Eight blood pressure loci identified by genome-wide association study of 34,433 people of European ancestry (original) (raw)
. Author manuscript; available in PMC: 2010 Sep 21.
Published in final edited form as: Nat Genet. 2009 May 10;41(6):666–676. doi: 10.1038/ng.361
Abstract
Elevated blood pressure is a common, heritable cause of cardiovascular disease worldwide. To date, identification of common genetic variants influencing blood pressure has proven challenging. We tested 2.5m genotyped and imputed SNPs for association with systolic and diastolic blood pressure in 34,433 subjects of European ancestry from the Global BPgen consortium and followed up findings with direct genotyping (N≤71,225 European ancestry, N=12,889 Indian Asian ancestry) and in silico comparison (CHARGE consortium, N=29,136). We identified association between systolic or diastolic blood pressure and common variants in 8 regions near the CYP17A1 (_P_=7×10−24), CYP1A2 (_P_=1×10−23), FGF5 (_P_=1×10−21), SH2B3 (_P_=3×10−18), MTHFR (_P_=2×10−13), c10orf107 (_P_=1×10−9), ZNF652 (_P_=5×10−9) and PLCD3 (_P_=1×10−8) genes. All variants associated with continuous blood pressure were associated with dichotomous hypertension. These associations between common variants and blood pressure and hypertension offer mechanistic insights into the regulation of blood pressure and may point to novel targets for interventions to prevent cardiovascular disease.
The World Health Organization estimated that, in 2005, the annual death toll from cardiovascular disease reached 17.5 million worldwide1–3. Increases in systolic and diastolic blood pressure (SBP, DBP), even within the normal range, have a continuous and graded impact on cardiovascular disease risk and are major contributors in half of all cardiovascular deaths 2,3. Lifestyle influences, including dietary sodium intake, alcohol excess, elevated body mass index and lack of exercise, are known to increase blood pressure4. Studies of familial aggregation suggest that there is also a substantial heritable component to blood pressure5. Studies of rare Mendelian disorders of hypertension and hypotension have produced the most significant progress toward understanding the heritable basis of blood pressure, showing that mutations in genes influencing renal salt handling can have a severe impact on blood pressure6. Detailed study of these genes has identified rare variants (minor allele frequency [MAF] <0.1%) that impact blood pressure in the general population7 and evolving evidence suggests a potential role for common variation in some of the same genes8–10.
Identification of common variants affecting blood pressure using genome-wide association studies (GWAS) has proved challenging, compared to other common complex disorders11,12. However, meta-analysis of multiple studies with large total sample sizes has the potential to facilitate detection of variants with modest effects. We therefore formed the Global Blood Pressure Genetics (Global BPgen) consortium and conducted meta-analysis of GWAS in 34,433 individuals of European ancestry with SBP and DBP measurements (stage 1), followed by large-scale direct genotyping (stage 2a) and in silico (stage 2b) analyses (Supplementary Figure 1). Our analyses identified eight loci demonstrating genome-wide significant association with systolic or diastolic blood pressure, with each locus also providing substantial evidence for association with hypertension.
RESULTS
Genome-wide association for blood pressure
Global BPgen includes 17 cohorts of European ancestry ascertained through population-based sampling or case-control studies. In our primary analysis (stage 1), we examined individuals aged ≤70 years from 13 population-based studies and from control groups from 4 case-control studies (Table 1). Individuals treated for hypertension were imputed to have 15 mm Hg higher SBP and 10 mm Hg higher DBP than the observed measurementsas this has been shown to reduce bias and improve statistical power13. SBP and (separately) DBP measures were each adjusted for age, age2, body mass index and any study-specific geographic covariates within cohort- and gender-specific regression analyses. Genome-wide SNP genotyping was performed on a variety of platforms and subjected to standard quality control measures (Methods, Supplementary Table 1). Genotypes for ~2.5M autosomal SNPs in the HapMap CEU sample were then imputed in each study and tested for association under an additive genetic model with SBP and DBP separately. Test statistics from association analysis of SBP and DBP from each cohort were adjusted using genomic control14 to avoid inflation of results due to inter-individual relatedness or residual population stratification, and to ensure good calibration of test statistics. Meta-analysis of results was performed using inverse variance weights. Test statistic inflation post-meta-analysis was modest (λGC = 1.08 SBP; λGC = 1.07 DBP); genomic control correction was applied again. The plots of test statistics against expectations under the null suggest an excess of extreme values (cohort-specific and meta-analysis quantile-quantile plots are presented in Supplementary Figure 2).
Table 1. Study sample characteristics.
Study characteristics are shown for cohort samples examined in stage 1 meta-analysis (population-based and controls from case-control studies), stage 2a (direct genotyping follow-up) and stage 2b (in silico follow-up with the CHARGE consortium). Population Cohorts: The Baltimore Longitudinal Study of Aging (BLSA), British 1958 Birth Cohort- Wellcome Trust Case Control Consortium (B58C-WTCCC), British 1958 Birth Cohort – Type 1 Diabetes Genetics Consortium (B58C-T1DGC), Cohorte Lausannoise (CoLaus), European Prospective Investigation of Cancer- Norfolk-Genome Wide Association Study (EPIC-Norfolk-GWAS), Fenland Study (Fenland), Invecchiare in Chianti (InCHIANTI), Kooperative Gesundheitsforschung in der Region Augsburg (KORA), Northern Finland Birth Cohort of 1966 (NFBC1966), SardiNIA, Study of Health in Pomerania (SHIP), Supplementation en Vitamines et Minéraux Antioxydants (SU.VI.MAX) and TwinsUK. Controls from case-control studies: Diabetes Genetics Initiative (DGI), Finland-United States Investigation of NIDDM Genetics (FUSION), the Myocardial Infarction Genetics Consortium (MIGen), the Precocious Coronary Artery Disease (PROCARDIS) study. Direct genotyping: The Utrecht Atherosclerosis Risk in Young Adults (AYRA), British Genetics of Hypertension study – hypertension cases (BRIGHT-HTN), BRIGHT study normotensive controls (BRIGHT-NT), EPIC-Italy, EPIC-Norfolk-Replication cohort (EPIC-Norfolk-REP), Finrisk97, FUSION stage 2 controls (FUSION2), London Life Sciences Population (LOLIPOP), Malmö Diet and Cancer Cardiovascular Cohort (MDC), Malmö Preventive Project (MPP), Prevention of REnal and Vascular ENd stage Disease (PREVEND), Metabolic Syndrome in Men Study (METSIM), Prospect-EPIC cohort, Utrecht Health Project (UHP). NA = not available. HTN = hypertension
| Study | N | % women | Age (SD) years | SBP (SD) mm Hg | DBP (SD) mm Hg | BMI (SD) kg/m2 | % HTN# | % anti-hypertensive therapy |
|---|---|---|---|---|---|---|---|---|
| Stage 1 – GWAS | ||||||||
| Population-based cohorts | ||||||||
| BLSA | 708 | 44 | 42.4 (13.2) | 119.5 (15.0) | 77.3(10.2) | 24.5(3.6) | 23.2 | 5.2 |
| B58C – T1DGC* | 2,580 | 51 | 44.3 (0.3) | 121.7 (15.3) | 79.4 (10.5) | 27.4 (4.9) | 20.5 | 4.7 |
| B58C – WTCCC* | 1,473 | 50 | 44.9 (0.4) | 126.7 (15.2) | 79.1(10.2) | 27.4(4.7) | 17.4 | 4.2 |
| CoLaus | 4,969 | 53 | 51.7 (9.5) | 127.3 (17.4) | 79.4(10.8) | 25.8(4.6) | 33.9 | 16 |
| EPIC- Norfolk - GWAS | 2,100 | 54 | 57.2 (7.8) | 136.7 (19.1) | 83.9(11.9) | 26.3(3.9) | 45.6 | 16 |
| Fenland | 1,401 | 56 | 45.0 (7.3) | 122.8 (16.3) | 75.5 (10.7) | 27.1 (4.9) | 18.8 | 5.5 |
| InCHIANTI | 562 | 55 | 56.9 (14.5) | 138.4 (20.1) | 81.4(10.1) | 27.1(4.2) | 59.6 | 23.7 |
| KORA | 1,644 | 51 | 52.5 (10.1) | 133.4 (18.5) | 81.8(10.9) | 27.3(4.1) | 20.9 | 17 |
| NFBC1966* | 4,761 | 52 | 31* | 125.2 (13.8) | 77.5(11.7) | 24.6(4.2) | 21.7 | 2 |
| SardiNIA | 3,998 | 57 | 40.8 (15.3) | 128.7 (28.4) | 79.7(17.3) | 25.1(4.6) | 29.5 | 10 |
| SHIP | 3,310 | 53 | 45.0 (13.9) | 133.1 (20.2) | 83.5(11.3) | 26.9 (4.7) | 40.9 | 16.3 |
| SUVIMAX | 1,823 | 60 | 50.5 (6.2) | 120.9 (12.3) | 78.0(8.1) | 23.5(3.3) | 19.0 | 0 |
| TwinsUK | 873 | 100 | 45.8 (11.9) | 122.9 (15.4) | 78.2(10.3) | 24.8(4.6) | 27.3 | 22 |
| Controls from case-control studies | ||||||||
| DGI controls | 1,277 | 51 | 56.1 (8.7) | 133.3 (18.4) | 80.1(10.0) | 26.7(3.8) | 41.4 | 18 |
| FUSION NGT controls | 1,038 | 49 | 58.2 (10.7) | 139.4 (19.3) | 81.5(10.3) | 27.1(4.0) | 51.8 | 21 |
| MIGen controls | 1,121 | 38 | 48.9 (8.3) | 127.1 (17.8) | 80.2 (11.6) | 27.1 (5.2) | 36.4 | 13.4 |
| PROCARDIS controls | 795 | 37 | 58.9 (6.9) | 134.7 (18.6) | 82.8(10.0) | 25.9(3.70) | 15.0 | 2 |
| Stage 2-follow up | ||||||||
| 2a. Cohorts with direct genotyping data | ||||||||
| ARYA | 736 | 52 | 57.0 (6.0) | 125.0 (13.0) | 72.0(8.0) | 25.0(4.0) | 15.8 | 1 |
| BRIGHT-HTN | 2,445 | 59 | 57.1 (10.8) | 153.9 (20.8) | 94.0 (11.0) | 27.4 (3.8) | 100 | 91.2 |
| BRIGHT-NT | 673 | 77 | 55.5 (8.5) | 111.1 (6.9) | 71.2 (6.6) | 24.4 (3.2) | 0 | 0 |
| EPIC-Italy | 3,909 | 37 | 49.0 (7.6) | 132.5 (15.5) | 83.7(9.0) | 26.0(3.6) | 43.1 | 12.7 |
| EPIC-Norfolk-REP | 15,858 | 48 | 56.2 (7.6) | 133.8 (17.5) | 82.3 (11.0) | 26.3 (3.8) | 44 | 15 |
| Finrisk97 | 7,023 | 51 | 47.1 (12.4) | 134.9 (19.4) | 82.3 (11.3) | 26.6 (4.5) | 45.5 | 12.4 |
| FUSION2 | 1,162 | 37 | 57.5 (6.8) | 138.2 (19.5) | 83.9 (10.1) | 26.8 (3.8) | 8.9 | 1 |
| Lolipop (Europeans) | 6,006 | 35 | 51.2 (10.3) | 130.4 (19.1) | 79.6 (10.6) | 27.5 (5.1) | 39.9 | 20 |
| Lolipop (Indian Asians) | 12,823 | 36 | 48.8 (9.9) | 129.9 (19.1) | 80.8 (10.8) | 27.4 (4.5) | 42.9 | 25 |
| MDC-CC | 5,330 | 58 | 57.4 (5.9) | 141.0 (19.0) | 87.0(9.5) | 25.7(4.0) | 63.8 | 17 |
| METSIM | 5,934 | 0 | 58.1 (6.0) | 142.0 (17.9) | 89.8 (10.2) | 27.3 (4.2) | 69.6 | 40.5 |
| MPP** | 14,249 | 34 | 45.3 (7.1) | 125.0 (14.0) | 83.0(9.1) | 24.4(3.4) | 34.8 | 4 |
| PREVEND | 7,272 | 51 | 47.5 (11.4) | 127.7 (19.3) | 73.6 (9.7) | 25.9 (4.2) | 22.0 | 13.7 |
| Prospect-EPIC | 1,680 | 100 | 57.0 (6.0) | 133.0 (20.0) | 79.0(11.0) | 26.0(4.0) | 42.4 | NA |
| Utrecht Health Project | 2,829 | 52 | 40.0 (12) | 128.0 (19.0) | 79.0 (11.0) | 25.0 (4.0) | 32.9 | NA |
| 2b. Cohorts with in silico data | ||||||||
| CHARGE*** | 29,136 |
On meta-analysis of results from 34,433 individuals in stage 1, we observed 11 independent signals with P < 10−5 for SBP and 15 for DBP, with two results attaining P < 5×10−8, corresponding to genome-wide significance when adjusting for ~1m independent common variant tests estimated for samples of European ancestry (Supplementary Figure 3)15.
Follow-up of strongest SBP and DBP signals in additional samples
To strengthen support for association we undertook two analyses. First, we selected 12 SNPs for follow-up genotyping in up to 71,225 individuals drawn from 13 cohorts of European ancestry and up to 12,889 individuals of Indian Asian ancestry from one cohort (stage 2a, Table 1, Supplementary Figure 1, Supplementary Table 2). Second, we performed a reciprocal exchange of association results for 10 independent signals each for SBP and DBP (stage 2b, Supplementary Figure 1, Supplementary Table 3) with colleagues from the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) blood pressure consortium who had recently meta-analyzed GWAS data for SBP and DBP in 29,136 individuals, independent of Global BPgen (Table 1). Meta-analysis of the stage 1 Global BPgen GWAS and stage 2a direct and stage 2b in-silico association results identified genome-wide significant (P < 5×10−8) associations at eight loci: 1p36 in MTHFR, 10q24 near CYP17A1 and 17q21 in PLCD3 with SBP, 4q21 near FGF5, 10q21 in C10orf107, 12q24 near SH2B3, 15q24 near CYP1A2, and 17q21 near ZNF652 with DBP (Table 2, Figure 1, Supplementary Table 2, Supplementary Table 3, Supplementary Figure 3). Three of these loci overlap with genome-wide significant loci identified in the CHARGE analyses (10q24 for SBP and 12q24 and 15q24 for DBP).
Table 2. Loci associated with blood pressure.
Shown is the top SNP for each independent locus associated with systolic or diastolic blood pressure (P < 5×10−7) on joint analysis in up to 134,258 individuals of European ancestry from Global BPgen GWAS (stage 1), follow up genotyping (stage 2a) and in silico exchange with the CHARGE consortium (stage 2b). The eight genome-wide significant loci (P<5×10−8) are shown in bold. For stage 1 and 2b results based on imputed genotypes, an effective sample size is estimated to be the sum of the cohort-specific products of the imputation quality metric and the sample size. The total sample size is the sum of the effective sample sizes and the direct genotyping sample size. Effect sizes are on the mm Hg scale for increasing copy of the coded (alphabetically higher) allele as estimated by the beta coefficient in linear regression. The proportion of variance explained by each SNP is shown (r2). Meta-analysis was performed using inverse variance weighting. Note that loci 10q21 and 15q24 show results for two SNPs selected for validation genotyping in an interim analysis (rs1530440, rs1378942) that were genome-wide significant on joint analysis of stage 1+2a+2b. These two SNPs are highly correlated with alternate SNPs at the locus (rs4590817, rs4886606, respectively) with slightly stronger significance in the final stage 1 meta-analysis. The originally selected SNPs are shown throughout the text for consistency.
| Chromosome | GenesNearby | BPTrait | SNP ID(pos NCBI35)function | Codedallele | Stage | Codedallelefreq | N | Beta (SE)mm Hg | P | Beta(SE) | P | Ntotal |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Joint analysis stages 1+2a+2b | ||||||||||||
| MTHFR | ||||||||||||
| 1p36 | CLCN6 | SBP | rs17367504 | G | 1 | 0.14 | 34,158 | −0.79 (0.17) | 1×10−5 | |||
| NPPA | ||||||||||||
| NPPB | (11,797,044) | 2a | 0.16 | 19,751 | −0.93 (0.22) | 2×10−5 | r2=0.07% | |||||
| AGTRAP | ||||||||||||
| intron MTHFR | 2b | 0.16 | 29,064 | −0.85 (0.20) | 3×10−5 | -0.85 (0.11) | 2×10−13 | 82,973 | ||||
| 10q24 | c10orf26 | SBP | rs11191548 | T | 1 | 0.91 | 33,123 | 1.17 (0.23) | 3×10−7 | |||
| CYP17A1 | ||||||||||||
| c10orf32 | ||||||||||||
| AS3MT | (104,836,168) | 2a | 0.91 | 71,225 | 1.19 (0.15) | 9×10−15 | r2=0.08% | |||||
| CNNM2 | ||||||||||||
| NT5C2 | intergenic CNNM2/NT5C2 | 2b | 0.92 | 28,204 | 1.05 (0.27) | 9×10−5 | 1.16 (0.12) | 7×10−24 | 132,552 | |||
| 17q21 | SBP | rs12946454 | T | 1 | 0.28 | 32,120 | 0.68 (0.15) | 4×10−6 | ||||
| PLCD3 | ||||||||||||
| ACBD4 | ||||||||||||
| HEXIM1 | (40,563,647) | 2a | 0.25 | 17,877 | 0.43 (0.21) | 0.045 | r2=0.04% | |||||
| HEXIM2 | ||||||||||||
| FMNL1 | ||||||||||||
| intron PLCD3 | 2b | 0.27 | 27,693 | 0.50 (0.17) | 0.004 | 0.57 (0.10) | 1×10−8 | 77,690 | ||||
| 3q26 | MDS1 | DBP | rs1918974 | T | 1 | 0.54 | 32,674 | −0.28 (0.09) | 1×10−3 | |||
| (170,648,590) | 2a | 0.55 | 26,910 | −0.18 (0.08) | 0.04 | r2=0.03% | ||||||
| intron | 2b | 0.53 | 28,307 | −0.35 (0.09) | 8×10−5 | -0.27 (0.05) | 8×10−8 | 87,891 | ||||
| 4q21 | DBP | rs16998073 | T | 1 | 0.21 | 26,106 | 0.65 (0.11) | 7×10−9 | ||||
| PRDM8 | ||||||||||||
| FGF5 | (81,541,520) | 2a | 0.29 | 53,508 | 0.50 (0.07) | 6×10−13 | r2=0.09% | |||||
| C4orf22 | ||||||||||||
| upstream FGF5 | 2b | 0.24 | 22,009 | 0.36 (0.12) | 0.003 | 0.50 (0.05) | 1×10−21 | 101,623 | ||||
| 10q21 | DBP | rs1530440 | T | 1 | 0.19 | 32,718 | −0.51 (0.11) | 3×10−6 | ||||
| c10orf107 | ||||||||||||
| TMEM26 | ||||||||||||
| RTKN2 | (63,194,597) | 2a | 0.18 | 19,884 | −0.21 (0.11) | 0.05 | r2=0.04% | |||||
| RHOBTB1 | ||||||||||||
| ARID5B | ||||||||||||
| intron c10orf107 | 2b | 0.19 | 27,651 | −0.44 (0.12) | 1×10−4 | -0.39 (0.06) | 1×10−9 | 87,273 | ||||
| 12q24 | DBP | rs653178 | T | 1 | 0.53 | 30,853 | −0.46 (0.09) | 1×10−7 | ||||
| SH2B3 | (110,470,476) | 2a | 0.54 | 19,689 | −0.40 (0.10) | 3×10−5 | r2=0.09% | |||||
| ATXN2 | ||||||||||||
| intron ATXN2 | 2b | 0.52 | 29,119 | −0.50 (0.09) | 2×10−8 | -0.46 (0.05) | 3×10−18 | 79,661 | ||||
| 15q24 | DBP | rs1378942 | C | 1 | 0.36 | 34,126 | 0.48 (0.09) | 6×10−8 | ||||
| CYP1A1 | ||||||||||||
| CYP1A2 | ||||||||||||
| CSK | (72,864,420) | 2a | 0.35 | 71,086 | 0.41 (0.06) | 2×10−12 | r2=0.07% | |||||
| LMAN1L | ||||||||||||
| CPLX3 | ||||||||||||
| ARID3b | intron CSK | 2b | 0.33 | 29,046 | 0.43 (0.09) | 3×10−6 | 0.43 (0.04) | 1×10−23 | 134,258 | |||
| 17q21 | DBP | rs16948048 | G | 1 | 0.39 | 34,052 | 0.40 (0.09) | 5×10−6 | ||||
| ZNF652 | (44,795,465) | 2a | 0.37 | 19,752 | 0.23 (0.10) | 0.02 | r2=0.04% | |||||
| PHB | ||||||||||||
| upstream ZNF652 | 2b | 0.37 | 28,637 | 0.29 (0.09) | 0.002 | 0.31 (0.05) | 5×10−9 | 82,441 |
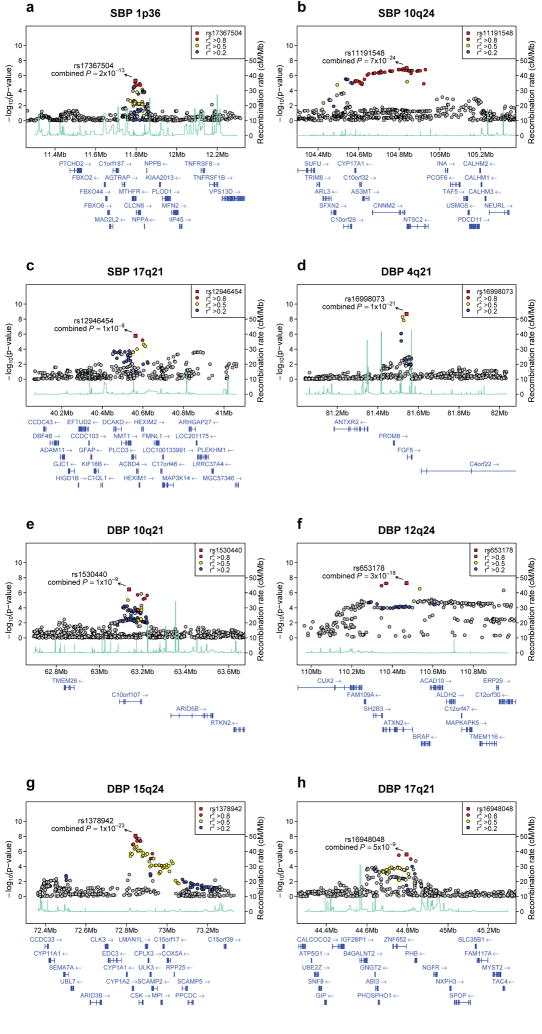
Figure 1.
Regional association plots of eight blood pressure loci. For each locus, we show the region extending to within 500kb of a SNP with P < 10−4 on either side. Statistical significance of associated SNPs at each locus are illustrated on the - log10(P) scale as a function of chromosomal position (NCBI Build 35). The sentinel SNP at each locus is shown in red. The correlation of the sentinel SNP to other SNPs at the locus is shown on a scale from minimal (gray and blue), to maximal (red). The meta-analysis result for stage 1 is shown with a red square. The joint analysis result (combined P) for stage 1+2a+2b is shown with an arrow. Fine-scale recombination rate from Myers et al50 is plotted in aqua.
For SBP, the strongest evidence for association was at 10q24 (rs11191548, MAF = 0.09, 1.16 mm Hg higher per major allele, P = 7×10−24, Table 2, Figure 1b). This SNP is part of a large cluster of associated SNPs spanning a ~430Kb region at 10q24 showing association in our GWAS meta-analysis. The locus includes six genes, most notably CYP17A1, which encodes the cytochrome P450 enzyme CYP17A1 (also known as P450c17) that mediates steroid 17α-hydroxylase and 17,20-lyase activity. The first enzymatic action is a key step in the biosynthesis of mineralocorticoids and glucocorticoids that affect sodium handling in the kidney and the second is involved in sex-steroid biosynthesis. Missense mutations in CYP17A1 cause one form of adrenal hyperplasia characterized by hypertension, hypokalemia, and reduced plasma renin and aldosterone levels16,17. None of the five other genes/transcripts in the region (Figure 1b) is an obvious candidate for blood pressure regulation.
The second locus associated with SBP was at 1p36 (rs17367504, MAF 0.14, 0.85 mm Hg lower SBP/minor allele, P = 2×10−13, Table 2, Figure 1a). This SNP is located in an intron of the MTHFR (methylenetetrahydrofolate reductase) gene in a region with many plausible candidate genes, including: MTHFR, CLCN6, NPPA, NPPB, and AGTRAP. The strongest signal in the locus is 6.4kb away from and uncorrelated with rs1801133 (C677T, A222V r2 CEU = 0.06), a coding variant that has been related to higher plasma homocysteine concentration18, pre-eclampsia19, and variably hypertension20. In Global BPgen rs1801133 was associated with 0.08 mm Hg higher SBP/T allele (P = 0.56), 0.24 mm Hg higher DBP (P = 0.01) and an odds ratio for hypertension of 1.00 (95% CI 0.94-1.05, P = 0.90).
The natriuretic peptides encoded by NPPA and NPPB, also located within the 1p36 associated interval, have vasodilatory and natriuretic properties and the NPPA knockout mouse has salt-sensitive hypertension21. A recent study found that the minor allele of rs5068 (43 kb from rs17367504, r2 CEU = 0.26), in the 3′ untranslated region of NPPA, is associated with higher plasma atrial natriuretic peptide and B-type natriuretic peptide, as well as lower SBP, DBP and odds of hypertension22. In the Global BPgen stage 1 meta-analysis we confirmed association of the minor allele of rs5068 with 0.97 mm Hg lower SBP (P = 3×10−4), 0.60 mm Hg lower DBP (P = 1 × 10−3) and 10% lower odds of hypertension (P = 0.04). Whether the associations of rs5068 and rs17367504 reflect the same or different underlying signals remains to be established. The less well-characterized gene CLCN6, also at the 1p36 locus, encodes a neuronally-expressed chloride channel that has not previously been implicated in blood pressure physiology, although rare mutations in other renally-expressed chloride channels have been associated with extremes of blood pressure23,24. Lastly, AGTRAP (encoding angiotensin II receptor-associated protein) negatively regulates angiotensin II signaling by interacting with the angiotensin II type 1 receptor, a critical component of the renin-angiotensin-aldosterone system and a target of antihypertensive therapy25.
The third locus associated with SBP was at 17q21 (rs12946454, MAF 0.28, 0.57 mm Hg higher SBP/minor allele, P = 1×10−8, Table 2, Figure 1c). This SNP is located in an intron in PLCD3 (phospholipase C-delta isoform), and is part of a cluster of associated SNPs. PLCD3 is a member of the phospholipase C family of enzymes; these are important in vascular smooth muscle signaling and are activated by the vasoactive peptides angiotensin II and endothelin26. Other genes of interest in the region include: HEXIM1 and HEXIM2 (encoding hexmethylene bis-acetamide inducible proteins 1 and 2). Both have been implicated in myocardial growth27, cardiac hypertrophy and inflammation28.
The DBP SNP with the strongest association evidence on joint analysis is rs1378942 (MAF = 0.36, 0.43 mm Hg higher/minor allele, P = 1×10−23, Table 2, Figure 1g), which is in an intron of CSK at 15q24. This is one of a cluster of associated SNPs spanning ~72kb. Genes in the region include CYP1A2 (cytochrome P450 enzyme), CSK (c-src tyrosine kinase), LMAN1L (lectin mannose-binding1 like) and ARID3b (encoding AT Rich Interacting Domain protein). Other nearby genes include CYP1A1 (~60kb) and C_YP11A1_ (~418kb). Cytochrome P450 enzymes are responsible for drug and xenobiotic chemical metabolism in the liver and cellular metabolism of arachidonic acid derivatives29, some of which influence renal function, peripheral vascular tone and blood pressure. CYP1A2 is widely expressed, representing 15% of CYP450 enzymes produced in the liver and mediating the metabolism of multiple medications (http://www.medicine.iupui.edu/Flockhart/table.htm). A correlated SNP, rs762551 (MAF = 0.31, r2 = 0.63, HapMap CEU) in an intron of CYP1A2 has been found to influence caffeine metabolism and recently association has been suggested between myocardial infarction risk and the allele associated with slow caffeine metabolism30. The ARID3B gene is embryonic lethal when knocked out in mouse, with branchial arch and vascular developmental abnormalities31, but is potentially interesting because of the presence of ARID5B at the 10q21 locus described below.
The second DBP SNP is rs16998073 (MAF = 0.21, 0.50 mm Hg higher/minor allele, P = 1×10−21, Table 2, Figure 1d) which lies 3.4kb upstream of FGF5 (fibroblast growth factor 5) on 4q21. The FGF5 protein is a member of the fibroblast growth factor (FGF) family that stimulates cell growth and proliferation in multiple cell types, including cardiac myocytes, and has been associated with angiogenesis in the heart32.
The third DBP SNP, rs653178 (MAF = 0.47, 0.46 mm Hg lower DBP/major allele, P = 3×10−18, Table 2, Figure 1f) at 12q24 is in an intron in the ATXN2 (Ataxin) gene. The SNP is in a cluster of strongly associated SNPs spanning 200kb. This SNP is perfectly correlated with a missense SNP in SH2B3 (rs3184504, R262W, r2 in CEU to rs653178 = 1.0, DBP P = 3×10−7 in stage 1 GWAS, change in log10(P) = 0.3 compared to rs653178). The minor allele of rs3184504, which is associated with higher DBP, has recently been associated with increased odds of type 1 diabetes33,34, celiac disease33,34, and most recently with eosinophil count, myocardial infarction, with a weak association with hypertension35. The SH2B3 protein (also known as lymphocyte-specific adapter protein, LNK) is one of a subfamily of SH2 domain-containing proteins and is implicated in growth factor, cytokine, and immunoreceptor signaling. In mice, it is primarily expressed in hematopoietic precursor cells, brain, testis and muscle36. There is some support for hypertension having an inflammatory component, possibly involving the adaptive immune system37. There are no prior studies linking blood pressure with type 1 diabetes or celiac disease. To explore this further, we looked up SNPs reported to be associated with T1D, celiac disease or myocardial infarction in the Global BPgen GWAS results and failed to find convincing association other than that for the SH2B3 missense SNP (data not shown). It is possible that the SH2B3 missense SNP impacts blood pressure through an action specific to cells outside of the immune system and that no direct link between blood pressure and autoimmune diseases exists.
The fourth DBP SNP, rs1530440 (MAF = 0.19, 0.39 mm Hg lower/minor allele, P = 1×10−9, Table 2, Figure 1e) at 10q21 is intronic and one of a cluster of SNPs in C10orf107, an open reading frame of unknown function. Nearby genes include ARID5B (AT rich interactive domain 5B (MRF1 like)), TMEM26 (transmembrane protein 26), RTKN2 (RhoA GTPase effector, rhotekin-2) and RHOBTB1 (RhoBTB GTPase). The Rho family of GTPases converts guanine triphosphate to inactive guanine diphosphate. The actions of other GTP-modulating enzymes may modulate salt-sensitive hypertension38,39. The ARID5B gene is a member of the AT-rich interaction domain family of transcription factors and is highly expressed in cardiovascular tissue and involved in smooth muscle cell differentiation40.
The fifth DBP SNP, rs16948048 (MAF 0.39, 0.34 mm Hg higher DBP/minor allele, P = 5×10−9, Table 2, Figure 1h) at 17q21 is upstream of ZNF652 (zinc finger protein 652) and PHB (prohibitin). Neither gene has previously been implicated in hypertension or other cardiovascular phenotypes.
We observed no significant interaction between the eight genome-wide significant SNPs and gender (P > 0.01, Supplementary Table 5). There was also no evidence of heterogeneity of effect across the samples examined for the eight SNPs (Q-statistic P > 0.05).
While we describe here promising candidates at each locus identified, the causal gene could be any of the genes around the association signal in each locus (Figure 1). Fine mapping and resequencing will be required to refine each association signal and to identify likely causal genetic variants which could be studied further in humans and in animal models.
All variants are related to both blood pressure traits
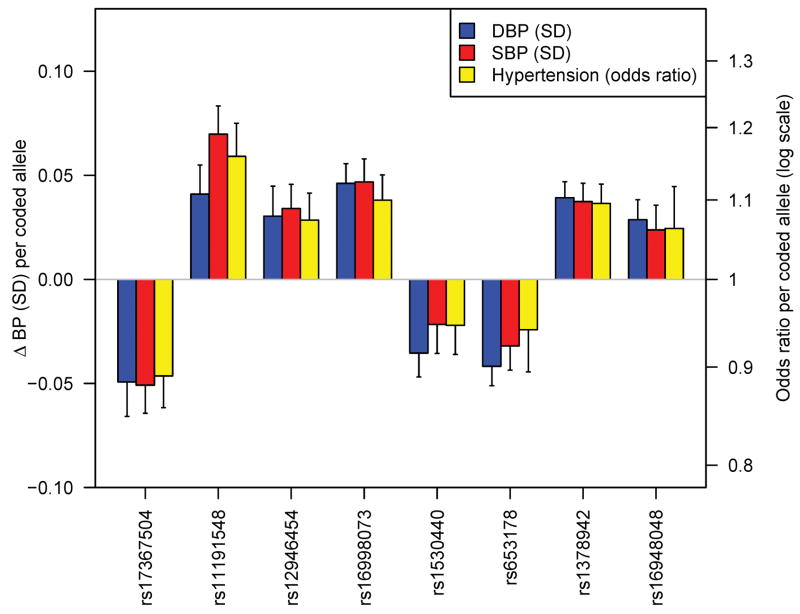
It remains to be clarified whether SBP or DBP is the better target for genetic investigation of blood pressure. The two traits are correlated and heritable, and both show strong increases with age, with DBP starting to plateau and in some individuals fall at ages above 60–65. Some have advocated the study of pulse pressure (SBP-DBP), which increases with advancing age, and is correlated positively with SBP and negatively with DBP and also shows evidence of heritability. In our GWAS and follow up, we chose a priori to consider SBP and DBP as separate traits. Thus, validation was only attempted for either SBP or DBP, according to the trait for which the stage 1 P value was lowest. Because SBP and DBP are correlated (r~0.50–0.70), it is perhaps not surprising to see that all eight genome-wide significant SNPs are associated with both SBP and DBP with the same directions of effect (Table 3, Figure 2). Thus, our presentation of results as SBP- or DBP-associated is somewhat arbitrary. The observation that each SNP shows stronger association with one trait or the other (typically by 1–2 orders of magnitude) could reflect sampling variation, small effect sizes or true differences in the underlying biologic basis of one trait or the other. A study designed to examine pulse pressure would be expected to show weaker (if any) association signals for the variants identified which all showed concordant effects on SBP and DBP.
Table 3. Relationship of SNPs at 8 genome-wide significant loci to both blood pressure traits.
For each of eight SNPs, the upper row shows association statistics for the blood pressure trait used for the analysis in which they were selected (SBP or DBP). The lower row (in bold) shows the equivalent association statistics for the alternate blood pressure trait. Results are shown for the 34,433 individuals in the stage 1 Global BPgen GWAS samples.
| SNP ID | Chr | Position (NCBI35) | Coded allele | Noncoded allele | Coded allele frequency | N (effective) | Trait | Beta mm Hg | SE | P |
|---|---|---|---|---|---|---|---|---|---|---|
| rs17367504 | 1 | 11,797,044 | G | A | 0.14 | 34,158 | SBP | −0.79 | 0.18 | 1×10−5 |
| DBP | −0.50 | 0.12 | 3×10−5 | |||||||
| rs11191548 | 10 | 104,836,168 | T | C | 0.91 | 33,123 | SBP | 1.17 | 0.22 | 3×10−7 |
| DBP | 0.56 | 0.15 | 2×10−4 | |||||||
| rs12946454 | 17 | 40,563,647 | T | A | 0.28 | 32,120 | SBP | 0.68 | 0.15 | 4×10−6 |
| DBP | 0.34 | 0.09 | 6×10−4 | |||||||
| rs16998073 | 4 | 81,541,520 | T | A | 0.21 | 26,106 | DBP | 0.65 | 0.11 | 7×10−9 |
| SBP | 0.74 | 0.17 | 1×10−5 | |||||||
| rs1530440 | 10 | 63,194,597 | T | C | 0.19 | 32,718 | DBP | −0.51 | 0.11 | 3×10−6 |
| SBP | −0.43 | 0.16 | 7×10−3 | |||||||
| rs653178 | 12 | 110,470,476 | T | C | 0.53 | 30,853 | DBP | −0.46 | 0.09 | 1×10−7 |
| SBP | −0.47 | 0.13 | 3×10−4 | |||||||
| rs1378942 | 15 | 72,864,420 | C | A | 0.36 | 34,126 | DBP | 0.48 | 0.09 | 6×10−8 |
| SBP | 0.62 | 0.13 | 2×10−6 | |||||||
| rs16948048 | 17 | 44,795,465 | G | A | 0.39 | 34,052 | DBP | 0.40 | 0.09 | 5×10−6 |
| SBP | 0.41 | 0.13 | 2×10−3 |
Figure 2.
Relationship of genome-wide significant loci to SBP, DBP and hypertension. Shown are the effects of each variant on continuous SBP and DBP and on the odds ratio for dichotomous hypertension compared to normotension (see Methods). For comparability, SBP and DBP effects are shown on the standard deviation scale (SBP SD = 16.6 mm Hg, DBP SD = 10.9 mm Hg). Alleles are coded as shown in Table 2.
All variants are related to hypertension
We did not perform a global GWAS of hypertension, which is expected to be underpowered to detect common variants of modest incremental effects on continuous blood pressure. For the eight SNPs that were genome-wide significant in continuous trait analysis, we examined the association with hypertension (SBP ≥ 140 mm Hg or DBP ≥ 90 mm Hg or antihypertensive medication use) compared to normotension (SBP ≤ 120 mm Hg and DBP ≤ 85 mm Hg and no antihypertensive medication use) in planned secondary analyses (N range = 57,410 – 99,802). All alleles associated with continuous blood pressure were also associated with odds of hypertension in directions consistent with the continuous trait effect (Table 4, Figure 2). The relative yields of the two approaches remain to be fully evaluated and will only become clearer upon completion of large ongoing GWA studies of dichotomous hypertension case-control samples. However, when we examined the hypertension association of each of the 8 SNPs genome-wide significantly associated with continuous SBP or DBP in just the stage 1 Global BPgen samples, 4 had 0.01 < P ≤ 0.10. These SNPs would not have been selected for follow-up genotyping had these tests been conducted as part of a hypertension GWAS. Thus, the study of continuous blood pressure allowed us to identify effects on risk of hypertension that would not have been readily discovered in a GWAS of hypertension drawn from these samples.
Table 4. Association of 8 SBP- and DBP-associated loci with hypertension.
Shown are the results for the top SNP from each genome-wide significant SBP or DBP locus from a logistic regression analysis of the odds of hypertension compared to normotension (see Methods). For comparison, the effect of the coded allele on the continuous blood pressure trait is shown. The inverse-variance-weighted meta-analysis results are shown. BP = blood pressure, OR = odds ratio.
| SNP ID | Chr | position (NCBI35) | Continuous Trait | Coded allele | Coded allele frequency | Continuous BP effect | HTN OR | HTN 95% CI | HTN P | N |
|---|---|---|---|---|---|---|---|---|---|---|
| rs17367504 | 1 | 11,797,044 | SBP | G | 0.14 | ↓ | 0.89 | 0.86 – 0.93 | 2×10−9 | 62,803 |
| rs11191548 | 10 | 104,836,168 | SBP | T | 0.91 | ↑ | 1.16 | 1.11 – 1.21 | 3×10−13 | 99,153 |
| rs12946454 | 17 | 40,563,647 | SBP | T | 0.28 | ↑ | 1.07 | 1.04 – 1.11 | 2×10−5 | 57,410 |
| rs16998073 | 4 | 81,541,520 | DBP | T | 0.19 | ↑ | 1.10 | 1.07 – 1.13 | 7×10−10 | 73,756 |
| rs1530440 | 10 | 63,194,597 | DBP | T | 0.19 | ↓ | 0.95 | 0.91 – 0.98 | 2×10−3 | 83,156 |
| rs653178 | 12 | 110,470,476 | DBP | T | 0.53 | ↓ | 0.93 | 0.91 – 0.96 | 8×10−7 | 60,030 |
| rs1378942 | 15 | 72,864,420 | DBP | C | 0.37 | ↑ | 1.10 | 1.07 – 1.12 | 2×10−14 | 99,802 |
| rs16948048 | 17 | 44,795,465 | DBP | G | 0.39 | ↑ | 1.06 | 1.03 – 1.09 | 1×10−4 | 62,411 |
Extension to non-European samples
To date, the majority of complex disease association signals reaching genome-wide significance have been concentrated in populations of European ancestry, and it remains unclear whether these findings will transfer to individuals with other genetic backgrounds. We genotyped all stage 2a SNPs (four of which were not confirmed in the European ancestry analyses) in a separate Indian Asian sample of up to 12,889 individuals. We replicated the association of the SNP at 4q21 near FGF5 (rs16998073, P = 5×10−4, Supplementary Table 2) and the SNP at 10q24 near CYP17A1 (rs11191548, P = 0.008, Supplementary Table 2). We did not replicate association of the SNP rs1378942 at CYP1A2 (P = 0.17, same direction), which could reflect limited power to detect the modest effect size, differences in linkage disequilibrium patterns in Indian Asians compared to Europeans, or simply lack of association in individuals of Indian Asian ancestry. The marked allele frequency differences between the European samples (C allele frequency ~0.35), the Indian Asian samples (0.77) and HapMap YRI (1.00) suggest distinct patterns of genetic variation at this locus across populations. A signal of positive selection has been suggested at the locus41 raising the potential functional importance of genetic variation in the region.
DISCUSSION
The eight loci described here and the additional loci reported by our colleagues in the CHARGE consortium are among the first confirmed associations between common genetic variants and blood pressure. Each association explains only a very small proportion of the total variation in SBP or DBP (~0.05–0.10%, approximately 1 mm Hg/allele SBP or 0.5 mm Hg/allele DBP, Table 2). However, the variants identified here have an aggregate effect on blood pressure, acting throughout the range of values (not just hypertensive), which has been shown to produce meaningful population changes in cardiovascular and stroke risk. For example, 2 mm Hg lower SBP, across the range of observed values, has been estimated to translate into 6% less stroke and 4% less coronary heart disease.42
Given the modest effects observed here and the limited power of this study to detect such effects, it is likely that many more common variants exist with weak effects upon blood pressure. This study illustrates the value of well-powered meta-analysis and follow-up genotyping, accompanied by in silico analysis, requiring the coordinated efforts of investigators across multiple studies, to establish definitively the relationship of these loci with blood pressure regulation in the general population.
In a companion paper, the CHARGE consortium reports as genome-wide significant 3 of the 8 loci that reached genome-wide significance in our Global BPgen joint analysis of stages 1+2. CHARGE also reports common variants at 5 additional genome-wide significant loci at: 11p15 (Global BPgen P = 0.009), 3p22 (P = 0.01), 12q21 (P = 0.008), 12q24 (P = 0.05), and 10p12 (P = 0.004, see companion CHARGE paper). While these SNPs did not appear among our top 10 SNPs for either blood pressure trait, the Global BPgen results from in silico exchange and for the same alleles are clearly consistent with the conclusions of the CHARGE investigators. Among the 10 SBP and 10 DBP loci at the top of the Global BPgen results, five loci were represented in the CHARGE top 10 results (Supplementary Table 3). With the modest effect sizes we observed, it is not surprising that the top 10 loci for each blood pressure trait would exhibit only partial overlap.
We acknowledge that some limitations apply to our study. The participants in the individual studies comprising Global BPgen and our follow-up cohorts were ascertained using diverse criteria, had their blood pressure measured in a variety of ways, and exhibited a broad range of age and treatment profiles. Even small differences in these factors could reduce power to detect the association of genetic variants with modest effect, although such heterogeneity should not increase the false-positive rate. Even though SBP and DBP are dynamic phenotypes resulting from multiple competing influences, estimates of the test-retest reliability of blood pressure measurements are approximately 0.65–0.75 in studies focused on blood pressure43,44. Moreover, a graded relationship between BP measures and cardiovascular risk has been consistently observed, despite variability in BP measures2. At the individual level, genetically-determined alteration of 1 mm Hg SBP or 0.5 mm Hg DBP would be difficult to detect in the clinic, but large sample sizes use group-level differences in means to detect small genetic effects.
Exposures such as dietary sodium and potassium intake or excessive alcohol use also contribute to inter-individual differences in blood pressure. These were measured in a minority of our samples and thus we could not meaningfully adjust for these in our study. Under the assumption that these do not alter blood pressure systematically by genotype, we would only expect this omission to reduce power slightly.
We chose a priori to adjust for body mass index, which explains ~6–8% of the total variation in SBP and DBP, with the goal of reducing potential non-genetic contributions to blood pressure variability. Genetic variants could influence blood pressure acting through BMI as an intermediate, but such variants are best identified through BMI GWA studies such as those recently reported by Loos et al45 and Willer et al46.
We adjusted for use of antihypertensive therapy by adding 15 mm Hg and 10 mm Hg to SBP and DBP, respectively. This approach has been shown to be superior to ignoring antihypertensive treatment or to excluding individuals on therapy13. However, it is clear that factors such as medication number and dosage, and variation in prescription patterns in different countries and time periods make this adjustment scheme an oversimplification. Again, such effects should generally bias our findings toward the null.
There are many classes of widely used therapies with strong antihypertensive effects. We examined the association of common variants at the loci extending 100kb on either side of the genes encoding the targets for thiazide diuretics (NCCT), loop diuretics (NKCC2), ACE inhibitors (ACE), angiotensin II receptor type 1 blockers (AGTR1), beta adrenoreceptor blockers (ADRB1, ADRB2), alpha adrenoreceptor antagonists (ADRA1A, ADRA1B, ADRA1D), calcium channel blockers (CACNA1S, CACNA1C, CACNA1D, CACNA1F), and aldosterone antagonists (CYP11B2). No results exceeded chance expectations. This does not exclude the existence of variants of weaker effects or variants that were missed because they were not covered by existing arrays. Obviously, it would be interesting to examine the impact of common variants in these genes on individual responses to therapies, which we have not done.
Moreover, the strength of association of variation in a gene with a trait (or lack thereof) says nothing about the potential strength of a drug designed to agonize or antagonize the product of that gene. For example, a common variant in HMGCR has only a modest effect on fasting lipids,47 yet statin therapy, which inhibits the HMGCR enzyme to lower LDL cholesterol, substantially lowers risk of cardiovascular disease. Thus, the implication of modest common variant genetic effects is not just a function of the ability to identify tendency toward higher or lower blood pressure in carriers of alternate alleles, but also the ability to recognize relevant targets for therapy that have defined in vivo relevance in human beings.
While targeted pharmacotherapy has theoretical appeal, clinical trials to demonstrate the utility and cost-effectiveness of such approaches will be required before such personalized medicine could be endorsed. The association signals identified here will need to be refined through fine mapping, and resequencing will be needed to define more fully the allelic spectrum of variants at each locus that contributes to inter-individual differences in blood pressure. Our findings offer initial insights into the genetic basis of a problem of global proportions and the potential for an improved understanding of blood pressure regulation. These loci may point to new targets for blood pressure reduction and ultimately additional opportunities to prevent the growing public health burden of cardiovascular disease.
METHODS
Overall study design
An expanded description of the methods is provided in the Supplementary Methods. The study comprised two staged analyses performed separately for SBP and DBP. Stage 1 was a meta-analysis of directly genotyped and imputed SNPs from individuals of European descent in 17 samples drawn from population-based or control samples in case-control studies in the Global BPgen consortium. In stage 2a, we selected 12 SNPs for genotyping in up to 71,225 individuals of European descent from 13 studies and up to 12,889 individuals of Indian Asian ancestry from one study. In stage 2b, we selected 20 SNPs (10 SBP, 10 DBP) for in silico analysis in 29,136 individuals of European descent from the CHARGE consortium (stage 2b, see Supplementary Figure 1).
Stage 1 samples
The Global BPgen consortium comprises 17 GWAS studies: the Baltimore Longitudinal Study of Aging (BLSA), British 1958 Birth Cohort (B58C-T1DGC and B58C-WTCCC), Cohorte Lausannoise (CoLaus), Diabetes Genetics Initiative (DGI), European Prospective Investigation of Cancer-Norfolk-Genome Wide Association Study (EPIC-Norfolk-GWAS), Fenland Study, Finland-United States Investigation of NIDDM Genetics (FUSION) study, Invecchiare in Chianti (InCHIANTI), Kooperative Gesundheitsforschung in der Region Augsburg (KORA), the Myocardial Infarction Genetics Consortium (MIGen), Northern Finland Birth Cohort of 1966 (NFBC1966), SardiNIA, Study of Health in Pomerania (SHIP), the Precocious Coronary Artery Disease (PROCARDIS), Supplementation en Vitamines et Mineraux Antioxydants (SU.VI.MAX), and TwinsUK. We excluded individuals >70 years of age and individuals ascertained on case status for type 1 or 2 diabetes (DGI, FUSION), coronary artery disease (MIgen, PROCARDIS) or hypertension (BRIGHT), leaving 34,433 individuals for analysis (Table 1). A detailed description of the study design and phenotype measurement for all cohorts can be found in the Supplementary Methods.
Genome-wide genotyping
Genotyping arrays and quality control filters are provided in Supplementary Table 1.
Imputation
Imputation of allele dosage of ungenotyped SNPs in HapMap CEU v21a or v22 was performed using MACH48 or IMPUTE49 with parameters and pre-imputation filters as specified in Supplementary Table 1. SNPs were excluded from analysis if the cohort-specific imputation quality as assessed by r2.hat (MACH) or .info (IMPUTE) metrics was <0.30. In total, up to 2,497,993 genotyped or imputed autosomal SNPs were analyzed.
Phenotype modeling
In individuals taking antihypertensive therapies, blood pressure was imputed by adding 15 mm Hg and 10 mm Hg for SBP and DBP, respectively13. Continuous SBP and DBP were adjusted for age, age2, body mass index, and any study-specific geographic covariates in gender-specific linear regression models. In FUSION and SardiNIA, which included family-based samples, gender-pooled linear regression was performed with the addition of gender as a covariate. Residuals on the mm Hg scale were used as univariate traits in genotype-phenotype analysis.
In secondary analyses, hypertension was defined by the presence of SBP ≥ 140 mm Hg or diastolic blood pressure ≥90 mm Hg or self-report of taking a medication for the treatment of hypertension. Normotensive controls were defined as individuals not taking any anti-hypertensives and having a SBP ≤120 mm Hg and a DBP ≤85 mm Hg.
Genotype-phenotype association analysis
Genotype-phenotype association of SBP and DBP residuals was performed under an additive model using software as specified in Supplementary Table 1. Analysis of hypertension for eight genome-wide significant continuous blood pressure loci was performed using logistic regression to adjust for age, age2, gender, body mass index.
Meta-analysis of stage 1 samples
All cohort-specific effect estimates and coded alleles were oriented to the forward strand of the NCBI35 reference sequence of the human genome, using the alphabetically higher allele as the coded allele. For example, for a G/T SNP coded GG=0, GT=1, TT=2, the coded allele would be T. To capture the power loss due to imperfect imputation, we estimated “N effective”, which was the sum of the sample-specific products of the imputation quality metric and the sample size. No filtering on minor allele frequency was used. Genomic control14 was performed on cohort- and gender-specific test statistics. Lambda estimates are given in Supplementary Table 1; quantile-quantile plots are shown in Supplementary Figure 2. Meta-analysis in stage 1 was performed using inverse variance weights. Stage 1 meta-analysis results were genomic controlled.
Selection of SNPs for stage 2
12 SNPs were selected for follow-up in stage 2a from among the results with P < 10−5 during interim analyses. For in silico exchange with the CHARGE consortium (stage 2b), we identified the top independent loci to select 10 SBP and 10 DBP SNPs. If a SNP in one top 10 list was also among the top 10 for the alternate blood pressure trait, we kept the locus with the lower p-value and went to the next locus on the list for the alternate blood pressure trait. Because a SNP at the 3q26 locus (MDS1) was selected in an interim analysis for direct genotyping, it was retained as the tenth locus for DBP even though its significance was reduced in the final stage 1 DBP GWAS analysis.
Stage 2a samples
We genotyped 12 SNPs in up to 71,225 individuals of European descent from 13 studies – Utrecht Atherosclerosis Risk in Young Adults (ARYA), British Genetics of Hypertension (BRIGHT), EPIC-Italy, EPIC-Norfolk-REP, Finrisk97, FUSION2, London Life Sciences Population (LOLIPOP), Malmö Diet and Cancer-Cardiovascular Cohort (MDC-CC), Metabolic Syndrome in Men (METSIM), Malmo Preventive Project (MPP), The Prevention of REnal and Vascular ENd stage Disease (PREVEND), Prospect-EPIC, and the Utrecht Health Project (UHP) – and in up to 12,889 individuals of Indian Asian ancestry from the LOLIPOP study. Summary demographics are shown in Table 1 and cohort information in the Supplementary Methods).
Stage 2a follow-up genotyping
For genotyping methods and platforms see Supplementary Methods.
Stage 2b in silico samples
We obtained results based on the analysis of the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) consortium, which comprises 29,136 samples from five population-based cohorts.
Pooled analysis of first and second stage samples
Meta-analysis of stage 1, 2a and 2b results was performed using inverse variance weighting. Standard errors were multiplied by the square root of the lambda estimate for genomic control and are presented throughout the text. Nominal P values after genomic control14 are presented. We considered associations genome-wide significant if they exceeded P = 5×10−8, a Bonferroni correction for the estimated 1M independent common variant tests in the human genome of European-derived individuals14,15.
Supplementary Material
1
2
Acknowledgments
The authors would like to thank the many colleagues who contributed to collection and phenotypic characterization of the clinical samples, as well as genotyping and analysis of the GWA data. They would also especially like to thank those who agreed to participate in the studies. Major funding for the work described in the paper comes from (alphabetically): Academy of Finland (124243, 129322, 129494, 118065), AGAUR (SGR 2005/00577), Albert Påhlsson Research Foundation, Alexander-von-Humboldt Foundation (V-Fokoop-1113183), American Diabetes Association, AstraZeneca AB, AVIS Torino blood donor organization, Barts and The London Charity, Biocenter of University of Oulu, Board of the UMC Utrecht, British Heart Foundation (PG02/128, FS/05/061/19501, SP/04/002), Burroughs Wellcome Fund, CamStrad, Cancer Research United Kingdom, CIBER Epidemiología y Salud Pública, Commissariat à l’Energie Atomique, Compagnia di San Paolo to the ISI Foundation (Torino, Italy), Conservatoire National des Arts et Métiers, Crafoord Foundation, Donovan Family Foundation, Doris Duke Charitable Foundation, Dutch Kidney Foundation (E033), Dutch College of Healthcare Insurance Companies, Dutch Ministry of Health, Dutch Organisation of Health Care Research, ENGAGE (HEALTH-F4-2007-201413), Ernhold Lundstroms Research Foundation, Estonian Ministry of Education and Science (0182721s06), EURO-BLCS, European Commission (QLG1-CT-2000-01643, LSHM-CT-2007-037273), European Commission-Europe Against Cancer (AEP/90/05), European Union (FP-6 LSHM-CT-2003-503041, FP-6 LSHM CT 2006 037697), European Society for the Study of Diabetes, Faculty of Biology and Medicine of Lausanne, Switzerland, Fannie E. Rippel Foundation, Finnish Foundation for Cardiovascular Research, FIS (CP05/00290), German Federal Ministry of Education and Research (01ZZ9603, 01ZZ0103, 01ZZ0403, 03ZIK012, 01EZ0874), German National Genome Research Network, German Research Center for Environmental Health, (Neuherberg, Germany), Giorgi-Cavaglieri Foundation, GlaxoSmithKline, Guy’s & St Thomas’ NHS Foundation Trust, Health Research and Development Council of the Netherlands (2100.0008, 2100.0042), Helmholtz Zentrum Munchen, Hulda and Conrad Mossfelt Foundation, Institut National de la Recherche Agronomique, Institut National de la Sante et de la Recherche Medicale, Italian Association for Research on Cancer, Italian Ministry of Health (110.1RS97.71), Italian National Research Council, Juvenile Diabetes Research Fund, King Gustaf V and Queen Victoria Foundation, King’s College London and King’s College Hospital NHS Foundation Trust, Knut and Alice Wallenberg Foundation, Lennart Hanssons Memorial Fund, LK Research Funds, Massachusetts General Hospital Cardiovascular Research Center and Department of Medicine, Medical Faculty of Lund University and Malmö University Hospital, Medical Research Council of the UK (G0000934, G0501942, G9521010D), Medical Research Council-GlaxoSmithKline (85374), MedStar Research Institute, Ministerio de Sanidad y Consumo, Instituto de Salud Carlos III (RD06/0009), Ministry of Cultural Affairs and Social Ministry (Federal State of Mecklenburg-West Pomerania), National Institute for Health Research (NIHR), National Institute for Health Research Cambridge Biomedical Research Centre, Novartis Institute for Biomedical Research, NWO VENI (916.76.170), Province of Utrecht, Region Skane, Siemens Healthcare (Erlangen, Germany), Sigrid Juselius Foundation, Stockholm County Council (562183), Support for Science Funding programme, Swedish Heart and Lung Foundation, Swedish Medical Research Council, Swedish National Research Council, Swedish Research Council (8691), Swiss National Science Foundation (33CSO-122661, 310030-112552, 3100AO-116323/1, PROSPER 3200BO-111362/1, 3233BO-111361/1), UNIL, University of Utrecht, US National Institutes of Health (U01DK062418, K23HL80025, DK062370, DK072193, U54DA021519, 1Z01HG000024, N01AG-916413, N01AG-821336, 263MD916413, 263MD821336, Intramural NIA, R01HL087676, K23HL083102, U54RR020278, R01HL056931, P30ES007033, R01HL087679, RL1MH083268, 263-MA-410953, NO1-AG-1-2109, N01-HD-1-3107), WCRF (98A04, 2000/30), Wellcome Trust (068545/Z/02, 076113/B/04/Z, 079895, 070191/Z/03/Z, 077016/Z/05/Z, WT088885/Z/09/Z).
Footnotes
Author contributions (alphabetical)
Project conception, design, management: ARYA: M.L.B., C.S.U.; BLSA: L.F.; B58C-T1DGC: D.P.S.; B58C-WTCCC: D.P.S.; BRIGHT: M.Brown, M.C., J.M.C., A. Dominiczak, M.F., P.B.M., N.J.S., J.W.; CoLaus: J.S.B., S.Bergmann, M.Bochud, V.M. (PI), P.Vollenweider (PI), G.W., D.M.W.; DGI: D.A., C.N.-C., L.G.; EPIC-Norfolk-GWAS: I.B., P.D., R.J.F.L., M.S.S., N.J.W., J.H.Z. EPIC-Italy: S. Polidoro, P.Vineis. Fenland Study: R.J.F.L., N.G. F., N.J.W.; Finrisk97: L.P., V.S.; FUSION: R.N.B., M Boehnke, F.S.C., K.L.M., L.J.S., T.V., J.T.; InCHIANTI: S. Bandinelli., L.F.; KORA: A. Döring, C.G., T.I., M.L., T.M., E.O., H.E.W. (PI); LOLIPOP: J.C.C., P.E., J.S.K. (PI); MDC-CC: G.B., O.M.; MPP: G.B., O.M.; MIGen: D.A., R.E., S.K., J.M., O.M., C.J.O., V.S., S.M.S., D.S.S.; METSIM: J.K., M.L.; NFBC1966: P.E., M.-R.J.; PREVEND: P.E.d.J. (PI), G.N., W.H.v.G.; PROCARDIS: R.C., M.F., A.H., J.F.P., U.S., G.T., H.W.(PI); PROSPECT-EPIC. N.C.O.-M., Y.T.v.d.S.; SardiNIA: E.G.L., D.S.; SHIP: M.D., S.B.F., G.H., R.L., T.R., R.R., U.V., H.V.; SUVIMAX: P.M.; TwinsUK: P.D., T.D.S. (PI); UHP: D.E.G., M.E.N.
Phenotype collection, data management: ARYA: M.L.B., C.S.U.; B58C-T1DGC: D.H., W.L.M., D.P.S.; B58C-WTCCC: D.H., K.P., D.P.S.; BRIGHT: M. Brown, M.C., J.M.C., A.Dominiczak, M.F., P.B.M., N.J.S., J.W.; CoLaus: G.W.; DGI: L.G., O.M.; EPIC-Italy: P. Vineis (PI); Finrisk97: P.J., M.P., V.S.;FUSION: J.T., T.V.; KORA: A. Doring, C.G., T.I.; MDC-CC: O.M.; MPP: O.M., P.N.; MIGen: D.A., R.E., S.K., J.M., O.M., C.J.O., S.M.S., D.S.S., V.S.; NFBC1966: A.-L.H., M.-R.J., A.P.; PREVEND: P.E.d.J., G.N., P.v.d.H., W.H.v.G.; PROCARDIS: R.C., A.H., U.S., G.T.; PROSPECT-EPIC. N.C.O.-M., Y.T.v.d.S.; SardiNIA: S.S.N., A.S.; SHIP: M.D., R.L., R.R., H.V.; SUVIMAX: P.G., S.H.; TwinsUK: F.M.W.; UHP: D.E.G., M.E.N.
Genome-wide, validation genotyping: B58C-T1DGC: W.L.M.; B58C-WTCCC: W.L.M.; DGI: D.A., O.M., M.O.-M.; EPIC-Norfolk-GWAS: I.B., P.D., N.J.W., J.H.Z.; EPIC-Norfolk-replication: S.A.B., K.-T.K., R.J.F.L., R.N.L., N.J.W.; EPIC-Italy: G.M.; EPIC-Italy: A.A., A.d.G., S.G., V.R.; Finrisk97: G.J.C., C.N.-C.; FUSION: L.L.B., M.A.M.; KORA: T.I., T.M., E.O., A.P.; MDC-CC: O.M., M.O.-M.; MPP: O.M., M.O.-M.; NFBC1966: P.E., N.B.F., M.-R.J., M.I.M., L.P. ; PREVEND: G.N., P.v.d.H. ; W.H.v.G.; PROCARDIS: S.C.H., G.M.L., A.-C.S.; SardiNIA: M.U.; SHIP: F.E., G.H., A.T., U.V.; SUVIMAX: I.G.G., S.C.H., G.M.L., D.Z.; TwinsUK: P.D.
Data analysis: BLSA: T.T.; B58C-T1DGC: D.H., S.H., D.P.S.; B58C-WTCCC: P.R.B., D.H., K.P., D.P.S, M.D.T.; B58C-T1DGC: D.H., S.H., D.P.S.; BRIGHT: S.J.N., C.W., E.Z.; CoLaus: S. Bergmann, M. Bochud, T.J., N.L., K.S., X.Y., DGI: O.M., C.N.-C., M.O.-M., B.F.V.; EPIC-Norfolk-GWAS: R.J.F.L., J.H.Z.; EPIC-Norfolk-replication: S.A.B., K.-T.K., R.J.F.L., R.N.L., N.J.W.; EPIC-Italy: S.G., G.M., S. Panico, S. Polidoro, F.R., C.S., P. Vineis; Fenland Study: J.L.; Finrisk97: C.N.-C.; FUSION: A.U.J., L.J.S., H.M.S., C.J.W.; InCHIANTI: T.T.; KORA: S.E., C.G., M.L., E.O.; LOLIPOP: J.C.C.; MDC-CC: O.M., M.O.-M.; MPP: O.M., M.O.-M.; MIGen: R.E., G.L., I.S., B.F.V.; NFBC1966: L.C., P.F.O.; PREVEND: H.S., P.v.d.H.; PROCARDIS: M.F., A.G., J.F.P.; SardiNIA: V.G., S.S., P.S.; SHIP: F.E., G.H., A.T., U.V.; SUVIMAX: S.C.H., T.J., P.M.; TwinsUK: N.S., F.Z., G.Z.
Analysis group. G.R.A., M.C., V.G., T.J., P.B.M., C.N.-C., M.D.T., L.V.W.
Writing group: G.R.A., M.C., P.E., V.G., T.J., P.B.M., C.N.-C., M.D.T.
COMPETING INTERESTS STATEMENT
The following authors declare the following potential conflicts of interest: N.L., V.M., K.S., D.M.W. and X.Y. are all full-time employees at GlaxoSmithKline. P. Vollenweider and G.W received financial support from GlaxoSmithKline to assemble the CoLaus study. No other authors reported conflicts of interest.
References
- 1.Ezzati M, Lopez AD, Rodgers A, Vander Hoorn S, Murray CJ. Selected major risk factors and global and regional burden of disease. Lancet. 2002;360:1347–60. doi: 10.1016/S0140-6736(02)11403-6. [DOI] [PubMed] [Google Scholar]
- 2.Lewington S, Clarke R, Qizilbash N, Peto R, Collins R. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360:1903–1913. doi: 10.1016/s0140-6736(02)11911-8. [DOI] [PubMed] [Google Scholar]
- 3.The world health report 2002. Reducing risks, promoting healthy life. World Health Organization; 2002. [DOI] [PubMed] [Google Scholar]
- 4.Whelton PK, et al. Primary prevention of hypertension: clinical and public health advisory from The National High Blood Pressure Education Program. Jama. 2002;288:1882–8. doi: 10.1001/jama.288.15.1882. [DOI] [PubMed] [Google Scholar]
- 5.Havlik RJ, et al. Blood pressure aggregation in families. Am J Epidemiol. 1979;110:304–12. doi: 10.1093/oxfordjournals.aje.a112815. [DOI] [PubMed] [Google Scholar]
- 6.Lifton RP, Gharavi AG, Geller DS. Molecular mechanisms of human hypertension. Cell. 2001;104:545–556. doi: 10.1016/s0092-8674(01)00241-0. [DOI] [PubMed] [Google Scholar]
- 7.Ji W, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–9. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newhouse SJ, et al. Haplotypes of the WNK1 gene associate with blood pressure variation in a severely hypertensive population from the British Genetics of Hypertension study. Hum Mol Genet. 2005;14:1805–14. doi: 10.1093/hmg/ddi187. [DOI] [PubMed] [Google Scholar]
- 9.Tobin MD, et al. Association of WNK1 gene polymorphisms and haplotypes with ambulatory blood pressure in the general population. Circulation. 2005;112:3423–9. doi: 10.1161/CIRCULATIONAHA.105.555474. [DOI] [PubMed] [Google Scholar]
- 10.Tobin MD, et al. Common variants in genes underlying monogenic hypertension and hypotension and blood pressure in the general population. Hypertension. 2008;51:1658–64. doi: 10.1161/HYPERTENSIONAHA.108.112664. [DOI] [PubMed] [Google Scholar]
- 11.Genome-wide association study of 14 000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy D, et al. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8(Suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–35. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 14.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 15.Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- 16.Martin RM, et al. P450c17 deficiency in Brazilian patients: biochemical diagnosis through progesterone levels confirmed by CYP17 genotyping. J Clin Endocrinol Metab. 2003;88:5739–46. doi: 10.1210/jc.2003-030988. [DOI] [PubMed] [Google Scholar]
- 17.Geller DH, Auchus RJ, Mendonca BB, Miller WL. The genetic and functional basis of isolated 17,20-lyase deficiency. Nat Genet. 1997;17:201–5. doi: 10.1038/ng1097-201. [DOI] [PubMed] [Google Scholar]
- 18.Kluijtmans LA, et al. Molecular genetic analysis in mild hyperhomocysteinemia: a common mutation in the methylenetetrahydrofolate reductase gene is a genetic risk factor for cardiovascular disease. Am J Hum Genet. 1996;58:35–41. [PMC free article] [PubMed] [Google Scholar]
- 19.Sohda S, et al. Methylenetetrahydrofolate reductase polymorphism and pre-eclampsia. J Med Genet. 1997;34:525–6. doi: 10.1136/jmg.34.6.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian X, Lu Z, Tan M, Liu H, Lu D. A meta-analysis of association between C677T polymorphism in the methylenetetrahydrofolate reductase gene and hypertension. Eur J Hum Genet. 2007;15:1239–45. doi: 10.1038/sj.ejhg.5201914. [DOI] [PubMed] [Google Scholar]
- 21.John SW, et al. Genetic decreases in atrial natriuretic peptide and salt-sensitive hypertension. Science. 1995;267:679–681. doi: 10.1126/science.7839143. [DOI] [PubMed] [Google Scholar]
- 22.Newton-Cheh C, et al. Association of Common Variants in NPPA and NPPB with Circulating Natriuretic Peptides and Blood Pressure. Nature Genetics. 2009 doi: 10.1038/ng.328. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simon DB, et al. Genetic heterogeneity of Bartter’s syndrome revealed by mutations in the K+ channel, ROMK. Nat Genet. 1996;14:152–6. doi: 10.1038/ng1096-152. [DOI] [PubMed] [Google Scholar]
- 24.Simon DB, et al. Gitelman’s variant of Bartter’s syndrome, inherited hypokalaemic alkalosis, is caused by mutations in the thiazide-sensitive Na-Cl cotransporter. Nat Genet. 1996;12:24–30. doi: 10.1038/ng0196-24. [DOI] [PubMed] [Google Scholar]
- 25.Daviet L, et al. Cloning and characterization of ATRAP, a novel protein that interacts with the angiotensin II type 1 receptor. J Biol Chem. 1999;274:17058–62. doi: 10.1074/jbc.274.24.17058. [DOI] [PubMed] [Google Scholar]
- 26.Suh PG, et al. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB Rep. 2008;41:415–34. doi: 10.5483/bmbrep.2008.41.6.415. [DOI] [PubMed] [Google Scholar]
- 27.Montano MM, et al. Mutation of the HEXIM1 gene results in defects during heart and vascular development partly through downregulation of vascular endothelial growth factor. Circ Res. 2008;102:415–22. doi: 10.1161/CIRCRESAHA.107.157859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dey A, Chao SH, Lane DP. HEXIM1 and the control of transcription elongation: from cancer and inflammation to AIDS and cardiac hypertrophy. Cell Cycle. 2007;6:1856–63. doi: 10.4161/cc.6.15.4556. [DOI] [PubMed] [Google Scholar]
- 29.Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat Rev Cancer. 2006;6:947–60. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- 30.Cornelis MC, El-Sohemy A, Kabagambe EK, Campos H. Coffee, CYP1A2 genotype, and risk of myocardial infarction. JAMA. 2006;295:1135–41. doi: 10.1001/jama.295.10.1135. [DOI] [PubMed] [Google Scholar]
- 31.Takebe A, et al. Microarray analysis of PDGFR alpha+ populations in ES cell differentiation culture identifies genes involved in differentiation of mesoderm and mesenchyme including ARID3b that is essential for development of embryonic mesenchymal cells. Dev Biol. 2006;293:25–37. doi: 10.1016/j.ydbio.2005.12.016. [DOI] [PubMed] [Google Scholar]
- 32.Vatner SF. FGF induces hypertrophy and angiogenesis in hibernating myocardium. Circ Res. 2005;96:705–7. doi: 10.1161/01.RES.0000164184.63158.6c. [DOI] [PubMed] [Google Scholar]
- 33.Todd JA, et al. Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet. 2007;39:857–64. doi: 10.1038/ng2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hunt KA, et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gudbjartsson DF, et al. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat Genet. 2009 doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- 36.Velazquez L, et al. Cytokine signaling and hematopoietic homeostasis are disrupted in Lnk-deficient mice. J Exp Med. 2002;195:1599–611. doi: 10.1084/jem.20011883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Iturbe B, Vaziri ND, Herrera-Acosta J, Johnson RJ. Oxidative stress, renal infiltration of immune cells, and salt-sensitive hypertension: all for one and one for all. Am J Physiol Renal Physiol. 2004;286:F606–16. doi: 10.1152/ajprenal.00269.2003. [DOI] [PubMed] [Google Scholar]
- 38.Du YH, Guan YY, Alp NJ, Channon KM, Chen AF. Endothelium-specific GTP cyclohydrolase I overexpression attenuates blood pressure progression in salt-sensitive low-renin hypertension. Circulation. 2008;117:1045–54. doi: 10.1161/CIRCULATIONAHA.107.748236. [DOI] [PubMed] [Google Scholar]
- 39.Zheng JS, et al. Gene transfer of human guanosine 5′-triphosphate cyclohydrolase I restores vascular tetrahydrobiopterin level and endothelial function in low renin hypertension. Circulation. 2003;108:1238–45. doi: 10.1161/01.CIR.0000089082.40285.C3. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe M, et al. Regulation of smooth muscle cell differentiation by AT-rich interaction domain transcription factors Mrf2alpha and Mrf2beta. Circ Res. 2002;91:382–9. doi: 10.1161/01.res.0000033593.05545.7b. [DOI] [PubMed] [Google Scholar]
- 41.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stamler J, et al. INTERSALT study findings. Public health and medical care implications. Hypertension. 1989;14:570–7. doi: 10.1161/01.hyp.14.5.570. [DOI] [PubMed] [Google Scholar]
- 43.Dyer AR, Shipley M, Elliott P. Urinary electrolyte excretion in 24 hours and blood pressure in the INTERSALT Study. I. Estimates of reliability. The INTERSALT Cooperative Research Group. Am J Epidemiol. 1994;139:927–39. doi: 10.1093/oxfordjournals.aje.a117099. [DOI] [PubMed] [Google Scholar]
- 44.Variability of blood pressure and the results of screening in the hypertension detection and follow-up program. J Chronic Dis. 1978;31:651–67. doi: 10.1016/0021-9681(78)90069-3. [DOI] [PubMed] [Google Scholar]
- 45.Loos RJ, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–75. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kathiresan S, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y, Abecasis GR. Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 49.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 50.Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–4. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2