209–216 [DOI] [PubMed] [Google Scholar]
- Landthaler M, Yalcin A, Tuschl T 2004. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol 14: 2162–2167 [DOI] [PubMed] [Google Scholar]
- Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF 2004. The multifactorial nature of HIV-1 latency. Trends Mol Med 10: 525–531 [DOI] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. 2003. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419 [DOI] [PubMed] [Google Scholar]
- Li J, Yang Z, Yu B, Liu J, Chen X 2005. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15: 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U 2004. Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Mangus DA, Evans MC, Jacobson A 2003. Poly(A)-binding proteins: Multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol 4: 223 doi: 10.1186/gb-2003-4-7-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Hannon GJ 2004. miRNAs on the move: miRNA biogenesis and the RNAi machinery. Curr Opin Cell Biol 16: 223–229 [DOI] [PubMed] [Google Scholar]
- Peterson S, Reid AP, Kim S, Siliciano RF 2007. Treatment implications of the latent reservoir for HIV-1. Adv Pharmacol 55: 411–425 [DOI] [PubMed] [Google Scholar]
- Ramachandran V, Chen X 2008. Degradation of microRNAs by a family of exoribonucleases in Arabidopsis. Science 321: 1490–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W 2006. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res 34: 4801–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD 2003. Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Sethi P, Lukiw WJ 2009. Micro-RNA abundance and stability in human brain: Specific alterations in Alzheimer's disease temporal lobe neocortex. Neurosci Lett 459: 100–104 [DOI] [PubMed] [Google Scholar]
- Shyu AB, Greenberg ME, Belasco JG 1989. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes & Dev 3: 60–72 [DOI] [PubMed] [Google Scholar]
- Song M, Kiledjian M 2007. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA 13: 2356–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Hajkova P, O'Carroll D, Lee C, Tarakhovsky A, Lao K, Surani MA 2008. MicroRNAs are tightly associated with RNA-induced gene silencing complexes in vivo. Biochem Biophys Res Commun 372: 24–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Day N, Trifillis P, Kiledjian M 1999. An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol 19: 4552–4560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ 2008. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456: 921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Hecht NB 2008. The DNA/RNA-binding protein, translin, binds microRNA122a and increases its in vivo stability. J Androl 29: 572–579 [DOI] [PubMed] [Google Scholar]
- Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X 2005. Methylation as a crucial step in plant microRNA biogenesis. Science 307: 932–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR 2003. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci 100: 9779–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W 2002. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]