Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome (original) (raw)
Abstract
In diploid mammalian genomes, parental alleles can exhibit different methylation patterns (allele-specific DNA methylation, ASM), which have been documented in a small number of cases except for the imprinted regions and X chromosomes in females. We carried out a chromosome-wide survey of ASM across 16 human pluripotent and adult cell lines using Illumina bisulfite sequencing. We applied the principle of linkage disequilibrium (LD) analysis to characterize the correlation of methylation between adjacent CpG sites on single DNA molecules, and also investigated the correlation between CpG methylation and single nucleotide polymorphisms (SNPs). We observed ASM on 23%∼37% heterozygous SNPs in any given cell line. ASM is often cell-type-specific. Furthermore, we found that a significant fraction (38%∼88%) of ASM regions is dependent on the presence of heterozygous SNPs in CpG dinucleotides that disrupt their methylation potential. This study identified distinct types of ASM across many cell types and suggests a potential role for CpG-SNP in connecting genetic variation with the epigenome.
DNA methylation is an epigenetic marker that plays a direct role in transcriptional regulation. DNA methylation patterns are tissue-specific. Embryonic stem cells undergoing differentiation show significant changes in DNA methylation patterns (Deng et al. 2009; Doi et al. 2009; Lister et al. 2009). In addition to DNA methylation pattern differences between cell lines, DNA methylation can also be allele-specific within a cell line and is thus linked to allele-specific gene expression (ASE). An example of allele-specific methylation (ASM) is genetic imprinting, which describes the parent-specific gene expression behavior of a small set of genes. The methylation pattern of imprinted genes is distinct; the inactive allele is significantly more methylated than the actively expressed allele. The number of currently known imprinting genes is suspected to be a small fraction of the total number of imprinted genes. Recent work has provided a large candidate list of imprinted genes (Luedi et al. 2007), though most of these candidates still remain to be validated. Even less is known is about the genes that fall into the broader ASM category. The biological importance of methylation is clear as disturbances of known methylation patterns are linked to disease phenotypes (Robertson 2005; Eggermann 2009).
In a recent survey of DNA methylation changes during nuclear reprogramming of human fibroblasts to induced pluripotent stem cells, 7.6% of CpG islands were found to be dominated by CpG sites that have intermediate levels of methylation (0.25–0.75, referred as fuzzy methylation), even though the samples used in the assay are polyclonal or monoclonal cell lines (Deng et al. 2009). A small fraction of fuzzily methylated CpG dinucleotides are related to X inactivation, and imprinting is unlikely the dominant effect that explains the other regions. ASM is another mechanism that could potentially explain fuzzy methylation. However, only a handful of ASM regions have been identified to date (Kerkel et al. 2008; Zhang et al. 2009b). Taking advantage of the high-resolution CpG methylation information generated from targeted bisulfite sequencing, we carried out a systematic study to characterize ASM and its role in fuzzy methylation (Supplemental Fig. 1).
Results
We reasoned that, if fuzzy methylation were due to unequal methylation levels between the two copies of the chromosomes in the same cells or the presence of heterogeneous epigenetic states among the cell population, the methylation levels on adjacent CpG sites of the same DNA molecules should be highly correlated. When performing Illumina sequencing on bisulfite-converted DNA, a sequencing read often contains multiple CpG sites, which can be treated as methylation haplotypes. Such methylation haplotypes are similar to SNP haplotypes, which allowed us to extend the concept of linkage disequilibrium (LD) analysis to characterizing the co-methylation of CpG sites on single DNA molecules. Specifically, we used the LD measurement _r_2, which indicates the fraction of variation (of methylation status) on a CpG site, A, that can be explained by the variation on another CpG site, B. Note that in this context LD is defined on a population of diploid cells.
Our analyses were based on targeted bisulfite sequencing data (41-bp reads) previously generated on 11 human pluripotent and adult cell lines (Deng et al. 2009), plus additional paired 36-bp Illumina sequencing reads on eight human cell lines (three cell lines were covered in both sets). The following regions were examined in this analysis: (1) all 2020 CpG islands on human chromosomes 12 and 20, (2) 237 promoters in eight ENCODE (the Encyclopedia of DNA Elements) regions, and (3) the 4-kb region centered around the transcription start sites (TSS) of 26 genes related to development or pluripotency. In addition to the Illumina short reads, Sanger sequencing data from cloned bisulfite PCR amplicons on a selected number of regions were also generated (Supplemental Tables 1–3). Expanding on the previously published read-mapping strategy (Deng et al. 2009), we used whole-genome mapping for Illumina data and developed methylation haplotype-identifying algorithms for Illumina and Sanger sequences. Regions with at least 10× read depth were used in our LD analysis.
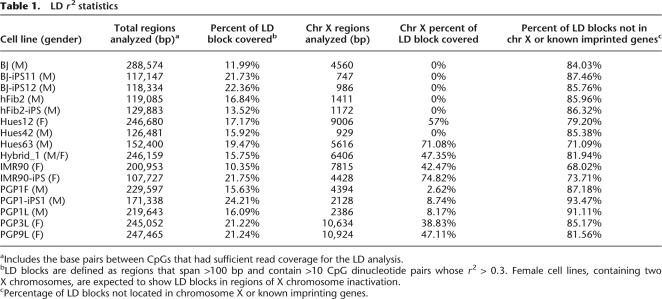
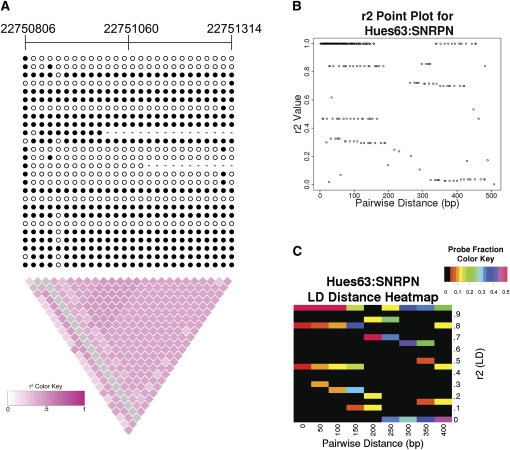
We first validated our LD analysis on known imprinted regions and female X chromosomes. In such regions we expected to observe high _r_2-values that extend over a long distance. We developed an algorithm to search for such regions (see Methods). Similar to the LD blocks in the human population, we called these regions “methylation LD blocks.” A block was first created between a pair of CpG sites with _r_2 > 0.3 and extended if another CpG pair with _r_2 > 0.3 was <100 bp away. Since we were most interested in examining extended organized methylatation regions, we filtered out LD blocks that spanned <100 bp or contained fewer than 10 CpG pairs with _r_2 > 0.3. We observed LD blocks in all 16 cell lines for the known imprinted genes SNRPN and GNAS (Luedi et al. 2007). NNAT, another known imprinted gene, was found to contain LD blocks in 13 cell lines. There were two other imprinted genes found, but the read coverage for these genes was much lower than for SNRPN, GNAS, and NNAT. IGF2AS was covered in only three cell lines and NDN was covered in only one cell line. No LD blocks were found within these two gene regions in their respective cell lines. In female cell lines, approximately 43%∼75% of the X chromosome regions included in our analysis were covered by LD blocks, which clearly distinguished them from the majority of male lines due to X chromosome inactivation (Table 1). A much lower fraction (0%∼9%) of X chromosome regions was within LD blocks for the male cell lines. The only exception is Hues63, a male human embryonic stem cell line, which exhibited extended LD on the X chromosome. This is likely due to the presence of a subpopulation of cells that was already in the early stage of differentiation (Supplemental Fig. 2). Bisulfite Sanger sequencing of a known imprinted gene, SNRPN, from Hues63 revealed a methylation LD block that extended over 500 bp. In such regions, LD does not decay over the physical distance (Fig. 1A–C). However, imprinted regions are not free of noise. We also observed many pairs of CpG sites that have little or no correlation (Fig. 1B,C).
Table 1.
LD _r_2 statistics
Figure 1.
Linkage disequilibrium (LD) analysis of CpG methylation haplotypes. (A) LD diagram of 5′ region of the imprinted gene SNRPN (chr15:22750806–22751314) from Hues63. Each row represents a Sanger sequence and each column represents a CpG dinucleotide. (Filled circles) Methylated CpGs; (open circles) unmethylated CpGs, (dashed lines) methylation state of the CpG could not be determined. Chromosomal coordinates are listed above. This region shows a methylation LD block spanned over 500 bp in Sanger reads. (B,C) CpG pairwise _r_2-value plots and heatmap for 5′ region of the SNRPN gene. While the majority of CpG pairs have high methylation correlation values (_r_2 > 0.3), some pairs of CpG sites have little or no correlation (_r_2 < 0.3). The pairwise distance represents the separation of the CpG dinucleotides used in the _r_2 calculation. The heatmap colors represent the probe fraction at a given pairwise distance (rounded down to the nearest 50 bp) that has the indicated _r_2-value. The probe fractions for each pairwise distance sum to one. The color scale saturates at 0.5, so that small probe fraction differences can be distinguished.
We next extended the LD analysis to the other autosomal regions. Various levels of LD were observed in all 16 cell lines included in this study (Supplemental Fig. 3). Pluripotent cell lines appear to exhibit more extended LD compared with the corresponding fibroblast lines, which can potentially be explained by the less compact chromatin structure in pluripotent cells such that _cis_-regulation can operate over a longer distance (Spivakov and Fisher 2007). Of the 108- to 289-kb genomic regions with sufficient read coverage for the LD analysis, 10%∼24% were within methylation LD blocks, which vary in both the number of CpG sites per block and the block size (Table 1). The majority of the methylation LD blocks (68%∼94%) are not known to be involved in genomic imprinting or X inactivation.
Many genes were found to contain LD blocks across several cell lines. Out of 309 genes that are associated with at least one LD block in at least one cell line, 100 genes (32%) contain LD blocks in at least five cell lines. Eighteen genes (5.9%) are found to have LD blocks in at least 12 cell lines (Supplemental Table 4). GNAS and SNRPN contain LD blocks in all 16 cell lines, and NNAT contains LD blocks in 13 cell lines. Overall, 31%∼50% of fuzzily methylated CpGs had strong LD (_r_2 > 0.3) with at least one adjacent site, and 14%∼36% of fuzzily methylated regions were found within methylation LD blocks. A fraction of fuzzily methylated CpGs has neighbors with correlated methylation patterns, but many of these patterns are too localized to meet our definition for a methylation LD block. The organized CpG methylation patterns within LD blocks cannot be explained by stochastic effects. They are evidence of either epigenetic heterogeneity across cells or preferential methylation of one particular parental chromosome copy.
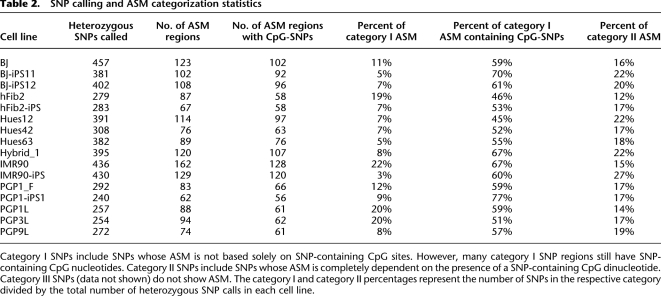
To explore whether the methylation LD blocks are due to heterogeneity of cell populations or unequal methylation of chromosome copies, we developed an algorithm for SNP calling from bisulfite sequencing reads (see Methods). Heterozygous SNPs allowed us to distinguish methylation haplotypes from the two parents, and to detect allele-specific methylation. If fuzzy methylation was simply due to the presence of heterogeneity in cell populations, we would not expect to observe allelic preference within a cell. Due to the reduced sequence complexity of the bisulfite-converted genome, we also adapted SAMtools to make SNP calls (Li et al. 2009a). We compiled a list of confident SNP calls based on the intersection between our own algorithm and SAMtools. In total, we identified 240–457 heterozygous SNPs and 197–391 homozygous SNPs in each cell line (Supplemental Table 5). The three cell lines PGP1L, PGPF, and PGP1-iPS were derived from the same individual (PGP1) in the Personal Genome Project, whose full genome was recently sequenced (Drmanac et al. 2010). We observed a high concordance between the SNPs called from our bisulfite sequencing data and those generated by whole-genome sequencing. For the PGP1 lines, 96%–97% of SNP calls made by Complete Genomics matched our SNP calls. As expected, cell lines of identical genetic background showed an expected higher number of pairwise overlapped SNP calls relative to the other cell lines (Supplemental Tables 6,7). From the bisulfite sequencing reads that contain both heterozygous SNPs and at least one CpG site, we identified methylation that significantly associates with one allele of a SNP using the Fisher's exact test (see Methods). We also required a minimum methylation frequency difference of 0.1 between the alleles for ASM categorization. In each cell line, 23%∼37% of heterozygous SNPs were found to associate with ASM (Table 2). Only a small fraction of ASM (6%) was consistent across all cell lines in which the heterozygous SNPs are present. The remaining cases are either cell-type-specific or individual-specific (Supplemental Fig. 4). We also compared the allelic methylation frequencies of 980 individual CpG sites that are linked to SNPs from two batches of IMR90 fibroblast cultures. Excellent correlation was observed between the biological replicates (Pearson correlation coefficient _r_2 = 0.90), which indicated that our observations were not due to technical artifacts or biological fluctuations.
Table 2.
SNP calling and ASM categorization statistics
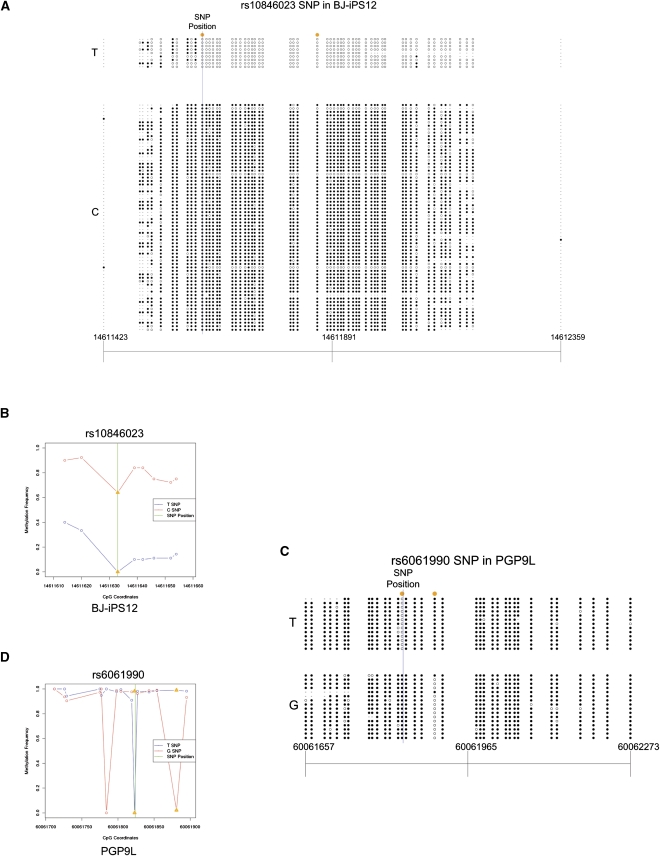
We validated 12 SNP sites by performing bisulfite PCR, cloning, and Sanger sequencing on one or more cell lines in which SNPs were called. A total of 21 SNP regions were amplified and Sanger sequenced. The Sanger regions that show ASM fall into two categories. In category I, more than one adjacent CpG site exhibit consistent bias in methylation. The known imprinted regions fall into this category. However, even in autosomal regions not known to be related to genetic imprinting, similar allelic preference can extend over 900 bp (Fig. 2A,B; Supplemental Fig. 1A,B). In category II, ASM is highly localized and restricted to only a very small number of CpG sites in a region (Fig. 2C,D; Supplemental Fig. 5). Seven sequences had inconsistent ASM classification between the Illumina and Sanger sequencing. Four of these inconsistencies were due to the inability to establish statistical ASM significance due to the low Sanger sequencing read depth. Two SNP sites predicted by the Illumina data were not found in the Sanger data (Supplemental Table 1), which are likely due to incorrect mapping of short bisulfite sequencing reads. One SNP site had inconsistent ASM behavior between Sanger and Illumina data. Overall, we found a Pearson correlation coefficient of _r_2 = 0.78 for the allelic methylation frequencies of CpGs shared between the Sanger sequencing and Illumina data sets.
Figure 2.
Allele-specific methylation. (A) Sanger reads from BJ-iPS12 show an extended ASM region in dbSNP 129 rs10846023 indexed region (chr12:14611423–14612359) in the intron of the RAB11FIP1 (also known as FLJ22622) gene. This ASM region includes two CpG dinucleotides that overlap with SNPs, but the ASM is not limited to these sites. (Orange circles) SNPs that overlap with CpG dinucleotides. (B) An allele-specific methylation frequency graph based on aligned Illumina data showing ASM at rs10846023 in BJ-iPS12 (chr12:14611616–14611654). (_Y_-axis) Methylation frequency, where a value of 1 indicates complete methylation at a CpG dinucleotide; (_x_-axis) chromosomal coordinates. (C) Sanger sequence data around SNP site rs6061990 (TAF4 intronic region, chr20:60061657–60062273) in PGP9L illustrates an example where ASM is solely dependent on SNPs at CpG dinucleotides. (D) An allele-specific methylation frequency graph based on Illumina data showing ASM at rs6061990 in PGP9L (chr20:60061712–60061895). (Green line) SNP position; (orange triangles) SNPs that overlap with CpG dinucleotides. Note that the Illumina data show ASM at a CpG (chr20:60061784) that is not supported by the Sanger data.
Next, we explored potential _cis_-regulatory mechanisms that account for ASM. We identified many ASM regions that contained at least one heterozygous SNP that overlapped with a CpG dinucleotide. As an example, the region containing the T/C SNP site rs10846023 in the BJ-iPS12 cell line shows ASM that spans over 500 bp. This SNP is present at a CG dinucleotide, and the T allele disrupts the CG site. However, the ASM behavior at this site is also exhibited in nearby sites (Fig. 2A,B), which suggests that some uncharacterized regulatory mechanisms determine the ASM and such regulation may be dependent on the local sequence context in cis. We also found examples of specific methylation behavior that did not correlate with the base identity of a nearby SNP (Supplemental Fig. 6). Given the presence of SNP-containing CpGs, we refined our ASM categories so that category I represented ASM that was not solely dependent on SNP-containing CpGs. Category II represented ASM that solely depended on SNP-containing CpGs. We found 45%–77% of category I SNP regions have SNPs at CpG dinucleotides, which suggests that SNPs on CpG sites may have an impact on differential methylation establishment if the SNPs are located in regions where methylation regulators are involved. In contrast are the regions where the ASM affects only CpG locations overlapping with SNPs. One allele of such SNPs eliminates the CpG sites, thus preventing them from being methylated. One example is the region around SNP site rs6061990 in PGP3L (Supplemental Fig. 5) and PGP9L (Fig. 2C,D). The PGP9L cell line shows ASM at two CpG dinucleotides while the region in PGP3L shows ASM at one CpG site. This example demonstrates that individual ASM sites can be dependent on sequence difference alone. A complete list of ASM categorizations of SNP regions is available on our supplemental website (http://genome-tech.ucsd.edu/public/ASM/) and in the Supplemental materials. Overall, 38%∼88% of ASM regions are solely due to the presence of SNPs at CpG dinucleotides, revealing that genetic variation at CpG sites is a dominating factor for ASM.
Finally, ASM regions that span across multiple CpG sites are likely regulated by other _cis_-regulatory mechanisms. We found many cases where the methylation state closely correlated with the alleles identified by a SNP. Such behavior is more likely explained by an allele-specific regulatory mechanism rather than cell subpopulations undergoing different epigenetic processes. For example, regions that exhibit reverse allelic preference in different cell lines of the same genetic background are observed (Supplemental Fig. 7), which can only be explained by a regulatory mechanism that involves more than one _cis_-regulator with opposite effects. Out of the category I ASM regions that are covered by our LD analysis, 8%–35% are within methylation LD blocks, demonstrating that ASM analysis uniquely identified new regions where methylation was due to allele specific _cis_-regulation. Finally, the two IMR90 biological replicates showed that 83% of ASM calls were consistent in both experiments (201 matched ASM SNP calls out of 242 total ASM SNP calls). Therefore, ASM is due to biological regulation instead of biological noise or technical artifacts.
Discussion
We set out in this analysis to investigate the basis of fuzzy methylation with two novel approaches: adapting linkage disequilibrium analysis to methylation data, and performing SNP calling on bisulfite sequencing reads. This study represents the largest survey of ASM in the human genome to date. Hundreds of methylation LD blocks were identified from over 2000 CpG islands in two human chromosomes and dozens of other regions. Roughly 30%∼48% of fuzzily methylated CpGs were found to have _r_2 > 0.3 and 14%∼36% of fuzzily methylated CpGs were found within LD blocks. This shows stochastic effects do not explain a significant amount of observed fuzzy methylation. Our SNP ASM analysis found that 23%–37% of heterozygous SNPs are associated with ASM. The frequency of ASM is similar to the frequency of allele-specific gene expression (ASE) observed in the human genome (Yan et al. 2002; Ge et al. 2009; Lee et al. 2009; Zhang et al. 2009a; Heap et al. 2010). ASE has been considered as an important indicator for the presence of functional _cis_-regulatory variants (Pastinen and Hudson 2004). ASE and ASM could be tightly coupled by the same _cis_-regulatory variants (Ghotbi et al. 2009; Milani et al. 2009). Similar to gene expression, methylation or ASM can be considered as quantitative traits for population genomic analysis. One important finding of this work is that a SNP could be a functional _cis_-regulatory variant by disrupting a CpG methylation site. According to the snp129 database, there are 225,659 known SNPs that locate on CpG sites. In addition, because CpG dinucleotides are highly mutable, there are many CpG rare variants in individual genomes (Li et al. 2009b). CpG SNPs are likely an important class of _cis_-regulatory polymorphisms that connects genetic variation to the individual variability of the epigenome.
Methods
Bisulfite targeted reseqeuncing with padlock probes
Bisulfite padlock capture and targeted resequencing were performed as described (Deng et al. 2009). Briefly, genomic DNA was extracted from frozen pellets of lymphocyte, fibroblast, iPS, or hES cells using Qiagen DNeasy columns, and bisulfite converted with Zymo DNA methylation Gold kit (Zymo research). The Cpg30k padlock probe library was annealed to the bisulfite-converted sample DNAs, circularized, and amplified by PCR. Random shotgun sequencing library was generated by USER/MmeI enzymatic fragmentation from the amplicons of captured targets, which then were subsequently sequenced by Illumina Genome Analyzer.
Bisulfite PCR and cloning for Sanger sequencing
Bisulfite PCR reactions were performed in 100-μL reactions including 50 ng of bisulfite-converted genomic DNA, 200 μM dNTP, 0.4 μM forward and reverse PCR primers, and 1× IQ PCR Supermix (Bio-Rad) for 2 min at 94°C; 45 cycles of 30 sec at 94°C, 1 min at 62°C, 1 min at 72°C; and finally 5 min at 72°C. Bisulfite PCR products were cloned in the pCR 2.1-TOPO vector (Invitrogen), and multiple clones were picked and sequenced at Agencourt. The primer sequences are in Supplemental Table 2.
Statistical analysis
Raw Illumina sequencing reads from the previously published data set and the new paired-end data set were combined to produce a data set that consisted of 16 cell lines (Deng et al. 2009). These combined reads were mapped to the in silico bisulfite-converted human genome sequence (hg18) via SOAP2 (Li et al. 2009c). Mates from paired-end reads were mapped independently. Reads were allowed to have up to two mismatches, and reads that mapped to multiple locations were excluded. Sanger reads were mapped analogously except that UCSC BLAT (Kent 2002) was used instead of SOAP to map these sequences to a reference template. Due to the longer length of Sanger sequences and their known position in the genome, Sanger sequence alignments were allowed to have multiple mismatches and gaps.
SNPs were called with an algorithm that assigned probabilities to each genotype. Bisulfite-converted strands were analyzed independently, and the probability of each genotype was assessed via the Fisher's exact test. SNP calls were limited to annotated dbSNP 129 sites, and quality score filtering was applied to filter out low-quality base calls. There was a 10× minimum read depth per strand requirement for SNP calling. SNP calls were also made with SAMtools, and the intersection of SNP calls between our algorithm and SAMtools was used in further analyses. SNP sites were grouped into three categories: (1) SNPs showing either CpG-specific or averaged ASM that is independent of SNPs at CG sites, (2) SNPs showing ASM that is dependent on the presence of a SNP CpG overlap, and (3) SNPs that do not show ASM. In order for a CpG site or SNP region to be classified as ASM in the Illumina data, it needed to have an allelic methylation frequency difference of at least 0.1. Sanger sequence ASM labeling used the same statistical test as described above without the allelic methylation frequency difference requirement due to the lower read coverage.
ASM was determined by creating a 2 × 2 contingency table where the two columns represented the two alleles identified by a SNP. The two rows represented the counts of methylated and unmethylated cytosines at CpG site(s) located on a SNP-containing read. Each CpG site was treated independently for CpG-specific ASM, and for average ASM the methylated and unmethylated cytosine counts were summed across CpG sites allele, specifically. If a SNP region contained SNPs at CpG sites, the average ASM calculation was repeated while excluding SNP-containing CpG sites.
Reads containing two or more CpG sites were used for the linkage disequilibrium _r_2 analysis. A CpG pair needed to have a minimum 10× read depth. LD _r_2-values were considered high if they were greater than 0.3. For the Illumina data, an LD block was created if a CpG pair had a high _r_2-value. This region was expanded if there was an overlapping CpG pair with a high _r_2-value or a CpG pair within 100 bp with a high _r_2-value. LD blocks span at least 100 bp and contain at least 10 CpG pairs with _r_2 > 0.3.
All raw data, SNP calls with ASM categorizations, LD analyses, Sanger diagrams, and methylation frequency data are available at http://genome-tech.ucsd.edu/public/ASM/ and in the Supplemental materials. The raw Illumina sequences are available at the NCBI Sequence Read Archive (accession no. SRA012435).
Acknowledgments
We thank the laboratories of George Church, George Daley, Kevin Eggan, Konrad Hochedlinger, and James Thomson for providing DNA samples, and the UCSD BioGem Core facility for assistance with Illumina sequencing. This study is supported by the UCSD new faculty startup fund, NIH/NIDA R01-DA025779 (K.Z.), and NIH R01GM072856 (W.W.). J.D. was sponsored by a CIRM post-doctoral fellowship.
Author contributions: K.Z. and W.W. oversaw the project. J.D. performed padlock bisulfite sequencing and various validation assays. R.S. performed algorithm development and bioinformatics analysis. R.S., J.D., W.W., and K.Z wrote the manuscript.
Footnotes
References
- Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol 27: 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. 2009. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet 41: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R, Sparks AB, Callow MJ, Halpern AL, Burns NL, Kermani BG, Carnevali P, Nazarenko I, Nilsen GB, Yeung G, et al. 2010. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327: 78–81 [DOI] [PubMed] [Google Scholar]
- Eggermann T 2009. Silver-Russell and Beckwith-Wiedemann syndromes: Opposite (epi)mutations in 11p15 result in opposite clinical pictures. Horm Res 71 (Suppl 2): 30–35 [DOI] [PubMed] [Google Scholar]
- Ge B, Pokholok DK, Kwan T, Grundberg E, Morcos L, Verlaan DJ, Le J, Koka V, Lam KC, Gagne V, et al. 2009. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet 41: 1216–1222 [DOI] [PubMed] [Google Scholar]
- Ghotbi R, Gomez A, Milani L, Tybring G, Syvanen AC, Bertilsson L, Ingelman-Sundberg M, Aklillu E 2009. Allele-specific expression and gene methylation in the control of CYP1A2 mRNA level in human livers. Pharmacogenomics J 9: 208–217 [DOI] [PubMed] [Google Scholar]
- Heap GA, Yang JH, Downes K, Healy BC, Hunt KA, Bockett N, Franke L, Dubois PC, Mein CA, Dobson RJ, et al. 2010. Genome-wide analysis of allelic expression imbalance in human primary cells by high throughput transcriptome resequencing. Hum Mol Genet 19: 122–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkel K, Spadola A, Yuan E, Kosek J, Jiang L, Hod E, Li K, Murty VV, Schupf N, Vilain E, et al. 2008. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat Genet 40: 904–908 [DOI] [PubMed] [Google Scholar]
- Lee JH, Park IH, Gao Y, Li JB, Li Z, Daley GQ, Zhang K, Church GM 2009. A robust approach to identifying tissue-specific gene expression regulatory variants using personalized human induced pluripotent stem cells. PLoS Genet 5: e1000718 doi: 10.1371/journal.pgen.1000718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 2009a. The Sequence Alignment/Map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Gao Y, Aach J, Zhang K, Kryukov GV, Xie B, Ahlford A, Yoon JK, Rosenbaum AM, Zaranek AW, et al. 2009b. Multiplex padlock targeted sequencing reveals human hypermutable CpG variations. Genome Res 19: 1606–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J 2009c. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25: 1966–1967 [DOI] [PubMed] [Google Scholar]
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. 2009. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ 2007. Computational and experimental identification of novel human imprinted genes. Genome Res 17: 1723–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L, Lundmark A, Nordlund J, Kiialainen A, Flaegstad T, Jonmundsson G, Kanerva J, Schmiegelow K, Gunderson KL, Lonnerholm G, et al. 2009. Allele-specific gene expression patterns in primary leukemic cells reveal regulation of gene expression by CpG site methylation. Genome Res 19: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen T, Hudson TJ 2004. _Cis_-acting regulatory variation in the human genome. Science 306: 647–650 [DOI] [PubMed] [Google Scholar]
- Robertson KD 2005. DNA methylation and human disease. Nat Rev Genet 6: 597–610 [DOI] [PubMed] [Google Scholar]
- Spivakov M, Fisher AG 2007. Epigenetic signatures of stem-cell identity. Nat Rev Genet 8: 263–271 [DOI] [PubMed] [Google Scholar]
- Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW 2002. Allelic variation in human gene expression. Science 297: 1143. [DOI] [PubMed] [Google Scholar]
- Zhang K, Li JB, Gao Y, Egli D, Xie B, Deng J, Li Z, Lee JH, Aach J, Leproust EM, et al. 2009a. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat Methods 6: 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Rohde C, Reinhardt R, Voelcker-Rehage C, Jeltsch A 2009b. Non-imprinted allele-specific DNA methylation on human autosomes. Genome Biol 10: R138 doi: 10.1186/gb-2009-10-12-r138 [DOI] [PMC free article] [PubMed] [Google Scholar]