A super-modular FHA/BRCT-repeat architecture mediates Nbs1 adaptor function in response to DNA-damage (original) (raw)
. Author manuscript; available in PMC: 2010 Jul 9.
SUMMARY
The Mre11/Rad50/Nbs1 (MRN) protein complex plays central enzymatic and signaling roles in the DNA-damage response. Nuclease (Mre11) and scaffolding (Rad50) components of MRN have been extensively characterized but the molecular basis of Nbs1 function has remained elusive. Here, we present a 2.3Å crystal structure of the N-terminal region of fission yeast Nbs1, revealing an unusual but conserved architecture in which the FHA and BRCT-repeat domains structurally coalesce. We demonstrate that di-phosphorylated pSer-Asp-pThr-Asp-like motifs, recently identified as multi-copy docking sites within human Mdc1, are evolutionarily conserved Nbs1 binding targets. Furthermore, we identify similar phospho-motifs within Ctp1, the fission yeast orthologue of the human tumor-suppressor, CtIP, and show that their interactions with the Nbs1 FHA domain are necessary for Ctp1-dependent resistance to DNA-damage. Finally, we establish that human Nbs1 interactions with Mdc1 can occur through both its FHA and BRCT-repeat domains, suggesting how their structural and functional inter-dependence may underpin Nbs1 adaptor functions in the DNA-damage response.
INTRODUCTION
Genomic integrity is constantly challenged by the generation of DNA lesions, of which double-stranded breaks (DSBs) are generally considered the most toxic. DSBs can arise through the actions of DNA-damaging chemicals and ionizing radiation, but also occur at sites of stalled DNA replication and as intermediates during programmed genomic rearrangements such as VDJ recombination, immunoglobulin class-switching and meiotic recombination. DNA-damage detection initiates a complex and orchestrated set of intracellular responses involving kinase-signaling cascades, cell-cycle checkpoint activation and the deployment of DNA-repair proteins (Riches et al., 2008). Central to these processes is a diverse group of adaptor and scaffold proteins that function as platforms for the assembly of multi-protein complexes through the combinatorial activities of phosphoserine/threonine binding protein modules, such as forkhead-associated (FHA), Brca1 C-terminal (BRCT)-repeat, Polo-box, WW and WD40-repeat domains (Reviewed in Seet et al., 2006) which interact with specific motifs following their phosphorylation by DNA-damage activated or cell-cycle responsive serine/threonine kinases.
Human Nbs1 (also known as Nibrin or p95) is a 754 amino-acid residue subunit of the Mre11-Rad50-Nbs1 (MRN) complex, which binds to DSBs and acts as a bridge to hold the DNA ends in close proximity and promote their rejoining (Williams et al., 2007). Nbs1 was originally identified as the gene mutated in Nijmegen breakage syndrome (NBS), a rare autosomal-recessive human disease characterized by immune disorders, microcephaly, growth retardation, hypersensitivity to ionizing radiation and predisposition to lymphoid cancers (Carney et al., 1998; Matsuura et al., 1998; Varon et al., 1998). Although homozygous-null Nbs1 mutations are embryonic lethal in mouse models (Zhu et al., 2001), these human diseases are characterized by a variety of hypomorphic mutations that can occur throughout the NBS1 gene.
Many NBS phenotypes overlap with those of another cancer-predisposition syndrome, ataxia-telangiectasia, which is caused by defects in the protein kinase ATM (ataxia-telangiectasia-mutated). It is now evident that Nbs1 functions in ATM activation, while also serving as a substrate for ATM-mediated phosphorylation on Ser-278 and Ser-343 (Difilippantonio and Nussenzweig, 2007). Nbs1 makes direct interactions with ATM and Mre11 through distinct motifs within its C-terminal region (Desai-Mehta et al., 2001; Falck et al., 2005; You et al., 2005) whereas the Nbs1 N-terminal region contains FHA and BRCT motifs that are characteristic of many other DNA-damage response proteins and which often mediate phospho-dependent protein-protein interactions. Functionally, Nbs1 seems to play key roles in most or all of the DNA-damage-checkpoint signaling functions of the MRN complex (D’Amours and Jackson, 2002) through interactions with a number of proteins, the best characterized being Mdc1 (mediator of the DNA-damage checkpoint 1) and ATM itself (Chapman and Jackson, 2008; Falck et al., 2005; Lukas et al., 2004; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008; You et al., 2005; Zhao et al., 2000). However, Nbs1 activities do not appear to be restricted to checkpoint signaling. Indeed, more direct contributions of Nbs1 to DSB repair are suggested by the observation that DNA-damage sequestration of CtIP, a molecule required for efficient DSB resection (Limbo et al., 2007; Sartori et al., 2007), is facilitated through direct interactions with Nbs1 in human cells (Chen et al., 2008).
In spite of the array of Nbs1 activities, little is currently known about how it mediates its diverse protein interactions at the molecular level. Here, we describe an integrated structural, biochemical, genetic and cell-biological analysis of the Nbs1 N-terminal FHA/BRCT-repeat region. This work reveals how the conserved architecture and phospho-dependent protein binding functions of the Nbs1 N-terminus play crucial roles in mediating Nbs1 adaptor functions in the DNA-damage response to promote DSB signaling and repair.
RESULTS & DISCUSSION
Structure of the Nbs1 N-terminal region reveals an unusual FHA/BRCT-repeat architecture
Following extensive screening of Nbs1 proteins from several species, we were able to express and purify 382 and 323 residue N-terminal fragments of human Nbs1 (hNbs1) and yeast Schizosaccharomyces pombe Nbs1 (spNbs1), respectively. The structure of the S. pombe protein was determined by single-wavelength anomalous diffraction methods at an initial resolution of 2.6Å, and then further refined against data extending to 2.3Å spacing collected from crystals of reductively methylated protein (Table S1). Representative electron density for the SAD-phased selenomethionine structure and the final refined methylated protein are shown in Figure S1.
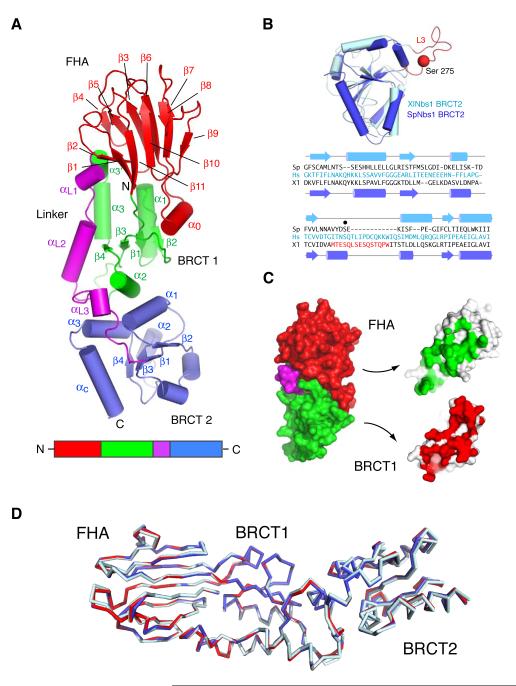
Overall, the spNbs1 N-terminus is extended, with approximate dimensions of 95 × 30 × 25 Å (Figure 1A). The N-terminal 114 residues adopt a β-sandwich fold characteristic of FHA domains: small phospho-dependent binding modules that function through recognition of phosphothreonine (pThr)-containing motifs in target proteins (Mahajan et al., 2008). Many functional studies of Nbs1 have been predicated on the assumption that only a single BRCT motif exists C-terminal to the Nbs1 FHA domain. Notably, the spNbs1 structure reveals the presence of tandem BRCT repeats, despite the lack of any significant homology between the second motif and other known BRCT domains. In the light of previous sequence analyses of various Nbs1 orthologues (Becker et al., 2006) and comparison of our spNbs1 structure with the NMR analysis of the second BRCT repeat from Xenopus laevis Nbs1 (Xu et al., 2008) (Figure 1B), it seems that the FHA/BRCT2 composition is universally conserved in eukaryotic Nbs1 proteins. Most significantly, our structure reveals an unprecedented structural fusion of the FHA domain and first BRCT repeat. Rather than being flexibly linked in a ‘beads-on-a-string’ arrangement as might be expected in a classical modular domain system, they are associated through a substantial interface within which 2150 Å2 of solvent-accessible surface is buried (Figure 1C). The bulk of the interfacial interactions occur between the base of the FHA domain and α1, α3, αL1 and α1/β2 loop of BRCT1. Additional stabilizing contacts are provided by the helical insertion in the FHA domain (α0) and the α1-β2/β2-β3 loops from BRCT1. Overall, the relative orientation of the FHA, BRCT1 and BRCT2 domains for three independent molecules (two methylated, one unmethylated) in the two crystal forms are almost identical, suggesting that the extensive FHA-BRCT1 interactions impose a rather rigid sub-structure (Figure 1D).
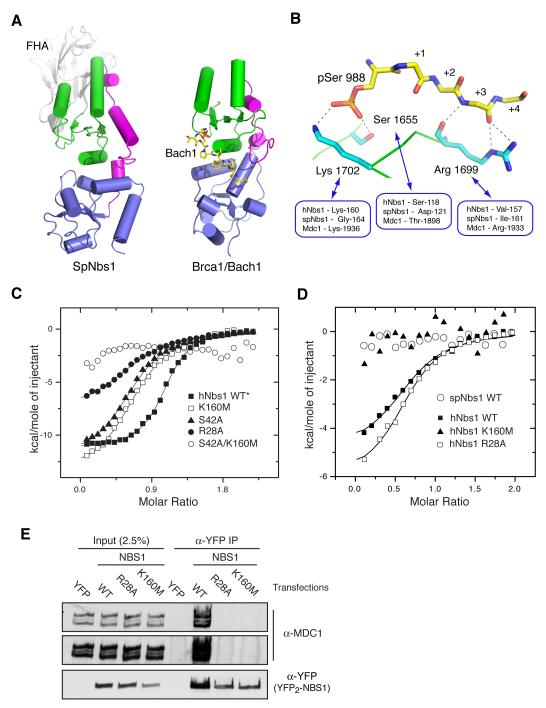
Figure 1. Structure of the S. pombe FHA/BRCT region.
(A) Ribbons representation of the spNbs1 FHA/BRCT region with β-strands shown as arrows and α-helices as tubes. The FHA, BRCT1, linker and BRCT2 domains are colored red, green, magenta and blue respectively with the major secondary structural elements labeled. All structural representations were generated using PyMOL (http://pymol.sourceforge.net/).
(B) Structural superposition (upper) shows a remarkable similarity between the second BRCT motif of spNbs1 and that of Xenopus laevis (PDBID: 2K2W) with a root mean square deviation of 2.0 Å for 65 matched Cα atoms from each domain. The ‘L3’ loop that contains a major site of ATM-phosphorylation corresponding to Ser-278 in human Nbs1 (Zhao et al., 2000), is replaced by a partially disordered helical loop in spNbs1. The resulting structure-based sequence comparison (lower) shows only a general conservation of non-polar residues that form the hydrophobic BRCT core.
(C) FHA and BRCT1 domain interactions are shown in an ‘open book’ representation. The contact areas of the FHA with BRCT1 (green) and BRCT1 with the FHA (red) represent >2000Å2 of total buried solvent-accessible surface.
(D) Least-squares superposition of the three crystallographically independent molecules from two crystal forms using the 114 Cα atoms from each FHA domain shows that the relative arrangements of FHA and BRCT1 are nearly identical although some small displacements of BRCT2 are evident.
The Nbs1 ‘super-modular’ architecture is conserved
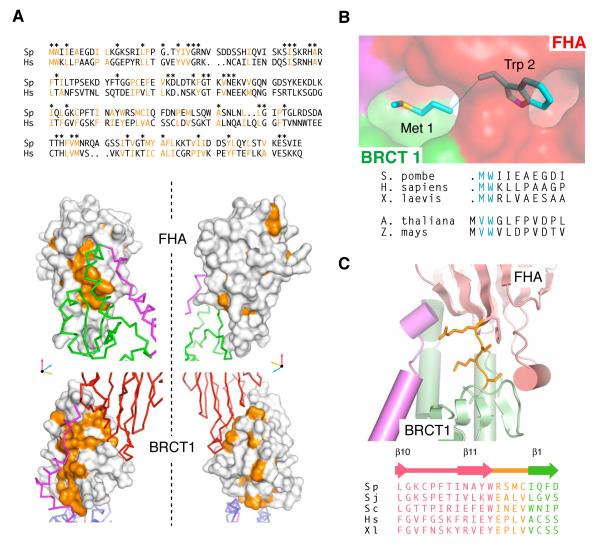
Several lines of evidence indicate that the overall architecture observed for spNbs1 is common to all eukaryotic Nbs1 orthologues. Thus, although limited sequence identity for interfacial residues is evident between spNbs1 and hNbs1, the non-polar character of amino-acid residues that contribute core FHA-BRCT1 interactions is conserved. (Figure 2A). In addition, a strip of conserved hydrophobic surface follows the path of the linker extending from the BRCT1-FHA interface to BRCT2. Notably, the N-terminal Met-Trp pair of spNbs1 is buried in the structure and forms an intrinsic part of the non-polar FHA-BRCT1 interface (Figure 2B). Trp-2 is highly conserved in animals and fungi and because of its bulk, it presumably insulates the initiator methionine from co-translational excision (Giglione et al., 2004). Plants, however appear to have followed a different evolutionary path to the same structural end-point whereby an extra valine residue is positioned to take the place of Met-1 and maintain the integrity of the FHA-BRCT1 interface following more efficient methionine-amino-peptidase cleavage expected when small hydrophobic residues are present in the second position (Martinez et al., 2008). Overall, the importance of the structure around Met-1/Trp-2 is exemplified by the insolubility/aggregation of recombinant Nbs1 expressed with either N-terminal GST or hexa-histidine affinity tags (data not shown). These observations, together with the absence of any significant ‘linker’ region between the C-terminus of the FHA domain and N-terminus of BRCT1 in all Nbs1 sequences (Becker et al., 2006) (Figure 2C), support our contention that the structural coupling and precise spatial apposition of the FHA and BRCT domains is likely to be crucial for Nbs1 functions in all eukaryotes.
Figure 2. A conserved FHA/BRCT1 architecture.
(A) Conserved hydrophobic residues (orange) within S. pombe and human Nbs1 proteins (top panel) mapped onto the spNbs1 crystal structure (lower panel) show that major, surface exposed non-polar patches (orange) common to both distantly related orthologues are located primarily at the FHA/BRCT1 and BRCT1-linker interfaces.
(B) Surface representation showing how the highly conserved N-terminal methionine/tryptophan motif is buried at the interface between the FHA domain (red surface) and BRCT1 (green surface).
(C) The FHA and BRCT1 domains (pink and green respectively) are connected through a short linker (orange) in all orthologues (Sp - S, pombe, Sj - S. japonicum, Sc - S. cerevisiae, Hs - Homo sapiens, Xl - Xenopus laevis)
Structural context of human disease-associated Nbs1 mutations
The apparent structural conservation between Nbs1 orthologues allows us to place in a molecular context various human disease-associated Nbs1 mutations that are located within the FHA/BRCT region. To date, around 90% of patients suffering from NBS are homozygous or heterozygous for a founder mutation, 657del5, a 5 bp deletion in exon 6 within the coding region for the BRCT1-BRCT2 linker that results in production of C- and N-terminally truncated proteins termed p26 and p70, respectively (Maser et al., 2001) (Figure S2A). In addition, a number of germ-line and somatic missense mutations in the Nbs1 N-terminal region have been noted in patients suffering from NBS and in a variety of cancers (http://www.nijmegenbreakagesyndrome.net) (Huang et al., 2008) (Figure S2B). Although the degree to which these mutations might contribute to disease is mostly unclear, we note that four (T148I, L150F, I171V and V210F) lie within a significant hydrophobic cluster in the BRCT1-domain core and the BRCT1-linker interface, suggesting that these lesions may result in structural perturbations, potentially contributing to Nbs1 protein instability in affected individuals (Figure S2C). Nonetheless, it remains possible that some mutations (such as V28I located immediately C-terminal to the highly conserved FHA phosphopeptide binding residue Arg-27) directly affect phospho-dependent and/or phospho-independent interaction surfaces on Nbs1.
Evolutionarily conserved binding of pSDpTD-like motifs to the Nbs1 FHA domain
The functional importance of the Nbs1 FHA/BRCT region is well established but the molecular basis for this remains obscure. Recruitment of the MRN complex to sub-nuclear foci at DSB sites has been proposed to involve a direct interaction between human Nbs1 (hNbs1) and γH2AX (Kobayashi et al., 2002), the ATM-phosphorylated form of variant histone H2A that serves as a major marker for damaged chromatin. However, by isothermal titration calorimetry (ITC), we were unable to detect any interactions of recombinant hNbs1 or spNbs1 proteins with peptides representing human or S. pombe γH2AX (Figure S3). This confirms the lack of binding of hNbs1 to γH2AX peptides observed previously by peptide pull-down assays (Chapman and Jackson, 2008) and strongly suggests that direct γH2AX-binding by Nbs1 is unlikely to contribute significantly to MRN localization to damaged chromatin.
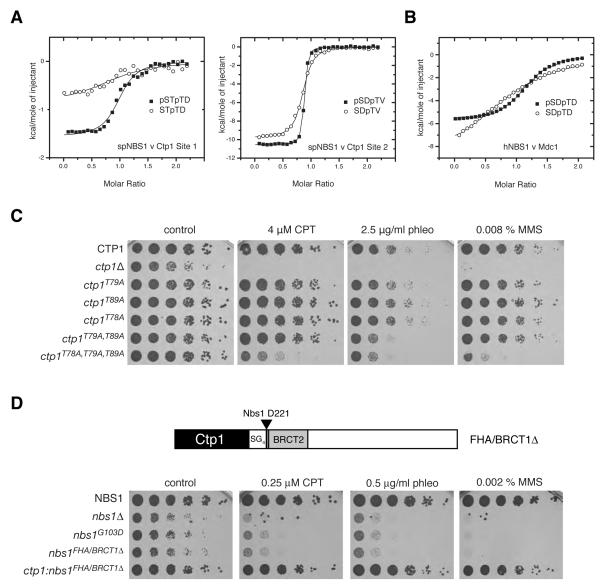
More recently, the mammalian MRN has been shown to be indirectly sequestered to DNA-damage sites through Nbs1 interactions with a cluster of six di-phosphorylated casein-kinase 2 (CK2) target sites within Mdc1, each with a core consensus sequence phospho-Ser, Asp, phospho-Thr, Asp (pSDpTD; (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008). This Mdc1/MRN/ATM complex is, in turn, tethered to damaged chromatin via interactions between the Mdc1 BRCT-repeat domain and γH2AX (Lee et al., 2005; Stucki et al., 2005). Consistent with the dependence of Mdc1-Nbs1 interactions on the Nbs1 FHA domain (Chapman and Jackson, 2008; Lukas et al., 2004; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008), we found that the hNbs1 fragment bound a synthetic Mdc1 pSDpTD motif peptide (Mdc1 residues 325-336) with an affinity of ~1 μM (Figure 3A, upper panel; Table S2) and a stoichiometry significantly greater than 1:1 (see below). In addition, and in spite of the absence of any clear Mdc1 orthologues in yeast, the purified spNbs1 protein bound the same Mdc1 phosphopeptide with a slightly higher affinity of 0.5 μM (Figure 3A, lower panel). By contrast, neither spNbs1 nor hNbs1 bound to a peptide of the same sequence in which the threonine was not phosphorylated, reflecting the acute pThr-selectivity of FHA domains.
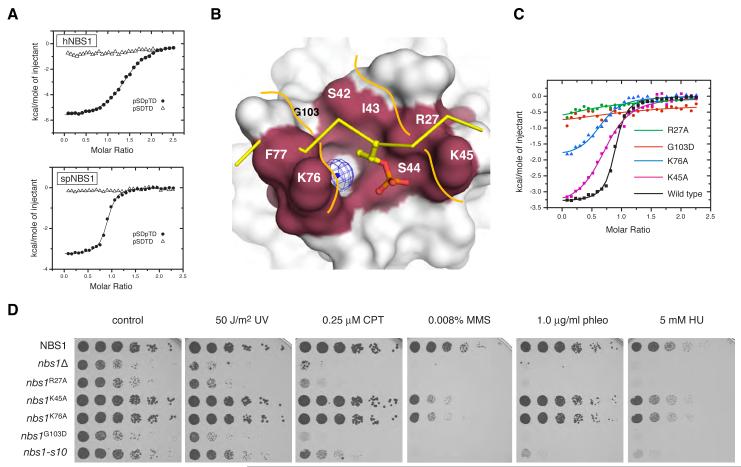
Figure 3. Yeast and human Nbs1 bind to Mdc1 pSDpTD-motif peptides.
(A) ITC experiments demonstrate that purified proteins of both hNbs1 (upper) and spNbs1 (lower) bind di-phosphorylated synthetic Mdc1 pSDpTD-motif peptides (closed circles). No binding of hNbs1 and spNbs1 to identical sequences containing a singly phosphorylated serine residue was detectable (open triangles).
(B) The phosphopeptide binding region of the FHA domain of spNbs1 is shown as a surface representation with conserved residues forming a potential binding channel highlighted. A superimposed phosphopeptide (yellow sticks) from the Chk2-FHA complex structure (Li et al., 2002) (PDB ID: 1GXC) highlights significant steric clashes with Phe-77 of spNbs1 and the phosphopeptide backbone.
(C) Binding contributions of residues that form the predicted ligand-binding surface of spNbs1 were assessed by mutagenesis and ITC and compared with wild-type spNbs1 binding to the Mdc1 pSDpTD di-phosphopeptide (black line).
(D) Nbs1 FHA-dependent phospho-interactions promote normal growth and resistance to DNA-damage. Five-fold serial dilutions of the indicated strains were plated on control medium (YEA) or medium supplemented with the indicated drugs, and then incubated for 3 or 5 days (HU) at 30°C (UV, ultra-violet light; phleo, phleomycin; CPT, camptothecin; MMS, methyl-methanesulphonate; HU, hydroxyurea). The additional sensitivity of the nbs1G103D mutant compared with nbs1-s10 may result from differences in genetic background.
Although the spNbs1 FHA domain is closely related to previously determined FHA-domain structures, superposition with a canonical Chk2 FHA-phosphopeptide complex (Li et al., 2002) predicts severe steric clashes of the spNbs1 FHA domain with the phosphopeptide. This results from phenylalanine/tyrosine substitutions in Nbs1 at a conserved position (Phe-77 in spNbs1) normally occupied by an asparagine in other FHA domains (Asn-166 in Chk2) (Figure S4A; Figure 3B). These observations suggest that bound phosphopeptide ligands follow a different path across the spNbs1 FHA binding surface, interacting with a surface groove formed by K76-F77 and S42-I-S-K45 (Figure 3B). These motifs form the β6-β7/β7-β8 loops that are more distant from one another in Nbs1 than in other FHA structures and are instead connected through a bridging water molecule. This generates a somewhat polar floor for the peptide-binding site and potentially provides for some structural flexibility in the relative juxtaposition of the peptide-binding loops. Lys-76 and Lys-45 each lie close to the pThr-binding site and potentially contact the phosphoryl moiety directly (Figure S4B), while Gly-103 and Ser-42 structure the rear of the pocket.
The above ideas were tested by site-directed mutagenesis. Significantly, of the three conserved basic residues located around the canonical FHA-domain binding site, the largest effect on binding to the Mdc1 pSDpTD peptide was observed for alanine substitution of the highly conserved Arg-27 residue (corresponding to Chk2 Arg-117; Figure S4) which reduced apparent binding affinity ~100-fold (Figure 3C; Table S2). More modest effects were seen for the K76A and K45A mutations, which reduced apparent affinity 7- and 5-fold, respectively. A G103D mutation has been implicated in the slow growth and DNA-damage hypersensitivity of the S. pombe nbs-s10 strain identified in a mutagenesis screen (Akamatsu et al., 2008). Given its structural context, we speculated that this substitution might directly affect phosphopeptide binding and, as shown in Figure 3C, this is clearly the case.
To explore the functional relevance of the somewhat unusual FHA-mediated binding mode implied by the above data, we introduced the same mutations into S. pombe cells and determined their effects on cell growth and sensitivity to various DNA-damaging agents. Consistent with previous findings (Akamatsu et al., 2008), a strain bearing the spNbs1 G103D mutation exhibited slow growth and hyper-sensitivity to a range of agents that cause DNA-damage and/or perturb DNA replication (Figure 3D). Moreover, a strain bearing the R27A mutation that severely reduced phosphopeptide binding in vitro, closely mimicked the slow-growth and DNA-damage hypersensitivity phenotypes of the nbs-s10 strain, implying that the G103D/s-10 phenotypes are not merely caused by improper Nbs1 folding. By contrast, strains with alanine substitutions of Lys-45 or Lys-76, which resulted in more modest phosphopeptide binding defects, were not slow-growing and exhibited DNA-damage sensitivity profiles close to those of the wild-type strain (Figure 3D). Nevertheless, these two strains showed significant hyper-sensitivity when exposed to increased concentrations of methyl-methane sulphonate, presumably reflecting the modest impairment of phospho-binding activity observed in ITC experiments. Collectively, these structural, biochemical and functional analyses thus provided strong evidence that FHA-dependent phosphopeptide interactions by spNbs1 are required for its roles in promoting normal growth and resistance to DNA-damaging agents.
Mdc1-like sites in S. pombe Ctp1 are phosphorylated in vivo
The above findings suggested that binding to pSDpTD-like motifs is an important, evolutionarily conserved function for Nbs1. In this regard, we noted that phospho-dependent interactions of budding yeast Nbs1/Xrs2 with the XRCC4 homologue, Lif1 (Matsuzaki et al., 2008) have previously been shown to require the Xrs2 FHA domain and two putative phosphorylation sites (Thr-387 and Thr-417) within Lif1 (Palmbos et al., 2008), both of which are clearly related to the Mdc1 SDTD motifs (Figure 4A). While S. pombe has no clear Mdc1 or Lif1 orthologues, we were intrigued by previous observations that the growth and DNA-repair defects of the nbs-s10 strain (harboring the functionally disruptive G103D FHA-domain mutation) are rescued by Ctp1 over-expression. We confirmed this and further showed that Ctp1 over-expression can suppress the impaired growth and DNA-damage hyper-sensitivity of a strain carrying the spNbs1 R27A FHA mutation that reduces phospho-binding function by ~100-fold in vitro (Figure 4B).
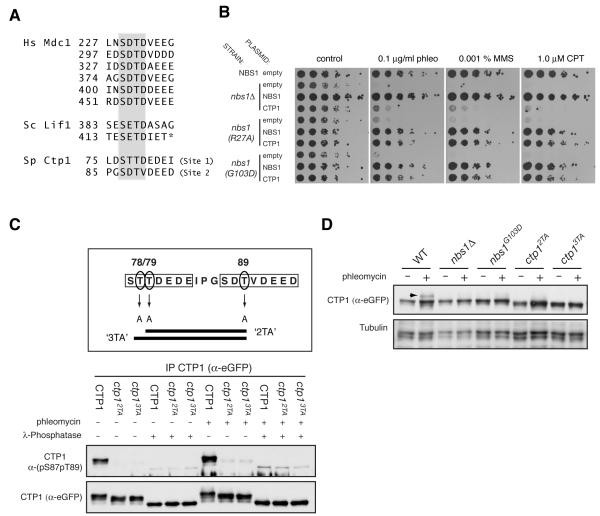
Figure 4. Conserved pSDpTD–like motifs in Ctp1 contribute to DNA-repair in S. pombe.
(A) Conservation of pSDpTD-related motifs between human Mdc1 and the yeast proteins Lif1 and Ctp1 (Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe). The core SDTD motif is highlighted. The asterisk denotes the C-terminus of the full-length Lif1 protein.
(B) Ctp1 over-expression suppresses the DNA damage sensitivity of the Nbs1 FHA phosphopeptide binding mutants. Indicated strains were transformed with a multicopy plasmid, pSP102 expressing either Nbs1 or Ctp1, or with an empty control plasmid. Five-fold serial dilutions of the indicated strains were plated on control medium (YPA-leu) or medium supplemented with the indicated drugs, and then incubated for 3 days at 30°C.
(C) Ctp1 SDTD-like sites are phosphorylated in vivo. eGFP-tagged wild-type or mutant Ctp1 was immunoprecipitated from extracts prepared from cultures of the indicated S. pombe strains, grown in the presence or absence of phleomycin (10 μg, 3 hours). Immunoprecipiates were either treated or mock treated with λ-phosphatase (NEB) before immunoblotting with the indicated antisera. The horizontal bars in the upper panel show the combination of alanine-substitutions in the 2TA and 3TA Ctp1 mutants.
(D) DNA damage-induced hyper-phosphorylation of Ctp1 is dependent on the integrity of Nbs1 and priming by CK2-like phosphorylation. Exponentially growing cultures of the indicated strains were treated as in (C), harvested and extracts immunoblotted with indicated antibodies; Arrow indicates hyper-phosphoryated form of Ctp1.
These data suggested to us that the spNbs1 FHA domain might promote interactions with Ctp1, and that spNbs1 FHA-domain defects can thus be circumvented by Ctp1 over-expression. In line with this idea, like its human counterpart CtIP (Chen et al., 2008; Sartori et al., 2007), S. pombe Ctp1 is a phospho-protein that is recruited to DSBs in a MRN-dependent fashion to promote DSB resection and DNA repair by homologous recombination (Akamatsu et al., 2008; Limbo et al., 2007). Moreover, while inspecting the Ctp1 sequence, we observed that it contains two motifs (Site 1 and Site 2) closely resembling the CK2-consensus, Nbs1-binding sites of Mdc1 and Lif1 (Figure 4A).
To investigate whether Ctp1 is phosphorylated on the Site 1 and 2 motifs in vivo, we generated a phospho-specific antibody directed against a di-phosphorylated peptide comprising Ctp1 Ser-87 and Thr-89 (pS87pT89). We then used this antibody to probe Ctp1-immunoprecipitates from extracts of S. pombe strains expressing epitope-tagged wild-type Ctp1 or Ctp1 mutants (‘2TA’ and ‘3TA’) in which the Thr residues in Site 1 or Site 2 were converted to non-phosphorylatable Ala residues (Figure 4C). Importantly, while the Ctp1 pS87pT89 antibody recognized wild-type Ctp1, its immunoreactivity was lost in the case of the mutant proteins, thus revealing that Ctp1 Site 2 (Ser-87/Thr-89) is phosphorylated in vivo. Consistent with this, we noted that the wild-type Nbs1 derivative migrated more slowly on SDS-polyacrylamide gels than the mutant proteins, while λ phosphatase treatment converted all proteins to a faster migrating form. Collectively, these findings indicated that Ctp1 is subject to multiple phosphorylations in vivo, some of which reside in the Site1/2 motifs. In accord with the apparent constitutive phosphorylation of CK2 motifs in mammalian Mdc1 (Chapman and Jackson, 2008; Melander et al., 2008; Spycher et al., 2008; Wu et al., 2008), we detected Ctp1 Ser-87/Thr-89 phosphorylation irrespective of whether the S. pombe cells had been treated with a DNA-damaging agent (Figure 4C). Significantly, we further noted that the DNA damage-dependent reduction in Ctp1 mobility previously attributed to phosphorylation by S. pombe orthologues of ATR and ATM, Tel1 and Rad3 (Akamatsu et al., 2008) was not observed in the Nbs1-deletion and FHA mutant strains, and was reduced or abolished in the 2TA and 3TA strains (Figure 4D). Together, these data suggested a mechanistic link between Nbs1 FHA-binding activity and priming phosphorylation of Ctp1 by CK2 as a pre-requisite for additional DNA-damage dependent Ctp1 phosphorylation by ATM/ATR-family kinases.
Ctp1 phosphosites bind to Nbs1 and promote resistance to DNA-damaging agents
The foregoing data prompted us to investigate whether Ctp1 phosphosites bind spNbs1 in vitro. This revealed tight spNbs1 binding to phosphorylated forms of both the Ctp1 Site1 and Site 2 motifs (Figure 5A; Table S2). Indeed, the Site 2 peptide containing both pSer-87 and pThr-89 bound some 30-fold tighter than a di-phosphorylated Site 1 peptide (pSer-77/pThr-79), with an apparent Kd of ~50 nM that is among the highest affinity interactions for any FHA domain reported to date. Significantly, interactions of spNbs1 with Site 1 or Site 2 peptides phosphorylated only on pThr-79 or pThr-89 were 8-fold and 10-fold weaker than those for the respective di-phosphorylated versions. This effect recapitulates the 17-fold reduced affinity observed for hNbs1 binding to the mono- versus di-phosphorylated Mdc1 peptide (Figure 5B), demonstrating a remarkably similar pattern of specificity for the fission yeast and human Nbs1 orthologs.
Figure 5. Phospho-dependent Nbs1 interactions with Ctp1 promote DNA-damage resistance.
(A) ITC titrations of spNbs1 with the pSDpTD-like motifs from Ctp1-Site 1 (upper left) and Ctp1-Site 2 (lower left) are depicted by closed squares. Each panel also shows the reduced affinities for singly-phosphorylated pThr peptides of the same length and sequence (open circles).
(B) ITC titrations of hNbs1 with mono-phosphorylated (SDpTD - closed squares) and di-phosphorylated (pSDpTD - open circles) Mdc1 motifs.
(C) Mutation of CK2-like sites in Ctp1 hypersensitizes cells to DNA double-strand break inducing agents. Five-fold serial dilutions of the indicated strains were plated on control medium (YPA) or medium supplemented with the indicated drugs, and then incubated for 3 days at 30°C.
(D) DNA-damage sensitivity due to deletion of the FHA domain and BRCT1 is suppressed by N-terminal fusion with Ctp1. The ctp1:nbs1FHA/BRCT1Δ strain was generated by replacing the nbs1 gene with a transgene encoding the chimeric fusion protein illustrated in the schematic. SG4 denotes a Ser-Gly poly-linker at the fusion site (Nbs1 Asp-221). Growth assays were performed as in Figure 5C.
To further investigate the potential functional significance of Ctp1 Sites 1 and 2 as binding epitopes for spNbs1, we generated mutant strains bearing alanine substitutions of Thr-79, Thr-89 or both. Perhaps surprisingly, both the individual and double site Ctp1 mutants largely corrected the slow-growth and DNA-damage hypersensitivity phenotypes of Ctp1-defective cells (Figure 5C). Nevertheless, while displaying little or no hypersensitivity towards low doses of the radio-mimetic drug phleomycin (data not shown), the double mutant did display some hypersensitivity to higher phleomycin levels. We thus speculated that phosphorylation of a third threonine, Thr-78 within Ctp1 Site 1, might also contribute to Ctp1 function and, accordingly, we found that the spNbs1 FHA/BRCT2 region bound tightly to a Ctp1 peptide containing singly phosphorylated Thr-78 with an apparent Kd of ~0.2 μM (Table S2). Moreover, while a mutant strain bearing the single T78A mutation displayed wild-type growth and drug-sensitivity profiles, combination of the T78A, T79A and T89A mutations yielded a strain that was hypersensitive to camptothecin, phleomycin, and methyl-methane sulphonate, albeit not to the same degree as the Ctp1-null strain (Figure 5A).
In order to investigate the specificity of spNbs1 interactions with the Site 1 and Site 2 Ctp1 motifs, we constructed a strain containing alanine mutations of Thr-78, 79, 89 and Thr-71 that is located immediately N-terminal to the Site 1 region, but not within a SDTD-like sequence. In this case, the additional mutation did not further increase damage sensitivity (Figure S5A), a lack of effect that correlated well with the absence of significant binding of spNbs1 FHA/BRCT2 to a pThr-71 phosphopeptide (Figure S5B; Table S2). These and additional data (Figure S6; Table S2) revealed an ‘extended’ specificity of the spNbs1 FHA domain beyond that normally observed for such modules. Binding absolutely depends on the presence of at least one pThr, and a further preference for glutamate/aspartate is evident at the pThr +3 position that is often a major determinant of FHA specificity (Durocher et al., 2000). For Thr-79 and Thr-89, additional serine phosphorylation in the -2 position increases affinity by ~10-fold. However, other positions, notably those in the C-terminal acidic stretch of the motif, contribute significantly to binding affinity reflecting a plasticity of the Nbs1 FHA domain that appears to have evolved to exploit and accommodate additional sequence requirements for Ctp1 phosphorylation.
Taken together, the above findings strongly suggested that S. pombe MRN, through the Nbs1 FHA domain, interacts directly with Ctp1 through redundant CK2 phosphorylation motifs. To test this idea, we first created an S. pombe strain containing Nbs1 in which the FHA-BRCT1 region had been deleted. As expected, these cells exhibited a severe hypersensitivity to low levels of DNA-damaging agents closely resembling the phenotypes of the G103D FHA mutant and Nbs1Δ strains (Figure 5D). Strikingly, however, hypersensitivity was completely rescued when the Nbs1 protein lacking the FHA/BRCT1 region was expressed as a fusion to full-length Ctp1 (Figure 5D). The ability of this fusion to circumvent a requirement for the FHA/BRCT1 domain thus provided strong support for Ctp1 binding as a primary function of the spNbs1 FHA/BRCT region.
The human Nbs1 BRCT-repeat domain contains a second phospho-binding site
Finally, we turned our attention to the role of the Nbs1 BRCT-repeat domain. Our spNbs1 structure revealed that the relative arrangement and packing of the tandem BRCT motifs resembles that previously observed in structures of other BRCT-repeat domains, many of which display phosphopeptide-binding functions (Figure 6A). SpNbs1 does not contain lysine and serine/threonine residues known to make direct phosphate contacts in other BRCT-phosphoprotein complexes (Figure 6B). They are, however, present in hNbs1 (Lys-160 and Ser-118) and although other positions involved in accessory main-chain peptide interactions and sequence-specific binding are not (Figure 6B), we nonetheless speculated that the Nbs1 N-terminus might possess dual phospho-recognition surfaces. In this regard, and in contrast to spNbs1, we noticed that titrations of wild-type hNbs1 with a variety of Mdc1-derived pSDpTD peptides consistently showed evidence of two interaction sites, manifested as binding stoichiometries significantly and reproducibly greater than 1:1 (Figure 6C; Table S2). Moreover, although mutation of the conserved phospho-binding residues in the hNbs1 FHA domain (Arg-28 or Ser-42), or in BRCT1 (Lys-160) had surprisingly small apparent effects on phosphopeptide-binding affinity (Figure 6C; Table S2), each of these reduced the stoichiometry by ~2-fold. These findings suggested that the effects of mutations in each individual domain were being partially obscured by interactions mediated by the other. Indeed, while a single FHA mutation (R27A) is sufficient to dramatically reduce phosphopeptide binding in spNbs1, we found that binding to hNbs1 was only abolished when both FHA and BRCT1 sites were disrupted in a S42A/K160M double mutant.
Figure 6. A phospho-binding function for the hNbs1 BRCT-repeat domain.
(A) Structural conservation of the BRCT-repeat domains from SpNbs1 (left) and the Brca1/Bach1 complex as an example of a BRCT-domain/ phosphopeptide interaction (right) (PDB ID: 1T15). BRCT1 and BRCT2 domains are colored green and blue respectively, while the BRCT-linker region is shown in magenta. The Bach1 phosphopeptide is drawn as a stick representation with carbon atoms colored yellow.
(B) Residues involved in Brca1 BRCT-repeat Bach1 phosphopeptide interactions shown in a stick representation, are partially conserved in hNbs1 but not in spNbs1.
(C) Mutation of potential phospho-binding residues in the hNbs1 FHA (R28A and S44A) and BRCT-domains (K160M) has a limited effect on binding affinities for Mdc1 pSDpTD motifs, but reduces the stoichiometry by ~2-fold in comparison to wild-type (see also Table S2). The double mutant, S44A/K160M, effectively abolishes binding.
(D) ITC measurements reveal that hNbs1 is able to bind to a pSDpSD variant of Mdc1 (closed squares) but not spNbs1 (open circles). Binding is abolished by the K160M (BRCT-repeat) mutation (closed triangles) but unaffected by R28A in the FHA domain (open squares).
(E) Interactions of hNbs1 and MDC1 requires the phosphopeptide-binding activity of both the hNbs1 FHA and BRCT domains. Immunoprecipitations were performed from 2mg of nuclease (Benzonase, Novagen)-treated whole-cell extracts derived from HEK 293 cells, transiently transfected with expression plasmids encoding YFP (pEYFP2-N1), YFP2-NBS1 (NBS1 pEYFP2-N1), or YFP2-NBS1 bearing either the R28A FHA point mutation, or the K160M BRCT1 point mutation. Samples were immunoblotted with the indicated antibodies. No interaction with Mdc1 is seen, even in longer exposures.
*Wild-type hNbs1 binding to the Mdc1 pSDpTD 326-336 peptide is shown in Figure 2D; here we used a similar di-phosphorylated peptide of the same length but with a core consensus of all six Mdc1 motifs (FIDpSDpTDVEEE).
To further explore the apparent dual phospho-binding mode of hNbs1, we attempted to ‘uncouple’ FHA and BRCT domain contributions by using a peptide synthesized with a variant pSDpSD motif that lacks a phosphothreonine and would not, therefore, be expected to interact with hNbs1 FHA domain. In line with the lack of ITC evidence for an additional binding site in spNbs1 and the absence of any of the characteristic BRCT phospho-binding residues in this protein, no interaction of spNbs1 with the pSDpSD peptide was detectable (Figure 6D). In marked contrast, significant binding of the pSDpSD peptide to both wild-type hNbs1 and the R28A hNbs1 FHA mutant was maintained but, crucially, no interaction was seen with the K160M BRCT1 mutant. Consistent with these and previous biochemical analyses (Chapman and Jackson, 2008), tagged wild-type hNbs1 expressed ectopically in human cells efficiently co-immunoprecipitated Mdc1, while Mdc1 was undetectable in immunoprecipitates of Nbs1 bearing either the R28A or the K160M point-mutation (Figure 6E). Thus, both FHA-dependent and BRCT-dependent phospho-specific interactions by Nbs1 are required for its stable interactions with Mdc1 in cell extracts.
Collectively, these data establish a clear in vitro phospho-dependent binding activity for the hNbs1 BRCT-repeat domain. Furthermore, the lack of hNbs1 interactions with the mono-phosphorylated pSDTD or γH2AX peptides containing a single phosphoserine (Figure 4A and Figure S3) implies a preference of the hNbs1 BRCT-domain for doubly-phosphorylated motifs that differs considerably from previously reported pS/TxxF/Y phospho-dependent BRCT specificities (Manke et al., 2003; Yu et al., 2003). Consistent with this model, hNbs1 lacks the highly conserved arginine corresponding to residue 1699 in Brca1 and residue 1933 in Mdc1 that mediates crucial contacts with main-chain atoms of bound phosphopeptides. Taken together, these observations point to a rather distinct overall phospho-motif binding mode for the hNbs1 BRCT-repeats, although further structural analysis of hNbs1 phosphopeptide complexes will clearly be necessary to ascertain the precise molecular basis for such interactions.
CONCLUSIONS
Here we have presented a combined structural, biochemical and cell-biological investigation of the N-terminal region of the key DSB repair and DNA damage-signaling protein, Nbs1. Our crystallographic analysis has revealed a conserved super-modular FHA-BRCT2 architecture that, along with our supporting biochemical data, explains the apparent functional inter-dependence of the Nbs1 FHA and BRCT motifs first noted some time ago (Cerosaletti and Concannon, 2003).
Through comparison of the in vitro binding activities of fission yeast and human Nbs1 fragments, we have identified a similar FHA domain specificity for pSDpTD-like CK2 motifs originally identified as hNbs1 binding sites in Mdc1. On this basis, we have identified related sequences in S. pombe Ctp1 that are phosphorylated in cells, bind in a phospho-dependent manner and with high affinity to the spNbs1 FHA domain in vitro and which, when mutated, impair Ctp1 function in promoting resistance to DNA-damaging agents in vivo. The resemblance of these sites to others located in budding yeast Lif1 further suggests that the reported interactions between Lif1 and Xrs2 (Palmbos et al., 2008) are likely to employ a closely related binding mode. Significantly, an increase in affinity for di-phosphorylated sequences as compared with singly phosphorylated motifs appears to be a characteristic of both human and yeast Nbs1. Taken together with the existence of multiple SDTD motifs in Mdc1, Ctp1 and Lif1 and the combinatorial effects of Site 1 and Site 2 mutations on Ctp1 function and its interactions with spNbs1, our data suggest that Nbs1 has evolved an ability to respond to varying levels of overall phosphorylation of its various interacting partners.
Overall, the intimate association of the Nbs1 FHA and BRCT-repeat domains observed in our structure suggests that the fixed spatial relationship between the two regions may be important in allowing a concerted mode of FHA and BRCT domain activity. Our cell-biological and biochemical approaches have shown a phosphorylation-dependent binding capability for the human Nbs1 BRCT-repeat domain that is exercised through interactions distinct from those previously observed for the BRCT-repeats of Brca1 and Mdc1. The apparent necessity for binding of the human Nbs1 BRCT-repeat domain to di-phosphorylated motifs is intriguing and contrasts the rather more accessory effect of the additional phosphoserine on interactions of the Nbs1 FHA domain. It is thus tempting to speculate that these binding properties confer a degree of flexibility to hNbs1 responses through the use of separate phospho-binding domains with distinct, but overlapping specificities. At the molecular level, this arrangement might, for example, accommodate binding and juxtaposition of different protein interaction partners, each primed through phosphorylation by upstream kinases, in a manner reminiscent of 14-3-3 function (Gardino et al., 2006). Alternatively, the FHA and BRCT-repeat domains might recognize dual phospho-dependent interaction sites within a single molecule such as Mdc1. In support of this, we have shown that individual mutation of phospho-binding residues in either the FHA or BRCT-repeat domains of hNbs1 is sufficient to severely compromise its interactions with Mdc1 in human cells, consistent with previously reports that intact FHA and BRCT-repeat domains of Nbs1 (Cerosaletti and Concannon, 2003; Wu et al., 2008) and more than a single Mdc1 pSDpTD motif (Spycher et al., 2008) are necessary for MRN localization to DNA-damage foci in human cells.
In contrast to the human protein, the role of the spNbs1 BRCT-repeat domain remains unclear and, consistent with the absence of residues analogous to those imparting direct phosphate interactions in Mdc1, Brca1 and also hNbs1, we have been unable to demonstrate a phospho-binding function for the spNbs1 BRCT-repeat domain. The presence of alternative interaction surfaces in this domain that might mediate phospho-binding cannot be discounted but it may also be that the spNbs1 BRCT-repeats function through as yet uncharacterized phospho-independent interactions. Indeed, the phospho-independent binding capability reported for FHA domains (Li et al., 2002; Nott et al., 2009) and previously seen in several BRCT-repeat domain complexes (Derbyshire et al., 2002; Doré et al., 2006; Joo et al., 2002; Sibanda et al., 2001) could potentially specify additional, evolutionarily conserved binding functions for the Nbs1 N-terminus, thus expanding the versatility of this region as a platform for promoting DNA repair and DNA-damage checkpoint signaling.
EXPERIMENTAL PROCEDURES
Expression and Purification
Synthetic genes encoding spNbs1 (1-324) and hNbs1 (1-382) were produced by GENEART AG (www.geneart.com) and expressed in pET22b as C-terminal (His)6-tag fusion proteins for purification utilizing a Ni-Sepharose affinity chromatography followed by Source 15Q ion-exchange and Superdex 75 size exclusion chromatography (GE Healthcare). Mutations were introduced using the QuikChange protocol (Strategene) and all constructs were verified by DNA sequencing. For preparation of seleno-methionine (SeMet) labelled spNbs1, plasmid was transformed into B834 (DE3) cells (Novagen). Protein expression was performed in supplemented M9 minimal media (Molecular Dimensions) and purified as for the native protein. SeMet incorporation was estimated to be > 98% by mass spectrometry.
Crystallization and Data collection
Growth of diffraction quality crystals of native and SeMet incorporated spNbs1 was aided by carboxypeptidase A digestion. Crystallization was carried out by sitting-drop vapour diffusion using an Oryx8 nano-dispensing robot (Douglas Instruments). Crystals grew at a protein concentration of 8 mg/ml against a reservoir containing 0.1 M MES pH 6.0, 0.1 M MgCl2 and 8% w/v PEG 6000. Diffraction data were collected at 100 K at a wavelength corresponding to the peak of the anomalous (f”) signal on beamline I04 at the Diamond Light Source, UK from crystals cryoprotected with 30% glycerol. Methods for crystal improvement by reductive methylation along with details of the structure solution and refinement and biochemical/cell biology procedures are provided in Supplemental Material.
Supplementary Material
Supplementary data
Acknowledgements
We thank Hiroshi Iwasaki for the kind gift of reagents, P. Walker for invaluable assistance with X-ray data collection, K. Rittinger for advice on ITC measurements and support from the NIMR large-scale laboratory. EH is supported by a CRUK grant C20600/A6620 and TC acknowledges funding by an MRC Program Grant G0600233. Research the SPJ laboratory is supported by grants from Cancer Research UK, the European Union (Integrated Project DNA repair, LSHG-CT-2005-512113, and Genomic Instability in Cancer and Precancer, GENICA, HEALTH-F2-2007-201630), and core infrastructure funding from Cancer Research UK and the Wellcome Trust. SJS is grateful to the Medical Research Council, UK for continuing support and Diamond Light Source, UK for synchrotron access and beamline support. Atomic coordinates have been deposited with the Protein Data Bank - accession numbers 1XXX and 1YYY.
Footnotes
The authors state that they have no competing financial interests.
References
- Akamatsu Y, Murayama Y, Yamada T, Nakazaki T, Tsutsui Y, Ohta K, Iwasaki H. Molecular characterization of the Schizosaccharomyces pombe nip1+/ctp1+ gene in DNA double strand break repair in association with the Mre11-Rad50-Nbs1 complex. Mol Cell Biol. 2008 doi: 10.1128/MCB.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker E, Meyer V, Madaoui H, Guerois R. Detection of a tandem BRCT in Nbs1 and Xrs2 with functional implications in the DNA damage response. Bioinformatics. 2006;22:1289–1292. doi: 10.1093/bioinformatics/btl075. [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Cerosaletti KM, Concannon P. Nibrin forkhead-associated domain and breast cancer C-terminal domain are both required for nuclear focus formation and phosphorylation. J Biol Chem. 2003;278:21944–21951. doi: 10.1074/jbc.M211689200. [DOI] [PubMed] [Google Scholar]
- Chapman JR, Jackson SP. Phospho-dependent interactions between NBS1 and MDC1 mediate chromatin retention of the MRN complex at sites of DNA damage. EMBO Rep. 2008 doi: 10.1038/embor.2008.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Nievera CJ, Lee AY, Wu X. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J Biol Chem. 2008;283:7713–7720. doi: 10.1074/jbc.M710245200. [DOI] [PubMed] [Google Scholar]
- D’Amours D, Jackson SP. The Mre11 complex: at the crossroads of dna repair and checkpoint signalling. Nat Rev Mol Cell Biol. 2002;3:317–327. doi: 10.1038/nrm805. [DOI] [PubMed] [Google Scholar]
- Derbyshire DJ, Basu BP, Serpell LC, Joo WS, Date T, Iwabuchi K, Doherty AJ. Crystal structure of human 53BP1 BRCT domains bound to p53 tumour suppressor. EMBO J. 2002;21:3863–3872. doi: 10.1093/emboj/cdf383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai-Mehta A, Cerosaletti KM, Concannon P. Distinct functional domains of nibrin mediate Mre11 binding, focus formation, and nuclear localization. Mol Cell Biol. 2001;21:2184–2191. doi: 10.1128/MCB.21.6.2184-2191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio S, Nussenzweig A. The NBS1-ATM Connection Revisited. Cell Cycle. 2007;6 doi: 10.4161/cc.6.19.4758. [DOI] [PubMed] [Google Scholar]
- Doré AS, Furnham N, Davies OR, Sibanda BL, Chirgadze DY, Jackson SP, Pellegrini L, Blundell TL. Structure of an Xrcc4-DNA ligase IV yeast ortholog complex reveals a novel BRCT interaction mode. DNA Repair (Amst) 2006;5:362–368. doi: 10.1016/j.dnarep.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Durocher D, Taylor IA, Sarbassova D, Haire LF, Westcott SL, Jackson SP, Smerdon SJ, Yaffe MB. The molecular basis of FHA domain:phosphopeptide binding specificity and implications for phospho-dependent signaling mechanisms. Mol Cell. 2000;6:1169–1182. doi: 10.1016/s1097-2765(00)00114-3. [DOI] [PubMed] [Google Scholar]
- Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature. 2005;434:605–611. doi: 10.1038/nature03442. [DOI] [PubMed] [Google Scholar]
- Gardino AK, Smerdon SJ, Yaffe MB. Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the X-ray crystal structures of all human 14-3-3 isoforms. Semin Cancer Biol. 2006;16:173–182. doi: 10.1016/j.semcancer.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Giglione C, Boularot A, Meinnel T. Protein N-terminal methionine excision. Cell Mol Life Sci. 2004;61:1455–1474. doi: 10.1007/s00018-004-3466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Grotzer MA, Watanabe T, Hewer E, Pietsch T, Rutkowski S, Ohgaki H. Mutations in the Nijmegen breakage syndrome gene in medulloblastomas. Clin Cancer Res. 2008;14:4053–4058. doi: 10.1158/1078-0432.CCR-08-0098. [DOI] [PubMed] [Google Scholar]
- Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev. 2002;16:583–593. doi: 10.1101/gad.959202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi J, Tauchi H, Sakamoto S, Nakamura A, Morishima K, Matsuura S, Kobayashi T, Tamai K, Tanimoto K, Komatsu K. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr Biol. 2002;12:1846–1851. doi: 10.1016/s0960-9822(02)01259-9. [DOI] [PubMed] [Google Scholar]
- Lee MS, Edwards RA, Thede GL, Glover JN. Structure of the BRCT repeat domain of MDC1 and its specificity for the free COOH-terminal end of the gamma-H2AX histone tail. J Biol Chem. 2005;280:32053–32056. doi: 10.1074/jbc.C500273200. [DOI] [PubMed] [Google Scholar]
- Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, Yaffe MB, Jackson SP, Smerdon SJ. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell. 2002;9:1045–1054. doi: 10.1016/s1097-2765(02)00527-0. [DOI] [PubMed] [Google Scholar]
- Limbo O, Chahwan C, Yamada Y, de Bruin RA, Wittenberg C, Russell P. Ctp1 is a cell-cycle-regulated protein that functions with Mre11 complex to control double-strand break repair by homologous recombination. Mol Cell. 2007;28:134–146. doi: 10.1016/j.molcel.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukas C, Melander F, Stucki M, Falck J, Bekker-Jensen S, Goldberg M, Lerenthal Y, Jackson SP, Bartek J, Lukas J. Mdc1 couples DNA double-strand break recognition by Nbs1 with its H2AX-dependent chromatin retention. EMBO J. 2004;23:2674–2683. doi: 10.1038/sj.emboj.7600269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Yuan C, Lee H, Chen ES, Wu PY, Tsai MD. Structure and function of the phosphothreonine-specific FHA domain. Science signaling. 2008;1:re12. doi: 10.1126/scisignal.151re12. [DOI] [PubMed] [Google Scholar]
- Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- Martinez A, Traverso JA, Valot B, Ferro M, Espagne C, Ephritikhine G, Zivy M, Giglione C, Meinnel T. Extent of N-terminal modifications in cytosolic proteins from eukaryotes. Proteomics. 2008;8:2809–2831. doi: 10.1002/pmic.200701191. [DOI] [PubMed] [Google Scholar]
- Maser RS, Zinkel R, Petrini JH. An alternative mode of translation permits production of a variant NBS1 protein from the common Nijmegen breakage syndrome allele. Nat Genet. 2001;27:417–421. doi: 10.1038/86920. [DOI] [PubMed] [Google Scholar]
- Matsuura S, Tauchi H, Nakamura A, Kondo N, Sakamoto S, Endo S, Smeets D, Solder B, Belohradsky BH, Der Kaloustian VM, et al. Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet. 1998;19:179–181. doi: 10.1038/549. [DOI] [PubMed] [Google Scholar]
- Matsuzaki K, Shinohara A, Shinohara M. FHA domain of yeast Xrs2, a homologue of human Nbs1, promotes non-homologous end joining through the interaction with a Ligase IV partner protein, Lif1. Genetics. 2008 doi: 10.1534/genetics.107.079236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melander F, Bekker-Jensen S, Falck J, Bartek J, Mailand N, Lukas J. Phosphorylation of SDT repeats in the MDC1 N terminus triggers retention of NBS1 at the DNA damage-modified chromatin. J Cell Biol. 2008;181:213–226. doi: 10.1083/jcb.200708210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott TJ, Kelly G, Stach L, Li J, Westcott SL, Patel D, Hunt DM, Howell S, Buxton RS, O’Hare H, Smerdon SJ. An intra-molecular switch regulates phospho-independent FHA domain interactions in Mycobacterium tuberculosis. Science signaling. 2009 doi: 10.1126/scisignal.2000212. [DOI] [PubMed] [Google Scholar]
- Palmbos PL, Wu D, Daley JM, Wilson TE. Recruitment of Saccharomyces cerevisiae Dnl4-Lif1 Complex to a Double-strand Break Requires Interactions with Yku80 and the Xrs2 FHA Domain. Genetics. 2008 doi: 10.1534/genetics.108.095539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riches LC, Lynch AM, Gooderham NJ. Early Events in the Mammalian Response to DNA Double-Strand Breaks. Mutagenesis. 2008 doi: 10.1093/mutage/gen039. [DOI] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- Sibanda BL, Critchlow SE, Begun J, Pei XY, Jackson SP, Blundell TL, Pellegrini L. Crystal structure of an Xrcc4-DNA ligase IV complex. Nat Struct Biol. 2001;8:1015–1019. doi: 10.1038/nsb725. [DOI] [PubMed] [Google Scholar]
- Spycher C, Miller ES, Townsend K, Pavic L, Morrice NA, Janscak P, Stewart GS, Stucki M. Constitutive phosphorylation of MDC1 physically links the MRE11-RAD50-NBS1 complex to damaged chromatin. J Cell Biol. 2008;181:227–240. doi: 10.1083/jcb.200709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005;123:1213–1226. doi: 10.1016/j.cell.2005.09.038. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanová E, Cooper PR, Nowak NJ, et al. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–520. doi: 10.1139/O07-069. [DOI] [PubMed] [Google Scholar]
- Wu L, Luo K, Lou Z, Chen J. MDC1 regulates intra-S-phase checkpoint by targeting NBS1 to DNA double-strand breaks. Proc Natl Acad Sci USA. 2008;105:11200–11205. doi: 10.1073/pnas.0802885105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Wu L, Cui G, Botuyan MV, Chen J, Mer G. Structure of a Second BRCT Domain Identified in the Nijmegen Breakage Syndrome Protein Nbs1 and its Function in an MDC1- Dependent Localization of Nbs1 to DNA Damage Sites. J Mol Biol. 2008 doi: 10.1016/j.jmb.2008.05.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol. 2005;25:5363–5379. doi: 10.1128/MCB.25.13.5363-5379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- Zhao S, Weng YC, Yuan SS, Lin YT, Hsu HC, Lin SC, Gerbino E, Song MH, Zdzienicka MZ, Gatti RA, et al. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- Zhu J, Petersen S, Tessarollo L, Nussenzweig A. Targeted disruption of the Nijmegen breakage syndrome gene NBS1 leads to early embryonic lethality in mice. Curr Biol. 2001;11:105–109. doi: 10.1016/s0960-9822(01)00019-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data