Genome-wide association studies identify novel loci associated with age at menarche and age at natural menopause (original) (raw)
. Author manuscript; available in PMC: 2010 Sep 20.
Published in final edited form as: Nat Genet. 2009 May 17;41(6):724–728. doi: 10.1038/ng.385
Abstract
Age at menarche and age at natural menopause are associated with causes of substantial morbidity and mortality such as breast cancer and cardiovascular disease. Studies have suggested both traits might be partially under genetic control. We performed a joint analysis of two genome-wide association studies of these two traits in a total of 17,438 women from the Nurses’ Health Study (NHS, N=2,287) and the Women’s Genome Health Study (WGHS, N=15,151). For age at menarche, we identified 10 associated SNPs (_P_=1×10-7-3×10-13) clustered at 6q21 (in or near the gene LIN28B) and 9q31.2 (in an intergenic region). For age at natural menopause, we identified 13 associated SNPs (_P_=1×10-7-1×10-21) clustered at 20p12.3 (in the gene MCM8), 19q13.42 (in or near the gene BRSK1), 5q35.2 (in or near genes RAP80 and HK3), and 6p24.2 (in the gene SYCP2L). These newly identified loci might expand understanding of the biological pathways regulating these two traits.
Age at menarche and age at natural menopause have important implications for the health of pre- and postmenopausal women. They mark the beginning and the end of the normal reproductive life. Early menarche and later menopause are well-established risk factors for breast cancer1 and endometrial cancer2. On the other hand, late menarche and early menopause increase risk of osteoporosis3 and cardiovascular disease4. Early natural menopause also implies a reduced span of fertility5. Twin and family studies have suggested high heritability for age at menarche (53-74%)6,7 and age at natural menopause (49-87%)8-10. However, these estimates need to be viewed with caution as some studies are small, and distinguishing between shared environment and true heritability is difficult for such environmentally influenced phenotypes. No correlation has been observed between the two traits, suggesting that they might be regulated differently8. Candidate gene association studies, focusing on the genes involved in steroid-hormone biosynthesis and metabolism pathways, have not yielded findings that have been consistently validated11. Recently, genome-wide linkage scans using microsatellite markers have identified chromosomal regions that might harbor genes for menarche12-15 and natural menopause16,17, but no genes have been identified.
To identify common genetic variants associated with normal variation in age at menarche and age at natural menopause, we performed a joint analysis of two genome-wide association (GWA) studies of these two traits in a total sample of 17,438 women from two prospective cohort studies, the Nurses’ Health Study (NHS, N=2,287) and the Women’s Genome Health Study (WGHS, N=15,151). The NHS sample consists of 1,145 women with postmenopausal invasive breast cancer and 1,142 matched controls. We evaluated 317,759 common SNPs (minor allele frequency [MAF] > 1%) across the two studies. The means and standard deviations (SDs) of these two traits are similar between the NHS and the WGHS (Supplementary Table 1 online). The mean age at menarche for the combined 17,406 women was 12.4 years (SD=1.43 years); while the mean age at natural menopause for the combined 9,112 women was 50.6 years (SD=3.58 years).
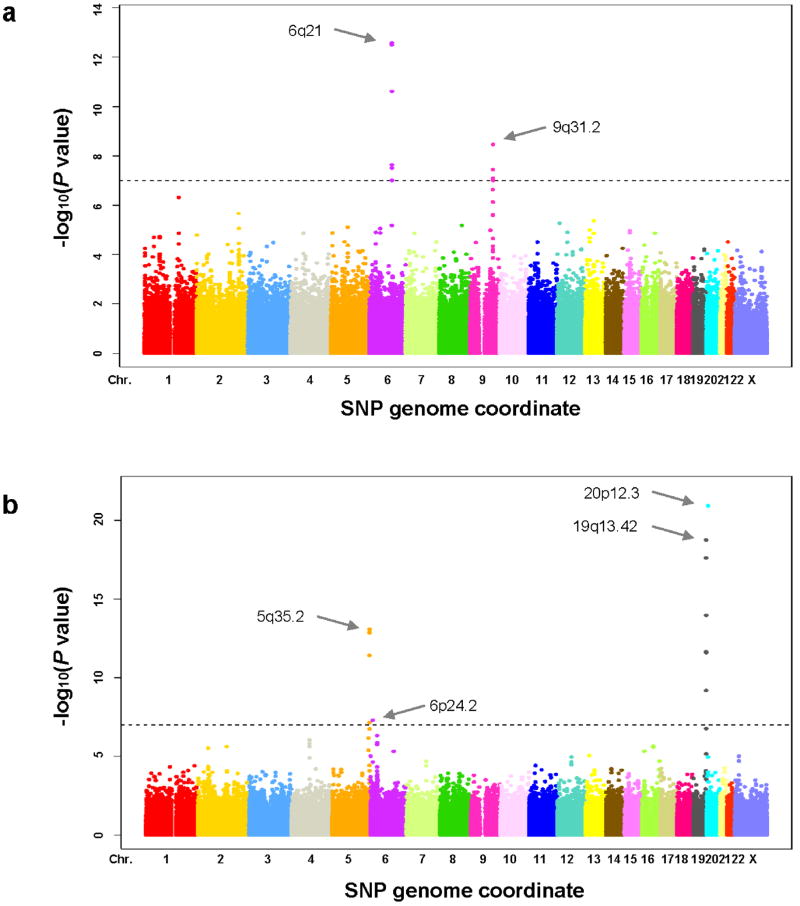
The distributions of the P values for the association tests across the SNPs tested showed little evidence of overall systematic bias or heterogeneity for age at menarche (genomic inflation factor for the combined analysis λ=1.074; Supplementary Figs. 1a and 2a online) or age at natural menopause (λ=1.026; Supplementary Figs. 1b and 2b online). The excess of low P values was consistent with the presence of true associations for each phenotype. In the joint analysis of the two GWA studies, ten and thirteen SNPs reached genome-wide significance (p< 1×10-7) for age at menarche (Fig. 1a) and age at natural menopause (Fig. 1b), respectively. Detailed results for these candidate SNPs are listed in Table 1. Although most of the SNPs associated with age at menarche showed no evidence of heterogeneity in effect estimates between the two studies, the effect estimates for two SNPs at 6q21 (rs314263 and rs369065) were significantly different (p for heterogeneity = 0.0037 and 0.0078, respectively). However, other genome-wide significant SNPs in this region (rs314277, rs314262, rs4946651 and rs314280) did not show evidence of statistical heterogeneity. Similarly, although one of the SNPs for age at natural menopause at 19q13.42 (rs1551562) showed some statistical evidence of heterogeneity (p for heterogeneity =0.04), five other genome-wide significant SNPs in this region (rs12611091, rs1172822, rs2384687, rs897798 and rs7246479) did not. We observed one SNP at 6q22.31 (rs12665088) that reached genome-wide significance for age at menarche in the NHS but not in the joint analysis (p=2.1×10-3), and one SNP at 16p13.13 (rs10852344) that reached genome-wide significance for age at natural menopause in the WGHS but not in the joint analysis (p=4.9×10-6). For each SNP that was genome-wide significant in the combined analysis, the differences in age at menarche across genotype are generally much smaller than that of age at natural menopause (Supplementary Table 2 online), consistent with the narrower standard deviation in age at menarche.
Figure 1.
Manhattan plots for 317,759 single-SNP tests of association in joint analysis with age at menarche (a) and age at natural menopause (b) by SNP position along chromosome. The dotted lines indicate the P value of 1×10-7 (genome wide significance level).
Table 1.
SNPs showing the strongest associations (combined P value < 1×10-7) with age at menarche or age at natural menopause in joint analysis
| SNP | CytologicalLocation | Position a | Nearby Genes b | Major,MinorAlleles | MAF c | NHS | WGHS | Joint analysis | Heterogeneity | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta d | S.E. | P value e | Beta d | S.E. | P value e | Beta d, f | S.E. | P value e, f | P value g | ||||||
| Age at Menarche | |||||||||||||||
| rs314277 | 6q21 | 105514355 | LIN28B | C, A | 0.14 | 0.087 | 0.059 | 0.14 | 0.17 | 0.024 | 3.4E-13 | 0.16 | 0.022 | 2.7E-13 | 0.18 |
| rs314263 | 6q21 | 105499438 | LIN28B | T, C | 0.31 | -0.0025 | 0.045 | 0.96 | 0.14 | 0.018 | 4.7E-15 | 0.12 | 0.017 | 3.2E-13 | 0.0037 |
| rs369065 | 6q21 | 105550751 | LIN28B | T, C | 0.33 | 0.00049 | 0.044 | 0.99 | 0.13 | 0.018 | 6.7E-13 | 0.11 | 0.016 | 2.4E-11 | 0.0078 |
| rs7861820 | 9q31.2 | 106016229 | T, C | 0.48 | -0.038 | 0.042 | 0.36 | -0.099 | 0.016 | 2.1E-09 | -0.090 | 0.015 | 3.4E-09 | 0.18 | |
| rs314280 | 6q21 | 105507530 | LIN28B | C, T | 0.45 | 0.044 | 0.041 | 0.28 | 0.093 | 0.017 | 2.3E-08 | 0.086 | 0.015 | 2.3E-08 | 0.27 |
| rs4946651 | 6q21 | 105476203 | LIN28B | G, A | 0.45 | 0.044 | 0.041 | 0.29 | 0.092 | 0.017 | 3.0E-08 | 0.085 | 0.015 | 3.1E-08 | 0.27 |
| rs12684013 | 9q31.2 | 106077669 | C, T | 0.27 | -0.086 | 0.046 | 0.063 | -0.096 | 0.019 | 2.1E-07 | -0.095 | 0.017 | 3.6E-08 | 0.84 | |
| rs4452860 | 9q31.2 | 106004944 | A, G | 0.28 | -0.082 | 0.045 | 0.072 | -0.093 | 0.018 | 4.2E-07 | -0.091 | 0.017 | 7.9E-08 | 0.83 | |
| rs7028916 | 9q31.2 | 106006623 | C, A | 0.28 | -0.077 | 0.045 | 0.089 | -0.093 | 0.018 | 4.1E-07 | -0.090 | 0.017 | 9.7E-08 | 0.75 | |
| rs314262 | 6q21 | 105501314 | LIN28B | T, C | 0.45 | 0.036 | 0.042 | 0.39 | 0.092 | 0.017 | 6.5E-08 | 0.084 | 0.016 | 9.7E-08 | 0.21 |
| Age at Natural Menopause | |||||||||||||||
| rs16991615 | 20p12.3 | 5896227 | TRMT6, MCM8 | G, A | 0.058 | 1.07 | 0.26 | 4.1E-05 | 1.07 | 0.12 | 7.6E-18 | 1.07 | 0.11 | 1.2E-21 | 1.00 |
| rs1172822 | 19q13.42 | 60511657 | BRSK1 | C, T | 0.37 | -0.63 | 0.12 | 3.4E-07 | -0.45 | 0.060 | 5.0E-14 | -0.49 | 0.054 | 1.8E-19 | 0.19 |
| rs2384687 | 19q13.42 | 60523000 | BRSK1 | T, C | 0.39 | -0.61 | 0.12 | 5.1E-07 | -0.43 | 0.059 | 4.5E-13 | -0.47 | 0.053 | 2.4E-18 | 0.18 |
| rs897798 | 19q13.42 | 60525566 | BRSK1, SUV420H2 | A, G | 0.47 | -0.49 | 0.12 | 4.0E-05 | -0.38 | 0.058 | 4.8E-11 | -0.40 | 0.052 | 1.1E-14 | 0.42 |
| rs365132 | 5q35.2 | 176311180 | RAP80 | G, T | 0.49 | 0.43 | 0.12 | 0.00027 | 0.38 | 0.058 | 7.6E-11 | 0.39 | 0.052 | 8.4E-14 | 0.70 |
| rs7718874 | 5q35.2 | 176290671 | RAP80 | A, G | 0.49 | 0.43 | 0.12 | 0.00027 | 0.38 | 0.058 | 1.1E-10 | 0.39 | 0.052 | 1.3E-13 | 0.68 |
| rs402511 | 5q35.2 | 176367046 | RAP80, ZNF346 | C, T | 0.49 | 0.43 | 0.12 | 0.00028 | 0.38 | 0.058 | 1.2E-10 | 0.39 | 0.052 | 1.4E-13 | 0.69 |
| rs7246479 | 19q13.42 | 60516144 | BRSK1 | G, T | 0.48 | 0.45 | 0.12 | 0.00012 | 0.34 | 0.058 | 3.2E-09 | 0.36 | 0.052 | 2.3E-12 | 0.40 |
| rs1551562 | 19q13.42 | 60506693 | BRSK1 | A, G | 0.23 | -0.69 | 0.14 | 8.8E-07 | -0.37 | 0.069 | 8.4E-08 | -0.43 | 0.062 | 2.6E-12 | 0.040 |
| rs691141 | 5q35.2 | 176255904 | HK3, RAP80 | C, T | 0.45 | 0.34 | 0.12 | 0.0039 | 0.37 | 0.058 | 2.9E-10 | 0.36 | 0.052 | 3.9E-12 | 0.85 |
| rs12611091 | 19q13.42 | 60492141 | HSPBP1, BRSK1 | T, C | 0.48 | 0.34 | 0.12 | 0.0043 | 0.32 | 0.059 | 4.5E-08 | 0.33 | 0.053 | 6.6E-10 | 0.88 |
| rs2153157 | 6p24.2 | 11005474 | GCM2, SYCP2L | C, T | 0.49 | 0.22 | 0.12 | 0.064 | 0.30 | 0.058 | 2.5E-07 | 0.29 | 0.052 | 5.1E-08 | 0.54 |
| rs2278493 | 5q35.2 | 176247040 | UNC5A, HK3 | G, A | 0.33 | -0.27 | 0.12 | 0.031 | -0.31 | 0.062 | 7.9E-07 | -0.30 | 0.056 | 7.2E-08 | 0.77 |
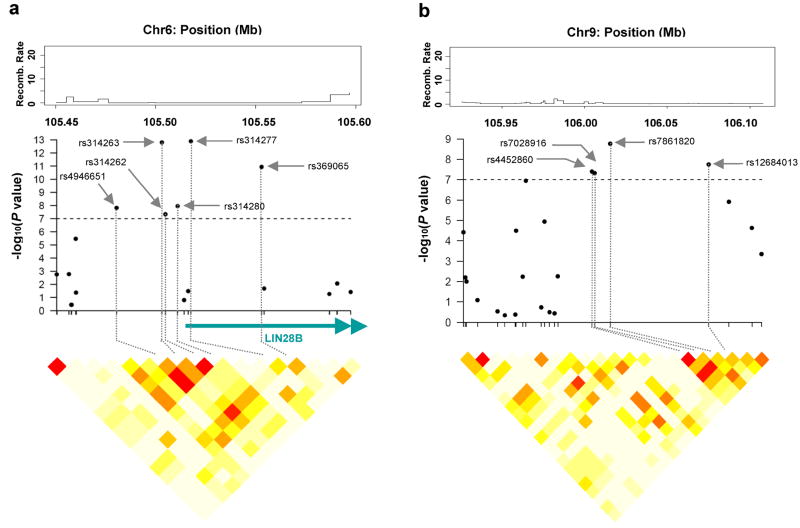
The ten genome-wide significant SNPs for age at menarche are clustered into two regions: six SNPs are at 6q21 (Fig. 2a); the other four are at 9q31.2 (Fig. 2b). In the 6q21 region, out of the four SNPs that are upstream of the LIN28B gene, three SNPs (rs4946651, rs314262 and rs314280) are in high linkage disequilibrium (LD) (pairwise r2>0.95; Supplementary Table 3 online); the other two SNPs (rs314277 and rs369065) are in intron 2 of the LIN28B gene. Three SNPs (rs7028916, rs4452860, rs12684013) in the 9q31.2 intergenic region are in high LD (pairwise r2>0.90; Supplementary Table 3 online). After adjusting for the most significant SNPs in each region (rs314263, rs314277, rs7861820), none of the remaining genome-wide significant SNPs was significantly associated with age at menarche (i.e. all had p>0.05). Together, these three most significant SNPs from the two independent regions explained 0.60% of variation in age at menarche in the combined sample.
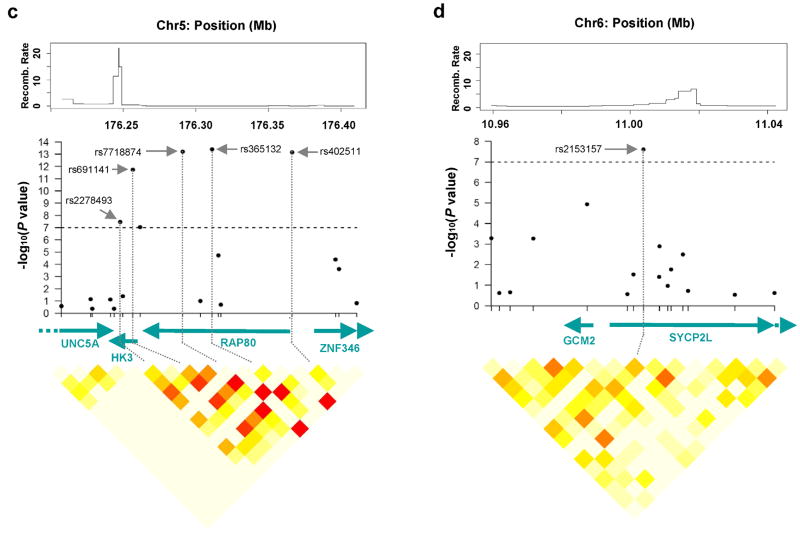
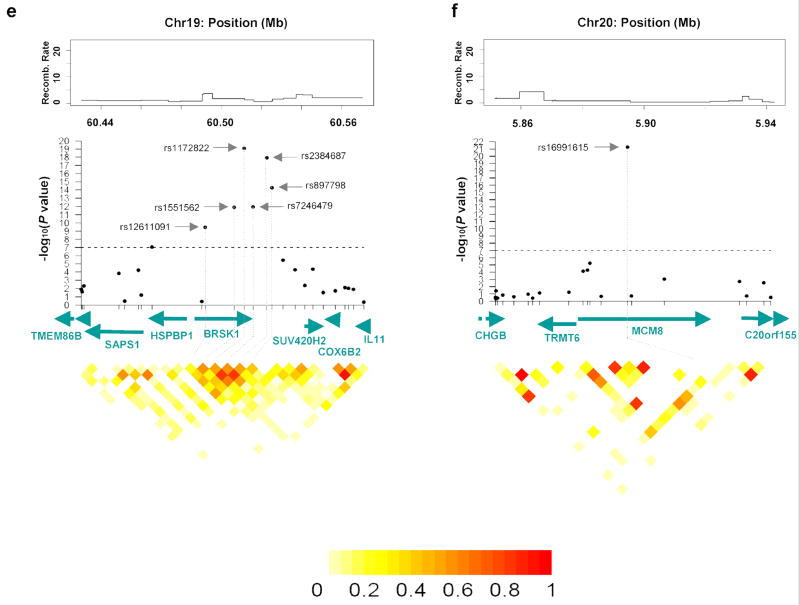
Figure 2.
Regional association analyses for age at menarche (a, b) and age at natural menopause (c, d, e, f) in joint analysis. The upper panels show the recombination rate for these regions estimated from HapMap data and P values for association testing from the joint analysis of two GWA studies in the NHS and the WHS, covering most strongly associated SNPs and approximately 50kb upstream and 50kb downstream. The lower panels show estimates of the square of the correlation coefficient (r2) calculated for each pairwise comparison of SNPs. The r2 values are color coded according to the scale at the bottom. The middle panels show gene name, their direction of transcription and corresponding genome position.
The thirteen genome-wide significant SNPs for age at natural menopause are located in four different genomic regions: 5q32.2, 6p24.2, 19q13.42 and 20p12.3. In the 5q32.2 region, the three most significant SNPs (rs7718874, rs365132 and rs402511) are in perfect LD (each pairwise r2=1) (Fig. 2c; Supplementary Table 3 online). Another SNP at 5q32.2, rs691141, is in relatively high LD with each of these SNPs (pairwise r2 =0.83). We note that rs365132 is a synonymous SNP in exon 9 of the RAP80 gene; the other genome-wide significant SNPs are distributed throughout the introns and flanking regions of RAP80 and HK3. In the 6p24.2 region, rs2153157 is in intron 4 of the SYCP2L gene (Fig. 2d). Six SNPs in the 19q13.42 region are either in or downstream of the BRSK1 gene (Fig. 2e). The two most significant SNPs (rs1172822, rs2384687) in this region are in high LD (r2=0.81; Supplementary Table 3 online) as are two other SNPs with genome-wide significance (rs897798 and rs7246479, r2=0.82). At 20p12.3, rs16991615 had the smallest P value (1.2×10-21) in the combined analysis for age at natural menopause (Fig. 2f) and it is a non-synonymous SNP in exon 9 of MCM8. After adjusting for one significant SNP in each region (rs7718874, rs2153157, rs1172822 and rs16991615), none of the remaining genome-wide significant SNPs was significantly associated with age at natural menopause (i.e. all had p>0.05). Together, these four significant SNPs from the four independent regions explained 2.69% of variation in age at natural menopause in the combined sample.
SNPs in the HapMap database with a pairwise correlation above 0.80 with the identified genome-wide significant SNPs in each region are listed in Supplemental Table 4 online.
In our results, the loci identified for age at menarche are different from those for age at natural menopause. Even at the nominal significance level (p<0.05), none of the genome-wide significant SNPs for age at menarche was associated with age at natural menopause and vice versa. This is consistent with previous studies8,18, suggesting that these traits might be regulated differently. We also observed fewer loci associated with age at menarche despite a larger sample size, which might be related to the fact that age at menarche has a more restricted age range (SD=1.43 years) compared to age at natural menopause (SD=3.58 years), and rounding reported age to the nearest year might increase the relative impact of measurement error and attenuate associations with age at menarche more than with age at natural menopause. Of note, none of the identified loci in this study contained previously reported candidate genes. We also did not observe any overlap between previously reported linkage peaks (LOD>1.5) and the loci we identified.
This study reports novel findings for women of European ancestry recruited using similar techniques in the NHS and the WGHS. We chose to combine evidence for association to optimize power and calculate a summary estimate across the two studies despite the differences in the sample sizes19 as we observed no evidence of systematic heterogeneity in genetic association with either trait. For age at natural menopause, three of the four chromosomal regions that were genome-wide significant (p<1.0×10-7) in the WGHS alone contained SNPs that had p<0.05 in the NHS. Direction and magnitude of the observed associations were also consistent between the two studies across all four regions. This result constitutes independent replication in the NHS of the findings in the WGHS for at least three of the four regions. For age at menarche, although none of the SNPs that reached genome-wide significance in the WGHS analysis was independently significant at the 0.05 level in the NHS, we note that in several instances the regression coefficients were very similar in magnitude and direction. There was no statistically significance evidence of heterogeneity for eight of the ten SNPs; for the other two SNPs, nearby SNPs in the region showed no significant heterogeneity. The reason we did not observe statistical significance in the NHS might be related to the modest absolute effects observed combined with the smaller sample size and the greater impact of measurement error on the analysis of age at menarche (see above). We cannot rule out the possibility that underlying differences in the two cohorts contribute to the observed discrepancy.
Our estimates of magnitude of association need to be confirmed in additional studies, as they might be overestimated due to the “Winner’s Curse”; on the other hand, it is probable the SNPs detected in this study are not the causal loci, but are in LD with the causal variants that might have stronger associations. Fine-mapping of the regions will be needed to pinpoint the variants that will be nominated for subsequent studies designed to understand the biological basis of the observed associations. The ages at menarche and ages at natural menopause for women in these studies are comparable to those of the US general population, suggesting that findings from this study might be generalizable to women of European ancestry in the US population.
For age at menarche, the most significant association is in a region that contains the LIN28B gene, which encodes a developmentally regulated RNA binging protein. This gene is highly expressed in most human hepatocellular carcinomas and embryonic stem cells. It functions as a negative regulator of microRNA (miRNA) by selectively blocking the processing of pri-let-7g miRNAs, and might play a critical role in stem cell fate determination and tumorigenesis20.
Several genes are present in the chromosomal regions identified for age at natural menopause. The MCM8 gene encodes a highly conserved mini-chromosome maintenance protein that is essential for genome replication21. The significant SNP (rs16991615) leads to an amino acid change from glutamic acid (Glu) to lysine (Lys) that might alter protein function. BRSK1 (also known as SAD1) is highly expressed in human brain at the presynaptic structure of neurons, and mutation or over-expression of the BRSK1 gene mislocates vesicles at the axonal terminals and thus affects vesicle transport and release at the axon terminals22,23. It is possible that BRSK1 gene might affect the secretion of gonadotropin-releasing hormone (GnRH) from the hypothalamus through this mechanism, and thus disturb the control of the hypothalamic-pituitary-ovary axis on menstrual cycles and influence onset of the natural menopause. The RAP80 (also known as UIMC1) gene is expressed in the ovary and the hypothalamus-pituitary-adrenal axis and can act as a transcriptional repressor. It has recently been identified to be in a complex with BRCA1, and is required for the localization of BRCA1 to DNA damage foci24. The HK3 gene is involved in carbohydrate metabolism and over-expressed in malignant follicular thyroid nodules25. Mouse SYCP2L is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. SYCP2L knockout mice exhibit sterility in male mice and subfertility with sharply reduced litter size in female mice26.
In summary, we identified two and four loci that have not been reported previously to be associated with age at menarche and age at natural menopause respectively. At these newly identified loci fine-mapping or sequencing might lead to identification of the causal variants, and thus expand our knowledge of the underlying physiology and biological regulation of these traits. Insights into the genetic factors influencing the timing of menarche and natural menopause might shed light on normal reproductive function and the prevention of the diseases associated with these two traits.
METHODS
Study populations
Detailed description of GWA study subjects in the NHS and the WGHS are available in the Supplementary Methods online. Briefly, the 2,287 NHS participants included in the present analysis were from the nested breast cancer case-control study in the NHS subcohort with blood sample collection between 1989-1990. The 15,151 women in the WGHS are participants in the ongoing Women’s Health Study (WHS) and provided a blood sample in 1993. The Institutional Review Board of Brigham and Women’s Hospital approved these studies.
Phenotype assessment
Age at menarche is defined as age at the first menstrual period (in years). This information was retrospectively ascertained by recall in the baseline questionnaires. In the NHS, the question was open-ended: “At what age did your menstrual periods begin?___ years of age”. In the WHS, the question asked, “At what age did your menstrual periods begin?” Response categories were: “9 or younger; 10; 11; 12; 13; 14; 15; 16; 17 or older”. We excluded women whose age at menarche was reported as 18 years or greater in the NHS from the analysis (N=2), as these women were more likely to have a pathological cause outside the spectrum of normal variation.
Age at natural menopause is defined as the age when menstrual periods ceased permanently and naturally (in years). Questions regarding menopause status were asked in baseline and subsequent questionnaires. The questions were: “Have your menstrual periods ceased permanently?” If yes, “At what age did your natural periods cease?” and “For what reason did your periods cease?” Response categories were: “Surgical; Radiation or Chemotherapy; Natural”. Age at natural menopause was assessed in the baseline questionnaire for postmenopausal women at baseline, and updated in subsequent questionnaires for premenopausal women at baseline. By the time we conducted this study, all women in the NHS and the WGHS had passed through menopause and age at menopause was recorded, either at baseline, or on subsequent questionnaires. We used this most updated information for age at natural menopause in the analysis. Women who had radiation/chemotherapy or surgically- induced menopause were excluded from the current study. We also excluded women who reported ages at menopause younger than 40 years or older than 60 years from the analysis as these extremes might reflect underlying pathologies.
Age at menarche and age at natural menopause are associated with breast cancer and other endpoints in both the NHS and the WHS with magnitudes and directions of association that are consistent with other published studies, attesting to the validity of the measurements. Further discussions about the validity of the measurement of age at menarche and age at natural menopause are provided in the Supplementary Methods online.
Genotyping
Genotyping in the NHS used the Illumina Infinium Sentrix HumanHap550 chip. Detailed methods related to the genotyping have been published previously27. Genotyping in the WGHS used either the Illumina HumanHap300 Duo-Plus chip or the combination of the HumanHap300 Duo and I-Select chips28. In the present analysis, the 317,759 SNPs shared between the NHS and the WGHS samples were almost entirely from the HumanHap300 panel.
Statistical analysis
In each of the NHS and the WGHS GWA studies separately, we performed linear regression to analyze the association between each of the SNPs (coded as counts of minor alleles) and age at menarche or age at natural menopause as continuous variables using PLINK software29. SNPs with low minor allele frequency (MAF) (<1%) in either the NHS or the WGHS samples were excluded from analysis. To control for potential confounding by population stratification, we adjusted for the top principal components of genetic variation chosen for each study after excluding any clearly non-Caucasian admixed individuals (see Supplementary Methods online). In the NHS analysis, controlling for breast cancer case-control status made no material difference to the GWAS results. To combine evidence for association with age at menarche or age at natural menopause and calculate a summary effect size across the NHS and the WGHS, we used fixed effect joint analysis; and heterogeneity in effect size across two studies was assessed using the Q- test30.
To estimate the proportion of variation in age at menarche and age at natural menopause that can be explained by the identified genome-wide significant SNPs, we pooled the original genotype and phenotype data from the NHS and the WGHS for the 23 genome-wide significant SNPs. After first adjusting the age at menarche or age at natural menopause for principal components of population substructure within each study separately, we performed a forward/backward stepwise selection of candidate SNPs with the BIC criteria in regression models that also included a study variable to select a minimal set of non-redundant SNPs for each phenotype. The proportion of variation explained (R2) in the combined phenotype was calculated by comparing the model containing the final selected SNPs and a study variable to the model containing a study variable alone.
Statistical power
Assuming a correlation (r2) between at least one tested SNP and any causal SNP is 0.80, our study has 80% power at a genome-wide significance level (P<1×10-7) to detect any causal SNP accounting for 0.27% of phenotypic variation in observed age at menarche, and 0.52% of phenotypic variation in observed age at natural menopause.
Supplementary Material
1
2
Acknowledgments
We thank J. Miletich and A. Parker as well as the technical staff at Amgen, Inc (Cambridge MA) for their collaboration and scientific support in performing the genotyping for the WGHS. The NHS GWAS was performed as part of the Cancer Genetic Markers of Susceptibility initiative of the NCI. We particularly acknowledge the contributions of R. Hoover, A. Hutchinson, K. Jacobs and G. Thomas. We thank Dr. J. Chen for discussion of gene functions. We thank H. Ranu and P. Soule for assistance. The WGHS is supported by HL 043851 and HL69757 from the National Heart Lung and Blood Institute and CA 047988 from the National Cancer Institute (Bethesda, MD), the Donald W Reynolds Foundation (Las Vegas, NV), the Fondation Leducq (Paris FR), with collaborative scientific support and funding for genotyping provided by Amgen, Inc. The NHS is supported by CA 40356 and U01-CA98233 from the National Cancer Institute. We acknowledge the study participants in the NHS and the WGHS for their contribution in making this study possible.
Footnotes
AUTHOR CONTRIBUTIONS C.H., C.C., P.K. and D.I.C. performed the primary GWAS analyses in each study. C.H., C.C., and P.K. performed the joint analysis and contributed to the graphics supporting the Figures. C.H., D.J.H. and D.I.C. wrote the manuscript with inputs from the other authors, especially S.J.C., P.K. and P.M.R. P.K., S.E.H. and D.J.H are investigators of the NHS. J.E.B., G.P., P.M.R and D.I.C. are investigators of the WGHS. All authors read and approved the final manuscript.
References
- 1.Kvale G. Reproductive factors in breast cancer epidemiology. Acta Oncol. 1992;31:187–94. doi: 10.3109/02841869209088901. [DOI] [PubMed] [Google Scholar]
- 2.Kaaks R, Lukanova A, Kurzer MS. Obesity, endogenous hormones, and endometrial cancer risk: a synthetic review. Cancer Epidemiol Biomarkers Prev. 2002;11:1531–43. [PubMed] [Google Scholar]
- 3.Paganini-Hill A, et al. Menstrual and reproductive factors and fracture risk: the Leisure World Cohort Study. J Womens Health (Larchmt) 2005;14:808–19. doi: 10.1089/jwh.2005.14.808. [DOI] [PubMed] [Google Scholar]
- 4.Cui R, et al. Relationships of age at menarche and menopause, and reproductive year with mortality from cardiovascular disease in Japanese postmenopausal women: the JACC study. J Epidemiol. 2006;16:177–84. doi: 10.2188/jea.16.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.te Velde ER, Pearson PL. The variability of female reproductive ageing. Hum Reprod Update. 2002;8:141–54. doi: 10.1093/humupd/8.2.141. [DOI] [PubMed] [Google Scholar]
- 6.Meyer JM, Eaves LJ, Heath AC, Martin NG. Estimating genetic influences on the age-at-menarche: a survival analysis approach. Am J Med Genet. 1991;39:148–54. doi: 10.1002/ajmg.1320390207. [DOI] [PubMed] [Google Scholar]
- 7.Sharma K. Genetic basis of human female pelvic morphology: a twin study. Am J Phys Anthropol. 2002;117:327–33. doi: 10.1002/ajpa.10055. [DOI] [PubMed] [Google Scholar]
- 8.Snieder H, MacGregor AJ, Spector TD. Genes control the cessation of a woman’s reproductive life: a twin study of hysterectomy and age at menopause. J Clin Endocrinol Metab. 1998;83:1875–80. doi: 10.1210/jcem.83.6.4890. [DOI] [PubMed] [Google Scholar]
- 9.de Bruin JP, et al. The role of genetic factors in age at natural menopause. Hum Reprod. 2001;16:2014–8. doi: 10.1093/humrep/16.9.2014. [DOI] [PubMed] [Google Scholar]
- 10.Murabito JM, Yang Q, Fox C, Wilson PW, Cupples LA. Heritability of age at natural menopause in the Framingham Heart Study. J Clin Endocrinol Metab. 2005;90:3427–30. doi: 10.1210/jc.2005-0181. [DOI] [PubMed] [Google Scholar]
- 11.Chanock SJ, et al. Replicating genotype-phenotype associations. Nature. 2007;447:655–60. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 12.Guo Y, et al. Genomewide linkage scan for quantitative trait loci underlying variation in age at menarche. J Clin Endocrinol Metab. 2006;91:1009–14. doi: 10.1210/jc.2005-2179. [DOI] [PubMed] [Google Scholar]
- 13.Rothenbuhler A, et al. Weight-adjusted genome scan analysis for mapping quantitative trait Loci for menarchal age. J Clin Endocrinol Metab. 2006;91:3534–7. doi: 10.1210/jc.2006-0150. [DOI] [PubMed] [Google Scholar]
- 14.Pan F, et al. Chromosomal regions 22q13 and 3p25 may harbor quantitative trait loci influencing both age at menarche and bone mineral density. Hum Genet. 2008;123:419–27. doi: 10.1007/s00439-008-0490-z. [DOI] [PubMed] [Google Scholar]
- 15.Anderson CA, et al. A genome-wide linkage scan for age-at-menarche in three populations of European descent. J Clin Endocrinol Metab. 2008 doi: 10.1210/jc.2007-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Asselt KM, et al. Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative trait loci influencing variation in human menopausal age. Am J Hum Genet. 2004;74:444–53. doi: 10.1086/382136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murabito JM, Yang Q, Fox CS, Cupples LA. Genome-wide linkage analysis to age at natural menopause in a community-based sample: the Framingham Heart Study. Fertil Steril. 2005;84:1674–9. doi: 10.1016/j.fertnstert.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 18.He LN, et al. Association study of the oestrogen signalling pathway genes in relation to age at natural menopause. J Genet. 2007;86:269–76. doi: 10.1007/s12041-007-0034-7. [DOI] [PubMed] [Google Scholar]
- 19.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 20.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinoshita Y, et al. Colocalization of MCM8 and MCM7 with proteins involved in distinct aspects of DNA replication. Microsc Res Tech. 2008;71:288–97. doi: 10.1002/jemt.20553. [DOI] [PubMed] [Google Scholar]
- 22.Inoue E, et al. SAD: a presynaptic kinase associated with synaptic vesicles and the active zone cytomatrix that regulates neurotransmitter release. Neuron. 2006;50:261–75. doi: 10.1016/j.neuron.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 23.Crump JG, Zhen M, Jin Y, Bargmann CI. The SAD-1 kinase regulates presynaptic vesicle clustering and axon termination. Neuron. 2001;29:115–29. doi: 10.1016/s0896-6273(01)00184-2. [DOI] [PubMed] [Google Scholar]
- 24.Yan J, et al. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–56. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooft L, et al. Hexokinase III, cyclin A and galectin-3 are overexpressed in malignant follicular thyroid nodules. Clin Endocrinol (Oxf) 2008;68:252–7. doi: 10.1111/j.1365-2265.2007.03031.x. [DOI] [PubMed] [Google Scholar]
- 26.Yang F, et al. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol. 2006;173:497–507. doi: 10.1083/jcb.200603063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunter DJ, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nat Genet. 2007;39:870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ridker PM, et al. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women’s Genome Health Study. Am J Hum Genet. 2008;82:1185–92. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2