Comparison of Self-Collected and Physician-Collected Vaginal Swabs for Microbiome Analysis (original) (raw)
Abstract
To our knowledge, no data are available on whether the microbial species composition and abundance sampled with self-collected vaginal swabs are comparable to those of swabs collected by clinicians. Twenty healthy women were recruited to the study during a routine gynecological visit. Eligible women were between 18 and 40 years old with regular menstrual cycles. Participants self-collected a vaginal swab using a standardized protocol and then were examined by a physician, who collected an additional five swabs from the lateral wall of the mid-vagina. In this study, the self-collected and three physician-obtained swabs were analyzed and compared using terminal restriction fragment length polymorphism and sequence analyses of the 16S rRNA genes. Vaginal microbial community comparative statistical analyses of both T-RFLP and 16S rRNA gene sequence datasets revealed that self-collected vaginal swabs sampled the same microbial diversity as physician collected swabs of the mid-vagina. These findings enable large-scale, field-based studies of the vaginal microbiome.
In recent years, “cultivation-independent” methods based on the analysis of 16S rRNA gene sequences directly extracted from biological samples are widely used to explore microbial diversity in various habitats (7, 27, 28). Using these methods, organisms are classified based on phylogenetic differences that are reflected in sequence polymorphisms of their 16S rRNA genes. The use of these approaches obviates the need to cultivate organisms, permits high-throughput analysis of samples, and provides precise and detailed information about the populations present. Using 16S rRNA gene analysis, the species (phylotype) composition and abundance in microbial communities can be readily determined, and similarities and differences among microbial communities can be quantitatively discerned. Despite difficulties in comparing data generated by different so-called “universal” PCR primer pairs, each introducing small biases (8, 36), this validated method has become the favored approach to characterizing the mutualistic microbial populations residing on and in the human body, including the gastrointestinal tract (4, 37), skin (4, 10), subgingival crevice (19), and vagina (4, 9, 15, 42). Importantly, the data obtained can be statistically analyzed to test the significance of changes that occur within individuals over time, or between individuals and treatment groups. These methods include terminal restriction fragment length polymorphisms (T-RFLP) (41) and high-throughput pyrosequencing of bar-coded 16S rRNA gene analysis (12, 22).
Surveys of vaginal microbial communities using cultivation-independent methods have been initiated under the NIH Roadmap Human Microbiome Project (14). Surveys of the vagina are important for a number of reasons. The beneficial effects of the endogenous microbiota on women's health in obstetric and gynecologic outcomes are numerous but poorly understood. A proper understanding of community membership, relative abundance, and variations therein are critical for recognizing potential pathogens and the physiological processes in microbial communities that are protective. Significant alterations or disruptions of the vagina's microbiota, such as in the clinical syndrome bacterial vaginosis (BV), may increase a woman's risk to invasion by infectious agents. The data from several prospective studies indicate that BV is a biological risk factor for adverse outcomes, including the acquisition and transmission of human immunodeficiency virus (HIV) (5) and the development of pelvic inflammatory disease (26).
To our knowledge, no data are available on the representation of microbial species composition and abundance in self-collected swabs compared to physician-collected swabs of the mid-vagina. A study of clinician-obtained samples reported by Kim et al. (17) showed heterogeneity in microbial population across the cervix, fornix, and outer vaginal canal within a woman. However, self-collected vaginal swabs of the mid-vagina are the only practical and financially feasible method to use for sampling in field-based longitudinal cohort studies. Frequent sampling by a clinician (weekly or more often) is not practicable, results in lower enrollment rates and higher rates of loss to follow-up. In addition, women often prefer self-collected over clinician-collected vaginal specimens (23). Several studies have shown that self-collection of vaginal swabs is sufficient for the detection of microbial pathogens (23, 29, 32, 40), diagnostic accuracy (31), and morphotype-specific validity in Gram staining (1, 25, 34). The present study sought to evaluate if self-collected swabs of the mid-vagina reflect the same microbial diversity as physician-collected swabs, thus enabling large-scale field-based microbiomic studies of the vaginal ecosystem.
MATERIALS AND METHODS
Sample collection and study design.
Women were recruited during a routine gynecological visit at Grady Memorial Hospital in Atlanta, GA. Eligible women were healthy, between 18 and 40 years old, and reported regular menstrual cycles between 21 to 35 days long. Women who were pregnant, had taken antibiotics or antifungal drugs in the past 30 days, had severe illnesses such as kidney failure, diabetes or HIV/AIDS, or who, in the 48 h prior to sample collection, were sexually active, had used douches, vaginal medications or suppositories, feminine sprays, genital wipes or contraceptive spermicides were ineligible for the study. The protocol was approved by the Institutional Review Board at Emory University School of Medicine, Grady Memorial Hospital and the University of Maryland School of Medicine. Guidelines of the universities were followed in the conduct of the clinical research. The study was registered at clinicaltrial.gov under identifier NCT00576797.
After giving written informed consent, participants completed a questionnaire on their medical history and sexual health. Standard pelvic examination was performed by the study physician. In total, six vaginal swabs were obtained from each participant using individual sterile Dacron swabs. The order of swab collection was as follows. First, participants were instructed by the study coordinator to insert the vaginal swab 1 to 2 in. into the vagina, twisting the swab to collect material on all sides of the tip, wipe in several full circles on the vaginal wall, keep the swab in the vagina for 20 s, and then carefully remove the swab and place it in a sterile vial. After the self-collection, the clinician conducted a pelvic exam and inserted a speculum lubricated with sterile saline. The clinician obtained five swabs from the lateral wall of the mid-vagina. All swabs were immediately placed in sterile cryovials (Sarstead tubes, 5 ml) containing 3 ml of sterile prereduced anaerobically sterilized medium (PRAS; Anaerobes Systems, Inc.). The swabs were placed immediately on dry ice and then stored at −80°C within 4 h. For the purpose of the present study, one self-collected and three physician-obtained swabs were analyzed and compared (total of 80 samples). The demographic characteristics and self-reported history of the subjects is shown in Table S1 in the supplemental material.
DNA extraction.
The swabs were thawed on ice before analysis and vortexed vigorously for 5 min to resuspend the cells. A 0.5-ml aliquot was transferred to a sterile 2.0-ml tube and stored on ice. Cell lysis was initiated by adding 50 μl of lysozyme (10 mg/ml), 6 μl of mutanolysin (25,000 U/ml; Sigma-Aldrich), 3 μl of lysostaphin (4,000 U/ml in sodium acetate; Sigma-Aldrich), and 41 μl of TE50 buffer (10 mM Tris-HCl, 50 mM EDTA [pH 8.0]). After a 1-h incubation at 37°C, 10 μl of proteinase K (20 mg/ml), 100 μl of 10% sodium dodecyl sulfate, and 20 μl of RNase A (20 mg/ml) were added to the mixture, followed by incubation for 1 h at 55°C. The sample was then transferred to a FastPrep lysing matrix B tube (Bio 101), and microbial cells were lysed by mechanical disruption using a bead beater (FastPrep instrument; Qbiogene) set at 6.0 m/s for 30 s. The lysate was processed by using a QIAamp DNA minikit (Qiagen) according to the manufacturer's recommendation but omitting the lysis steps. The samples were eluted with 50 μl of hot molecular biology-grade water (56°C).
T-RFLP analysis of 16S rRNA genes.
For the analysis of T-RFLPs of 16S rRNA genes, internal regions of 16S rRNA genes in each sample were amplified in two separate reactions using fluorescently labeled primer pairs, 8fm-926r and 49f-926r (based on the Escherichia coli position). Primer 8fm was labeled with VIC, 49f was labeled with NED and 926r was labeled with 6-FAM (Applied Biosystems, Foster City, CA). DNA amplification and T-RFLP analysis was performed as previously described (42). T-RFLP profiles were determined by using an ABI 3730xl DNA analyzer and GeneScan software (Applied Biosystems).
Pyrosequencing of bar-coded 16S rRNA gene amplicons.
The two universal primers 27F and 338R were used for PCR amplification of the V1-V2 hypervariable regions of the 16S rRNA gene (12). The 338R primer included a unique sequence tag to bar-code each sample. The primers used were 27F (5′-GCCTTGCCAGCCCGCTCAGTCAGAGTTTGATCCTGGCTCAG-3′) and 338R (5′- GCCTCCCTCGCGCCATCAGNNNNNNNNCATGCTGCCTCCCGTAGGAGT-3′), where the underlined sequences are the 454 Life Sciences FLX sequencing primers B and A in 27F and 338R, respectively, and the boldfacing denotes the universal 16S rRNA primers 27F and 338R. The 8-bp bar code within primer 338R is denoted by 8 Ns. 16S rRNA genes was amplified in 96-well microtiter plates by using AmpliTaq DNA polymerase (Applied Biosystems), and 50 ng of template DNA in a total reaction volume of 50 μl. Reactions were run in a PTC-100 thermal controller (MJ Research) using the following cycling parameters: 5 min of denaturation at 95°C; followed by 20 cycles of 30 s at 95°C (denaturing), 30 s at 56°C (annealing), and 90 s at 72°C (elongation); with a final extension at 72°C for 7 min. Negative controls without a template were included for each reactions. The presence of amplicons was confirmed by gel electrophoresis on a 2% agarose gel and staining with Sybr green. PCR products were quantified by using a GelDoc quantification system (Bio-Rad), and equimolar amounts (∼100 ng) of the PCR amplicons were mixed in a single tube. Amplification primers and reaction buffer were removed by processing the amplicons mixture with the AMPure kit (Agencourt). The purified amplicon mixtures were sequenced by 454 FLX pyrosequencing using 454 Life Sciences primer A by the Genomics Resource Center at the Institute for Genome Sciences, University of Maryland School of Medicine, using protocols recommended by the manufacturer as amended by the Center.
Statistical analyses of T-RFLP patterns.
Analyses were performed on individual fragment length values to observe whether self-collected samples affect intrasubject agreement in peak area values. This approach permits investigation of whether differences in self-collected samples can lead to false identification of microbial species. One-way analyses of variance (ANOVA) were performed separately for the peak area values for each fragment length with subjects as the treatment factor. The self-collected and clinician-collected samples were combined for each subject in the analyses, so if self-collected samples differed from clinician-collected samples, an insignificant ANOVA would tend to result. For each ANOVA, the intraclass correlation (ICC) for subjects was calculated (18). The ICC can be used as a measure of how well profiles of subjects can be distinguished based on the presence and abundance of a given fragment. For fragment lengths with an ICC near 1, it is expected that self-collected samples will agree with clinician-collected samples, since the subjects differ from each other. Lower ICC values are of particular interest for trying to detect lack of agreement between self-collected and clinician-collected samples, so peak areas for fragment lengths with low ICC values were examined for possible disagreement. Power calculations were also performed to quantify the ability to detect differences between self-collected and clinician-collected samples from these data.
16S rRNA gene sequence community composition comparative analysis.
Sequences were binned by samples using the sample-specific bar-code sequences and trimmed (removal of the bar-code and primer sequences). We used criteria previously described to assess the quality of sequence reads. Briefly, to pass, a sequence read (i) included a perfect match to the sequence tag (bar code) and the 16S rRNA gene primer, (ii) was at least 200 bp in length, (iii) had no more than two undetermined bases, and (iv) had a least a 60% match to a previously determined 16S rRNA gene sequence. Genus level taxonomic assignments were performed by using the RDP Classifier (2), and further species level assignments for Lactobacillus sp. were done using 127 HMM Lactobacillus species models followed by clustering analysis. The relative abundance of taxa within a sample were used to test whether or not self-collected and physician-collected samples belonged to the same distribution. The samples of self-collected (SC) and physician-collected (PC) relative taxa abundances came from the same distribution if the distances d(SC,SC), d(PC,PC), and d(SC,PC) came from the same distribution. Since only one self-collected sample per woman was available, we could not draw any inference about the distribution of d(SC,SC). Therefore, we tested the hypothesis that d(PC,PC) and d(SC,PC) were drawn from the same distribution by applying the Kolmogorov-Smirnov test (3). To boost the power of the test, we pooled the distances of physician-collected and self-collected across all women. If the Kolmogorov-Smirnov test rejects the null of equal distributions, then either SC and PC do not come from the same distribution or the covariances of these samples for different woman were substantially different and pooling of the corresponding distances would be responsible for the test failure. However, if the test failed to reject the null hypothesis, i.e., that pooled d(PC,PC) and d(SC,PC) samples came from the same distribution, then the result of the test was evidence supporting the hypothesis that SC and PC came from the same distribution and that the variance of physician-collected samples and covariance of self-collected versus physician-collected samples were not significantly different across all women.
Clustering of vaginal microbial communities based on taxa composition and abundance was performed by using the Ward hierarchical clustering method (39).
RESULTS
T-RFLP analysis.
ANOVA and ICC analyses were done to compare the T-RFLP profiles of SC and PC samples. A total of 1,450 fragment lengths were present in the data set that included four samples from 20 subjects. Of these 1,450 fragment lengths, 772 had discordant within-subject peak areas as measured by an ICC value less than or equal to zero. Of these 772 fragment lengths, typically the zero or negative ICC value resulted from having nearly every sample with a zero fragment length but a few that were nonzero (see Fig. S1 in the supplemental material). We examined whether an excess of these nonzero outlier values were due to the self-collected samples. The 772 discordant fragment lengths had a total of 1,482 nonzero fragment length values, of which 312 were from self-collected samples. Thus, 21% of these values were from self-collected samples, less than the 25% than would be expected by chance (z = −3.51, P < 0.0001). These results demonstrate that self-collected samples do not contribute disproportionally to outlier samples that are observed for peak area values and support the conclusion that vaginal communities sampled using self-collection are similar to those collected by a physician. Power calculations were conducted (21) based on the number of discordant fragment lengths. Since two primer pairs were used, a sample size of 1,482/2 = 741 was used. For a sample size of 741, a test of a binomial proportion with a null value of 0.25 will have a power of 0.87 to detect a true proportion of ≥0.295. Thus, the number of distinct fragment lengths observed gives high power for detecting small differences from the expected 0.25 fraction of discordant lengths due to self-collected samples.
Microbial communities species compositions and abundances comparative analysis.
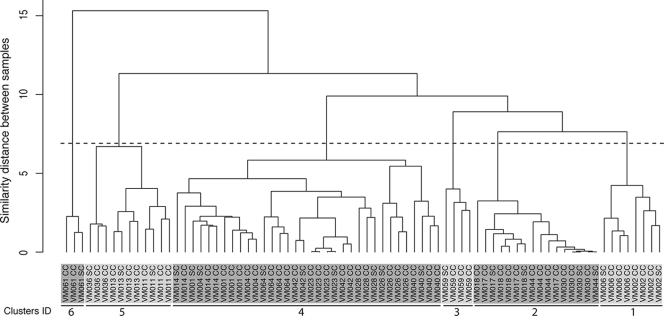
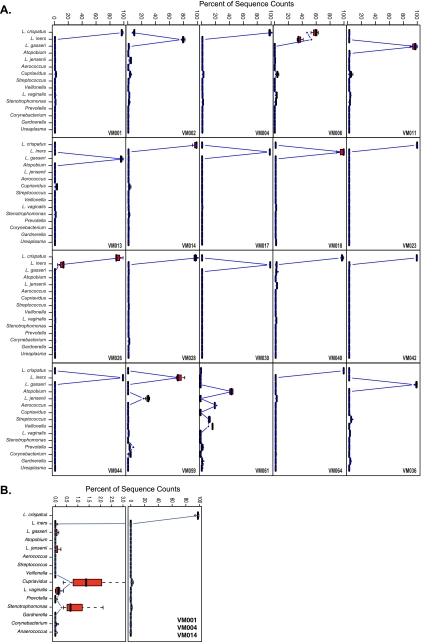
A total of 77 vaginal swabs from 20 women were analyzed by pyrosequencing of bar-coded 16S rRNA genes. An average of 1,650 16S rRNA gene sequences were obtained per sample. The taxonomic assignments to the reads obtained for each sample are summarized in Table 1, where taxon abundance (as represented by the total number of 16S rRNA gene sequences assigned to each taxa) is indicated as the percentage of total reads obtained for each sample. The species composition and abundances of microbial communities fell into six groups, which is comparable to what has been previously reported (42). Communities in groups 1 to 5 were dominated by one or two Lactobacillus sp., while group 6 included a diverse set of taxa belonging to the genera Atopobium, Veillonella, Aerococcus, Prevotella, Gardnerella, Dialister, and Streptococcus spp. among others (Table 1). A Ward hierarchical clustering analysis (Fig. 1) showed that self-collected samples clustered with the corresponding physician-collected samples, indicating that the diversity that is sampled in both sets of swabs is identical. Clusters 4 and 2 are the largest clusters and comprise vaginal microbial communities that are dominated (>90%) by L. crispatus and L. iners, respectively. The high level of similarity between subjects was reflected by subclusters composed of mixed samples from two or more subjects (Fig. 2). Statistical analysis of the distributions of microbial communities composition and abundance sampled by self-collection and physician collection using the Kolmogorov-Smirnov test resulted in a P value of 0.5446, supporting the null hypothesis that the distributions d(PC,PC) and d(SC,PC) came from the same distribution. These analyses further support the T-RFLP analysis and confirm that the vaginal community microbial species compositions and abundances sampled by each method were similar. Detailed comparisons of the microbial communities sampled by physician collected or self-collected for each women are shown in Fig. 2.
TABLE 1.
Microbial community composition as percentage of the top 18 most abundant taxa present in each of three swabs collected by a physician (CC1, CC2, and CC3) and the sample self-collected by the participant (SC)
FIG. 1.
Ward hierarchical clustering of vaginal microbial community taxa composition and abundance characterized by 16S rRNA gene sequencing in three clinical collected (CC) and one self-collected (SC) swabs from each of 20 participants. The clusters were established using the cutoff indicated by the dotted black horizontal line. Clusters are numbered 1 to 6 and colored accordingly.
FIG. 2.
(A) Detailed comparisons of microbial community composition of samples collected by self-collection (gray dot) and by physician (red boxplot, average from three samples) for each of the 20 women enrolled in the study. (B) Similar profiles for three women combined (VM001, VM004, and VM0014), with the region 0 to 3% enlarged.
DISCUSSION
Our study demonstrates that self-collected vaginal swabs of the mid-vagina reflect the same microbial diversity as physician-collected vaginal specimens. This finding enables large scale, field-based longitudinal microbiomic studies of the vaginal ecosystem.
The microbiota of the human vagina is a dynamic and complex example of microbial mutualistic symbiosis, the regulation of which is not fully understood (20). The vagina and its unique microbiota form a finely balanced ecosystem. It is hypothesized that the physical and chemical environment control the microbial types present and that the microbiota in turn control the vaginal environment, thus forming a dynamic equilibrium (30). Previous studies have established that several distinct kinds of vaginal communities with markedly different species composition occur in white and black women in North America (41, 42), and Japanese women in Tokyo, Japan (43). Six of these microbial community compositions were observed in our study of 20 women.
Since vaginal bacterial communities differ in species composition (15, 35, 38, 42), they are likely to differ in how they respond to disturbances. Conceptually, this is important since vaginal communities continually experience various kinds of chronic and acute disturbances caused by human behaviors such as the use of antibiotics, hormonal contraceptives and other methods of birth control, sexual intercourse, vaginal lubricants, douching, and many others. In addition, the structure and composition of vaginal microbial communities are known to be influenced by natural changes in normal healthy women, including aging, time in the menstrual cycle, menstruation, pregnancy, and stress (6, 11, 16, 24). Very little is known about the biotic or abiotic factors that control the dynamics of the vaginal microbiota. Ultimately, resilience to disruptions of the ecological equilibria may be an important factor in protection against infectious agents, sexually transmitted infections and development of adverse obstetric outcomes, including preterm delivery of low-birth-weight infants.
Daily fluctuations in the composition of the vaginal microbiota have been previously documented by microscopy and Gram's staining (13, 16, 33). These changes are not reflected in cross-sectional studies; hence, longitudinal studies with frequent sampling are needed to capture the dynamics of vaginal communities. For example, in a 6-week, daily self-collection longitudinal study (33), it was observed that in 75% of the participants, the vaginal microbiota composition fluctuated over the time course of the menstrual cycle. The epidemiological study concluded that hygiene and sexual behavior were associated with the observed fluctuations. However, because molecular analyses of the vaginal communities were not performed, it was not possible to assess the importance of inherent differences in vaginal community composition. Similarly, Hay et al. noted the rapid onset and resolution (as established by Nugent scores) of BV in a study of 18 women with recurrent BV who self-collected vaginal swabs daily for up to 10 months (13). These studies highlight the dynamic nature of vaginal microbial communities (34) and reveal a potential weakness of cross-sectional sampling, since shifts in the vaginal microbial communities composition can be missed if samples are collected weekly, monthly, or less frequently. In order to observe the frequent fluctuations in vaginal microbiota, field-based cohort studies must rely on self-collection of samples since frequent clinician-obtained samples would not be practical or financially feasible. These studies further highlight the need for prospective field-based studies so that we can better understand the biotic or abiotic factors driving the dynamics of the vaginal microbiota and their association with adverse outcomes.
The present study demonstrates that self-collected vaginal swabs of the mid-vagina reflect identical microbial diversity as physician-collected swabs by culture-independent methodologies. Microbial surveys of microbiota, as performed by 16S rRNA gene analyses, require sampling to be quantitative and qualitative, and we provide the evidence that self-collection quantitatively samples the vaginal microbiota. In the present study, we further demonstrate that the insertion of a speculum by a physician is not necessary for the collection of reliable quantitative vaginal swabs from the mid-vagina. Importantly, participant's instructions for the collection of the vaginal swabs were no different than those given to women in other studies evaluating self-collection as a sampling method (1, 23, 25, 29, 31, 32, 34, 40). We believe that self-sampling can be easily implemented for daily or frequent sampling in a home-based setting. Samples can be stored at room temperature or in a freezer depending on the transport medium used for the swab sample, and participants could mail-in or drop off samples weekly to the enrolling center. The results of the present study not only validate self-collection for prospective longitudinal studies of the vaginal microbiota but also enable such studies by minimizing the burden on the participants by obviating the need for clinician-collected sampling.
Acknowledgments
This study is based on work funded by grant AI-U01070921 from the National Institutes of Health, National Institute for Allergies and Infectious Diseases.
Footnotes
▿
Published ahead of print on 3 March 2010.
REFERENCES
- 1.Boskey, E. R., S. A. Atherly-Trim, P. J. O'Campo, D. M. Strobino, and D. P. Misra. 2004. Acceptability of a self-sampling technique to collect vaginal smears for Gram stain diagnosis of bacterial vaginosis. Womens Health Issues 14**:**14-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33**:**D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conover, W. J. 1971. Practical nonparametric statistics. John Wiley & Sons, Inc., New York, NY.
- 4.Costello, E. K., C. L. Lauber, M. Hamady, N. Fierer, J. I. Gordon, and R. Knight. 2009. Bacterial community variation in human body habitats across space and time. Science 326**:**1694-1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cu-Uvin, S., J. W. Hogan, A. M. Caliendo, J. Harwell, K. H. Mayer, and C. C. Carpenter. 2001. Association between bacterial vaginosis and expression of human immunodeficiency virus type 1 RNA in the female genital tract. Clin. Infect. Dis. 33**:**894-896. [DOI] [PubMed] [Google Scholar]
- 6.Eschenbach, D. A., S. S. Thwin, D. L. Patton, T. M. Hooton, A. E. Stapleton, K. Agnew, C. Winter, A. Meier, and W. E. Stamm. 2000. Influence of the normal menstrual cycle on vaginal tissue, discharge, and microflora. Clin. Infect. Dis. 30**:**901-907. [DOI] [PubMed] [Google Scholar]
- 7.Forney, L. J., X. Zhou, and C. J. Brown. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7**:**210-220. [DOI] [PubMed] [Google Scholar]
- 8.Frank, J. A., C. I. Reich, S. Sharma, J. S. Weisbaum, B. A. Wilson, and G. J. Olsen. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74**:**2461-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fredricks, D. N., T. L. Fiedler, and J. M. Marrazzo. 2005. Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med. 353**:**1899-1911. [DOI] [PubMed] [Google Scholar]
- 10.Grice, E. A., H. H. Kong, S. Conlan, C. B. Deming, J. Davis, A. C. Young, G. G. Bouffard, R. W. Blakesley, P. R. Murray, E. D. Green, M. L. Turner, and J. A. Segre. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324**:**1190-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guise, J. M., S. M. Mahon, M. Aickin, M. Helfand, J. F. Peipert, and C. Westhoff. 2001. Screening for bacterial vaginosis in pregnancy. Am. J. Prev. Med. 20**:**62-72. [DOI] [PubMed] [Google Scholar]
- 12.Hamady, M., J. J. Walker, J. K. Harris, N. J. Gold, and R. Knight. 2008. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat. Methods 5**:**235-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hay, P. E., A. Ugwumadu, and J. Chowns. 1997. Sex, thrush, and bacterial vaginosis. Int. J. STD AIDS 8**:**603-608. [DOI] [PubMed] [Google Scholar]
- 14.Hsiao, W. W., and C. M. Fraser-Liggett. 2009. Human Microbiome Project: paving the way to a better understanding of ourselves and our microbes. Drug Discov. Today 14**:**331-333. [DOI] [PubMed] [Google Scholar]
- 15.Hyman, R. W., M. Fukushima, L. Diamond, J. Kumm, L. C. Giudice, and R. W. Davis. 2005. Microbes on the human vaginal epithelium. Proc. Natl. Acad. Sci. U. S. A. 102**:**7952-7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keane, F. E., C. A. Ison, and D. Taylor-Robinson. 1997. A longitudinal study of the vaginal flora over a menstrual cycle. Int. J. STD AIDS 8**:**489-494. [DOI] [PubMed] [Google Scholar]
- 17.Kim, T. K., S. M. Thomas, M. Ho, S. Sharma, C. I. Reich, J. A. Frank, K. M. Yeater, D. R. Biggs, N. Nakamura, R. Stumpf, S. R. Leigh, R. I. Tapping, S. R. Blanke, J. M. Slauch, H. R. Gaskins, J. S. Weisbaum, G. J. Olsen, L. L. Hoyer, and B. A. Wilson. 2009. Heterogeneity of vaginal microbial communities within individuals. J. Clin. Microbiol. 47**:**1181-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirk, R. E. 1995. Experimental design: procedures for the behavioral sciences, 3rd ed. Brooks/Cole, Pacific Grove, CA.
- 19.Kumar, P. S., E. J. Leys, J. M. Bryk, F. J. Martinez, M. L. Moeschberger, and A. L. Griffen. 2006. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J. Clin. Microbiol. 44**:**3665-3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsen, B., and G. R. Monif. 2001. Understanding the bacterial flora of the female genital tract. Clin. Infect. Dis. 32**:**e69-e77. [DOI] [PubMed] [Google Scholar]
- 21.Lenth, R. V. 2009. Java applets for power and sample size.
- 22.McKenna, P., C. Hoffmann, N. Minkah, P. P. Aye, A. Lackner, Z. Liu, C. A. Lozupone, M. Hamady, R. Knight, and F. D. Bushman. 2008. The Macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 4**:**e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer, B. M., M. C. Taylor, J. L. Fricke, V. S. Baselski, and B. M. Sibai. 1995. The accuracy and patient preference for self-collected group B Streptococcus cultures. Am. J. Obstet. Gynecol. 173**:**1325-1328. [DOI] [PubMed] [Google Scholar]
- 24.Nansel, T. R., M. A. Riggs, K. F. Yu, W. W. Andrews, J. R. Schwebke, and M. A. Klebanoff. 2006. The association of psychosocial stress and bacterial vaginosis in a longitudinal cohort. Am. J. Obstet. Gynecol. 194**:**381-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson, D. B., S. Bellamy, T. S. Gray, and I. Nachamkin. 2003. Self-collected versus provider-collected vaginal swabs for the diagnosis of bacterial vaginosis: an assessment of validity and reliability. J. Clin. Epidemiol. 56**:**862-866. [DOI] [PubMed] [Google Scholar]
- 26.Ness, R. B., K. E. Kip, S. L. Hillier, D. E. Soper, C. A. Stamm, R. L. Sweet, P. Rice, and H. E. Richter. 2005. A cluster analysis of bacterial vaginosis-associated microflora and pelvic inflammatory disease. Am. J. Epidemiol. 162**:**585-590. [DOI] [PubMed] [Google Scholar]
- 27.Nocker, A., M. Burr, and A. K. Camper. 2007. Genotypic microbial community profiling: a critical technical review. Microb. Ecol. 54**:**276-289. [DOI] [PubMed] [Google Scholar]
- 28.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276**:**734-740. [DOI] [PubMed] [Google Scholar]
- 29.Polaneczky, M., C. Quigley, L. Pollock, D. Dulko, and S. S. Witkin. 1998. Use of self-collected vaginal specimens for detection of Chlamydia trachomatis infection. Obstet. Gynecol. 91**:**375-378. [DOI] [PubMed] [Google Scholar]
- 30.Pybus, V., and A. B. Onderdonk. 1996. The effect of pH on growth and succinate production by Prevotella bivia. Microb. Ecol. Health Dis. 9**:**19-22. [Google Scholar]
- 31.Rompalo, A. M., C. A. Gaydos, N. Shah, M. Tennant, K. A. Crotchfelt, G. Madico, T. C. Quinn, R. Daniel, K. V. Shah, J. C. Gaydos, and K. T. McKee, Jr. 2001. Evaluation of use of a single intravaginal swab to detect multiple sexually transmitted infections in active-duty military women. Clin. Infect. Dis. 33**:**1455-1461. [DOI] [PubMed] [Google Scholar]
- 32.Safaeian, M., M. Kiddugavu, P. E. Gravitt, J. Ssekasanvu, D. Murokora, M. Sklar, D. Serwadda, M. J. Wawer, K. V. Shah, and R. Gray. 2007. Comparability of self-collected vaginal swabs and physician-collected cervical swabs for detection of human papillomavirus infections in Rakai, Uganda. Sex. Transm. Dis. 34**:**429-436. [DOI] [PubMed] [Google Scholar]
- 33.Schwebke, J. R., C. M. Richey, and H. L. Weiss. 1999. Correlation of behaviors with microbiological changes in vaginal flora. J. Infect. Dis. 180**:**1632-1636. [DOI] [PubMed] [Google Scholar]
- 34.Strauss, R. A., B. Eucker, D. A. Savitz, and J. M. Thorp, Jr. 2005. Diagnosis of bacterial vaginosis from self-obtained vaginal swabs. Infect. Dis. Obstet. Gynecol. 13**:**31-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sundquist, A., S. Bigdeli, R. Jalili, M. L. Druzin, S. Waller, K. M. Pullen, Y. Y. El-Sayed, M. M. Taslimi, S. Batzoglou, and M. Ronaghi. 2007. Bacterial flora-typing with targeted, chip-based Pyrosequencing. BMC Microbiol. 7**:**108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tringe, S. G., and P. Hugenholtz. 2008. A renaissance for the pioneering 16S rRNA gene. Curr. Opin. Microbiol. 11**:**442-446. [DOI] [PubMed] [Google Scholar]
- 37.Turnbaugh, P. J., and J. I. Gordon. 2009. The core gut microbiome, energy balance, and obesity. J. Physiol. 587**:**4153-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Verhelst, R., H. Verstraelen, G. Claeys, G. Verschraegen, J. Delanghe, L. Van Simaey, C. De Ganck, M. Temmerman, and M. Vaneechoutte. 2004. Cloning of 16S rRNA genes amplified from normal and disturbed vaginal microflora suggests a strong association between Atopobium vaginae, Gardnerella vaginalis, and bacterial vaginosis. BMC Microbiol. 4**:**16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward, J. J. H. 1963. Hierarchical grouping to optimize an objective function. J. Am. Statist. Assoc. 58**:**236-244. [Google Scholar]
- 40.Wiesenfeld, H. C., D. L. Lowry, R. P. Heine, M. A. Krohn, H. Bittner, K. Kellinger, M. Shultz, and R. L. Sweet. 2001. Self-collection of vaginal swabs for the detection of chlamydia, gonorrhea, and trichomoniasis: opportunity to encourage sexually transmitted disease testing among adolescents. Sex. Transm. Dis. 28**:**321-325. [DOI] [PubMed] [Google Scholar]
- 41.Zhou, X., S. J. Bent, M. G. Schneider, C. C. Davis, M. R. Islam, and L. J. Forney. 2004. Characterization of vaginal microbial communities in adult healthy women using cultivation-independent methods. Microbiology 150**:**2565-2573. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, X., C. J. Brown, Z. Abdo, C. C. Davis, M. A. Hansmann, P. Joyce, J. A. Foster, and L. J. Forney. 2007. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1**:**121-133. [DOI] [PubMed] [Google Scholar]
- 43.Zhou, X., M. A. Hansmann, C. C. Davis, H. Suzuki, C. J. Brown, U. Shütte, J. D. Pierson, and L. J. Forney. 2009. The vaginal bacterial communities of Japanese women resemble those of women in other racial groups. FEMS Immunol. Med. Microbiol. 58**:**169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]