AtEXO70A1, a member of a family of putative exocyst subunits specifically expanded in land plants, is important for polar growth and plant development (original) (raw)
. Author manuscript; available in PMC: 2010 May 10.
Summary
The exocyst is a hetero-oligomeric protein complex involved in exocytosis and has been extensively studied in yeast and animal cells. Evidence is now accumulating that the exocyst is also present in plants. Bioinformatic analysis of genes encoding plant homologs of the exocyst subunit, Exo70, revealed that three Exo70 subgroups are evolutionarily conserved among angiosperms, lycophytes and mosses. Arabidopsis and rice contain 22 and approximately 39 EXO70 genes, respectively, which can be classified into nine clusters considered to be ancient in angiosperms (one has been lost in Arabidopsis). We characterized two independent T-DNA insertional mutants of the At_EXO70A1_ gene (exo70A1-1 and exo70A1-2). Heterozygous EXO70A1/exo70A1 plants appear to be normal and segregate in a 1:2:1 ratio, suggesting that neither male nor female gametophytes are affected by the EXO70A1 disruption. However, both exo70A1-1 and exo70A1-2 homozygotes exhibit an array of phenotypic defects. The polar growth of root hairs and stigmatic papillae is disturbed. Organs are generally smaller, plants show a loss of apical dominance and indeterminate growth where instead of floral meristems new lateral inflorescences are initiated in a reiterative manner. Both exo70A1 mutants have dramatically reduced fertility. These results suggest that the putative exocyst subunit EXO70A1 is involved in cell and organ morphogenesis.
Keywords: exocyst, evolution, Arabidopsis, root hairs, cell polarity, EXO70
Introduction
Only a few components of the plant secretory pathway have been identified by direct biochemical studies, whereas others have been identified on the basis of forward genetic screens. Fortunately, the core components of the secretory pathway, e.g. SNARE proteins, rat brain Ras related GTPase (RAB) and ADP-ribosylation factor (ARF) guanosine triphosphatases (GTPases), or coat complexes, appear to be highly conserved across eukaryotes (Dacks and Field, 2004; Sanderfoot and Raikhel, 2003). Rapidly accumulating expressed sequence tags (ESTs) and complete genome sequences for diverse plant species offer a straightforward way to find, on the basis of sequence homology, candidate plant secretory genes for reverse genetic and further experimental analyses.
Genetic analysis of secretion in the yeast Saccharomyces cerevisiae led to the identification of a series of SEC genes (Novick et al., 1980). Six out of 10 so-called late SEC genes encode components of a multimeric complex, which was denoted the exocyst because of its role in exocytosis (TerBush et al., 1996; recently reviewed in Hsu et al., 2004). Based on sequence homology, the mammalian exocyst was subsequently characterized (Kee et al., 1997). In both yeast and mammals, the exocyst complex consists of eight subunits called Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70 and Exo84 (Guo et al., 1999; Matern et al., 2001). The exocyst is involved in spatially regulated exocytosis, and is thought to tether secretory vesicles to the plasma membrane prior to the assembly of the SNARE complex leading to membrane fusion. In yeasts, the exocyst localizes at sites of extensive polarized vesicle exocytosis (Hsu et al., 1999): e.g. at the tip and neck of the budding yeast (TerBush and Novick, 1995), and at the furrow of dividing fission yeast (Fielding et al., 2005). In polarized animal epithelial cells, the exocyst is responsible for targeting proteins to the basolateral membrane, and localizes to the region of tight junctions (Yeaman et al., 2004); in neuronal cells, the exocyst is present at the tip of the growing neurite (Vega and Hsu, 2001). Numerous proteins interacting with the exocyst and regulating its localization and function have been described, many of them belonging to the superfamily of Ras-like GTPases (Lipschutz and Mostov, 2002).
Interaction of the exocyst with Rho GTPases, in particular, may define sites of landing and assembly of the tethering complex at the plasma membrane. In S. cerevisiae, the Exo70 subunit is known to interact with Rho3 GTPase, and may act together with Sec3, the Rho1-interacting exocyst subunit, as a spatial marker at the plasma membrane (Boyd et al., 2004; Roumanie et al., 2005). In the fission yeast Schizosaccharomyces pombe, the interaction of Rho3 with Exo70 is instrumental in the last step of cytokinesis called abscission (Wang et al., 2003). In animals, the Exo70 subunit was also recognized as the spatial landmark of the exocyst (Matern et al., 2001). In adipocytes and muscle cells, the assembly of the exocyst and the polarization of the glucose membrane transporter 4 (GLUT4) transporter at the plasma membrane was shown to occur as Exo70 interacts with the Rho GTPase TC10 during insulin-dependent glucose transport (Inoue et al., 2003).
Plant homologs of all eight subunits could be identified in silico (Cvrckova et al., 2001; Elias et al., 2003; Jurgens and Geldner, 2002). However, it had not yet been demonstrated whether exocyst subunits assemble in a similar complex and share the same functions in plants as they do in yeast and animals. Recently, an electron tomographic analysis of cell plate formation during cytokinesis of somatic cells and pollen development in Arabidopsis (Otegui and Staehelin, 2004; Segui-Simarro et al., 2004) uncovered the existence of a 24-nm long structure, which tethers membrane vesicles and resembles the mammalian exocyst as observed in the electron microscope (Hsu et al., 1998). In addition, a maize roothairless1 mutation, which is manifested by the failure of root hair primordia to elongate properly, was shown to result from a transposon insertion into the SEC3 gene (Wen et al., 2005). Phenotypic analysis of a series of T-DNA insertion mutants in the Arabidopsis SEC8 gene revealed that the putative Sec8 exocyst subunit is necessary for pollen tube germination (Cole et al., 2005). These findings suggest that the exocyst is conserved in plants and may be involved in cytokinesis and polarized exocytosis.
As reported previously (Elias et al., 2003), a striking feature of the Arabidopsis and rice genomes is the presence of a large family of genes coding for multiple isoforms of a putative Exo70 subunit. Here we provide a detailed phylogenetic analysis of the plant EXO70 family, establishing a phylogeny-based nomenclature of plant EXO70 paralogs. We characterized mutant Arabidopsis plants with a disruption of the EXO70A1 gene. Homozygous exo70A1-1 and exo70A1-2 mutants are small, sterile and demonstrate decreased apical dominance. Inflorescences are recurrently branched as a result of the ectopic initiation of lateral inflorescences instead of flowers. Additionally, the polar growth of root hairs and stigmatic papillae is altered, consistent with the expected function of EXO70A1 as a subunit of the putative plant exocyst complex.
Results
Plant species possess a large family of EXO70 genes
We identified 23 annotated Arabidopsis genes that are significantly similar to either the human or the yeast Exo70 (Table 1). TBLASTN searches of all Arabidopsis sequence data available (incl. ESTs) did not reveal any additional un-annotated EXO70 genes. Each of the 23 genes is described as an ‘exocyst subunit EXO70 family protein’ in the current annotation of the Arabidopsis genome (TAIR6.0), and animal and yeast Exo70 entries are the top-most non-plant hits. The Arabidopsis EXO70 family with the same 23 members has been also found independently by Cannon et al. (2004). One of these genes, here referred to as At_EXO70A3_ (At5g52350), is probably non-functional because it seems to be truncated at the N-terminus, and there is no evidence for its transcription in either ESTs or in experiments with the Arabidopsis whole-genome tiling array (Yamada et al., 2003) or ATH1 array (Affymetrix) (Zimmermann et al., 2004).
Table 1.
EXO70 genes in Arabidopsis thaliana
| Gene name | AGI gene symbol | No. exons | E-value (H.s./S.c.) | Duplication history | No. aa residues | Predicted m.w. (kDa) |
|---|---|---|---|---|---|---|
| At_EXO70A1_ | At5g03540 | 12 | 3e-54/3e-10 | 638 | 72.3 | |
| At_EXO70A2_ | At5g52340* | 11 | 1e-46/3e-08 | 631 | 72.1 | |
| At_EXO70A3_ | At5g52350* | 9 | 9e-23/8e-08 | 586 | 67.6 | |
| At_EXO70B1_ | At5g58430 | 1 | 1e-19/6e-06 | 624 | 70.6 | |
| At_EXO70B2_ | At1g07000 | 2 | 6e-12/0.070 | 599 | 67.7 | |
| At_EXO70C1_ | At5g13150 | 1 | 1e-17/9e-11 | 653 | 75.0 | |
| At_EXO70C2_ | At5g13990 | 1 | 1e-20/1e-04 | 695 | 79.8 | |
| At_EXO70D1_ | At1g72470 | 1 | 3e-19/5e-04 | Segmental duplication with At_EXO70D2_/D3 (round β) | 633 | 71.5 |
| At_EXO70D2_ | At1g54090 | 1 | 9e-19/3e-04 | Segmental duplication with At_EXO70D3_ (round α); segmental duplication with At_EXO70D1_ (round β) | 622 | 70.8 |
| At_EXO70D3_ | At3g14090 | 1 | 4e-20/9e-05 | Segmental duplication with At_EXO70D2_ (round α); segmental duplication with At_EXO70D1_ (round β) | 623 | 70.5 |
| At_EXO70E1_ | At3g29400 | 1 | 8e-15/8e-05 | 658 | 75.2 | |
| At_EXO70E2_ | At5g61010 | 1 | 1e-13/0.002 | 639 | 73.5 | |
| At_EXO70F1_ | At5g50380 | 1 | 2e-22/2e-11 | 637 | 72.0 | |
| At_EXO70G1_ | At4g31540 | 1 | 1e-17/0.001 | 686 | 77.6 | |
| At_EXO70G2_ | At1g51640 | 1 | 8e-05/0.51 | 660 | 76.2 | |
| At_EXO70H1_ | At3g55150 | 1 | 6e-05/1e-05 | Segmental duplication with At_EXO70H2_ (round α); segmental duplication with At_EXO70H3_/H4 (round β); segmental duplication with At_EXO70H5_/H6/H7/H8 (round γ) | 636 | 72.0 |
| At_EXO70H2_ | At2g39380 | 1 | 1e-05/8e-07 | Segmental duplication with At_EXO70H1_ (round α); segmental duplication with At_EXO70H3_/H4 (round β); segmental duplication with At_EXO70H5_/H6/H7/H8 (round γ) | 637 | 72.2 |
| At_EXO70H3_ | At3g09530 | 1 | 7e-05/0.034 | Tandem duplication with At_EXO70H4_; segmental duplication with At_EXO70H1_/H2 (round β); segmental duplication with At_EXO70H5_/H6/H7/H8 (round γ) | 637 | 72.1 |
| At_EXO70H4_ | At3g09520 | 1 | 0.005/3e-06 | Tandem duplication with At_EXO70H3_; segmental duplication with At_EXO70H1_/H2 (round β); segmental duplication with At_EXO70H5_/H6/H7/H8 (round γ) | 628 | 71.0 |
| At_EXO70H5_ | At2g28640 | 2 | 8e-12/9e-05 | Segmental duplication with At_EXO70H6_ (round α); segmental duplication with At_EXO70H7_ (round β); segmental duplication with At_EXO70H1_/H2/H3/H4 (round γ) | 605 | 68.8 |
| At_EXO70H6_ | At1g07725* | 1 | 4e-10/1e-05 | Segmental duplication with At_EXO70H5_ (round α); segmental duplication with At_EXO70H7_ (round β); segmental duplication with At_EXO70H1_/H2/H3/H4 (round γ) | 615 | 68.4 |
| At_EXO70H7_ | At5g59730 | 1 | 3e-09/12 | Segmental duplication with At_EXO70H5_/H6/H8 (round β); segmental duplication with At_EXO70H1_/H2/H3/H4 (round γ) | 634 | 71.2 |
| At_EXO70H8_ | At2g28650 | 1 | 1e-12/0.066 | Tandem duplication with At_EXO70H5_ (after round β but prior round α); segmental duplication with At_EXO70H7_ (round β); segmental duplication with At_EXO70H1_/H2/H3/H4 (round γ) | 573 | 64.8 |
We wanted to know if a large family of EXO70 genes is also characteristic for other plant species. Indeed, our analysis of the complete rice genome sequence (International Rice Genome Sequencing Project, 2005) revealed an even larger EXO70 family (Table S1). The actual number of rice EXO70 genes is not entirely clear, as in several cases it is difficult to distinguish functional EXO70 genes and pseudogenes. In the Oryza sativa ssp. japonica subspecies the total number of likely functional EXO70 genes reaches 39, with the EXO70H1a–EXO70H1b and EXO70H3a–EXO70H3b pairs coding for identical proteins. In the Oryza sativa ssp. indica rice the number may differ, as some presumed japonica pseudogenes seem to be functional genes in indica and vice versa (Table S1).
The first assembly of the poplar (Populus trichocarpa) genome has been released but not yet annotated. We therefore relied on TBLASTN searches with various Arabidopsis EXO70 sequences as queries and found 26 EXO70 genes and a probable pseudogene (Table S2). Recently, the first genome sequence of a non-angiosperm land plant, the lycophyte Selaginella moellendorffii, has become available (http://selaginella.genomics.purdue.edu/). Using tblastn, we identified five Selaginella EXO70 genes (Table S3). The whole-genome shotgun (WGS) sequencing project is well advanced for the moss Physcomitrella patens (see http://www.jgi.doe.gov/sequencing/why/CSP2005/physcomitrella.html), but the genome assembly has not yet been released. We were able to assemble and annotate 13 Physcomitrella EXO70 genes from available WGS reads, representing most if not all EXO70 genes in this species (Table S4). Finally, we identified a single EXO70 gene in the genomes of the chlorophytes Chlamydomonas reinhardtii and Volvox carteri.
We found that the exon–intron structure of some of the Arabidopsis and rice EXO70 genes and the Chlamydomonas EXO70 gene have been incorrectly predicted in the current database annotations (see Table 1, Table S1 and Appendix S1). For some rice EXO70 genes alternative splicing is supported by either full-length cDNAs or ESTs.
Embryophyte EXO70 proteins typically comprise 600–700 amino acid residues, with a predicted molecular weight mostly in the range between 70 and 80 kDa. This fits well the size of the human (684 aa/78.1 kDa) and S. cerevisiae (623 aa/71.3 kDa) Exo70. Recently, the structure of most of the yeast Exo70 protein (except the very N-terminus) has been determined (Dong et al., 2005; Hamburger et al., 2006). The protein forms a rod composed of contiguous α-helical bundles comprising 19 helices separated by loops (Dong et al., 2005). Multiple alignment of plant EXO70s revealed that regions corresponding to helices in the yeast Exo70 are generally well conserved, but that loops and the region N-terminal to the helix 1 of the solved structure are poorly conserved in both length and sequence. The N-terminal region (approximately helices 1–4) exhibits great sequence divergence, and a few predicted plant EXO70 proteins (a subset of the rice EXO70FX group, see below) completely lack the region N-terminal to the helix 2. Nevertheless, it is very likely that the plant EXO70 proteins assume a structure similar to that of the yeast Exo70, with possible differences in the N-terminal region.
Nine evolutionarily conserved clusters of EXO70 genes are present in angiosperms
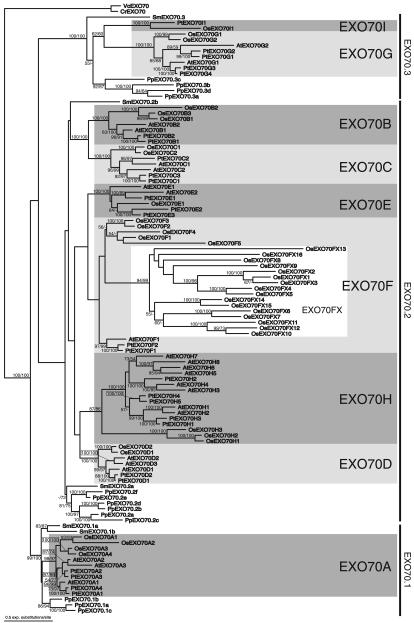
A broad phylogenetic analysis of EXO70 protein sequences showed that all land plant EXO70s form a monophyletic group to the exclusion of Exo70s from other eukaryotes (data not shown). We restricted further analyses on embryophyte and green algal EXO70 sequences, allowing for more reliably aligned positions. Figure 1 represents a maximum likelihood (ML) tree of Arabidopsis, poplar, rice, Selaginella and Physcomitrella EXO70s rooted with sequences from chlorophytes (Chlamydomonas and Volvox). We also constructed a Fitch–Margoliash tree based on Γ-corrected ML distances; this tree was largely congruent with the ML tree.
Figure 1. Protein maximum-likelihood phylogeny of the plant EXO70 family.
The tree was constructed with ProML as described in Experimental procedures. Bootstrap values (in %) based on PhyML/BIONJ analyses are shown for nodes that received support over 50%. Nine angiosperm EXO70 clusters (EXO70A-I) are highlighted with shading. Species abbreviations: At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; Os, Oryza sativa; Pp, Physcomitrella patens; Pt, Populus trichocarpa; Sm, Selaginella moellendorffii; Vc, Volvox carteri. All rice sequences are from O. sativa ssp. japonica cultivar Nipponbare, except EXO70F5 and EXO70FX12, which are from O. sativa ssp. indica, because the corresponding japonica sequences represent pseudogenes (see Table S1).
All angiosperm EXO70s analyzed group into nine clusters, each comprising sequences from both the monocot rice and the eudicots. We designated these clusters EXO70A–EXO70I (Figure 1). Seven of these clusters receive a strong support in bootstrap analysis, and the Exo70H cluster is moderately supported. In EXO70A, EXO70B, EXO70C, EXO70D, EXO70G and EXO70H clusters, rice and eudicot sequences form independent branches, implying a single ancestral gene for each of these clusters in the common ancestor of monocots and eudicots, and in the subsequent lineage-specific multiplications. In the EXO70E cluster, the single rice EXO70E gene branches off among eudicot sequences, but weak bootstrap support for this topology does not rule out a simpler history, with a single ancestral angiosperm EXO70E subsequently multiplicated in the eudicot lineage. A single EXO70I gene was also probably present in the monocot and eudicot common ancestor, but has been lost from the Arabidopsis lineage.
The EXO70F cluster is the only one lacking bootstrap support. It is not recovered in bootstrap consensus trees because the chlorophyte EXO70s branch within the EXO70F cluster, disrupting its monophyly. This is probably a result of the distantly related chlorophyte sequences being artificially attracted by either the highly divergent OsExo70F5 or by a rice-specific subgroup of the EXO70F cluster, here denoted as EXO70FX.
We were interested in probable evolutionary mechanisms yielding the huge number of EXO70 genes in angiosperms. Unequal crossing-over events could explain the tandemly arranged pairs EXO70A2–EXO70A3, EXO70H3–EXO70H4 and EXO70H5–XO70H8 in Arabidopsis and EXO70A1–EXO70A2, EXO70B1–EXO70B2 and EXO70FX14–EXO70FX15 in rice. Some other rice EXO70FX genes are arranged in groups of linked loci comprising several intact genes accompanied with deteriorating _EXO70_-like pseudogenes and fragments (see Table S1).
The origin of some other Arabidopsis and rice EXO70 genes can be clearly associated with presumed whole-genome duplication (WGD) events that occurred independently in the monocot and eudicot lineages (Bowers et al., 2003; Paterson et al., 2004). For example, EXO70D2 and EXO70D3 appear to result from the latest (α) WGD round, whereas the precursor of EXO70D2/EXO70D3 and EXO70D1 were generated by an earlier (β) round of duplication. An even more complex history can also be deduced for the Arabidopsis EXO70H cluster, including a duplication that can be traced to the most ancient γ WGD round (Table 1). The possible role of WGD events in the evolution of other EXO70 clusters cannot be excluded, but is not apparent in our present understanding of the duplication history of the Arabidopsis genome. On the other hand, the involvement of the presumed WGD that occurred in the ancestor of grasses, and resulted in the duplication of genes in most EXO70 clusters in rice, is readily evident (Table S1).
The rice EXO70H3a and EXO70H3b, which are 99% identical at the nucleotide level, map to regions of the chromosome 12 and 11, respectively, which represent a recent segmental duplication estimated to occur at 7.7 mya (Rice Chromosomes 11 And 12 Sequencing Consortia and Messing, 2005). EXO70H1a and EXO70H1b are completely identical at the nucleotide level and represent two independent loci on the chromosome 11, probably resulting from an even more recent duplication.
At least three EXO70 genes were present in the common ancestor of mosses and vascular plants
Our phylogenetic analysis further revealed that there are three principal EXO70 groups conserved across land plants (Figure 1). All three groups can probably be traced to three ancient EXO70 genes present in the common ancestor of mosses and vascular plants, which have been duplicated independently in the moss, lycophyte and angiosperm lineages.
The first group, further referred to as EXO70.1, is represented by the angiosperm EXO70A cluster, and two and three sequences from Selginella and Physcomitrella, respectively. The whole grouping is strongly supported by bootstrap analysis, but the relationship among EXO70A, and Selaginella and Physcomitrella EXO70.1 is not resolved. Contrary to the expectation based on the known relationship among embryophyte groups, Physcomitrella EXO70.1 sequences do not form the basal-most branch within the EXO70.1 group in the ML tree (Figure 1), but bootstrap support is lacking and the expected topology has been recovered in the Fitch–Margoliash tree.
The second major embryophyte EXO70 group, herein referred to as EXO70.2, comprises the angiosperm EXO70B, EXO70C, EXO70D, EXO70E, EXO70F and EXO70H clusters, a moderately supported cluster of six Physcomitrella sequences, and two Selaginella sequences. The EXO70.2 group lacks bootstrap support, but the reason for this is the same as for the EXO70F cluster (see above). Indeed in the ML tree constructed without the EXO70FX sequences, the monophyly of the EXO70.2 group is recovered with moderate support (76%). Apart from strong support for the sisterhood of the EXO70B and EXO70C clusters, the interrelationship within the EXO70.2 group remains unresolved. The affinities of the Selaginella EXO70.2a and EXO70.2b are especially unclear. It is likely that they represent a lycophyte-specific duplication independent of the duplication events that gave rise to the six angiosperm EXO70.2 clusters, but further analyses are needed to corroborate this hypothesis.
All the remaining EXO70 sequences, i.e. a single Selaginella representative, a strongly supported cluster of four Physcomitrella sequences, and the angiosperm EXO70G and EXO70I clusters, which branch sister to each other with weak bootstrap support, are included in the EXO70.3 group. Similarly to the EXO70.1 group, the EXO70.3 group is strongly supported but the internal topology of the group in the ML tree does not follow the expected basal-most position for Physcomitrella EXO70.3 sequences. However, the branching order is not well supported by bootstrap analysis, and a common ancestry of angiosperm and Selaginella EXO70.3 sequences to the exclusion of Physcomitrella EXO70.3 cannot be ruled out.
We investigated how the exon–intron structure of EXO70 genes correlates with the reconstructed phylogeny. Genes of the EXO70.1 group share 11 introns at conserved positions, with the exception of EXO70A2 and EXO70A3 from Arabidopsis and poplar, which have lost the corresponding 10th intron. The probable pseudogene At_EXO70A3_ in addition misses the first two exons. Most genes belonging to the EXO70.2 and EXO70.3 groups, especially in vascular plants, consist of only a single coding exon. No clear picture of the evolutionary history of the rare introns in the EXO70.2 and EXO70.3 genes can be drawn with the present data, but several independent intron gains (e.g. in At_EXO70B2_, At_EXO70H5_, Os_EXO70F5_ and in the EXO70FX subgroup) are probable given their largely non-conserved positioning. On the other hand, the intron conserved in the EXO70I cluster and the intron conserved in a subset of Physcomitrella EXO70.2 group genes seem to be ancient, as corresponding introns are present in the EXO70 genes from chlorophyte algae (Figure S1).
Expression patterns revealed a major role for the EXO70A1 gene
To gain insights into the developmental and organ-specific regulation of the expression of individual Arabidopsis EXO70 genes, we analyzed the publicly available Affymetrix ATH1 Arabidopsis genome array data in the Genevestigator database (Zimmermann et al., 2004). Except for EXO70A3, EXO70H4 and EXO70H6, all other 19 EXO70 genes are detectable at least in one microarray experiment (Figure 2). Probes for the EXO70H6 gene are missing on the ATH1 array but its expression is documented by several ESTs. No ESTs are available for EXO70H4 but the gene is transcriptionally active according to data from the Arabidopsis whole-genome tiling array (Yamada et al., 2003). For EXO70A3 alone there is no experimental evidence for transcription; as mentioned above, EXO70A3 is very likely to be an inactive pseudogene.
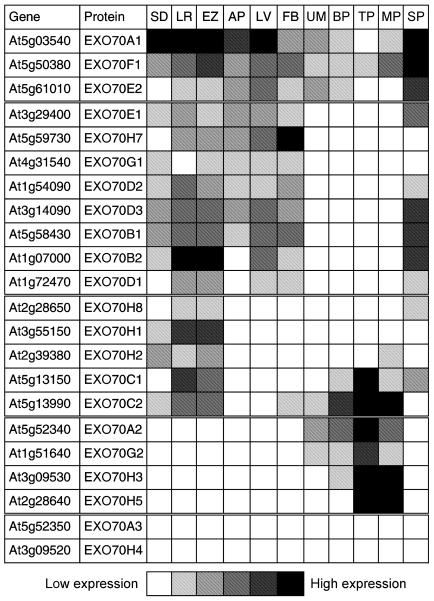
Figure 2. Expression analysis of Arabidopsis EXO70 genes.
Microarray data for Arabidopsis EXO70 genes were retrieved from the Genevestigator database. Sources of RNA for the expression analysis: seedlings (SD), lateral roots (LR), elongation zone of roots (EZ), aerial parts (AP), leaves (LV), flower buds (FB), uninucleate microspores (UM), bicellular pollen (BP), tricellular pollen (TP), mature pollen (MP) and suspension (SP). The particular microarray experiments (chips) included in this analysis are listed in Table S5.
According to their expression pattern, the expressed EXO70 genes can be roughly divided into four classes. EXO70A1, E2 and F1 are expressed in most organs and cells. Expression of EXO70B1, B2, D1, D2, D3, E1, G1 and H7 is detectable in sporophytic tissues and organs, but not during male gametophytic development. Complementary, EXO70A2, G2, H3 and H5 are only active during pollen development. The last group of genes, EXO70H1, H2, H8, C1 and C2, show a more restricted activity, being predominantly expressed in roots. EXO70A1 is the most strongly expressed gene except during pollen development.
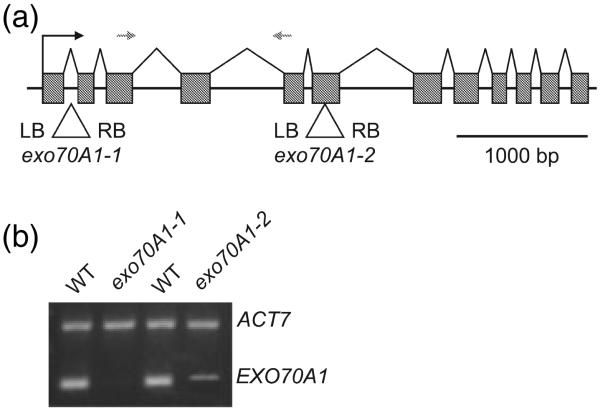
In order to functionally characterize the EXO70A1 gene, we obtained two different mutant lines (exo70A1-1 and exo70A1-2) with T-DNA insertions in the first intron and in the sixth exon respectively (Figure 3a). In addition to mutants in EXO70A1, we analyzed insertional mutants for five other EXO70 genes (EXO70B2, D3, F1, G1 and H7) that are mostly highly expressed. However, only exo70A1-1 and exo70A1-2 mutants exhibited a discernible phenotype (Figure S2).
Figure 3. EXO70A1 mRNA level analysis by RT-PCR.
(a) Two T-DNA insertional mutants, exo70A1-1 and exo70A1-2, were used in this study. exo70A1-1 and exo70A1-2 insertions interrupt the EXO70A1 gene (At5g03540) in the first intron and in the sixth exon, respectively (exons are boxed). Positions of primers used for EXO70A1 mRNA level analysis are indicated with grey arrows. LB, left border; RB, right border of T-DNA.
(b) Total RNA from wild-type Col-0 and exo70A1 plants was reverse-transcribed and PCR amplified using primers against EXO70A1. Primers specific to ACT7 (constitutively transcribed gene) were used as an internal control.
Heterozygotes EXO70A1/exo70A1-1 and EXO70A1/exo70A1-2 are indistinguishable from wild-type plants, and their offspring segregates in a Mendelian ratio 1:2:1. Homozygous exo70A1-1 and exo70A1-2 plants exhibit identical developmental alterations. No transcripts were detected in exo70A1-1 by semiquantitative RT-PCR (Figure 3b), indicating that exo70A1-1 is a null allele. In exo70A1-2, which has the T-DNA positioned in the middle of the EXO70A1 open reading frame, a weak level of transcription could be detected (Figure 3b). The phenotype identical to exo70A1-1 suggests either that the transcript does not allow for the sufficient production of EXO70A1 or that a non-functional protein is produced. Consequently, exo70A1-2 is probably also a null allele.
We did not analyze the expression levels of EXO70B2, D3, F1, G1 and H7 in the respective insertional mutants mentioned above (i.e. those without phenotypic deviations from the wild type).
EXO70A1 is required for hypocotyl elongation and tip growth of root hairs
Both exo70A1-1 and exo70A1-2 exhibit identical phenotypes (Figures 4-8). Mutant seedlings germinate normally, and their cotyledons reach the same size and shape as wild-type ones (data not shown).
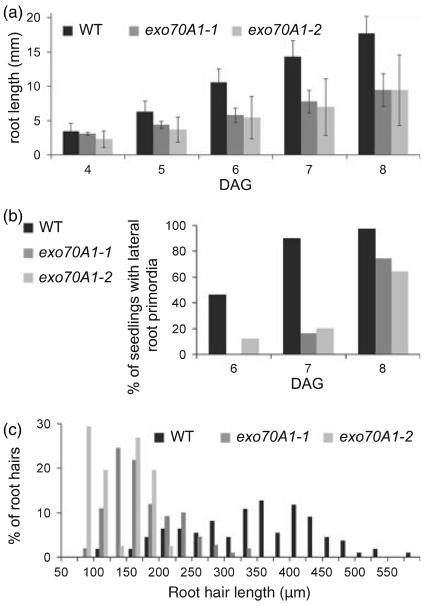
Figure 4. Quantification of root growth and root hair characteristics of exo70A1 mutants.
(a) Root growth of exo70A1-1 (grey) and exo70A1-2 (light grey) seedlings grown on solid MS medium supplemented with 4.5% sucrose grow slower than wild-type Col-0 (black). _T_-tests for exo70A1-1 to wild type: day 4, P = 0.064; days 5–8, P < 0.01. _T_-tests for exo70A1-2 to wild type: days 4–8, P < 0.01. DAG, days after germination.
(b) Lateral root initiation of exo70A1-1 and exo70A1-2 is similarly delayed in comparison with wild-type Col-0 seedlings. DAG – days after germination.
(c) Distribution of mature root hair lengths of exo70A1-1 and exo70A1-2 mutants and wild-type Col-0. Lengths of individual root hairs were measured in 7-day-old seedlings grown on solid MS medium without sucrose.
Figure 8. Phenotypes of generative organs of exo70A1 mutants.
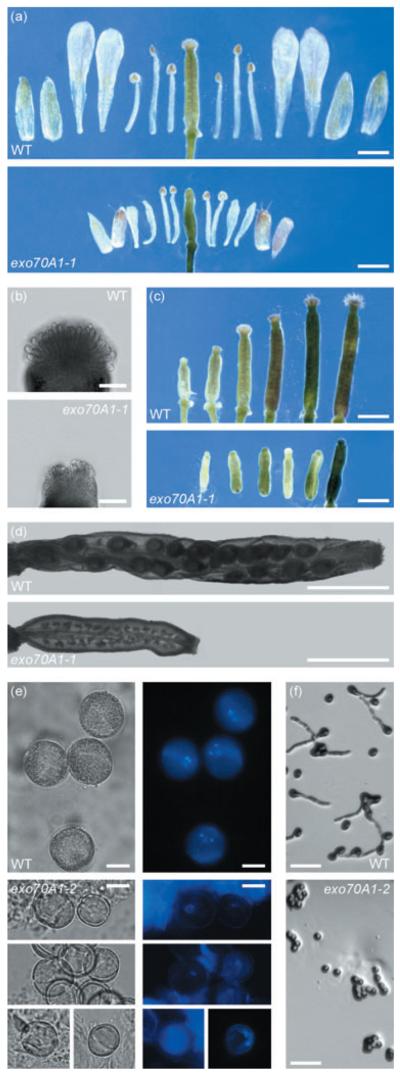
(a) Comparison of floral organ sizes in dissected wild-type Col-0 and exo70A1-1 flowers. All floral organs are shorter and the number of stamen is frequently reduced in mutant flowers. Bars = 1 mm.
(b) Stigmas after the opening of flowers. Bars = 0.1 mm.
(c) Pistil development in wild-type Col-0 and exo70A1-1. The two left pistils represent stages before anthesis, the third at and the last three after anthesis. Nearly no papillae elongation occurs after pollination in mutant flowers. Bars = 1 mm.
(d) Siliques 1 week after anthesis. exo70A1-1 siliques typically do not develop embryos. Bars = 1 mm.
(e) Pollen grains isolated from wild-type Col-0 and exo70A1-2 flowers after anthesis. Mutant pollen grains are smaller and mostly uninuclear as revealed by DAPI staining. Bars = 10 μm.
(f) In vitro germination of wild-type Col-0 and exo70A1-2. Pollen of homozygous mutants does not germinate under these conditions. Bars = 0.1 mm.
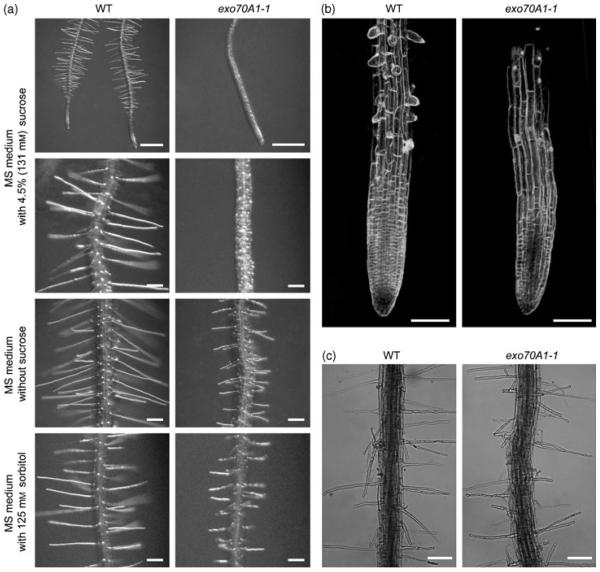
However, root growth is slower after 4 days in exo70A1 mutants, resulting in shorter roots (Figure 4a), and lateral root initiation is also retarded (Figure 4b). The length of root epidermal cells indicates that epidermal cell expansion is not significantly affected in exo70A1-1 (exo70A1-1, 121.4 ± 38.3 μm; wild type, 116.7 ± 42.8 μm; _t_-test _P_-value = 0.73).
We also measured etiolated hypocotyls of 7-day-old seedlings grown on medium supplemented with 4.5% sucrose. Both exo70A1 mutants develop shorter hypocotyls (exo70A1-1, 11.1 ± 1.8 mm, n = 16; exo70A1-2, 11.4 ± 1.9 mm, n = 18) than in wild type (16.3 ± 1.6 mm, n = 35). This difference is highly significant (_t_-test _P_-values <0.01). The average epidermal cell length in _exo70A1-1_ is not significantly different from wild type (_exo70A1-1_, 593.3 ± 107.7; wild type, 479.3 ± 38.8; _t_-test _P_-value = 0.073). However, the class of longest cells (>800 μm) present in wild type is missing (Figure S3). Moreover, the number of cells per cell file is reduced in exo70A1-1 (exo70A1-1, 18.3 ± 3.0, n = 9; wild type, 24.0 ± 2.1, n = 9; _t_-test _P_-value = 0.015). These results show that EXO70A1 is involved in both cell division and cell elongation in etiolated hypocotyls.
Addition of 5 μm 1-aminocyclopropane-1-carboxylic acid (ACC), an ethylene precursor, to the medium causes an inhibition of cell elongation of exo70A1-1 and exo70A1-2 hypocotyls (exo70A1-1, 3.7 ± 0.8 mm, n = 15; exo70A1-2, 4.1 ± 0.7 mm, n = 13; wild type, 6.0 ± 0.6 mm, n = 39). Thus, mutants respond to ACC treatment comparably with the wild type. The hook formation is also unaffected (Figure S4), so we conclude that both exo70A1 mutants are sensitive to ethylene and exhibit the triple response.
Furthermore, we observed a conditional tip growth defect of root hairs in exo70A1 mutants. Whereas on medium with 4.5% sucrose exo70A1-1 and exo70A1-2 initiate only bulges, which do not elongate and result in ‘naked’ roots (Figure 5a,b), on medium without sucrose root hairs do elongate, but are significantly shorter than the wild-type ones (Figures 4c and 5a). To reveal if this conditionality depends on either the osmotic or the metabolic properties of sucrose, we cultivated exo70A1-1 mutants on medium supplemented with sorbitol in concentration with a comparable osmotic potential. As seen in Figure 5a, the root hairs develop in the same manner as those on medium without sucrose. A similar result was obtained using mannitol in appropriate concentrations (data not shown). These results indicate that the inhibition of root hair elongation depends on the presence of sucrose, and is independent of the surrounding osmotic potential. When exo70A1-1 seedlings were grown in liquid medium between two slides, a higher percentage of branched root hairs was observed for homozygous mutants (56%, n = 101) than for wild types and heterozygotes (22%, n = 123; Figure 5c). All of these observations are indicative of polar growth defects.
Figure 5. Root phenotypes of exo70A1 mutants.
(a) Ten-day-old exo70A1-1 seedlings grown on solid MS medium supplemented with 4.5% sucrose initiate bulges, but do not elongate root hairs. On solid MS medium either without sucrose or with 125 mm sorbitol exo70A1-1 seedlings produce short root hairs. Detailed images were taken at a position where root hairs do not further elongate. Bars = 500 μm (first row) and 100 μm.
(b) CLSM images of root tips of 10-day-old seedlings grown on solid MS medium supplemented with 4.5% sucrose. The cell outlines were visualized by FM4-64 labeling. Bars = 100 μm.
(c) Root hairs 24 h after the transfer from solid MS medium without sucrose to liquid medium of similar composition. Under these conditions, a much higher percentage of exo70A1-1 root hairs are branched. Bars = 100 μm.
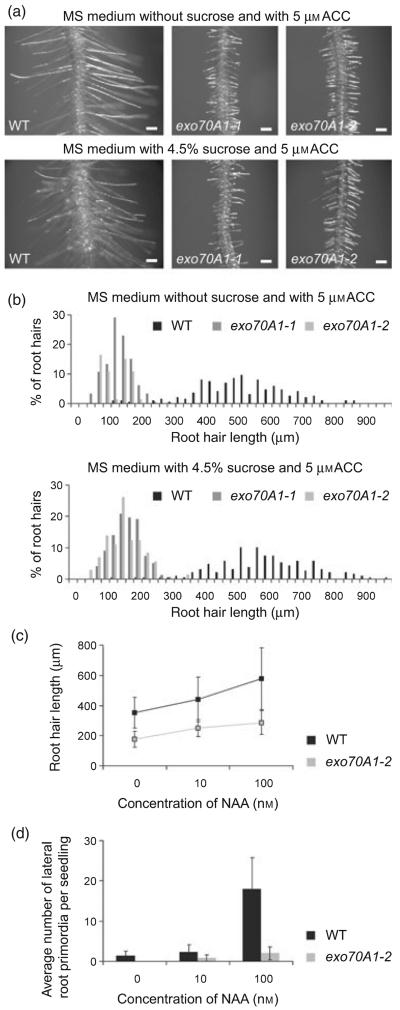
We asked whether the root hair growth could be rescued by treatment with ACC. ACC induces ectopic root hairs and enhances root hair elongation (Masucci and Schiefelbein, 1996; Pitts et al., 1998). In exo70A1-1 and exo70A1-2, the sucrose-dependent inhibition of root hair development was rescued by 5 μm ACC (Figure 6a), but root hairs in the mutants were still unable to reach the length of wild-type ones (Figure 6b).
Figure 6. Aminocyclopropane carboxylic acid (ACC) and naphtyl acetic acid (NAA) treatments.
(a) ACC overcomes the inhibition of root hair elongation in 7-day-old exo70A1 mutants on solid MS medium supplemented with 4.5% sucrose and 5 μm ACC. Bars = 100 μm.
(b) Distribution of root hair lengths of wild-type Col-0 (black), exo70A1-1 (grey) and exo70A1-2 (light grey) seedlings grown on solid MS medium supplemented with 5 μm ACC.
(c) Average lengths of mature root hairs of 7-day-old wild-type Col-0 and exo70A1-2, respectively, grown on solid MS medium supplemented with different concentrations of NAA. exo70A1-2 responds to NAA comparably with the wild type. _t_-test _P_-values <0.01 for all concentrations. (At least 100 root hairs per each data point were measured.)
(d) Average number of lateral root primordia per seedling of 7-day-old wild-type Col-0 and exo70A1-2, respectively, grown on solid MS medium supplemented with different concentrations of NAA. Initiation of lateral roots is retarded in exo70A1-2. (Eight exo70A1-2 and 20 wild-type seedlings, respectively, were inspected for each value).
Auxin is also known to promote root hair growth (Cernac et al., 1997; Leyser et al., 1996; Pitts et al., 1998; Wilson et al., 1990). Root hairs of exo70A1-2 cultivated on a medium containing either 10 nm or 100 nm of naphtyl acetic acid (NAA) elongated in a similar way as the wild-type ones (Figure 6c). However, the initiation of lateral roots was not enhanced by NAA (Figure 6d). Therefore, we concluded that exo70A1-2 is partly insensitive to auxin.
exo70A1 plants are smaller, exhibit reduced apical dominance and show an indeterminate growth
As described above, exo70A1 seedlings germinate normally and their cotyledons are indistinguishable from the wild-type ones. However, after approximately 2 weeks, and generally upon the formation of true leaves, the shoot phenotype becomes evident and results in semidwarf exo70A1-1 and exo70A1-2 plants with much smaller rosette leaves (Figure 7a). Nevertheless, the number of rosette leaves is similar to this of wild type (exo70A1-1, 7.3 ± 1.0, n = 31; wild type, 7.7 ± 0.8, n = 35). Comparison of the size and number of epidermal cells of rosette leaves shows that the cell size is not changed (Figure 7c), indicating that the smaller leaves are a result of fewer cell divisions.
Figure 7. General phenotypes of exo70A1 mutants.
(a) Three-week-old plants cultivated in vitro. Bar = 1 cm.
(b) Rosette leaves of 4-week-old plants. Bar = 5 mm.
(c) Epidermal cells of abaxial surface of cauline leaves have the same size and shape in exo70A1-1 as in wild-type Col-0. Bars 50 = μm.
(d) Six-week-old plants in soil. Bar = 1 cm.
(e) Inflorescences with cauline leaves of exo70A1-1 and exo70A1-2 exhibit a repetitive pattern. Wild-type Col-0 inflorescence is displayed for comparison. Bars = 1 mm.
(f) Four-month-old exo70A1-1 exhibiting indeterminate highly branched inflorescence and delayed senescence. Bar = 1 cm.
Although mutants develop secondary inflorescences earlier, the bolting time is similar to that of wild type (data not shown). However, the most dramatic phenotypical change occurs in the inflorescence architecture. Instead of initiating floral meristems at the flanks of the apical meristem, exo70A1-1 and exo70A1-2 also develop lateral secondary inflorescence meristems, resulting in an indeterminate highly branched inflorescence with cauline leaves (Figure 7e). This repetitive pattern of secondary inflorescences correlates with a significantly delayed senescence. The lifespan of exo70A1-1 and exo70A1-2 is approximately 5 months, i.e. more than twice the lifespan of wild-type plants. Whereas wild-type plants reach a maximum height of approximately 30 cm, exo70A1 mutants reach at most 12 cm (Figure 7f).
exo70A1 mutants display impaired flower development and are nearly sterile
Although a large number of flowers develop during the indeterminate growth, the exo70A1-1 and exo70A1-2 mutants are nearly sterile. Only four out of >100 propagated mutants produced in total 10 siliques with 63 seeds. All germinated, gave rise to plants that exhibited the mutant phenotype and were indeed shown to be homozygous mutants by PCR genotyping, thus excluding cross-pollination by wild-type pollen.
To explain the diminished fertility of the mutant we examined the development of floral organs precisely. Mature mutant flowers reach about half the size of wild-type flowers and have a variable number of stamens, which is reduced to either four or five in 33% of exo70A1-2 flowers. As the stamens reach the stigma, heterostyly cannot be the reason for the infertility (Figure 8a).
However, the stigmatic papillae are not fully developed and resemble the early stages of wild-type stigma development before pollination (Figure 8b). Nearly no elongation of papillae occurs unlike that seen in the wild type (Figure 8c). Siliques usually start to form but they typically do not contain seeds. After a short phase of elongation they stop their development at a length of only 2.1 ± 0.2 mm (n = 40; Figure 8d). Manual pollination of mutant stigmas by either mutant or wild-type pollen always failed, indicative of defective stigmatic function.
We also found defects in anther development. Anthesis is remarkably delayed and does not happen until the petals start to fall off and the pistil has become slightly elongated. All anthers examined contained fewer pollen grains compared with the wild type. Tetrads are formed, and microspores are released from the tetrads, but only very rarely do pollen grains resemble wild-type mature pollen grains (Figure 8e). They contain either only one or two nuclei and their average diameter is distinctively smaller (exo70A1-2, 16.2 ± 1.4 μm; n = 62, wild type, 19.8 ± 1.3 μm, n = 37). Pollen of exo70A1-2 was unable to germinate in vitro (Figure 8f) and to pollinate wild-type flowers. To test the hypotheses that either the pollen defect depends on the sporophyte or is cell autonomous, we analyzed the pollen of heterozygotes. Neither decrease in germination efficiency with respect to the wild type nor dimorphic pollen was observed in EXO70A1-2/exo70A1-2, suggesting that the pollen defect depends on the sporophyte. Mutant pollen of heterozygous plants is able to fertilize, and the stigma is fully functional, leading to a segregation ratio that is not significantly different from the expected ratio 1:2:1 (n = 208, χ2 = 0.952, _P_-value = 0.621). Slower growing mutant pollen tubes result in an unequal distribution of mutant seeds within the resultant silique (Goubet et al., 2003). To reveal whether exo70A1-2 pollen tubes either develop slower or are shorter we analyzed EXO70A1-2/exo70A1-2 siliques after self-pollination. However, the percentage of homozygous mutant seeds in the top half (30%, n = 221, χ2 = 2.929, _P_-value = 0.087) and in the bottom half (23%, n 186, χ2 0.581, _P_-value = 0.446) corresponds to a random distribution. We concluded that the almost complete sterility of homozygous exo70A1 mutants is caused by a combination of underdeveloped stigmatic papillae and disturbed anther/pollen development resulting from sporophytic dysfunction.
Discussion
The vastly expanded EXO70 family is a unique feature of land plants
The exocyst has been extensively studied in yeast and animal cells, but there has been no systematic survey of the exocyst subunit homologs in other eukaryotes. We searched sequences from diverse eukaryotes to see whether the large EXO70 family is an exclusive feature of land plants. Only a single EXO70 gene occurs in genomes of animals, fungi, Dictyostelium, Entamoeba, diatoms or Phytophthora, and some organisms (e.g. kinetoplastids or ciliates) are even devoid of a detectable EXO70 homolog. The only known organism apart from land plants with more that one EXO70 gene is the parabasalid Trichomonas vaginalis with three closely related paralogs. Hence, just a single EXO70 gene has been retained in most organisms including vertebrates whose ancestor passed through two rounds of whole-genome duplications (Dehal and Boore, 2005), and even in teleost fish, which encountered another round of a whole-genome duplication (Volff, 2005). The so-called balance hypothesis argues that the duplication of genes encoding subunits of complexes may lead to deleterious changes in stoichiometry, and is therefore under negative selection (Papp et al., 2003).
Interestingly, some of the remaining exocyst subunits in plants are also encoded by multiple paralogous genes, but their number is not comparable with the extent of the Exo70 set. For example, Arabidopsis has two SEC3, SEC5 and SEC15 paralogs and three EXO84 paralogs (Elias et al., 2003), whereas for EXO70 this number reaches to 22 likely functional genes. In rice, the difference is even more pronounced (M. Elias, unpublished data). In the light of the balance hypothesis, this discrepancy strengthens a notion that sequence homology to the yeast and animal Exo70 exocyst subunit may not necessarily indicate a homologous cellular function. It is possible that only some of the plant EXO70 proteins serve as subunits of the plant exocyst complex, whereas others have been recruited to perform a function independent of the remaining exocyst subunits. We suggest that the EXO70.1 proteins (including the angio-sperm EXO70A cluster) are the best candidates for genuine exocyst subunits, because EXO70.1 protein sequences are more similar than other plant EXO70s to yeast and especially mammalian Exo70s (see E-values in Table 1). Indeed, shorter branches of EXO70.1 sequences in the phylogenetic tree (Figure 1), as compared with EXO70.2 and EXO70.3 branches, indicate that they diverged less from the ancestral EXO70 sequence and are therefore more likely to retain the ancestral function.
However, we cannot discount the possibility that EXO70s in plants do not obey the implications of the balance hypothesis, and that most or even all of them can really incorporate into the exocyst complex. In which case the plant cell would be endowed with a number of different forms of the exocyst, each with a specific EXO70 subunit and potentially a different function. Biochemical studies are necessary to answer the puzzle of the large plant EXO70 family.
EXO70 genes were duplicated many times independently during the evolution of land plant phyla
Based on our phylogenetic analysis we inferred that at least three EXO70 genes were already present in the common ancestor of mosses and vascular plants. Given the presence of only one EXO70 gene in the chlorophytes Chlamydomonas and Volvox, and in most other eukaryotes, it is very likely that a single EXO70 gene is an ancestral condition for green plants (Viridiplantae). The expansion of the EXO70 family thus started somewhere in the lineage leading to mosses and vascular plants after its divergence from the chlorophyte lineage, but sequences from charophyte green algae, hornworts and liverworts are required to pinpoint when this happened.
Reconstruction of the evolutionary history of the EXO70 family in angiosperms based on genomes of three species uncovered nine conserved clusters that can be traced to nine ancestral genes in the common ancestor of monocots and eudicots. EXO70 genes identified in partial genome sequences from other angiosperms (Medicago truncatula, Lotus japonicus and maize; data not shown) can all be unambiguously placed into one of the nine clusters, suggesting that we have already grasped the essence of the angiosperm EXO70 diversity. We would need data from moniliforms (ferns and horsetails) and gymnosperms to determine more precisely how the ancestral EXO70.2 gene has given rise to the nine angiosperm EXO70 clusters.
The moss lineage represents an independent history of EXO70 gene proliferation (Figure 1). Our survey of the Selaginella genome suggests that the lycophyte lineage has been less obsessed with inflating the EXO70 family, but data from other lycophytes are needed to ascertain whether Selaginella is a typical representative of its phylum.
The evolution of the EXO70 family in plants is not only a story of diversification but also of gene losses, as exemplified by the absence of an EXO70I gene from Arabidopsis. EXO70I orthologs are present in rice and poplar, and are evident in the expanding genomic databases for maize and Medicago, suggesting that EXO70I is widespread in angiosperms and was clearly present already in the common ancestor of monocots and eudicots. Poplar and Medicago represent the fabid (or eurosid I) lineage belonging to rosids, which comprise also the malvid (or eurosid II) clade with Arabidopsis as a notorious representative (Judd and Olm-stead, 2004). We may therefore infer that the absence of an EXO70I gene in Arabidopsis is a result of a gene loss that most likely occurred not earlier than in the ancestor of the malvid lineage. It would be interesting to define the role of EXO70I in either rice or Medicago, and then to examine whether the corresponding process has been either simplified or lost altogether from Arabidopsis.
The reduced organ size of exo70A1 mutants is the result of organ-specific reduction of cell division
We started to characterize the function of the EXO70 family in Arabidopsis using a reverse genetic approach. Among the insertional mutants obtained, only T-DNA insertions in the EXO70A1 gene resulted in a discernible phenotype. Despite the fact that most of the other genes studied (EXO70B2, D3, F1, G1 and H7) are highly expressed, the respective insertional mutants (all of which have T-DNA insertions positioned in exons) showed no obvious phenotype, suggesting that EXO70 genes may be functionally redundant. However, as we did not analyze the expression levels of EXO70B2, D3, F1, G1 and H7 in the respective insertional mutants, we also cannot exclude the possibility that some of these mutant loci are still functional.
In all organs from the exo70A1-1 and exo70A1-2 investigated, cell morphology generally does not appear to be affected. Epidermis of leaves and flowers consists of cells that have the same size as in wild type. Organ size is, however, strongly diminished. The number of stamens is often reduced in mutant flowers, pointing to the possibility of reduced cell number in floral primordia. Thus we suggest that cell division may be affected, resulting in a decreased total number of cells and consequently in small plants. Alternatively, the small stature of exo70A1-1 and exo70A1-2 might be explained by insufficient nutrition because of underdeveloped root hairs. However, we do not choose this hypothesis because several root hair mutants, which grow well and cannot be distinguished from wild type in any other way, have been characterized (rhd2 and rhd4, for example; Schiefelbein and Somerville, 1990).
In eukaryotes, cell division depends on targeting exocytic vesicles, e.g. either to the cell plate in plants or to the cleavage furrow in animals (Guertin et al., 2002), and a direct role of the exocyst in this process has been demonstrated in S. cerevisiae (Dobbelaere and Barral, 2004; VerPlank and Li, 2005) and mammals (Fielding et al., 2005; Gromley et al., 2005). Interestingly, there is now indication that the cytokinetic role of the exocyst may also be conserved in plants, because exocyst-like particles were observed by electron tomography to tether vesicles during the formation of cell plate in Arabidopsis meristematic cells (Segui-Simarro et al., 2004) and developing pollen (Otegui and Staehelin, 2004). Therefore, we hypothesize that cell division is dependent on EXO70A1, and that the loss of function of this crucial exocyst subunit leads to reduced cell division.
EXO70A1 is involved in polarized cell growth
Cells featuring polarized cell growth such as tip-growing root hairs, stigmatic papillae and hypocotyl cells show reduced length in exo70A1 mutants. One of the most conspicuous phenotypes is the lack of root hairs on plants grown in medium supplemented with 4.5% sucrose. However, bulges are formed, suggesting that there is a defect in the transition to tip growth. Omitting sucrose from the medium allows the roots to develop root hairs, but they are significantly shorter compared with wild-type plants, indicating a defect in tip growth under these conditions. Furthermore, the higher frequency of branched root hairs in exo70A1 mutants observed in liquid medium without sucrose indicates that not only the tip growth itself is affected, but that the polarity is also poorly established and/or maintained.
Genetic analysis of root hair growth has led to the characterization of a number of Arabidopsis root hair mutants (Grierson et al., 1997; Masucci and Schiefelbein, 1994; Schiefelbein and Somerville, 1990; Schiefelbein et al., 1993). Defects in three phases of root hair development can be recognized: (i) initiation of a site for hair growth and formation of a bulge, (ii) transition to slow tip growth, and (iii) rapid tip growth, which starts when the hair is between 20 and 40 lm long (Dolan et al., 1994). The exo70A1 mutants appear to be defective in rapid tip growth, but their phenotype does not match any of the previously described mutants.
Among known root hair mutants, rhd3 and tip1 resemble the exo70A1 mutants in that the stature and architecture of the plant is also affected (Schiefelbein and Somerville, 1990; Schiefelbein et al., 1993; Wang et al., 1997). RHD3 encodes a putative GTP-binding protein that controls anterograde vesicle trafficking between the endoplasmic reticulum and the Golgi complex (Wang et al., 1997; Zheng et al., 2004). TIP1 codes for a protein with palmitoyl transferase activity, but its exact cellular role has not been defined (Hemsley et al., 2005). Phenotypes of the rhd3, tip1 and exo70A1 mutants are generally similar. In contrast to exo70A1 mutants, where cell number is reduced and cell dimension is less prominently decreased, in the rhd3 and tip1 mutants cells are smaller.
Importantly, a phenotype very similar to that of exo70A1 mutants has been recently reported for the maize roothairless1 (rth1) mutant bearing a transposon insertion in a gene coding for a homolog of the Sec3 exocyst subunit (Wen et al., 2005). Based on the Arabidopsis exo70A1 and maize rth1 phenotypes, we suggest that both the Arabidopsis EXO70A1 and maize RTH1 (SEC3) proteins function in vesicle delivery during tip growth of root hairs as subunits of the plant exocyst complex.
The Arabidopsis Ras homology (RHO) GTPase ROP2 acts as a positive regulator of root hair initiation and elongation (Jones et al., 2002). In both yeast and mammalian cells the Exo70 subunit serves as an effector of GTPases of the RHO family (Adamo et al., 1999; Inoue et al., 2003). Taken together, it is tempting to speculate that EXO70A1 could equivalently interact with ROP2 to mediate its regulatory role in root hair development.
The inhibitory effects of sucrose on the transition to tip growth in root hairs in exo70A1 mutants is not an osmotic effect of sucrose, and it is rescued by ACC treatment. Sucrose is known to influence ethylene signaling (Gazzarrini and McCourt, 2001; Gibson, 2004), which is required for root hair elongation (Pitts et al., 1998). It is possible that in exo70A1 mutants the ethylene signaling pathway is more sensitive to sucrose than in the wild type.
The exact mechanism of extension of stigmatic papillae has, to our knowledge, not yet been established. However, the similarity between the failure of stigmatic papillae elongation and the defect in root hair tip growth in exo70A1 mutants points to a possibility that papillae elongation might also proceed via tip growth.
In contrast, growth of pollen tubes harboring the exo70A1 mutation is unaffected. We could not evaluate the growth of the mutant pollen tube directly, because pollen of homozygous mutants did not germinate in vitro. However, several lines of evidence suggest that mutations in EXO70A1 do not disturb pollen tube function. First, mutant alleles do not exhibit reduced transmission, as indicated by the expected segregation ratio of 3:1 in offspring of heterozygous plants. Second, the percentage of homozygous mutant seeds in the top and bottom half of siliques of EXO70A1-2/exo70A1-2 plants did not differ significantly, indicating that the in vivo growth rate of mutant pollen tubes is not reduced compared with wild-type pollen tubes. Recently, the putative SEC8 exocyst subunit was shown to be involved in pollen germination and tube growth in Arabidopsis (Cole et al., 2005). Null alleles of SEC8 completely abrogate pollen germination, whereas hypomorphic alleles lead to a reduced pollen tube growth rate. These results are consistent with the expected role of the exocyst complex in polar cell growth. Why do we not observe a similar phenotype for the exo70A1-1 and exo70A1-2 mutants? Although the EXO70A1 gene is expressed at least in some stages of pollen development (Figure 2), there is in Arabidopsis a closely related paralog EXO70A2, the expression of which is restricted to pollen and seems to be much stronger compared with that of EXO70A1. We therefore suggest that EXO70A2 can fully compensate for the loss of function of the EXO70A1 gene in pollen.
The almost complete sterility of homozygous exo70A1 mutants results from a combination of underdeveloped stigmatic papillae and aberrant pollen development caused by sporophytic defects in the anther function. A couple of mutants have been described where the defective tapetum, which is important for the nutrition of developing pollen, results in the arrest of pollen development. In this respect, exo70A1 mutants resemble, e.g. the ms1 (male sterility) Arabidopsis mutant, where pollen development is arrested at the uninuclear stage (Wilson et al., 2001).
EXO70A1 affects shoot meristem function
The exo70A1 mutants also display an array of phenotypic defects that are more difficult to explain in the perspective of the known EXO70 function in fungi and animals. The most dramatic phenotypic alterations in exo70A1 mutants occur in the inflorescence architecture and lifespan. In spite of normal bolting time, secondary inflorescences are initiated earlier. Mutants produce highly branched inflorescence with cauline leaves as a result of the recurrent initiation of new ectopic lateral inflorescences at the positions of the wild-type flower initiation and formation. This might be partly similar to the process of reversion of flower primordia to inflorescence primordia, which seldom occurs in the Arabidopsis Ler ecotype. Such reversions are very frequent in either lfy-6 or ag-1 mutants because of the weakening of the repression of inflorescence development and the reprogramming of the original flower primordia (Okamuro et al., 1996; Tooke et al., 2005). Upon overexpression of AGL24, flower primordia are also reverted to inflorescence primordia (Yu et al., 2004). It is unclear how and whether EXO70A1 relates to the function of these genes. However, it is interesting that a role for the exocyst complex in cell fate determination was reported recently in Drosophila (Jafar-Nejad et al., 2005).
The longevity of exo70A1 mutants is certainly linked to the indeterminate growth based on reiterative initiation of new inflorescences, along with the lack of fruit production. A similar phenotype is well documented in different mutants including ms1 (discussed above) and mutants with floral meristem reversion (Tooke et al., 2005; Wilson et al., 2001). It is improbable that the longevity results from decreased sensitivity to ethylene because exo70A1 mutants exhibit normal triple response and their root hair development is sensitive to ethylene.
Some aspects of the exo70A1 mutant phenotype, mainly the reduced auxin sensitivity in root hair elongation, delayed lateral root initiation and reduced root growth, point to the possibility that either auxin transport or signalling is partially compromised. In an attempt to build a mechanistic model of polarized auxin transport, Muday and Murphy (2002) suggested using the well-studied mechanisms of establishing the membrane localization of GLUT4 in response to insulin as a paradigm for speculations about mechanisms and proteins regulating polarized localization of PINFORMED (PIN) protein. Interestingly, the exocyst complex and its EXO70 subunit interacting with the TC10 Rho-related GTPase is required for the delivery of the GLUT4 transporter to specific domains of the plasma mebrane (Inoue et al., 2003, 2006). It is therefore possible that the EXO70A1 protein, probably as a subunit of the exocyst complex, might be involved in localizing PIN proteins to the plasma membrane, and that the phenotypic defects caused by EXO70A1 disruption are caused by disturbed auxin transport resulting from less efficient polarized PIN proteins localization. We are currently testing this hypothesis experimentally.
This report along with previously published genetic analyses of the function of putative exocyst subunits SEC3 (Wen et al., 2005) and SEC8 (Cole et al., 2005) establishes an essential base for further studies on the functions of exocyst subunits (and putative exocyst complex) in the plant cell and organ morphogenesis of plants.
Experimental procedures
Bioinformatic identification of EXO70 genes
Arabidopsis EXO70 proteins were identified by psi-blast (Altschul et al., 1997) with default settings searching against the non-redundant protein database (including all proteins from the TAIR6.0 annotation of the Arabidopsis genome) at the National Center for Biotechnology Information (NCBI, http://www.ncbi.nlm.nih.gov/BLAST/). Searches with the human Exo70 as a query converged with the third iteration. The highest but still significantly low E-value (2 × 10−58) had a match with a protein entry representing a fragment of At1g07000 (the corresponding full-length protein matched the query with E-value 7 × 10−154). Only one additional protein (NP_196133.3) was found with an E-value (0.76) lower than the default threshold of 10, but it is obviously not related to Exo70.
Genomic, cDNA, EST and protein sequences from Arabidopsis thaliana and Oryza sativa were searched using the blast server at NCBI. Annotated protein sequences from the tigr release three assembly of the rice genome were searched with blast at tigr rice genome annotation database (http://www.tigr.org/tdb/e2k1/osa1/). We also checked the latest assembly (release 4) of the rice genome generated by The International Rice Genome Sequencing Project by blast, but no additional EXO70 gene was found. A draft genome sequence (version 1.0) of P. trichocarpa was searched with tblastn at the DOE Joint Genome Institute (http://genome.jgi-psf.org/Poptr1/Poptr1.home.html). A draft genome sequence (Purdue contigs 12/28/05) of S. moellendorffii was searched with tblastn at the Selaginella Genomics web-site (http://selaginella.genomics.purdue.edu/). A draft genome sequence and predicted proteome (version 3.0) of C. reinhardtii were downloaded from DOE Joint Genome Institute and searched with stand-alone blast downloaded from ftp://ftp.ncbi.nlm.nih.gov/blast/executables/.
Sequence reads from whole-genome shotgun sequencing projects for P. patens (≈6 500 000 reads) and V. carteri (≈2 900 000 reads) were downloaded from the Trace Archive maintained at NCBI (http://www.ncbi.nlm.nih.gov/Traces/) and searched with stand-alone blast. Reads identified by tblastn searches with plant EXO70 sequences were subsequently used as queries in blastn searches against the read databases, and those reads matching the query with at least 94% were assembled to create a contig. The contig was then extended by repeated blast searches as needed to assemble a complete gene. The coverage of both genomes represented by the reads available for our analysis was already high enough to allow for complete and reliable assembly of all EXO70 genes detected by the blast searches.
Either prediction or correction of the exon–intron structure of EXO70 genes was performed manually with the aid of ESTs and cDNAs (when available) and homology comparison, additionally taking advantage of the conservation of exon–intron structures among some plant EXO70 genes. In subsequent analyses we used the corrected sequences.
Phylogenetic analysis
Protein sequences were aligned with clustal × 1.83 (Thompson et al., 1997). The multiple alignment was manually adjusted using GeneDoc (http://www.psc.edu/biomed/genedoc/). Unreliably aligned regions and regions with extensive gaps were removed, resulting in 297 aligned positions of 106 land plant EXO70 sequences and two chlorophyte sequences. Phylogeny was inferred by the ML method using the ProML program in the PHYLIP 3.6 package (Felsenstein, 2005) with the JTT matrix, Γ-corrected among site–rate variation (the α parameter estimated using TREE-PUZZLE, Schmidt et al., 2002), and a randomized input order of sequences with two jumbles and global rearrangements. A distance tree was computed with Fitch–Margoliash (as implemented in the Fitch program in the PHYLIP 3.6 package) using a ML distance matrix produced with TREE-PUZZLE (the JTT matrix, Γ-corrected among-site rate variation with eight rate site categories plus a category for invariable sites, all parameters estimated from the data), global rearrangements and randomized input order of sequences with 10 jumbles. Bootstrap analysis was performed with replicates generated by SEQBOOT (in PHYLIP 3.6). One hundred bootstrap trees were inferred using PhyML and a JTT + Γ + I model with parameters as estimated for the ML tree computed with PhyML tree from the original data (eight rate site categories plus a category for invariable sites, all parameters estimated from the data). Γ-corrected distances were calculated for 500 bootstrap replicates using protdist in the phylip package (α parameter estimated with TREE-PUZZLE from the original data). bionj (Gascuel, 1997) was then used to infer trees from the distance matrices. Bootstrap values were calculated by consense in the PHYLIP package.
Expression analysis
Public expression data from experiments using the Affymetrix ATH1 Arabidopsis genome array were downloaded from the Genevestigator database (http://www.genevestigator.ethz.ch; Zimmermann et al., 2004). We selected only experiments on different organs/tissues from wild-type plants grown under physiological conditions (Table S5). Average values of normalized signal intensities were determined for parallel chips after the exclusion of non-significant values (P > 0.05). For representational purposes we grouped expression values into six categories: 0–400, 401–800, 801–1200, 1201–1600, 1601–2000, >2000.
Plant material and growth conditions
Lines of Columbia-0 ecotype of A. thaliana L. Heynh with T-DNA insertions were obtained from the SALK Institute (Alonso et al., 2003) and Syngenta (Sessions et al., 2002) collections as follows: exo70A1-1 – SALK_014826; exo70A1-2 – SALK_135462; exo70B2 – Garlic_339_D07; exo70D3 – Garlic_175_D08; exo70F1 – SALK_036927; exo70G1 – SALK_099892; exo70H7 – SALK_072673.
Seeds were surface-sterilized (either 3 min in 70% ethanol, with two times 5 min in 10% v/v commercial bleach or in 5% hypochlorite v/v with a few drops of Tween-20; as described in Hauser et al., 1995; and finally rinsed three times with sterile distilled water) and dispersed onto Petri plates with growing medium: 1× MS-salts (Sigma, St Louis, MO, USA), either 0% or 4.5% (w/v) sucrose (Fluka, Buchs, Switzerland), 1% (w/v) agar (Duchefa, Haarlem, the Netherlands), buffered to pH 5.7. Stratification was performed at 4°C for 3 days. Seedlings for root hair observations and measurement were grown vertically on 1.6% agar. Plants were grown in a climate chamber at 22°C under long-day conditions (16 h of light per day). Seedlings were transferred from plates to Magenta jars 10 days after germination and subsequently to soil after 14 more days.
For microscopy of root hair morphology 6-day-old seedlings picked from vertical agar (MS medium without sucrose) were cultivated vertically between two slides for 24 h in liquid MS medium of the same composition.
Elongation of etiolated hypocotyls was measured 7 days after germination in complete darkness. Seedlings were cultivated vertically on MS medium supplemented with 4.5% sucrose either with or without 5 μm ACC.
Genotype analysis
T-DNA insertions in EXO70 genes were followed by a PCR genotyping of individual plants using primers annealing to the left border of the T-DNA sequence (either LBb1 or LB3, recommended by either salk or Syngenta, respectively) and a set of gene-specific primers. The presence of T-DNA insertions elsewhere in the genome was probed by primers specific to nptII (kanamycin resistance) gene. All primer sequences are listed in Table S6.
DNA was extracted from young flower buds by alkali treatment as described Klimyuk et al. (1993).
mRNA level analysis by RT-PCR
Samples of leaves and shoots of 3-week-old plants were harvested and immediately frozen in liquid nitrogen. Total RNA was extracted using RNeasy Plant Mini Kit (Qiagen, Germantown, MD, USA) according to the manufacturer's instructions. RNA (3 μg) was converted to cDNA by the AMV First Strand cDNA Synthesis Kit (Bio Basic Inc., Markham, Ontario, Canada) according to the manufacturer's recommendations using an oligo-dT primer. The cDNA was amplified by PCR using a set of primers specific to the EXO70A1 gene. An equal quantity of PCR product was loaded on 0.8% agarose gel. PCR amplification of the constitutively expressed actin gene ACT7 (At5g09810) was used as an internal amplification and template control. (All primer sequences are listed in Table S6).
Pollen germination and labeling
Mature anthers were picked and pollen was brushed on slides with a thin layer of nutrient agar (25% sucrose, 8 mm H3BO3, 2 mm Ca(NO3)2 × 4 H2O, 1 mm MgSO4 × 7 H2O, 0.7 mm CaCl2 × 2 H2O, 0.26 mm K2HPO4, 3% agar, pH adjusted to 7.4 with KOH). Slides were cultivated in a wet chamber at 26°C for 10–12 h. The pollen grains were considered as germinated if the pollen tube was longer than one-grain diameter.
For labeling of pollen grains by 4′,6-diamidino-2-phenylindol, either anthers or whole flowers were mounted in 5% DMSO, 1% Tween 20 and 1 mm DAPI (Sigma). Flowers were squeezed by pressure applied on the cover slip and labeled for 1 h at room temperature (approximately 22°C).
Microscopy and image analysis
Microscopic pictures were acquired with a DP50 camera (Olympus Europa, GmbH) attached to Olympus BX51 microscope. Details of inflorescence, leaves, roots and dissected flowers were captured with a Stemi 2000C (Carl Zeiss AG, Oberkochen, Germany) stereo-microscope equipped with Minolta x-300 camera (KONICA MINOLTA HOLDINGS INC., Tokyo, Japan). Images of seedlings and whole plants were taken by a C-4040Zoom (Olympus) digital camera. Confocal laser scanning microscopy pictures of FM4-64 dye-labeled root meristems were obtained with a Leica TCS SP2 confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany). For FM4-64 staining, seedlings were incubated for 1 min in 16.4 μm FM4-64 (Molecular Probes, Eugene, OR, USA) solution, washed and mounted in water.
For root and hypocotyl lengths measurement, images of seedlings on plates were taken by a C-4040Zoom (Olympus) digital camera, lengths were measured with analysis® (Soft Imaging System GmbH, Münster, Germany) and the results were analyzed with Microsoft Excel 2000.
For root hair measurements, microscopical images of roots were taken, root hairs that were in focus throughout their length were measured with analysis® and the results were analyzed with Microsoft Excel 2000.
Supplementary Material
Figure S1
Exon-intron structure of EXO70 genes.
Figure S2
Homozygous exo70 mutants.
Figure S3
Epidermal cell length of etiolated hypocotyls.
Figure S4
Apical hook formation of etiolated seedlings.
Tables S1-S6, Text S1
Acknowledgements
The sequence data from P. trichocarpa, S. moellendorffii, P. patens, C. reinhardtii and V. carteri were produced by the US Department of Energy Joint Genome Institute http://www.jgi.doe.gov/. We are highly indebted to Hana Soukupová and Petra Žižková for their help with propagating and maintaining the plants, David Honys for helpful discussions on the expression analysis, and Fatima Cvrčková, John Fowler and Rex Cole for their critical reading of the manuscript. We are also grateful to the two anonymous reviewers for important and constructive suggestions.
This work was supported by the GAAV ČR grant A6038410 and the EU RTN project TIPNET HPRN-CT-2002-00265. Nicole Schlager and Marie-Theres Hauser were supported by the Austrian Science Fund (P16410).
Footnotes
Supplementary Material
The following supplementary material is available for this article online:
Table S1. Rice EXO70 genes
Table S2. Poplar EXO70 genes
Table S3. EXO70 genes from Selaginella moellendorffii
Table S4. EXO70 genes from Physcomitrella patens
Table S5. Experiments included in the expression analysis
Table S6. Primers for genotyping and mRNA level analysis
Appendix S1. Corrected and newly predicted EXO70 sequences
Figure S1. Exon-intron structure of EXO70 genes.
Figure S2. Homozygous exo70 mutants.
Figure S3. Epidermal cell length of etiolated hypocotyls.
Figure S4. Apical hook formation of etiolated seedlings.
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Adamo JE, Rossi G, Brennwald P. The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol. Biol. Cell. 1999;10:4121–4133. doi: 10.1091/mbc.10.12.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped blast and psi-blast:a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers JE, Chapman BA, Rong J, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Boyd C, Hughes T, Pypaert M, Novick P. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell. Biol. 2004;167:889–901. doi: 10.1083/jcb.200408124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernac A, Lincoln C, Lammer D, Estelle M. The SAR1 gene of Arabidopsis acts downstream of the AXR1 gene in auxin response. Development. 1997;124:1583–1591. doi: 10.1242/dev.124.8.1583. [DOI] [PubMed] [Google Scholar]
- Cole RA, Synek L, Zarsky V, Fowler JE. SEC8, a subunit of the putative Arabidopsis exocyst complex, facilitates pollen germination and competitive pollen tube growth. Plant Physiol. 2005;138:2005–2018. doi: 10.1104/pp.105.062273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvrckova F, Elias M, Hala M, Obermeyer G, Zarsky V. Small GTPases and conserved signalling pathways in plant cell morphogenesis: from exocytosis to Exocyst. In: Geitmann A, Cresti M, editors. Cell Biology of Plant and Fungal Tip Growth. IOS Press; Amsterdam: 2001. pp. 105–122. [Google Scholar]
- Dacks JB, Field MC. Eukaryotic cell evolution from a comparative genomic perspective: the endomembrane system. In: Hirt RP, Horner DS, editors. Organelles, Genomes and Eukaryote Phylogeny: An Evolutionary Synthesis in the Age of Genomics. CRC Press; Boca Raton: 2004. pp. 309–334. [Google Scholar]
- Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005;3:e314. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- Dolan L, Duckett CM, Grierson C, Linstead P, Schneider K, Lawson E, Dean C, Poethig S, Roberts K. Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development. 1994;120:2465–2474. [Google Scholar]
- Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat. Struct. Mol. Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- Elias M, Drdova E, Ziak D, Bavlnka B, Hala M, Cvrckova F, Soukupova H, Zarsky V. The exocyst complex in plants. Cell Biol. Int. 2003;27:199–201. doi: 10.1016/s1065-6995(02)00349-9. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Version 3.6. Department of Genome Sciences, University of Washington; Seattle: 2005. Distributed by the author. [Google Scholar]
- Fielding AB, Schonteich E, Matheson J, Wilson G, Yu X, Hickson GR, Srivastava S, Baldwin SA, Prekeris R, Gould GW. Rab11-FIP3 and FIP4 interact with Arf6 and the Exocyst to control membrane traffic in cytokinesis. EMBO J. 2005;24:3389–3399. doi: 10.1038/sj.emboj.7600803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P. Genetic interactions between ABA, ethylene and sugar signalling pathways. Curr. Opin. Plant Biol. 2001;4:387–391. doi: 10.1016/s1369-5266(00)00190-4. [DOI] [PubMed] [Google Scholar]
- Gibson SI. Sugar and phytohormone response pathways: navigating a signalling network. J. Exp. Bot. 2004;55:253–264. doi: 10.1093/jxb/erh048. [DOI] [PubMed] [Google Scholar]
- Goubet F, Misrahi A, Park SK, Zhang Z, Twell D, Dupree P. AtCSLA7, a cellulose synthase-like putative glycosyl-transferase, is important for pollen tube growth and embryo-genesis in Arabidopsis. Plant Physiol. 2003;131:547–557. doi: 10.1104/pp.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson CS, Roberts K, Feldmann KA, Dolan L. The COW1 locus of arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol. 1997;115:981–990. doi: 10.1104/pp.115.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromley A, Yeaman C, Rosa J, Redick S, Chen C, Mirabelle S, Guha M, Sillibourne J, Doxsey SJ. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Trautmann S, McCollum D. Cytokinesis in eukaryotes. Microbiol. Mol. Biol. Rev. 2002;66:155–178. doi: 10.1128/MMBR.66.2.155-178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Grant A, Novick P. Exo84p is an exocyst protein essential for secretion. J. Biol. Chem. 1999;274:23558–23564. doi: 10.1074/jbc.274.33.23558. [DOI] [PubMed] [Google Scholar]
- Hamburger ZA, Hamburger AE, West AP, Weis WI. Crystal structure of the S. cerevisiae exocyst component Exo70p. J. Mol. Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- Hauser M-T, Morikami A, Benfey PN. Conditional root expansion mutants of Arabidopsis. Development. 1995;121:1237–1252. doi: 10.1242/dev.121.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley PA, Kemp AC, Grierson CS. The TIP GROWTH DEFECTIVE1 S-acyl transferase regulates plant cell growth in Arabidopsis. Plant Cell. 2005;17:2554–2563. doi: 10.1105/tpc.105.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu SC, Hazuka CD, Foletti DL, Heuser J, Scheller RH. Subunit composition, protein interactions and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron. 1998;20:1111–1122. doi: 10.1016/s0896-6273(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Hazuka CD, Foletti DL, Scheller RH. Targeting vesicles to specific sites on the plasma membrane: The role of the sec6/8 complex. Trends Cell Biol. 1999;9:150–153. doi: 10.1016/s0962-8924(99)01516-0. [DOI] [PubMed] [Google Scholar]
- Hsu SC, TerBush D, Abraham M, Guo W. The exocyst complex in polarized exocytosis. Int. Rev. Cytol. 2004;233:243–265. doi: 10.1016/S0074-7696(04)33006-8. [DOI] [PubMed] [Google Scholar]
- Inoue M, Chang L, Hwang J, Chiang SH, Saltiel AR. The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature. 2003;422:629–633. doi: 10.1038/nature01533. [DOI] [PubMed] [Google Scholar]
- Inoue M, Chiang SH, Chang L, Chen XW, Saltiel AR. Compartmentalization of the exocyst complex in lipid rafts controls Glut4 vesicle tethering. Mol. Biol. Cell. 2006;17:2303–2311. doi: 10.1091/mbc.E06-01-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Jafar-Nejad H, Andrews HK, Acar M, Bayat V, Wirtz-Peitz F, Mehta SQ, Knoblich JA, Bellen HJ. Sec15, a component of the exocyst, promotes notch signaling during the asymmetric division of Drosophila sensory organ precursors. Dev. Cell. 2005;9:351–363. doi: 10.1016/j.devcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Jones MA, Shen JJ, Fu Y, Li H, Yang Z, Grierson CS. The Arabidopsis Rop2 GTPase is a positive regulator of both root hair initiation and tip growth. Plant Cell. 2002;14:763–776. doi: 10.1105/tpc.010359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd WS, Olmstead RG. A survey of tricolpate. (eudicot) phylogenetic relationships. Am. J. Bot. 2004;91:1627–1644. doi: 10.3732/ajb.91.10.1627. [DOI] [PubMed] [Google Scholar]
- Jurgens G, Geldner N. Protein secretion in plants: from the trans-Golgi network to the outer space. Traffic. 2002;3:605–613. doi: 10.1034/j.1600-0854.2002.30902.x. [DOI] [PubMed] [Google Scholar]
- Kee Y, Yoo JS, Hazuka CD, Peterson KE, Hsu SC, Scheller RH. Subunit structure of the mammalian exocyst complex. Proc. Natl Acad. Sci. USA. 1997;94:14438–14443. doi: 10.1073/pnas.94.26.14438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimyuk VI, Carroll BJ, Thomas CM, Jones JD. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 1993;3:493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- Leyser HM, Pickett FB, Dharmasiri S, Estelle M. Mutations in the AXR3 gene of Arabidopsis result in altered auxin response including ectopic expression from the SAUR-AC1 promoter. Plant J. 1996;10:403–413. doi: 10.1046/j.1365-313x.1996.10030403.x. [DOI] [PubMed] [Google Scholar]
- Lipschutz JH, Mostov KE. Exocytosis: the many masters of the exocyst. Curr. Biol. 2002;12:R212–R214. doi: 10.1016/s0960-9822(02)00753-4. [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. The rhd6 mutation of Arabidopsis thaliana alters root-hair initiation through an auxinand ethylene-associated process. Plant Physiol. 1994;106:1335–1346. doi: 10.1104/pp.106.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell. 1996;8:1505–1517. doi: 10.1105/tpc.8.9.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matern HT, Yeaman C, Nelson WJ, Scheller RH. The Sec6/8 complex in mammalian cells: characterization of mammalian Sec3, subunit interactions, and expression of subunits in polarized cells. Proc. Natl Acad. Sci. USA. 2001;98:9648–9653. doi: 10.1073/pnas.171317898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muday GK, Murphy AS. An emerging model of auxin transport regulation. Plant Cell. 2002;14:293–299. doi: 10.1105/tpc.140230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R. Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell. 1980;21:205–215. doi: 10.1016/0092-8674(80)90128-2. [DOI] [PubMed] [Google Scholar]
- Okamuro JK, den Boer BG, Lotys-Prass C, Szeto W, Jofuku KD. Flowers into shoots: photo and hormonal control of a meristem identity switch in Arabidopsis. Proc. Natl Acad. Sci. USA. 1996;93:13831–13836. doi: 10.1073/pnas.93.24.13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otegui MS, Staehelin LA. Electron tomographic analysis of post-meiotic cytokinesis during pollen development in Arabidopsis thaliana. Planta. 2004;218:501–515. doi: 10.1007/s00425-003-1125-1. [DOI] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc. Natl Acad. Sci. USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J. 1998;16:553–560. doi: 10.1046/j.1365-313x.1998.00321.x. [DOI] [PubMed] [Google Scholar]
- Rice Chromosomes 11 and 12 Sequencing Consortia and Messing The sequence of rice chromosomes 11 and 12, rich in disease resistance genes and recent gene duplications. BMC Biol. 2005;3:20. doi: 10.1186/1741-7007-3-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roumanie O, Wu H, Molk JN, Rossi G, Bloom K, Brennwald P. Rho GTPase regulation of exocytosis in yeast is independent of GTP hydrolysis and polarization of the exocyst complex. J. Cell Biol. 2005;170:583–594. doi: 10.1083/jcb.200504108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderfoot AA, Raikhel NV. The secretory system of Arabidopsis. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. American Society of Plant Biologists; Rockville: 2003. pp. 1–24. [Google Scholar]
- Schiefelbein JW, Somerville C. Genetic-control of root hair development in Arabidopsis thaliana. Plant Cell. 1990;2:235–243. doi: 10.1105/tpc.2.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Galway M, Masucci J, Ford S. Pollen-tube and root hair tip growth is disrupted in a mutant of Arabidopsis thaliana. Plant Physiol. 1993;103:979–985. doi: 10.1104/pp.103.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- Segui-Simarro JM, Austin JR, II, White EA, Staehelin LA. Electron tomographic analysis of somatic cell plate formation in meristematic cells of Arabidopsis preserved by high-pressure freezing. Plant Cell. 2004;16:836–856. doi: 10.1105/tpc.017749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions A, Burke E, Presting G, et al. A high-throughput Arabidopsis reverse genetics system. Plant Cell. 2002;14:2985–2994. doi: 10.1105/tpc.004630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. Blast 2 sequences - a new tool for comparing protein and nucleotide sequences. FEMS Microbiol. Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- TerBush DR, Novick P. Sec6, Sec8, and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J. Cell Biol. 1995;130:299–312. doi: 10.1083/jcb.130.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TerBush DR, Maurice T, Roth D, Novick P. The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J. 1996;15:6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The clustal_x windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke F, Ordidge M, Chiurugwi T, Battey N. Mechanisms and function of flower and inflorescence reversion. J. Exp. Bot. 2005;56:2587–2599. doi: 10.1093/jxb/eri254. [DOI] [PubMed] [Google Scholar]
- Vega IE, Hsu SC. The exocyst complex associates with microtubules to mediate vesicle targeting and neurite outgrowth. J. Neurosci. 2001;21:3839–3848. doi: 10.1523/JNEUROSCI.21-11-03839.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank L, Li R. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol. Biol. Cell. 2005;16:2529–2543. doi: 10.1091/mbc.E04-12-1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volff JN. Genome evolution and biodiversity in teleost fish. Heredity. 2005;94:280–294. doi: 10.1038/sj.hdy.6800635. [DOI] [PubMed] [Google Scholar]
- Wang HY, Lockwood SK, Hoeltzel MF, Schiefelbein JW. The ROOT HAIR DEFECTIVE3 gene encodes an evolutionarily conserved protein with GTP-binding motifs and is required for regulated cell enlargement in Arabidopsis. Genes Dev. 1997;11:799–811. doi: 10.1101/gad.11.6.799. [DOI] [PubMed] [Google Scholar]
- Wang H, Tang X, Balasubramanian MK. Rho3p regulates cell separation by modulating exocyst function in Schizosaccharomyces pombe. Genetics. 2003;164:1323–1331. doi: 10.1093/genetics/164.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen TJ, Hochholdinger F, Sauer M, Bruce W, Schnable PS. The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol. 2005;138:1637–1643. doi: 10.1104/pp.105.062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AK, Pickett FB, Turner JC, Estelle M. A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 1990;222:377–383. doi: 10.1007/BF00633843. [DOI] [PubMed] [Google Scholar]
- Wilson ZA, Morroll SM, Dawson J, Swarup R, Tighe PJ. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001;28:27–39. doi: 10.1046/j.1365-313x.2001.01125.x. [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- Yeaman C, Grindstaff KK, Nelson WJ. Mechanism of recruiting Sec6/8 (exocyst) complex to the apical junctional complex during polarization of epithelial cells. J. Cell Sci. 2004;117:559–570. doi: 10.1242/jcs.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Wellmer F, Meyerowitz EM. Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat. Genet. 2004;36:157–161. doi: 10.1038/ng1286. [DOI] [PubMed] [Google Scholar]
- Zheng H, Kunst L, Hawes C, Moore I. A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 2004;37:398–414. doi: 10.1046/j.1365-313x.2003.01969.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Exon-intron structure of EXO70 genes.
Figure S2
Homozygous exo70 mutants.
Figure S3
Epidermal cell length of etiolated hypocotyls.
Figure S4
Apical hook formation of etiolated seedlings.
Tables S1-S6, Text S1