Identification of two evolutionarily conserved genes regulating processing of engulfed apoptotic cells (original) (raw)
. Author manuscript; available in PMC: 2011 Feb 1.
Published in final edited form as: Nature. 2010 Mar 21;464(7289):778–782. doi: 10.1038/nature08853
Abstract
Engulfment of apoptotic cells occurs throughout life in multi-cellular organisms. Impaired apoptotic cell clearance (due to defective recognition/internalization or degradation) results in autoimmune disease1, 2. One fundamental challenge in understanding how defects in corpse removal translate into diseased states is the identification of critical players orchestrating the different stages of engulfment. Here, through genetic, cell biological and molecular studies in C. elegans and mammalian cells, we identify SAND-1 and its partner CCZ-1 as new players in corpse removal. In _sand-1_- and ccz-1 deficient worms, apoptotic cells are internalized and the phagosomes recruit the small GTPase RAB-5, but fail to progress to the subsequent RAB-7(+) stage. The mammalian orthologues of SAND-1, Mon1a and Mon1b, were similarly required for phagosome maturation. Mechanistically, Mon1 interacts with GTP-bound Rab5, identifying Mon1 as a novel Rab5 effector. Moreover, a Mon1:Ccz1 complex (but not either protein alone) could bind Rab7 and also influence Rab7 activation, suggesting Mon1:Ccz1 as an important link in progression from Rab5+ to Rab7+ stages of phagosome maturation. Collectively, these data identify SAND-1(Mon1), and CCZ-1(Ccz1) as critical and evolutionarily conserved players regulating the processing of ingested apoptotic cell copses.
The nematode C. elegans represents a powerful genetic tool for the study of programmed cell death3; in the adult gonad, germ cell corpses are rapidly recognized and internalized by the phagocytic somatic sheath cells, which encase the germ line4. Two evolutionarily conserved signaling pathways (comprised of CED-1/CED-6 and CED-7, and CED-2/CED-5/CED-12), both of which function upstream of the small GTPase CED-10(Rac1), mediate the recognition and internalization of apoptotic cells5, 6. The subsequent steps in degradation of the ingested apoptotic cells, termed ‘phagosome maturation’, have only recently begun to be defined.
Previous studies have identified the sequential recruitment and activation of the small GTPases RAB-5 and RAB-7 to apoptotic cell-containing phagosomes; however, many of the players required for these steps remain unidentified and mechanistic steps are not well understood. We had previously conducted a targeted reverse genetic screen in the worm to identify candidate RAB-5 activators/effectors required for phagosome maturation (Ref. 7 and data not shown). Although no requirement for many of the known regulators of RAB-5 function (such as EEA-1, HGRS-1/Hrs and RABS-5/Rabenosyn7–9) was seen in apoptotic cell processing, this screen identified one candidate gene, sand-1, which has been proposed to function upstream of RAB-7 activation/function10. SAND-1 is the nematode orthologue of S. cerevisiae Mon1p and has been implicated in fluid-phase uptake in coelomocytes; how sand-1 (or Mon1p) functioned in corpse removal or its mechanism of action were not known.
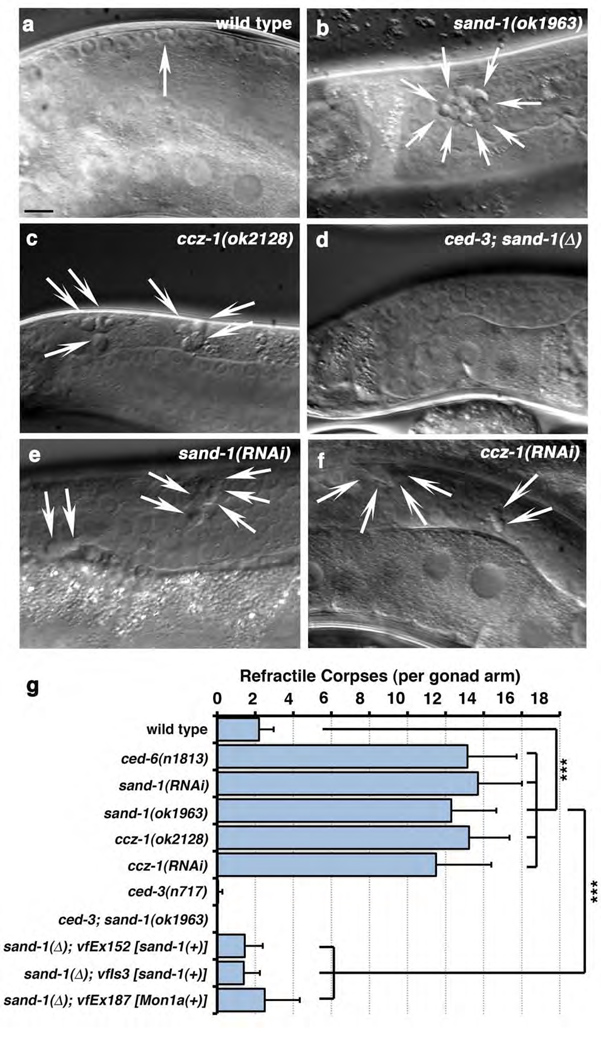
When cells die in the nematode, they gradually condense to generate ‘refractile’ apoptotic bodies that can be visualized by DIC microscopy4, 11. Analysis of a sand-1(ok1963) deletion mutant, denoted sand-1(Δ), revealed increased numbers of refractile bodies in the gonad (Figure 1, a vs. b quantitated in g and Table 1a). Refractile corpses in sand-1(Δ) worms arose due to apoptotic cell death, as sand-1(Δ); ced-3(n717) double mutant worms (lacking the executioner caspase CED-33) did not accumulate refractile corpses in the gonad (Figure 1d, e, quantitated in g). Overexpression of YFP::SAND-1 under the ced-1 promoter was sufficient to rescue defects in sand-1(Δ) mutant worms [transgenics referred to as _sand-1_(Δ); _vfEx152_ or _sand-1_(Δ); _vfIs3_, Figure 1, g], suggesting that defects in sand-1(Δ) mutant worms are due to lesions within sand-1 and not a closely linked mutation. The ced-1 promoter targets expression to the phagocytic somatic sheath cells but not apoptotic germ cells12, suggesting SAND-1 expression within the phagocyte is sufficient for function. This raised the possibility that defects in sand-1(lf) worms arose due to impairment of uptake, or the subsequent processing of the ingested apoptotic cell. We addressed both of these possibilities as detailed below.
Figure 1. SAND-1 and CCZ-1 are required for removal of apoptotic cell corpses in the adult hermaphrodite gonad.
(a–c) Refractile corpses in the adult hermaphrodite gonad wild-type (a), sand-1(ok1963) (b) and ccz-1(ok2128) (c) mutant animals. Size bar, 10µm. (d) Lack of refractile bodies in sand-1(ok1963), ced-3(n717) double mutants. (e–f) Accumulation of refractile corpses in the gonad after RNAi-mediated knockdown of sand-1 (e) or ccz-1 (f). Size bar, 10µm. (g) Quantitation of corpse defects in these conditions at the 12-hour adult stage (see Supplementary Methods). Data shown represents mean ± s.d., with details of corpse numbers and n shown in Supplemental Table 1a, ***, p<0.001.
Acidification is an important marker for ‘maturation’ of the phagosome13–15. In the nematode, Acridine Orange (AO) or Lysotracker red (LTR) can be used as markers of acidic organelles7, 16; phagosomes begin to acidify following the acquisition of RAB-5 and grow progressively more acidic as they mature through the RAB-7(+) stage7, 17. Worms deficient in dyn-1 or vps-34 (which are required for RAB-5 recruitment to nascent phagosomes14), or rab-5 (Supplementary Figure S1, Table 1b) show phagosomes arrested without LTR or AO staining7; in comparison, rab-7 RNAi results in delayed acidification17, though the majority of corpses still stain with AO or LTR (Supplementary Figure S1, Table 1b). In sand-1(Δ) mutant worms, refractile corpses stained brightly with both LTR (Supplementary Figure S1) or AO (quantitated in Table S1b), suggesting that apoptotic cells were internalized but arrested at a late stage of degradation.
To further rule out a corpse internalization defect in sand-1 mutants, we took two approaches. By transmission electron microscopy (TEM), apoptotic cells in sand-1 mutant worms appear phagocytosed (Supplementary Figure S2), often with multiple apoptotic cells per phagosome (see below). Second, we expressed YFP::Actin as a transgene to visualize cells undergoing internalization in real-time (Supplementary Figure S2). sand-1(lf) worms show similar numbers of actin halos as controls, ruling out increased germ cell death or defects in corpse internalization, and suggesting increased corpse number arose from defects in apoptotic cell degradation.
To identify the stage at which phagosomes in sand-1(Δ) worms were arrested, we examined the localization/recruitment of different markers to the phagosome. To date, recruitment of DYN-1 is the earliest available maturation-specific marker of apoptotic cell-containing phagosomes7, 17, 18, occurring at a similar time as actin polymerization and apoptotic cell uptake. We found no defect in DYN-1 recruitment or release from the phagosome in sand-1(Δ) mutant worms (Table S1c), suggesting that SAND-1 might function downstream of DYN-1. Since DYN-1 recruitment requires phagocytic uptake7, these results further confirmed that sand-1(lf) worms have no defect in corpse internalization.
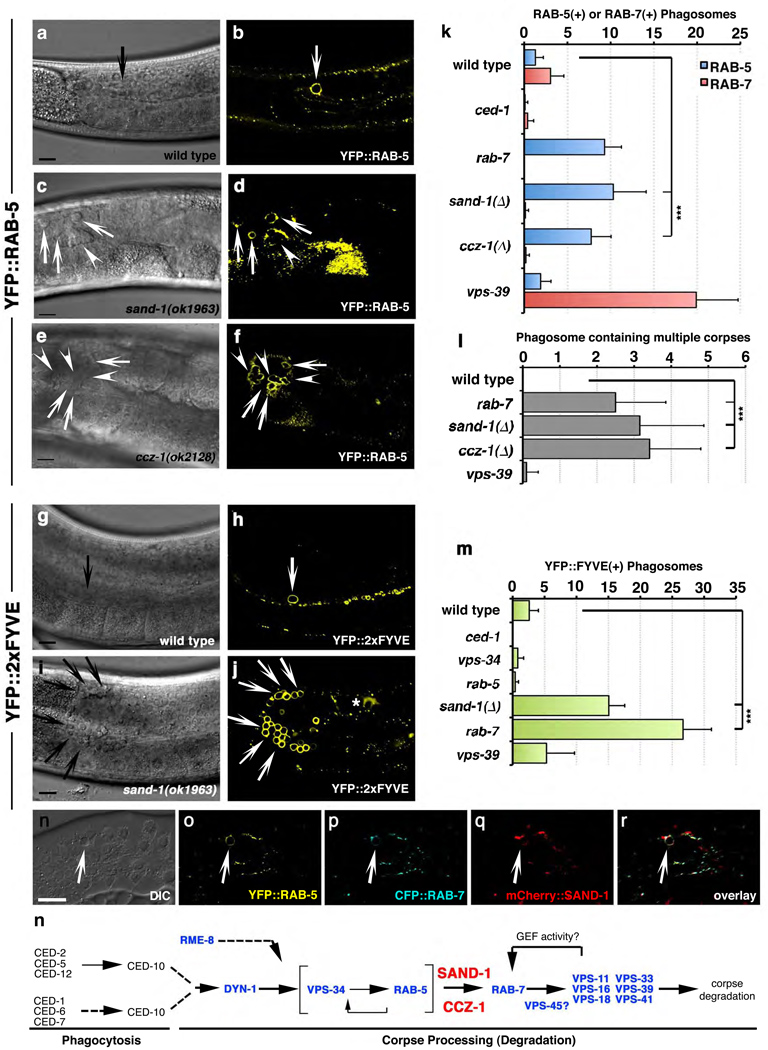
Following apoptotic cell internalization, the GTPases RAB-5 and RAB-7 are sequentially recruited to the surface of the phagosome7, 19. In sand-1(Δ) mutant worms, we found a substantial increase in the number of RAB-5(+) phagosomes (Figure 2a–d, quantitated in k, Table S1d, see Supplementary Figure S3 for widefield images) with a concomitant decrease in the number of RAB-7(+) phagosomes (Figure 2k). This suggested a defect in transition from the RAB-5(+) stage to the RAB-7(+) stage. A marker for the ‘mature’ phagolysosome, LMP-117, was also decreased on phagosomes in sand-1(Δ) worms (Table S1e, Supplementary Figure S4). Interestingly, arrest at the RAB-5(+) stage in sand-1(Δ) or rab-7(RNAi) worms resulted in phagosomes containing multiple apoptotic cells (Figure 2d, asterisks, quantitated in l). Since multi-corpse phagosomes are rarely seen in wild-type worms or vps-39(lf), where phagosomes are arrested at the RAB-7(+) stage (Figure 2k, l), this might represent fusion events between RAB-5(+) structures, as described in other contexts20.
Figure 2. Phagosomes are arrested at the RAB-5(+) stage in sand-1 mutant worms.
(a–j) Compared to wild-type gonads (a,b), sand-1(ok1963) (c,d) and ccz-1(ok2128) (e,f) mutant worms accumulate apoptotic cells within RAB-5(+) (a–f) PtdIns(3)(+) (g–j) phagosomes. Size bar, 10µm. (k–m) Quantitation of RAB-5(+) or RAB-7(+) phagosome (k), phagosomes containing multiple apoptotic cells (l) or PtdIns(3)P(+) phagosomes (m). ***, p<0.001. Data shown represents mean ± s.d., n is shown in Supplementary Table S1. (n–r) Nematodes co-expressing YFP::RAB-5 (o), CFP::RAB-7 (p) and mCherry::SAND-1 (q) (from the same array) were assayed for simultaneous localization to phagosomes containing apoptotic cells. DIC shown in (n), and overlay in (r). (s) Schematic of the genetic pathway for phagosome maturation in the nematode denoting the position of SAND-1 and CCZ-1.
Activation of RAB-5 and subsequent VPS-34/PI3K activation result in the generation of PtdIns(3)P on the phagosome surface7, 21, which can be detected using a YFP::2xEEA-1(FYVE) fusion protein. In sand-1(Δ) mutant worms, phagosomes were enriched in PtdIns(3)P (Figure 2g–j, quantitated in m), suggesting that recruitment/activation of RAB-5 and VPS-34 occurred normally on the phagosome surface. Collectively, these data suggested that loss of SAND-1 caused a block in phagosome maturation after RAB-5 recruitment, but before RAB-7 recruitment to the phagosome (see genetic pathway, Figure 2s).
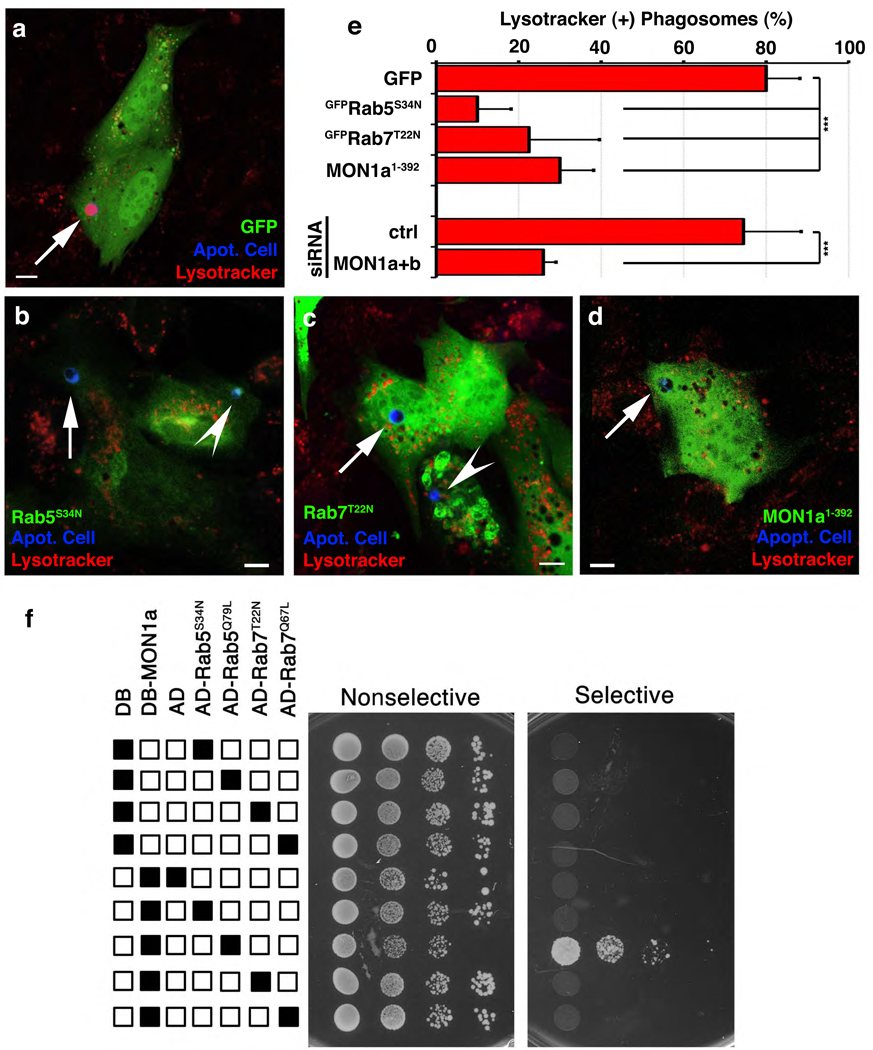
We next asked whether SAND-1 and its function are evolutionarily conserved. Mammals possess two orthologues of SAND-1, namely Mon1a and Mon1b. Several lines of evidence suggested that the function of SAND-1/Mon1 during apoptotic cell clearance is evolutionarily conserved: first, transgenic expression of mouse Mon1a (as YFP::Mon1a) in sand-1(Δ) mutant worms efficiently rescued the corpse clearance defect [Figure 1g, _sand-1(Δ); vfEx187_]. Second, expression of Mon1a1–392 in mammalian phagocytes, which matched the predicted deletion mutation in sand-1(Δ), blocked phagosome acidification (Lysotracker Red staining, see Methods) (Figure 3d vs. a, quantitated in e); this block was analogous to the block in phagosome acidification that results from overexpression of dominant negative mutants of Rab5 and Rab7 (GFP-Rab5S34N or GFP-Rab7T22N, respectively) (Figure 3b, c and quantitated in e, see Supplementary Figure S5 for individual panels). Third, siRNA-mediated knockdown of Mon1a and Mon1b (denoted Mon1a+b) caused a significant decrease in acidified phagosomes (Figure 3e, see Supplementary Figure S6 for quantitation of knockdown). It is noteworthy that in the above experiments, there were no defects in corpse recognition or internalization (Supplementary Figure S6). Finally, we could detect localization of YFP-Mon1a on the phagosome (Supplementary Figure S7), supporting a functional role for Mon1 on the phagosome. Collectively, these data suggested that SAND-1/Mon1 plays an evolutionarily conserved role in phagosome maturation.
Figure 3. Mon1 is required for phagosome acidification and is recruited to phagosomes containing apoptotic cells.
(a–e) NIH/3T3 fibroblasts transfected with plasmids coding for GFP only (a), Mon11–392 (b), GFP-Rab5S34N (c), or GFP-Rab7T22N (d), or Mon1a/Mon1b siRNA (e) were assessed for Lyostracker Red(+) phagosomes. Arrows indicate apoptotic cells (blue) or Lysotracker Red(+) phagosomes. Quantification of these data are shown in e as mean ± s.d. from four experiments. _n_>30 cells per condition, ***, p<0.001. (f) Yeast expressing indicated GAL4 DNA binding domain (DB) alone, VP16 activation domain (AD) alone, DB-Mon1a or AD-Rab proteins were assessed for protein interaction in a yeast two-hybrid assay via growth on selective and non-selective plates (plated in progressive 2-fold dilutions).
To elucidate a mechanism of SAND-1/Mon1 action, we asked whether Mon1 might interact with Rab5. Interestingly, this was the case in two different read-outs. First, Mon1a bound to Rab5 when expressed in 293T cells, with enhanced interaction between Mon1a and Rab5Q79L, the preferentially GTP-associated version of Rab5 (Supplementary Figure S8). Second, in a yeast two-hybrid interaction assay, we observed a specific interaction between Mon1a and Rab5Q79L, but not with the preferentially GDP-associated Rab5S34N or Rab7 (neither the active Q79L, nor the inactive T22N mutants) (Figure 3f); interestingly, a truncated version of Mon1a (Mon1a1–392), mimicking the sand-1(Δ) mutant allele, failed to interact with Rab5Q79L (Supplementary Figure S9). These data suggested Mon1a as a new class of Rab5 effector lacking the canonical FYVE domain architecture found in other Rab5 effectors such as EEA1, Rabenosyn-5 or Hrs9.
Although SAND-1 was identified in a high molecular weight complex with both RAB-5 and RAB-710, the role of SAND-1, if any, in linking RAB-5 and RAB-7 was not known. We first tested whether SAND-1, RAB-5 and RAB-7 could be detected simultaneously on the apoptotic cell-containing phagosomes: transgenic worms coexpressing YFP::RAB-5, CFP::RAB-7 and mCherry::SAND-1, SAND-1 was present on ~75% of phagosomes that were positive for both RAB-5 and RAB-7 (n=5 gonads, Figure 2, n–r), suggesting a possible role for SAND-1 during the transition from the RAB-5(+) to the RAB-7(+) phagosome. One intriguing possibility was that SAND-1 might act as a ‘physical bridge’ between Rab5 and Rab7 thereby linking Rab5 activation to Rab7 recruitment. However, we could not detect an interaction between Mon1a and Rab7 (despite a robust interaction with Rab5, Figure 3f). This prompted us to ask whether Mon1, perhaps via a Mon1 interacting protein(s), might mediate an interaction between Rab5 and Rab7.
To date, one binding partner for the yeast Mon1p has been described, Ccz1p22; however, Ccz1p orthologues have not been characterized in worms or mammals. A ccz-1(ok2128) deletion mutant or ccz-1(RNAi) showed increased numbers of internalized refractile cell corpses in the gonad (Figure 1c, f, quantitated in Table S1). Upon further analysis, we found that in ccz-1(ok2128) worms, phagosomes containing apoptotic cells were arrested at the RAB-5(+) stage (Figure 2, e, f, quantitated in k). The phenocopy of the defects among sand-1(Δ) and ccz-1(ok2128) mutant animals suggested that SAND-1 and CCZ-1 proteins likely function at the same step of phagosome maturation.
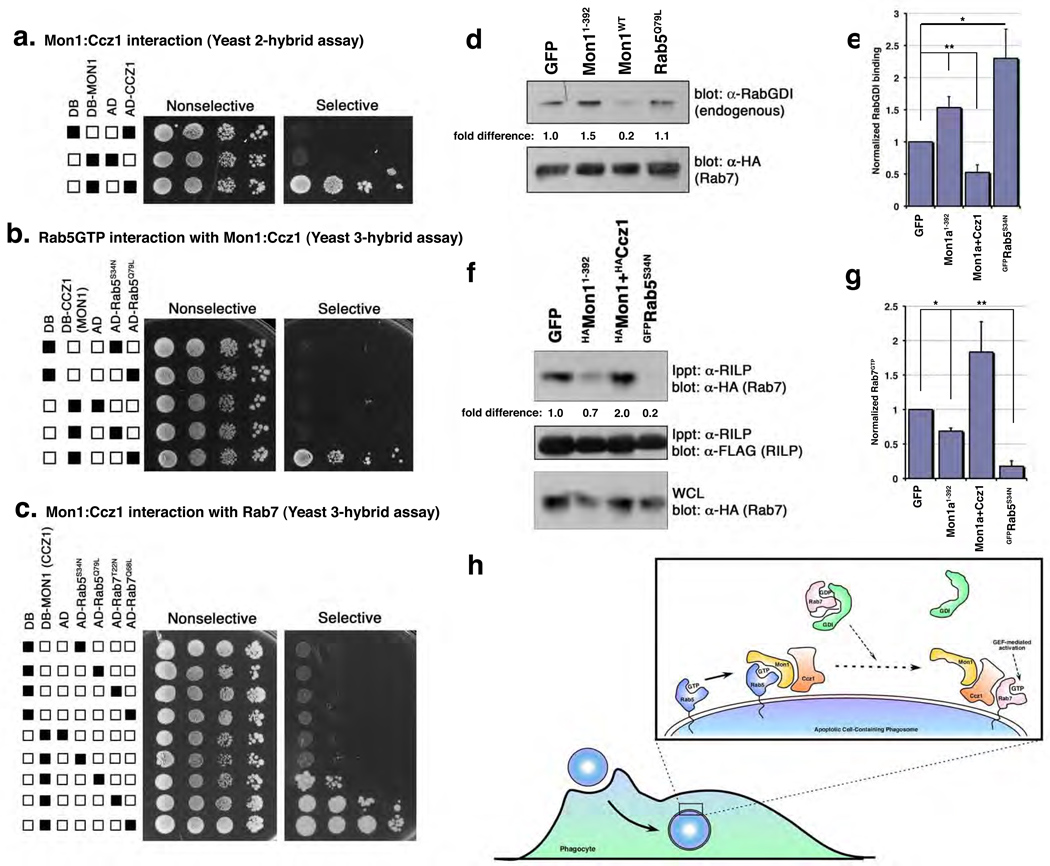
An interesting possibility was that Ccz1 might interact with Rab7, providing a link between Rab5 activation and recruitment of Rab7 to the phagosome. To test this, we needed to establish a few baseline parameters. First, mammals contain one CCZ-1 orthologue, which we named Ccz1. Mammalian Mon1a and Mon11–392 interacted with mammalian Ccz1, both by immunoprecipitation (Supplementary Figure S9) and in the yeast two-hybrid assay (Figure 4a). The region of Mon1a responsible for binding Ccz1 and Rab5 are distinct (with Rab5 and Ccz1 binding to the C-terminal and N-terminal regions of Mon1a, respectively) (Supplementary Figure S9). Furthermore, in a yeast three-hybrid assay (see Methods), Mon1a could simultaneously associate with both Ccz1 and Rab5GTP (Figure 4b). It is noteworthy that under these conditions Ccz1 itself does not directly interact with Rab5 (Supplementary Figure S10).
Figure 4. A Mon1/Ccz1 complex is a novel Rab5 effector that links activated Rab5 to Rab7 recruitment and GDI displacement.
(a–c) Interaction between MON1a and CCZ1 (a), NLSMon1:DBCcz1 and ADRab5 (b) or DBMon1:NLSCcz1 and ADRab5/ADRab7 (c) was assayed by growth of yeast on selective plates. (d, e) Mon1:Ccz1 affect Rab7 recruitment/activation by enhancing GDI displacement. 293T cells were transiently transfected with the indicated constructs, lysed and immunoprecipitated as indicated. Co-precipitation of endogenous RabGDI by HARab7 (d) or Rab7GTP by the Rab7 effector FLAGRILP (f) was assessed by immunoblotting. A representative experiment for each is shown. Graphs (e, g) represent mean densitometry values from multiple independent experiments for GFP, Mon11–392(_n_=6), S34N (_n_=5), Mon1+Ccz1 (_n_=4) (e), or _n_=3 (g). Error bars represent s.e.m. (e) *, p=0.01, **, p=0.009 or (g) **, p= 0.02, * p=0.04. (f) Working model of Mon1:Ccz1 function during phagosome maturation.
We next tested whether Ccz1 might bind Rab7; however, we failed to detect a direct interaction between Ccz1 and Rab7 (or Rab5) (Supplementary Figure S10), which prompted us to ask whether a ‘Mon1:Ccz1 complex’ might generate (or open up) a Rab7 interaction site, thereby potentially bridging Rab5GTP and the recruitment of Rab7. Remarkably, coexpression of both Mon1a and Ccz1 resulted in interaction with Rab7 (Figure 4c) in the yeast three-hybrid assay, suggesting that the Mon1:Ccz1 complex could potentially act as a protein:protein interaction bridge linking active Rab5 and Rab7.
We took two further approaches to address this whether Mon1:Ccz1 might regulate Rab7 activity following recruitment. RabGDIs (Rab Guanine Nucleotide Dissociation Inhibitors) bind to GDP-bound forms of Rab GTPases and function to inhibit basal dissociation of GDP, keeping GTPases in the inactive form. We asked whether Mon1:Ccz1 may facilitate the dissociation of GDI from Rab7 and in turn, promote GTP loading of Rab7. Interestingly, the association of endogenous RabGDI with Rab7 was decreased when MON1a:Ccz1 were overexpressed in 293T cells (Figure 4e, f, see Supplementary Figure S11 for protein expression controls). In contrast, expression of Mon1a1–392 blocked dissociation of RabGDI from Rab7, similar to dominant-negative Rab5S34N, consistent with a role for Mon1:Ccz1 at the Rab5-to-Rab7 transition.
We also assessed GTP binding to Rab7 under these conditions by coexpressing the Rab7 effector RILP23, which specifically binds Rab7GTP (Supplementary Figure S12). Overexpression of Mon1a1–392 decreased the levels of GTP-bound Rab7 in cells (Figure 4f, g see Supplementary Figure S12 for protein expression controls). These data suggest a possible model wherein the Mon1:Ccz1 complex facilitates Rab7 recruitment as well as RabGDI release, thereby promoting GTP loading of Rab7 during maturation of apoptotic cell-containing phagosomes (Figure 4h).
In summary, this work has identified two new players, SAND-1/Mon1 and CCZ-1/Ccz1, having an evolutionarily conserved role in processing of apoptotic cell-containing phagosomes. Mechanistically, SAND-1/Mon1 appears to be a new type of Rab5 effector that can link Rab5 activation to Rab7 recruitment, with the Mon1:Ccz1 complex (but not the individual proteins) being able to bind/recruit Rab7. Lastly, the Mon1:Ccz1 complex appears to facilitate the displacement of GDI from Rab7, an important step in Rab7 activation, although the details of this process remain to be defined. Since deficiencies in RabGDI have been associated with mental retardation both in humans and in mouse models24, the possible role of Mon1:Ccz1 in GDI displacement from Rab7 may have broader implications.
Mon1:Ccz1 function identified here in the context of apoptotic cell clearance may also be relevant for vesicular trafficking, such as receptor mediated endocytosis and its pathological alterations (e.g. tumorigenesis)25,26 Finally, genes required for maturation of apoptotic cell-containing phagosomes may be relevant in processing of antigens derived from apoptotic cells (which requires acidification-dependent cathepsin activation27) and in contributing to immune tolerance; pathogens have also been shown to modify phagosome maturation to evade the immune response28. Thus, the identification of genes and pathways related to phagosome maturation could impact a number of different cellular processes.
Methods Summary
C. elegans imaging
C. elegans feeding RNAi was performed as described7 (see Online Methods for detail). Worms were synchronized by picking hermaphrodites at the L4 larval stage (Christmas tree vulva) then incubated for 24h at 20°C and scored for persistent cell corpses and fluorescent protein localization where appropriate.
To score, worms were placed on 2% agarose pads and anaesthetized with 3–5mM levamisole (Sigma) and mounted under a cover slip for observation using a Zeiss Axiovert 200, AxioImager Z2 (with deconvolution) or LSM 510 confocal microscope equipped with DIC (Nomarski) optics. Staining of worms with Acridine Orange or Lysotracker Red (Invitrogen) were performed as previously described7, 29.
Mammalian phagocytosis assays
Cells were incubated overnight in Lipofectamine 2000 as previously described, or transfected using Amaxa program U-30 and Kit R for NIH/3T3 cells (Amaxa, Germany) with an siRNA SMARTpool containing 4 siRNAs targeting mouse Mon1a (Dharmacon cat # M-049528-01), Mon1b (M-055500-01) or a noncoding SMARTpool (Dharmacon cat # D-001206-13) using 1.2 µg of total siRNA (0.3 µg of each individual siRNA) as previously described7, then incubated 48h to recover. Images were acquired using a Zeiss 510 laser scanning confocal microscope with 405, 488, 543, and 633nm lasers (Zeiss AG, Germany).
Apoptotic thymocytes were generated as previously described7; apoptotic thymocytes (5 × 105 cells per condition) were added to NIH/3T3 cells in 4-well Labtek II culture; thymocytes were allowed engulfed for 30 minutes, then unbound apoptotic thymocytes were gently washed off with DMEM + 10% FBS, and then subsequently incubated for 2h in DMEM + 10% FBS containing 1/10,000 dilution of Lysotracker Red. Cells were then fixed with 3% paraformaldehyde (Sigma) in PBS for 30 minutes, permeabilized with 0.1% Triton X-100 (Sigma) and blocked with 5% milk that had been clarified by high speed centrifugation. Antibody staining was then done as previously described30.
Supplementary Material
01
Acknowledgements
The authors would like to thank Jim Casanova, Cynthia Grimsley and members of the Ravichandran lab for helpful conversations, the Caenorhabditis Genetics Consortium (CGC) for nematode strains, A. Wandinger-Ness for Rab7 expression constructs and Jan Redick and Stacey Guillot of the Advanced Microscopy Facility for preparation of EM specimens.
Footnotes
Author contributions: J.M.K performed all the experiments; J.M.K and K.S.R. planned and analyzed the experimental results wrote the manuscript.
Competing Interests Statement
The authors declare they have no competing financial interests.
References
- 1.Albert ML. Death-defying immunity: do apoptotic cells influence antigen processing and presentation? Nat Rev Immunol. 2004;4:223–231. doi: 10.1038/nri11308. [DOI] [PubMed] [Google Scholar]
- 2.Ravichandran KS, Lorenz U. Engulfment of apoptotic cells: signals for a good meal. Nat Rev Immunol. 2007;7:964–974. doi: 10.1038/nri2214. [DOI] [PubMed] [Google Scholar]
- 3.Lettre G, Hengartner MO. Developmental apoptosis in C. elegans: a complex CEDnario. Nat Rev Mol Cell Biol. 2006;7:97–108. doi: 10.1038/nrm1836. [DOI] [PubMed] [Google Scholar]
- 4.Gumienny TL, Lambie E, Hartwieg E, Horvitz HR, Hengartner MO. Genetic control of programmed cell death in the Caenorhabditis elegans hermaphrodite germline. Development. 1999;126:1011–1022. doi: 10.1242/dev.126.5.1011. [DOI] [PubMed] [Google Scholar]
- 5.Kinchen JM, Hengartner MO. Tales of cannibalism, suicide, and murder: Programmed cell death in C. elegans. Current Topics in Developmental Biology. 2005;65:1–45. doi: 10.1016/S0070-2153(04)65001-0. [DOI] [PubMed] [Google Scholar]
- 6.Mangahas PM, Zhou Z. Clearance of apoptotic cells in Caenorhabditis elegans. Semin Cell Dev Biol. 2005;16:295–306. doi: 10.1016/j.semcdb.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Kinchen JM, et al. A pathway for phagosome maturation during engulfment of apoptotic cells. Nat Cell Biol. 2008;10:556–566. doi: 10.1038/ncb1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gengyo-Ando K, et al. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 2007;8:152–157. doi: 10.1038/sj.embor.7400882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103:11821–11827. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poteryaev D, Fares H, Bowerman B, Spang A. Caenorhabditis elegans SAND-1 is essential for RAB-7 function in endosomal traffic. EMBO J. 2007;26:301–312. doi: 10.1038/sj.emboj.7601498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoeppner DJ, et al. eor-1 and eor-2 are required for cell-specific apoptotic death in C. elegans. Dev Biol. 2004;274:125–138. doi: 10.1016/j.ydbio.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 12.Zhou Z, Hartwieg E, Horvitz HR. CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 13.Huynh KK, et al. LAMP proteins are required for fusion of lysosomes with phagosomes. Embo J. 2007;26:313–324. doi: 10.1038/sj.emboj.7601511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinchen JM, Ravichandran KS. Phagosome maturation: going through the acid test. Nat Rev Mol Cell Biol. 2008;9:781–795. doi: 10.1038/nrm2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hackam DJ, et al. Regulation of phagosomal acidification. Differential targeting of Na+/H+ exchangers, Na+/K+-ATPases, and vacuolar-type H+-atpases. J Biol Chem. 1997;272:29810–29820. doi: 10.1074/jbc.272.47.29810. [DOI] [PubMed] [Google Scholar]
- 16.Lettre G, et al. Genome-wide RNAi identifies p53-dependent and -independent regulators of germ cell apoptosis in C. elegans. Cell Death Differ. 2004;11:1198–1203. doi: 10.1038/sj.cdd.4401488. [DOI] [PubMed] [Google Scholar]
- 17.Yu X, Lu N, Zhou Z. Phagocytic receptor CED-1 initiates a signaling pathway for degrading engulfed apoptotic cells. PLoS Biol. 2008;6:e61. doi: 10.1371/journal.pbio.0060061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu X, Odera S, Chuang CH, Lu N, Zhou Z. C. elegans Dynamin mediates the signaling of phagocytic receptor CED-1 for the engulfment and degradation of apoptotic cells. Dev Cell. 2006;10:743–757. doi: 10.1016/j.devcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Lu Q, et al. C. elegans Rab GTPase 2 is required for the degradation of apoptotic cells. Development. 2008;135:1069–1080. doi: 10.1242/dev.016063. [DOI] [PubMed] [Google Scholar]
- 20.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 21.Li W, et al. C. elegans Rab GTPase activating protein TBC-2 promotes cell corpse degradation by regulating the small GTPase RAB-5. Development. 2009;136:2445–2455. doi: 10.1242/dev.035949. [DOI] [PubMed] [Google Scholar]
- 22.Wang CW, Stromhaug PE, Kauffman EJ, Weisman LS, Klionsky DJ. Yeast homotypic vacuole fusion requires the Ccz1-Mon1 complex during the tethering/docking stage. J Cell Biol. 2003;163:973–985. doi: 10.1083/jcb.200308071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, et al. Mycobacterium bovis BCG disrupts the interaction of Rab7 with RILP contributing to inhibition of phagosome maturation. J Leukoc Biol. 2007;82:1437–1445. doi: 10.1189/jlb.0507289. [DOI] [PubMed] [Google Scholar]
- 24.D'Adamo P, et al. Mutations in GDI1 are responsible for X-linked non-specific mental retardation. Nat Genet. 1998;19:134–139. doi: 10.1038/487. [DOI] [PubMed] [Google Scholar]
- 25.Jekely G, Sung HH, Luque CM, Rorth P. Regulators of endocytosis maintain localized receptor tyrosine kinase signaling in guided migration. Dev Cell. 2005;9:197–207. doi: 10.1016/j.devcel.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 26.Mosesson Y, Mills GB, Yarden Y. Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer. 2008;8:835–850. doi: 10.1038/nrc2521. [DOI] [PubMed] [Google Scholar]
- 27.Rock KL, Shen L. Cross-presentation: underlying mechanisms and role in immune surveillance. Immunol Rev. 2005;207:166–183. doi: 10.1111/j.0105-2896.2005.00301.x. [DOI] [PubMed] [Google Scholar]
- 28.Gruenberg J, van der Goot FG. Mechanisms of pathogen entry through the endosomal compartments. Nat Rev Mol Cell Biol. 2006;7:495–504. doi: 10.1038/nrm1959. [DOI] [PubMed] [Google Scholar]
- 29.Kinchen JM, et al. Two pathways converge at CED-10 to mediate actin rearrangement and corpse removal in C. elegans. Nature. 2005;434:93–99. doi: 10.1038/nature03263. [DOI] [PubMed] [Google Scholar]
- 30.Grimsley CM, Lu M, Haney LB, Kinchen JM, Ravichandran KS. Characterization of a novel interaction between ELMO1 and ERM proteins. J Biol Chem. 2006;281:5928–5937. doi: 10.1074/jbc.M510647200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01