Two small ncRNAs jointly govern virulence and transmission in Legionella pneumophila (original) (raw)
. Author manuscript; available in PMC: 2010 Sep 1.
Summary
To transit from intra- to extracellular environments, L. pneumophila differentiates from a replicative/non-virulent to a transmissive/virulent form using the two-component system LetA/LetS and the global repressor protein CsrA. While investigating how both regulators act coordinately we characterized two ncRNAs, RsmY and RsmZ that link the LetA/LetS and CsrA regulatory networks. We demonstrate that LetA directly regulates their expression and show that RsmY and RsmZ are functional in E. coli and are able to bind CsrA in vitro. Single mutants have no (Δ_rsmY_) or a little (Δ_rsmZ_) impact on virulence, but the Δ_rsmYZ_ strain shows a drastic defect in intracellular growth in Acanthamoeba castellanii and THP-1 monocyte-derived macrophages. Analysis of the transcriptional programs of the Δ_letA_, Δ_letS_ and Δ_rsmYZ_ strains revealed that the switch to the transmissive phase is partially blocked. One major difference between the Δ_letA_, Δ_letS_ and Δ_rsmYZ_ strains was that the latter synthesizes flagella. Taken together, LetA activates transcription of RsmY and RsmZ, which sequester CsrA and abolish its post-transcriptional repressive activity. However, the RsmYZ-CsrA pathway appears not to be the main or only regulatory circuit governing flagella synthesis. We suggest that rather RpoS and LetA, by influencing LetE and probably cyclic-di-GMP levels, regulate motility in L. pneumophila.
Keywords: Legionella pneumophila, RsmY, RsmZ, non-coding RNA, CsrA, transcriptome, pathogenicity, Acanthamoeba castellanii
Introduction
Legionella pneumophila is the causative agent of the pneumonia-like Legionnaires’ disease. The bacterium’s survival and spread depends on its ability to replicate inside eukaryotic phagocytic cells like Acanthamoeba castellanii, Hartmanella sp. or Naeglaria sp. It is thought that the interaction of L. pneumophila with aquatic protozoa has generated a pool of virulence traits, which also allow Legionella to infect human cells and to cause disease. Once internalized, L. pneumophila is able to survive and replicate by manipulating host cell functions to reprogram the endosomal-lysosomal degradation pathway of the phagocytic cell (for reviews see (Albert-Weissenberger et al., 2007; Steinert et al., 2002; Swanson and Hammer, 2000; Vogel and Isberg, 1999)).
For successful infection of host cells, L. pneumophila needs to ensure a precise timing of its life cycle. Signal transduction pathways, such as two-component systems (TCS) are known to be involved in these regulatory processes. TCS comprise a sensor histidine kinase that senses specific signals leading to autophosphorylation of the sensor and activation of the response regulators, which then activate or repress the transcription of target genes. In L. pneumophila strain Paris, 13 histidine kinases and 14 response regulators were identified (Cazalet et al., 2004); however, only a few of these systems have been studied to date. Examples are CpxA/CpxR that regulate icm and dot virulence genes (Altman and Segal, 2008; Gal-Mor and Segal, 2003) or the PmrA/PmrS TCS that was shown to be a global regulator implicated in intracellular growth of L. pneumophila and in regulation of the Dot/Icm type IV secretion system (Al-Khodor et al., 2008; Zusman et al., 2007). An important TCS in L. pneumophila, described previously, is LetA/LetS (Legionella transmission activator and sensor, respectively) that induces traits necessary for efficient host transmission and survival in the environment (Hammer et al., 2002). letA and letS mutants are non-motile, non-cytotoxic, sodium sensitive, and less proficient in infecting macrophages; however, letA mutants still multiply in macrophage host cells (Gal-Mor and Segal 2003; Hammer et al., 2002). Furthermore, letA mutants are more sensitive to oxidative and acid stress than the wild type (Lynch et al., 2003) and infectivity of A. castellanii is reduced (Hammer et al., 2002; Lynch et al., 2003; Molofsky and Swanson, 2004). Recently it was shown that the LetA/LetS TCS belongs to a family of signal-transducing proteins that employ a four-step phosphorelay to regulate gene expression. Histidine 307 of the LetS protein is the primary site of phosphorylation required to activate LetA (Edwards et al., 2009). Additionally, a threonine substitution at position 311 of LetS generated a L. pneumophila mutant with an intermediate phenotype (Edwards et al., 2009), in which gene expression of the flagellar regulon and numerous other loci was delayed when compared to wild type bacteria. Thus, histidine 307 and threonine 311 are necessary for the complete function of LetS (Edwards et al., 2009).
There are orthologous systems to LetA/LetS in many other Gram-negative bacteria, such as Salmonella enterica (BarA/SirA), Erwinia carotovora (ExpA/ExpS), Vibrio cholerae (VarA/VarS), Pseudomonas spp (GacA/GacS), Photorhabdus luminescens (UvrY/BarA) and E. coli (UvrY/BarA). These TCS are part of global regulatory circuits that control a variety of functions such as carbon metabolism, biosynthesis of secondary metabolites, cell motility, biofilm formation, quorum sensing, and, in pathogenic bacteria, virulence traits (Babitzke and Romeo, 2007; Cui et al., 2001; Kay et al., 2005; Krin et al., 2008; Lenz et al., 2005; Suzuki et al., 2002). These regulatory networks comprise small non-coding RNAs (ncRNAs) that bind proteins of the CsrA (carbon storage regulator)/RsmA (repressor of secondary metabolites) family and antagonize their activity by sequestering CsrA (Babitzke and Romeo, 2007; Lapouge et al., 2008; Liu et al., 1997; Romeo, 1998). CsrA-mediated repression involves the binding of CsrA to the ribosome-binding site of target transcripts, thereby blocking ribosome access to the mRNA; activation by CsrA seems to be due to mRNA stabilization (Babitzke and Romeo, 2007; Pernestig et al., 2003). The best-studied example is the BarA/UvrY system of E. coli, in which a few CsrA targets have been identified and studied so far. These are glgC (glycogen synthesis), cstA (peptide transport) and pgaA (synthesis of biofilm adhesin). CsrA of E. coli also binds to the untranslated region of the flhDC transcript (regulator of flagella biosynthesis) (Wei et al., 2001) and has recently been shown to control cyclic di-GMP metabolism by directly binding to the mRNA leaders of different GGDEF/EAL protein encoding genes (Jonas et al., 2008).
Legionella encodes a CsrA protein homologous to CsrA of E. coli (Fettes et al., 2001; Molofsky and Swanson, 2003) that is able to complement an E. coli csrA mutant (Fettes et al., 2001) as well as three CsrA homologues (Cazalet et al., 2004). CsrA of L. pneumophila was shown to repress the production of virulence traits during the exponential (E) growth phase (replicative /non-virulent bacteria) (Bachman and Swanson, 2004b; Molofsky and Swanson, 2003). In the post exponential (PE) growth phase (transmissive/virulent bacteria), it appears that the LetA/LetS system is necessary to overcome post-transcriptional repression of transmissive traits by CsrA (Molofsky and Swanson, 2003, , 2004). However, the link between LetA/LetS regulation and the CsrA-system was missing; it was unknown whether the regulatory process in Legionella involved small ncRNAs like in E. coli and Pseudomonas sp.
In order to understand the regulation of the switch from replicative/non-virulent to transmissive/virulent Legionella and to elucidate the link between the LetA/LetS system and the CsrA regulatory network we took a genomic-level approach to analyze LetA/LetS regulation and to assess the role of two putative ncRNAs that were predicted in L. pneumophila by a bioinformatics approach (Kulkarni et al., 2006). Here we present evidence that these ncRNAs, RsmY and RsmZ of L. pneumophila, are the missing link between LetA/LetS signaling and CsrA regulation. The genome-wide transcriptional profiles of letS, letA and rsmYZ double mutants showed the global effect of these different regulators on gene expression and their role in regulating virulence traits and transmission.
Results
The LetA/LetS regulatory system comprises two small ncRNAs - RsmY and RsmZ
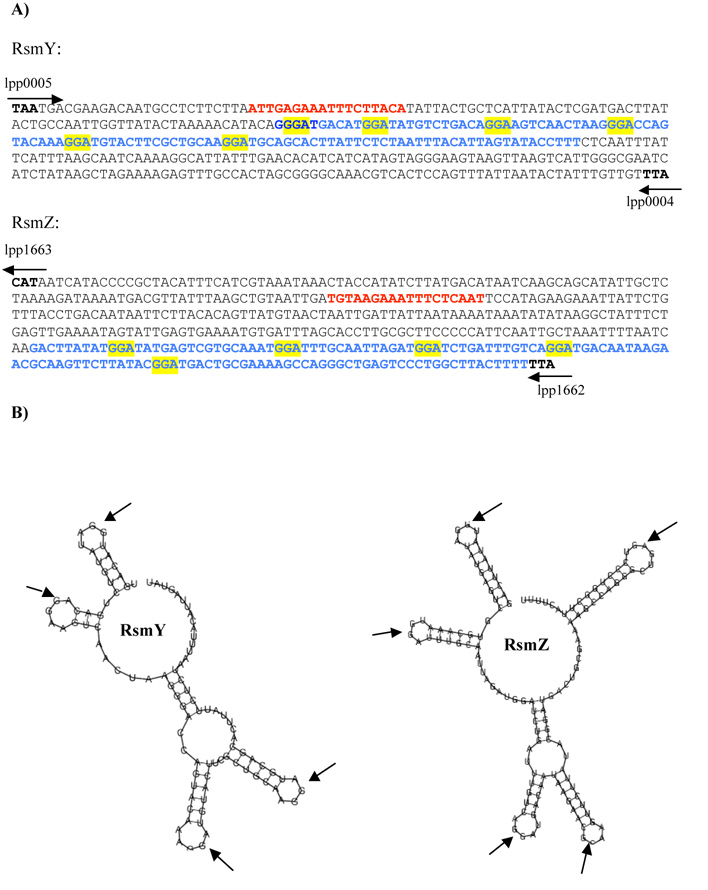
A recent bioinformatics study searching for putative CsrB/C or RsmY/Z-like RNAs in a variety of bacterial genomes predicted two ncRNAs in the genome of L. pneumophila. One was designated RsmY and is present in the intergenic region between gyrB (lpp0004) and lpp0005 (predicted 5’ end at nucleotide position 6,714), while the other is located in the intergenic region between lpp1662 and lpp1663 (position 1,870,666) and was designated RsmZ (Kulkarni et al., 2006) (Figure 1A). Repeated GGA motifs in the loop regions of the secondary structure are a crucial element in small RNAs that regulate CsrA and its homologs (Dubey et al., 2005). Analysis of the putative structure of RsmY and RsmZ using the program MFOLD (Zucker, 2003) indeed predicted loops in which conserved GGA motifs were located (Figure 1B). To verify whether these two regions were transcribed, real-time PCR was performed. This showed that RsmY and RsmZ are expressed in a growth phase-dependent manner, with an mRNA transcript increase of 3.1 and 6.8-fold for rsmY and rsmZ in the PE (OD 4.3) compared to the E growth phase (OD 2.5), respectively. The predicted size of the rsmY transcript is 110 nts, and that of rsmZ 132 nts; which was confirmed by Northern blot analysis (data not shown). The LetA/LetS TCS is predicted to regulate expression of RsmY and RsmZ. To gain further insight into this regulatory cascade, we also constructed a letA and a letS deletion mutant in L. pneumophila strain Paris and compared their roles in gene regulation with that of the two ncRNAs RsmY and RsmZ. In contrast to the wt, in the letA and the letS mutants no up-regulation of rsmY or rsmZ in the PE phase was observed. Thus, the LetA/S TCS is essential for the growth phase dependent expression of rsmY and rsmZ.
Figure 1. Two small ncRNAs, named RsmY and RsmZ are located in intergenic regions.
A) Sequence of the small RNAs and the flanking regions; Blue letters, sequence of the small RNA. Red letters, LetA binding site; yellow boxes, GGA motifs representing the CsrA binding sites; arrows indicate flanking genes in the L. pneumophila strains Paris genome. B) Predicted structure of the RsmY and RsmZ of Legionella pneumophila predicted by the program Mfold (http://mfold.burnet.edu.au/). The typical GGA (arrows) motifs are located in the loop regions
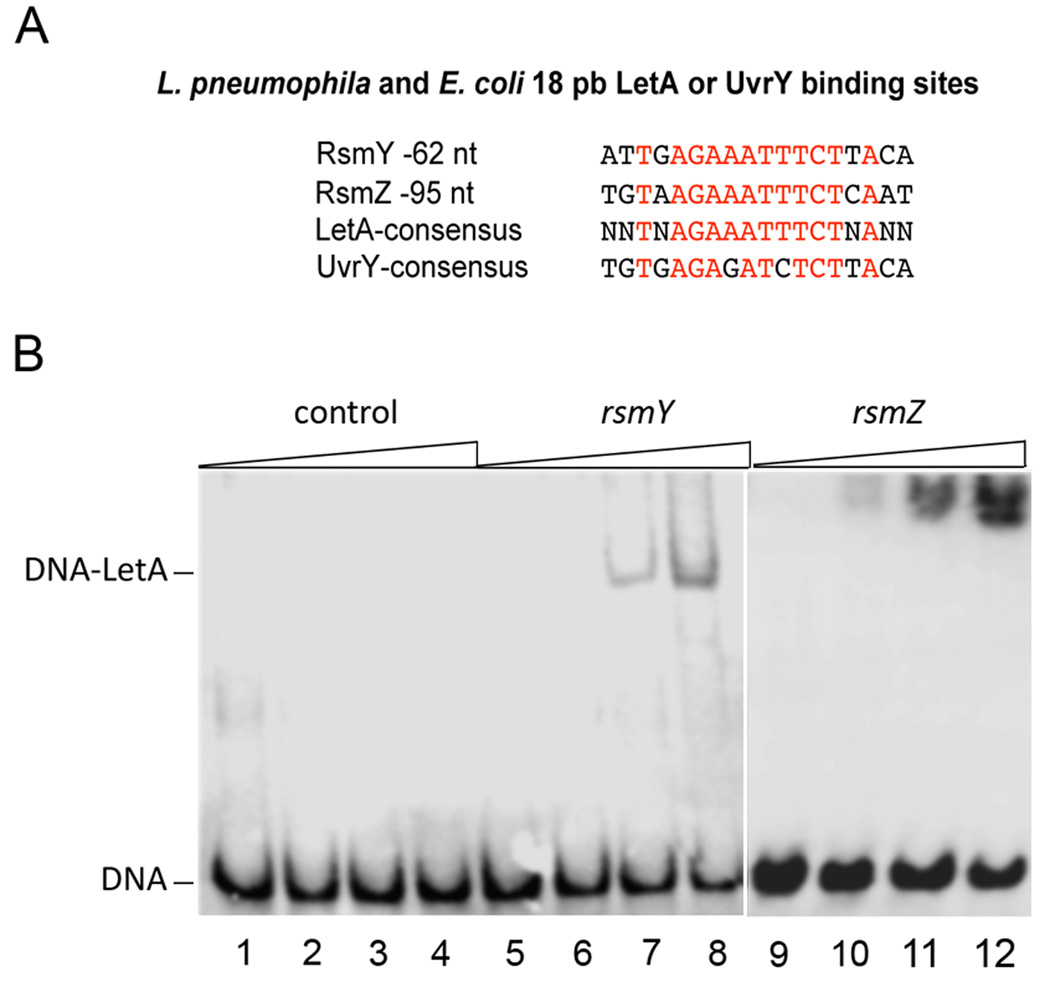
To determine if LetA directly regulates rsmY and rsmZ expression by binding to their promoter regions, we searched for a putative LetA-binding site upstream rsmY and rsmZ. Indeed, 62 nts and 95 nts upstream these genes, respectively, a single, conserved motif similar to the E. coli UvrY binding site was identified (Figure 2A). A consensus motif of the likely LetA-binding site is the palindromic sequence TNAGAAATTTCTNA. To confirm this site we performed mobility shift assays with purified, recombinant LetA and a 35bp fragment containing this predicted site. A band shift was observed for both rsmY and rsmZ upstream regions (Figure 2B), demonstrating that LetA binds specifically upstream of these loci. A stringent pattern search (no mismatch with respect to the consensus sequence) in the genome sequence of the L pneumophila strain Paris genome did not identify additional LetA-binding sites, suggesting that rsmY and rsmZ might be the only direct targets of LetA. Thus LetA binds to the upstream regions of rsmY and rsmZ and regulates their expression.
Figure 2. LetA binds specifically upstream of rsmY and rsmZ.
Electrophoretic Mobility Shift Assay with Strep-tag purified LetA protein and 35 bp fragment of HPLC-purified rsmY and rsmZ oligonucleotides containing the predicted LetA binding site and a 35 bp control DNA fragment without LetA binding site each with a biotin tag at the 5’ends A) Identified LetA-binding site and the L. pneumophila consensus sequence as compared to the E. coli consensus binding site for UvrY. B) Lane 1–4: 50ng control DNA; Lane 5–8: 50ng 35bp DNA fragment upstream rsmY containing the predicted LetA binding site, incubated with 0, 2, 4, 6 µM LetA; Lane 9–12: 50ng 35bp DNA fragment upstream rsmZ containing the predicted LetA binding site incubated with 0, 2, 4, 6 µM LetA, respectively.
RsmY and RsmZ of L. pneumophila induce premature glycogen synthesis and biofilm formation in E. coli
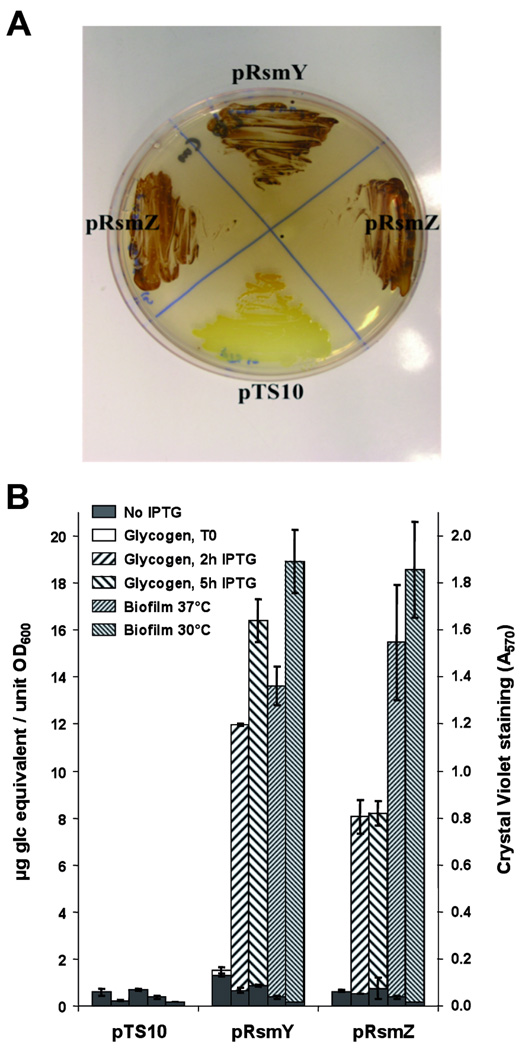
Known targets of CsrA in E.coli are mRNAs of genes involved in glycogen (glgCAP) and polysaccharide synthesis (pgaABCD). Inhibition of translation of these genes is effected by CsrA binding to these mRNAs during exponential growth (Wang et al., 2005; Yang et al., 1996). As it was shown that a functional similarity between the CsrA system of E. coli and L. pneumophila exists, and that CsrA of L. pneumophila can repress glycogen accumulation in E. coli (Fettes et al., 2001), we hypothesized that RsmY and RsmZ of L. pneumophila might also be capable of interacting with CsrA of E. coli. In order to test our assumption and to verify the in vivo functionality of both small ncRNAs of L. pneumophila, we cloned rsmY and rsmZ into the IPTG-inducible vector pMMB207C (Chen et al., 2004) generating the plasmid constructs p_rsmY_ and p_rsmZ_, respectively. Overexpression of rsmY and rsmZ in E. coli led to drastic changes in two reporter phenotypes known to be controlled by CsrA/CsrB in E. coli, namely glycogen storage and biofilm formation. After induction with IPTG of exponentially growing E. coli cells, glycogen quickly accumulated (<2 hours) and reached a plateau 10-times higher than that of the control strain transformed with the empty vector pTS10 (Figure 3A, B). Similarly, biofilm formation was enhanced by a factor of 100 (Figure 3B) showing that RsmY and RsmZ of L. pneumophila can mimic the action of CsrB and CsrC in E. coli.
Figure 3. RsmY and RsmZ of L. pneumophila induce glycogen accumulation and biofilm formation in E. coli.
A) Escherichia coli wild-type (BL21) containing the empty plasmid pTS10 and E. coli producing L. pneumophila RsmY (p_rsmY_) or RsmZ (p_rsmZ_), respectively, were streaked onto Kornberg medium containing 1% glucose and 100 mg.ml-1 Ampicillin, then stained with iodine vapour to visualize glycogen accumulation. B) Quantification of glycogen accumulation expressed in µg equivalent glucose per unit of OD600 measures and biofilm formation of E. coli carrying RsmY and RsmZ of L. pneumophila as judged from OD570 measures after solubilization of cristal violet.
CsrA binds RsmY and RsmZ in vitro
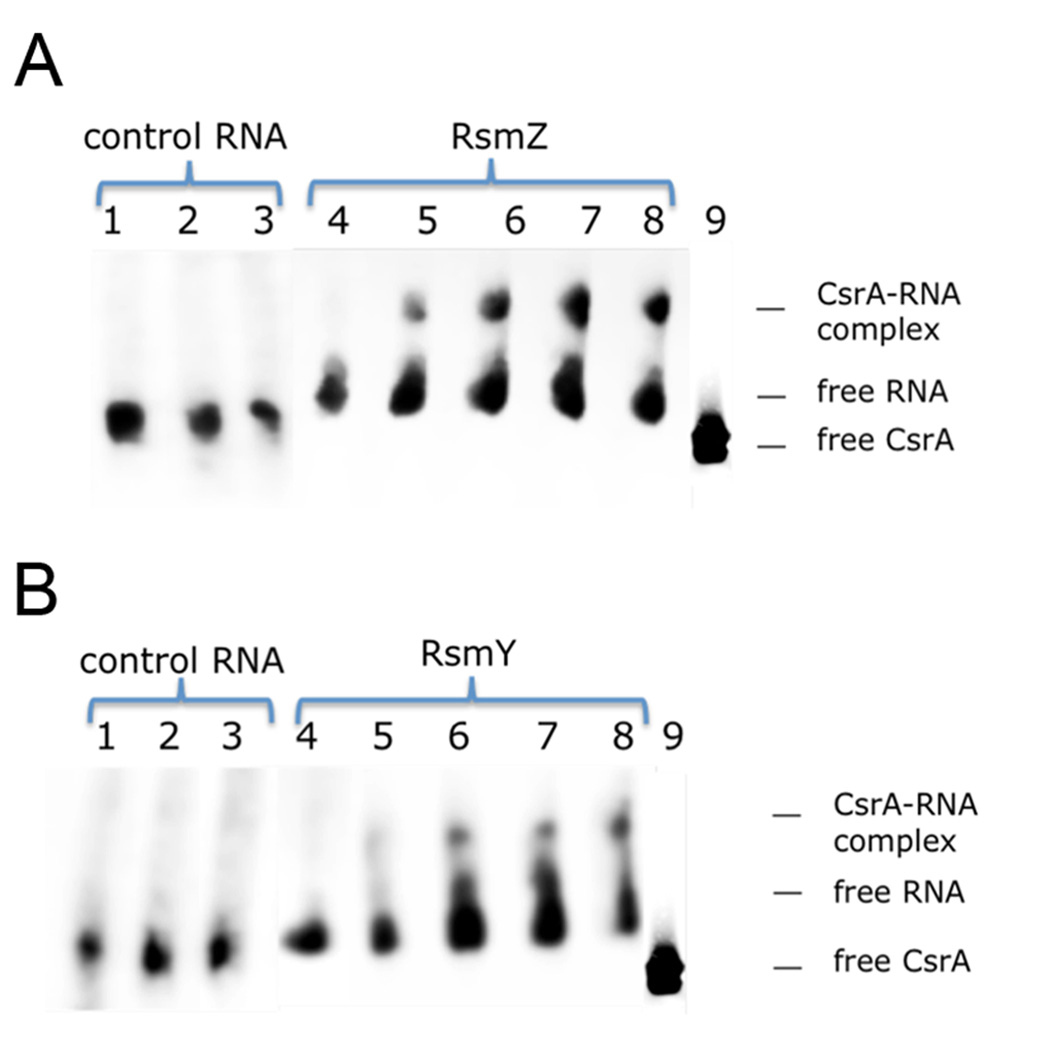
To determine whether RsmY and RsmZ are able to interact with CsrA of L. pneumophila, we prepared both ncRNA by in vitro transcription and performed mobility shift assays with Ni-NTA purified His-tagged L. pneumophila -CsrA, heterologously expressed in E.coli BL21 cells. At RNA concentrations of 100nM, a shift of RsmZ could be observed in the presence of 500nM of CsrA (Figure 4A). This additional band reached maximal intensity at 1µM CsrA; above this level (up to 5µM CsrA) no additional increase in the intensity of the shifted band was detectable. For RsmY, protein concentrations of 1µM CsrA were necessary to see a retarded band on the gel and the intensity of the band seemed to increase with higher CsrA concentrations up to 5µM (Figure 4B). The affinity for CsrA thus seems higher for RsmZ than for RsmY, suggesting that RsmZ is the major suppressor of CsrA in the PE phase. As controls, similar amounts of RNA of comparable size but lacking putative CsrA-binding structures did not lead to a mobility shift, indicating that CsrA bound specifically to RsmY and RsmZ in vitro. Thus, RsmY and RsmZ are two small ncRNAs that are able to bind CsrA. Altogether, our data suggest that LetA-dependent formation of small ncRNAs is required to dissociate RNA-binding proteins like CsrA from recognition sequences present in the target RNAs.
Figure 4. RsmY and RsmZ bind CsrA in vitro.
Electromobility shift assay with 100nM of RNA combined with varying concentrations of purified CsrA-His. A) RsmZ RNA and recombinant CsrA in 6% Blue Native Bis-Tris/Tricine Gel. Lane 1: no CsrA, 0.1 µM control RNA; lane 2: 2.0 µM CsrA + 0.1 µM control RNA, lane 3: 5.0 µM CsrA + 0.1 µM control RNA, lane 4: no CsrA, 0.1 µM RsmZ, lane 5: 0.5 µM CsrA + 0.1 µM RsmZ; lane 6: 1.0 µM CsrA+ 0.1 µM RsmZ; lane 7: 2.0 µM CsrA+ 0.1 µM RsmZ; lane 8: 5.0 µM CsrA+ 0.1 µM RsmZ; lane 9: 1µg CsrA (silver-stained). B) RsmY RNA and recombinant CsrA in 6% Blue Native Bis-Tris/Tricine Gel. Lane 1: no CsrA, 0.1 µM control RNA; lane 2: 2.0 µM CsrA + 0.1 µM control RNA, lane 3: 5.0 µM CsrA + 0.1 µM control RNA, lane 4: no CsrA, 0.1 µM RsmY, lane 5: 0.5 µM CsrA + 0.1 µM RsmY; lane 6: 1.0 µM CsrA+ 0.1 µM RsmY; lane 7: 2.0 µM CsrA+ 0.1 µM RsmY; lane 8: 5.0 µM CsrA+ 0.1 µM RsmY; lane 9: 1µg CsrA (silver-stained)
The rsmYZ double mutant is dramatically affected in infection and intracellular replication and is impaired in sodium sensitivity and pigment production
In order to gain further insight into the physiological role of these two small ncRNAs in L. pneumophila, we constructed Δ_rsmY_ and Δ_rsmZ_ single mutants. Their in vitro growth (either liquid or solid BYE) was not notably affected except for an elongated lag-phase (data not shown). The single knockout mutants have no (Δ_rsmY_) or little (Δ_rsmZ_) impact on virulence in the infection of A. castellanii (Figure 5A). In addition, the transcriptome of the two mutants showed no significant differences as compared to that of the wild type (data not shown). These observations are consistent with the fact that overproduction of both RsmY and RsmZ in E. coli and L. pneumophila led to similar phenotypic changes, indicating that their expression probably triggers an identical response and that they can functionally replace the other one. In contrast, a double mutant strain should have a detectable phenotype.
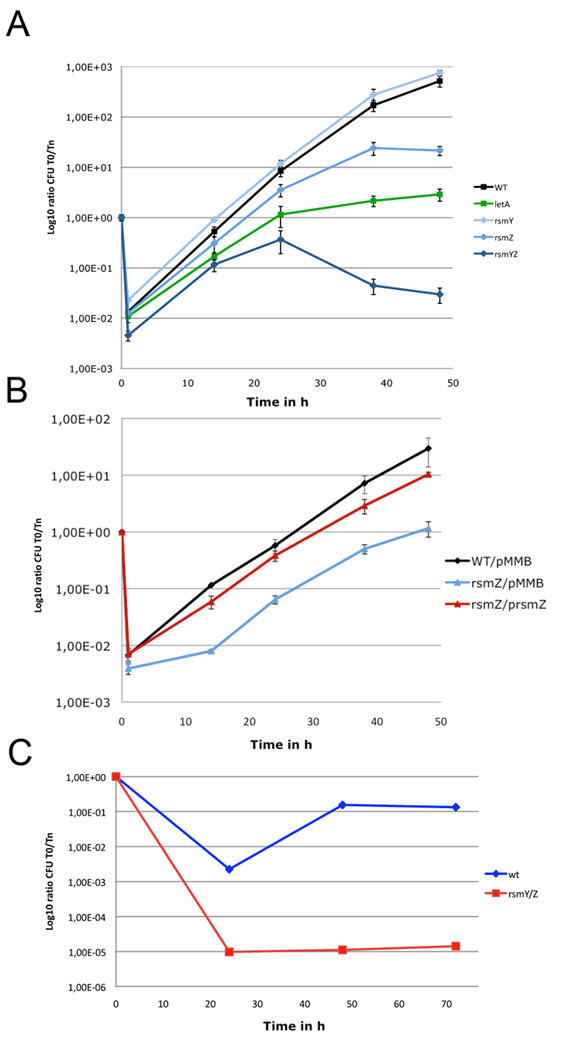
Figure 5. Intracellular replication of L. pneumophila in Acanthamoeba castellanii and THP-1 macrophages is dependent on functional RsmY and RsmZ.
A) A. castellani infection; Black, wild type L. pneumophila strain Paris; light blue, Δ_rsmY L. pneumophila_ strain Paris; blue, Δ_rsmZ L. pneumophila_ strain Paris; dark blue, Δ_rsmYZ L. pneumophila_ strain Paris; green Δ_letA L. pneumophila_ strain Paris. B) A. castellani infection; Black, wild type L. pneumophila strain Paris; light blue, Δ_rsmZ L. pneumophila_ strain Paris carrying the empty plasmid pMMB; red, Δ_rsmZ L. pneumophila_ strain Paris carrying plasmid p_rsmZ_ with an IPTG inducible promoter; For the complementation experiment all strains were grown in presence of IPTG. C) Infection of THP-1 human macrophages with Legionella pneumophila Paris wild type and Δ_rsmYZ L. pneumophila_ strain Paris. The number of viable bacteria within amoebae was evaluated by the standard plate count assay. Results are expressed as Log10 ratio CFU Tn/T0. Each time point represents the mean of +/− SD of two independent experiments. Infections were performed at 37°C.
To test this hypothesis we constructed a Δ_rsmYZ_ double mutant. The double mutant infected A. castellanii less effectively than the wild type, and was not able to replicate efficiently. Moreover, after 24h of infection, when bacteria normally transit to the next host cell, the bacterial count of the Δ_rsmYZ_ mutant declined, indicating that it was either not able to infect a new host or was degraded inside the amoeba (Figure 5A). Complementation of the Δ_rsmZ_ mutant with plasmid p_rsmZ in trans_ rescued it in amoeba infection (Figure 5B). Deletion of rsmY and rsmZ as compared to the deletion of letA, had a more pronounced impact on intracellular growth (Figure 5A). In infection of THP-1 monocyte-derived macrophages the Δ_rsmYZ_ double mutant was also strongly impaired in intracellular growth and infection of a new host (Figure 5C).
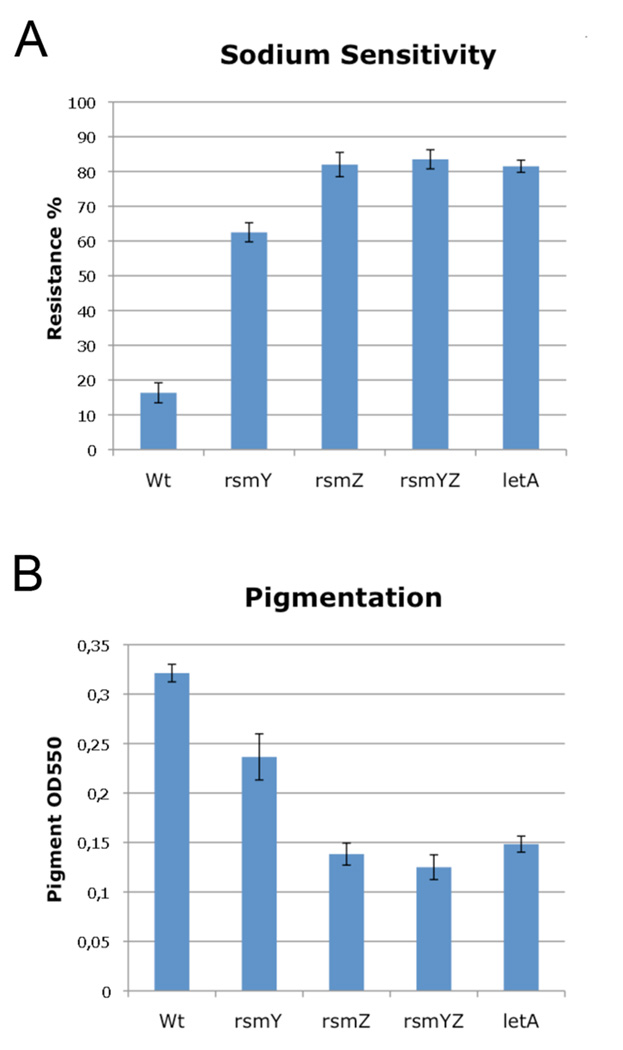
Wild type L. pneumophila are sodium sensitive as they grow poorly on agar containing 100mM NaCl (Catrenich and Johnson, 1989) and accumulate substantial pigment during growth (Wiater et al., 1994). However, Δ_letA_ or Δ_letS_ mutants are sodium resistant and accumulate little pigment during the PE growth (Hammer et al., 2002). Here we show that the double (Δ_rsmYZ_) and the single knockout mutants (Δ_rsmY_, Δ_rsmZ_) were sodium resistant (Figure 6A) and accumulate little pigment (Figure 6B) comparable to a Δ_letA_ mutant. This suggests that RsmY and RsmZ, through the CsrA system, are essential for intracellular replication of L. pneumophila and are involved in regulating pigment production and sodium sensitivity.
Figure 6. Sodium sensitivity is repressed and pigment production is activated by LetA, RsmY and RsmZ.
A) PE phase Δ_rsmY_, Δ_rsmZ_, Δ_rsmYZ_ and Δ_letA_ mutants of strain Paris are not sensitive to NaCl as compared to PE phase wild type strain Paris. B) RsmZ, RsmY, RsmYZ and LetA activate pigment production in Legionella
The over expression of rsmY or rsmZ in L. pneumophila wt induces expression of transmissive genes during exponential growth
Provided that RsmY and RsmZ interact with and sequester CsrA protein in the PE phase to allow the transition of wild-type cells to the transmissive phase, early expression (i.e. in the E phase) of these RNAs in L. pneumophila should abolish the post-transcriptional repression of transmissive traits by CsrA in the E phase. To verify this prediction p_rsmZ_, was transformed into L. pneumophila Paris and cells were grown to OD 1.0. After 90 min of induction of rsmZ transcription cells were harvested and microarray analyses were performed to determine gene expression differences between the recombinant strain and wild-type strain harboring an empty plasmid pMMB207C. Differences in mRNA transcript levels could indeed be observed. Eighty-three genes were down- and 380 genes were significantly up-regulated. Among the down-regulated genes were typical replicative genes, while among the up-regulated genes were genes characteristic of the transmissive stage, as defined previously (Brüggemann et al., 2006b) (the complete dataset is available at http://genoscript.pasteur.fr). For example, the strongest down-regulated gene was lpp0706 (3.4-fold repressed) and the adjacent lpp0707, both typical replicative genes expected to be up-regulated at OD 1 (Brüggemann et al., 2006b). These genes, together with a third repressed gene, lpp1860, code for proteins that are members of a subfamily of the major facilitator superfamily, previously described as phagosomal transporters (Pht) of L. pneumophila that may be involved in the transport of amino acids (Chen et al., 2008). It was shown that one mutant, phtA, prematurely differentiates to the transmissive phase (Sauer et al., 2005). Other down-regulated genes were lpp0153 (glyceraldehyde 3-phosphate dehydrogenase) and lpp0529 (acetyl-CoA biotin carboxyl carrier protein), lpp2756 and lpp2757 similar to stringent starvation protein A and B, respectively, or genes related to ATP synthesis like lpp3052 and lpp3053. These are again classically seen as replicative genes.
Several typical transmissive genes were already found to be up-regulated at OD 1. Those are genes encoding for proteins containing a GGDEF/EAL domain (lpp1170) or an EAL motif (lpp0351), respectively, and the superoxide-dismutase precursor (lpp2297) possibly related to stress response as well as several Legionella specific proteins with yet unknown function (e.g. lpp1112 or lpp1340). Other strongly up-regulated genes in the RsmZ overexpressing strain with known or predicted functions were lpp2788, encoding the response regulator LqsR, a protein involved in phagocytosis and cytotoxicity (Tiaden et al., 2007), lpp1324, coding a protein similar to a DNA-binding protein Fis, and lpp2675 which exhibits similarity to Papain-like cysteine peptidases. Weak induction of fleQ (lpp0915), coding for the regulator of flagella synthesis FleQ, lpp0969, coding for the anti-sigma28 factor, a negative regulator of flagellin synthesis, and lpp0541, coding a putative sigma-54 modulation protein as well as two flagellar genes fliE (lpp1725) and flgA (lpp0970) was observed.
We also overexpressed rsmY by transforming plasmid p_rsmY_ into L. pneumophila strain JR32, since transformation of strain Paris was not successful. Comparison of microarray analyses at OD 0.5 and OD 1 showed similar results for RsmY on L. pneumophila gene expression as described above for RsmZ (the complete dataset is available at http://genoscript.pasteur.fr).
The LetA/LetS two-component system regulates rsmY and rsmZ expression and many genes involved in virulence formation of L. pneumophila
To compare the role of the LetA/LetS TCS in gene regulation with that of the two ncRNAs RsmY/RsmZ, we sought to identify all genes controlled by these systems through global gene expression profiling. Microarray analyses were performed with cells harvested in E and PE growth phases. No significant differences in gene expression between mutant and wild type strains were observed during the E phase (ODs 1 and 2.5), underlining the previous assumption that LetA/LetS senses a signal generated at the onset of the PE phase (Hammer et al., 2002; Molofsky and Swanson, 2004). In contrast, drastic changes in gene expression of Δ_letA_ and Δ_letS_ were observed in the PE phase (OD 4.3), with 425 significantly down- and 314 significantly up-regulated genes (fold-change >1.5) as compared to the wild type (Supplementary material Table S1 and Table S2). The expression of rsmY and rsmZ was reduced three and two fold, respectively. However, expression was not completely abolished, suggesting that additional factors might regulate the expression of these small ncRNAs or that a basal level of RsmY and RsmZ is constitutively expressed. Direct comparison of the gene expression pattern of Δ_letA_ and Δ_letS_ identified no significant differences (data not shown), indicating that LetS has no additional function besides phosphorylating and activating LetA, and that no cross talk of LetA with other sensor kinases exists in these conditions.
Analyses of the transcriptome of the mutants revealed that up-regulated genes in the PE phase are those previously shown to be typical replicative traits (Brüggemann et al., 2006b), such as protein biosynthesis (e.g. ribosomal structure and biogenesis), energy transfer (e.g. NADH dehydrognenase - lpp2830-lpp2835; NADH-quinone oxidoreductase - lpp2827, lpp2828; ATP synthetase - lpp3052-lpp3059), or proteins involved in carbohydrate metabolism (e.g. pyruvate dehydrogenase - lpp1517; phosphoenolpyruvate synthase – lpp0867) (Table 1). Furthermore, in contrast to the wild type, in letA and letS mutants the biosynthesis of the lipopolyssacharide (LPS) capsule and of legioaminic acid appears to continue also in the PE phase as several genes encoding proteins involved in LPS-biosynthesis (lpp0814, hisF/lpp0815, hisH/lpp0816, neuA/lpp0817, neuB/lpp0818, neuC/lpp0819, hisF, hisH, lpp2026) are induced (Table 1 and the complete dataset is available at http://genoscript.pasteur.fr).
Table 1.
Selection of PE phase induced genes in the letA and rsmYZ mutants
| Gene name | Annotation | Δ_letA/wt_ | Δ_rsmYZ/wt_ |
|---|---|---|---|
| lpp3059 | AtpB (H+-transporting ATP synthase chain a) | 2.45 | - |
| lpp3058 | AtpE (H+-transporting ATP synthase chain c) | 1.57 | - |
| lpp3057 | AtpF (H+-transporting ATP synthase chain b) | 1.55 | - |
| lpp3056 | AtpH (H+-transporting ATP synthase chain delta) | 1.78 | - |
| lpp3055 | AtpA (H+-transporting ATP synthase chain alpha) | 1.21 | - |
| lpp3054 | AtpG (H+-transporting ATP synthase chain gamma) | 2.01 | - |
| lpp3053 | AtpD (H+-transporting ATP synthase beta chain) | 2.47 | - |
| lpp3052 | AtpC (H+-transporting ATP synthase epsilon chain) | 2.47 | - |
| lpp3039 | SodB (Superoxide dismutase) | - | 3.11 |
| lpp3038 | GlrA (Glutaredoxin-like protein) | 2.46 | 4.34 |
| lpp3037 | AhpC (Alkyl hydroperoxide reductase) | 2.61 | 3.21 |
| lpp3033 | Major outer membrane protein precursor | 3.41 | 7.74 |
| lpp3032 | Major outer membrane protein | 4.22 | 7.52 |
| lpp2835 | NADH dehydrogenase I chain B | 3.84 | 2.37 |
| lpp2831 | NADH dehydrogenase I chain F | 2.35 | 1.69 |
| lpp2830 | NADH dehydrogenase I chain G | 3.14 | - |
| lpp2828 | NADH-quinone oxidoreductase chain I | 1.99 | - |
| lpp2827 | NADH-quinone oxidoreductase chain J | 2.42 | - |
| lpp2768 | 50S ribosomal protein L35 | 3.06 | 2.88 |
| lpp2689 | 30S ribosomal subunit protein S2 | 3.33 | 1.78 |
| lpp2026 | Peptidoglycan-associated lipoprotein precursor | 1.79 | 4.52 |
| lpp1958 | Major outer membrane protein | 2.16 | 6.57 |
| lpp1773 | Similar to long-chain fatty acid transport protein | 3.07 | 3.16 |
| lpp1731 | Thioredoxin reductase | 2.77 | 2.63 |
| lpp1726 | FleR | - | 3.01 |
| lpp1673 | AstB (Succinylarginine dihydrolase) | 2.08 | 1.86 |
| lpp1672 | AstD (Succinylglutamic semialdehyde dehydrogenase) | 2.46 | 1.96 |
| lpp1671 | AstA (Arginine N-succinyltransferase- beta chain) | 2.16 | 2.21 |
| lpp1517 | Pyruvate dehydrogenase E2 component | 3.74 | 2.09 |
| lpp1516 | Pyruvate dehydrogenase- (E1 beta subunit) | 2.23 | 1.67 |
| lpp1346 | 50S ribosomal subunit protein L32 | 2.39 | 2.46 |
| lpp1247 | RpoS (RNA polymerase sigma factor) | - | 3.11 |
| lpp1223 | Oxygen-dependent coproporphyrinogen III oxidase | 3.14 | 2.58 |
| lpp1204 | CydA (Cytochrome d ubiquinol oxidase subunit I) | 3.43 | 2.69 |
| lpp1205 | CydB (Cytochrome d ubiquinol oxidase subunit II) | 1.74 | 1.71 |
| lpp1202 | HisG | 3.40 | - |
| lpp1201 | HisD | 3.13 | - |
| lpp1200 | HisC | 8.50 | 2.05 |
| lpp1199 | HisB | 8.65 | 1.85 |
| lpp1198 | HisH | 4.74 | 1.70 |
| lpp1197 | HisA | 7.79 | 1.68 |
| lpp1196 | HisF | 4.00 | - |
| lpp1195 | HisI | 2.76 | 1.55 |
| lpp1117 | ChiA | 4.88 | 2.23 |
| lpp0867 | PpsA (phosphoenolpyruvate synthase) | 2.93 | 1.89 |
| lpp0855 | Mip (Macrophage infectivity potentiator) | 2.51 | 3.91 |
| lpp0819 | NeuC (N-acylglucosamine 2-epimerase) | 1.65 | - |
| lpp0818 | NeuB (N-acetylneuraminic acid conden. enzyme) | 2.01 | - |
| lpp0817 | NeuA (CMP-N-acetlyneuraminic acid synthetase) | 2.34 | - |
| lpp0816 | HisH (Imidazole glycerol phosphate synthase) | 2.91 | - |
| lpp0815 | HisF (Imidazole glycerol phosphate synthase) | 3.55 | 2.00 |
| lpp0814 | Similar to LPS biosynthesis protein | 2.56 | 2.29 |
| lpp0602 | LetE (Transmission trait enhancer protein) | 2.71 | 1.60 |
| lpp0570 | OmpH (Outer membrane protein) | 3.08 | 3.54 |
| lpp0532 | ProA (Zinc metalloproteinase precursor) | 3.01 | 3.07 |
| lpp0493 | similar to Cold shock-like protein CspD | 2.84 | 5.88 |
| lpp0413 | 50S ribosomal subunit protein L15 | 2.41 | - |
| lpp0384 | 50S ribosomal protein L1 | 2.53 | - |
| lpp0383 | 50S ribosomal protein L11 | 1.98 | - |
| lpp0845 | Global regulator CsrA | 1.66 | - |
Conversely, down-regulated genes of both single mutants in the PE phase as compared to the wild type comprise functions associated with transmissive traits such as motility, virulence, and factors involved in host-cell manipulation (Table 2). Several substrates of the Dot/Icm type IV secretion system (T4SS), which are necessary at the onset of infection, were down regulated, such as RalF (lpp1932), PatA/VipD (lpp2888), LidA (lpp1002), the SidE homologs SdeA, SdeC (lpp2096, lpp2092), SidF (lpp2637) or SidC (lpp2579). T4SS-independent genes were also down-regulated, such as the structural toxin protein RtxA (lpp0699), the global stress protein GspA (lpp2141), the enhanced entry protein EnhB (lpp2693), EnhA homologs (lpp0972, lpp1290), and EnhC homologs (lpp1310, lpp2174). The highest repressed gene cluster in both Δ_letS_ and Δ_letA_ mutants is lpp0726–0730. This operon is of unknown function but lpp0726 is predicted to encode a patatin-like esterase and lpp0727, lpp0728 and lpp0730 encode proteins with characteristics of a membrane-bound NADH dehydrogenase, an acetoacetate decarboxylase and an adenylate cyclase, respectively (Table 2 and the complete dataset is available at http://genoscript.pasteur.fr). The expression of ten selected genes (lpp0952, flaA, mip, ralF, rpoS, rsmZ, lpp1170, lpp0351, fis2, csrA) was also analyzed by qRT-PCR. These results were in perfect agreement with the mircoarray data (supplementary material, Figure S2). Thus, the LetA/LetS TCS is clearly implicated in regulating genes important for the switch from the replicative to transmissive phase.
Table 2.
Selection of PE phase genes down-regulated in the letA and rsmYZ mutants
| Gene name | Known or predicted product | Δ_letA/wt_ | Δ_rsmYZ/wt_ |
|---|---|---|---|
| lpp0728 | Similar to acetoacetate decarboxylase | 0.04 | 0.67 |
| lpp0972 | Similar to enhanced entry protein EnhA | 0.05 | 0.15 |
| lpp0729 | Similar to protein | 0.07 | 0.64 |
| lpp1294 | FlaA | 0.08 | - |
| lpp1293 | FlaG | 0.08 | - |
| lpp2096 | SdeA | 0.10 | 0.18 |
| lpp0727 | NADH-ubiquinone oxidoreductase | 0.11 | - |
| lpp1900 | Unknown | 0.11 | 0.41 |
| lpp1942 | Unknown | 0.11 | 0.34 |
| lpp0197 | Similar to adenine specific DNA methylase | 0.13 | - |
| lpp2327 | Legionella specific protein | 0.13 | - |
| lpp0730 | Similar to adenylate cyclase | 0.14 | - |
| lpp0075 | Similar to L. pneumophila LvrA protein | 0.14 | - |
| lpp1162 | Legionella specific protein | 0.14 | 0.16 |
| lpp1291 | FliS | 0.14 | - |
| lpp1292 | FliD | 0.14 | - |
| lpp1453 | SidE homolog | 0.14 | 0.22 |
| lpp1641 | Putative alpha amylase | 0.14 | 0.23 |
| lpp2200 | Unknown | 0.15 | 0.23 |
| lpp1233 | FlgK | 0.16 | 1.80 |
| lpp1615 | SidE homolog | 0.16 | 0.19 |
| lpp2357 | unknown | 0.16 | - |
| lpp1341 | Some similarity with EnhA protein | 0.17 | 0.17 |
| lpp2174 | Similar to EnhC | 0.17 | 0.47 |
| lpp2578 | SdcA | 0.17 | 0.26 |
| lpp1002 | LidA | 0.18 | - |
| lpp1932 | RalF | 0.18 | 0.26 |
| lpp2092 | SdeC | 0.18 | - |
| lpp1290 | Similar to EnhA | 0.19 | 0.40 |
| lpp1479 | PilB | 0.19 | 0.29 |
| lpp2485 | Some similarities with eukaryotic proteins | 0.19 | 0.42 |
| lpp2579 | SidC | 0.20 | 0.31 |
| lpp0952 | Regulatory protein (GGDEF and EAL) | 0.21 | - |
| lpp0969 | FlgM (Anti-sigma-28 factor) | 0.23 | - |
| lpp1234 | FlgL | 0.23 | 1.61 |
| lpp0686 | FimU | 0.26 | 0.22 |
| lpp0683 | PilX | 0.27 | 0.30 |
| lpp2267 | MotB2 | 0.27 | - |
| lpp0726 | Similar to patatin-like esterase | 0.28 | 0.67 |
| lpp1480 | PilC | 0.28 | 0.26 |
| lpp1227 | FlgE | 0.29 | 3.13 |
| lpp2487 | Some similarity with eukaryotic proteins | 0.29 | 0.35 |
| lpp1228 | FlgF | 0.30 | 4.02 |
| lpp1925 | PatE | 0.30 | 0.34 |
| lpp2161 | Unknown | 0.30 | 0.47 |
| lpp2265 | PatB | 0.30 | 0.53 |
| lpp2362 | Chemiosmotic efflux system B protein B | 0.30 | - |
| lpp2363 | Chemiosmotic efflux system B protein C | 0.30 | - |
| lpp1324 | Similar to DNA-binding protein Fis | 0.31 | 6.75 |
| lpp2266 | MotA2 | 0.31 | 0.62 |
| lpp2888 | PatA/VipD | 0.31 | 0.23 |
| lpp0351 | Regulatory protein (EAL domain) | 0.34 | - |
| lpp0352 | Regulatory protein (GGDEF domain) | 0.34 | 0.42 |
| lpp1745 | MotA | 0.34 | - |
| lpp1723 | FliG | 0.35 | 2.01 |
| lpp1657 | FliK | 0.36 | - |
| rsmY | small ncRNA rsmY | 0.36 | 0.05 |
| lpp2141 | Global stress protein GspA | 0.37 | 2.67 |
| lpp1170 | Regulatory protein (GGDEF and EAL) | 0.37 | - |
| lpp0809 | Regulatory protein (GGDEF domain) | 0.38 | - |
| lpp2071 | Regulatory protein (GGDEF domain) | 0.38 | 0.38 |
| lpp2355 | Regulatory protein (GGDEF domain) | 0.38 | - |
| lpp0220 | Regulatory protein (EAL domain) | 0.39 | 0.39 |
| lpp1744 | MotB | 0.40 | - |
| lpp0440 | Regulatory protein (GGDEF domain) | 0.41 | 0.32 |
| lpp2477 | Regulatory protein (GGDEF domain) | 0.41 | 0.40 |
| lpp1754 | FliO | 0.43 | 2.53 |
| lpp0029 | Regulatory protein (GGDEF and EAL) | 0.44 | 0.36 |
| lpp1310 | EnhC homolog | 0.45 | - |
| lpp1747 | FleN | 0.45 | 2.92 |
| lpp2635 | FlhB | 0.46 | - |
| lpp0166 | LvrA | 0.47 | - |
| lpp1755 | FliN | 0.47 | 2.76 |
| lpp2637 | SidF | 0.48 | 0.44 |
| lpp2082 | Ankyrin repeat and a F-box | 0.49 | 0.61 |
| lpp0699 | RtxA | 0.50 | 0.37 |
| lpp0150 | SdhB | 0.51 | 0.34 |
| lpp1475 | Regulatory protein (GGDEF and EAL) | 0.51 | 0.53 |
| lpp1748 | FlhF | 0.51 | 2.54 |
| lpp2693 | EnhB | 0.51 | 1.74 |
| lpp0304 | SidE | 0.52 | 0.36 |
| lpp1746 | FliA (Sigma factor 28) | 0.52 | - |
| lpp2788 | LqsR (Response regulator) | 0.52 | 1.74 |
| lpp1722 | FliH | 0.53 | 1.98 |
| rsmZ | small ncRNA rsmZ | 0.54 | 0.01 |
| lpp2095 | SdeB | 0.55 | 0.47 |
| lpp1232 | FlgJ | 0.57 | 2.16 |
| lpp0542 | RpoN (sigma-54 factor) | 0.58 | - |
| lpp2546 | SdbB | 0.58 | 0.38 |
| lpp1231 | FlgI | 0.60 | 2.47 |
| lpp1226 | FlgD | 0.61 | 3.66 |
| lpp0009 | Similar to host factor-1 protein Hfq | 0.65 | 1.67 |
| lpp2692 | EnhC | 0.68 | - |
LetA/LetS governs flagella biosynthesis largely independently of RsmY and RsmZ
Consistent with the lack of a functional flagellum in letA and letS mutants (Figure 7), the flagellin-encoding gene (flaA, lpp1294) was one of the strongest down-regulated genes in the dataset. Levels of flaA mRNA were 13-fold lower in Δ_letA_ as compared to the wild-type (Table 3). Western blot analysis with anti-FlaA antibodies confirmed that no flagellin was present in Δ_letA_ strains (data not shown). Other flagellum encoding genes were significantly repressed as well, such as flgEF (lpp1226–1228), flgKL (lpp1231–1234), flgMNO (lpp1754–1756), the flaA_-associated operon containing flaG, fliDS (lpp1291-93), and the gene encoding the alternative sigma factor 28 (fliA, lpp1746) and its antagonist (flgM, lpp0969). Thus, LetA/LetS regulation seems to be hierarchically upstream of RpoN/FleQ, the master regulator of flagella biosynthesis genes (Albert-Weissenberger et al., in preparation), since most flagellar genes are down-regulated in Δ_letS and Δ_letA_ strains.
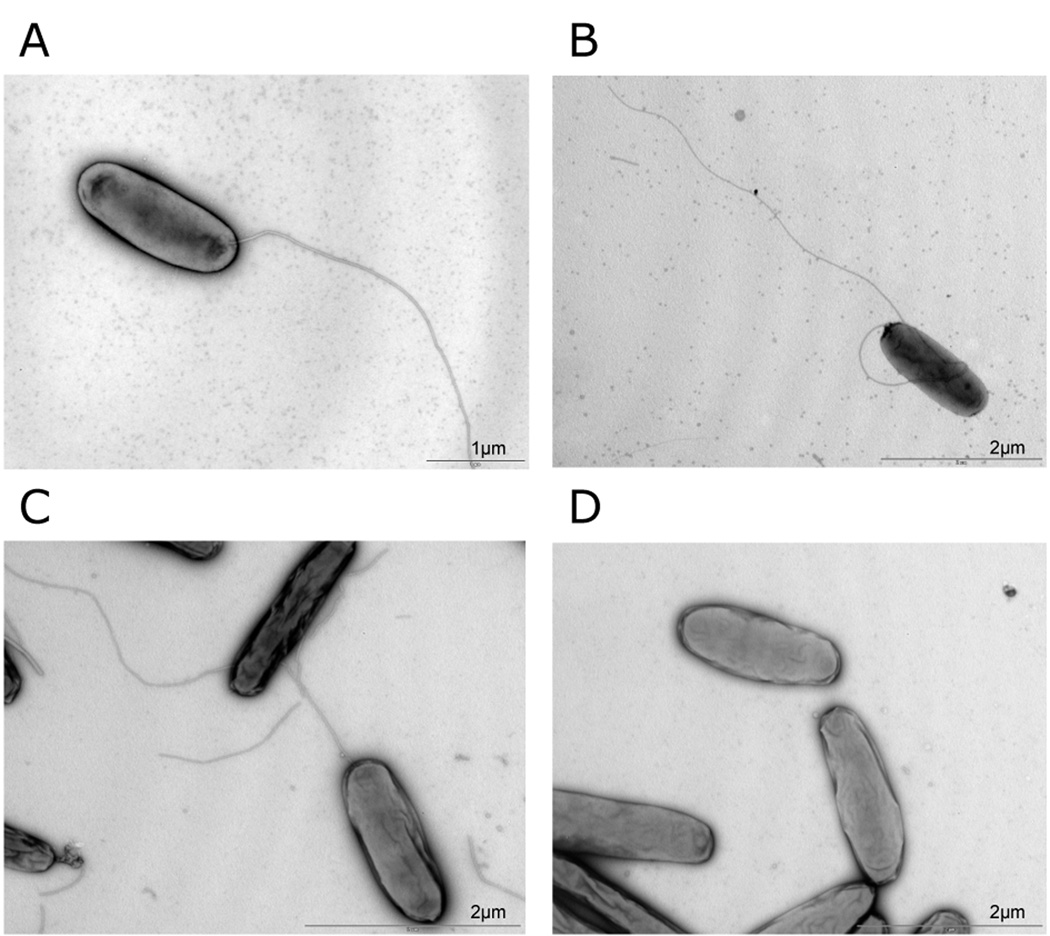
Figure 7. Electron microscopy of PE phase Legionella pneumophila wild type, Δ_letA_, Δ_rsmZ_ and Δ_rsmYZ_.
In contrast to the letA mutant, mutants in the small ncRNAs synthesize flagella in PE-growth phase. A) wild type, B) Δ_rsmZ, C) ΔrsmYZ, D) ΔletA_.
Table 3.
Flagella gene expression of letA and rsmYZ mutants as compared to wild type L. pneumophila strain Paris
| Gene name | Lpp | Known or predicted product | Δ_letA/ wt_ | Δ_rsmYZ/ wt_ | Δ_let/Δ_rsmYZ | |
|---|---|---|---|---|---|---|
| rpoN | lpp0542 | RNA polymerase sigma-54 factor | I | 0.58 | - | - |
| fleQ | lpp0915 | Transcriptional regulator FleQ | I | - | - | - |
| flgN | lpp0968 | Unknown | III | 0.35 | - | - |
| flgM | lpp0969 | Anti-sigma-28 factor | III | 0.23 | - | 7.16 |
| flgA | lpp0970 | Flagellar basal body P-ring biosynthesis | IIb | 0.66 | - | - |
| flgB | lpp1224 | Flagellar basal-body rod protein FlgB | IIb | - | 4.17 | - |
| flgC | lpp1225 | Flagellar basal-body rod protein FlgC | IIb | - | 3.23 | - |
| flgD | lpp1226 | Flagellar basal-body rod modification | IIb | 0.61 | 3.66 | 3.75 |
| flgE | lpp1227 | Flagellar hook protein FlgE | IIb | 0.29 | 3.13 | 19.7 |
| flgF | lpp1228 | Flagellar biosynthesis protein FlgF | IIb | 0.30 | 4.02 | 14.8 |
| flgG | lpp1229 | Flagellar biosynthesis protein FlgG | IIb | - | 5.20 | - |
| flgH | lpp1230 | Flagellar L-ring protein precursor FlgH | IIb | - | 4.62 | 5.50 |
| flgI | lpp1231 | Flagellar P-ring protein precursor FlgI | IIb | 0.60 | 2.47 | 4.59 |
| flgJ | lpp1232 | Flagellar biosynthesis protein FlgJ | IIb | 0.57 | 2.16 | 21.6 |
| flgK | lpp1233 | Flagellar hook-associated protein 1 | IIb | 0.16 | 1.80 | 16.4 |
| flgL | lpp1234 | Flagellar hook-associated protein FlgL | IIb | 0.23 | 1.61 | 5.50 |
| fliS | lpp1291 | Flagellar protein FliS | IV | 0.14 | - | 11.3 |
| fliD | lpp1292 | Flagellar hook-associated protein 2 | IV | 0.14 | - | 13.0 |
| flaG | lpp1293 | Unknown | IV | 0.08 | - | 20.6 |
| flaA | lpp1294 | Flagelline | IV | 0.08 | - | 18.8 |
| fliK‘ | lpp1657 | Flagellar hook-length control protein FliK | IIb | 0.36 | - | 2.54 |
| fliJ | lpp1720 | Flagellar protein FliJ | IIa | - | - | - |
| fliI | lpp1721 | Flagellum-specific ATP synthase FliI | IIa | - | - | - |
| fliH | lpp1722 | Polar flagellar assembly protein FliH | IIa | 0.53 | 1.98 | - |
| fliG | lpp1723 | Flagellar motor switch protein | IIa | 0.35 | 2.01 | 2.50 |
| fliF | lpp1724 | Flagellar M-ring protein | IIa | 0.42 | 2.23 | 2.99 |
| fliE | lpp1725 | Flagellar hook-basal body complex protein | IIa | - | 2.58 | - |
| fleR | lpp1726 | Two-component response regulator | IIa | - | 3.00 | - |
| fleS | lpp1727 | Sensor histidine kinase | IIa | - | - | - |
| motB | lpp1744 | Chemotaxis MotB protein | III | 0.40 | - | 2.78 |
| motA | lpp1745 | Flagellar motor protein MotA | III | 0.34 | - | 4.32 |
| fliA | lpp1746 | Sigma factor 28 | III | 0.52 | - | - |
| fleN | lpp1747 | Flagellar synthesis regulator | IIb | 0.45 | 2.92 | 3.61 |
| flhF | lpp1748 | Flagellar biosynthesis protein FlhF | IIb | 0.51 | 2.54 | - |
| flhA | lpp1749 | Flagellar biosynthesis protein FlhA | IIa | - | - | - |
| flhB | lpp1750 | Flagellar biosynthetic protein FlhB | IIa | 0.46 | - | - |
| fliR | lpp1751 | Flagellar biosynthetic protein FliR | IIa | - | - | - |
| fliQ | lpp1752 | Flagellar biosynthetic protein FliQ | IIa | - | - | - |
| fliP | lpp1753 | Flagellar biosynthetic protein FliP | IIa | - | 1.52 | - |
| fliO | lpp1754 | Flagellar protein FliO | IIa | 0.43 | 2.53 | - |
| fliN | lpp1755 | Flagellar motor switch protein FliN | IIa | 0.47 | 2.76 | 3.53 |
| fliM | lpp1756 | Flagellar motor switch protein FliM | IIa | 0.47 | 2.37 | 3.30 |
| motA2 | lpp2266 | Proton conductor component of motor, | III | 0.31 | - | 3.14 |
| motB2 | lpp2267 | Flagellar motor protein | III | 0.27 | - | 5.86 |
| flhB‘ | lpp2635 | Putative part of flagellar apparatus | III | 0.46 | - | - |
| motY | lpp3034 | Sodium-type flagellar protein MotY | IV | - | - | - |
Analysis of the transcription results of the Δ_letA_ strain and the Δ_rsmYZ_ double mutant as well as direct comparison between the two mutants in the PE phase showed, as expected, only slight differences in the expression patterns and transcript levels of most of the genes (the complete dataset is available at http://genoscript.pasteur.fr). Nevertheless, microarray analyses highlighted some genes and gene clusters which were expressed in Δ_rsmYZ_ but strongly down-regulated in Δ_letA_ (Table 2). Most surprisingly, these comprised the genes necessary for flagellar biosynthesis as they were expressed or even induced in the Δ_rsmYZ_ but not in Δ_letA_ as compared to the wild type strain (e.g. flgB-L, fleN, flgM, flaA) (Table 3). To further analyze these unexpected results we undertook western blot experiments with anti-FlaA antibodies. These revealed that FlaA protein was indeed expressed in wt and Δ_rsmYZ_, but not in Δ_letA_ (data not shown) indicating that, in contrast to Δ_letA_ strains, Δ_rsmYZ_ synthesizes a flagellum. We observed the Δ_letA_, Δ_rsmZ_ and Δ_rsmYZ_ strains under the electron microscope; this confirmed that the different rsm mutants did indeed synthesize flagella (Figure 7).
Furthermore, rpoS (lpp1247), lqsR (lpp2788) and 6 GGDEF/EAL protein-coding genes (lpp0351, lpp0352, lpp0809, lpp0952, lpp1170 and lpp2355) showed significantly lower expression in Δ_letA_ as compared to a Δ_rsmYZ_ strain, with all of these genes encoding proteins putatively implicated in regulating L. pneumophila motility. Another protein previously implicated in influencing flagella expression, letE, encoding the Legionella transmission enhancer protein LetE (Bachman and Swanson, 2004b; Hammer et al., 2002), seems also to play an important role as letE expression was induced nearly 3 fold in the Δ_letA_ strain suggesting that LetA directly or indirectly represses letE expression, and that the repression of flagellum synthesis in a Δ_letA_ strain may be linked to repression of letE.
Discussion
Bacterial regulatory RNAs participate in a broad range of cellular processes, including carbon storage and utilization, response to oxidative stress, quorum sensing, transition to stationary phase and virulence. Two major classes of ncRNAs are described. They can interact with 5’ leader regions of target mRNAs by base paring, facilitated by the RNA chaperone Hfq or they have high affinity to RNA binding proteins of the RsmA/CsrA family (for a review see (Repoila and Darfeuille, 2009)). Small RNAs that bind RsmA/CsrA are typically produced under the positive control of TCS like GacA/GacS of P. aeruginosa. In this way, the small RNAs expand the scope of genetic control exerted by the TCS.
In L. pneumophila no ncRNAs were characterized to date. However, L. pneumophila encodes a CsrA homologue of E. coli and a TCS homologous to GacA/GacS of Pseudomonas or BarA/UvrY of E. coli, whose regulatory activity is linked through ncRNAs in these bacteria. The present study was thus initiated to decipher the regulatory cascade under the control of the LetA/LetS two-component system (TCS) and the CsrA signaling pathway, to learn whether L. pneumophila also employs ncRNAs to link the two systems, and to identify the functions controlled by the L. pneumophila CsrA-pathway. Although many Gram-negative bacteria possess the Gac/Rsm signaling pathway, the target genes that are translationally regulated by this system vary considerable. In L. pneumophila, both systems had previously been shown to be implicated in regulating differentiation from a replicative/non-virulent form to a transmissve/virulent form (Forsbach-Birk et al., 2004; Hammer et al., 2002; Lynch et al., 2003; Molofsky and Swanson, 2003, , 2004). LetA/LetS was shown to influence the formation of the virulent phenotype of L. pneumophila, since it was reported to control the progression into the transmissive phase (Gal-Mor and Segal 2003; Hammer et al., 2002; Lynch et al., 2003). In contrast, CsrA activates replication and represses transmissive traits, as it was reported to repress motility, pigmentation, sodium resistance, cytotoxicity and efficient macrophage infection (Fettes et al., 2001; Molofsky and Swanson, 2003).
Here we showed that L. pneumophila indeed employs ncRNAs, RsmY and RsmZ, two small RNAs predicted by a bioinformatics approach (Kulkarni et al., 2006) that link this signaling pathway. At the onset of the transmissive phase, LetA induces the expression of rsmY and rsmZ by binding to an upstream LetA binding site with the proposed consensus sequence TNAGAAATTTCTNA, as shown by in vitro binding assays of purified LetA to a DNA fragment containing the predicted binding site (Figure 2). Subsequently RsmY and RsmZ bind CsrA, as evidenced by the in vitro binding of these two small ncRNAs to purified CsrA of L. pneumophila. The small ncRNAs of L. pneumophila are able to functionally complement the CsrA-regulating small RNAs of E. coli. Indeed, our study revealed that over-expression of the L. pneumophila rsmY and rsmZ gene in E. coli induces premature glycogen synthesis and increased biofilm formation, two known target traits of CsrA in E. coli (Wang et al., 2005; Yang et al., 1996). This suggests that L. pneumophila RsmY and RsmZ are able to interact with E. coli CsrA. However, the E. coli and L. pneumophila ncRNAs belong to two different classes of CsrA interacting RNAs. The ones similar to CsrB binding to E. coli CsrA are around 300 bp and those similar to RsmY, RsmZ, RsmX binding to RsmA of e.g. P. fluorescence are about 100 bp (Babitzke and Romeo, 2007). They differ also in the number of GGA motifs present mainly in the loops. It has been shown that mutation of the GGA motifs leads to substantial reduction in CsrA binding affinity (Dubey et al., 2005). There are 22 GGA motifs present in CsrB of E. coli but only 6 and 5 in RsmY and RsmZ of L. pneumophila, respectively. Still, RsmY and RsmZ of L. pneumophila are able to mimic CsrB or CsrC activity in E. coli, showing that the Csr-like systems in the different bacteria function by very similar mechanisms.
To further understand the impact of RsmY and RsmZ on virulence and its influence on the progression of L. pneumophila to the transmissive phase, single and double knockout mutants were constructed and analyzed. The double mutant was drastically affected in virulence and appeared unable to transit to a new host (Figure 5A). Interestingly, the intracellular replication defect was much stronger in the Δ_rsmYZ_ double mutant than in the Δ_letA_ mutant strain (Figure 5A). This might be due to the fact that a basal level of RsmY and RsmZ expression still remained in a Δ_letA_ strain as judged from transcriptome analysis, but these two small RNAs were completely missing in the Δ_rsmYZ_ double mutant. Thus, RsmY and RsmZ are crucial regulators of the transmissive/virulent phenotype of L. pneumophila and are indispensible for virulence of L. pneumophila.
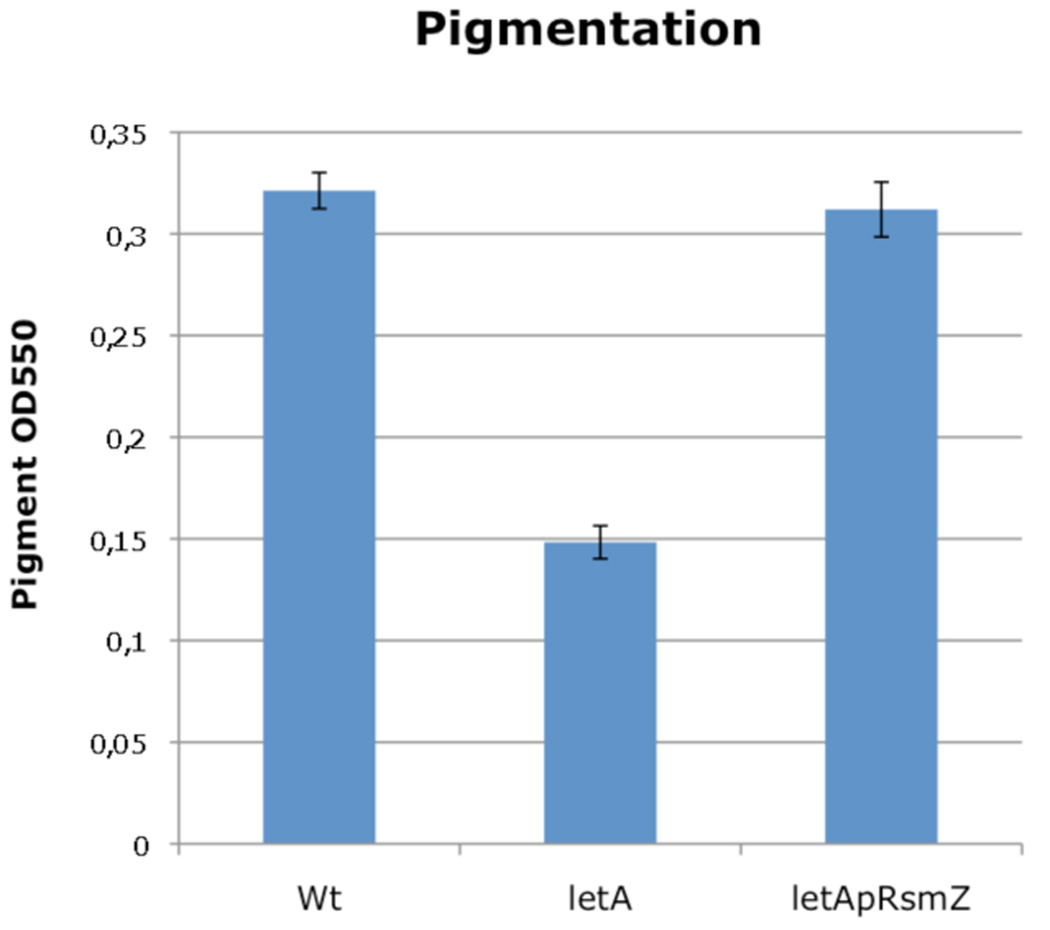
Direct regulation by CsrA is post-transcriptional and occurs by CsrA binding to the mRNA of target genes (Babitzke and Romeo, 2007). Thus the target genes for which CsrA affects translation but not transcript levels will not be detected in standard microarray experiments. We therefore undertook a bioinformatics search for the occurrence of multiple GGA motifs in the vicinity of the Shine-Dalgarno sequence, a known feature of CsrA targets in other bacteria combined with a structural analysis of the identified sequences. By this approach 42 putative targets of CsrA were predicted (supplementary material Figure S1). Among those are LqsR, a recently identified pleiotropic regulator whose expression was reported to be influenced by RpoS and, to a lesser extent, also by LetA (Tiaden et al., 2007; Spirig et al., 2008), but also several flagella biosynthesis genes like fleQ, fliI, fliH, flgE, fliQ and fliK indicating that some influence of CsrA on flagella biosynthesis might exist. The bioinformatics approach did not predict lly, coding legiolysin, responsible for the pigmentation phenotype, as a target. However, when overexpressing RsmZ in the Δ_letA_ mutant strain, pigmentation was restored, suggesting that lly may be a target of CsrA (Figure 8). Another target of CsrA might be Hfq, as shown for E. coli and suggested previously for L. pneumophila (McNealy et al., 2005).
Figure 8. RsmZ restores pigment production in a letA mutant strain.
Wilde type Legionella pneumophila and a letA mutant strain carrying p_rsmZ_ activate pigment production in PE growth phase, in contrast to a letA mutant strain.
Comparison of the effect of the mutations of rsmYZ, letA and letS as compared to wt L. pneumophila by whole genome transcriptional profiling in different growth phases, revealed many similar but also different traits. The transcriptional changes induced by the LetA/LetS TCS were in line with phenotypes previously reported (Hammer et al., 2002) and also observed in this study, like lack of flagella (Figure 7), reduced pigmentation and sodium sensitivity (Figure 6), or reduced infectivity and less efficient intracellular replication (Figure 5). In particular genes defined previously as typical replicative traits (Brüggemann et al., 2006b) were expressed in all mutants in the PE (transmissive) phase, whereas several genes coding proteins characteristic of the transmissive/virulent phenotype were down-regulated. The genes affected by these changes have been shown to play central roles in the virulence of L. pneumophila. For example, SidF contributes to apoptosis resistance in _L. pneumophila_-infected cells by specifically interacting and neutralizing the effects of two pro-apoptotic members of the Bcl2 protein family (Banga et al., 2007). SidC is a phosphatidylinositol-4 phosphate-binding type IV substrate that recruits endoplasmic reticulum vesicles to a replication-permissive vacuole (Weber et al., 2006b; Ragaz et al., 2008). The patatin-like protein PatA/VipD was reported to assist in the establishment of a vacuole suitable for bacterial replication by interfering with multi-vesicular body formation at the late endosome and ER to Golgi transport (Shohdy et al., 2005; VanRheenen et al., 2006). SdeA is secreted early during macrophage infection, suggesting that it is important in the initial formation of the replicative vacuole (Bardill et al., 2005). Taken together, our results from gene expression profiling of regulatory mutants identify the genes implicated in the observed phenotype of letA and letS mutants and confirm as suggested previously that the TCS LetA/LetS regulates the switch from replicative/non-virulent to transmissive/virulent L. pneumophila (Hammer et al., 2002). Furthermore, we show here for the first time, that this regulation is dependent on two small ncRNAs. Thus, LetA/LetS is part of a Csr-like regulatory system in Legionella as previously proposed (Molofsky and Swanson, 2003).
If the LetA/LetS system only regulates RsmY and RsmZ, as suggested by a genome wide stringent search for additional LetA binding sites, a rsmYZ double mutant and a letA or letS mutant should show the same transcriptional profile. Indeed, in the transmissive phase the rsmYZ double mutant had a strong global impact on gene expression, very similar to the changes seen for a letA or a letS mutant. However, we observed one significant difference. The expression of the genes necessary for flagella biosynthesis were repressed in the letA mutant but induced or unchanged in the rsmYZ double mutant with respect to the wild type (Table 3). This result was unexpected, as it had been postulated that flagella biosynthesis in L. pneumophila was regulated through the LetA/LetS–CsrA signaling pathway (Fettes et al., 2001; Heuner and Steinert, 2003; Molofsky and Swanson, 2003, 2004). However, we demonstrate here that although letA mutants do not synthesize flagella, rsmY, rsmZ and rsmYZ mutants synthesize flagella as judged by electron microscopy (Figure 7) and transcriptional profiling and flagellin is produced as assessed by western blot analysis with anti-FlaA antibodies (data not shown). Thus our results suggest that flagella expression in L. pneumophila is largely independent of the RsmYZ-CsrA pathway but is clearly dependent on the LetA/LetS TCS. This is different from what is seen in E. coli where CsrA positively regulates motility (Wei et al., 2001) but similar to Photorhabdus luminescens, where flagella synthesis is reported to be independent of the CsrA pathway but dependent on BarA/UvrY, a homologous system to LetA/LetS (Krin et al., 2008).
Our transcriptome data also showed that expression of letE, encoding the LetA/LetS enhancer protein previously reported to enhance flagella synthesis (Bachman and Swanson, 2004b; Hammer et al., 2002) is 3-fold induced in a Δ_letA_ strain but not in a ΔrsmYZ mutant strain. Previous reports show that letE is mainly expressed during the replicative phase (Bachman and Swanson, 2004b), a stage where indeed no flagellum is synthesized. This might suggest, that in a Δ_letA_ strain the direct or indirect repression of letE expression by LetA is abolished when Legionella progresses into the transmissive phase. Thus, in a Δ_letA_ mutant, LetE may be further produced and may continue to repress flagella biosynthesis genes like in replicative phase. In contrast, in the presence of LetA letE is repressed in the transmissive phase and flagella repression is released.
Furthermore, according to the transcriptional profile, LetA, but not RsmYZ, influences the expression of genes coding the pleiotropic regulator LqsR and several GGDEF/EAL proteins. GGDEF/EAL proteins have been shown to repress motility in various bacteria (Romling, 2005). In contrast, the transcript level of the gene coding the sigma factor RpoS is strongly induced only in the ΔrsmYZ mutant during the PE phase. Furthermore it was recently shown that RpoS also influences rsmY and rsmZ expression (Hovel-Mine et al., 2009), suggesting that a feedback loop between the two systems exists. Thus RsmY and RsmZ seem to have some influence on RpoS expression, a regulator known to positively influence flagella expression. But not only RpoS also the other regulators discussed above are all known to influence motility and virulence in Legionella (Bachman and Swanson, 2004a; Hickman and Harwood, 2008; Tiaden et al., 2007; Weber et al., 2006a). Therefore, our results suggest that rsmYZ and LetA may directly or indirectly regulate the transcription of other regulators or regulatory elements important for flagella regulation and, moreover, alter the cyclic-diGMP level, which may influence motility by controlling the amount of specific GGDEF/EAL proteins.
However, impact of the CsrA pathway on flagella biosynthesis cannot be completely ruled out as a constitutively expressing CsrA strain was reported to have reduced motility in PE growth phase and it shows only 30% flagellin expression as compared to a wt strain (Molofsky and Swanson, 2003; Fettes et al., 2001). Thus, another, yet undiscovered small ncRNA, e.g. like RsmX of Pseudomonas fluorescence (Kay et al., 2005) could be present also in L. pneumophila. In P. fluorescence it was shown, that only a triple rsmX, rsmY, rsmZ mutation caused the same phenotypic effect as those observed in a gacA or gacS mutant (Kay et al., 2005). This arrangement may allow a more efficient regulatory response via a gene dosages effect and may allow fine-tuning of the LetA/LetS system in response to different stimuli. Furthermore, in other systems where small RNAs play a role, like in the control of quorum sensing in Vibrio harvey it has been shown that the action of small RNAs can be additive (Tu and Bassler, 2007). Search for this putative third small ncRNA and further analysis of CsrA and the different regulators involved in the CsrA signaling pathway are under way to shed light on this issue.
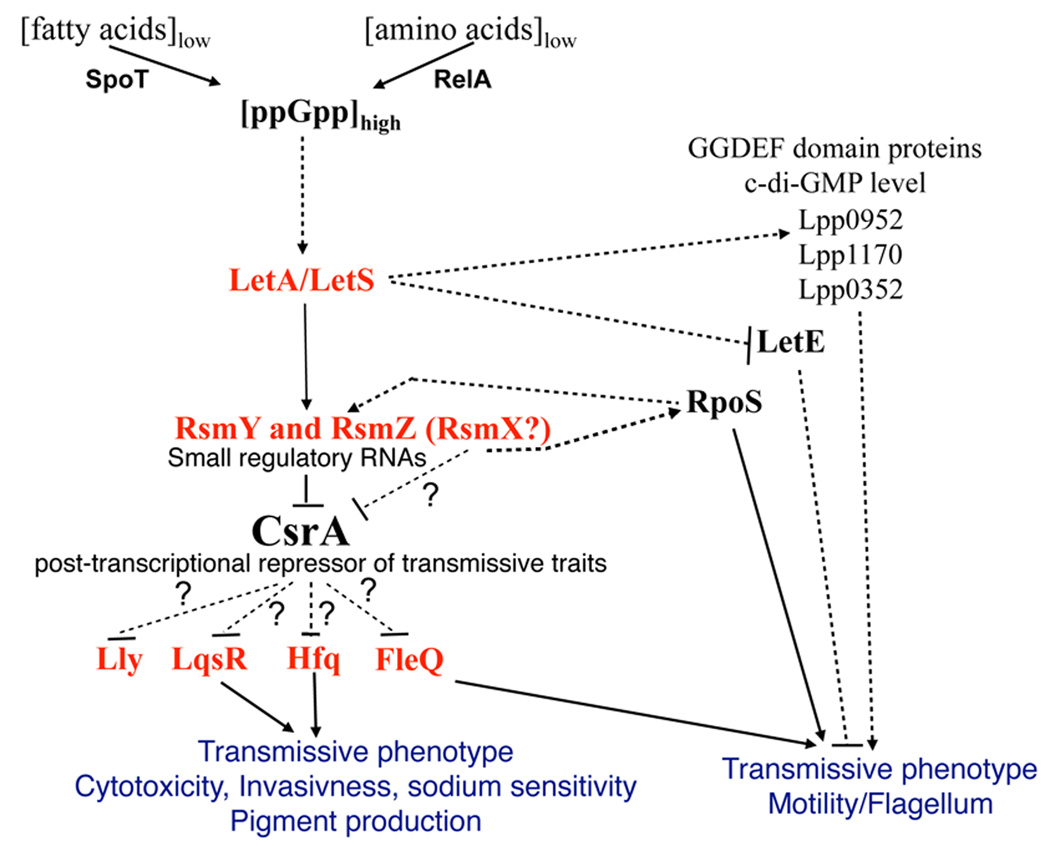
On the basis of the data presented here and of previously published work (reviewed in Molofsky and Swanson, 2004) we propose a refined model of this signaling network (Figure 9). Upon entering the PE phase LetS is auto-phosphorylated after having received a yet unknown signal (presumably a starvation signal related to the formation of ppGpp (Edwards et al., 2009; Hammer and Swanson, 1999) and then, in turn, phosphorylates LetA. Subsequently, activated LetA binds to a LetA-binding site in close proximity to the promoter region of the regulatory sRNA genes rsmY and rsmZ and therewith induces their expression. The RNA transcripts form a distinctive secondary structure, which is able to bind to and to sequester CsrA. Thus, CsrA is released from mRNA transcripts of its targets, which then allows either their translation or their decay. In L. pneumophila, this results in the production of transmissive phase traits, and probably also in the decay of mRNAs of replicative traits, thus allowing the cell to prepare itself for the last stage of the infectious cycle, the evasion from the host cell and the infection of a new host. Flagella biosynthesis appears to be independent of the RsmYZ-CsrA signaling pathway and seems mainly to be regulated by LetA, LetE, RpoS and probably specific levels of cyclic-diGMP controlled through the regulation of specific GGDEF/EAL proteins.
Figure 9. Model of the regulatory network governing differentiation of L. pneumophila from replicative/non virulent to transmissive/virulent.
During the transmissive phase, amino acid starvation triggers the ppGpp synthetase RelA and fatty acid starvation triggers the ppGpp synthetase/hydrolase SpoT to produce the alarmone (p)ppGpp. (p)ppGpp may be sensed by the sensor kinase LetS which then phosphorylates LetA. Phosphorylated LetA binds upstream of the small ncRNAs RsmY and RsmZ and activates their transcription. CsrA that is bound near the ribosomal binding site of its mRNAs targets (e.g. probably LqsR, FleQ and Hfq) inhibits their translation. The presence of RsmY and RsmZ titrate CsrA away from its targets, which then enables translation of the mRNAs, leading to expression of transmissive phenotypes. In contrast flagella biosynthesis does not seem to be dependent on the RsmYZ-CsrA-pathway but is controlled by the LetA/LetS TCS, probably via LetE repression. Furthermore, LetA/LetS influences directly or indirectly ci-di-GMP levels which also may influence motility.
However, it is also possible that the CsrA pathway has more influence on flagella biosynthesis as evidenced here, if other not yet identified ncRNAs are implicated in the LetA/CsrA signaling pathway. Thus, determination whether other ncRNAs are part of this regulatory signaling cascade or not, is an important question for the future. Equally important is the elucidation of direct targets of CsrA, which would greatly enhance our understanding of this regulatory network in L. pneumophila.
Materials and Methods
Strains, media, growth conditions and A. castellanii and THP-1 infection assay
L. pneumophila strains Paris and Philadelphia JR32 were cultured in _N_-(2-acetamido)-2-aminoethanesulfonic acid (ACES)-buffered yeast extract broth or on ACES-buffered charcoal-yeast (BCYE) extract agar at 37°C. A. castellanii ATCC50739 was cultured in PYG 712 medium (2% proteose peptone, 0.1% yeast extract, 0.1 M glucose, 4 mM MgSO4, 0.4 M CaCl2, 0.1% sodium citrate dihydrate, 0.05 mM Fe(NH4)2(SO4)2 x 6H2O, 2.5 mM NaH2PO3, 2.5 mM K2HPO3) at 20°C. THP-1 human monocytes were grown in RPMI 1640 GlutaMAX medium (GIBCO) supplemented with 10% foetal bovine serum at 37°C, 5% CO2.
For in vivo growth of L. pneumophila Paris and its derivatives in A. castellanii we followed a protocol previously described (Brüggemann et al., 2006b). In brief, three days old cultures of A. castellanii were washed in infection buffer (PYG 712 medium without proteose peptone, glucose, and yeast extract) and adjusted to 105−106 cells per ml. Stationary phase Legionella grown on BCYE agar, diluted in water were mixed with A. castellanii at a MOI of 0.01. After allowing invasion for 1 hour at 37°C the A. castellanii layer was washed twice with infection buffer (start point of time-course experiment). Intracellular multiplication was monitored using a 300µl sample, which was centrifuged (14500 rpm) and vortexed to break up amoeba. 72 hours before the infection cells were seeded into 12 well tissue culture trays (Falcon) at a density of 3 × 105 cells/well and pretreated with 10−8 M phorbol 12-myristate 13-acetate (SIGMA) to induce differentiation into macrophage-like cells. Stationary phase Legionella were resuspended in RPMI 1640 serum free medium and added to THP-1 cell monolayers at a MOI of 10. After 1 hour of incubation cells were treated with 100 ug ml−1 gentamycin for 1 hour to kill extracellular bacteria. Infected cells were then washed with PBS before incubation with serum-free medium. At 24, 48 and 72 THP-1 were lysed with 0.1% tritonX-100. The number of colony forming units (CFU) of Legionellae was determined by plating on BCYE agar. Each infection was carried out in duplicates.
Plasmids and L. pneumophila mutant construction
Mutant strains of L. pneumophila were constructed as described previously (Brüggemann et al., 2006b; Heuner et al., 2002). In brief, the gene of interest was inactivated by introduction of a kanamycin resistance (kanR) cassette into the chromosomal gene. To construct the letA mutant strain, the chromosomal region containing the letA gene was PCR-amplified with the primers LetA_for TTGATGTGGATCCGGAAAAT and LetA_rev TAGTACGGTGCCTTCCTCGT. The product was cloned into the pGEM vector (Promega), the resulting plasmid was digested with _Xho_I and ligated to the kanamycin resistance cassette, which was amplified via PCR from the plasmid pGEM-KanR, using primers containing _Xho_I restriction sites at the ends (Kan_XhoI_for AGCTCGAGAAAGCAGGTAGCTTGCAG, Kan_XhoI_rev AGCTCGAGCCGCTGCTGGTTTCCTGG). A second letA mutant strain (in strain Philadelphia, kindly provided by M. Swanson, (Hammer et al., 2002)) was also used in this study. For the letS mutant, the chromosomal region for letS was amplified using the primers LetS_for CCGAGTTTACCATGAACGCA and LetS_rev GGATGACACCACAAGCTGAT and the resulting 2.9 kb PCR fragment was cloned into the pGEM T easy vector (Promega). The resulting plasmid was digested with HindIII and the HindIII-kanamycin resistance cassette was inserted as described before.
To construct the rsmY mutant, the corresponding intergenic region between lpp0004 and lpp0005 plus ca. 400 bp up- and downstream sequence of rsmY was PCR-amplified with the primers rsmY_for AGGAAGATCCTAACTACCATCGCTT and rsmY_rev ATAGAGGAAGGCGCCTATATTCAAC and cloned into pGEM T easy vector (Promega). On this template, an inverse PCR was performed using the primers rsmY_inv_HindIII_for AGAGAAGCTTATTCTCTAATTTACATTAGTATACC and rsmY_inv_HindIII_rev AGAGAAGCTTAGTTGACTTCCTGTCAGACATAT. These primers contained a HindIII restriction site, and amplified the pGEM backbone and the 400 bp flanking regions of rsmY. The resulting PCR product was HindIII-digested and ligated to the kanR cassette (amplified via PCR from the plasmid pGEM-KanR (Kanamycin resistance cassette (kindly provided by Klaus Heuner) inserted into a pGEM vector), using primers containing HindIII restriction sites at the ends (Kan_HindIII_for CTAAGCTTGCAGTGGGCTTACATG, Kan_HindIII_rev CTAAGCTTCGGGGGCATGGATGTGCGGATA). For chromosomal recombination, the construct (i.e. PCR fragment containing the kanR cassette with flanking regions of the gene of interest) was introduced into L. pneumophila by transformation.
For complementation experiments, pRsmZ was introduced into the rsmZ single mutant and A. castellanii infection assay were performed at presence of 0.25mM IPTG. As control L. pneumophila Paris wild type and the rsmZ mutant containing the empty plasmid pTS-10 were used, respectively. All mutants and plasmids used in this study are listed in Table 4.
Table 4.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties1 | Reference |
|---|---|---|
| E. coli | ||
| DH5α | Invitrogen | |
| BL21 | Invitrogen | |
| L. pneumophila | ||
| CIP 107629 | Virulent L. pneumophila serogroup 1, strain Paris | [5] |
| JR32 | Virulent L. pneumophila serogroup 1, strain Philadelphia | [65] |
| Δ_letA_ | Paris letA::Km | This study |
| Δ_letS_ | Paris letS::Km | This study |
| Δ_rsmY_ | Paris rsmY::Cm | This study |
| Δ_rsmZ_ | Paris rsmZ::Km | This study |
| Δ_rsmYZ_ | Paris rsmZY::Km::Cm | This study |
| Plasmids | ||
| pGEM-T Easy | Cloning of PCR products, Ap | Promega |
| pMMB207C | Legionella expression vector, Δ_mobA_ | [35] |
| pMMB207C–RBS-lcsC | Legionella expression vector for LcsC (with ribosome binding site) | [55] |
| pTS-10 | pMMB207C–RBS | [40] |
| p_rsmY_ | pMMB207C, rsmY | This study |
| p_rsmZ_ | pMMB207C, rsmZ | This study |
| p_letA__Strep | pMMB207C, letA with C-terminal Strep-Tag | This study |
| pIVEX2.3d | Expression of C-terminal hexa-histidine tag fusions | Roche Diagnostics |
| pIVEX2.3d–csrA | pIVEX2.3d, csrA | This study |
RsmY and RsmZ overexpression in L. pneumophila
For overexpression of RsmZ in L. pneumophila we used the plasmid pMMB207C–RBS-lcsC (derived from pTS-10; kindly provided by H. Hilbi, see (Weber et al., 2006b)). The lcsS gene was removed by cutting the plasmid with EcoRI and BamHI, and replaced with the rsmZ gene, which was amplified from chromosomal DNA with primers containing _EcoR_I and _BamH_I restriction sites (rsmZ_EcoRI_for GGCGAATTCGACTTATATGGATATGAGTC, rsmZ_BamHI_rev GCGGGATCCAAAAGTAAGCCAGGGACTC), respectively. The resulting plasmid (pRsmZ) and the control plasmid pTS-10 were introduced via electroporation into L. pneumophila strain Paris. For overexpression of RsmY in L. pneumophila the same plasmid and cloning strategy was used. For amplification of the rsmY genes primers rsmY_EcoRI_for GGCGAATTCTGACATGGATATGTCTGACAGGA and rsmY_BamHI_rev GCGGGATCCATACTAATGTAAATTAGAGAATAAGTGC, respectively were used. The resulting plasmid (pRsmY) and the control plasmid pTS-10 were introduced via electroporation into L. pneumophila strain JR32. For overexpression IPTG (100µM) was added to the plasmid-carrying strains at ODs of 0.5 and OD 1. Samples for RNA extraction were taken after 90 min of induction.
RsmY/RsmZ overexpression in E. coli
RsmY and RsmZ were amplified and cloned into pGEM (Promega), by using the following primers: rsmY_for AGGAAGATCCTAACTACCATCGCTT, rsmY_rev ATAGAGGAAGGCGCCTATATTCAAC and rsmZ_for CCAGCTATAGCACAAACC, rsmZ_rev GTTACCCAGACAAACACATG. The resulting plasmids (pGEM-rsmY, pGEM-rsmZ) were introduced into E. coli BL21 by electroporation.
Determination of E. coli intracellular glycogen content
Cell samples (about 2–5×108 cells) were collected by centrifugation (4000_g_, 4 min, 4°C), washed once with 1 ml cold water and the pellet stored at −20°C until further use. Cells were resuspended in 250 µL 0.25 M Na2CO3 and heated at 95°C for 2 h with occasional stirring. The suspension was adjusted to pH 5.2 with 150 µL of 1 M acetic acid and 600 µL of 0.2 M sodium acetate (pH 5.2). This mixture was incubated overnight at 57 °C with continuous shaking in the presence of 100 µg of α-amyloglucosidase (Boehringer, 208464), freshly prepared as a 10 mg mL−1 stock solution made in 0.2 M sodium acetate (pH 5.2). The released glucose was determined with the glucose oxidase reagent (Sigma, kit 510-A) in 96-well microplates and glycogen content was expressed in µg equivalent glucose per unit of OD600.
Qualitative assessment of the E. coli glycogen content was carried out using the iodine-staining method (Govons et al., 1969). Briefly, cells were grown overnight in Luria-Bertani (LB) liquid medium. An LB agar plate containing 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was then streaked with large amounts of bacteria, which were grown for 7 hours at 37°C. Then a iodine crystal was positioned in the lid of the Petri dish positioned upside down. In 10 min, iodine vapor stained the cells according to the amount of glycogen they contained.
Biofilm formation assay
The initial steps of biofilm formation were assayed by determining the ability of cells to adhere to the wells of 96-well polyvinyl chloride (PVC) microtiter dishes (Falcon) as described previously (Roux et al., 2005). The medium (LB for E. coli, BYE for L. pneumophila, 100 µl/well) was inoculated with a 1:100 dilution from an overnight culture. After inoculation, the plates were incubated at 37°C for 24 h (72 h in the case of L. pneumophila) and then rinsed and 125 µl of a 1% solution of crystal violet was added to each well. The plates were incubated at room temperature for 15 min and rinsed. For analyzing biofilm formation, the crystal violet was solubilized by addition of 200 µL of ethanol-acetone (80:20), and the OD570 was determined. The results are averages of four replicate wells from three independent experiments.
Pigmentation and sodium sensitivity
For quantifying pigment accumulation, 1 ml samples - obtained from 5 day old broth cultures grown at 37°C - were centrifuged at 16,000 × g for 10 min and supernatants measured at OD550nm. Sodium sensitivity was tested by plating dilutions of PE broth cultures on BYE agar containing or lacking 100 mM NaCl. CFUs were counted and percentage of sodium sensitive bacteria were calculated.
LetA overexpression, purification and Electrophoretic Mobility Shift Assay (EMSA)
To detect binding of LetA (Lpp2699) protein to the upstream regions of the rsmY and rsmZ genes we performed gel shift assays using the LightShift Chemiluminescent EMSA kit (Pierce, Rockford). For purification of the LetA protein, the letA gene was cloned into the vector pMMB207C using the primers letA_for TCTAGAATGATTAAAGTATTAATTGTTGATGAC and letA_Strep_rev CTGCAGTTACTTCTCAAATTGAGGATGTGACCAATCATCTTGTGACAAATCATTGG. The resulting plasmid (pletA_Strep) was sequenced and introduced into E. coli BL21, and the LetA Strep-tag fusion protein was over expressed and purified with a Strep-Tactin sepharose column according to the instructions of the manufacturer (IBA, Göttingen).
A set of complementary pairs of 35mers HPLC-purified rsmY and rsmZ oligonucleotides were used as substrates containing the predicted LetA binding site and a biotin tag at the 5’end: rsmY_upstream_for CTCTTCTTAATTGAGAAATTTCTTACATATTACTGC, rsmY_upstream_rev GCAGTAATATGTAAGAAATTTCTCAATTAAGAAGAG; rsmZ_upstream_for GTAATTGATGTAAGAAATTTCTCAATTCCATAGA, rsmZ_upstream_rev TCTATGGAATTGAGAAATTTCTTACATCAATTAC. As control peptide a random sequence containing motifs of the rsmY and rsmZ oligonucleotides, but without LetA binding site was used (control_for GTAATTGCAATTCCATAGATATTACTGCCATTCAGA, control_rev TCTGAATGGCAGTAATATCTATGGAATTGCAATTAC). Binding assays of purified LetA protein with biotin-labeled DNA probes were performed at room temperature for 30 min. Binding reactions contained: 0.2 µM biotin end-labeled target DNA, LetA (0–6 µM), 1 x binding buffer, 5% glycerol, 5 mM MgCl2. As nonspecific competitor 50 ng/µl poly(dI-dC) was used; as specific competitor unlabeled target DNA was added (4pmol). Subsequently, the binding reaction was loaded on an 8% native polyacrylamide gel, followed by electrophoretic transfer to a Nylon membrane. Cross-linking was performed under UV-light at 120 mJ/cm2 for 60 sec. The biotin-labeled DNA probe was visualized on thy nylon membrane by chemiluminescence using a streptavidin-horseradish peroxidase conjugate and the chemiluminescence substrate of the manufacturer
CsrA overexpression and purification
Full-length cDNAs encoding CsrA (Lpp0845) were amplified by PCR using primer CsrA-for ATACCATGGTGATTTTGACTCGGCGTATAG and CsrA-rev CCCCCCGGGTACTGCTTGCGAATCAGATTC. Fragments were cloned in frame into the expression vector pIVEX2.3d (Roche Diagnostics), which adds a hexa-histidine tag to the C-terminus of the protein; positive clones in E. coli DH5α were selected on 50 µg/ml ampicillin and sequences were verified. E. coli BL21 (Invitrogen) cells were used for protein overexpression. The cells were grown in M9 minimal medium, containing 1 mM MgSO4, 0.1 mM CaCl2, 0.2% (w/v) glucose, 50 µg ml−1 ampicilin, at 16°C. Protein production was induced by adding 0.5 mM IPTG at A600 ∼0.5, and cells were harvested late exponential phase by centrifugation at 4°C. Cells from one liter of culture were resuspended in 1 ml buffer A – PBS buffer (120 mM NaCl, 4 mM KH2PO4, 16 mM Na2HPO4, pH 7.8), 2 mM MgCl2, 5 mM DTT, 5% (v/v) glycerol and a cocktail of protease inhibitors (Sigma) at the concentration recommended by the manufacturer. Cells were disrupted by sonication, centrifuged (18,000 × g, 30 min, 4°C) and the supernatant was applied to a Ni-NTA affinity column (GE Healthcare) equilibrated with buffer A including 40 mM imidazole. The column was washed with the same buffer containing 100 mM imidazole then the enzyme was eluted with 500 mM imidazole in buffer A. Fractions were dialyzed against buffer A and concentrated by centrifugation (Microcon, 3 kDa cut-off, Millipore) to a final concentration of 0.1 mg protein/ml. Proteins were quantified according to Bradford using BSA as standard. Aliquots of 20 µl were frozen in liquid N2 and kept at −80°C until further use. The quality of the purification was determined after SDS-PAGE analysis (4% stacking gel and 12% running gel) and staining with Brilliant Blue G - Colloidal Concentrate (Sigma).
In vitro transcription of RsmY/RsmZ-RNA and EMSA gel mobility assay
RsmY and RsmZ was amplified from bacterial DNA with the primer pairs RsmY-for TGACATGGATATGTCTGAC/RsmY-rev AAAGGTATACTAATGTAAATTAG and RsmZ-for GACTTATATGGATATGAGTC/RsmZ-rev AAAAGTAAGCCAGGGACTC, respectively, and cloned under the control of a T7 promoter into pGEMT-easy vector. Insertion was verified by sequencing and correct inserts were used in MEGAscript T7 Kit (Ambion) to produce in vitro RNA; 2µM of Biotin-16-UTP was added into the reaction mix for later detection. As negative control, empty pGEMT-easy vector cut with NdeI was used as template to produce in vitro RNA without CsrA binding motif. The reaction was incubated at 37°C for 12h and RNA was purified by Phenol/Chloroform extraction according to the supplier’s protocol. RNA concentration was estimated by UV absorption at 260nm. For 10µl interaction assays, 100nM of RNA was combined with varying concentrations of purified CsrA-His (0–5µM) and incubated in buffer A in presence of 10ng tRNA yeast (Invitrogen) and 1U Rnase Out (Invitrogen) for 30min. Subsequently, samples were fractionated under non-denaturing conditions on Blue-Native PAGE (Schägger and von Jagow, 1991), 1991) and blotted to Imobilon-P transfer membranes (Millipore). Membranes were blocked in PBS buffer containing 0.1% Tween-20 and 1% BSA for 1h at RT and, afterwards, incubated for 1h in the same buffer including streptavidin-horseradish peroxidase conjugate (Invitrogen). RNA-bands were visualized with ECL Plus Western blotting solutions (GE Healthcare) and detected with Chemi-Smart 5000 (Vilber Loumert).
RNA Isolation, labeling and microarray hybridization
Total RNA was extracted as previously described (Milohanic et al., 2003). Paris wild type and mutant strains were grown in AYE medium, and harvested for RNA isolation at the exponential (OD1 or OD 2.5) and post-exponential growth phase (OD. 4.3). RNA was prepared in triplicates (three independent cultures) and each RNA sample was hybridized twice to the microarrays (dye swap). RNA was reverse-transcribed with Superscript indirect cDNA kit (Invitrogen) and labeled with Cy5 or Cy3 (Amersham Biosciences) according to the supplier’s instructions. The design of microarrays containing gene-specific 70mer oligonucleotides based on all predicted genes of the genome of L. pneumophila strain Paris (CR628336) and its plasmid (CR628338) was previously described (Brüggemann et al., 2006a). Hybridization was performed following the manufacturers’ recommendations (Corning) using 250 pmol of Cy3 and Cy5 labeled cDNA. Slides were scanned on a GenePix 4000A scanner (Axon Instruments). Laser power and/or PMT were adjusted to balance the two channels and the resulting files were analyzed using Genepix Pro 4.0 software. Spots were excluded from analysis in case of high local background fluorescence, slide abnormalities, or weak intensity.
Data analysis and statistics
Data normalization and differential analysis were conducted using the R software (http://www.R-project.org). No background subtraction was performed, but a careful graphical examination of all the slides was conducted to ensure a homogeneous, low-level background in both channels. A loess normalization (Yang et al., 2002) was performed on a slide-by-slide basis (BioConductor package marray; http://www.bioconductor.org/packages/bioc/stable/src/contrib/html/marray.html). Differential analysis was carried out separately for each comparison between two time points, using the VM method (VarMixt package (Delmar et al., 2005)), together with the Benjamini and Yekutieli (Reiner et al., 2003) p-value adjustment method. If not stated otherwise, only differently expressed genes with 2-fold-changes were taken into consideration. Empty and flagged spots were excluded from the data set, and only genes with no missing values for the comparison of interest were analyzed. The complete dataset is available at http://genoscript.pasteur.fr in a MIAME compliance public database maintained at the Institut Pasteur and was submitted to the ArrayExpress database maintained at http://www.ebi.ac.uk/microarray-as/ae/ under the Acc. No A-MEXP-1539.
Real-time PCR
Real-time PCR was performed as described previously (Brüggemann et al., 2006b) at cDNA concentrations from 100 pg to 0.1pg. Two biological repetitions were carried out and each transcript was quantified twice. The genes tested were lpp0952, ralF, rpoS, rsmZ, lpp1170, lpp0351, fis2, csrA (for letA mutant flaA and mip in addition). Primers used are listed in supplementary material, Table S2. As control lpp1040 and csrA were used. Both were confirmed by microarray data to be stably expressed housekeeping genes under these conditions. Values of relative induction were calculated according to (Pfaffl, 2001), by group-wise comparison of induced samples versus control samples.
Acknowledgements
We like to thank T Romeo for providing CsrA antibodies, P Glaser for his interest and critical comments, SV Gordon for critical reading of the manuscript, ZD Dalebroux and M Swanson for helpful discussions, S Rousseau and I Moszer (Pasteur Genopole Ile-de-France -- PF4 Genome analysis and Integration) for curating and updating the transcriptome database, O Sismeiro and JY Coppée for spotting of arrays (Pasteur Genopole Ile-de-France – PF2 DNA microarrays), MC Prevost for electron microscopy (Ultrastructural Microscopy). Financial support was received from the Institut Pasteur, from the Network of Excellence “Europathogenomics” LSHB-CT-2005–512061 and NIH Grant 2 R01 AI44212. H. Brüggemann was holder of a fellowship of the German Academy of Natural Scientists Leopoldina (funding number BMBF-LPD9901/8–101), M. Jules received support from PTR 185 (Institut Pasteur –Biorad), M. Lomma is holder of a Marie Curie fellowship (Early stage training in infectious diseases) financed by the European Commission in the framework of the INTRAPTH project MEST-CT-2005–020715 and C. Albert-Weissenberger was holder of a research Fellowship of the Bavarian Research Foundation (Bayerische Forschungsstiftung)
References
- Al-Khodor S, Kalachikov S, Morozova I, Price CT, Abu Kwaik Y. The PmrA/B two component system of Legionella pneumophila is a global regulator required for Intracellular Replication within macrophages and protozoa. Infect Immun. 2009;77:374–386. doi: 10.1128/IAI.01081-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albert-Weissenberger C, Cazalet C, Buchrieser C. Legionella pneumophila - a human pathogen that co-evolved with fresh water protozoa. Cell Mol Life Sci. 2007;64:432–448. doi: 10.1007/s00018-006-6391-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman E, Segal G. The response regulator CpxR directly regulates expression of several Legionella pneumophila icm/dot components as well as new translocated substrates. J Bacteriol. 2008;90:1985–1996. doi: 10.1128/JB.01493-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol. 2007;10:156–163. doi: 10.1016/j.mib.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Bachman MA, Swanson MS. Genetic evidence that Legionella pneumophila RpoS modulates expression of the transmission phenotype in both the exponential phase and the stationary phase. Infect Immun. 2004a;72:2468–2476. doi: 10.1128/IAI.72.5.2468-2476.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman MA, Swanson MS. The LetE protein enhances expression of multiple LetA/LetS-dependent transmission traits by Legionella pneumophila. Infect Immun. 2004b;72:3284–3293. doi: 10.1128/IAI.72.6.3284-3293.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banga S, Gao P, Shen X, Fiscus V, Zong WX, Chen L, Luo ZQ. Legionella pneumophila inhibits macrophage apoptosis by targeting pro-death members of the Bcl2 protein family. Proc Natl Acad Sci U S A. 2007;104:5121–5126. doi: 10.1073/pnas.0611030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardill JP, Miller JL, Vogel JP. IcmS-dependent translocation of SdeA into macrophages by the Legionella pneumophila type IV secretion system. Mol Microbiol. 2005;56:90–103. doi: 10.1111/j.1365-2958.2005.04539.x. [DOI] [PubMed] [Google Scholar]
- Brüggemann H, Cazalet C, Buchrieser C. Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. Curr Opin Microbiol. 2006a;9:86–94. doi: 10.1016/j.mib.2005.12.009. Epub 2006 Jan 2006. [DOI] [PubMed] [Google Scholar]
- Brüggemann H, Hagman A, Jules M, Sismeiro O, Dillies M, Gouyette C, Kunst F, Steinert M, Heuner K, Coppée J, Buchrieser C. Virulence strategies for infecting phagocytes deduced from the in vivo transcriptional program of Legionella pneumophila. Cell Microbiol. 2006b;8:1228–1240. doi: 10.1111/j.1462-5822.2006.00703.x. [DOI] [PubMed] [Google Scholar]
- Catrenich CE, Johnson W. Characterization of selective inhibition of growth of virulent Legionella pneumophila by supplemented Mueller-Hinton Medium. Infect Immun. 1989;57:1862–1864. doi: 10.1128/iai.57.6.1862-1864.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalet C, Rusniok C, Bruggemann H, Zidane N, Magnier A, Ma L, Tichit M, Jarraud S, Bouchier C, Vandenesch F, Kunst F, Etienne J, Glaser P, Buchrieser C. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- Chen DE, Podell S, Sauer JD, Swanson MS, Saier MHJ. The phagosomal nutrient transporter (Pht) family. Microbiology. 2008;154:42–53. doi: 10.1099/mic.0.2007/010611-0. [DOI] [PubMed] [Google Scholar]
- Chen J, de Felipe KS, Clarke M, Lu H, Anderson OR, Segal G, Shuman HA. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303:1358–1361. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- Cui Y, Chatterjee A, Chatterjee AK. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp.carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and harpinEcc. Mol Plant Microbe Interact. 2001;14:516–526. doi: 10.1094/MPMI.2001.14.4.516. [DOI] [PubMed] [Google Scholar]
- Dalebroux ZD, Edwards RL, Swanson MS. SpoT governs Legionella pneumophila differentiation in host macrophages. Mol Microbiol. 2009;71:640–658. doi: 10.1111/j.1365-2958.2008.06555.x. [DOI] [PubMed] [Google Scholar]
- Delmar P, Robin S, Daudin JJ. VarMixt: efficient variance modelling for the differential analysis of replicated gene expression data. Bioinformatics. 2005;21:502–508. doi: 10.1093/bioinformatics/bti023. [DOI] [PubMed] [Google Scholar]
- Dubey AK, Baker CS, Romeo T, Babitzke P. RNA sequence and secondary structure participate in high-affinity CsrA-RNA interaction. RNA. 2005;11:1579–1587. doi: 10.1261/rna.2990205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R, Jules M, Buchrieser C, Swanson M. The multi-step design of the LetA/LetS two-component system increases Legionella pneumophila versatility. in preparation. 2009 [Google Scholar]
- Fettes PS, Forsbach-Birk V, Lynch D, Marre R. Overexpresssion of a Legionella pneumophila homologue of the E.coli regulator csrA affects cell size, flagellation, and pigmentation. Int J Med Microbiol. 2001;291:353–360. doi: 10.1078/1438-4221-00141. [DOI] [PubMed] [Google Scholar]
- Forsbach-Birk V, McNealy T, Shi C, Lynch D, Marre R. Reduced expression of the global regulator protein CsrA in Legionella pneumophila affects virulence-associated regulators and growth in Acanthamoeba castellanii. Int J Med Microbiol. 2004;294:15–25. doi: 10.1016/j.ijmm.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Gal-Mor O, Segal G. Identification of CpxR as a positive regulator of icm and dot virulence genes of Legionella pneumophila. J Bacteriol. 2003;185:4908–4919. doi: 10.1128/JB.185.16.4908-4919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal-Mor O, Segal G. The Legionella pneumophila GacA homolog (LetA) is involved in the regulation of icm virulence genes and is required for intracellular multiplication in Acanthamoeba castellanii. Microb Pathog. 2003;34:187–194. doi: 10.1016/s0882-4010(03)00027-5. [DOI] [PubMed] [Google Scholar]
- Govons S, Vinopal R, Ingraham J, Preiss J. Isolation of mutants of Escherichia coli B altered in their ability to synthesize glycogen. J Bacteriol. 1969;97:970–972. doi: 10.1128/jb.97.2.970-972.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer BK, Swanson MS. Co-ordination of Legionella pneumophila virulence with entry into stationary phase by ppGpp. Mol Microbiol. 1999;33:721–731. doi: 10.1046/j.1365-2958.1999.01519.x. [DOI] [PubMed] [Google Scholar]
- Hammer BK, Tateda ES, Swanson MS. A two-component regulator induces the transmission phenotype of stationary-phase Legionella pneumophila. Mol Microbiol. 2002;44:107–118. doi: 10.1046/j.1365-2958.2002.02884.x. [DOI] [PubMed] [Google Scholar]
- Heuner K, Dietrich C, Skriwan C, Steinert M, Hacker J. Influence of the alternative sigma(28) factor on virulence and flagellum expression of Legionella pneumophila. Infect Immun. 2002;70:1604–1608. doi: 10.1128/IAI.70.3.1604-1608.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuner K, Steinert M. The flagellum of Legionella pneumophila and its link to the expression of the virulent phenotype. Int J Med Microbiol. 2003;293:133–143. doi: 10.1078/1438-4221-00259. [DOI] [PubMed] [Google Scholar]
- Hickman JW, Harwood CS. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69:376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovel-Miner G, Pampou S, Faucher SP, Clarke M, Morozova I, Morozov P, Russo JJ, Shuman HA, Kalachikov S. {sigma}S controls multiple pathways associated with intracellular multiplication of Legionella pneumophila. J Bacteriol. [Epub ahead of print] 2009 doi: 10.1128/JB.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas K, Edwards AN, Simm R, Romeo T, Römling U, Melefors O. The RNA binding protein CsrA controls c-di-GMP metabolism by directly regulating the expression of GGDEF proteins. Mol Microbiol. 2008;70:236–257. doi: 10.1111/j.1365-2958.2008.06411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay E, Dubuis C, Haas D. Three small RNAs jointly ensure secondary metabolism and biocontrol in Pseudomonas fluorescens CHA0. Proc Natl Acad Sci U S A. 2005;102:17136–17141. doi: 10.1073/pnas.0505673102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krin E, Derzelle S, Bedard K, Adib-Conquy M, Turlin E, Lenormand P, Hullo MF, Bonne I, Chakroun N, Lacroix C, Danchin A. Regulatory role of UvrY in adaptation of Photorhabdus luminescens growth inside the insect. Environ Microbiol. 2008;10:1118–1134. doi: 10.1111/j.1462-2920.2007.01528.x. [DOI] [PubMed] [Google Scholar]
- Kulkarni PR, Cui X, Williams JW, Stevens AM, Kulkarni RV. Prediction of CsrA-regulating small RNAs in bacteria and their experimental verification in Vibrio fischeri. Nucleic Acids Res. 2006;34:3361–3369. doi: 10.1093/nar/gkl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapouge K, Schubert M, Allain FH, Haas D. Gac/Rsm signal transduction pathway of gamma-proteobacteria: from RNA recognition to regulation of social behaviour. Mol Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- Lenz DH, Miller MB, Zhu J, Kulkarni RV, Bassler BL. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol Microbiol. 2005;58:1186–1202. doi: 10.1111/j.1365-2958.2005.04902.x. [DOI] [PubMed] [Google Scholar]
- Liu MY, Gui G, Wei B, Preston JFr, Oakford L, Yüksel U, Giedroc DP, Romeo T. he RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- Lynch D, Fieser N, Gloggler K, Forsbach-Birk V, Marre R. The response regulator LetA regulates the stationary-phase stress response in Legionella pneumophila and is required for efficient infection of Acanthamoeba castellanii. FEMS Microbiol Lett. 2003;219:241–248. doi: 10.1016/S0378-1097(03)00050-8. [DOI] [PubMed] [Google Scholar]
- Milohanic E, Glaser P, Coppee JY, Frangeul L, Vega Y, Vazquez-Boland JA, Kunst F, Cossart P, Buchrieser C. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol. 2003;47:1613–1625. doi: 10.1046/j.1365-2958.2003.03413.x. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Legionella pneumophila CsrA is a pivotal repressor of transmission traits and activator of replication. Mol Microbiol. 2003;50:445–461. doi: 10.1046/j.1365-2958.2003.03706.x. [DOI] [PubMed] [Google Scholar]
- Molofsky AB, Swanson MS. Differentiate to thrive: lessons from the Legionella pneumophila life cycle. Mol Microbiol. 2004;53:29–40. doi: 10.1111/j.1365-2958.2004.04129.x. [DOI] [PubMed] [Google Scholar]
- Pernestig AK, Georgellis D, Romeo T, Suzuki K, Tomenius H, Normark S, Melefors O. The Escherichia coli BarA-UvrY two-component system is needed for efficient switching between glycolytic and gluconeogenic carbon sources. J Bacteriol. 2003;185:843–853. doi: 10.1128/JB.185.3.843-853.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragaz C, Pietsch H, Urwyler S, Tiaden A, Weber SS, Hilbi H. The Legionella pneumophila phosphatidylinositol-4 phosphate-binding type IV substrate SidC recruits endoplasmic reticulum vesicles to a replication-permissive vacuole. Cell Microbiol. 2008;210:2416–2433. doi: 10.1111/j.1462-5822.2008.01219.x. [DOI] [PubMed] [Google Scholar]
- Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol Cell. 2009;101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- Romling U. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci. 2005;62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Beloin C, Ghigo JM. Combined inactivation and expression strategy to study gene function under physiological conditions: application to identification of new Escherichia coli adhesins. J Bacteriol. 2005;187:1001–1013. doi: 10.1128/JB.187.3.1001-1013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadosky AB, Wiater LA, Shuman HA. Identification of Legionella pneumophila genes required for growth within and killing of human macrophages. Infect Immun. 1993;61:5361–5373. doi: 10.1128/iai.61.12.5361-5373.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer JD, Bachman MA, Swanson MS. The phagosomal transporter A couples threonine acquisition to differentiation and replication of Legionella pneumophila in macrophages. Proc Natl Acad Sci U S A. 2005;102:9924–9929. doi: 10.1073/pnas.0502767102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991;199:223–231. doi: 10.1016/0003-2697(91)90094-a. [DOI] [PubMed] [Google Scholar]
- Shohdy N, Efe JA, Emr SD, Shuman HA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci U S A. 2005;102:4866–4871. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig T, Tiaden A, Kiefer P, Buchrieser C, Vorholt JA, Hilbi H. The Legionella autoinducer synthase LqsA produces an alpha-hydroxyketone signaling molecule. J Biol Chem. 2008;283:18113–18123. doi: 10.1074/jbc.M801929200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert M, Hentschel U, Hacker J. Legionella pneumophila: an aquatic microbe goes astray. FEMS Microbiol Rev. 2002;26:149–162. doi: 10.1111/j.1574-6976.2002.tb00607.x. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Wang X, Weilbacher T, Pernestig AK, Melefors O, Georgellis D, Babitzke P, Romeo T. Regulatory circuitry of the CsrA/CsrB and BarA/UvrY systems of Escherichia coli. J Bacteriol. 2002;184:5130–5140. doi: 10.1128/JB.184.18.5130-5140.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson MS, Hammer BK. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu Rev Microbiol. 2000;54:567–613. doi: 10.1146/annurev.micro.54.1.567. [DOI] [PubMed] [Google Scholar]
- Tiaden A, Spirig T, Weber SS, Brüggemann H, Bosshard R, Buchrieser C, Hilbi H. The Legionella pneumophila response regulator LqsR promotes virulence as an element of the regulatory network controlled by RpoS and LetA. Cell Microbiol. 2007;9:2903–2920. doi: 10.1111/j.1462-5822.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- Tu KC, Bassler BL. Multiple small RNAs act additively to integrate sensory information and control quorum sensing in Vibrio harveyi. Genes Dev. 2007;21:221–233. doi: 10.1101/gad.1502407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRheenen SM, Luo ZQ, O’Connor T, Isberg RR. Members of a Legionella pneumophila family of proteins with ExoU (phospholipase A) active sites are translocated to target cells. Infect Immun. 2006;74:3597–3606. doi: 10.1128/IAI.02060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JP, Isberg RR. Cell biology of Legionella pneumophila. Curr Opin Microbiol. 1999;2:30–34. doi: 10.1016/s1369-5274(99)80005-8. [DOI] [PubMed] [Google Scholar]
- Wang X, Dubey AK, Suzuki K, Baker CS, Babitzke P, Romeo T. CsrA post-transcriptionally represses pgaABCD, responsible for synthesis of a biofilm polysaccharide adhesin of Escherichia coli. Mol Microbiol. 2005;56:1648–1663. doi: 10.1111/j.1365-2958.2005.04648.x. [DOI] [PubMed] [Google Scholar]
- Weber H, Pesavento C, Possling A, Tischendorf G, Hengge R. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol Microbiol. 2006a;62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- Weber SS, Ragaz C, Reus K, Nyfeler Y, Hilbi H. Legionella pneumophila exploits PI(4)P to anchor secreted effector proteins to the replicative vacuole. PLoS Pathog. 2006b;2:e46. doi: 10.1371/journal.ppat.0020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei BL, Brun-Zinkernagel AM, Simecka JW, Prüss BM, Babitzke P, Romeo T. Positive regulation of motility and flhDC expression by the RNA-binding protein CsrA of Escherichia coli. Mol Microbiol. 2001;40:245–256. doi: 10.1046/j.1365-2958.2001.02380.x. [DOI] [PubMed] [Google Scholar]
- Wiater LA, Sadosky AB, Shuman HA. Mutagenesis of Legionella pneumophila using Tn903 dlllacZ: identification of a growth-phase-regulated pigmentation gene. Mol Microbiol. 1994;11 doi: 10.1111/j.1365-2958.1994.tb00343.x. 641-353. [DOI] [PubMed] [Google Scholar]
- Yang H, Liu MY, Romeo T. Coordinate genetic regulation of glycogen catabolism and biosynthesis in Escherichia coli via the CsrA gene product. J Bacteriol. 1996;178:1012–1017. doi: 10.1128/jb.178.4.1012-1017.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 2002;30:e15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman T, Aloni G, Halperin E, Kotzer H, Degtyar E, Feldman M, Segal G. The response regulator PmrA is a major regulator of the icm/dot type IV secretion system in Legionella pneumophila and Coxiella burnetii. Mol Microbiol. 2007;63:1508–1523. doi: 10.1111/j.1365-2958.2007.05604.x. [DOI] [PubMed] [Google Scholar]