Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control TH2 responses (original) (raw)
. Author manuscript; available in PMC: 2010 May 5.
Published in final edited form as: Nature. 2009 Feb 1;458(7236):351–356. doi: 10.1038/nature07674
Abstract
In the course of infection or autoimmunity, particular transcription factors orchestrate the differentiation of TH1, TH2 or TH17 effector cells, the responses of which are limited by a distinct lineage of suppressive regulatory T cells (Treg). Treg cell differentiation and function are guided by the transcription factor Foxp3, and their deficiency due to mutations in Foxp3 results in aggressive fatal autoimmune disease associated with sharply augmented TH1 and TH2 cytokine production1–3. Recent studies suggested that Foxp3 regulates the bulk of the Foxp3-dependent transcriptional program indirectly through a set of transcriptional regulators serving as direct Foxp3 targets4,5. Here we show that in mouse Treg cells, high amounts of interferon regulatory factor-4 (IRF4), a transcription factor essential for TH2 effector cell differentiation, is dependent on Foxp3 expression. We proposed that IRF4 expression endows Treg cells with the ability to suppress TH2 responses. Indeed, ablation of a conditional Irf4 allele in Treg cells resulted in selective dysregulation of TH2 responses, IL4-dependent immunoglobulin isotype production, and tissue lesions with pronounced plasma cell infiltration, in contrast to the mononuclear-cell-dominated pathology typical of mice lacking Treg cells. Our results indicate that Treg cells use components of the transcriptional machinery, promoting a particular type of effector CD4+ T cell differentiation, to efficiently restrain the corresponding type of the immune response.
Treg cell deficiency results in activation and expansion of CD4+ and CD8+ T cells, dendritic cells, granulocytes and macrophages, and greatly increased production of a wide range of cytokines including interleukin (IL)-2, TH1 and TH2 cytokines6,7. Expression of Foxp3 is required for the establishment and maintenance of Treg lineage identity and suppressor function8–11. Our recent study suggested that in Treg cells Foxp3 might regulate expression of IRF4 (refs 12–14) a transcription factor that is indispensable for TH2 effector cell differentiation15,16. Furthermore, a recent study suggested a prominent role for IRF4 in TH17 differentiation17. Thus, we decided to examine a role for IRF4 in Treg cell differentiation and function.
Foxp3 binding within the promoter region of Irf4 in Treg cells4 was confirmed by chromatin immunoprecipitation (ChIP)-coupled quantitative PCR (qPCR) (Supplementary Fig. 1a, b). Irf4 messenger RNA was increased in thymic and peripheral Foxp3+ Treg cells in comparison to CD25− Foxp3− CD4+ T cells (data not shown)8. Furthermore, Foxp3 knockdown using a retrovirally encoded Foxp3-specific short hairpin RNA resulted in a marked diminution in Irf4 mRNA (Supplementary Fig. 1c), suggesting that Foxp3 directly regulates IRF4 expression in Treg cells.
Next we induced deletion of a conditional Irf4 allele (Irf4fl) in Treg cells by crossing Irf4fl mice to Foxp3Cre mice expressing yellow fluorescent protein (YFP)-Cre recombinase fusion protein under the Foxp3 locus control18,19. The Irf4fl allele has a ‘built-in’ reporter capacity in that Cre-mediated recombination results in the deletion of the Irf4 promoter and the exon containing the translational start, and the concomitant expression of green fluorescent protein (GFP)18. The specificity of the Irf4fl deletion was examined by flow cytometric analysis of Foxp3 expression in a sorted GFP+ CD4+ T cell population that underwent Cre-mediated recombination, and in a GFP− CD4+ T cell population that did not. Essentially all GFP+ cells expressed Foxp3, whereas GFP− cells lacked Foxp3 expression (Fig. 1a). Flow-cytometric analysis of control Irf4fl/+ Foxp3Cre mice showed that IRF4 expression was markedly increased in all peripheral Treg cells, but only modestly in Foxp3+ CD4+ thymocytes, whereas IRF4 levels were undetectable in corresponding Foxp3− cell subsets. On Cre-mediated deletion in Irf4fl/flFoxp3Cre mice, IRF4 protein became undetectable in both peripheral and thymic Foxp3+ cells (Fig. 1b). Irf4fl/flFoxp3Cre and _Irf4fl/_− Foxp3Cre mice harbouring an IRF4-deficient Treg subset were born at the expected Mendelian ratio and were indistinguishable from their wild-type or heterozygous littermates during the first month of life. However, by 6–8 weeks of age Irf4fl/flFoxp3Cre and _Irf4fl/_− Foxp3Cre mice manifested identical autoimmune disease—including lymphadenopathy, weight loss, blepharitis and dermatitis—and succumbed to disease at 3–4 months of age (Fig. 1c, d and data not shown). Histopathological evaluation of diseased mice showed massive infiltration in the pancreas, lung and stomach, whereas control littermates did not show any noticeable pathology. For comparison to a complete Treg cell deficiency, we analysed tissue lesions in Foxp3DTR knock-in mice (expressing human diphtheria receptor, DTR under control of Foxp3 locus) that were subjected to chronic ablation of a Treg cell subset caused by diphtheria toxin treatment, starting from birth. These mice showed analogous lesions in the pancreas and stomach, and much more severe lesions in the lung in comparison to mice harbouring an IRF4-deficient Treg subset (Fig. 1e and data not shown). In contrast to the massive liver lesions observed after Treg depletion, livers in mice containing an IRF4-deficient Treg subset were unaffected, but kidneys showed the opposite trend (Supplementary Fig. 2 and data not shown). Furthermore, flow cytometric analysis showed expansion and activation of peripheral T cells but not dendritic cells in Irf4fl/flFoxp3Cre mice, in contrast to the increased numbers of activated dendritic cells observed in Treg-deficient mice (Fig. 2a, b and data not shown).
Figure 1. Ablation of IRF4 in Treg cells results in autoimmune lymphoproliferative disease.
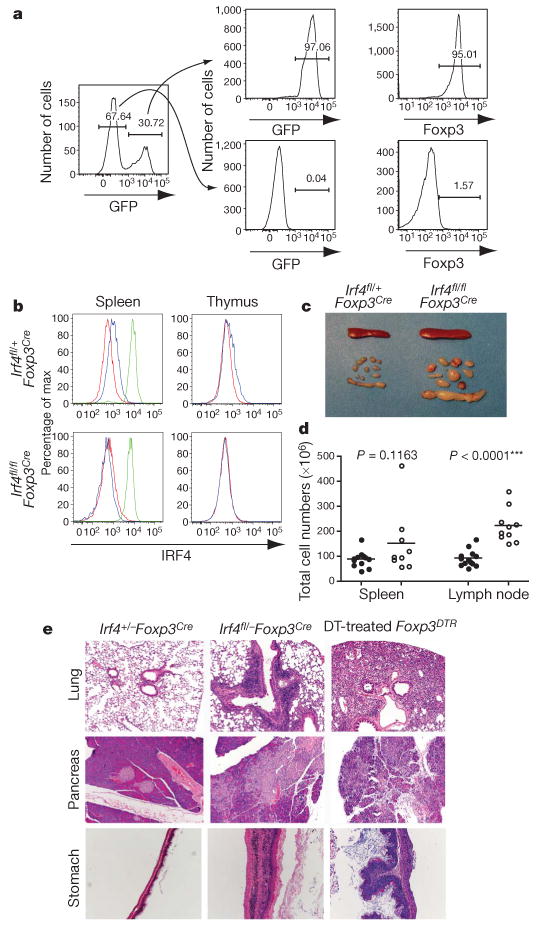
a, _Foxp3Cre_-mediated deletion of Irf4 is restricted to Treg cells. Spleen and lymph node CD4+ T cells from _Irf4fl/− Foxp3Cre mice were FACS-sorted into GFP+ and GFP− cells containing a recombined and unrecombined Irf4fl allele, respectively, and stained for Foxp3. The post-sorting purity of GFP+ and GFP− population was greater than 97%. b, Intracellular staining of IRF4 in splenic and thymic CD4+ Foxp3− (red) and CD4+ Foxp3+ (blue) cells and splenic B220− CD138+ (green) plasma cells (positive control) in Irf4fl/+ Foxp3Cre (top panels) and Irf4fl/flFoxp3Cre mice (bottom panels). c, Splenomegaly and lymphadenopathy in Irf4fl/flFoxp3Cre mice. d, Spleen and lymph node cellularity in Irf4fl/+ Foxp3Cre (filled circles) and Irf4fl/flFoxp3Cre (open circles) mice. e, Histopathology induced after IRF4 ablation in Treg cells. Representative haematoxylin and eosin (H&E)-stained tissue sections from 8-week-old Irf4+/_− Foxp3Cre and Irf4fl/-Foxp3Cre mice and diphtheria-toxin-treated Foxp3DTR mice. Note pronounced infiltrates in the lung, pancreas and stomach of _Irf4fl/_− Foxp3Cre mice and severe lesions in tissues from diphtheria-toxin-treated Foxp3DTR mice (n = 3–5). Original magnification (c, e), ×10.
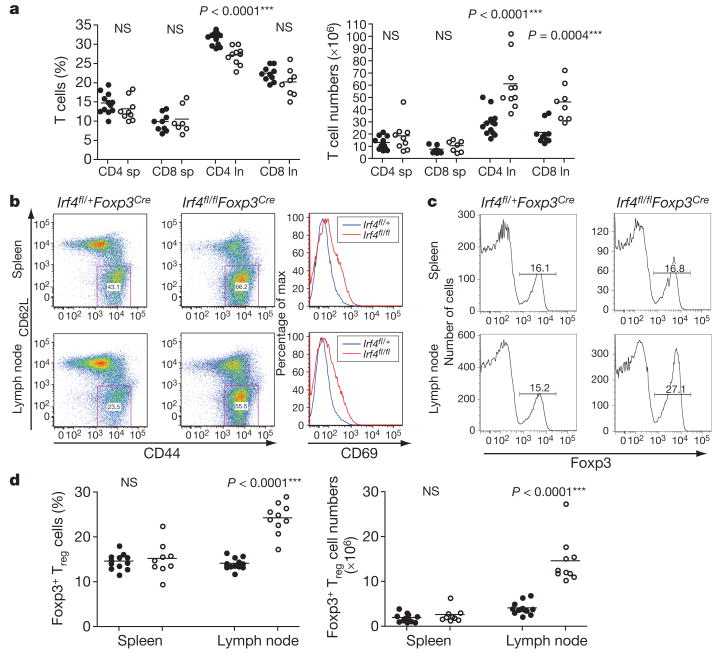
Figure 2. Increased numbers and activation of CD4+ T cells in mice harbouring IRF4-deficient Tregcells.
a, CD4+ and CD8+ T cell numbers in the spleen (sp) and lymph nodes (ln) of Irf4fl/flFoxp3Cre (open circles) mice and Irf4fl/+ Foxp3Cre (filled circles) littermate control mice. NS, not significant. b, Flow cytometric analysis of CD44, CD62L and CD69 expression on CD4+ T cells in 8-week-old Irf4fl/flFoxp3Cre mice and Irf4fl/+ Foxp3Cre littermates. A representative of three independent experiments is shown. c, d, Increased Foxp3+ Treg cell subset in Irf4fl/flFoxp3Cre mice. Flow cytometric analyses of spleen and lymph node cells from Irf4fl/+ Foxp3Cre (filled circles) and Irf4fl/flFoxp3Cre (open circles) mice. A representative of three independent experiments is shown.
The aforementioned phenotypic differences between mice lacking Treg cells and mice containing IRF4-deficient Treg cells could be due to a numerical decrease in the Treg cells in the absence of IRF4. However, the IRF4-deficient Treg subset was markedly increased in diseased Irf4fl/flFoxp3Cre mice compared to Irf4fl/+ Foxp3Cre littermate controls, probably in response to T cell activation (Fig. 2c, d). IRF4 deficiency did not change Foxp3 protein expression on a per cell basis (Fig. 2c), excluding the possibility of down-modulation of Foxp3 in Treg cells in the absence of IRF4 as a reason for their functional incompetence. These results pointed to impairment in a particular aspect of Treg function on IRF4 ablation.
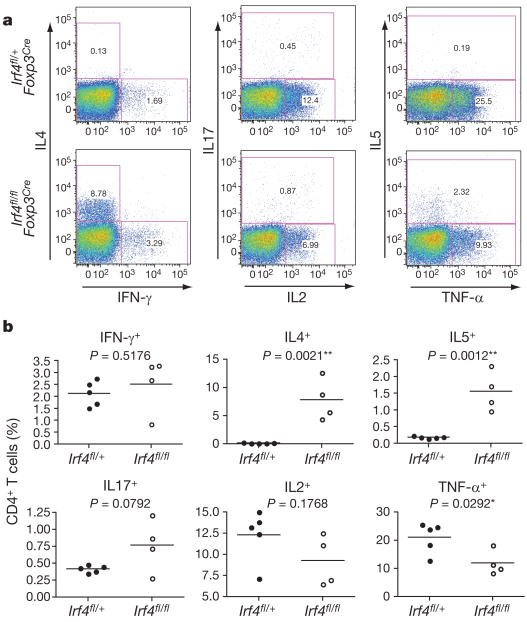
Indeed, analysis of cytokine production by CD4+ T cells from Irf4fl/flFoxp3Cre mice showed a notable increase in the numbers of IRF4-sufficient Foxp3− CD4+ T cells producing the TH2 cytokines IL-4 and IL-5 (Fig. 3a, b), whereas IRF4-deficient Foxp3+ Treg cells, unlike their IRF4-sufficient counterparts, were unable to produce effector cytokines (Supplementary Fig. 3). The production of IL-2 and the TH1 cytokine IFN-γ by CD4+ T cells was largely unchanged, but production of the TH17 cytokine IL-17 was marginally increased in Irf4fl/flFoxp3Cre mice compared to Irf4fl/+ Foxp3Cre controls (Fig. 3a, b and data not shown). In agreement with these results, enzyme-linked immunosorbent assay (ELISA) showed high amounts of the TH2 cytokines IL-5, IL-13 and IL-10 in the supernatants of CD3-stimulated splenic Irf4fl/flFoxp3Cre T cell cultures (Supplementary Fig. 4). In contrast, a lack of Treg cells in _Foxp3_− mice led to sharply increased production of IFN-γ and IL-4 by CD4+ T cells (Supplementary Fig. 5). Massive increases in IFN-γ and IL-2 production were also observed after ablation of Treg cells in Foxp3DTR mice7 (J. M. Lund and A.Y.R., unpublished observations). Thus, TH2 effector CD4+ T cell responses seem to be selectively dysregulated in mice containing IRF4-deficient Treg cells. Notably, the augmented TH2 responses appeared in Irf4fl/flFoxp3Cre mice only by 3–4 weeks of age, whereas both TH2 and TH1 cytokine production in 11-day-old Irf4fl/flFoxp3Cre and Irf4fl/+ Foxp3Cre mice was low (data not shown).
Figure 3. IRF4 deficiency in Treg cells results in a selective failure to control TH2 responses.
a, b, Flow cytometric analysis of cytokine production by splenic CD4+ T cells from Irf4fl/+ Foxp3Cre mice and Irf4fl/flFoxp3Cre littermates. Splenocytes were stimulated with CD3 (5 μg ml−1) and CD28 (5 μg ml-1) antibodies in the presence of Golgi-Plug (1 μg ml−1) for 5 h before staining for CD4, CD8 and the indicated cytokines. A representative of three independent experiments is shown.
Because IL-4 is an important cytokine that promotes IgG1 and IgE class-switch recombination, we measured levels of serum immunoglobulin isotypes in diseased 8-week-old Irf4fl/flFoxp3Cre and littermate control Irf4fl/+ Foxp3Cre mice. As a positive control, we also analysed 3–4-week-old Treg-deficient _Foxp3_− mice. Consistent with an uncontrolled TH2 response, IL-4-dependent IgG1 and IgE serum concentrations reached very high levels (10–30 mg ml−1 and 0.75–1.5 mg ml−1, respectively) in Irf4fl/flFoxp3Cre mice, whereas serum IgM and IgA concentrations were only modestly increased in comparison to control mice. However, the amounts of the remaining immunoglobulin isotypes, including IFN-γ-dependent IgG2a, were diminished in the serum of afflicted mice (Fig. 4a). Unlike Irf4fl/flFoxp3Cre mice, _Foxp3_− mice showed in addition to increased IgG1 and IgE, sharply increased amounts of IgG2a consistent with the increased IFN-γ production in these mice (Fig. 4a). Furthermore, we observed ubiquitous germinal centre formation and increased numbers of plasma cells in the spleens of diseased Irf4fl/flFoxp3Cre mice, but not in the littermate controls (Fig. 4b and data not shown). In Irf4fl/flFoxp3Cre mice, 15% and 67% of plasma cells produced IgE and IgG1, respectively, in contrast to 3% IgG1-producing and undetectable numbers of IgE-producing plasma cells in control mice (Supplementary Fig. 6). Furthermore, examination of diseased mice harbouring an IRF4-deficient Treg cell subset revealed a predominance of cells with a characteristic plasma cell morphology infiltrating the pancreas, kidney and stomach (Fig. 4c and data not shown). In contrast, the pancreatic and other tissue infiltrates in Foxp3DTR mice subjected to chronic Treg ablation, or in _Foxp3_− mice, consisted predominantly of macrophages, lymphocytes and neutrophils with very few plasma cells (Fig. 4c and data not shown). We also tested whether the dysregulated TH2 response in the presence of IRF4-deficient Treg cells promoted autoantibody production. Indeed, serum IgG1 from affected Irf4fl/flFoxp3Cre mice showed robust reactivity with several tissue antigens, whereas minimal reactivity was found in control animals (Supplementary Fig. 7).
Figure 4. Increased serum IgG1 and IgE concentration, germinal centre formation, and plasma cell tissue infiltration caused by IRF4 deficiency in Treg cells.
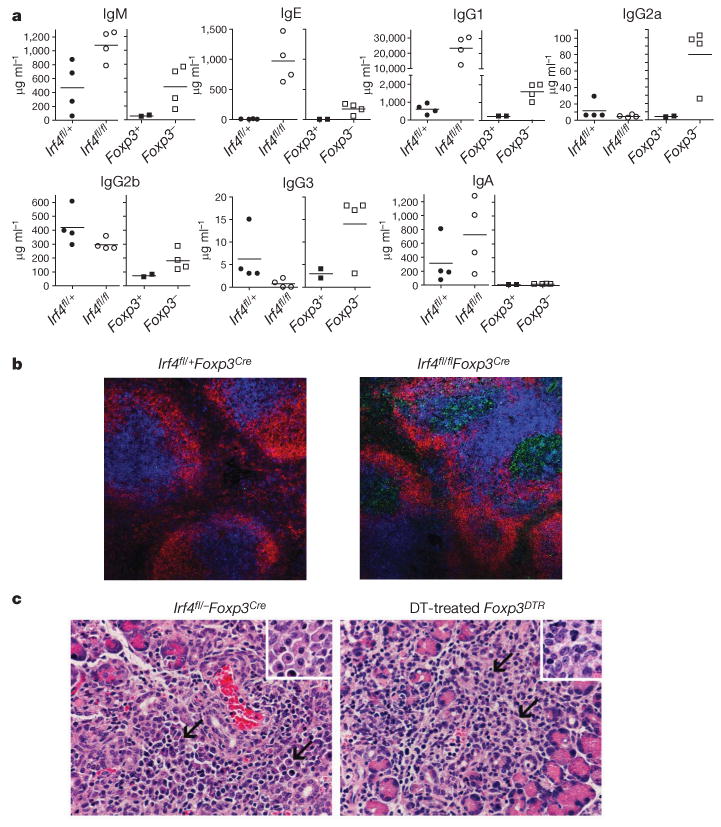
a, Analysis of immunoglobulin isotype amounts in sera of 8-week-old Irf4fl/flFoxp3Cre mice and Irf4fl/+ Foxp3Cre littermates, and of 3–4-week-old _Foxp3_− and Foxp3+ littermates. b, Immunofluorescent staining of germinal centre B cells (GL7+, green), follicular B cells (IgD+, red), and CD4+ T cells (blue) in spleens of Irf4fl/flFoxp3Cre mice and Irf4fl/+ Foxp3Cre littermates. Original magnification, ×20. c, Histological sections of H&E-stained pancreas from 8-week-old _Irf4fl_− Foxp3Cre mice and Treg-deficient diphtheria-toxin-treated Foxp3DTR mice. The _Irf4fl/_− Foxp3Cre pancreas is infiltrated primarily by plasma cells (arrows), that is, distinct round cells containing an eccentric nucleus with a cartwheel chromatin appearance and perinuclear clearing (inset; original magnification, ×60). In contrast, the pancreatic infiltrates of diphtheria-toxin-treated Foxp3DTR mice contained principally macrophages (arrows), that is, large cells with abundant eosinophilic cytoplasm, reniform to oval nuclei, and indistinct cell borders (inset; original magnification, ×60). Original magnification for both panels, ×40. Representative sections are shown.
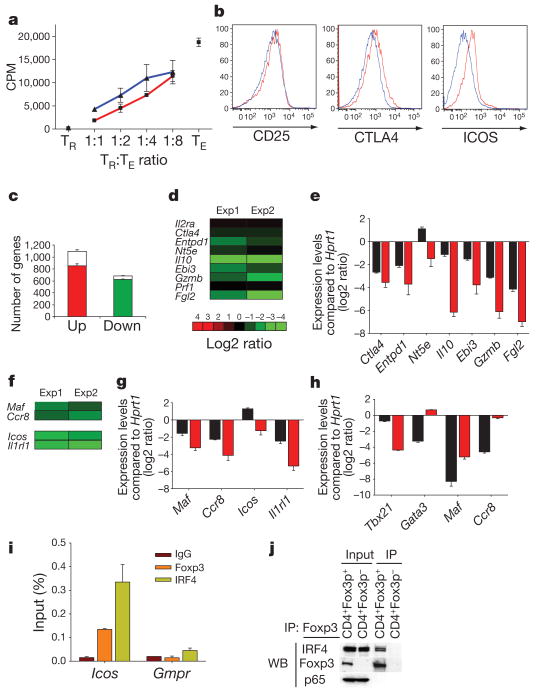
The distinct immune activation profile and tissue lesions in diseased mice harbouring an IRF4-deficient Treg cell subset were in agreement with the previously mentioned idea that only a part of the suppressor program was impaired in IRF4-deficient Treg cells. In further support of this notion, the in vitro suppressor capacity of IRF4-deficient Treg cells isolated from Irf4fl/flFoxp3Cre mice was largely intact (Fig. 5a). Furthermore, cell surface amounts of two putative Treg suppressor effector molecules CTLA4 and CD73 (also known as NT5E) were not altered in IRF4-deficient Treg cells. The levels of CD25 (IL2RA) and GITR (TNFRSF18) were also unchanged (Fig. 5b and data not shown). However, ICOS expression was decreased, suggesting that IRF4 regulates expression of a distinct subset of functionally important genes in Treg cells.
Figure 5. IRF4 interacts with Foxp3 and diminished expression of a subset of suppressor effector and TH2 specific genes in IRF4-deficient Treg cells.
a, IRF4-sufficient (Irf4+ TR, red) and -deficient (_Irf4_− TR, blue) Treg cells from 5-week-old Irf4+/+ Foxp3Cre mice and Irf4fl/flFoxp3Cre littermates suppress in vitro proliferative response of Foxp3− CD4+ T cells (TE) from B6 mice. A representative of two independent experiments is shown. b, Flow cytometric analysis of CD25, CTLA4 and ICOS expression by IRF4-sufficent (red) and -deficient (blue) Treg cells. c, Numbers of IRF4-independent genes that were up- (red bars) or downregulated (green bars), respectively, in Treg cells compared to naive CD25− Foxp3− CD4+ T cells9. Open bars represent genes in which expression was changed by twofold or more in the absence of IRF4. d, Decreased expression of genes with a presumed role in Treg suppressor function in IRF4-deficient compared to IRF4-sufficient Treg cells. The data (c, d) represent average of two independent microarray experiments (exp) performed using YFP-Cre+ Treg cells FACS-purified from healthy _Irf4fl/_− Foxp3Cre/wt and Irf4+/− Foxp3Cre littermates. e, qPCR analysis of relative expression of genes shown in d in Irf4+ Treg (black) and _Irf4_− Treg (red). f, The decreased expression of TH2-specific or functionally important genes in IRF4-deficient in comparison to IRF4-sufficient Treg cells (two independent microarray experiments as above). g, h, qPCR analysis of relative expression of the TH2-specific gene set in Irf4+ Treg (black) and _Irf4_− Treg (red) (g), and in in vitro differentiated TH1 (black) and TH2 cells (red) (h). Data in e, g and h represent mean and s.d. of the expression of genes relative to Hprt1 in two independent experiments using three replicates each. i, Both IRF4 and Foxp3 bind to the promoter region of the Icos gene. qPCR analysis of Foxp3- and IRF4-bound chromatin isolated from wild-type Treg cells using primer set corresponding to the Icos promoter region. IgG ChIP and qPCR using primers corresponding to the promoter region of Gmpr was used as a specificity controls. j, Western blot (WB) analysis of IRF4 in nuclear lysates of wild-type Foxp3+ Treg cells and total Foxp3− CD4+ T cells (control) (lanes 1 and 2), and in Foxp3 complexes immunoprecipitated (IP) from the nuclear lysates using Foxp3 antibody. Transcription factor p65 was a negative control. IRF4 signal in control nuclear lysates is due to the presence of activated IRF4+ CD25+ Foxp3− T cells.
An unbiased assessment of the effect of IRF4 deficiency on Treg transcriptome demonstrated that the expression of up to 20% of ‘Treg-specific’ genes decreased in the absence of IRF4, whereas approximately 7% were increased (Fig. 5c). Recent gene targeting studies demonstrated that Treg cells use several suppressor modalities19–25. Examination of the IRF4-dependent subset of ‘Treg’ genes and the subsequent independent confirmation by qPCR showed significantly decreased expression of several genes (Fgl2, Il10 and Gzmb) encoding putative suppressor effector molecules; a few (Ebi3 and Entpd1) were only marginally decreased, whereas others (Tgfb1 and Ctla4) did not change (Fig. 5d, e and data not shown). It is likely that changes in expression of a combination of genes, but not a single gene, account for the impaired suppressor capacity of IRF4-deficient Treg cells. In this regard, we recently showed a role for IL10 in Treg-mediated suppression20. In addition, ICOS-deficient Treg cells were inferior in comparison to their wild-type counterparts in limiting expansion of effector T cells in lymphopenic hosts (Y.Z., unpublished observations).
The observation that ICOS—the expression of which is critically important for TH2 differentiation26—was expressed in Treg cells in an IRF4-dependent manner suggested that IRF4 might be involved in the regulation of a functionally important transcriptional module shared by both Treg and TH2 effector T cells. To test this hypothesis, we cross-referenced the IRF4-dependent gene data set derived from our Treg cell studies to an effector TH2-specific gene data set. This comparison and independent qPCR analysis showed that in addition to Icos, the expression of two important TH2 genes Maf and Ccr8 was compromised in IRF4-deficient Treg cells, and the expression Il1rl — another essential player in TH2 differentiation—was also decreased (Fig. 5f–h)27–29.
Next, using Icos as a model gene, we sought to investigate the possibility that in Treg cells IRF4 and Foxp3 co-operate in transcriptional regulation. Using CLOVER, a transcription factor binding site prediction algorithm30, we identified a putative IRF4 binding site within the Icos promoter corresponding to the Foxp3 binding sites4 (Supplementary Fig. 8), and confirmed binding of both Foxp3 and IRF4 by ChIP-coupled qPCR (Fig. 5i). These results raised the possibility that Foxp3 physically interacts with IRF4. In fact, this interaction was confirmed by IRF4 and Foxp3 co-immunoprecipitation from Treg cell nuclear lysates using a Foxp3-specific antibody (Fig. 5j). These results indicate that IRF4 binds to Foxp3 and the resulting complexes affect expression of certain target genes such as Icos.
Our results indicate that Foxp3 induces IRF4 expression in Treg cells. A transcriptional module downstream of IRF4 is probably modified after IRF4 interaction with Foxp3 to facilitate efficient TH2 suppression by Treg cells. Uniformly increased IRF4 protein expression in peripheral Treg cells seems to suggest that they are equally poised to suppress TH2 responses. However, Treg cell populations may still be heterogeneous in this regard, if signalling-dependent post-translational modifications of IRF4 or recruitment of other nuclear factors are needed for Treg-mediated TH2 suppression. We propose that Treg cells might hijack certain components of transcriptional machinery promoting a particular effector T cell differentiation to efficiently control the corresponding type of the immune response.
Methods Summary
Mice
Mice containing Irf4fl, _Irf4_−, Foxp3Cre and Foxp3DTR alleles were previously described7,18,19. The Irf4fl/flFoxp3Cre and Irf4fl/− Foxp3Cre mice were indistinguishable in regards to the IRF4 deletion efficiency in Treg cells and to the immunological and clinical manifestations of autoimmunity, and so were all three types of healthy littermate control mice, Irf4+/+ _Foxp3_Cre, _Irf4_fl/+ _Foxp3_Cre and Irf4+/− _Foxp3_Cre.
Flow cytometric and serum immunoglobulin analyses
Rat anti-mouse IRF4 monoclonal antibody (clone 3E4) was raised against a glutathione _S_-transferase (GST)-fusion protein containing the carboxy-terminal 65-amino-acid sequence of murine IRF4. FACS data were acquired on a FACSCanto flow cytometer (Becton Dickinson) and analysed using FlowJo software package (Tri-Star).
Serum IgM, IgG1, IgG2a, IgG2b, IgG3 and IgA concentrations were measured using SBA Clonotyping System (Southern Biotech). IgE ELISA was performed using biotinylated anti-IgE antibody (BD Pharmingen) and streptavidin-conjugated HRP.
ChIP and qPCR
ChIP with Foxp3 and IRF4 antibodies (Santa Cruz Biotechnology) were performed using MACS-purified CD4+ CD25+ Treg cells as previously described4. qPCRs were performed using primers listed in Supplementary Table 1.
Methods
Mice
C57BL/6 (B6) mice were purchased from the Jackson Laboratory. Mice containing Irf4fl, _Irf4_−, Foxp3Cre and Foxp3DTR alleles were previously described7,18,19. The Irf4fl/flFoxp3Cre mice were indistinguishable from _Irf4fl/_− Foxp3Cre mice in regards to the efficiency of IRF4 deletion in Treg cells, and to the immunological and clinical manifestations of autoimmunity, as were all three types of healthy littermate control mice, Irf4+/+ _Foxp3_Cre, _Irf4_fl/+ _Foxp3_Cre and Irf4+/− _Foxp3_Cre. Mice were housed under specific pathogen-free conditions and used according to the guidelines of the Institutional Animal Care Committee at the University of Washington.
Antibodies and FACS analysis
Fluorescent-dye-conjugated antibodies were purchased from BD Pharmingen and eBioscience. Rat anti-mouse IRF4 monoclonal antibody (clone 3E4) was raised against a GST-fusion protein containing the C-terminal 65-amino-acid sequence of murine IRF4. FACS data were acquired on a FACSCanto flow cytometer (Becton Dickinson) and analysed using FlowJo software package (Tri-Star). Intracellular staining of Foxp3 and IRF4 was conducted using Foxp3 Mouse Regulatory T cell Staining Kit (eBioscience). For flow cytometric analysis of cytokine- and immunoglobulin-secreting cells, cell populations were first stained with antibodies against the indicated cell surface markers, followed by permeabilization in Fix/Perm buffer, and intracellular staining in Perm/Wash buffer (BD Pharmingen).
Immunofluorescence microscopy
Frozen tissue sections were fixed in cold acetone for 20 min, washed twice and pre-blocked in PBS containing 5% normal rat serum, 5% normal rabbit serum and 5% BSA for 30 min. Slides were then incubated in primary antibodies (anti-GL7-FITC, anti-IgD-biotin, anti-CD4-AF647) for 45 min, washed twice, and incubated with secondary antibodies (anti-FITC-AF488, StrepAv-AF555) for 45 min and fluorescence was examined using a Leica SL confocal microscope.
In vitro suppression assay
For in vitro suppression assays, Treg cells and ‘effector’ T cells were purified by positive selection using CD4-specific MACS beads (Miltenyi), followed by sorting on a FACSAria cell sorter (Becton Dickinson). Antigen-presenting cells were prepared from wild-type B6 splenocytes by T-cell depletion using Thy1-specific MACS beads. Effector T cells (2 × 104 cells well−1) were co-cultured with Treg cells at indicated ratios in the presence of irradiated (30 Gy) antigen-presenting cells (1 × 105 cells well−1) in 96-well plates in complete RPMI1640 medium supplemented with 10% FBS and CD3 antibody (1 μg ml−1) for 60 h. One microcurie 3H-thymidine was added into the cultures for a further 8–12 h and cell proliferation in triplicate cultures was measured using a scintillation counter.
Serum immunoglobulin ELISA
Serum IgM, IgG1, IgG2a, IgG2b, IgG3 and IgA concentrations were measured using SBA Clonotyping System (Southern Biotech). For IgE, ELISA was performed using biotinylated anti-IgE detection antibody (BD Pharmingen) and streptavidin-conjugated HRP.
Affymetrix microarray and qRT–PCR
Total RNA was extracted with RNA Stat-60 reagent (Iso-Tex Diagnostics) from IRF4-sufficient and -deficient CD4+ GFP/YFP-Cre+ Treg cells FACS-purified from healthy Irf4+/− Foxp3Cre and _Irf4fl/_− Foxp3Cre/wt mice, respectively. Complementary DNA was synthesized using Superscript II Reverse Transcriptase (Invitrogen), and amplified twice and labelled using MessageAmp II aRNA Amplification kit (Ambion). Biotinylated complementary RNA was fragmented and hybridized to Affymetrix GeneChip Mouse Genome 430 2.0 arrays at the Stanford PAN Facility. All data analyses were performed by using Bioconductor for the statistical software R (http://www.r-project.org). Expression values were background corrected, normalized and summarized by using the default settings of the gcrma package. From the resulting data sets we extracted a list of genes with a more than twofold change in expression and cross-referenced this list to the previously published set of Treg-specific genes9. Effector TH2-specific gene data set was extracted by subtracting gene expression values for CD4 T cells activated in vitro under TH1 skewing conditions or without skewing (TH0) from those for T cells activated under TH2 skewing conditions (K. Hilgner and K. Murphy, unpublished observations).
To verify expression array data, independently prepared Treg cell samples were used to generate cDNA. qPCR was performed using Power SYBR Green PCR master mix (Applied Biosystems). PCR primer sequences are listed in Supplementary Table 1.
Chromatin immunoprecipitation and qPCR
Foxp3 and IRF4 antibody (Santa Cruz Biotechnology) ChIPs were performed using MACS purified CD4+ CD25+ Treg cells as previously described4. qPCRs were performed using primers listed in Supplementary Table 1.
Immunoprecipitation and western blot analysis
Nuclear extracts from sorted CD4+ Foxp3+ and CD4+ Foxp3− cells were prepared in nuclear lysis buffer (Active Motif) according to the manufacturer's protocol. Immunoprecipitation was carried out using anti-Foxp3-coated magnetic beads followed by western blotting with Foxp3, IRF4 or p65 antibodies.
Supplementary Material
Supplementary Data
Acknowledgments
We thank K. Hilgner and K. Murphy for providing TH1, TH2 and TH0 gene expression data sets, B. Sullivan, R. Locksley, S. Quezada and J. Allison for critical reagents, K. Forbush and L. Karpik for expert technical assistance and mouse colony management, and R. Dalla-Favera for discussions. This work was supported by grants from the National Institutes of Health (A.Y.R.). Y.Z.and J.M.K. were supported by the CRI-Irvington Institute postdoctoral fellowship. A.Y.R. is an investigator with the Howard Hughes Medical Institute.
Footnotes
References
- 1.Ramsdell F. Foxp3 and natural regulatory T cells: key to a cell lineage? Immunity. 2003;19:165–168. doi: 10.1016/s1074-7613(03)00207-3. [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi S, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 3.Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nature Immunol. 2007;8:457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 4.Zheng Y, et al. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- 5.Marson A, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanangat S, et al. Disease in the scurfy (sf) mouse is associated with overexpression of cytokine genes. Eur J Immunol. 1996;26:161–165. doi: 10.1002/eji.1830260125. [DOI] [PubMed] [Google Scholar]
- 7.Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nature Immunol. 2007;8:191–197. doi: 10.1038/ni1428. [DOI] [PubMed] [Google Scholar]
- 8.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 9.Gavin MA, et al. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 10.Lin W, et al. Regulatory T cell development in the absence of functional Foxp3. Nature Immunol. 2007;8:359–368. doi: 10.1038/ni1445. [DOI] [PubMed] [Google Scholar]
- 11.Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nature Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- 12.Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 13.Mittrucker HW, et al. Requirement for the transcription factor LSIRF/IRF4 for mature B and T lymphocyte function. Science. 1997;275:540–543. doi: 10.1126/science.275.5299.540. [DOI] [PubMed] [Google Scholar]
- 14.Iida S, et al. Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nature Genet. 1997;17:226–230. doi: 10.1038/ng1097-226. [DOI] [PubMed] [Google Scholar]
- 15.Lohoff M, et al. Dysregulated T helper cell differentiation in the absence of interferon regulatory factor 4. Proc Natl Acad Sci USA. 2002;99:11808–11812. doi: 10.1073/pnas.182425099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rengarajan J, et al. Interferon regulatory factor 4 (IRF4) interacts with NFATc2 to modulate interleukin 4 gene expression. J Exp Med. 2002;195:1003–1012. doi: 10.1084/jem.20011128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brustle A, et al. The development of inflammatory TH-17 cells requires interferon-regulatory factor 4. Nature Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 18.Klein U, et al. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nature Immunol. 2006;7:773–782. doi: 10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 19.Rubtsov YP, et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity. 2008;28:546–558. doi: 10.1016/j.immuni.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 20.Collison LW, et al. The inhibitory cytokine IL-35 contributes to regulatory T-cell function. Nature. 2007;450:566–569. doi: 10.1038/nature06306. [DOI] [PubMed] [Google Scholar]
- 21.Li MO, Wan YY, Flavell RA. T cell-produced transforming growth factor-β1 controls T cell tolerance and regulates Th1- and Th17-cell differentiation. Immunity. 2007;26:579–591. doi: 10.1016/j.immuni.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 22.Takahashi T, et al. Immunologic self-tolerance maintained by CD25+ CD4+ regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J Exp Med. 2000;192:303–310. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobie JJ, et al. T regulatory and primed uncommitted CD4 T cells express CD73, which suppresses effector CD4 T cells by converting 5′-adenosine monophosphate to adenosine. J Immunol. 2006;177:6780–6786. doi: 10.4049/jimmunol.177.10.6780. [DOI] [PubMed] [Google Scholar]
- 24.Deaglio S, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med. 2007;204:1257–1265. doi: 10.1084/jem.20062512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao X, et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity. 2007;27:635–646. doi: 10.1016/j.immuni.2007.08.014. [DOI] [PubMed] [Google Scholar]
- 26.Nurieva RI, et al. Transcriptional regulation of Th2 differentiation by inducible costimulator. Immunity. 2003;18:801–811. doi: 10.1016/s1074-7613(03)00144-4. [DOI] [PubMed] [Google Scholar]
- 27.Ho IC, Lo D, Glimcher LH. c-maf promotes T helper cell type 2 (Th2) and attenuates Th1 differentiation by both interleukin 4-dependent and -independent mechanisms. J Exp Med. 1998;188:1859–1866. doi: 10.1084/jem.188.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chensue SW, et al. Aberrant in vivo T helper type 2 cell response and impaired eosinophil recruitment in CC chemokine receptor 8 knockout mice. J Exp Med. 2001;193:573–584. doi: 10.1084/jem.193.5.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohning M, et al. T1/ST2 is preferentially expressed on murine Th2 cells, independent of interleukin 4, interleukin 5, and interleukin 10, and important for Th2 effector function. Proc Natl Acad Sci USA. 1998;95:6930–6935. doi: 10.1073/pnas.95.12.6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frith MC, et al. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data