Smoking-dependent Reprogramming of Alveolar Macrophage Polarization: Implication for Pathogenesis of COPD (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 15.
Published in final edited form as: J Immunol. 2009 Jul 27;183(4):2867–2883. doi: 10.4049/jimmunol.0900473
Abstract
When exposed to specific microenvironment, macrophages acquire either M1- or M2-polarized phenotypes associated with inflammation and tissue remodeling, respectively. Alveolar macrophages (AM) directly interact with environmental stimuli such as cigarette smoke, the major risk factor for chronic obstructive pulmonary disease (COPD), a disease characterized by lung inflammation and remodeling. Transcriptional profiling of AM obtained by bronchoalveolar lavage of 24 healthy nonsmokers, 34 healthy smokers and 12 COPD smokers was performed to test the hypothesis whether smoking alters AM polarization resulting in a disease-relevant activation phenotype. The analysis revealed that AM of healthy smokers exhibited a unique polarization pattern characterized by substantial suppression of M1-related inflammatory/immune genes and induction of genes associated with various M2-polarization programs relevant to tissue remodeling and immunoregulation. Such reciprocal changes progressed with the development of COPD with M1-related gene expression being most dramatically down-regulated (p<0.0001 vs healthy nonsmokers, p<0.002 vs healthy smokers), results confirmed with TaqMan real-time PCR and flow cytometry. Among progressively down-regulated M1-related genes were those encoding type I chemokines CXCL9, CXCL10, CXCL11, and CCL5. Progressive activation of M2-related program was characterized by induction of tissue remodeling and immunoregulatory genes such as MMP2, MMP7 and ADORA3. Principal component analysis revealed that differential expression of polarization-related genes has substantial contribution to global AM phenotypes associated with smoking and COPD. In summary, the data provides transcriptome-based evidence that AM likely contribute to COPD pathogenesis in non-inflammatory manner due to their smoking-induced reprogramming towards M1-deactivated, partially M2-polarized macrophages.
Introduction
Mononuclear phagocytes are heterogeneous population of cells with significant phenotypic plasticity (1). Depending on the microenvironment, they undergo distinct activation programs acquiring polarized phenotypes and different functional capacities that together provide an armamentarium that helps to protect, repair and sometimes damage tissues (2–4). Mononuclear phagocyte “M1 polarization”, also referred to as the “classical activation” program, is induced by signals generated during Th1-mediated immune response such as interferon (IFN) γ and by exposure to pathogen components such as bacterial lipopolysaccharide (LPS) (2–4). The M1 polarization response is characterized by up-regulation of genes relevant to inflammation and cell-mediated immunity. In contrast, mononuclear phagocyte “M2 polarization”, induced upon exposure to the Th2 cytokines IL-4 and IL-13 (referred to as “alternative activation”) or immunoregulatory signals such as IL-10 (also called “deactivation”) and glucocorticoids, is highlighted by induction of expression of receptors with scavenger functions, anti-inflammatory cytokines and molecules implicated in tissue remodeling (1–4).
Although considerable evidence has accumulated regarding the reprogramming of human blood monocytes and murine macrophages in vitro depending on the environment to which they are exposed, little attention has been paid in defining how the in vivo environment modifies the global polarization program of human macrophages in health and disease. Alveolar macrophages (AM), the pulmonary representatives of the mononuclear phagocyte system, play a central role in defending the lung against pathogens and other environmental challenges, as well as in mediating damage and repair in the lung parenchyma (5,6). AM are unique among mononuclear phagocytes in that AM mostly reside on the respiratory epithelial surface, and thus are exposed directly to the outside environment. One of the most common of these environmental exposures is cigarette smoking, the major risk factor for the development of chronic obstructive pulmonary disease (COPD) (7) that is currently a leading cause of morbidity and mortality worldwide (7,8). Studies in experimental animals and humans have led to the concept that AM play a central role in the pathogenesis of COPD as a major source of mediators that derange the normal lung structure (6,9). In humans, AM numbers are increased in the lung of healthy smokers and individuals with COPD, AM accumulate in areas of lung destruction, and there is a correlation between the AM numbers, airway obstruction and severity of COPD (6,9–12).
Based on studies in murine transgenic models and several human studies that suggest that IFNγ-dependent inflammation is responsible for the development of smoking-induced lung disease (10,13–16), and reports indicating up-regulation of genes related to scavenger function, anti-inflammatory cytokines and remodeling mediators in AM of smokers (17,18), we hypothesized that, compared to healthy nonsmokers, AM of healthy smokers demonstrate an altered polarization program, and that this polarization pattern progresses with the development of COPD. To assess this hypothesis, global transcriptional profiles were used to assess the M1 and M2 polarization-related genes in AM of 24 healthy nonsmokers, 34 healthy and 12 COPD smokers using Affymetrix microarrays with TaqMan real-time PCR and FACS confirmation of the phenotypic changes. The data demonstrates that cigarette smoke does indeed alter the steady-state polarization status of human AM in vivo, including induction of several genes representing M2 sub-phenotypes. Surprisingly, however, rather than up-regulating the M1 polarization program as expected, cigarette smoking induces in AM of healthy smokers the opposite phenotype, characterized by a substantial down-regulation of the majority of the genes associated with M1 polarization. The overall expression of M1-related genes was progressively further down-regulated in COPD smokers accompanied with a gradual up-regulation of some M2-related genes, suggesting that the transcriptome of AM in COPD smokers is characterized by progressive reciprocal dysregulation of M1- and M2-polarization patterns. The data supports the concept that AM contribute to tissue remodeling during the development of smoking-induced lung disease. However, the data also suggests that it is unlikely that AM play a significant role as inflammatory cells in the early pathogenesis of smoking-induced COPD, a departure from the concept that AM-mediated inflammation participates in the early derangements of the lung induced by smoking.
Methods
Study Population
A total of 70 subjects were assessed including, healthy never-smokers with normal lung function (n=24, referred to as “healthy nonsmokers”), healthy smokers with normal lung function (n=34, “healthy smokers”), and COPD smokers (GOLD classification, n=12, Table I). No COPD smokers with current exacerbation were included in the study. The study was approved by the Weill Cornell Medical College Institutional Review Board and written informed consent was obtained from each individual before enrollment in the study. Subjects were evaluated at the Weill Cornell NIH General Clinical Research Center and Department of Genetic Medicine Clinical Research Facility based on clinical history, physical examination, routine blood screening tests, urinalysis, chest X-ray, ECG and pulmonary function tests. Current smoking status was confirmed by history, venous carboxyhemoglobin levels, and urinalysis for nicotine levels and its derivative cotinine.
Table I.
Demographic Characteristics of the Study Population and Biologic Samples1, 2
| Parameter | Healthynonsmokers | Healthysmokers | COPDsmokers |
|---|---|---|---|
| Number | 24 | 34 | 12 |
| Sex (male / female) | 18/6 | 26/8 | 10/2 |
| Age (yr) | 40.3 ± 8.2 | 41.3 ± 7.9 | 54.7 ± 9.3*, ** |
| Ancestry (B/W/H3) | 15/6/3 | 20/10/4 | 1/8/3 |
| Smoking history (pack-yr) | 0 | 26.8 ± 17.7 | 49.8 ± 28.3** |
| Urine nicotine (ng/ml) | negative | 864 ± 1053 | 1217 ± 1301 |
| Urine cotinine | negative | 1211 ± 1034 | 1243 ± 639 |
| Venous carboxyhemoglobin (%)4 | 0.5 ± 0.8 | 3.0 ± 2.1 | 3.2 ± 2.1 |
| Pulmonary function5 | |||
| FVC | 106.1 ± 11.2 | 107.0 ± 10.8 | 105.2 ± 24.6 |
| FEV1 | 102.9 ± 11.8 | 108.0 ± 13.2 | 85.2 ± 22.9*, ** |
| FEV1/FVC | 81.5 ± 4.7 | 81.9 ± 4.9 | 64.2 ± 4.4*, ** |
| TLC | 96.8 ± 8.1 | 99.0 ± 12.6 | 112.8 ± 21.6*, **** |
| DLCO | 94.3 ± 8.5 | 96.0 ± 11.2 | 75.4 ± 17.0*, ** |
| GOLD Stage (I/II/III)6 | -- | -- | 8/3/1 |
Alveolar Macrophages
AM were collected by bronchoalveolar lavage as previously described (19). The total volume used per site was typically 100 ml and a maximum of 3 sites were lavaged per individual. Recovery of the infused volume ranged from 45 to 65%. BAL fluid was filtered with gauze and centrifuged at 1,200 rpm for 5 min, at 4°C. Cells were washed twice in RPMI 1640 containing 10% fetal bovine serum, 50 U/ml penicillin, 50 U/ml streptomycin and 2 mM glutamine (Invitrogen, Carlsbad, CA), suspended in 10 ml medium. Cell viability was assessed by trypan blue exclusion and expressed as a percentage of the total cells recovered. Total cell number was determined by counting on a hemocytometer. Differential cell count was performed on sedimented cells prepared by cytocentrifugation (Cytospin 3; Shandon Instruments, Pittsburgh, PA) stained with DiffQuik reagents (Dade Behring, Newark, NJ) and performed by counting 500 cells on each slide. For the microarray analysis, the remaining cells were seeded in six-well plastic culture dishes (2 × 106 per 2 ml/well) and the AM purified by adherence at 37°C, 2 hr in a 5% CO2 humidified incubator, removing any nonadherent cells by washing with RPMI 1640 before RNA extraction. Light microscopy was used to assess the morphological features of the cells. After the adherence step, all samples were >98% AM. For FACS analysis, the cells were processed immediately after isolation as described below.
cDNA Preparation and Affymetrix Microarrays
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) followed by RNeasy (Qiagen, Valencia, CA) to remove residual DNA, yielding 2 to 4 µg RNA per 106 cells. RNA samples were stored in RNA Secure (Ambion, Austin, TX) at −80°C. RNA integrity was determined by running an aliquot of each RNA sample on an Agilent Bioanalyzer (Agilent Technologies, Palo Alto, CA). A NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE) was used to determine the concentration of RNA. Double stranded cDNA was synthesized from 3 µg of total RNA using the GeneChip One-Cycle cDNA Synthesis Kit, followed by cleanup with GeneChip Sample Cleanup Module, in vitro transcription reaction using the GeneChip IVT Labeling Kit, and cleanup and quantification of the biotin-labeled cRNA yield by spectrophotometric analysis (all kits from Affymetrix, Santa Clara, CA). Hybridizations to test chips and to the HG-U133 Plus 2.0 array (54,000 probe sets representing approximately 47,000 full-length human transcripts) were performed according to Affymetrix protocols, processed by the Affymetrix GeneChip Fluidics Station 450 and scanned with an Affymetrix GeneChip Scanner 2500. Overall microarray quality was verified by the following criteria: (1) RNA Integrity Number (RIN) >=7.0; (2) 3'/5' ratio for GAPDH <=3; (3) scaling factor <=10.0; and (4) expression level for all 100 housekeeping genes (as defined by Affymetrix, www.affymetrix.com) with coefficient of variation of <40%. The captured image data from the HG-U133 Plus 2.0 arrays was processed using MAS5 algorithm (Affymetrix Microarray Suite Version 5 software). MAS5-processed data was normalized using GeneSpring version 7.3.1 (Agilent technologies) by setting measurements <0.01 to 0.01, per array, by dividing the raw data by the 50th percentile of all measurements, and per gene, by dividing the raw data by the median expression level for all the genes across all arrays in a data set.
Genes that were significantly modified between 2 groups were identified according to the following criteria: (1) P call of “Present” in ≥20% of samples; (2) magnitude of fold change in average expression value for 2 comparative groups ≥1.5; and (3) p<0.05 between the groups with a Benjamini-Hochberg correction to limit the false positive rate (20). To exclude the effect of adherence step of AM purification on global transcriptional changes associated with smoking, we compared our results with the signature of significant smoking-responsive genes identified in a previous study in which AM were isolated by flow cytometry (17).
Characterization of M1 and M2-related Gene Expression
The genes representing the M1 and M2 polarization patterns were chosen based on the literature regarding in vitro studies of human and murine monocytes and macrophages (2–4,21–32). A gene was classified as “M1-related” based on the current definition of M1 polarization (classical activation) of mononuclear phagocytes as a molecular pattern that may be induced in macrophages and/or monocytes upon stimulation with IFNγ and / or LPS (2–4,23,24,26,29,30). A gene was classified as “M2-related” if it can be induced either by Th2 cytokines IL-4 and IL-13 (alternative activation) (2–4,27,29), IL-10 (deactivation) (22,25,28), transforming growth factor-beta (TGF-β) (21,22,32), glucocorticoids (31), or macrophage colony stimulating factor (M-CSF) (29). A gene was also classified as M2 if it has been observed to be down-regulated upon exposure to M1-polarization stimuli (33). Genes that can be induced during both M1 and M2 macrophage polarization (for example, HLA-related genes (3)) were excluded from the analysis. Based on these criteria, 41 M1-related (Table II) and 33 M2-related (Table III) genes were selected for analysis.
Table II.
M1-related Gene Expression in Alveolar Macrophages of Healthy Nonsmokers and Healthy Smokers and COPD-smokers1, 2
| P call (%) | Fold-change | p value | M profile3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene name | Ref | Symbol | Probeset ID | NS | S | COPD-S | S _vs_NS | COPD-S_vs_ NS | COPD-S_vs_ S | S vs NS | COPD-S_vs_ NS | COPD-S_vs_ S | S _vs_NS | COPD-S vs NS | COPD-S vs S |
| Membrane receptors | |||||||||||||||
| Fc fragment of IgG, low affinity IIIa, receptor (CD16a) | (2–4,23) | FCGR3A | 204006_s_at | 100 | 100 | 100 | −1.1 | −1.8 | −1.5 | N.S. | N.S. | N.S. | - | - | - |
| Fc fragment of IgG, low affinity IIa, receptor (CD32) | (2–4,23) | FCGR2A | 203561_at | 100 | 100 | 100 | −1.5 | −1.4 | 1 | N.S. | N.S. | N.S. | - | - | - |
| 1565674_at | 29 | 27 | 42 | −1.2 | 1.3 | 1.5 | N.S. | N.S. | N.S. | ||||||
| Fc fragment of IgG, high affinity Ia, receptor (CD64) | (2–4,23) | FCGR1A | 216950_s_at | 100 | 100 | 100 | −1.2 | −1.2 | 1 | N.S. | N.S. | N.S. | - | - | - |
| Interleukin 15 receptor, alpha | (29) | IL15RA | 207375_s_at | 100 | 100 | 100 | −1.6 | −1.7 | −1.1 | <0.02 | <0.02 | N.S. | ↓ | ↓ | - |
| Complement component 3a receptor 1 | (23,26) | C3AR1 | 209906_at | 100 | 100 | 100 | −1.8 | −1.7 | 1.1 | <0.03 | N.S. | N.S. | ↓ | - | - |
| CD69 antigen (p60, early T-cell activation antigen) | (23) | CD69 | 209795_at | 83 | 27 | 8 | −4.8 | -10 | −2.1 | <0.0003 | <0.01 | N.S. | ↓ | ↓ | - |
| CD80 antigen (CD28 antigen ligand 1, B7-1 antigen) | (2–4,23) | CD80 | 1554519_at | 100 | 100 | 100 | −1.9 | −1.6 | 1.2 | <0.03 | N.S. | N.S. | ↓ | - | - |
| 1555689_at | 96 | 47 | 92 | −3.3 | −1.3 | 2.5 | <0.002 | N.S. | <0.03 | ||||||
| CD86 antigen (CD28 antigen ligand 2, B7-2 antigen) | (2–4,23) | CD86 | 205686_s_at | 100 | 100 | 100 | 1.2 | 1 | −1.2 | N.S. | N.S. | N.S. | - | - | - |
| Toll-like receptor 2 | (4) | TLR2 | 204924_at | 100 | 100 | 100 | −1.6 | −1.8 | −1.1 | N.S. | N.S. | N.S. | - | - | - |
| Toll-like receptor 4 | (4) | TLR4 | 1552798_a_at | 100 | 100 | 100 | 1.8 | 1 | −1.5 | <0.04 | N.S. | <0.03 | ↑ | - | ↓ |
| 221060_s_at | 100 | 100 | 100 | 1.5 | −1.1 | −1.6 | <0.05 | N.S. | N.S. | ||||||
| Intercellular adhesion molecule 1 (CD54) | (26) | ICAM1 | 215485_s_at | 96 | 88 | 92 | −1.4 | −3.3 | −2.4 | N.S. | <0.005 | <0.02 | - | ↓ | ↓ |
| Cytokines and chemokines | |||||||||||||||
| Interleukin 1, beta | (2) | IL1B | 205067_at | 100 | 100 | 100 | −2.3 | −3.3 | −1.4 | <0.03 | <0.01 | N.S. | ↓ | ↓ | - |
| 39402_at | 100 | 100 | 100 | −2.3 | −3.1 | −1.3 | <0.02 | <0.01 | N.S. | ||||||
| Interleukin 6 (interferon, beta 2) | (2,29) | IL6 | 205207_at | 75 | 56 | 67 | −2.2 | −1.7 | 1.3 | N.S. | N.S. | N.S. | - | - | - |
| Interleukin 12B (natural killer cell stimulatory factor 2, p40) | (2,3,29) | IL12B | 207901_at | 21 | 3 | 17 | −2.1 | −1.2 | 1.7 | N.S.* | N.S.* | N.S.* | - | - | - |
| Interleukin 18 (interferon-gamma-inducing factor) | (85) | IL18 | 206295_at | 100 | 100 | 100 | −1.9 | −1.7 | 1.1 | <0.0007 | N.S. | N.S. | ↓ | - | - |
| Interleukin 23, alpha subunit p19 | (4) | IL23 | 220054_at | 42 | 27 | 33 | −1.6 | −2.1 | −1.4 | N.S. | N.S. | N.S. | - | - | - |
| Interleukin 32 | (86) | IL32 | 203828_s_at | 13 | 6 | 0 | −5.2 | −17.1 | −3.3 | <0.003* | <0.0002* | <0.05* | ↓ | ↓ | ↓ |
| Tumor necrosis factor (TNF superfamily, member 2) | (2,29) | TNF | 207113_s_at | 100 | 100 | 100 | −1.2 | −2.1 | −1.8 | N.S. | N.S. | N.S. | - | - | - |
| Tumor necrosis factor (ligand) superfamily, member 10 | (29) | TNFSF10 | 202687_s_at | 96 | 68 | 42 | −2.9 | −3.7 | −1.3 | <0.02 | <0.02 | N.S. | ↓ | ↓ | - |
| 202688_at | 96 | 91 | 92 | −3.2 | −3.4 | −1.1 | <0.02 | <0.02 | N.S. | ||||||
| 214329_x_at | 83 | 50 | 92 | −3.4 | −2.2 | 1.6 | <0.02 | N.S. | N.S. | ||||||
| Tumor necrosis factor, alpha-induced protein 6 | (29) | TNFAIP6 | 206025_s_at | 100 | 100 | 92 | −4.6 | −7.9 | −1.7 | <0.01 | <0.02 | N.S. | ↓ | ↓ | - |
| 206026_s_at | 100 | 100 | 100 | −4.9 | −5.4 | −1.1 | <0.004 | <0.03 | N.S. | ||||||
| Chemokine (C-X-C motif) ligand 1 | (23) | CXCL1 | 204470_at | 100 | 100 | 100 | −3.1 | −2.6 | 1.2 | <0.03 | N.S. | N.S. | ↓ | - | - |
| Chemokine (C-X-C motif) ligand 9 | (4,29) | CXCL9 | 203915_at | 100 | 62 | 75 | −4 | −6.5 | −1.6 | <0.02 | <0.0005 | N.S. | ↓ | ↓ | - |
| Chemokine (C-X-C motif) ligand 10 | (4,29) | CXCL10 | 204533_at | 83 | 47 | 17 | −3.8 | −5.3 | −1.4 | N.S. | <0.05 | N.S. | - | ↓ | - |
| Chemokine (C-X-C motif) ligand 11 | (4,29) | CXCL11 | 211122_s_at | 63 | 18 | 8 | −7.7 | −11.5 | −1.5 | <0.01 | <0.01 | N.S. | ↓ | ↓ | - |
| 210163_at | 63 | 21 | 8 | −4.1 | −5 | −1.2 | <0.01 | <0.05 | N.S. | ||||||
| Chemokine (C-C motif) ligand 4 | (4,26) | CCL4 | 204103_at | 100 | 100 | 100 | −3.3 | −3.7 | −1.1 | <0.02 | N.S. | N.S. | ↓ | - | - |
| Chemokine (C-C motif) ligand 5 | (4,29) | CCL5 | 1405_i_at | 92 | 68 | 50 | −3.8 | −4.7 | −1.2 | <0.01 | <0.01 | N.S. | ↓ | ↓ | - |
| CCL5 | 1555759_a_at | 75 | 35 | 25 | −2.8 | −3.5 | −1.3 | <0.01 | <0.008 | N.S. | |||||
| 204655_at | 92 | 65 | 75 | −2.8 | −2.5 | 1.1 | <0.005 | <0.05 | N.S. | ||||||
| Chemokine (C-C motif) ligand 20 | (4,29) | CCL20 | 205476_at | 100 | 85 | 83 | −2.5 | −5.8 | −2.3 | N.S. | N.S. | N.S. | - | - | - |
| Signaling related proteins | |||||||||||||||
| Guanylate binding protein 1, interferon-inducible, | (23,26) | GBP1 | 202270_at | 100 | 100 | 100 | −2.2 | −2 | 1.1 | <0.02 | N.S. | N.S. | ↓ | - | - |
| 67 kDa | 231577_s_at | 100 | 100 | 100 | −1.9 | −2.5 | −1.3 | N.S. | N.S. | N.S. | |||||
| 231578_at | 67 | 50 | 75 | −2 | −1.2 | 1.7 | N.S. | N.S. | N.S. | ||||||
| Guanylate binding protein 2, interferon-inducible | (23,26) | GBP2 | 202748_at | 100 | 100 | 100 | −1.9 | −2.7 | −1.5 | <0.02 | <0.0005 | <0.04 | ↓ | ↓ | ↓ |
| 242907_at | 100 | 100 | 100 | −2.2 | −2.1 | 1 | <0.002 | <0.03 | N.S. | ||||||
| Guanylate binding protein 3 | (26) | GBP3 | 223434_at | 100 | 100 | 100 | −1.5 | −1.4 | 1.1 | N.S. | N.S. | N.S. | - | - | - |
| Guanylate binding protein 4 | (26) | GBP4 | 235175_at | 96 | 56 | 25 | −2.1 | −3 | −1.4 | <0.01 | <0.002 | N.S. | ↓ | ↓ | - |
| 235574_at | 79 | 44 | 0 | −2 | −6 | −3.1 | N.S. | <0.0006 | <0.02 | ||||||
| Guanylate binding protein 5 | (26) | GBP5 | 229625_at | 96 | 68 | 42 | −2.7 | −3.4 | −1.3 | <0.05 | <0.003 | N.S. | ↓ | ↓ | - |
| 238581_at | 88 | 50 | 58 | −3.1 | −3 | 1 | <0.01 | <0.01 | N.S. | ||||||
| Immunoresponsive 1 homolog | (23,26) | IRG1 | 240287_at | 79 | 24 | 17 | −4.1 | −4.2 | 1 | <0.001 | <0.004 | N.S. | ↓ | ↓ | - |
| Interferon regulatory factor 1 | (23,26,29) | IRF1 | 202531_at | 100 | 100 | 100 | −1.3 | −1.9 | −1.5 | N.S. | N.S. | N.S. | - | - | - |
| Interferon regulatory factor 7 | (23,26,29) | IRF7 | 208436_s_at | 100 | 100 | 100 | −1.3 | −1.7 | −1.3 | N.S. | <0.02 | N.S. | - | ↓ | - |
| Suppressor of cytokine signaling 3 | (87) | SOCS3 | 227697_at | 100 | 97 | 100 | −2.7 | −3.1 | −1.2 | <0.02 | <0.03 | N.S. | ↓ | ↓ | - |
| 206359_at | 50 | 35 | 0 | −1.9 | −6 | −3.1 | N.S. | <0.02 | <0.05 | ||||||
| Enzymes and other proteins | |||||||||||||||
| Nitric oxide synthase 2A (inducible) | (3,23,26) | NOS2A | 210037_s_at | 0 | 0 | 0 | N.D.** | N.D.** | N.D.** | N.S.* | N.S.* | N.S.* | - | - | - |
| Phosphodiesterase 4B, cAMP-specific | (23) | PDE4B | 203708_at | 100 | 97 | 92 | −3.7 | −3.7 | 1 | <0.0002 | <0.05 | N.S. | ↓ | ↓ | - |
| 211302_s_at | 100 | 100 | 100 | −3.2 | −4.5 | −1.4 | <0.002 | <0.02 | N.S. | ||||||
| 215671_at | 54 | 15 | 33 | −3.8 | −2.6 | 1.5 | <0.02 | N.S. | N.S. | ||||||
| 222326_at | 83 | 44 | 50 | −4.4 | −3.1 | 1.4 | <0.01 | N.S. | N.S. | ||||||
| Apolipoprotein L, 3 | (29) | APOL3 | 221087_s_at | 100 | 100 | 92 | −1.8 | −2 | −1.2 | <0.03 | <0.003 | N.S. | ↓ | ↓ | - |
| B-factor, properdin (complement factor B) | (26) | CFB | 202357_s_at | 96 | 85 | 50 | −2.7 | −3.9 | −1.4 | <0.005 | <0.0005 | N.S. | ↓ | ↓ | - |
Table III.
M2-related Gene Expression in AM of Healthy Nonsmokers and Healthy Smokers1, 2
| P call (%) | Fold-change | p value | M profile3 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene name | Ref | Symbol | Probe set ID | NS | S | COPD-S | S vs NS | COPD-S_vs_ NS | COPD-S_vs_ NS | S vs NS | COPD-S_vs_ NS | COPD-S_vs_ NS | S vs NS | COPD-S_vs_ NS | COPD-S_vs_ NS |
| Membrane receptors | |||||||||||||||
| Macrophage scavenger receptor 1 | (3,29) | MSR1 | 208422_at | 100 | 100 | 100 | 1.1 | −1.7 | −1.8 | N.S. | N.S. | N.S. | - | - | - |
| 208423_s_at | 100 | 100 | 100 | 1 | −1.1 | −1.2 | N.S. | N.S. | N.S. | ||||||
| Mannose receptor, C type 1 | (2,3,29) | MRC1 | 204438_at | 100 | 100 | 100 | 1 | 1.1 | 1 | N.S. | N.S. | N.S. | - | - | - |
| Mannose receptor, C type 2 | (2,3) | MRC2 | 37408_at | 13 | 47 | 25 | 1.5 | 1.9 | 1.3 | N.S. | N.S. | N.S. | - | - | - |
| Chemokine (C-X-C motif) receptor 4 | (4,29) | CXCR4 | 217028_at | 100 | 100 | 100 | 1.1 | 1.4 | 1.3 | N.S. | N.S. | N.S. | - | - | - |
| 211919_s_at | 96 | 100 | 100 | 1.2 | 1 | −1.2 | N.S. | N.S. | N.S. | ||||||
| Chemokine (C-C motif) receptor 5 | (3,71) | CCR5 | 206991_s_at | 100 | 100 | 100 | 1.9 | 1.6 | −1.2 | <0.02 | N.S. | N.S. | ↑ | - | - |
| C-type lectin domain family 7A | (2,88) | CLEC7A | 221698_s_at | 100 | 100 | 100 | −1.2 | 1 | 1.2 | N.S. | N.S. | N.S. | - | - | - |
| 241098_at | 42 | 50 | 92 | 1.3 | 2.3 | 1.7 | N.S. | N.S. | N.S. | ||||||
| 1554406_a_at | 100 | 100 | 100 | 1 | −1.2 | −1.2 | N.S. | N.S. | N.S. | ||||||
| 1555214_a_at | 88 | 100 | 92 | 1.1 | −1.6 | −1.8 | N.S. | N.S. | N.S. | ||||||
| 1555756_a_at | 100 | 100 | 100 | 1.1 | 1 | −1.2 | N.S. | N.S. | N.S. | ||||||
| Purinergic receptor P2Y, G-protein coupled, 5 | (29) | P2RY5 | 218589_at | 100 | 100 | 100 | 1.5 | 2.1 | 1.3 | N.S. | <0.02 | N.S. | - | ↑ | - |
| Stabilin 1 | (89) | STAB1 | 38487_at | 13 | 53 | 67 | 1.8 | 2.1 | 1.2 | N.S. | N.S. | N.S. | - | - | - |
| CD9 antigen (p24) | (33) | CD9 | 233317_at | 4 | 38 | 25 | 3.9 | 1.9 | −2 | <0.01 | N.S. | N.S. | ↑ | - | - |
| CD36 (collagen type I receptor) | (29,70) | CD36 | 241929_at | 88 | 100 | 100 | 2.9 | 3 | 1 | <0.02 | <0.03 | N.S. | ↑ | ↑ | - |
| 228766_at | 100 | 100 | 100 | 1.9 | 2.2 | 1.1 | <0.05 | N.S. | N.S. | ||||||
| 236923_x_at | 100 | 100 | 100 | 1.7 | 2.5 | 1.4 | N.S. | <0.01 | N.S. | ||||||
| 206488_s_at | 100 | 100 | 100 | 1.5 | 1.6 | 1.1 | N.S. | N.S. | N.S. | ||||||
| 209554_at | 8 | 27 | 17 | −1.1 | 1.2 | 1.4 | N.S. | N.S. | N.S. | ||||||
| 209555_s_at | 100 | 100 | 100 | 1.6 | 1.4 | -1.1 | N.S. | N.S. | N.S. | ||||||
| 242197_x_at | 42 | 85 | 83 | 2.9 | 2.3 | -1.3 | <0.01 | N.S. | N.S. | ||||||
| CD163 antigen | (2,3) | CD163 | 203645_s_at | 100 | 100 | 100 | 1 | 1.2 | 1.1 | N.S. | N.S. | N.S. | - | - | – |
| 215049_x_at | 100 | 100 | 100 | 1.2 | 1.2 | 1.2 | N.S. | N.S. | N.S. | ||||||
| 216233_at | 83 | 91 | 92 | 1.2 | 1.5 | 1.2 | N.S. | N.S. | N.S. | ||||||
| c-mer proto-oncogene tyrosine kinase | (31) | MERTK | 206028_s_at | 54 | 91 | 100 | 2.7 | 4.7 | 1.7 | <0.04 | <0.004 | N.S. | ↑ | ↑ | - |
| 211913_s_at | 54 | 91 | 100 | 2.7 | 3 | 1.1 | <0.006 | <0.03 | N.S. | ||||||
| Adenosine A3 receptor | (31) | ADORA3 | 223660_at | 13 | 56 | 83 | 2.7 | 4.6 | 1.7 | N.S. | <0.01 | N.S. | - | ↑ | - |
| 206171_at | 21 | 53 | 50 | 2.1 | 3.1 | 1.5 | N.S. | N.S. | N.S. | ||||||
| Fc fragment of IgE, low affinity II,receptor for (CD23) | (2,3) | FCER2 | 206759_at | 0 | 0 | 0 | N.D.** | N.D.** | N.D.** | N.S.* | N.S.* | N.S.* | - | - | - |
| Interleukin 4 receptor | (3) | IL4R | 203233_at | 100 | 100 | 92 | −1.1 | −1.2 | −1.1 | N.S. | N.S. | N.S. | - | - | - |
| 242743_at | 58 | 82 | 67 | 1.3 | 1.4 | 1.1 | N.S. | N.S. | N.S. | ||||||
| Cytokines and chemokines | |||||||||||||||
| Interleukin 10 | (2–4) | IL10 | 207433_at | 54 | 50 | 58 | 1 | 1.7 | 1.7 | N.S. | N.S. | N.S. | - | - | - |
| Interleukin 1 receptor antagonist | (2–4) | IL1RN | 212657_s_at | 100 | 100 | 100 | 1 | −1.4 | −1.3 | N.S. | N.S. | N.S. | - | - | - |
| 212659_s_at | 100 | 100 | 100 | 1.3 | −2.6 | −3.5 | N.S. | N.S. | N.S. | ||||||
| 216243_s_at | 100 | 100 | 100 | 1.4 | −3.3 | −4.4 | N.S. | N.S. | <0.006 | ||||||
| Transforming growth factor, beta 1 | (3) | TGFB1 | 203085_s_at | 100 | 100 | 100 | 1.4 | −1.2 | −1.7 | N.S. | N.S. | N.S. | |||
| Chemokine (C-C motif) ligand 17 | (4) | CCL17 | 207900_at | 25 | 0 | 0 | −1.2 | N.D.** | N.D.** | N.S.* | N.S. | N.S. | - | - | - |
| Chemokine (C-C motif) ligand 18 | (2–4,29) | CCL18 | 32128_at | 100 | 100 | 100 | −1.2 | 1.1 | 1.3 | N.S. | N.S. | N.S. | - | - | - |
| 209924_at | 100 | 100 | 100 | −1.1 | 1.1 | 1.2 | N.S. | N.S. | N.S. | ||||||
| Chemokine (C-C motif) ligand 22 | (4) | CCL22 | 207861_at | 54 | 50 | 42 | 1.4 | −1.1 | −1.6 | N.S. | N.S. | N.S. | - | - | - |
| Chemokine (C-C motif) ligand 23 | (29,40) | CCL23 | 210548_at | 100 | 97 | 92 | −2.5 | −3.9 | −1.6 | N.S. | <0.02 | N.S. | - | ↓ | - |
| 210549_s_at | 100 | 97 | 92 | −2.4 | −3.9 | −1.6 | N.S. | <0.03 | N.S. | ||||||
| Chemokine (C-C motif) ligand 24 | (4) | CCL24 | 221463_at | 54 | 68 | 8 | 1.2 | −2.1 | −2.5 | N.S. | N.S. | N.S. | - | - | - |
| Signaling-related proteins | |||||||||||||||
| Regulator of G-protein signaling 1 | (32) | RGS1 | 216834_at | 100 | 100 | 100 | 1 | 2.6 | 2.5 | N.S. | <0.005 | <0.008 | - | ↑ | - |
| Growth arrest-specific 7 | (29) | GAS7 | 202191_s_at | 92 | 100 | 100 | 2.1 | 3.3 | 1.6 | N.S. | <0.02 | N.S. | - | ↑ | - |
| 211067_s_at | 75 | 82 | 100 | 1.8 | 1.9 | 1.1 | N.S. | N.S. | N.S. | ||||||
| Enzymes and other proteins | |||||||||||||||
| Arginase, liver | (2,3,38) | ARG1 | 206177_s_at | 17 | 24 | 0 | 1 | N.D.** | N.D.** | N.S.* | N.S.* | N.S.* | - | - | - |
| Matrix metallopeptidase 2 (gelatinase A, 72 kDa gelatinase) | (72) | MMP2 | 201069_at | 63 | 88 | 100 | 2.7 | 6.2 | 2.2 | <0.03 | <0.002 | N.S. | ↑ | ↑ | - |
| 1566678_at | 29 | 21 | 42 | −1.4 | 1 | 1.4 | N.S. | N.S. | N.S. | ||||||
| Matrix metallopeptidase 7 (matrilysin, uterine) | (27) | MMP7 | 204259_at | 25 | 50 | 58 | 2 | 3.9 | 2 | N.S. | <0.009 | N.S. | - | ↑ | - |
| Matrix metallopeptidase 9 (gelatinase B, 92 kDa gelatinase) | (3) | MMP9 | 203936_s_at | 100 | 100 | 100 | 1.4 | 1.7 | 1.2 | N.S. | N.S. | N.S. | - | - | - |
| Heparan sulfate (glucosamine) 3-0- sulfotransferase 1 | (29) | HS3ST1 | 205466_s_at | 42 | 71 | 67 | 2.2 | 3.2 | 1.4 | N.S. | N.S. | N.S. | - | - | - |
| Heparan sulfate (glucosamine) 3-0- sulfotransferase 2 | (29) | HS3ST2 | 219697_at | 83 | 79 | 100 | 1.5 | 5.2 | 3.6 | N.S. | <0.006 | <0.02 | - | ↑ | ↑ |
| Collagen, type VI, alpha 2 | (90) | COL6A2 | 209156_s_at | 38 | 50 | 42 | 1.6 | 1.1 | −1.4 | N.S. | N.S. | N.S. | - | - | - |
| Fibronectin | (29,91) | FN1 | 214701_s_at | 100 | 100 | 92 | 2.4 | 1.3 | 1.8 | N.S. | N.S. | N.S. | - | - | - |
The M1 and M2 polarization patterns were determined by comparing, for each probe, the significance of the fold-change in the level of expression among groups. If more than 1 probe from the same gene was on the array, the gene was considered significantly up- or down-regulated in one group compared to another if ≥50% of the probes were significantly changed and/or the change was confirmed by TaqMan analysis.
The polarization pattern of AM associated with cigarette smoking was determined based on the analysis of expression of M1- and M2-related genes in AM of healthy smokers as compared to healthy nonsmokers. M1- and/or M2 genes were determined to be smoking responsive if their expression changed significantly in healthy smokers compared to healthy nonsmokers.
The polarization pattern of AM of COPD smokers was assessed by comparing the expression of M1- and M2-related genes of COPD smokers with that of healthy nonsmokers and healthy smokers. The global transcriptional changes related to M1- and M2-polarization of healthy smokers compared to healthy nonsmokers as well as COPD smokers compared to both healthy nonsmokers and healthy smokers were analyzed by assessing the mean normalized expression levels of all M1-related and M2-related probe sets for healthy nonsmokers, healthy smokers and COPD smokers, using all (i.e., significantly changed as well as those not significantly changed) M1- and M2-related probe sets with P calls ≥20%. To assess whether changes in M1- and M2-gene expression in AM of COPD smokers were related to smoking, an additional comparative analysis was performed using the M1- and M2-related probe sets significantly differently expressed between COPD smokers and healthy nonsmokers.
Unsupervised hierarchical clustering of the healthy nonsmokers vs healthy smokers was carried out for the significantly changed M1- and M2-related genes using the MAS5-analyzed data with the Spearman correlation as similarity measure and the complete linkage clustering algorithm using GeneSpring software. To visualize the differences in expression of the M1- and M2-related genes among the 3 groups (healthy nonsmokers, healthy smokers, COPD-smokers), principal component analysis (PCA) was performed using all M1- and M2-related probe sets with P calls >20% for each group. For comparison, all probe sets on the array were independently assessed with the identical individuals. These analyses were carried out using GeneSpring by mean centering of microarray normalized intensity values of all subjects or the average for each of the three groups in order to assign the general variability in the data to a reduced set of principal components (34). The first 3 principal components containing most of the variance-based information were visualized in 3-dimensional space. To evaluate the relative contribution of the differences in M1- and M2-gene expression to the global transcriptional differences between the study groups, the variability (as measured by the first 3 principal components) determined by PCA of M1- and M2-related probe sets was compared to that revealed by PCA of the same study groups using all 26,959 probe sets with a P call ≥20%. To exclude the possible influence of differences in age between COPD smokers and other groups as well as differences in pack-yr between COPD smokers and healthy smokers, nonparametric Spearman correlation analysis has been used to evaluate the correlation of COPD-relevant M1- and M2-related gene expression with age in healthy smokers and smokers with COPD as well as with pack-yr in smokers with COPD.
Real-time PCR Confirmation of Microarray Data
TaqMan real-time RT-PCR was used to confirm differential expression of a subset of genes found to be differentially expressed by microarray, using the same RNA samples that were used for the microarray analysis. cDNA was synthesized from 2 µg RNA isolated from AM in a 100 µl reaction volume, using the TaqMan Reverse Transciptase Reaction Kit (Applied Biosystems, Foster City, CA) with random hexamers as primers. Two dilutions of 1:10 and 1:100 were made from each sample and duplicate wells were run with each dilution. TaqMan PCR reactions were carried out using pre-made kits from Applied Biosystems and 2 µl of cDNA were used in each 25 µl reaction volume. The PCR reactions were run in an Applied Biosystems Sequence Detection System 7500 and relative expression levels were determined using the ΔΔCt method using β-actin as endogenous control. TaqMan gene expression assays (all from Applied Biosystems) were assessed for selected M1-related genes [CD80 (Hs00175478_m1), IL1B (Hs00174097_s1), TNFSF10 (Hs00234356_m1), CXCL9 (Hs00171065_m1), CXCL11 (Hs00171138_m1), GBP5 (Hs00369472_m1), CCL5 (Hs00174575_m1)], and selected M2-related genes [CCR5 (Hs99999149_s1), CD36 (Hs01567186_m1), ADORA3 (Hs00252933_m1), MMP2 (Hs00165949_m1), and MMP7 (Hs01042795_m1)].
Flow Cytometry
To assess AM expression of M1 and M2-related genes at the protein level, freshly isolated cells recovered by lavage (~2 × 106) were washed in PBS containing 2% bovine serum albumin (BSA), resuspended in PBS, 2% BSA and 5% inactivated human serum for 20 min, at 4°C to block nonspecific antibody binding. The cells were stained for surface antigens with the following mouse monoclonal anti-human antibodies: fluorescein (FITC)-conjugated antibodies against CD206 (macrophage mannose receptor, to verify the macrophage phenotype), CD3 (to identify contaminating T cells), phycoerythrin (PE)-conjugated antibodies against CD56 (to identify contaminating NK cells), CD15 (to identify contaminating granulocytes), TNFSF10 and CD80 (for validation of M1 polarization); and CCR5 and CD36 (for validation of M2 polarization; all from BD Biosciences). Appropriate isotype controls were used for each antibody. The incubation with the antibodies was for 30 min on ice in dark according to the manufacturer’s instructions. After subsequent washing, cells were fixed and permeabilized with Cytofix/Cytoperm reagent (BD Biosciences) for 20 min on ice and washed with Perm/Wash solution. For intracellular staining, cells were fixed and permeabilized with Cytofix/Cytoperm reagent (BD Biosciences) for 20 min, 4°C, and washed twice with Perm/Wash solution prior to incubation with the following mouse monoclonal anti-human antibodies: FITC-conjugated antibodies against CCL5 or CXCL9, and PE-conjugated IL1B (all from R&D Systems). All samples were prepared and analyzed with and without quenching with 0.2% crystal violet (1 min, 4°C; Polyscientific, Bayshore, NY) to reduce the autofluorescence (35). After washing, cells were analyzed by a FACScalibur cytometer (BD Biosciences, Pharmingen) using Cell Quest software. Thirty thousand events were collected for each sample. FACS data were analyzed using WinMDI 2.8 software (The Scripps Institute, La Jolla, CA) and expressed as a percentage increase of mean fluorescence for a given antibody over isotype control.
Statistical Analysis
Statistical comparisons for microarray data were calculated using GeneSpring software and associated two-tailed t test with unequal variance. Differences with a fold-change >1.5 and p<0.05 with a Benjamini-Hochberg correction were considered statistically significant. For other experiments, comparisons between groups were analyzed by 2-tailed t test, or ANOVA for experiments with more than two subgroups, and values are displayed as mean ± standard deviation.
Deposition of Data
All gene expression data has been deposited at the Gene Expression Omnibus (GEO) site (http://www.ncbi.nlm.nih.gov/geo/). Accession number is as follows: HG-U133 Plus 2.0 GSE13896.
Results
Biologic Samples
The total numbers of cells recovered by lavage for healthy nonsmokers was 12.7 ± 8.1 × 106, healthy smokers 25.5 ± 17.0 × 106 (p<0.05 compared to healthy nonsmokers) and COPD smokers 12.9 ± 8.1 × 106 (p<0.05 compared to healthy nonsmokers, p>0.05 compared to healthy smokers). After purification, the AM populations were >98% pure in all groups. The cell viability ranged from 94 to 97%. To exclude the effect of adherence step of AM purification on global transcriptional changes associated with smoking, we compared the expression of smoking-responsive genes identified in a previous study in which AM were isolated by flow cytometry (17) with our results. Such analysis is relevant for interpretation of data since it has been shown that macrophage may change expression of a set of genes following in vitro culture (36). The data shows that the method of AM purification does not significantly modify the differential expression of smoking-responsive genes: around 46% gene probe sets found to be significantly up- or down-regulated in smokers in a study with flow cytometry-based AM isolation had similar significant change in our study (Supplemental Figure 1A) and there is significant positive correlation of smoking-responsive probe set expression detected in these two studies (r=0.71, p<0.0001; Supplemental Figure 1B).
Deactivation of the Steady-state M1 Polarization Program in AM of Healthy Smokers
Since an altered inflammatory response of AM to cigarette smoking is thought to play a central role in the development of pathological changes leading to chronic lung disease (9,10) and IFNγ, an inducer of M1 macrophage polarization, has been identified as important mediator of smoking-induced disease in animal models (13–15), we first asked whether chronic cigarette smoking skews human AM activation toward M1 polarization in vivo? To evaluate this question, the expression of the set of M1-related genes by AM of healthy smokers was compared to that of healthy nonsmokers. Surprisingly, the data showed that the majority of genes associated with M1 polarization were down-regulated in AM of healthy smokers as compared to healthy nonsmokers (Figure 1A, Table II). Of the 41 M1 genes evaluated, 23 genes (56%) were down-regulated in healthy smokers.
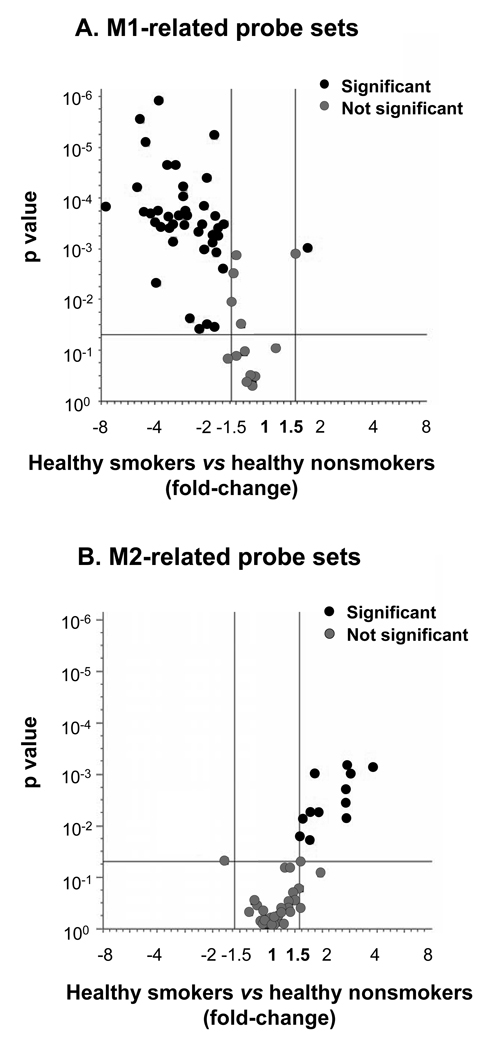
Figure 1.
Smoking-mediated reciprocal induction of M1 and M2 polarization programs of human alveolar macrophages. A. Volcano plot of M1-related probe sets significantly differently expressed between healthy nonsmokers (n=24) and healthy smokers (n=34). B. Volcano plot for the M2-related gene probe sets comparing the same groups. For both panels, the x-axis corresponds to the fold-change and the y-axis corresponds to p value. Red dots represent significant differentially expressed probe sets, grey dots represent probe sets with no significant difference between healthy smokers and healthy nonsmokers. The changes in gene expression were considered statistically significant based on the criteria of fold-change ≥1.5, p<0.05 with Benjamini-Hochberg correction.
Many of the M1-related genes down-regulated in AM by smoking encode T-cell-recruiting chemokines. Among these were IFNγ-inducible type 1 chemokines C-X-C chemokine ligand (CXCL) 11 (the most down-regulated gene, fold-change 7.65 vs healthy nonsmokers; p<0.01), CXCL9 (p<0.02), C-C chemokine ligand (CCL)5 (p<0.01), CCL4 (p<0.02), as well as a family of proteins implicated in IFN signaling such as guanylate binding proteins (GBP) 1, 2, 4, 5. Among other down-regulated M1-related genes were IFNγ-dependent gene CD69 (p<0.0003), costimulatory molecule CD80 (p<0.002), inflammasome-related cytokines IL-1 (p<0.02) and IL-18 (p<0.0006), TNF-related proteins TNFAIP6 (p<0.01) and TNFSF10 (p<0.02), as well as complement-related genes such as complement factor B and complement component 3a receptor. The TLR4, a LPS receptor, was the only M1-related gene up-regulated in AM of healthy smokers (fold-change 1.5, p<0.05). Interestingly, the IL-12 gene, typical for M1 polarization (2–4), was very weakly expressed in AM, there was a tendency (albeit not significant) for its down-regulation in AM of healthy smokers. Another typical M1-related gene, inducible nitric oxide synthase, was not expressed in AM regardless of smoking status, consistent with previous observations in human macrophages (37).
Interestingly, the decreased average expression of many M1-related genes was associated with a marked increase in the number of subjects in the group of healthy smokers in which these genes were not detected at all. For example, only 27% of healthy smokers expressed CD69 as compared to 83% of healthy nonsmokers (p<10−6); less than half of healthy smokers (47%) expressed CD80 gene vs 96% of healthy nonsmokers (p<10−6; Table II). A dramatic smoking-related decrease of P calls was also detected for several chemokine genes including CXCL11 (18% in healthy smokers vs 63% in healthy nonsmokers; p<0.001) and CXCL10 (47% in healthy smokers vs 83% in healthy nonsmokers; p<0.005).
The differential expression of several M1-related genes between healthy smokers and healthy nonsmokers was further confirmed by TaqMan real-time PCR (Supplemental Figure 2). For example, the M1-related genes CD80, TNFSF10, IL1B, CXCL11, CXCL9, and CCL5 were all observed to be down-regulated by both microarray and TaqMan PCR. Consistent with these observations, M1-related gene expression was also observed to be down-regulated at the protein level (Supplemental Figure 3). FACS analysis demonstrated that CD80, TNFSF10, IL1B, CXCL9, and CCL5 are down-regulated in AM of healthy smokers compared to healthy nonsmokers AM (p<0.05, all comparisons).
Unusual M2-like Polarization Program Induced in AM of Healthy Smokers
The current concept of macrophage polarization programs suggests their reciprocal regulation, implying that M2 polarization develops when IFNγ, the inducer of macrophage activation towards M1-polarization, does not dominate over alternative group of stimuli such as IL-4, IL-13, IL-10, TGFβ, glucocorticoids, and M-CSF and others (3,4). We sought, therefore, to determine whether down-regulation of M1-related gene expression pattern in human AM by chronic cigarette smoking is accompanied by induction of M2 polarization program?. To assess this question, the expression of M2-related genes was analyzed in AM of healthy smokers in comparison to healthy nonsmokers. Interestingly, by contrast to M1-related genes, the expression of all of the genes associated with M2-like polarization that were significantly modified by smoking were up-regulated in healthy smokers (Table II, Table III, Figure 1B).
However, unlike the broad and distinct pattern of M2-polarization observed with some stimuli, smoking was not associated with a uniform induction of genes related to particular M2 polarization pathway. For example, the genes known to be induced during alternative, IL-4 driven, macrophage activation in vitro such as those encoding mannose receptor, macrophage scavenger receptor 1, DECTIN1, and CD163 (2,3) were not significantly modulated by smoking (Table III). Further, expression of other M2-typical genes, such as arginase-1 and YM1 (not shown), were not detected in AM of either group, consistent with previous studies of human macrophages (38). Overall, the panel of smoking-induced M2-related probe sets in AM represented a mixture of different subtypes of M2-like macrophage polarization with simultaneous up-regulation of genes related to IL-4-induced (CD36; p<0.02), glucocorticoid-induced (MERTK; p<0.006), IL-10-induced (C-C chemokine receptor (CCR)5; p<0.02), M-CSF-induced (MMP2; p<0.03, and CD36; p<0.02), as well as IFNγ-down-regulated (CD9; p<0.01) M2-like polarization programs (Table III).
The expression of several M2-related genes was analyzed using TaqMan real-time PCR (Supplemental Figure 2). As examples, MMP2, CCR5 and CD36 were up-regulated in both the microarray and TaqMan analysis, confirming the microarray data regarding their differential expression in AM of healthy smokers vs healthy nonsmokers. Furthermore, the M2 genes up-regulated at the mRNA level were also up-regulated at the protein level. For example, FACS analysis showed that CCR5 and CD36 were found to be significantly up-regulated at the protein level in AM of healthy smokers vs healthy nonsmokers (p<0.05 and p<0.01, respectively; Supplemental Figure 3).
Biologic Segregation of Healthy Smokers and Healthy Nonsmokers Based on Expression of M1- and M2-related Genes in AM
Based on the observation of reciprocal dysregulation of M1 and M2 polarization programs in human AM by cigarette smoking, we asked whether the expression of M1- and M2-related genes may serve as a biologic discriminator between healthy smokers and healthy nonsmokers and what is the inter-individual variability of M1- and M2- gene expression among these groups? To address this issue, the normalized expression data of all probe sets for M1- and M2-related genes was analyzed at the personalized level, i.e., for every healthy smoker and healthy nonsmoker assessed in the study. There was a coordinate down-regulation of M1-related gene expression in the majority, but not all healthy smokers, similar to coordinate relatively high expression levels of the same probe sets in the majority, but not all healthy nonsmokers (Figure 2A). The difference between healthy smokers and healthy nonsmokers based on the comparative quantitative evaluation of the average normalized expression levels of all M1-related probe sets in both groups was highly significant (p<10−15; Figure 3A). In contrast, the personalized M2-related gene expression profile demonstrated a tendency towards higher overall expression in healthy smokers (Figure 2B). The difference in the overall expression of all M2-related probe sets between the healthy nonsmokers and healthy smokers was significant, albeit less so than for the M1 gene (p<0.001; Figure 3C).
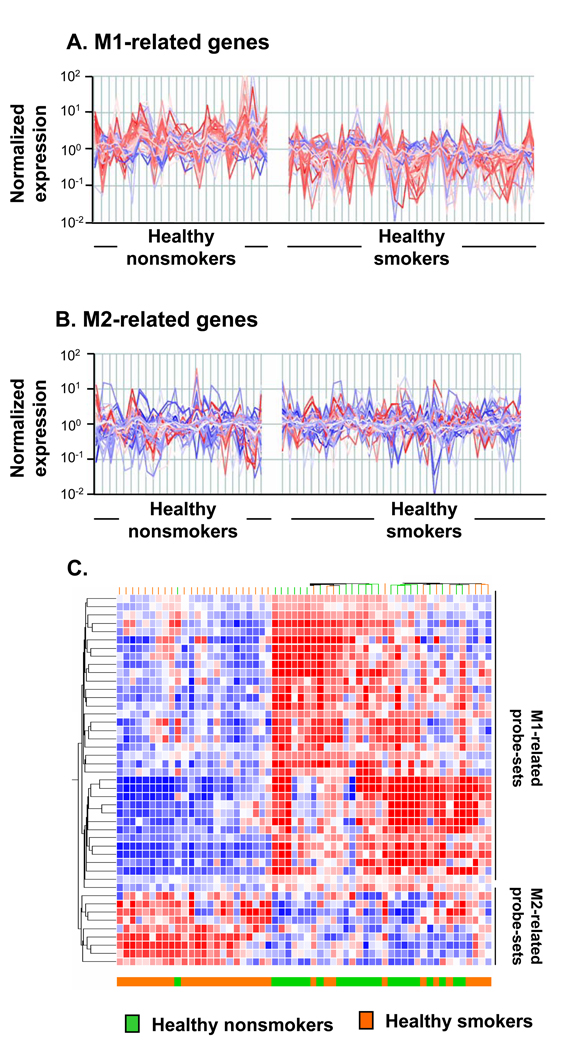
Figure 2.
Biologic phenotypes of healthy smokers compared to healthy nonsmokers based on alveolar macrophage M1- and M2-related gene expression. A. Expression of all M1-related probe sets in AM of healthy nonsmokers (n=24; left panels) compared to that of healthy smokers (n=34; right panels). See Table II for a list of all M1-related probes. B. Expression of all M2-related probe sets in AM comparing the same groups (healthy nonsmokers, left; healthy smokers, right). See Table III for a list of all M2-related probes. For both A and B, the y-axis indicates normalized relative expression levels for the probe sets, the x-axis shows the individuals belonging to each group randomly ordered but of similar order in A and B. Red = gene probe sets down-regulated in healthy smokers as compared to healthy nonsmokers; blue = gene probe sets up-regulated in healthy smokers as compared to healthy smokers. Intensity of color indicates the degree of down- or up-regulation. Note that overall, the M1-related genes tend to be down-regulated in the healthy smokers compared to the healthy nonsmokers. The opposite is observed among the M2-related genes, but not to the same extent as the down-regulation of the M1 genes. C. Non-supervised hierarchical cluster analysis of AM M1- and M2-related gene expression of healthy nonsmokers and healthy smokers. The analysis is based on healthy smokers, the differential expression of M1- and M2- related genes of the same groups of healthy nonsmokers (n=24) and healthy smokers (n=34) using Spearman correlation as a similarity measure and an average linkage as a clustering algorithm. Statistically significant differentially expressed M1- and M2-gene probe sets were used as input data set. Genes expressed above average are represented in red, below average in blue, and average in white. The genes are represented vertically, and individual subjects horizontally at the bottom. Healthy nonsmokers are indicated by green, healthy smokers by orange. Although, there is variability within each group, compared to the healthy nonsmokers, the general tendency for the healthy smokers is for the M1 genes to be down-regulated and the M2 genes up-regulated.
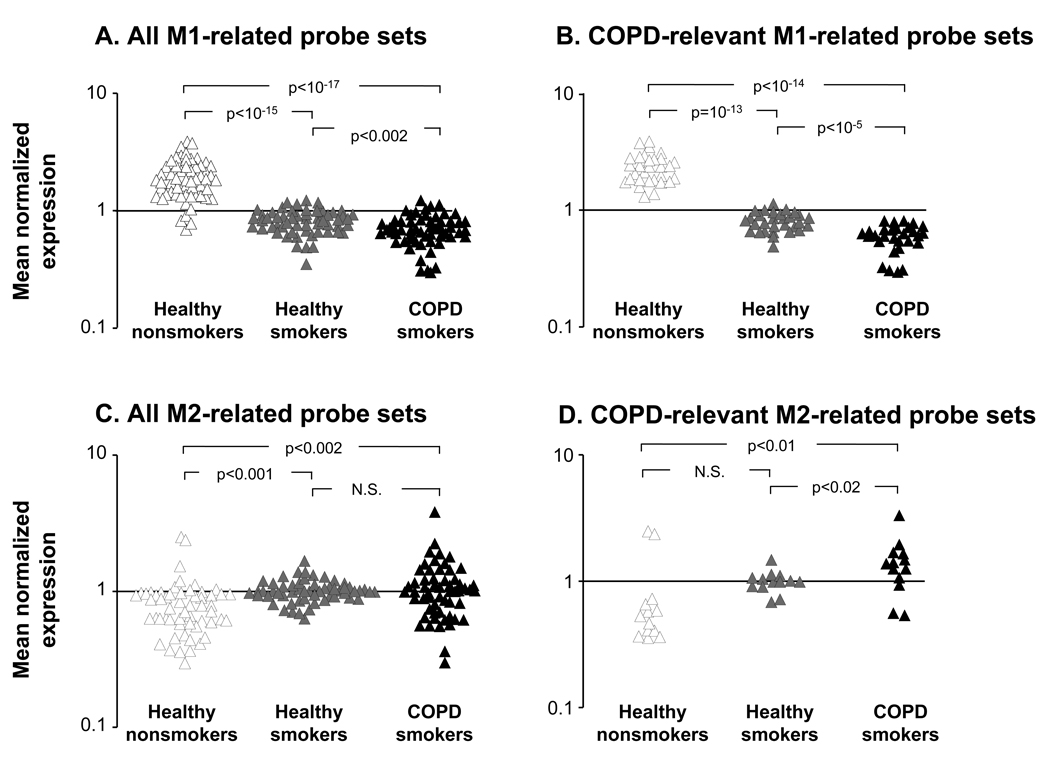
Figure 3.
Progressive reciprocal alteration of M1- and M2-related gene expression in alveolar macrophages with the development of COPD. A. Normalized expression levels of all M1-related probe sets; B. M1-related probe sets significantly differentially expressed in AM of COPD smokers vs healthy nonsmokers; C. All M2-related probe sets; and D. M2-related probe sets significantly differentially expressed in AM of COPD smokers vs healthy nonsmokers. The data is based on healthy nonsmokers (n=24), healthy smokers (n=34) and COPD smokers (n=12). The y-axis indicates mean normalized, expression levels for the probe sets in each group; the x-axis indicates the groups. p values represent differences among groups as indicated.
To analyze this in a greater detail, we performed hierarchical clustering of healthy smokers and healthy nonsmokers using Spearman correlation analysis and significantly regulated M1- and M2-related gene probe sets as input data set (Figure 2C). This analysis revealed that healthy smokers as a group could be quite effectively segregated from healthy nonsmokers based on expression of smoking-sensitive M1- and M2-related gene probe sets in AM, with only 1 of 24 healthy non-smokers similar to healthy smokers. Remarkable heterogeneity was observed among healthy smokers based on the expression of genes related to macrophage polarization. Indeed, 24 of 34 healthy smokers (71%) exhibited an expression pattern strictly different from healthy nonsmokers, characterized by almost coordinate reciprocal alteration of M1- and M2-related gene expression (Figure 2C). Interestingly, the remaining (29%) of healthy smokers were clustered together with healthy nonsmokers. Together, this suggests that the expression profile of M1- and M2-related genes in AM not only discriminates among healthy smokers and healthy nonsmokers, but also identifies different biologic phenotypes within each group.
Progressive Deactivation of M1-Gene Expression Program in AM with the Development of Smoking-induced Lung Disease
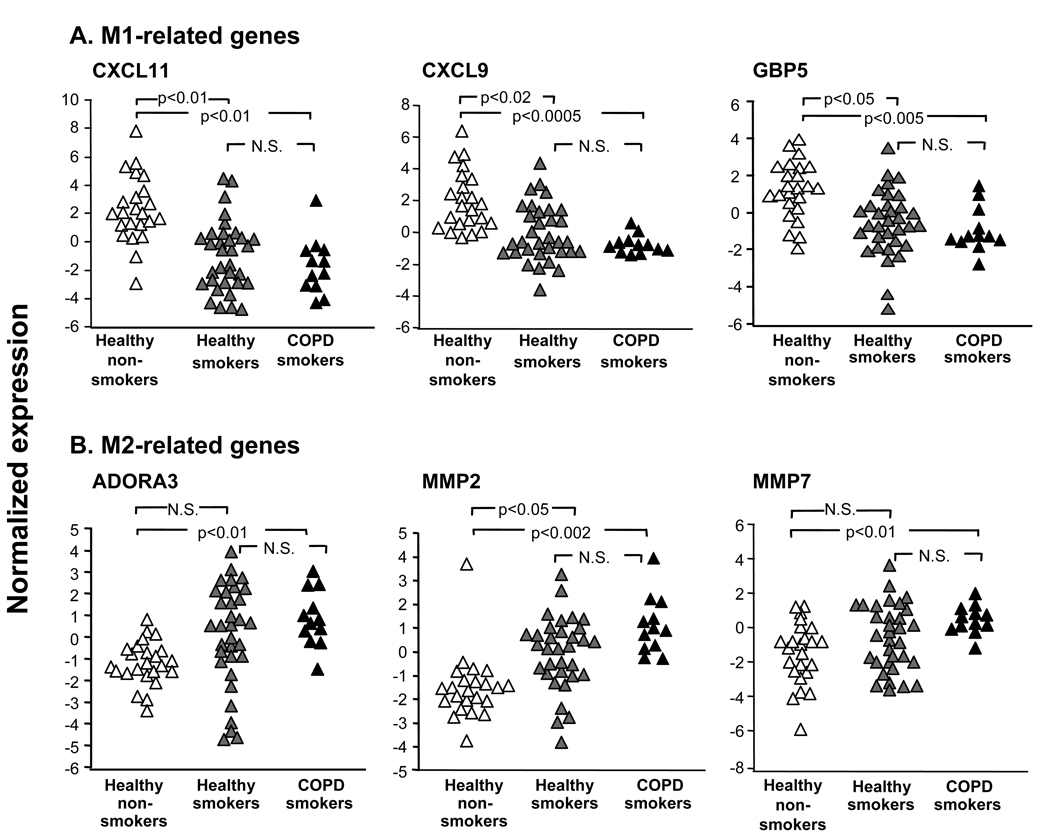
Based on these observations of alteration in M1- and M2-related polarization changes in AM of healthy smokers vs healthy nonsmokers, we assessed whether transcriptional changes induced in AM of healthy individuals by chronic cigarette smoking progresses with the development of smoking-induced lung disease. First, we asked whether there is a shift in AM polarization from M1-deactivated to M1-polarized phenotype when smokers develop COPD, a disease hypothesized to be dependent on cigarette smoking-induced IFNγ-mediated inflammatory response in the lung (10,13–16,39). To answer this question, expression of all M1-related gene probe sets in AM of COPD smokers was compared to that of healthy smokers and healthy nonsmokers. In the COPD smokers, the overall average expression of M1-related gene probe sets was substantially suppressed as compared to healthy nonsmokers (p<10−17; Figure 3A). Further, COPD smokers exhibited a further down-regulated M1-related gene expression pattern, significantly suppressed compared to healthy smokers (p<0.002). The down-regulation of M1-related genes appeared to be rather smoking-dependent than disease-dependent, i.e., there were more M1-related genes that were significantly down-regulated in healthy smokers vs healthy nonsmokers than COPD smokers vs healthy smokers (Table II). Notably, all M1-related genes significantly down-regulated in COPD smokers vs healthy nonsmokers were suppressed to some extend in healthy smokers as compared to nonsmokers. Thus, the earliest changes in the M1 polarization pattern relevant to COPD are already present in smokers without disease, with a progression of deactivation of M1-related gene expression with the development of smoking-induced lung disease. Among the genes progressively down-regulated with the development of smoking-induced lung disease were the M1-related IFNγ-inducible genes CXCL11, CXCL9, and GBP5 (Figure 4A), a phenotypic pattern that was confirmed by TaqMan real-time PCR (Supplemental Figure 4A). The similar trend was characteristic for other M1-related genes including CXCL10, IL1B, and IRF7 (Table II). Consistent with the concept of progressive suppression of expression of the M1-related genes with the development of disease, many M1-related genes were not detected in AM of a considerable portion of COPD smokers. For example, CXCL10 was expressed in AM of only 2 of 12 (17%) COPD smokers whereas it was detected in 92% of healthy nonsmokers and 50% of healthy smokers (Table II). Similar progressive decrease of P calls was noted for other M1-related genes including CXCL11, CD69, CAMP, GBP4, GBP5 (Table II).
Figure 4.
Examples of AM expression of M1 and M2 polarization-related genes demonstrating progressive differences with the development of COPD. A. M1-related genes; and B. M2-related genes. Log2-transformed normalized expression levels for selected M1-related genes [C-X-C chemokine ligand 11 (CXCL11); C-X-C chemokine ligand 9 (CXCL9); guanylate binding protein (GBP 5)]; and M2-related genes [adenosine A3 receptor (ADORA3); matrix metalloprotease 2 (MMP2); and matrix metalloproteinase 7 (MMP7), are plotted for all healthy nonsmokers (n=24; green triangles), healthy smokers (n=32; orange triangles), and COPD smokers (n=12; blue triangles). p values are indicated. N.S., non-significant.
Since COPD smokers are significantly older than healthy nonsmokers and smokers (Table I), it is essential to confirm that observed changes in M1-related gene expression are due to smoking but not the older age. If expression of COPD-relevant M1-related genes decreases with age independently on smoking, such tendency should be seen in a general population, such as healthy non-smokers. To exclude this, we analyzed correlation between the expression of COPD-relevant M1-probesets and the age in healthy nonsmokers using Spearman correlation analysis. We found that 9 of 30 (30%) M1-related probe sets have significant positive correlation with age (Supplemental Figure 5A), an opposite direction to expected if assume that down-regulation of M1-related genes in COPD smokers is due to their older age. Only one M1-related gene (TNFAIP6) had a significant negative correlation with age in healthy nonsmokers, however it did not correlate significantly with age in COPD smokers (Supplemental Figure 5B). Only one out of 30 COPD-relevant M1-related probe sets (IRG1) significantly negatively correlated with age (Supplemental Figure 5B). In addition, we excluded that progressive down-regulation of M1-related probe sets in COPD smokers is not due to longer pack-years as compared to healthy smokers (Table II). Only one M1-related probe set (SOCS3) significantly negatively correlated with pack-years in smokers with COPD (Supplemental Figure 7A). Thus, global down-regulated expression of M1-related genes in smokers with COPD is unlikely due to their older age or longer smoking experience.
Progressive Induction of M2-related Gene Expression Program in AM with the Development of Smoking-induced Lung Disease
We then asked whether M2-like transcriptional pattern of AM observed in healthy smokers progresses with the development of COPD in parallel with a progressive deactivation of M1 polarization program? To answer this question, expression of all M2-related probe sets was analyzed in all three groups. Reciprocal to the observations with the M1-related genes, the overall expression of M2-related gene probe sets was progressively increased with the development of COPD (Figure 3C; p<0.002 vs healthy nonsmokers). Similar to alterations in M1-related gene expression, the up-regulation of M2-related probe sets in AM of COPD smokers is likely initiated by smoking and not disease itself, since the earliest changes in expression of M2-related probe sets relevant to COPD, albeit not significant, were already observed in smokers without disease (Figure 3D; Table III). Among progressively up-regulated M2-related genes were those encoding adenosine A3 receptor (ADORA3), matrix metalloproteases MMP2 and MMP7 (Figure 4B; Table III), whose expression pattern was confirmed by TaqMan real-time PCR (Supplemental Figure 4B). Interestingly, CCL23, one of markers of IL-4-driven alternative macrophage activation (29,40), was the only significantly down-regulated M2-related gene in AM of COPD smokers (Table III), suggesting that the M2-like phenotype of AM induced by smoking is complex, and differs from that described for alternatively activated macrophages based on the in vitro studies.
The possible effect of older age and longer pack-yr on M2-related changes in AM of COPD smokers was excluded similarly as for M1-related genes. Only 1 of 13 COPD-relevant M2-related probe sets (MMP7) has a significant negative correlation with age in healthy nonsmokers (Supplemental Figure 6A), an opposite direction to expected if assume that induction of M2-related probe sets in COPD smokers is due to general increase of their expression with ageing. In the COPD group, none of M2-related COPD-relevant probe sets significantly correlated with age (Supplemental Figure 6B). Only one M2-related probe set (MMP2) significantly correlated with pack-yr in smokers with COPD (Supplemental Figure 7B, respectively). Interestingly, COPD smokers with longer pack-years had significantly lower MMP2 expression (Supplemental Figure 7B), what is opposite to expectation based on assumption that increased expression of M2-related genes in COPD smokers is due to their higher pack-year values. Thus, global up-regulated expression of M2-related genes similarly to down-regulated expression of M1-related genes in smokers with COPD is unlikely due to their older age or longer smoking experience.
Overall Differences in M1- and M2-Related Gene Expression in AM of Healthy Smokers and COPD Smokers
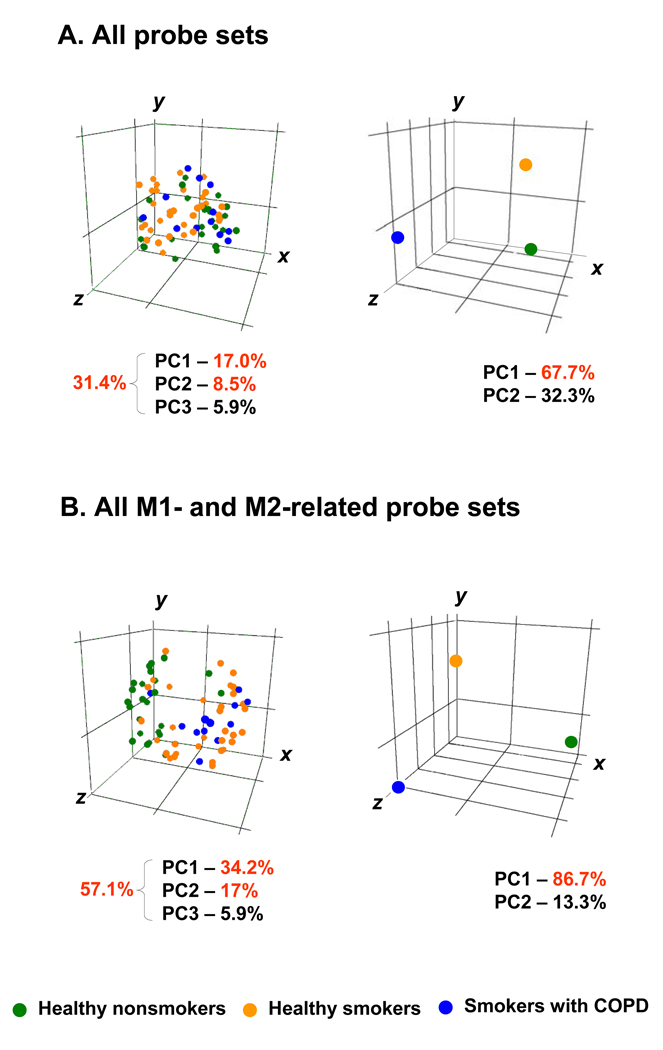
To determine how changes in expression of both M1- and M2-related genes collectively contribute to differences between healthy nonsmokers, healthy smokers and COPD smokers, principal component analysis was first applied to all 26,959 probe sets with P call≥20% compared to the experimental data set of all 114 M1- and M2-related gene probe sets. In the global 26,959-probe set space, subjects of different groups were not clearly separated from each other, indicating heterogeneity within the study groups based on the global transcriptome patterns, with only 31% of variation in the system represented by the first three principal components (Figure 5A, left panel). However, when the analysis was limited to the expression of M1- and M2-polarization related genes in AM, the first three principal components in this 114- probe set space captured 57% variation of gene expression among the subjects (Figure 5B, left panel). In the 3-dimensional space generated based on expression of M1- and M2-related probe sets, healthy nonsmokers and COPD smokers formed two separate clusters. Consistent with results of hierarchical clustering, principal component analysis on polarization-related genes revealed a substantial heterogeneity of healthy smokers, with some clustering with healthy nonsmokers, while others were similar to COPD smokers (Figure 5B, left panel).
Figure 5.
Principle component analysis (PCA) comparison of alveolar macrophage gene expression patterns of healthy smokers, healthy smokers and COPD smokers. A. PCA of all 26,959 probe sets expressed in AM of at least 20% of subjects. B. PCA of all 114 M1- and M2-related probe sets expressed in AM of at least 20% of subjects (see Tables I, II). In both analyses, all samples within each group were centered (left panels) or averaged and then the means were centered (right panels) in the three-dimensional space based on the expression pattern. In left panels, each circle represents an individual sample; in right panels, each circle represents an averaged sample for each group. Healthy nonsmokers, n=24, green; healthy smokers, n=34, orange; and COPD smokers, n=12, blue). The percentage contributions of the first three (left panels) or two (right panels) principal components (PC) to the observed variability between the groups are indicated.
Next, the averaged expression patterns for each group were subjected to principal component analysis. When analyzed as groups, healthy nonsmokers, healthy smokers and smokers with COPD were clearly segregated from each other in the global 26,959-probe set principal component space with a 67.7% of variation detected by the first component (Figure 5A, right panel), suggesting that the global average AM transcriptomes in these three groups are indeed quite different. However, when the averaged representatives for each group were plotted in the 3-dimensional principal component space based on the expression of all M1- and M2-related probe sets, the variation among the groups captured by the first component increased to 86.7% (Figure 5B, right panel). Based on visualization of principal components, gene expression of the AM of COPD smokers were clearly different from AM of healthy nonsmokers, with healthy smokers being between these two groups, consistent with the data showing that smoking-associated reprogramming of AM polarization progresses with the development of COPD.
Discussion
In this study, we employed the concept of macrophage polarization to help understand the role of AM in smoking-induced lung disease. The complex pathogenesis of COPD, a human disease associated with cigarette smoking, is clearly environment-dependent, similar to polarization phenotypes that are acquired by mononuclear phagocytes depending on particular environmental settings. The two major macrophage activation programs, referred to as M1- and M2-polarization, define the ability of macrophages to play a role in inflammation and regulation of tissue integrity, respectively (2–4). Both of these macrophage functions are altered in COPD, a disease in which the remodeling of the airways and lung parenchyma is accompanied by, and thought to be dependent on, an abnormal inflammatory response-induced by cigarette smoking (7,9–12,39,41–43). In the present study, we applied a global transcriptional profiling to assess the polarization pattern of AM in healthy smokers and COPD smokers as compared to healthy nonsmokers. The data demonstrates that cigarette smoking, indeed, alters the steady-state polarization program in human AM in vivo. However, contrary to the widespread concept that non-related inflammation is a central driver in the early pathogenesis of COPD, smoking was associated with a substantial down-regulation of genes related to M1 macrophage polarization, i.e., there is an overall hypoinflammatory gene expression pattern in AM of healthy smokers. Deactivation of M1 polarization pattern was accompanied by the induction of an unusual phenotype characterized by up-regulation of genes associated with different M2 polarization programs. The data further demonstrates that, with the development of COPD, there is progression of suppression of the M1 program and, to a lesser extent, enhanced expression of some M2-related genes.
Deactivation of M1 Polarization Program in AM by Cigarette Smoking
A major observation of the present study is that AM of healthy smokers exhibit a coordinate down-regulation of a considerable number of genes typical for M1 polarization, a distinct activation program of inflammatory and host defense genes induced in mononuclear phagocytes in vitro by IFNγ and LPS (2–4). Among these genes are those encoding type 1 chemokines CXCL11, CXCL9, CCL4, CCL5, inflammasome-related cytokines IL-1β and IL-18, costimulatory molecule CD80, complement-related proteins, a number of proteins involved in IFNγ signaling. This observation leads to several conclusions.
First, the smoking-induced suppression of the M1 polarization program of AM, and of the inflammation program of AM in general, has implications for understanding the role of AM as inflammatory cells in the pathogenesis of the early events in smoking-induced lung disease. The current concepts of COPD suggest that the disease develops as a result of abnormal inflammatory response of the lung to cigarette smoke or other noxious gases and particles, and this inflammatory response mediates small airways derangements and alveolar destruction (7,9–12,42,43). The lungs of smokers contain increased numbers of AM, which accumulate in the sites of alveolar wall destruction (6,9–12). While there is clear evidence that inflammatory and immune mechanisms play a role in mediating lung damage in established COPD (7,9,10,16,41–43), the effect of smoking on the pro-inflammatory properties of individual cell types in the human lung in vivo before the manifestation of the disease has not been studied in detail. The present study suggests that, at least for the role of alveolar macrophages, the early events in the pathogenesis of smoking-induced COPD mediated by AM in humans is unlikely inflammatory. Consistent with our observation of suppression of the M1 polarization pattern of human AM with smoking, other studies have observed decreased levels of transcripts for selected inflammatory cytokines in the BAL of smokers. For example, IL-6, CCL4, CCL5, CCL20 have been noted to be down-regulated in cells recovered by lavage of healthy smokers, although the cellular source of transcripts was not identified (44). Decreased expression of pro-inflammatory cytokines such as IL-6 and TNF-α as well as some surface molecules related to immune response in AM of healthy smokers has also been described (45–48). However, in general, these observations have been ignored in the context that exaggerated inflammatory and immune processes dominate in the lungs of patients with established COPD (7,9–12,41–43). The global gene expression analysis of AM carried out in the present study provides a transcriptome-based evidence that AM unlikely contribute to augmented production of inflammatory mediators in response to smoking, and is consistent with the basic concepts of macrophage polarization that not all forms of macrophage activation are pro-inflammatory (1–4).
Compatible with our observations, two previous studies in which the transcriptional pattern of AM of smokers was compared to nonsmokers have revealed that cigarette smoking is associated with induction of several genes related to tissue remodeling without activation of inflammatory program in AM, although the expression of macrophage polarization-related genes was not been addressed specifically (17,18). With regard to M1/M2-related genes, there is considerable overlap between these and our studies. Of 28 smoking-responsive M1- and M2-related genes identified in our study, 25 (89%) were detected in the study of Woodruff et al. (17) and 21 (75%) in the study of Heguy et al. (18). Among detected M1- and M2-related genes, 20 genes (80%) in the study of Woodruff et al (17) and 10 genes (47%) in the study of Heguy et al. (18) had the same direction of change as in our study (not shown). The overlapping genes, i.e. M1- and M2-related genes with the same direction of change, in these two studies and our study include M1-related genes TNFSF10, GBP1, PDE4B, and M2-related genes CCR5, CD9, and CD36. However, a direct gene-to-gene comparison of these studies is difficult due to different microarray analyzes used in these studies. Of interest, one of these studies demonstrated that AM of smokers displayed expression pattern distinct from those evoked in murine models of emphysema (17).
Second, from a host defense perspective, a broad suppression of inflammatory/immune genes in smokers is consistent with the epidemiologic data that smokers and COPD smokers are more susceptible to respiratory tract infection than nonsmokers (49). Accordant with this concept, AM of healthy smokers exhibit a decreased ability to kill intracellular bacteria Listeria monocytogenes (50) and clearance of Pseudomonas aeruginosa is impaired in mice following chronic exposure to cigarette smoke (51). A decreased host defense potential of M1-deactivated AM resulting from smoking may, at least in part, explain why smokers who develop lung disease have an increased airway bacterial load and are frequently infected with respiratory viruses (52). A growing body of clinical evidence suggests that a variety of viruses such as rhinovirus, influenza virus, and respiratory syncytial virus are common causes of COPD exacerbations (52–54). It is, therefore, possible that in smokers with advanced COPD, persistent lung infection, which might develop due to deactivation of M1 polarization program in AM, initiates compensatory inflammatory responses, which are not directly induced by smoking. In support of this concept, cigarette smoking and viral components synergistically stimulate innate immune signaling in the mouse lung (55).
An important question that arises from these observations is how cigarette smoking might interfere with the M1 polarization program in AM? The direct immunosuppressive effect of various cigarette smoke constituents such as nicotine is well established (56). AM express the nicotinic acetylcholine receptor; when stimulated by nicotine, the AM support enhanced replication of intracellular bacteria and exhibit down-regulation of inflammatory cytokine production (57). Cigarette smoke extract inhibits the activation of nuclear factor kappa B (NF-kB), a master regulator of multiple inflammatory and immune processes in human macrophages (58). Acrolein, an aldehyde present in cigarette smoke, has been shown to decrease NF-kB activation in human AM (59). In contrast to the effects of chronic smoking on AM, acute exposure of macrophages to cigarette smoking extract, triggers the release of IL-8 and TNFα (60). The acute effect of cigarette smoking components on AM in vitro is likely very different from the effect of chronic smoking within the complex in vivo environment of human lung, involving multiple cell types. In this regard, the function of T cells, the major source of the M1-inducing cytokine IFNγ, is directly suppressed by cigarette smoke (56), and levels of IFNγ-producing T cells were found to be depressed in the lung of healthy smokers (61), consistent with decreased expression of IFNγ transcripts in cells recovered by lavage of smokers (44). Further, there are increased numbers of CD4+CD25+ regulatory T cells on the epithelial surface of healthy smokers (62,63). These regulatory T cells likely maintain an immunosuppressive microenvironment due to production of IL-10 and TGFβ (64), both inducing macrophage deactivation (3,21,22,65). In support of this scenario, the gene encoding integrin αE, important for induction of regulatory T cells in peripheral tissues (66), is up-regulated in AM of smokers (18). Another possibility is that chronic exposure to LPS, found in cigarette smoke at concentrations 120 times higher than levels found in smoke-free indoor air (67), induces inflammatory paralysis in AM, a phenomenon known as endotoxin tolerance (68). Consistent with this concept, stimulation of AM of smokers with LPS does not induce release of inflammatory cytokines comparable to that of AM of nonsmokers (69). Thus, a direct effect of smoke on AM as well as various changes in the cellular and cytokine microenvironment may account for cigarette smoking-induced deactivation of M1 polarization program in AM.
Induction of an Unusual M2 Polarization Program in AM by Cigarette Smoking
A broad deactivation of M1-polarization program in AM of healthy smokers was accompanied by induction of an unusual pattern of M2 polarization. This pattern was unique, in that the panel of up-regulated genes was not typical for any single M2 polarization program observed in vitro, but rather represented a mixture of different M2-related phenotypes. Smoking was associated with up-regulation of genes encoding scavenger receptors CD36, that can be induced in macrophages by IL-4 (29,70) or M-CSF (70), and MERTK, a receptor for apoptotic cells, that can be induced by glucocorticoids (31). AM of healthy smokers exhibited an increased expression of CCR5, a gene induced by IL-10 (71) and associated with the development of smoking-induced emphysema in mice (13,14). In addition, smoking increased expression of MMP2, a gene associated with M-CSF-induced M2-like polarization program (72) and recently linked to the pathogenesis of COPD (73). In agreement with deactivated M1 gene expression pattern, the expression of CD9, a gene negatively regulated by IFNγ (33), was significantly increased in the AM of healthy smokers. Interestingly, induction of CD36 and CCR5 have been noted in two previous human studies in which the AM transcriptomes of healthy nonsmokers and smokers were compared (17,18).
Thus, cigarette smoking induces in AM a unique kind of M2-like polarization characterized by deactivation of M1 polarization program and induction of a diverse set of M2 polarization patterns, in a manner similar to M2-inducing immunosuppressive factors such as IL-10, TGFβ, and glucocorticoids, that do not only down-regulate the expression of M1-related genes, but also induce a special patterns of non-classical activation (3). However, the polarization state of AM in smokers was unique in that a broad M1 deactivation was accompanied by simultaneous activation of genes typical for various kinds of M2 polarization. Such pattern has not been described yet for any of known inducers of M2 macrophage phenotype. In the context that the M2 gene list used in our study was based on in vitro studies in which single cell-types (macrophages or monocytes) have been exposed to a defined, usually single, stimulus, our results suggest that the complex in vivo microenvironment generated in the lung of smokers dictates a novel macrophage polarization phenotype, distinct from those described in vitro.
It is not certain, however, whether induction of a such unusual M2-like phenotype in AM is due to abundance of known activators of M2-polarization program in the alveolar microenvironment of smokers. Although increased expression of IL-4 has been detected in human emphysematous lungs (42) and overexpression of both IL-4 and IL-13 in mice result in emphysema (13,74), decreased levels of IL-13 have been found in lavage fluid recovered from healthy smokers (44). While there are higher percentage of T cells expressing IL-4 and IL-13 in COPD smokers, these changes have not been observed in healthy smokers (75). Whether smoking modulates the levels of immunoregulatory cytokines IL-10 and TGFβ in the lung is also not certain, but the observation of increased numbers of regulatory T cells in the BAL of smokers (62,63) suggests such a possibility.
Heterogeneity of AM Responses to Smoking
Personalized assessment of AM transcriptomes revealed a diversity of biological phenotypes based on the response of AM polarization-related genes to smoking. Healthy smokers appeared to be a heterogeneous group in terms of expression of polarization-related genes; while ~70% cluster separately from nonsmokers, others expressed a pattern similar to healthy nonsmokers. These observations are consistent with epidemiological studies demonstrating that only a subset of smokers develop COPD (76) and lead to a hypothesis for future studies that subgroups of healthy smokers with a higher risk for the development of smoking-induced lung disease might be identified at the biologic level prior to the development of lung disease. Furthermore, the observed heterogeneity of AM responses to smoking suggests that the early pathogenetic mechanisms mediated by AM during the development of lung disease may be different in various subgroups of smokers, possibly resulting in distinct clinical phenotypes of the disease (77,78). Additional studies are necessary to determine whether healthy smokers whose AM are M1-deactivated will progress into a hypoinflammatory clinical phenotype of COPD. Indeed, it is well documented that anti-inflammatory therapy with corticosteroids is not effective in modifying the natural course of disease in subsets of patients with COPD (8–10).
Progressive Alteration of AM Polarization During the Development of COPD
The results of population-based comparative analysis utilized in our study indicate that transcriptional changes induced in AM by smoking have a progressive character, since M1-related genes were further down-regulated and M2-related genes were further up-regulated in AM of COPD smokers when compared to smokers without disease. In AM of COPD smokers, there was advanced down-regulation of many host defense genes, including those encoding IFNγ-inducible chemokines CXCL9, CXCL10, CXCL11, and CCL5. The overall M1-related gene expression pattern in AM of COPD smokers was profoundly inhibited as compared to healthy nonsmokers and further down-regulated as compared to healthy smokers. The progressive suppression of the host defense and M1 polarization program was accompanied by progressive induction of several M2-related genes including MMP2, MMP7, and ADORA3. The tissue remodeling potential of MMPs (6,9,13) and MMP2, in particular, has already been implicated in the pathogenesis of COPD (73). Recent studies have shown that signaling through ADORA3, the glucocorticoid-inducible anti-inflammatory receptor, increases metalloprotease activity of macrophages (79) and activates TGF-β-dependent pro-fibrotic pathway in the lung (80).
Taken together, these results of the present study suggest that chronic cigarette smoking reprograms the steady-state AM polarization toward M1-deactivated, partially M2-activated macrophages with increased tissue remodeling potential but decreased expression of genes related to inflammation and immunity. This observation differs from those obtained in animal studies, in which induction of M2-like proteases in AM during the development of smoking-associated emphysema, a major component of COPD, has been found to occur in a concert with inflammatory response, as, for example, in transgenic mice overexpressing IFNγ (13,81). Paradoxically, chemokines CCL5, CXCL9, CXCL10, and CXCL11 shown to be progressively down-regulated at the transcriptional level in healthy smokers and COPD smokers in our study, have been recognized as key mediators of emphysema development in different mouse models (13–15).
There may be several explanations for these mice-human differences. First, although IFNγ does induce emphysema in the murine lung, it seems to be not necessary for emphysema development in mice, because emphysema can also develop in mice that lack functional T cells (82), a major source of IFNγ, and mice overexpressing IL-13, a cytokine inducing M2 macrophage polarization (3), also develop emphysema (74). Second, it is known that physiology of respiratory and immune systems in rodents have a number of significant differences from human and animal models may not always reflect the features of human disease (42). Consistent with this, there are substantial differences among the AM gene signatures of human smokers and two transgenic models of emphysema (17).
Finally, there are several other cell types than AM in the lung capable of producing inflammatory mediators (9,10,39). In this regard, several lines of evidence suggest that lung T cells activated during the development of smoking-induced lung disease may serve as source of IFNγ-inducible chemokines, such as CXCL9, CXCL10 and CXCL11(15,16), known to be associated with M1 macrophage polarization (4,29) and found to be suppressed in AM of healthy smokers and COPD smokers in our study. Elevated levels of CCL5, CXCL9, CXCL10, CXCL11 detected in the induced sputum of COPD smokers in one recent study correlated positively with neutrophil but negatively with macrophage numbers (83) and increased expression of CXCL10 was found in the bronchiolar epithelium and pulmonary arteries in smokers with COPD (84). Studies in transgenic mice and lung cells obtained from COPD patients have revealed that these chemokines are responsible for inducing protease activity in lung macrophages increasing thereby their tissue damaging potential (13,15,16). However, it remains unclear why such IFNγ-dependent mechanism does not induce M1 polarization program in AM. It is possible that inflammatory mechanisms unraveled in that studies are more characteristic for advanced stages of COPD pathogenesis. The results of the present study suggest that early mechanisms of smoking-induced lung disease in humans are likely highlighted by a complex suppression of various aspects of immune response in the lung, including deactivation of AM inflammatory and host defense function, and development of tissue remodeling processes.
Supplementary Material
01
Acknowledgments
We thank Tina Raman, Howard Lou, David Dang and Matthew Teater for excellent technical assistance; and N Mohamed for help in preparing this manuscript.
Footnotes
*
R01 HL074326, P50 HL084936 and UL1-RR024996. This publication was made possible by Grant Number U54RR024385 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and the NIH Roadmap for Medical research.
Publisher's Disclaimer: This is an author-produced version of a manuscript accepted for publication in The Journal of Immunology (The JI). The American Association of Immunologists, Inc. (AAI), publisher of The JI, holds the copyright to this manuscript. This version of the manuscript has not yet been copyedited or subjected to editorial proofreading by The JI; hence, it may differ from the final version published in The JI (online and in print). AAI (The JI) is not liable for errors or omissions in this author-produced version of the manuscript or in any version derived from it by the U.S. National Institutes of Health or any other third party. The final, citable version of record can be found at www.jimmunol.org.
References
- 1.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat. Rev. Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 2.Goerdt S, Orfanos CE. Other functions, other genes: alternative activation of antigen-presenting cells. Immunity. 1999;10:137–142. doi: 10.1016/s1074-7613(00)80014-x. [DOI] [PubMed] [Google Scholar]
- 3.Gordon S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 4.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 5.Bezdicek P, Crystal RG. Pulmonary macrophages. In: Crystal RG, West JB, Weibel ER, Barnes PJ, editors. The Lung: Scientific Foundations. Philadelphia: Lippincott-Raven Publishers; 1997. p. 859. [Google Scholar]
- 6.Shapiro SD. The macrophage in chronic obstructive pulmonary disease. Am. J Respir. Crit Care Med. 1999;160:S29–S32. doi: 10.1164/ajrccm.160.supplement_1.9. [DOI] [PubMed] [Google Scholar]
- 7.Rabe KF, Hurd S, Anzueto A, Barnes PJ, Buist SA, Calverley P, Fukuchi Y, Jenkins C, Rodriguez-Roisin R, van WC, Zielinski J. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit Care Med. 2007;176:532–555. doi: 10.1164/rccm.200703-456SO. [DOI] [PubMed] [Google Scholar]
- 8.Barnes PJ. Chronic obstructive pulmonary disease: a growing but neglected global epidemic. PLoS. Med. 2007;4:e112. doi: 10.1371/journal.pmed.0040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnes PJ, Shapiro SD, Pauwels RA. Chronic obstructive pulmonary disease: molecular and cellular mechanisms. Eur. Respir. J. 2003;22:672–688. doi: 10.1183/09031936.03.00040703. [DOI] [PubMed] [Google Scholar]
- 10.Barnes PJ. Immunology of asthma and chronic obstructive pulmonary disease. Nat. Rev. Immunol. 2008;8:183–192. doi: 10.1038/nri2254. [DOI] [PubMed] [Google Scholar]
- 11.Finkelstein R, Fraser RS, Ghezzo H, Cosio MG. Alveolar inflammation and its relation to emphysema in smokers. Am. J. Respir. Crit Care Med. 1995;152:1666–1672. doi: 10.1164/ajrccm.152.5.7582312. [DOI] [PubMed] [Google Scholar]
- 12.Hogg JC, Chu F, Utokaparch S, Woods R, Elliott WM, Buzatu L, Cherniack RM, Rogers RM, Sciurba FC, Coxson HO, Pare PD. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N. Engl. J Med. 2004;350:2645–2653. doi: 10.1056/NEJMoa032158. [DOI] [PubMed] [Google Scholar]
- 13.Elias JA, Kang MJ, Crothers K, Homer R, Lee CG. State of the art. Mechanistic heterogeneity in chronic obstructive pulmonary disease: insights from transgenic mice. Proc. Am. Thorac. Soc. 2006;3:494–498. doi: 10.1513/pats.200603-068MS. [DOI] [PubMed] [Google Scholar]
- 14.Ma B, Kang MJ, Lee CG, Chapoval S, Liu W, Chen Q, Coyle AJ, Lora JM, Picarella D, Homer RJ, Elias JA. Role of CCR5 in IFN-gamma-induced and cigarette smoke-induced emphysema. J. Clin. Invest. 2005;115:3460–3472. doi: 10.1172/JCI24858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maeno T, Houghton AM, Quintero PA, Grumelli S, Owen CA, Shapiro SD. CD8+ T Cells are required for inflammation and destruction in cigarette smoke-induced emphysema in mice. J. Immunol. 2007;178:8090–8096. doi: 10.4049/jimmunol.178.12.8090. [DOI] [PubMed] [Google Scholar]
- 16.Grumelli S, Corry DB, Song LZ, Song L, Green L, Huh J, Hacken J, Espada R, Bag R, Lewis DE, Kheradmand F. An immune basis for lung parenchymal destruction in chronic obstructive pulmonary disease and emphysema. PLoS. Med. 2004;1:e8. doi: 10.1371/journal.pmed.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woodruff PG, Koth LL, Yang YH, Rodriguez MW, Favoreto S, Dolganov GM, Paquet AC, Erle DJ. A distinctive alveolar macrophage activation state induced by cigarette smoking. Am. J. Respir. Crit Care Med. 2005;172:1383–1392. doi: 10.1164/rccm.200505-686OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heguy A, O'Connor TP, Luettich K, Worgall S, Cieciuch A, Harvey BG, Hackett NR, Crystal RG. Gene expression profiling of human alveolar macrophages of phenotypically normal smokers and nonsmokers reveals a previously unrecognized subset of genes modulated by cigarette smoking. J. Mol. Med. 2006;84:318–328. doi: 10.1007/s00109-005-0008-2. [DOI] [PubMed] [Google Scholar]
- 19.Russi TJ, Crystal RG. Use of bronchoalveolar lavage and airway brushing to investigate the human lung. In: Crystal RG, West JB, Weibel ER, Barnes PJ, editors. The Lung: Scientific Foundations. Philadelphia: Lippincott-Raven Publishers; 1997. p. 371. [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;B57:289–300. [Google Scholar]
- 21.Tsunawaki S, Sporn M, Ding A, Nathan C. Deactivation of macrophages by transforming growth factor-beta. Nature. 1988;334:260–262. doi: 10.1038/334260a0. [DOI] [PubMed] [Google Scholar]
- 22.Bogdan C, Nathan C. Modulation of macrophage function by transforming growth factor beta, interleukin-4, and interleukin-10. Ann. N. Y. Acad. Sci. 1993;685:713–739. doi: 10.1111/j.1749-6632.1993.tb35934.x. [DOI] [PubMed] [Google Scholar]
- 23.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 24.Kopydlowski KM, Salkowski CA, Cody MJ, van RN, Major J, Hamilton TA, Vogel SN. Regulation of macrophage chemokine expression by lipopolysaccharide in vitro and in vivo. J. Immunol. 1999;163:1537–1544. [PubMed] [Google Scholar]
- 25.Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- 26.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 27.Nagorsen D, Deola S, Smith K, Wang E, Monsurro V, Zanovello P, Marincola FM, Panelli MC. Polarized monocyte response to cytokine stimulation. Genome Biol. 2005;6:R15. doi: 10.1186/gb-2005-6-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Park-Min KH, Antoniv TT, Ivashkiv LB. Regulation of macrophage phenotype by long-term exposure to IL-10. Immunobiology. 2005;210:77–86. doi: 10.1016/j.imbio.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J. Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 30.Sharif O, Bolshakov VN, Raines S, Newham P, Perkins ND. Transcriptional profiling of the LPS induced NF-kappaB response in macrophages. BMC. Immunol. 2007;8:1. doi: 10.1186/1471-2172-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrchen J, Steinmuller L, Barczyk K, Tenbrock K, Nacken W, Eisenacher M, Nordhues U, Sorg C, Sunderkotter C, Roth J. Glucocorticoids induce differentiation of a specifically activated, anti-inflammatory subtype of human monocytes. Blood. 2007;109:1265–1274. doi: 10.1182/blood-2006-02-001115. [DOI] [PubMed] [Google Scholar]
- 32.Gratchev A, Kzhyshkowska J, Kannookadan S, Ochsenreiter M, Popova A, Yu X, Mamidi S, Stonehouse-Usselmann E, Muller-Molinet I, Gooi L, Goerdt S. Activation of a TGF-beta-specific multistep gene expression program in mature macrophages requires glucocorticoid-mediated surface expression of TGF-beta receptor II. J. Immunol. 2008;180:6553–6565. doi: 10.4049/jimmunol.180.10.6553. [DOI] [PubMed] [Google Scholar]
- 33.Wang XQ, Evans GF, Alfaro ML, Zuckerman SH. Down-regulation of macrophage CD9 expression by interferon-gamma. Biochem. Biophys. Res. Commun. 2002;290:891–897. doi: 10.1006/bbrc.2001.6293. [DOI] [PubMed] [Google Scholar]
- 34.Jolliffe IT. Principal component analysis. New York: Springer; 2002. [Google Scholar]
- 35.Hodge SJ, Hodge GL, Holmes M, Reynolds PN. Flow cytometric characterization of cell populations in bronchoalveolar lavage and bronchial brushings from patients with chronic obstructive pulmonary disease. Cytometry B Clin. Cytom. 2004;61:27–34. doi: 10.1002/cyto.b.20020. [DOI] [PubMed] [Google Scholar]
- 36.Chiang CS, Chen FH, Hong JH, Jiang PS, Huang HL, Wang CC, McBride WH. Functional phenotype of macrophages depends on assay procedures. Int. Immunol. 2008;20:215–222. doi: 10.1093/intimm/dxm137. [DOI] [PubMed] [Google Scholar]
- 37.Schneemann M, Schoedon G, Hofer S, Blau N, Guerrero L, Schaffner A. Nitric oxide synthase is not a constituent of the antimicrobial armature of human mononuclear phagocytes. J. Infect. Dis. 1993;167:1358–1363. doi: 10.1093/infdis/167.6.1358. [DOI] [PubMed] [Google Scholar]
- 38.Raes G, Van den Bergh R, De BP, Ghassabeh GH, Scotton C, Locati M, Mantovani A, Sozzani S. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J. Immunol. 2005;174:6561–6562. doi: 10.4049/jimmunol.174.11.6561. [DOI] [PubMed] [Google Scholar]
- 39.Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin. Invest. 2008;118:3546–3556. doi: 10.1172/JCI36130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Novak H, Muller A, Harrer N, Gunther C, Carballido JM, Woisetschlager M. CCL23 expression is induced by IL-4 in a STAT6-dependent fashion. J. Immunol. 2007;178:4335–4341. doi: 10.4049/jimmunol.178.7.4335. [DOI] [PubMed] [Google Scholar]
- 41.Cosio MG, Majo J, Cosio MG. Inflammation of the airways and lung parenchyma in COPD: role of T cells. Chest. 2002;121:160S–165S. doi: 10.1378/chest.121.5_suppl.160s. [DOI] [PubMed] [Google Scholar]
- 42.Taraseviciene-Stewart L, Voelkel NF. Molecular pathogenesis of emphysema. J. Clin. Invest. 2008;118:394–402. doi: 10.1172/JCI31811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cosio MG, Guerassimov A. Chronic obstructive pulmonary disease. Inflammation of small airways and lung parenchyma. Am. J. Respir. Crit Care Med. 1999;160:S21–S25. doi: 10.1164/ajrccm.160.supplement_1.7. [DOI] [PubMed] [Google Scholar]
- 44.Meuronen A, Majuri ML, Alenius H, Mantyla T, Wolff H, Piirila P, Laitinen A. Decreased cytokine and chemokine mRNA expression in bronchoalveolar lavage in asymptomatic smoking subjects. Respiration. 2008;75:450–458. doi: 10.1159/000114855. [DOI] [PubMed] [Google Scholar]
- 45.Soliman DM, Twigg HL., III Cigarette smoking decreases bioactive interleukin-6 secretion by alveolar macrophages. Am. J. Physiol. 1992;263:L471–L478. doi: 10.1152/ajplung.1992.263.4.L471. [DOI] [PubMed] [Google Scholar]
- 46.McCrea KA, Ensor JE, Nall K, Bleecker ER, Hasday JD. Altered cytokine regulation in the lungs of cigarette smokers. Am. J. Respir. Crit Care Med. 1994;150:696–703. doi: 10.1164/ajrccm.150.3.8087340. [DOI] [PubMed] [Google Scholar]
- 47.Lofdahl JM, Wahlstrom J, Skold CM. Different inflammatory cell pattern and macrophage phenotype in chronic obstructive pulmonary disease patients, smokers and non-smokers. Clin. Exp. Immunol. 2006;145:428–437. doi: 10.1111/j.1365-2249.2006.03154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hodge S, Hodge G, Ahern J, Jersmann H, Holmes M, Reynolds PN. Smoking alters alveolar macrophage recognition and phagocytic ability: implications in chronic obstructive pulmonary disease. Am J Respir. Cell Mol. Biol. 2007;37:748–755. doi: 10.1165/rcmb.2007-0025OC. [DOI] [PubMed] [Google Scholar]
- 49.Murin S, Bilello KS. Respiratory tract infections: another reason not to smoke. Cleve. Clin. J. Med. 2005;72:916–920. doi: 10.3949/ccjm.72.10.916. [DOI] [PubMed] [Google Scholar]
- 50.King TE, Jr, Savici D, Campbell PA. Phagocytosis and killing of Listeria monocytogenes by alveolar macrophages: smokers versus nonsmokers. J. Infect. Dis. 1988;158:1309–1316. doi: 10.1093/infdis/158.6.1309. [DOI] [PubMed] [Google Scholar]
- 51.Drannik AG, Pouladi MA, Robbins CS, Goncharova SI, Kianpour S, Stampfli MR. Impact of cigarette smoke on clearance and inflammation after Pseudomonas aeruginosa infection. Am. J. Respir. Crit Care Med. 2004;170:1164–1171. doi: 10.1164/rccm.200311-1521OC. [DOI] [PubMed] [Google Scholar]
- 52.Sethi S, Murphy TF. Infection in the Pathogenesis and Course of Chronic Obstructive Pulmonary Disease. N. Engl. J Med. 2008;359:2355–2365. doi: 10.1056/NEJMra0800353. [DOI] [PubMed] [Google Scholar]
- 53.Seemungal T, Harper-Owen R, Bhowmik A, Moric I, Sanderson G, Message S, Maccallum P, Meade TW, Jeffries DJ, Johnston SL, Wedzicha JA. Respiratory viruses, symptoms, and inflammatory markers in acute exacerbations and stable chronic obstructive pulmonary disease. Am. J. Respir. Crit Care Med. 2001;164:1618–1623. doi: 10.1164/ajrccm.164.9.2105011. [DOI] [PubMed] [Google Scholar]
- 54.McManus TE, Marley AM, Baxter N, Christie SN, O’Neill HJ, Elborn JS, Coyle PV, Kidney JC. Respiratory viral infection in exacerbations of COPD. Respir. Med. 2008;102:1575–1580. doi: 10.1016/j.rmed.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang MJ, Lee CG, Lee JY, Dela Cruz CS, Chen ZJ, Enelow R, Elias JA. Cigarette smoke selectively enhances viral PAMP- and virus-induced innate immune and remodeling responses in mice. J. Clin. Invest. 2008;118:2771–2784. doi: 10.1172/JCI32709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sopori M. Effects of cigarette smoke on the immune system. Nat. Rev. Immunol. 2002;2:372–377. doi: 10.1038/nri803. [DOI] [PubMed] [Google Scholar]
- 57.Matsunaga K, Klein TW, Friedman H, Yamamoto Y. Involvement of nicotinic acetylcholine receptors in suppression of antimicrobial activity and cytokine responses of alveolar macrophages to Legionella pneumophila infection by nicotine. J. Immunol. 2001;167:6518–6524. doi: 10.4049/jimmunol.167.11.6518. [DOI] [PubMed] [Google Scholar]
- 58.Birrell MA, Wong S, Catley MC, Belvisi MG. Impact of tobacco-smoke on key signaling pathways in the innate immune response in lung macrophages. J. Cell Physiol. 2008;214:27–37. doi: 10.1002/jcp.21158. [DOI] [PubMed] [Google Scholar]
- 59.Li L, Hamilton RF, Jr, Holian A. Effect of acrolein on human alveolar macrophage NF-kappaB activity. Am. J. Physiol. 1999;277:L550–L557. doi: 10.1152/ajplung.1999.277.3.L550. [DOI] [PubMed] [Google Scholar]
- 60.Yang SR, Chida AS, Bauter MR, Shafiq N, Seweryniak K, Maggirwar SB, Kilty I, Rahman I. Cigarette smoke induces proinflammatory cytokine release by activation of NF-kappaB and posttranslational modifications of histone deacetylase in macrophages. Am. J. Physiol Lung Cell Mol. Physiol. 2006;291:L46–L57. doi: 10.1152/ajplung.00241.2005. [DOI] [PubMed] [Google Scholar]
- 61.Hagiwara E, Takahashi KI, Okubo T, Ohno S, Ueda A, Aoki A, Odagiri S, Ishigatsubo Y. Cigarette smoking depletes cells spontaneously secreting Th(1) cytokines in the human airway. Cytokine. 2001;14:121–126. doi: 10.1006/cyto.2001.0860. [DOI] [PubMed] [Google Scholar]
- 62.Smyth LJ, Starkey C, Vestbo J, Singh D. CD4-regulatory cells in COPD patients. Chest. 2007;132:156–163. doi: 10.1378/chest.07-0083. [DOI] [PubMed] [Google Scholar]
- 63.Barcelo B, Pons J, Ferrer JM, Sauleda J, Fuster A, Agusti AG. Phenotypic characterisation of T-lymphocytes in COPD: abnormal CD4+CD25+ regulatory T-lymphocyte response to tobacco smoking. Eur. Respir. J. 2008;31:555–562. doi: 10.1183/09031936.00010407. [DOI] [PubMed] [Google Scholar]
- 64.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 65.Bogdan C, Vodovotz Y, Nathan C. Macrophage deactivation by interleukin 10. J. Exp. Med. 1991;174:1549–1555. doi: 10.1084/jem.174.6.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larsson L, Szponar B, Pehrson C. Tobacco smoking increases dramatically air concentrations of endotoxin. Indoor. Air. 2004;14:421–424. doi: 10.1111/j.1600-0668.2004.00290.x. [DOI] [PubMed] [Google Scholar]
- 68.Cavaillon JM, Adib-Conquy M. Bench-to-bedside review: endotoxin tolerance as a model of leukocyte reprogramming in sepsis. Crit Care. 2006;10:233. doi: 10.1186/cc5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen H, Cowan MJ, Hasday JD, Vogel SN, Medvedev AE. Tobacco smoking inhibits expression of proinflammatory cytokines and activation of IL-1R–associated kinase, p38, and NF-kappaB in alveolar macrophages stimulated with TLR2 and TLR4 agonists. J. Immunol. 2007;179:6097–6106. doi: 10.4049/jimmunol.179.9.6097. [DOI] [PubMed] [Google Scholar]
- 70.Yesner LM, Huh HY, Pearce SF, Silverstein RL. Regulation of monocyte CD36 and thrombospondin-1 expression by soluble mediators. Arterioscler. Thromb. Vasc. Biol. 1996;16:1019–1025. doi: 10.1161/01.atv.16.8.1019. [DOI] [PubMed] [Google Scholar]
- 71.Houle M, Thivierge M, Le GC, Stankova J, Rola-Pleszczynski M. IL-10 up-regulates CCR5 gene expression in human monocytes. Inflammation. 1999;23:241–251. doi: 10.1023/a:1020273903224. [DOI] [PubMed] [Google Scholar]
- 72.Bonfield TL, Swaisgood CM, Barna BP, Farver CF, Kavuru MS, Thomassen MJ. Elevated gelatinase activity in pulmonary alveolar proteinosis: role of macrophage-colony stimulating factor. J. Leukoc. Biol. 2006;79:133–139. doi: 10.1189/jlb.0805447. [DOI] [PubMed] [Google Scholar]
- 73.Baraldo S, Bazzan E, Zanin ME, Turato G, Garbisa S, Maestrelli P, Papi A, Miniati M, Fabbri LM, Zuin R, Saetta M. Matrix metalloproteinase-2 protein in lung periphery is related to COPD progression. Chest. 2007;132:1733–1740. doi: 10.1378/chest.06-2819. [DOI] [PubMed] [Google Scholar]
- 74.Zheng T, Zhu Z, Wang Z, Homer RJ, Ma B, Riese RJ, Jr, Chapman HA, Jr, Shapiro SD, Elias JA. Inducible targeting of IL-13 to the adult lung causes matrix metalloproteinase- and cathepsin-dependent emphysema. J. Clin. Invest. 2000;106:1081–1093. doi: 10.1172/JCI10458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Barcelo B, Pons J, Fuster A, Sauleda J, Noguera A, Ferrer JM, Agusti AG. Intracellular cytokine profile of T lymphocytes in patients with chronic obstructive pulmonary disease. Clin. Exp. Immunol. 2006;145:474–479. doi: 10.1111/j.1365-2249.2006.03167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lokke A, Lange P, Scharling H, Fabricius P, Vestbo J. Developing COPD: a 25 year follow up study of the general population. Thorax. 2006;61:935–939. doi: 10.1136/thx.2006.062802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Celli BR. Roger s. Mitchell lecture. Chronic obstructive pulmonary disease phenotypes and their clinical relevance. Proc. Am. Thorac. Soc. 2006;3:461–465. doi: 10.1513/pats.200603-029MS. [DOI] [PubMed] [Google Scholar]
- 78.Rennard SI, Vestbo J. The many "small COPDs": COPD should be an orphan disease. Chest. 2008;134:623–627. doi: 10.1378/chest.07-3059. [DOI] [PubMed] [Google Scholar]
- 79.Velot E, Haas B, Leonard F, Ernens I, Rolland-Turner M, Schwartz C, Longrois D, Devaux Y, Wagner DR. Activation of the adenosine-A3 receptor stimulates matrix metalloproteinase-9 secretion by macrophages. Cardiovasc. Res. 2008;80:246–254. doi: 10.1093/cvr/cvn201. [DOI] [PubMed] [Google Scholar]
- 80.Morschl E, Molina JG, Volmer JB, Mohsenin A, Pero RS, Hong JS, Kheradmand F, Lee JJ, Blackburn MR. A3 adenosine receptor signaling influences pulmonary inflammation and fibrosis. Am. J. Respir. Cell Mol. Biol. 2008;39:697–705. doi: 10.1165/rcmb.2007-0419OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang Z, Zheng T, Zhu Z, Homer RJ, Riese RJ, Chapman HA, Jr, Shapiro SD, Elias JA. Interferon gamma induction of pulmonary emphysema in the adult murine lung. J. Exp. Med. 2000;192:1587–1600. doi: 10.1084/jem.192.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.D'hulst AI, Maes T, Bracke KR, Demedts IK, Tournoy KG, Joos GF, Brusselle GG. Cigarette smoke-induced pulmonary emphysema in scid-mice. Is the acquired immune system required? Respir. Res. 2005;6:147. doi: 10.1186/1465-9921-6-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Costa C, Rufino R, Traves SL, Lapa E Silva, JR, Barnes PJ, Donnelly LE. CXCR3 and CCR5 chemokines in induced sputum from patients with COPD. Chest. 2008;133:26–33. doi: 10.1378/chest.07-0393. [DOI] [PubMed] [Google Scholar]
- 84.Saetta M, Mariani M, Panina-Bordignon P, Turato G, Buonsanti C, Baraldo S, Bellettato CM, Papi A, Corbetta L, Zuin R, Sinigaglia F, Fabbri LM. Increased expression of the chemokine receptor CXCR3 and its ligand CXCL10 in peripheral airways of smokers with chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2002;165:1404–1409. doi: 10.1164/rccm.2107139. [DOI] [PubMed] [Google Scholar]
- 85.Stienstra R, Duval C, Keshtkar S, van der Laak J, Kersten S, Muller M. Peroxisome proliferator-activated receptor gamma activation promotes infiltration of alternatively activated macrophages into adipose tissue. J. Biol. Chem. 2008;283:22620–22627. doi: 10.1074/jbc.M710314200. [DOI] [PubMed] [Google Scholar]
- 86.Netea MG, Azam T, Lewis EC, Joosten LA, Wang M, Langenberg D, Meng X, Chan ED, Yoon DY, Ottenhoff T, Kim SH, Dinarello CA. Mycobacterium tuberculosis induces interleukin-32 production through a caspase- 1/IL-18/interferon-gamma-dependent mechanism. PLoS. Med. 2006;3:e277. doi: 10.1371/journal.pmed.0030277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, Stewart KN, Bishop E, Marek CJ, Kluth DC, Rees AJ, Wilson HM. Unique expression of suppressor of cytokine signaling 3 is essential for classical macrophage activation in rodents in vitro and in vivo. J. Immunol. 2008;180:6270–6278. doi: 10.4049/jimmunol.180.9.6270. [DOI] [PubMed] [Google Scholar]
- 88.Willment JA, Lin HH, Reid DM, Taylor PR, Williams DL, Wong SY, Gordon S, Brown GD. Dectin-1 expression and function are enhanced on alternatively activated and GM-CSF-treated macrophages and are negatively regulated by IL-10, dexamethasone, and lipopolysaccharide. J Immunol. 2003;171:4569–4573. doi: 10.4049/jimmunol.171.9.4569. [DOI] [PubMed] [Google Scholar]
- 89.Kzhyshkowska J, Workman G, Cardo-Vila M, Arap W, Pasqualini R, Gratchev A, Krusell L, Goerdt S, Sage EH. Novel function of alternatively activated macrophages: stabilin-1-mediated clearance of SPARC. J. Immunol. 2006;176:5825–5832. doi: 10.4049/jimmunol.176.10.5825. [DOI] [PubMed] [Google Scholar]
- 90.Schnoor M, Cullen P, Lorkowski J, Stolle K, Robenek H, Troyer D, Rauterberg J, Lorkowski S. Production of type VI collagen by human macrophages: a new dimension in macrophage functional heterogeneity. J. Immunol. 2008;180:5707–5719. doi: 10.4049/jimmunol.180.8.5707. [DOI] [PubMed] [Google Scholar]
- 91.Gratchev A, Guillot P, Hakiy N, Politz O, Orfanos CE, Schledzewski K, Goerdt S. Alternatively activated macrophages differentially express fibronectin and its splice variants and the extracellular matrix protein betaIG-H3. Scand. J Immunol. 2001;53:386–392. doi: 10.1046/j.1365-3083.2001.00885.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01