From microscopes to microarrays: dissecting recurrent chromosomal rearrangements (original) (raw)
. Author manuscript; available in PMC: 2010 Apr 22.
Published in final edited form as: Nat Rev Genet. 2007 Nov;8(11):869–883. doi: 10.1038/nrg2136
Abstract
Submicroscopic chromosomal rearrangements that lead to copy-number changes have been shown to underlie distinctive and recognizable clinical phenotypes. The sensitivity to detect copy-number variation has escalated with the advent of array comparative genomic hybridization (CGH), including BAC and oligonucleotide-based platforms. Coupled with improved assemblies and annotation of genome sequence data, these technologies are facilitating the identification of new syndromes that are associated with submicroscopic genomic changes. Their characterization reveals the role of genome architecture in the aetiology of many clinical disorders. We review a group of genomic disorders that are mediated by segmental duplications, emphasizing the impact that high-throughput detection methods and the availability of the human genome sequence have had on their dissection and diagnosis.
The sequencing of the human genome has facilitated efforts to define a wide spectrum of human genetic disorders. Variation at the nucleotide or sequence level has been extensively characterized and utilized for multiple purposes, including whole-genome association studies. This field has been galvanized by the availability of robust high-throughput array-based SNP detection, allowing major advances in defining the loci associated with age-related macular degeneration1, childhood and adult obesity2,3 and myocardial infarction4, among others. These approaches have been successfully used to examine genetic factors associated with common complex traits and population variation, providing a wealth of information for human genetic and evolutionary studies5,6. On a larger scale, variation in chromosome structure has been appreciated since the advent of chromosome banding. Examination of karyotypes has not only provided diagnostic information on genetic syndromes, but has also revealed microscopically detectable structural differences in the general population. More recently, emphasis has been placed on characterizing variations of the genome that fall in the range between the single nucleotide and visible chromosomal changes — submicroscopic structural variants (those that involve less than 5 Mb). Several recent studies have focused on identifying copy-number changes that have now been shown to exist in apparently normal individuals7–12. These copy-number variations (CNVs) include insertions and deletions as well as more complex changes that involve gains and losses at the same locus. In addition, there are multilocus complex CNVs, which have been further characterized and classified11. Depending on the resolution of the technique used, as many as 90 interindividual CNVs comprising regions of several kilobases to megabases can be identified11. Readily accessible, comprehensive databases contain much of the data from these studies (see the web site for The Sick Kids Centre for Applied Genomics and the UCSC Genome Browser).
The recent findings that large (>1 kb) insertions and deletions of DNA segments contribute significantly to human genome variation have focused the attention of human geneticists on the role of CNVs in the aetiology of human genetic disease. There is a strong correlation between segmental duplications and copy-number variants8. Segmental duplications (discussed further below) are large highly homologous low-copy repeats that appear to constitute approximately 5% of the genome13. However, analysis of the content of CNVs showed that about 30% of sequence within deletion CNVs consisted of segmental duplications, a sixfold enrichment with respect to the genome average8. The same study found that duplications showed an even greater enrichment for segmental duplications. Although it is unclear whether this type of variation contributes directly to abnormal clinical phenotypes per se, as a group it is a major contributor to genome variation12,14,15. CNVs occur in genomic regions that are associated with known disorders, providing a substrate and a risk for genomic rearrangement.
Several chromosomal regions are characterized by the presence of chromosome-specific segmental duplications. These segmental duplications share a high level of sequence identity, predisposing the regions to aberrant non-allelic homologous recombination16, which can result in deletions, duplications or inversions. Segmental duplications have been shown to flank genomic regions that are associated with known human syndromes. Thus, these features of genome architecture have been regarded as the underlying basis for many genomic disorders15–17.
The term genomic disorder18 is typically used to describe a gain (duplication) or loss (deletion) of a specific chromosomal region associated with a clinical genetic syndrome that may present with congenital anomalies or with impairment in neurological and cognitive function. In general, the clinical features are believed to reflect changes in the normal copy number or dosage of the genes that are contained in a given genomic interval, one or more of which contribute to the resulting phenotype. Less frequently, the aberrant phenotype might be the result of gene disruption, creation of a fusion gene, a position effect or the unmasking of a recessive gene mutation. Technological improvements, coupled with higher-quality sequence and better annotation, have uncovered new syndromes19–23 that are associated with copy-number changes, revealing several underlying mechanistic aetiologies. Using a group of genomic disorders that are mediated by segmental duplications (FIG. 1), we discuss the impact that new technologies based on the human genome sequence have had on our understanding of the mechanisms of genomic rearrangements.
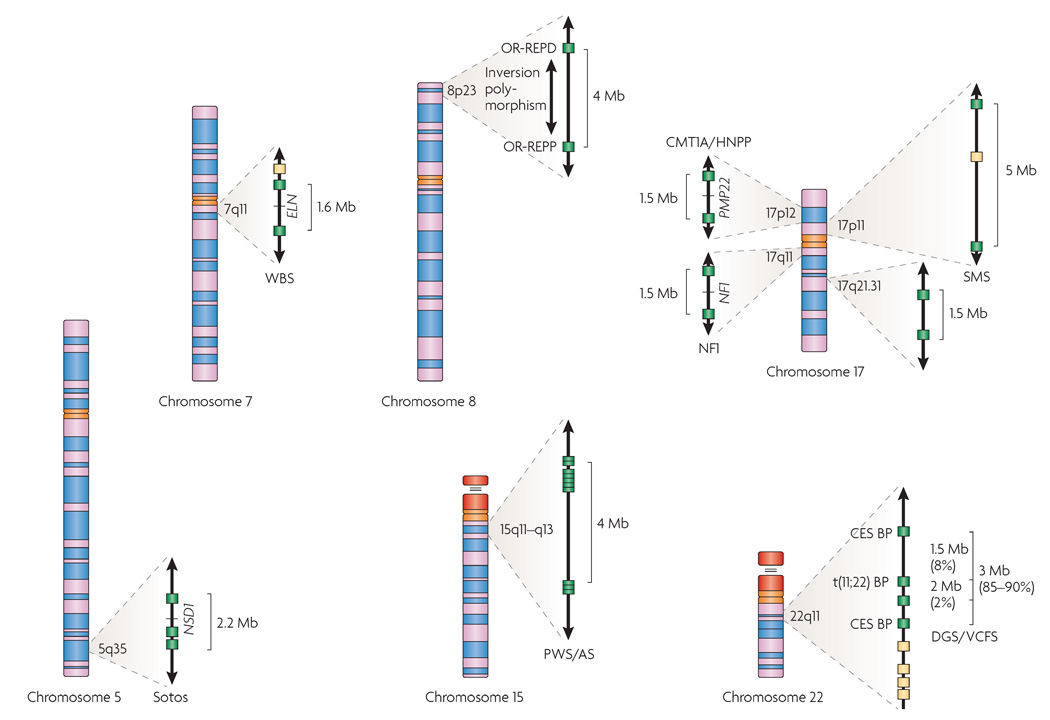
Figure 1. Chromosomal rearrangements mediated by segmental duplications.
The chromosomal regions involved in segmental-duplication-mediated rearrangements described in this manuscript are shown expanded. Segmental duplications are shown as filled boxes: green boxes indicate segmental duplications that are known to be involved in recurrent chromosomal rearrangements, whereas yellow boxes indicate copies that are less frequently involved in mediating known rearrangements. On chromosome 5q, the region associated with Sotos syndrome deletions is indicated along with the causative gene, nuclear-receptor-binding SET-domain protein 1 gene (NSD1). On chromosome 7q11, the deletion associated with Williams–Beuren syndrome (WBS) is shown along with the elastin gene (ELN), the main gene responsible for the supravalvular aortic stenosis of WBS. Chromosome 8 is shown with the proximal (OR-REPP) and distal (OR-REPD) olfactory-receptor gene clusters that are associated with inverted duplications of 8p and the t(4;8). Also indicated on 8p is the polymorphic inversion of the region. The region on chromosome 15q11–q13 that is deleted in Prader–Willi and Angelman syndromes (PWS/AS) is shown with its numerous duplicated hect domain and RLD 2 (HERC2) gene segments. On chromosome 17, the region on that is 17p12 duplicated or deleted in Charcot–Marie–Tooth disease Type 1A and hereditary neuropathy with pressure palsies (CMT1A/HNPP) is shown with peripheral myelin protein 22 (PMP22), the gene that is known to be involved in their aetiology. Also on 17p11 is the region involved in Smith–Magenis syndrome (SMS) deletions and the reciprocal duplication. On 17q11 is the region involved in neurofibromatosis type 1 (NF1) deletions. A recently identified segmental duplication-mediated microdeletion syndrome at 17q21.3 is indicated. On chromosome 22, recurrent deletions associated with DiGeorge and velocardiofacial syndromes (DGS/VCFS) are indicated. The proximal and distal breakpoints (BP) for the marker chromosomes in cat eye syndrome (CES) and the t(11;22) BP region are also shown.
Detecting rearrangements
Under ideal conditions, conventional cytogenetic banding techniques allow for the visualization of segmental aneusomy for chromosomal regions of 2–10 Mb of duplicated or deleted DNA24–26. The introduction of molecular cytogenetic approaches such as chromosomal fluorescence in situ hybridization (FISH) into the repertoire of clinical testing and genetic investigation in the 1990s led to an explosion of information about the prevalence and variety of genomic rearrangements. The ability to locate specific DNA segments on metaphase chromosomes and in interphase nuclei identified gains, losses and rearrangements of DNA sequences associated with microdeletion and microduplication syndromes. The sensitivity of FISH is limited both by the size of the probes that are used and the fact that only the genomic region recognized by the specific probes can be queried. The information and resources provided by the Human Genome Project have made it possible to query any part of the genome. For example, a set of FISH probes were designed specifically to interrogate the integrity of human telomeres27. Their use revealed that submicroscopic subtelomeric rearrangements are a significant cause of mental retardation and multiple congenital anomalies28,29. As they are commercially available, these probes are now used routinely for research and clinical diagnostic testing.
Once the involvement of genomic alterations in numerous genetic syndromes became clear, additional genome-wide approaches were developed to detect copy-number differences. Comparative genomic hybridization (CGH) is one such technique that was first described for the detection of copy-number changes in solid tumours30. Originally, copy-number variation was assessed by the differential labelling of DNAs from a test (for example, tumour) genome and from a reference genome (from a normal individual) by hybridizing them to metaphase chromosome spreads. The ratio of the test and reference hybridization signals was then used to determine the relative copy number of sequences in the test genome when compared with the reference genome. Chromosomal CGH is robust for the identification of large-scale imbalances25; however, the use of metaphase chromosomes makes it difficult to detect events that involve regions that are less than 20 Mb of the genome, and limits the resolution of closely spaced aberrations and the ability to link ratio changes to genomic or genetic markers31.
More recent technological improvements have resulted in a shift towards microarray-based formats for CGH analysis — array CGH31,32. DNA probes are arrayed on a chip and CGH is used to test for increased or decreased dosage of chromosomal regions of interest. With a single test, array CGH can detect genomic errors for disorders that are usually identified by cytogenetic analysis and multiple FISH tests33. Array CGH has been widely used in the detection of chromosomal imbalances in solid tumours34–38, mental retardation39–41, subtelomeric rearrangements42–44 and other constitutional chromosomal abnormalities45–50. The detection limits of copy-number differences by array CGH depend on the probe density and the resolution of the platform used. The DNA probes range from genomic clones, most often BAC clones (80–200 kb), to oligonucleotides (25–85 bp). If genomic clones are used as the hybridization target, the size of each individual target and the distance between targets defines the size limits of what is detectable. For example, if BACs are spaced at 1-Mb intervals, any copy-number change that occurs between the BACs and is smaller than 1 Mb will not be detected. Spacing BACs at 0.5-Mb intervals results in a twofold improvement in the ability of the array to detect changes.
Oligonucleotide arrays can detect gains or losses of shorter stretches of the genome, and if oligonucleotides for a given region are densely arrayed, the sensitivity for detecting alterations of that region is greatly enhanced. Commercially available platforms that typically utilize oligonucleotides representing from tens of thousands to more than 1 million SNPs distributed across the genome are now frequently being used to assess copy number. These arrays or chips are used for genotyping studies, but groups of adjacent SNPs can be interrogated to determine copy number for a given chromosomal region. Studies using oligonucleotide arrays have shown that known microdeletion syndromes and previously undiagnosed rearrangements can be detected in patients with mental retardation and/or multiple congenital anomalies51–53. In addition, when combined with appropriate analytical tools, SNP-based arrays can detect other types of genomic alteration. For example, regions of copy-number neutral loss of heterozygosity could be encountered in uniparental disomy (UPD) of a genomic interval54.
In general, array-CGH-based methods are currently focused on detecting copy-number changes; rearrangements such as balanced translocations or inversions are not detectable using this methodology. In addition, given the frequency of CNVs in the genome, higher-density oligonucleotide arrays will require careful assessment of probe design for accurate copy-number genotyping of complex regions of the genome55. Although array CGH is proving robust and provides an exceptional level of resolution from a diagnostic perspective, the main difficulty with the current interpretation of array-CGH results lies in assigning causality and clinical significance to the alterations that are detected. Towards this end, the availability of databases with information on normal variation in multiple ethnic populations, and testing of unaffected parents, remain the standard approaches to discerning whether a copy-number change that has been detected by array CGH is likely to be disease-causing.
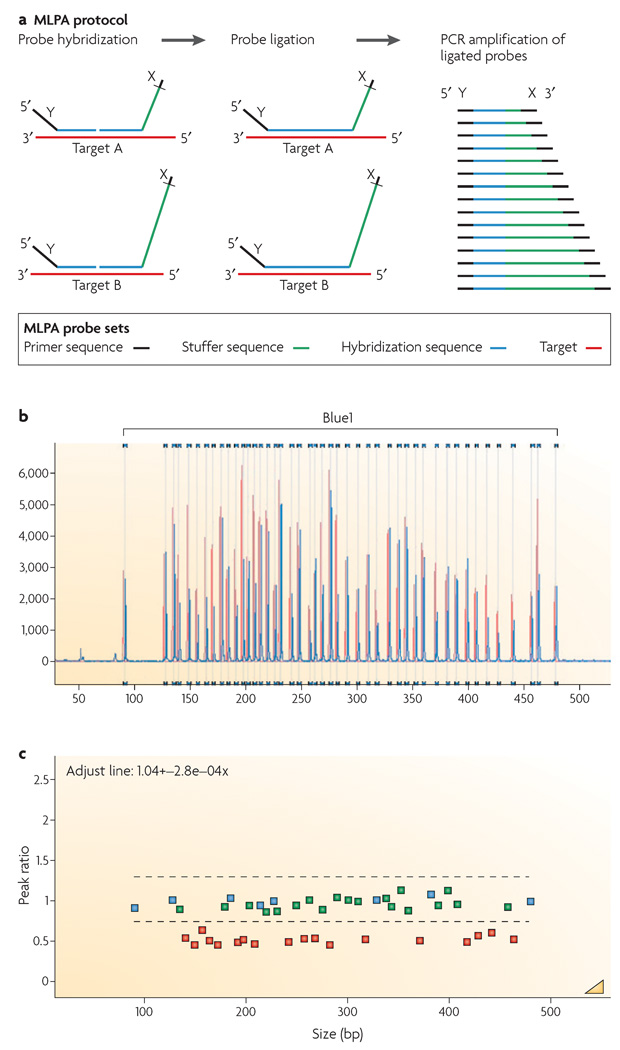
Other technologies that have been used to determine or confirm copy-number changes associated with genomic disorders include quantitative or real-time PCR, multiplex amplifiable probe hybridization (MAPH)56 and multiplex ligation-dependent probe amplification (MLPA). In traditional PCR, the amount of product is often not related to the amount of input DNA. By contrast, real-time or quantitative PCR is a kinetic approach that assesses the products in the early linear stages of the reaction. Thus, the higher the starting copy number of the nucleic acid target, the shorter the time required for a significant increase in fluorescent product to be observed. MLPA, similar to MAPH, allows the relative quantification of up to 45 different target DNA sequences in a single reaction based on the quantification of PCR products of varied lengths (FIG. 2). This sensitive diagnostic screening method57–58 is also faster, less expensive and less labour-intensive than FISH or array CGH. For both quantitative PCR and MLPA, copy-number detection is strictly limited to the regions that the amplification primers have been chosen to assess.
Figure 2. The multiplex ligation-dependent probe amplification (MLPA) reaction.
a | The figure shows two MLPA hemiprobes for each target. Each hemiprobe has a universal primer on one end. One hemiprobe is synthetic and the second, the one with the stuffer sequence (shown in green), is M13-derived. Each of the M13-derived hemiprobes has a different stuffer sequence. During the MLPA reaction, each pair of hemiprobes hybridizes to its adjacent target sequences to be enzymatically ligated. The ligation products are PCR amplified using a single primer pair (indicated as X and Y). Amplification products for each target locus have unique lengths. b | Products are separated by capillary electrophoresis. Relative probe signal strength depends on the relative amounts of target sequence that are present. c | Comparison of a test DNA sample to the copy number for control DNAs allows for calculation of the ratio. DNA from a patient with a chromosome 22 deletion indicating a 0.5 ratio for the deleted probes is used as an example. Part a modified with permission from REF. 57 © (2002) Oxford University Press.
Anatomy of genomic rearrangements
There are several types of repeat structure in the human genome, a feature that is thought to underlie its capacity for evolution. Smaller repeats, such as Alu repeats, long interspersed nuclear elements (LINES) and short interspersed nuclear elements (SINES) have already been implicated in causing human disease59–60. Large (> 1 kb) low-copy repeats, or segmental duplications, are also an important structural element. Segmental duplications can be complex, consisting of modular segments, and are characterized by a high degree of sequence identity, often 96–100% over large stretches of the repeat13. Paralogues can therefore be distinguished by examining sequence variation. Segmental duplications are more frequently found in the pericentromeric and subtelomeric regions of chromosomes. This bias is also mirrored by the concentration of segmental duplications in regions that are associated with genomic disorders (FIG. 1). Array CGH was recently used to further investigate regions that are prone to potential genomic rearrangements, based on the duplication architecture of the genome, leading to the identification of a novel genomic disorder in chromosome 17q21.31 (REF. 19). Additionally, a significant number of CNVs in control individuals8,11,13 and visible structural variants in chromosomes have been shown to cluster in these same chromosomal regions15, underscoring the involvement of segmental duplications in perpetuating the dynamic nature of the human genome.
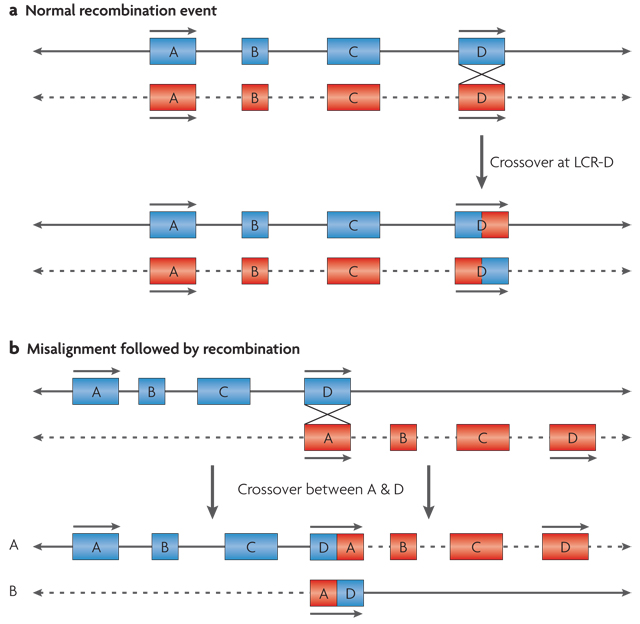
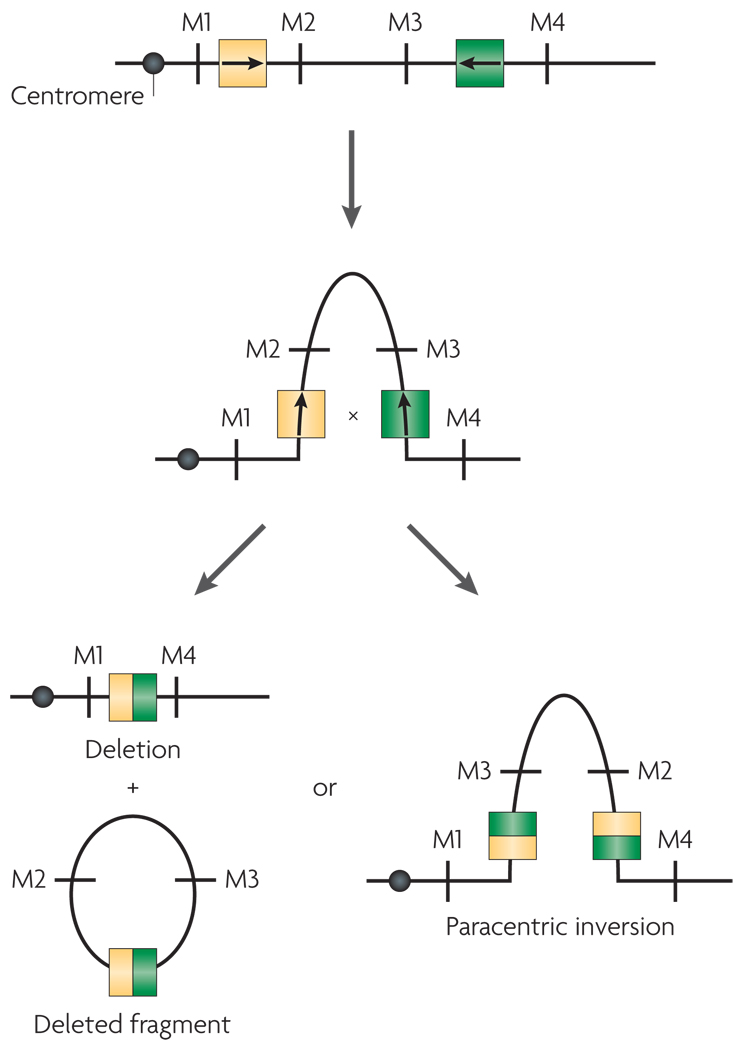
Segmental duplications mediate aberrant recombination by misalignment, or non-allelic homologous recombination16, which can occur either between homologues (interchromosomally) (FIG. 3), or intra-chromosomally by looping out within a single homologue (FIG. 4). These large, highly similar repeats can include sequences that are shared among different chromosomes, or can contain paralogous sequence from within a given chromosome. The orientation of segmental duplications might also contribute to genomic rearrangements17,18,61. For example, direct repeats are more likely to mediate deletions and duplications of a given region, whereas those that are arranged in an inverse orientation are likely to be associated with inversions of the intervening regions17 (FIG. 4).
Figure 3. Model for interchromosomal recombination leading to formation of a deletion and a duplication.
Chromosomes are shown as lines; solid and dotted lines are used to distinguish between the two homologues. a | Segmental duplications or low-copy repeats (LCRs) are shown as blue or red boxes with arrows to indicate the orientation of the shared modules within them. They are depicted during a normal recombination event between two properly aligned segmental duplications, A and D. b | Misalignment of segmental duplications that share sequence homology in the same orientation results in interchromosomal recombination between the two homologues of a chromosome. This results in a reciprocal duplication on homologue A and a deletion on homologue B.
Figure 4. Intrachromosomal recombination between segmental duplications on a single chromosome.
Chromosomes are shown as lines with markers M1, M2, M3 and M4 to indicate marker order. Arrows within the segmental duplications indicate the orientation of the shared sequence elements within them. Segmental duplications that are oriented in opposite orientation to one another pair with one another based on sequence homology and loop out. A recombination event within the segmental duplications leads to either a deletion with a deleted fragment or a paracentric inversion.
Genomic disorders
The presence of multiple congenital anomalies in a patient can be associated with a rearrangement of the genome18, which can occur through multiple mechanisms17. Some of these rearrangements are not associated with changes in dosage or copy number, such as balanced translocations of chromosomal material or chromosomal inversions. In these cases, it is thought that the rearrangement affects the expression of a gene or genes in the interval by causing a breakage within the coding region or by disrupting the regulatory regions, leading to a phenotype. Microdeletion syndromes are genomic disorders that have been referred to as “contiguous gene-deletion syndromes”62 because the commonly deleted chromosomal regions contain multiple genes. Microduplication syndromes are associated with a gain in copy number of a group of contiguous genes. Although only some patients have cytogenetically visible chromosomal losses or gains, molecular studies frequently reveal submicroscopic changes. A group of segmental-duplication-mediated rearrangements, many of which have been extensively characterized both phenotypically and molecularly, is discussed below, with an emphasis on their aetiological and mechanistic features. See also FIG. 1.
Chromosome 5q35
Translocations of the long arm of chromosome 5 associated with Sotos syndrome led to the identification of the associated gene, NSD1 (REF. 63). Sotos syndrome is characterized by overgrowth, craniofacial differences, hypotonia and variable mental retardation. Haploinsufficiency of NSD1, which lies on chromosome 5q35 (chromosome 5, long arm, region 3, band 5) and encodes a nuclear receptor SET-binding-domain protein, which confers these phenotypic features. NSD1 is a putative co-regulator that interacts with steroid receptors and DNA response elements to regulate transcription. Approximately 45% of the initial cases studied were caused by microdeletions that included this gene64. Subsequently, 83% of cases in a large study in the United Kingdom were shown to carry intragenic deletions and mutations of NSD1 (REF. 65). A common 1.9-Mb deletion encompassing the entire gene that is mediated by flanking segmental duplications has subsequently been identified64,66. The prevalence of these structural elements is variable, which might account for the variable frequency of microdeletions in different populations.
The reciprocal duplication of this same region has been identified by array CGH and MLPA-based techniques67 in a small cohort of patients; their decreased growth, including small stature and microcephaly, suggests that gene dosage is directly related to the distinct growth phenotypes that are associated with microdeletions and microduplications of this locus68.
Disorders of chromosome 7
The pericentromeric region of chromosome 7q contains intrachromosomal segmental duplications that give rise to recurrent constitutional genomic rearrangements. The well-known micro-deletion syndrome Williams–Beuren syndrome (WBS) is associated with this region. WBS is characterized by cardiovascular abnormalities, hypertension, intermittent hypercalcaemia, growth retardation, facial dysmorphology and mental retardation with a unique, outgoing personality. The vast majority of patients have a micro-deletion of about 1.5 Mb from chromosome 7q11, which contains 17 genes including the elastin gene (ELN). Haploinsufficiency of ELN underlies a specific cardiac defect — supravalvular aortic stenosis — that is often seen in WBS. Further, mutations in ELN are associated with isolated autosomal dominant supravalvular aortic stenosis, which has been described in families69. Two of the genes that are located near the typical breakpoints of the WBS microdeletion, neutrophil cytosolic factor 1 gene (NCF1) and GTF2I-repeat-domain-containing 2 (GTF2IRD2), have been studied for their association with hypertension, a prevalent feature of WBS. Studies of gene expression within and around the deletion demonstrated that not only the hemizygous genes but also intact genes flanking the deletion showed reduced expression levels, presumably because their transcription control elements are affected by the deletion70.
Up to one-third of unaffected parents of WBS children have inversions of chromosome 7q11 (REF. 71). The inversion variant is present in up to 5% of the general population, and is thought to confer a genetic risk for a carrier to have a child with WBS72, by predisposing the chromosome to aberrant meiotic recombination. This recombination is mediated by the complex, modular ~400-kb segmental duplications that lie in the vicinity of the WBS deletion breakpoints. Although similar predisposing inversions are not typical of all the well-known segmental-duplication-mediated microdeletion syndromes, for WBS these inversions seem to have a significant effect on its aetiology.
A chromosome 7q11 duplication syndrome, which is the reciprocal of the WBS deletion, is associated with characteristic facial features67, learning and speech-acquisition delays, and behavioural features that fall within the autism spectrum73. The severe problem with expressive speech in particular is in striking contrast to the unusual fluency that is observed in WBS, suggesting that dosage of specific genes in 7q11.2 is important for human language and visual–spatial ability74. It also illustrates the point that phenotypic manifestations of microduplications and microdeletions of the same genomic region can be quite different.
Chromosome 8 rearrangements
Olfactory-receptor (OR) genes represent the largest superfamily in the mammalian genome75. They frequently cluster and are present on most human chromosomes, providing evidence for the evolutionary processes — gene duplication and conversion — that are responsible for their genome-wide expansion76. The estimated copy number is up to 1,000 ORs per haploid genome. Two OR gene clusters reside on chromosome 8p, providing a substrate for the formation of several recurrent intrachromosomal rearrangements. Among them are the inverted duplication or (inv dup(8p))77, del(8)(p22)78–80 and small marker chromosome der(8)(p23–pter)81 (the derivative 8 is an inverted duplication of the material on the short arm of chromosome 8 starting from region 2 band 3 and extending to the terminus of the short arm of chromosome 8). Some of these recurrent 8p rearrangements occur as a consequence of an inversion polymorphism in the parent who has transmitted the disease-related chromosome.
The inverted duplications on chromosome 8p23 have a consistently maternal origin, presumably owing to the differences between male and female meiosis. Studies using probes located between the two OR clusters on 8p in phenotypically normal mothers of individuals with inv dup(8p) revealed that these mothers were heterozygous for a submicroscopic inversion of chromosome 8p (REF. 82). The inversion is flanked by the 8p OR gene clusters and is present, in a heterozygous state, in 26% of a population of European descent. The presence of the inversion was suggested to result in a predisposition to aberrant recombination, leading to the formation of the inv dup(8p) or its reciprocal 8p23 deletion product83.
The recurrent deletions of 8p23 have been associated with cardiac defects and congenital diaphragmatic hernia84,85. Furthermore, heterozygous submicroscopic inversions of this region were detected in the transmitting progenitor of many deletion-carrying probands. Here again, non-allelic homologous recombination between segmental duplications creates aberrant exchanges in meiosis.
Similarly, another OR gene cluster resides at 4p16 and provides the partner region for the recurrent t(4;8)(p16;p23) — a translocation between chromosomes 4 and 8 with breakpoints at 4p16 and 8p23. In the unbalanced form this translocation has been reported multiple times in association with congenital anomalies. Individuals with an unbalanced karyotype containing the der(4) show typical features of Wolf–Hirschhorn syndrome, a well-known deletion syndrome86,87, whereas those with the der(8) show a milder dysmorphic syndrome83. Double-heterozygous inversion polymorphisms in non-homologous chromosomes that are generated by OR-gene clusters have been shown to mediate some of these interchromosomal rearrangements.
Disorders of chromosome 15
The 15q11–q13 region has an extremely complex organization and is susceptible to rearrangements. The acrocentric morphology of chromosome 15 and the presence of several large low-copy repeats in the region near the centromere facilitate rearrangements including deletions, duplications and small marker chromosomes. These segmental duplications flank the typical recurrent breakpoints and lead to segmental aneusomy of the 15q11–q13 region. Because this region is subject to imprinting, the phenotypes that are associated with the disorders depend on the parental origin of the rearranged chromosome.
Two distinct disorders are associated with interstitial deletions of chromosome 15q11: Prader–Willi syndrome (PWS) and Angelman syndrome. The regions that are deleted in these two conditions often physically overlap, although PWS occurs only when the deletion is on the paternally inherited chromosome88–90, whereas Angelman syndrome occurs when the deletion is on the maternally derived chromosome, emphasizing the role of imprinting in the aetiology of these human diseases. Recent work has demonstrated that there is significant heterogeneity in the mechanisms that lead to loss of imprinted genes in these syndromes91,92.
Sixty to seventy percent of PWS cases are caused by a 15q11–q13 deletion. Most are sporadic, de novo deletions, although in a few patients deletions are secondary to familial translocations93. The common deletions in PWS encompass an ~5-Mb region flanked by chromosome-15-specific segmental duplications, which mediate the rearrangements94,95. These segmental duplications consist of complex modular subunits, and are composed of duplications of the HECT (also known as HERC5) domain and RLD2 (also known as HERC2), a conserved gene of unknown function96. Importantly, the deleted region contains several paternally imprinted genes97. To date, four protein-coding genes that are transcribed only from the paternal chromosome (makorin, ring finger protein, 3 gene (MKRN3), MAGE-like 2 (MAGEL2), necdin homologue (NDN), and SNRPN upstream reading frame (SNURF-SNRPN)) have been identified, but their precise contribution to the PWS phenotype is unclear. Further, several other imprinted transcripts and genes have emerged, and their role in the phenotype is under investigation98,99. SNRPN (small nuclear ribonucleoprotein polypeptide N) has been a leading candidate gene for the PWS phenotype, although some patients who carry interrupted SNRPN do not show all features of the disorder100. Recurrence risks for PWS vary depending on the underlying molecular mechanism — it is low with de novo deletions and uniparental disomy and increased with translocations involving this region. Additionally, several rare familial cases have been identified in which there is an imprinting-centre defect. In these families, the risk for recurrence is significantly greater. Many of these infrequent imprinting-centre defects appear to result from SNRPN exon 1 deletions, as this region seems to be essential for the imprinting-centre function91. Like PWS, the genetic aetiology of Angelman syndrome is complex. Approximately 65–70% of patients have a de novo segmental-duplication-mediated ~5-Mb deletion of maternal 15q11–q13; these cases involve the same segmental duplications that are associated with PWS on the paternal 15q homologue. Fewer patients (3–5%), those without deletions, have paternal uniparental disomy for chromosome 15. Another 3–5% of patients have an imprinting defect associated with microdeletions of a region that lies 35–40 kb upstream of SNRPN exon 1.
Another mechanism associated with Angelman sydrome involves mutations in UBE3A. UBE3A maps to the critical region for Angelman syndrome and encodes an E6-AP ubiquitin-protein ligase, which catalyses the transfer of activated ubiquitin to protein substrates, thereby earmarking them for degradation by the protea-some. Mutations that disrupt normal patterns of DNA methylation at the imprinting centre occur in 5–10% of patients, accounting for a significant proportion of familial cases101.
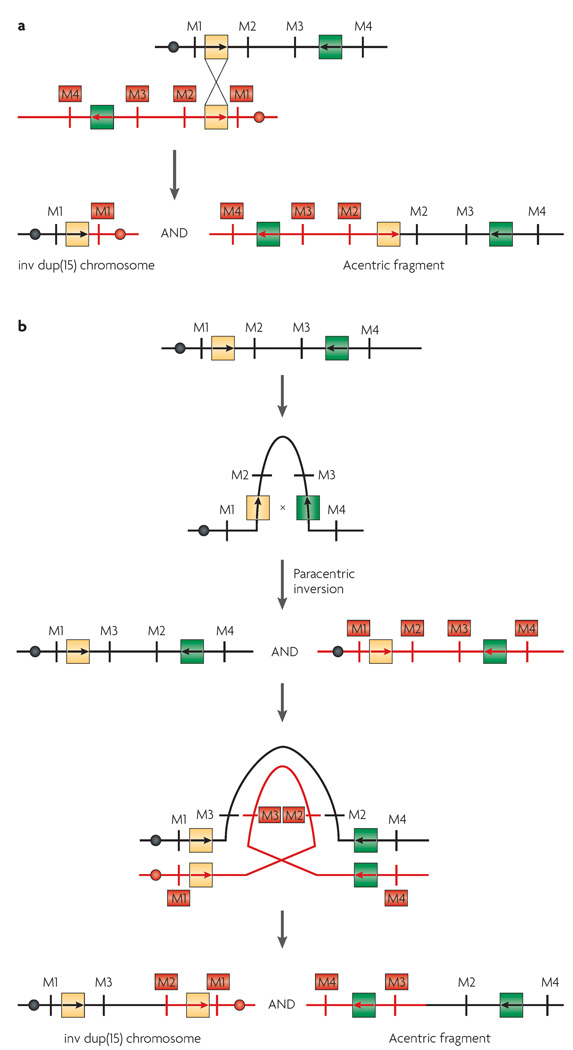
Inv dup(15) is the most common supernumerary marker chromosome seen in humans, accounting for approximately 40% of all such cases. It has been suggested that a predisposing mechanism to inv dup(15) formation is inherent in the complex structure of proximal chromosome 15. Several regions in which breakage occurs have frequently been described. With the use of array CGH, at least four different distal breakpoints have been detected indicating that there are symmetrical and asymmetrical inv dup(15q) chromosomes102. Because the involved regions (between 15q11 and 15q13) contain a large number of segmental duplications, it has been proposed that non-allelic homologous recombination between these repeats is likely to be involved in the formation of the marker chromosomes102 (FIG. 5).
Figure 5. Models for formation of the inv dup(15) or the cat eye syndrome marker chromosomes.
Chromosomes are shown as lines; black and red are used to distinguish between the two homologues. Filled circles indicate centromeres. Segmental duplications are shown as yellow and green boxes with internal arrows to indicate the orientation of sequence blocks with high homology. a | Interchromosomal recombination between the two homologues of a particular chromosome leads to the formation of a bisatellited marker chromosome and an acentric fragment. b | Paracentric inversion within one homologue of a chromosome followed by recombination within an inversion loop leads to the formation of a bisatellited marker chromosome. Formation of an asymmetrical inv dup chromosome is shown.
Inv dup(15) chromosomes are dicentric, bisatellited and consist of two inverted copies of the short (p) arm, the centromere and the proximal long (q) arm of chromosome 15, which are fused at the proximal long arm. This rearrangement results in tetrasomy of 15p and partial tetrasomy of 15q (REFS 102–104). Phenotypically, the individuals with the extra chromosome are characterized by mental retardation, seizures, autism or autistic-like behaviour, abnormal dermatoglyphics and strabismus. Several studies have shown a correlation between the extent of the duplication and the resulting phenotype105–108. In fact, Inv dup(15) chromosomes have been reported in three groups of subjects: normal individuals, individuals with mental retardation and other anomalies, and some patients with PWS and Angelman syndrome.
Disorders of chromosome 17
Both arms of chromosome 17 have been shown to harbour intrachromosomal segmental duplications. There are distinct, well-characterized disorders associated with the rearrangements that are mediated by the segmental duplications, most of which have been studied at the molecular level109.
Smith–Magenis syndrome (SMS)97,98 is typically associated with a 3.7-Mb deletion of chromosome 17p11.2 (REFS 110,111). Segmental duplications that contain sequences that are specific to chromosome 17, or repeats known as SMS-REPs112, flank the microdeletion interval. Duplications of the same region lead to clinical features that include hypotonia, mental retardation, structural cardiac defects, autistic features and sleep apnoea, collectively known as Potocki–Lupski syndrome113, providing another example of distinct phenotypes associated with loss versus gain of the same genomic interval. Intrachromosomal and interchromosomal non-allelic homologous recombination, without apparent parental origin bias, underlies both the deletions and duplications of this region114. An 8-kb recombination hotspot has been described115, and alternative low-copy repeats seem to serve as the substrates for non-allelic homologous recombination, resulting in larger, atypical deletions (5 Mb) that encompass the SMS region114,116.
Duplication of a more telomeric region of 17p, of approximately 1.5 Mb lying adjacent to the region that is missing in SMS, causes Charcot–Marie–Tooth disease type 1A (CMT1A)109. This polyneuropathy is characterized by a progressive weakness and atrophy of the distal limb muscles, which usually begins in the legs and feet. The duplication of this region of 17p contains the peripheral myelin protein 22 gene (PMP22). PMP22 is a major component of myelin that is found in all myelinated fibres in the peripheral nervous system, and is produced predominantly by Schwann cells. The reciprocal deletion of the same 1.5-Mb region is associated with hereditary neuropathy and liability to pressure palsies (HNPP) — an episodic neuropathy109.
Two copies of the CMT1A repeat (CMT1A-REP), each of 24 kb, show greater than 98% sequence identity117. They flank the 17p12 region and mediate the interstitial duplications and deletions that are associated with CMT1A and HNPP, respectively. Recombination breakpoints for both disorders cluster around a 1.7-kb hotspot that contains recombination-promoting motifs and a mariner transposon element118, although the functional relationship between these elements and the hotspot is not well defined. The exchanges appear to take place in a 557-bp region within the breakpoint cluster interval.
Microdeletions of chromosome 17q11.2 can result in neurofibromatosis type 1, if they encompass the gene encoding neurofibromin119. The segmental duplications that mediate these deletions are complex NF1-repeats. They are approximately 15–100 kb in size, and are associated with interchromosomal exchanges causing deletions that are primarily of maternal origin. In addition, recombinogenic sequences similar to those seen in Escherichia coli, which can instigate aberrant recombination, have been reported in the vicinity of these microdeletions120. Recombination-promoting motifs, such as polymerase-arrest sites, are associated with both homologous and non-homologous recombination leading to genomic rearrangements121,122. These motifs have been previously identified at or near deletion and translocation breakpoints123,124, and near deletions on other chromosomes61.
Predictions based on genome architecture and the location of segmental duplications have recently led to the identification of a novel recurrent genomic disorder characterized by microdeletions of chromosome 17q21.3 (REFS 19–21). Its clinical features include mental retardation, hypotonia and facial dysmorphia. The disorder is associated with a common inversion polymorphism20 that encompasses the microtubule-associated protein tau gene (MAPT), which has previously been associated with neurological disorders21. This is one of the first genomic disorders to be identified primarily using array-CGH-based techniques.
Disorders of chromosome 22
There are several recurrent, constitutional abnormalities on chromosome 22q. Among them are translocations and deletions associated with DiGeorge syndrome (DGS) and velocardiofacial syndrome (VCFS), the recently described interstitial duplications of 22q11, the bisatellited marker chromosome of cat eye syndrome (CES), and the translocations that give rise to the recurrent t(11;22) supernumerary der(22) syndrome (Emanuel syndrome). All of the rearrangement breakpoints cluster around the same chromosome-specific segmental duplications of proximal 22q11, indicating that they are involved in the aetiology of these disorders96.
The 22q11.2 deletion syndrome — the most common human microdeletion syndrome — is a genomic disorder that encompasses the clinically defined DiGeorge syndrome, velocardiofacial syndrome and conotruncal anomaly face syndrome (CTAF). Affected individuals show a wide range of phenotypic features, including cardiac defects, palatal anomalies, thymic, parathyroid, craniofacial, developmental and behavioural manifestations125– 127. However, there is significant variation, even within families.
Although smaller, nested recurrent 1.5-Mb deletions have been associated with the syndrome, 80–90% of patients carry the same deletion of approximately 3 Mb in size. However, the phenotype of those patients with the large and small deletions is indistinguishable, making genotype–phenotype correlations difficult128. The 3-Mb deletion contains approximately 35 genes, including TBX1. TBX1, a member of the T-box family of transcription factor genes, was identified as the leading candidate gene for the syndrome when several 22q11.2 deletion-associated phenotypes were modelled in the mouse129–131. Point mutations in TBX1 were subsequently identified in individuals who did not carry deletions, but who had the main physical, but not behavioural and cognitive, features that are seen in the disorder132. More recently, studies using mouse models attributed the behavioural defects of the syndrome to Tbx1 (REF. 133). The same authors also identified a frameshift mutation of TBX1 in a family with phenotypic and psychiatric features of the 22q11.2 deletion.
Most 22q11 deletions occur as de novo lesions. Studies of crossover events in affected probands confirmed that increased interchromosomal exchanges between homologous chromosomes in the regions containing low-copy repeats are associated with the deleted chromosome134,135. By contrast, normal chromosomes 22 show a pattern of recombination that is consistent with more telomeric exchanges136, as is predicted by standard recombination maps. Although this region of 22q11.2 does not seem to be a recombination hotspot, the presence of segmental duplications predisposes the region to atypical recombination events.
The reciprocal duplications of 22q11 have been identified less frequently than anticipated. This is probably due to under-ascertainment caused by insensitivity of the standard FISH technique used for detection, and also due to the indeterminate phenotype. In fact, the patients who have been identified to date seem to be phenotypically diverse, ranging from individuals with multiple defects to those who are phenotypically normal137–140.
A supernumerary, bisatellited marker chromosome comprising portions of chromosome 22 has been associated with CES — a malformation syndrome associated with a highly variable pattern of congenital anomalies. It is named for the ocular defect or pupil malformation that is seen in half of the affected individuals. The acrocentric marker chromosome of CES contains two copies of 22pter→q11.2 (chromosome 22 starting at the terminus of the short arm and ending on the long arm at region 1 band 1.2), so affected individuals carry four copies of the genes that are located in this region. CES marker chromosomes vary in size and can be asymmetrical with regard to the region that is duplicated141. The breakpoints of the CES chromosome cluster in two intervals, as shown in FIG. 1. Although the critical region for CES lies in the 2-Mb region centromeric to the VCFS and DGS region, which contains at least ten genes, the extra material in the larger CES chromosomes can extend across the 3-Mb region that is usually deleted in VCFS and DGS.
The t(11;22) translocation and Emanuel Syndrome
The t(11;22)(q23;q11.2) is the most frequently occurring non-Robertsonian, constitutional translocation in humans142,143. Carriers of the constitutional translocation t(11;22) are phenotypically normal, and are often identified after the birth of abnormal offspring with the der(22) as a supernumerary chromosome — (47,XX or XY,+der(22)t(11;22) — or upon treatment for infertility. Only one type of unbalanced karyotype is seen in live-born individuals in association with this translocation: 47,XX(Y),+der(22)t(11;22)(q11.2;q23). This is referred to as the supernumerary der(22)t(11;22) syndrome (or Emanuel syndrome).
Patients with Emanuel syndrome have a distinctive phenotype that includes severe mental retardation, facial malformations, heart defects and genital abnormalities in males. Presumably, unbalanced karyotypes with 46 chromosomes and the der(11) or the der(22) are not viable, as meiotic analysis in males has shown that the other segregants do arise144,145. The minimal overall recurrence risk for producing offspring with an unbalanced karyotype is estimated to be 1.8–5.6% (see the Gene Reviews web site). Risks vary depending on whether the mother or father of a proband is the carrier of the balanced translocation, with a slightly greater risk for meiotic malsegregation in female than in male carriers143.
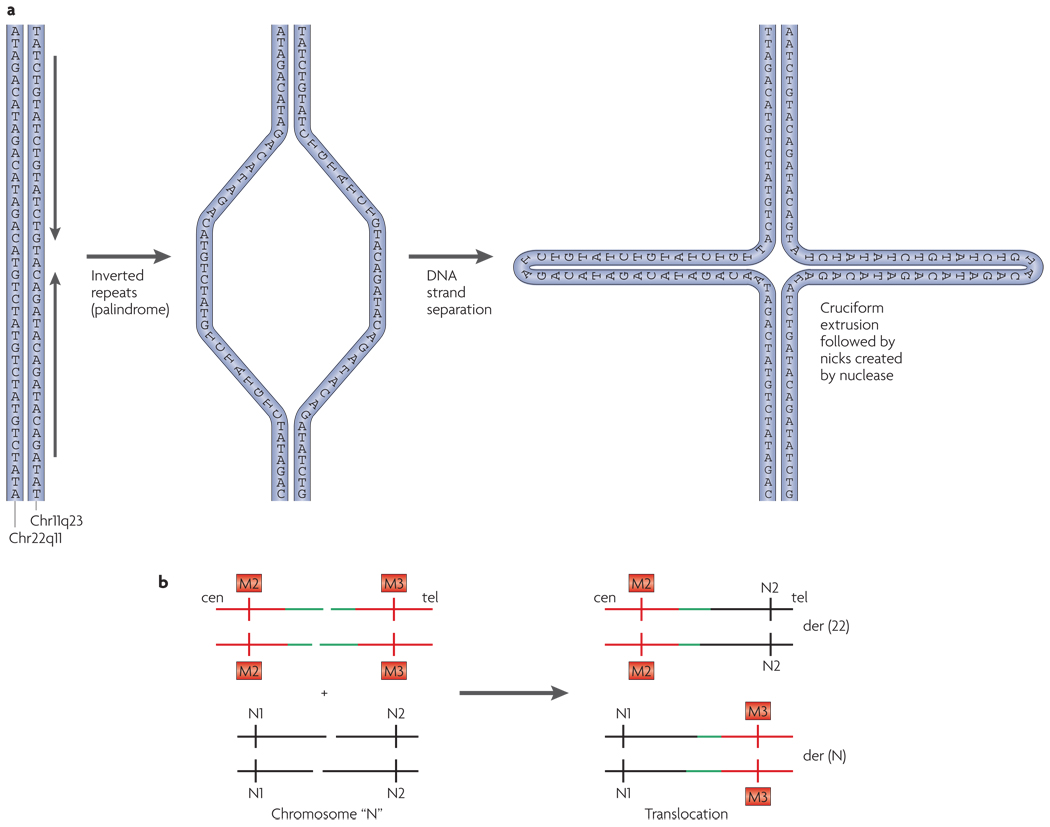
The translocation breakpoints on both chromosomes 11 and 22 are tightly clustered in multiple unrelated families137,146. Nearly identical breakpoints have been identified within palindromic AT-rich repeat regions (PATRRs) on chromosomes 11q23 and 22q11, suggesting that genomic instability is the mechanism that underlies the rearrangement146. Palindrome-mediated double-strand breaks in meiosis are thought to result in non-homologous end joining between the two chromosomes, resulting in this recurrent translocation (FIG. 6). The PATRR on chromosome 11 is polymorphic in healthy individuals; this variation seems to alter an individual’s susceptibility to producing de novo t(11;22) translocations in meiosis147. The recurrent translocation is facilitated by physical proximity between 11q23 and 22q11 during meiosis136.
Figure 6. Model for the translocation between 11q23 and 22q11.
a | The palindromic AT-rich repeat on chromosome 22q11 or 11q23 is shown as dsDNA with the arrows indicating the head-to-head inverted repeats. DNA strand separation occurs followed by cruciform extrusion. Palindromic sequences (head-to-head arrows) are predicted to form a hairpin or cruciform structure on chromosome 11, and a similar one on chromosome 22. The tips of the cruciforms may be prone to nicking by nucleases. b | Chromosome 22 with double-strand breaks within the palindrome (coloured green) could recombine with 11q23 or with another chromosome (chromosome ‘N’) that has similar double-strand breaks. This would lead to a translocation between chromosome 22 and chromosome 11 or chromosome N, leading to the formation of der(22) and a der(11) or der(N). Most often, N is chromosome 11. cen, centromere; tel, telomere.
The genomic architecture of the pericentromeric region of chromosome 22q predisposes it to recurrent deletions, duplications and translocations (FIG. 7). Recognizable phenotypes are associated with copy-number variation of the chromosome 22q11.21 region: hemizygous loss reflected by one copy in DiGeorge syndrome, and gains reflected by three copies (Emanuel syndrome) and four copies (large cat eye chromosome) each display a distinct set of clinical features, even when the same genomic interval is involved. The genomic instability of 22q11 is attributed to its large (200–500 kb), highly homologous (98–99% sequence identity), modular segmental duplications that facilitate misalignment before meiotic exchange61,96,135,137,148. BCRL modules that contain sequences related to the BCR (breakpoint cluster region) gene exist within most of the 22q11 segmental duplications149. It has recently been shown that deletion endpoints for distal deletions of 22q11.2 lie within BCRL modules61. The BCR-like sequences might serve as the substrate for many of the genomic rearrangements associated with this chromosome’s pericentromeric region. In addition, there is evidence that structural variation, such as polymorphic Alu repeats, might have a role in the non-allelic homologous recombination150 that is associated with deletions on this chromosome in particular61.
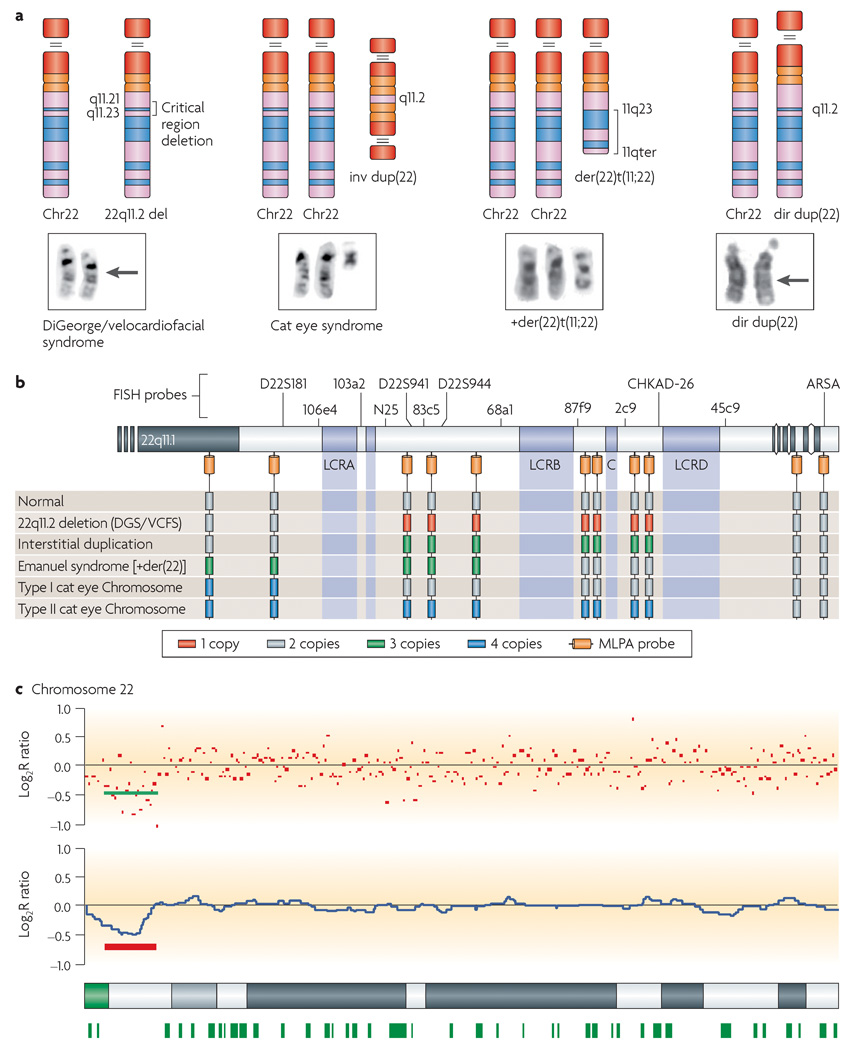
Figure 7. Ideograms and partial karyotypes of chromosome 22 abnormalities.
a | The deletion of chromosome 22q11.21–11.23 (indicated by an arrow) is associated with DiGeorge and velocardiofacial syndromes. The inv dup(22) is associated with the cat eye syndrome, the +der(22)t(11;22) — a derivative chromosome 22 that is generated by the translocation between chromosomes 11 and 22 — is associated with Emanuel syndrome. The interstitial direct duplication is not visible cytogenetically. b | A copy-number diagram for the above disorders. Multiplex ligation-dependent probe amplification (MLPA) and fluorescence in situ hybridization (FISH) probes as well as copy number for the disorders shown in panel a are indicated. c | Graphical output for copy number of SNP-based probes on chromosome 22 on the Affymetrix 50K Xba Mapping GeneChip as computed by copy-number analysis software CNAG155. Red dots represent raw log2 ratio values for each SNP. Blue curves represent copy-number inferences based on local mean analysis for ten consecutive SNPs. Heterozygous SNP calls are shown as green bars below the chromosome 22 ideogram at the bottom of the figure. For probes that are normal in copy number, the signal intensity ratio of the subject versus controls is expected to be 1 and the log2 ratio should be around 0.0 (log2 = 0). The deletion that has been detected in this patient with DiGeorge and velocardiofacial syndromes (DGS/VCFS) based on log2 ratio is underlined in red. Loss of copy number due to a deletion results in a negative log2 ratio (mean log2 ratio ~ −0.5).
Aberrant recombination and genomic disorders
Segmental duplications that flank specific regions of the genome have been shown to mediate misalignment and aberrant recombination in meiosis. Evidence for unequal meiotic exchanges in genomic regions that contain segmental duplications has been provided for a number of human deletion and reciprocal duplication syndromes including WBS (chromosome 7q11.2), PWS and Angelman syndrome (chromosome 15q11.2), SMS (chromosome 17p11.2), neurofibromatosis type I (chromosome 17q11.2), and DiGeorge syndrome and VCFS (22q11.2) (see REF. 96 for a review). A recent study of a large group of patients with WBS, PWS, Angelman syndrome, DiGeorge syndrome and VCFS found that most of the rearrangements were interchromosomal, with approximately equal numbers of maternal and paternal deletions151. The proportions of interchromosomal deletions varied among the groups, with chromosome 22q11 having the most and chromosome 15q having the least.
Studies of other genomic disorders have demonstrated the presence of chromosomal inversions involving regions that are flanked by segmental duplications. Inversions are thought to predispose a chromosome to rearrangement in meiosis17. For example, one-third of transmitting progenitors of WBS have an inversion flanked by the segmental duplications on chromosome 7q11.2 (REF. 71). Inversions involving the segmental duplications on chromosome 5q35 associated with Sotos syndrome were detected in fathers whose children carried a paternal deletion of the region66.
Regions in which recurrent and varied types of rearrangements occur show an unusual degree of genomic instability, and are presumably susceptible to double-strand breaks. Synaptonemal complexes and sites of exchange might form in response to these breaks. The presence of palindromic sequences on 22q, for example, might have a major role in susceptibility to double-strand breaks. Palindromic sequences have been implicated in double-strand breaks that mediate translocations of 22q11, including the t(11;22).
Conclusions
Segmental duplications have arisen in the primate genome, driving the process of chromosome evolution152,153. Evolutionary studies showed that the segmental duplications involved in Sotos syndrome apparently occurred before the divergence of Old World monkeys66, and those mediating the 22q deletion of DiGeorge syndrome arose around the time of the divergence of Old and New World monkeys148. WBS repeats were demonstrated to have rapidly evolved during the divergence of primates154. In general, species-specific duplications and rearrangements are associated with significant differences among the different species of primates studied. In the hominoid species, Alu elements150 were found at the ends of segmental duplications, suggesting that they have a role in facilitating the rapid evolution of these regions. The current focus on studying genome structure should significantly improve our understanding of its evolution. However, in addition to creating a dynamic, evolvable genome, these same structural elements (segmental duplications) result in instability, rearrangement and disease.
The ability to detect subtle chromosomal abnormalities has escalated progressively in recent years. With the advent of high-resolution G-banding, routine karyotyping has allowed for the ascertainment of changes that are greater than 5–10 Mb. The introduction of FISH led to the detection of many microdeletion and microduplication syndromes, as cosmid and BAC-based probes were developed and targeted to numerous known regions flanked by segmental duplications, as well as to the sub-telomeric regions of human chromosomes. The development of chromosomal CGH provided the capability to assess the entire genome for copy-number differences in a single experiment, adding to the wealth of information about genomic disorders and copy-number imbalances. The development of array CGH with BAC or oligonucleotide targets has revolutionized the ability to detect small copy-number changes, leading to a higher rate of detection of clinically relevant chromosomal abnormalities and the description of new chromosomal disorders.
In addition, the surprising degree of variability of large segments of the genome has been exposed. Application of these array-based techniques will result in the identification of new genetic syndromes and the discovery of multiple loci that cause phenotypic abnormalities when their expression levels are modified by numerous mechanisms, including duplication or deletion. Thus, array CGH has the potential to replace the light microscope as the diagnostic tool of choice in the identification of clinically relevant chromosomal and genomic changes.
Note added in proof
A recent article by Korbel, Urban and Affourtit et al. describes a large-scale sequencing approach to identify mapping structural variants in the genome156. It utilizes high-throughput and extensive paired-end mapping methods. Its ability to detect variants at a 3-kb resolution offers an improvement over the resolution of array CGH, and might ultimately allow for the identification of new genomic disorders.
DATABASES
Entrez Gene
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=gene
BCR | ELN | GTF2IRD2 | HECT | MAGEL2 | MAPT | MKRN3 | NCF1 | NDN | neurofibromin | NSD1 | PMP22 | RLD2 | SNURF-SNRPN | TBX1 | UBE3A
OMIM
http://www.ncbi.nlm.nih.gov/entrez/query. fcgi?db=OMIM
Angelman syndrome | cat eye syndrome | Charcot–Marie–Tooth Disease type 1A | conotruncal anomaly face syndrome | DiGeorge syndrome | Emanuel syndrome | hereditary neuropathy and liability to pressure palsies | neurofibromatosis type 1 | Potocki–Lupski syndrome | Prader–Willi syndrome | Smith–Magenis syndrome | Sotos syndrome | velocardiofacial syndrome | Williams–Beuren syndrome | Wolf–Hirschhorn syndrome
FURTHER INFORMATION
Gene Reviews
http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=gene.chapter.emanuel
The Sick Kids Centre for Applied Genomics
UCSC Genome Browser
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
Acknowledgements
These studies were supported in part by HL74731, CA39926, a research grant from the National Organization for Rare Disorders (NORD) and funds from the Charles E.H. Upham chair (B.S.E.). S.C.S. was supported by HL04487 and HL080637. The authors wish to thank T. Shaikh for helpful discussions and assistance with artwork. Additional assistance with the figures was provided by R. Jalali, as well as A. Hacker and R. O’Connor.
Glossary
Constitutional abnormalities
Chromosomal abnormalities that are present at birth in the somatic cells of an individual.
Uniparental Disomy
A condition whereby both members of a chromosome pair are contributed by one parent rather than one from each parent.
Translocation
The fusion or exchange of material between chromosomes. When there is no gain or loss of material, the translocation is said to be balanced; when there is a gain or loss, resulting in trisomy or monosomy for a particular chromosome segment, it is said to be unbalanced.
Pericentromeric
The region surrounding the centromere, the chromosomal region where two sister chromatids are joined.
Haploinsufficiency
The inactivation or deletion of one of two alleles in diploid cells such that insufficient protein is produced.
Hypercalcaemia
A condition that refers to an elevated calcium level in the blood.
Unbalanced karyotype
A gain or loss of genetic material, resulting in trisomy or monosomy for a particular chromosome or chromosomal segment.
Acrocentric chromosome
A chromosome in which the centromere lies very close to one end. The short (p) arms are very short and usually have small dot-like appendages on stalks, known as satellites. In humans, chromosomes 13,14,15, 21 & 22 are acrocentric.
Imprinting
The differential expression of genes depending on whether they were inherited maternally or paternally.
Supernumerary marker chromosome
A marker chromosome present in a karyotype that consists of more than 46 chromosomes.
Bisatellited marker chromosome
A marker chromosome is usually small, of unknown origin and unidentifiable from its G-banding pattern. Bisatellited markers have small round appendages that are attached by fine stalks to both ends of the marker chromosome.
Non-Robertsonian
A reciprocal translocation of material between two chromosome arms, not at the centromere of an acrocentric chromosome.
Constitutional translocations
Translocations that are present at birth in the somatic cells of an individual.
Palindrome
A DNA sequence that reads the same in both directions.
Synaptonemal complex
The structure seen in electron micrographs of paired chromosomes at the pachytene stage of meiosis. The name is derived from the word synapsis, which has been used to designate chromosome pairing. Originally it referred to the protein structure aggregating the two chromosomes.
References
- 1.Klein RJ, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dina C, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nature Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 4.Helgadottir A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 5.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conrad DF, Hurles ME. The population genetics of structural variation. Nature Genet. 2007;39:S30–S36. doi: 10.1038/ng2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iafrate AJ, et al. Detection of large-scale variation in the human genome. Nature Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 8.Sebat J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. A landmark study demonstrating genome-wide analysis of copy-number polymorphisms or variants in normal individuals. It provided an initial glimpse of the surprising amount of large-scale variation and its implications for human diversity.
- 9.Tuzun E, et al. Fine-scale structural variation of the human genome. Nature Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 10.McCarroll SA, et al. Common deletion polymorphisms in the human genome. Nature Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 11.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. The first CNV map of the human genome. This study utilized oligonucleotide arrays and hundreds of reference samples from several different human populations to show a remarkable number of variable regions in ‘normal’ individuals, underscoring the need to catalogue CNVs comprehensively.
- 12.Scherer SW, et al. Challenges and standards in integrating surveys of structural variation. Nature Genet. 2007;39:S7–S15. doi: 10.1038/ng2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey JA, et al. Recent segmental duplications in the human genome. Science. 2002;297:1003–1007. doi: 10.1126/science.1072047. This work analysed public sequence databases to detect large intervals of high sequence identity that were localized to regions of genomic instability associated with human disease and primate evolution.
- 14.Cooper GM, Nickerson DA, Eichler EE. Mutational and selective effects on copy-number variants in the human genome. Nature Genet. 2007;39:S22–S29. doi: 10.1038/ng2054. [DOI] [PubMed] [Google Scholar]
- 15.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nature Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. A comprehensive review of genome-scanning technologies and the discovery of large-scale genomic variation. Provides an excellent overview of structural variants and their contribution to genomic diversity and disease.
- 16.Stankiewicz P, Lupski JR. Genome architecture, rearrangements, and genomic disorders. Trends Genet. 2002;18:74–82. doi: 10.1016/s0168-9525(02)02592-1. [DOI] [PubMed] [Google Scholar]
- 17.Shaffer LG, Lupski JR. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- 18.Lupski JR. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 1998;14:415–420. doi: 10.1016/s0168-9525(98)01555-8. The paper that coined the phrase ‘genomic disorders’ based on the concept that structural features of the genome can lead to DNA rearrangements and human disease traits.
- 19.Sharp AJ, et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nature Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- 20.Koolen DA, et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nature Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- 21.Shaw-Smith C, et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nature Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- 22.Sharp AJ, et al. Characterization of a recurrent 15q24 microdeletion syndrome. Hum. Mol. Genet. 2007;16:567–572. doi: 10.1093/hmg/ddm016. [DOI] [PubMed] [Google Scholar]
- 23.Ballif BC, et al. Discovery of a novel microdeletion syndrome on 16p11.2p12.2. Nature Genet. 2007;39:1071–1073. doi: 10.1038/ng2107. [DOI] [PubMed] [Google Scholar]
- 24.Ledbetter DH, Ballabio A. In: The metabolic and molecular bases of inherited disease. Scriver CR, Beaudet AL, Sly WS, Valle D, editors. Vol. 1. New York: McGraw–Hill; 1995. pp. 811–839. [Google Scholar]
- 25.Levy B, Dunn TM, Kaffe S, Kardon N, Hirschhorn K. Clinical applications of comparative genomic hybridization. Genet. Med. 1998;1:4–12. doi: 10.1097/00125817-199811000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Feenstra I, Brunner HG, van Ravenswaaij CM. Cytogenetic genotype–phenotype studies: improving genotyping, phenotyping and data storage. Cytogenet. Genome Res. 2006;115:231–239. doi: 10.1159/000095919. [DOI] [PubMed] [Google Scholar]
- 27.Knight SJ, et al. An optimized set of human telomere clones for studying telomere integrity and architecture. Am. J. Hum. Genet. 2000;67:320–332. doi: 10.1086/302998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Vries BB, et al. Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J. Med. Genet. 2001;38:145–150. doi: 10.1136/jmg.38.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight SJ, Flint J. The use of subtelomeric probes to study mental retardation. Methods Cell Biol. 2004;75:799–831. doi: 10.1016/s0091-679x(04)75035-9. [DOI] [PubMed] [Google Scholar]
- 30.Kallioniemi A, et al. Comparative genomic hybridization for molecular cytogenetic analysis of solid tumors. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 31.Pinkel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nature Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 32.Snijders AM, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nature Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 33.Shaffer LG, Bejjani BA. Medical applications of array CGH and the transformation of clinical cytogenetics. Cytogenet. Genome Res. 2006;115:303–309. doi: 10.1159/000095928. [DOI] [PubMed] [Google Scholar]
- 34.Fritz B, et al. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res. 2002;62:2993–2998. [PubMed] [Google Scholar]
- 35.Wilhelm M, et al. Array-based comparative genomic hybridization for the differential diagnosis of renal cell cancer. Cancer Res. 2002;62:957–960. [PubMed] [Google Scholar]
- 36.Snijders AN, et al. Genome-wide array based comparative genomic hybridization reveals genetic homogeneity and frequent copy number increases encompassing CCNE1 in Fallopian tube carcinoma. Oncogene. 2003;22:4281–4286. doi: 10.1038/sj.onc.1206621. [DOI] [PubMed] [Google Scholar]
- 37.Squire JA, et al. High-resolution mapping of amplifications and deletions in pediatric osteosarcoma by use of CGH analysis of cDNA microarrays. Genes Chromosomes Cancer. 2003;38:215–225. doi: 10.1002/gcc.10273. [DOI] [PubMed] [Google Scholar]
- 38.Albertson DG, Collins C, McCormick F, Gray JW. Chromosome aberrations in solid tumors. Nature Genet. 2003;34:369–376. doi: 10.1038/ng1215. [DOI] [PubMed] [Google Scholar]
- 39.Vissers LE, et al. Array-based comparative genomic hybridization for the genomewide detection of submicroscopic chromosomal abnormalities. Am. J. Hum. Genet. 2003;73:1261–1270. doi: 10.1086/379977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Menten B, et al. Emerging patterns of cryptic chromosomal imbalance in patients with idiopathic mental retardation and multiple congenital anomalies: a new series of 140 patients and review of published reports. J. Med. Genet. 2006;43:625–633. doi: 10.1136/jmg.2005.039453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Vries BB, et al. Diagnostic genome profiling in mental retardation. Am. J. Hum. Genet. 2005;77:606–616. doi: 10.1086/491719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Veltman JA, et al. High-throughput analysis of subtelomeric chromosome rearrangements by use of array-based comparative genomic hybridization. Am. J. Hum. Genet. 2002;70:1269–1276. doi: 10.1086/340426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu W, et al. Development of a comparative genomic hybridization microarray and demonstration of its utility with 25 well-characterized 1p36 deletions. Hum. Mol. Genet. 2003;12:2145–2152. doi: 10.1093/hmg/ddg230. [DOI] [PubMed] [Google Scholar]
- 44.Ballif BC, et al. The clinical utility of enhanced subtelomeric coverage in array CGH. Am. J. Med. Genet. A. 2007;143:1850–1857. doi: 10.1002/ajmg.a.31842. [DOI] [PubMed] [Google Scholar]
- 45.Rauen KA, Albertson DG, Pinkel D, Cotter PD. Additional patient with del(12)(q21.2q22): further evidence for a candidate region for cardio-facio-cutaneous syndrome? Am. J. Med. Genet. 2002;110:51–56. doi: 10.1002/ajmg.10478. [DOI] [PubMed] [Google Scholar]
- 46.Veltman JA, et al. Definition of a critical region on chromosome 18 for congenital aural atresia by arrayCGH. Am. J. Hum. Genet. 2003;72:1578–1584. doi: 10.1086/375695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gunn SR, et al. Molecular characterization of a patient with central nervous system dysmyelination and cryptic unbalanced translocation between chromosomes 4q and 18q. Am. J. Med. Genet. 2003;120:127–135. doi: 10.1002/ajmg.a.20026. [DOI] [PubMed] [Google Scholar]
- 48.Hoffman JD, et al. Array based CGH and FISH fail to confirm duplication of 8p22–p23.1 in association with Kabuki syndrome. J. Med. Genet. 2005;42:49–53. doi: 10.1136/jmg.2004.024372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prescott K, et al. A novel 5q11.2 deletion detected by microarray comparative genomic hybridisation in a child referred as a case of suspected 22q11 deletion syndrome. Hum. Genet. 2005;116:83–90. doi: 10.1007/s00439-004-1195-6. [DOI] [PubMed] [Google Scholar]
- 50.Lu X, et al. Clinical implementation of chromosomal microarray analysis: summary of 2513 postnatal cases. PLoS ONE. 2007;2:e327. doi: 10.1371/journal.pone.0000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedman J, et al. Oligonucleotide microarray analysis of genomic imbalance in children with mental retardation. Am. J. Hum. Genet. 2006;79:500–513. doi: 10.1086/507471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ming JE, et al. Rapid detection of submicroscopic chromosomal rearrangements in children with multiple congenital anomalies using high density oligonucleotide arrays. Hum. Mutat. 2006;27:467–473. doi: 10.1002/humu.20322. [DOI] [PubMed] [Google Scholar]
- 53.Shaikh TH, et al. Oligonucleotide arrays for high-resolution analysis of copy number alteration in mental retardation/multiple congenital anomalies. Genet. Med. 2007;9:617–625. doi: 10.1097/gim.0b013e318148bb81. [DOI] [PubMed] [Google Scholar]
- 54.Wagenstaller J, et al. Copy-number variations measured by single-nucleotide-polymorphism oligonucleotide arrays in patients with mental retardation. Am. J. Hum. Genet. 2007;81:768–779. doi: 10.1086/521274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sharp AJ, et al. Optimal design of oligonucleotide microarrays for measurement of DNA copy number. Hum. Mol. Genet. 2007 August 28; doi: 10.1093/hmg/ddm234. (doi:10.1093/hmg/ddm234) [DOI] [PubMed] [Google Scholar]
- 56.Armour JA, Sismani C, Patsalis PC, Cross G. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res. 2000;28:605–609. doi: 10.1093/nar/28.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schouten JP, et al. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vorstman JA, et al. MLPA: a rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum. Mutat. 2006;27:814–821. doi: 10.1002/humu.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deininger PL, Batzer MA. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 60.Babushok DV, Kazazian HH., Jr Progress in understanding the biology of the human mutagen LINE-1. Hum. Mutat. 2007;28:527–539. doi: 10.1002/humu.20486. [DOI] [PubMed] [Google Scholar]
- 61.Shaikh TH, et al. Low copy repeats mediate distal chromosome 22q11.2 deletions: sequence analysis predicts breakpoint mechanisms. Genome Res. 2007;17:482–491. doi: 10.1101/gr.5986507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmickel RD. Contiguous gene syndromes: a component of recognizable syndromes. J. Pediatr. 1986;109:231–241. doi: 10.1016/s0022-3476(86)80377-8. The paper that introduced the concept of a non-traditional pattern of inheritance involving duplication or deletion of contiguous genes.
- 63.Kurotaki N, et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nature Genet. 2002;30:365–366. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 64.Kurotaki N, Stankiewicz P, Wakui K, Niikawa N, Lupski JR. Sotos syndrome common deletion is mediated by directly oriented subunits within inverted Sos-REP low-copy repeats. Hum. Mol. Genet. 2005;14:535–542. doi: 10.1093/hmg/ddi050. [DOI] [PubMed] [Google Scholar]
- 65.Tatton-Brown K, et al. Multiple mechanisms are implicated in the generation of 5q35 microdeletions in Sotos syndrome. J. Med. Genet. 2005;42:307–313. doi: 10.1136/jmg.2004.027755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Visser R, et al. Identification of a 3.0-kb major recombination hotspot in patients with Sotos syndrome who carry a common 1.9-Mb microdeletion. Am. J. Hum. Genet. 2005;76:52–67. doi: 10.1086/426950. A detailed analysis of breakpoint junctions within segmental duplications for Sotos syndrome. This paper identified the influence of both sequence and orientation on hotspots for mediating aberrant recombination.
- 67.Kirchhoff M, Bisgaard AM, Bryndorf T, Gerdes T. MLPA analysis for a panel of syndromes with mental retardation reveals imbalances in 5.8% of patients with mental retardation and dysmorphic features, including duplications of the Sotos syndrome and Williams–Beuren syndrome regions. Eur. J. Med. Genet. 2007;50:33–42. doi: 10.1016/j.ejmg.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 68.Chen CP, et al. Molecular cytogenetic analysis of de novo dup(5)(q35.2q35.3) and review of the literature of pure partial trisomy 5q. Am. J. Med. Genet. A. 2006;140:1594–1600. doi: 10.1002/ajmg.a.31329. [DOI] [PubMed] [Google Scholar]
- 69.Tassabehji M, Urban Z. Congenital heart disease: molecular diagnostics of supravalvular aortic stenosis. Methods Mol. Med. 2006;126:129–156. doi: 10.1385/1-59745-088-X:129. [DOI] [PubMed] [Google Scholar]
- 70.Merla G, et al. Submicroscopic deletion in patients with Williams–Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am. J. Hum. Genet. 2006;79:332–341. doi: 10.1086/506371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Osborne LR, et al. A 1.5 million-base pair inversion polymorphism in families with Williams–Beuren syndrome. Nature Genet. 2001;29:321–325. doi: 10.1038/ng753. The first study to demonstrate the presence of inversion polymorphisms in regions flanked by segmental duplications. This suggested that genomic polymorphism is an important contributor to disease-associated chromosomal rearrangements, which were previously thought to be random.
- 72.Scherer SW, et al. Observation of a parental inversion variant in a rare Williams–Beuren syndrome family with two affected children. Hum. Genet. 2005;117:383–388. doi: 10.1007/s00439-005-1325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.DePienne C, et al. Autism, language delay and mental retardation in a patient with 7q11 duplication. J. Med. Genet. 2007;44:452–458. doi: 10.1136/jmg.2006.047092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Somerville MJ, et al. Severe expressive-language delay related to duplication of the Williams–Beuren locus. N. Engl. J. Med. 2005;353:1694–1701. doi: 10.1056/NEJMoa051962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mombaerts P. Odorant receptor genes in humans. Curr. Opin. Genet. Dev. 1999;9:315–320. doi: 10.1016/s0959-437x(99)80047-1. [DOI] [PubMed] [Google Scholar]
- 76.Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc. Natl Acad. Sci. USA. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Floridia G, et al. The same molecular mechanism at the maternal meiosis I produces mono- and dicentric 8p duplications. Am. J. Hum. Genet. 1996;58:785–796. [PMC free article] [PubMed] [Google Scholar]
- 78.Devriendt K, et al. Delineation of the critical deletion region for congenital heart defects, on chromosome 8p23.1. Am. J. Hum. Genet. 1999;64:1119–1126. doi: 10.1086/302330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pehlivan T, et al. GATA4 haploinsufficiency in patients with interstitial deletion of chromosome region 8p23.1 and congenital heart disease. Am. J. Med. Genet. 1999;83:201–206. [PubMed] [Google Scholar]
- 80.Giglio S, et al. Deletion of a 5-cM region at chromosome 8p23 is associated with a spectrum of congenital heart defects. Circulation. 2000;102:432–437. doi: 10.1161/01.cir.102.4.432. [DOI] [PubMed] [Google Scholar]
- 81.Ohashi H, et al. A stable acentric marker chromosome: possible existence of an intercalary ancient centromere at distal 8p. Am. J. Hum. Genet. 1994;55:1202–1208. [PMC free article] [PubMed] [Google Scholar]
- 82.Giglio S, et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms, and common chromosome rearrangements. Am. J. Hum. Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Giglio S, et al. Heterozygous submicroscopic inversions involving olfactory receptor-gene clusters mediate the recurrent t(4;8)(p16;p23) translocation. Am. J. Hum. Genet. 2002;71:276–285. doi: 10.1086/341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Faivre L, et al. Prenatal diagnosis of an 8p23.1 deletion in a fetus with a diaphragmatic hernia and review of the literature. Prenat. Diagn. 1998;18:1055–1060. doi: 10.1002/(sici)1097-0223(1998100)18:10<1055::aid-pd405>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 85.Lopez I, et al. Prenatal diagnosis of de novo deletions of 8p23.1 or 15q26.1 in two fetuses with diaphragmatic hernia and congenital heart defects. Prenat. Diagn. 2006;26:577–580. doi: 10.1002/pd.1468. [DOI] [PubMed] [Google Scholar]
- 86.Zollino M, et al. Mapping the Wolf–Hirschhorn syndrome phenotype outside the currently accepted WHS critical region and defining a new critical region, WHSCR-2. Am. J. Hum. Genet. 2003;72:590–597. doi: 10.1086/367925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rodríguez L, et al. The new Wolf–Hirschhorn syndrome critical region (WHSCR-2): a description of a second case. Am. J. Med. Genet. 2005;136:175–178. doi: 10.1002/ajmg.a.30775. [DOI] [PubMed] [Google Scholar]
- 88.Butler MG, Meaney FJ, Palmer CG. Clinical and cytogenetic survey of 39 individuals with Prader–Labhart–Willi syndrome. Am. J. Med. Genet. 1986;23:793–809. doi: 10.1002/ajmg.1320230307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nicholls RD, Knoll JH, Butler MG, Karam S, Lalande M. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader–Willi syndrome. Nature. 1989;342:281–285. doi: 10.1038/342281a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Robinson WP, et al. Molecular, cytogenetic, and clinical investigations of Prader–Willi syndrome patients. Am. J. Hum. Genet. 1991;49:1219–1234. [PMC free article] [PubMed] [Google Scholar]
- 91.Lalande M, Calciano MA. Molecular epigenetics of Angelman syndrome. Cell. Mol. Life Sci. 2007;64:947–960. doi: 10.1007/s00018-007-6460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sahoo T, et al. Identification of novel deletions of 15q11q13 in Angelman syndrome by array-CGH: molecular characterization and genotype–phenotype correlations. Eur. J. Hum. Genet. 2007;15:943–949. doi: 10.1038/sj.ejhg.5201859. [DOI] [PubMed] [Google Scholar]
- 93.Smeets DF, et al. Prader–Willi syndrome and Angelman syndrome in cousins from a family with a translocation between chromosomes 6 and 15. N. Engl. J. Med. 1992;326:807–811. doi: 10.1056/NEJM199203193261206. [DOI] [PubMed] [Google Scholar]
- 94.Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH. Large genomic duplicons map to sites of instability in the Prader–Willi/Angelman syndrome chromosome region (15q11–q13) Hum. Mol. Genet. 1999;8:1025–1037. doi: 10.1093/hmg/8.6.1025. [DOI] [PubMed] [Google Scholar]
- 95.Amos-Landgraf JM, et al. Chromosome breakage in the Prader–Willi and Angelman syndromes involves recombination between large, transcribed repeats at proximal and distal breakpoints. Am. J. Hum. Genet. 1999;65:370–386. doi: 10.1086/302510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Emanuel BS, Shaikh TH. Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nature Rev. Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- 97.Lee S, Wevrick R. Identification of novel imprinted transcripts in the Prader–Willi syndrome and Angelman syndrome deletion region: further evidence for regional imprinting control. Am. J. Hum. Genet. 2000;66:848–858. doi: 10.1086/302817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chai JH, et al. Identification of four highly conserved genes between breakpoint hotspots BP1 and BP2 of the Prader–Willi/Angelman syndromes deletion region that have undergone evolutionary transposition mediated by flanking duplicons. Am. J. Hum. Genet. 2003;73:898–925. doi: 10.1086/378816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Buiting K, et al. C15orf2 and a novel noncoding transcript from the Prader–Willi/Angelman syndrome region show monoallelic expression in fetal brain. Genomics. 2007;89:588–595. doi: 10.1016/j.ygeno.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 100.Schulze A, et al. Exclusion of SNRPN as a major determinant of Prader–Willi syndrome by a translocation breakpoint. Nature Genet. 1996;12:452–454. doi: 10.1038/ng0496-452. [DOI] [PubMed] [Google Scholar]
- 101.Jiang Y, Lev-Lehman E, Bressler J, Tsai TF, Beaudet AL. Genetics of Angelman syndrome. Am. J. Hum. Genet. 1999;65:1–6. doi: 10.1086/302473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang NJ, Liu D, Parokonny AS, Schanen NC. High-resolution molecular characterization of 15q11–q13 rearrangements by array comparative genomic hybridization (array CGH) with detection of gene dosage. Am. J. Hum. Genet. 2004;75:267–281. doi: 10.1086/422854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wandstrat AE, Schwartz S. Isolation and molecular analysis of inv dup(15) and construction of a physical map of a common breakpoint in order to elucidate their mechanism of formation. Chromosoma. 2000;109:498–505. doi: 10.1007/s004120000103. [DOI] [PubMed] [Google Scholar]
- 104.Wandstrat AE, Leana-Cox J, Jenkins L, Schwartz S. Molecular cytogenetic evidence for a common breakpoint in the largest inverted duplications of chromosome 15. Am. J. Hum. Genet. 1998;62:925–936. doi: 10.1086/301777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Robinson WP, et al. Uniparental disomy explains the occurrence of the Angelman or Prader–Willi syndrome in patients with an additional small inv dup(15) chromosome. J. Med. Genet. 1993;30:756–760. doi: 10.1136/jmg.30.9.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheng SD, Spinner NB, Zackai EH, Knoll JH. Cytogenetic and molecular characterization of inverted duplicated chromosomes 15 from 11 patients. Am. J. Hum. Genet. 1994;55:753–759. [PMC free article] [PubMed] [Google Scholar]
- 107.Leana-Cox J, et al. Molecular cytogenetic analysis of inv dup(15) chromosomes, using probes specific for the Prader–Willi/Angelman syndrome region: clinical implications. Am. J. Hum. Genet. 1994;54:748–755. [PMC free article] [PubMed] [Google Scholar]
- 108.Mignon C, et al. Clinical heterogeneity in 16 patients with inv dup 15 chromosome: cytogenetic and molecular studies, search for an imprinting effect. Eur. J. Hum. Genet. 1996;4:88–100. doi: 10.1159/000472176. [DOI] [PubMed] [Google Scholar]
- 109.Lupski JR. Genomic rearrangements and sporadic disease. Nature Genet. 2007;39:S43–S47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 110.Smith AC, et al. Interstitial deletion of (17)(p11.2p11.2) in nine patients. Am. J. Med. Genet. 1986;24:393–414. doi: 10.1002/ajmg.1320240303. [DOI] [PubMed] [Google Scholar]
- 111.Stratton RF, et al. Interstitial deletion of (17)(p11.2p11.2): report of six additional patients with a new chromosome deletion syndrome. Am. J. Med. Genet. 1986;24:421–432. doi: 10.1002/ajmg.1320240305. [DOI] [PubMed] [Google Scholar]
- 112.Chen KS, et al. Homologous recombination of a flanking repeat gene cluster is a mechanism for a common contiguous gene deletion syndrome. Nature Genet. 1997;17:154–163. doi: 10.1038/ng1097-154. [DOI] [PubMed] [Google Scholar]
- 113.Potocki L, et al. Characterization of Potocki–Lupski syndrome (dup(17)(p11.2p11.2)) and delineation of a dosage-sensitive critical interval that can convey an autism phenotype. Am. J. Hum. Genet. 2007;80:633–649. doi: 10.1086/512864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shaw CJ, Withers MA, Lupski JR. Uncommon deletions of the Smith–Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates. Am. J. Hum. Genet. 2004;75:75–81. doi: 10.1086/422016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bi W, et al. Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am. J. Hum. Genet. 2003;73:1302–1315. doi: 10.1086/379979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shaw CJ, Lupski JR. Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum. Mol. Genet. 2004;13:R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- 117.Reiter LT, et al. A recombination hotspot responsible for two inherited peripheral neuropathies is located near a mariner transposon-like element. Nature Genet. 1996;12:288–297. doi: 10.1038/ng0396-288. Erratum in: Nature Genet. 19, 303 (1998). [DOI] [PubMed] [Google Scholar]
- 118.Reiter LT, et al. Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am. J. Hum. Genet. 1998;62:1023–1033. doi: 10.1086/301827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dorschner MO, Sybert VP, Weaver M, Pletcher BA, Stephens K. NF1 microdeletion breakpoints are clustered at flanking repetitive sequences. Hum. Mol. Genet. 2000;9:35–46. doi: 10.1093/hmg/9.1.35. [DOI] [PubMed] [Google Scholar]
- 120.Lopez-Correa C, et al. Recombination hotspot in NF1 microdeletion patients. Hum. Mol. Genet. 2001;10:1387–1392. doi: 10.1093/hmg/10.13.1387. [DOI] [PubMed] [Google Scholar]
- 121.Stary A, Sarasin A. Molecular analysis of DNA junctions produced by illegitimate recombination in human cells. Nucleic Acids Res. 1992;20:4269–4274. doi: 10.1093/nar/20.16.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hyrien O. Mechanisms and consequences of replication fork arrest. Biochimie. 2000;82:5–17. doi: 10.1016/s0300-9084(00)00344-8. [DOI] [PubMed] [Google Scholar]
- 123.Badge RM, Yardley J, Jeffreys AJ, Armour JA. Crossover breakpoint mapping identifies a subtelomeric hotspot for male meiotic recombination. Hum. Mol. Genet. 2000;9:1239–1244. doi: 10.1093/hmg/9.8.1239. [DOI] [PubMed] [Google Scholar]
- 124.Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN. Translocation and gross deletion breakpoints in human inherited disease and cancer I: nucleotide composition and recombination-associated motifs. Hum. Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- 125.Gerdes M, et al. Cognitive and behavioral profile of preschool children with chromosome 22q11.2. Am. J. Med. Genet. 1999;85:127–133. [PubMed] [Google Scholar]
- 126.McDonald-McGinn DM, et al. The 22q11.2 deletion: screening for deletion, diagnostic workup, and outcome of results. Report on 181 patients. Genet. Test. 1997;1:99–108. doi: 10.1089/gte.1997.1.99. [DOI] [PubMed] [Google Scholar]
- 127.Swillen A, Vogels A, Devriendt K, Fryns JP. Chromosome 22q11 deletion syndrome: update and review of the clinical features, cognitive-behavioral spectrum, and psychiatric complications. Am. J. Med. Genet. 2000;97:128–135. doi: 10.1002/1096-8628(200022)97:2<128::aid-ajmg4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 128.Emanuel BS, McDonald-McGinn D, Saitta SC, Zackai EH. The 22q11.2 deletion syndrome. Adv. Pediatr. 2001;48:39–73. [PubMed] [Google Scholar]
- 129.Jerome LA, Papaioannou VE. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nature Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 130.Lindsay EA, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 131.Merscher S, et al. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 132.Yagi H, et al. Role of TBX1 in human del22q11.2 syndrome. Lancet. 2003;362:1366–1373. doi: 10.1016/s0140-6736(03)14632-6. [DOI] [PubMed] [Google Scholar]
- 133.Paylor R, et al. Tbx1 haploinsufficiency is linked to behavioral disorders in mice and humans: implications for 22q11 deletion syndrome. Proc. Natl Acad. Sci. USA. 2006;103:7729–7734. doi: 10.1073/pnas.0600206103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Baumer A, et al. High level of unequal meiotic crossovers at the origin of the 22q11.2 and 7q11.23 deletions. Hum. Mol. Genet. 1998;7:887–894. doi: 10.1093/hmg/7.5.887. [DOI] [PubMed] [Google Scholar]
- 135.Saitta SC, et al. Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum. Mol. Genet. 2004;13:417–428. doi: 10.1093/hmg/ddh041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ashley T, et al. Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22) Am. J. Hum. Genet. 2006;79:524–538. doi: 10.1086/507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Edelmann L, et al. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum. Mol. Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. A demonstration of the role of segmental duplications in promoting multiple different rearrangements of chromosome 22, including the first 22q11.2 duplications.
- 138.Ensenauer RE, et al. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am. J. Hum. Genet. 2003;73:1027–1040. doi: 10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yobb TM, et al. Microduplication and triplication of 22q11.2: a highly variable syndrome. Am. J. Hum. Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Alberti A, et al. 1.5 Mb de novo 22q11.21 microduplication in a patient with cognitive deficits and dysmorphic facial features. Clin. Genet. 2007;71:177–182. doi: 10.1111/j.1399-0004.2007.00750.x. [DOI] [PubMed] [Google Scholar]
- 141.Fraccaro M, Lindsten J, Ford CE, Iselius L. The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum. Genet. l980;56:2l–5l. doi: 10.1007/BF00281567. [DOI] [PubMed] [Google Scholar]
- 142.Mears AJ, et al. Molecular characterization of the marker chromosome associated with cat eye syndrome. Am. J. Hum. Genet. 1994;55:134–142. [PMC free article] [PubMed] [Google Scholar]
- 143.Zackai EH, Emanuel BS. Site specific reciprocal translocation, t(11;22)(q23;q11) in several unrelated families with 3:l meiotic disjunction. Am. J. Med. Genet. l980;7:507–52l. doi: 10.1002/ajmg.1320070412. [DOI] [PubMed] [Google Scholar]
- 144.Martin RH, et al. Analysis of human sperm chromosome complements from a male heterozygous for a reciprocal translocation t(11;22)(q23;q11) Clin. Genet. 1984;25:357–361. doi: 10.1111/j.1399-0004.1984.tb02004.x. [DOI] [PubMed] [Google Scholar]
- 145.Armstrong SJ, Goldman AS, Speed RM, Hulten MA. Meiotic studies of a human male carrier of the common translocation, t(11;22), suggests postzygotic selection rather than preferential 3:1 MI segregation as the cause of liveborn offspring with an unbalanced translocation. Am. J. Hum. Genet. 2000;67:601–609. doi: 10.1086/303052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kurahashi H, et al. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 147.Kato T, et al. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. The first study to demonstrate that genomic sequence variation has an important role in influencing the frequency of chromosomal rearrangements.
- 148.Shaikh TS, et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. This study provides detailed analysis of the complex modular segmental duplication that mediate the most frequently occurring microdeletion syndrome in humans.
- 149.Budarf M, Canaani E, Emanuel BS. Linear order of the four BCR-related loci in 22q11. Genomics. 1988;3:168–171. doi: 10.1016/0888-7543(88)90149-8. [DOI] [PubMed] [Google Scholar]
- 150.Bailey JA, Liu G, Eichler EE. An Alu transposition model for the origin and expansion of human segmental duplications. Am. J. Hum. Genet. 2003;73:823–834. doi: 10.1086/378594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Thomas NS, et al. Parental and chromosomal origins of microdeletion and duplication syndromes involving 7q11.23, 15q11–q13 and 22q11. Eur. J. Hum. Genet. 2006;14:831–837. doi: 10.1038/sj.ejhg.5201617. [DOI] [PubMed] [Google Scholar]
- 152.Bailey JA, Eichler EE. Primate segmental duplications: crucibles of evolution, diversity and disease. Nature Rev. Genet. 2006;7:552–564. doi: 10.1038/nrg1895. A recent review of the associations among segmental duplications, genomic instability and large-scale variation. The authors discuss evidence for distinct waves of duplication during primate evolution and the creation of novel gene families.
- 153.Johnson ME, et al. Recurrent duplication-driven transposition of DNA during hominoid evolution. Proc. Natl Acad. Sci. USA. 2006;103:17626–17631. doi: 10.1073/pnas.0605426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Antonell A, de Luis O, Domingo-Roura X, Pérez-Jurado LA. Evolutionary mechanisms shaping the genomic structure of the Williams–Beuren syndrome chromosomal region at human 7q11.23. Genome Res. 2005;15:1179–1188. doi: 10.1101/gr.3944605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Nannya, et al. A robust algorithm for copy number detection using high-density oligonucleotide single nucleotide polymorphism genotyping arrays. Cancer Res. 2005;65:6071–6079. doi: 10.1158/0008-5472.CAN-05-0465. [DOI] [PubMed] [Google Scholar]
- 156.Korbel JO, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007 September 27; doi: 10.1126/science.1149504. (doi:10.1126/science.1149504) [DOI] [PMC free article] [PubMed] [Google Scholar]