Host-Interactive Genes in Amerindian Helicobacter pylori Diverge from Their Old World Homologs and Mediate Inflammatory Responses (original) (raw)
Abstract
Helicobacter pylori is the dominant member of the gastric microbiota and has been associated with an increased risk of gastric cancer and peptic ulcers in adults. H. pylori populations have migrated and diverged with human populations, and health effects vary. Here, we describe the whole genome of the _cag_-positive strain V225d, cultured from a Venezuelan Piaroa Amerindian subject. To gain insight into the evolution and host adaptation of this bacterium, we undertook comparative H. pylori genomic analyses. A robust multiprotein phylogenetic tree reflects the major human migration out of Africa, across Europe, through Asia, and into the New World, placing Amerindian H. pylori as a particularly close sister group to East Asian H. pylori. In contrast, phylogenetic analysis of the host-interactive genes vacA and cagA shows substantial divergence of Amerindian from Old World forms and indicates new genotypes (e.g., VacA m3) involving these loci. Despite deletions in CagA EPIYA and CRPIA domains, V225d stimulates interleukin-8 secretion and the hummingbird phenotype in AGS cells. However, following a 33-week passage in the mouse stomach, these phenotypes were lost in isolate V225-RE, which had a 15-kb deletion in the cag pathogenicity island that truncated CagA and eliminated some of the type IV secretion system genes. Thus, the unusual V225d cag architecture was fully functional via conserved elements, but the natural deletion of 13 cag pathogenicity island genes and the truncation of CagA impaired the ability to induce inflammation.
Helicobacter pylori is a microaerophilic bacterium of the Epsilonproteobacteria that has colonized the stomach since early in human evolution (45) and diverged with ancient human migrations (24, 45, 92). Thus, several major H. pylori populations, such as hpAfrica1, hpEurope, hspEAsia, and hspAmerind, whose names indicate their original geographic associations (45, 51), have been defined. In particular, similarities between the hspAmerind and hspEAsia populations suggest that the first colonizers of the New World brought H. pylori with them (24, 28). With recent mixing of human groups, H. pylori populations are also mixing and competing, with an apparent dominance by the hpEurope population at least in Latin America (19).
H. pylori usually does not cause illness, but colonization with strains bearing the cag (cytotoxin-associated gene) pathogenicity island (cag PAI) (3, 7, 25, 52, 57, 61, 63) is associated with an increased risk of noncardia gastric adenocarcinoma and peptic ulcer disease (56, 64). Nonetheless, a high prevalence of _cag_-positive H. pylori strains occurs concurrently with low gastric cancer rates in Africa (40) and some regions in Latin America, such as the Venezuelan savannas and Amazonas (29, 53). Moreover, clinical and epidemiological data provide evidence for an inverse relationship between H. pylori colonization and the prevalence of certain metabolic disorders, esophageal diseases, asthma and allergic disorders, and acute infectious diseases, as well as a direct relationship with improved nutritional status of rural children (3, 14, 34, 37, 49, 68). That the host interaction with an indigenous gastric microbe provides some health benefits to the host is not unexpected given the well-established role of gastrointestinal microflora in maintaining gastroenteric homeostasis (8).
The most thoroughly studied H. pylori proteins that interact with human cells are CagA and VacA. CagA is an effector protein injected into gastric epithelial cells by a type IV secretion system encoded by the cag PAI (10, 12, 15, 83). VacA is initially secreted from the bacterial cell by an autotransporter mechanism (16). Both proteins have multiple effects on host cells. Inside the host cell, phosphorylation of CagA on EPIYA repeats in the phosphotyrosine (PY) region (73) induces cellular elongation known as the hummingbird phenotype (72). CagA may also induce secretion of interleukin-8 (IL-8) (11), a process commonly attributed to NF-κB, and disrupt the barrier function of the tight junctions in polarized epithelial cells, leading to a loss of adhesion (1, 5). Other motifs in the PY region promote phosphorylation-independent effects (79). In addition, cagA may be considered an oncogene (60), since transgenic expression of cagA in mice leads to gastric epithelial hyperplasia through aberrant epithelial cell signaling and gastric carcinogenesis (60, 62). In contrast, VacA is a multifunctional protein with several activities in epithelial and immune cells (16). VacA induces cell vacuolation (43), alters mitochondrial membrane permeability (27, 41, 90), and increases epithelial monolayer permeability. VacA also activates several signal transduction pathways that are important in immune and epithelial cells, including the mitogen-activated protein (MAP) kinase and p38/ATF-2-mediated signal pathways (9, 55).
Genomic analysis provides insights into the evolution of H. pylori strains and their relation with their human hosts and may be useful for the development of diagnostic tools and novel therapies. To date, there are six published complete H. pylori genomes, mostly from the hpEurope population (see Table SA1 in the supplemental material). Here, we report the whole genome of a newly characterized hspAmerind strain, V225d, and assess its genetic structure in comparison to those of Old World H. pylori strains through a comprehensive multiprotein phylogenetic analysis, as well as through single-gene examination of cagA and vacA, revealing clues to the evolution and migration of this strain into the New World and the implications for human health. We also present the results of functional and genomic studies using gastric epithelial cells demonstrating that V225d can induce an inflammatory host response, an effect that was lost following passage through the mouse stomach.
MATERIALS AND METHODS
Strain information and DNA extraction and characterization.
H. pylori strain V225d was isolated from a gastric antral biopsy specimen from a Piaroa Amerindian who underwent a gastroscopy and was found to have acute superficial gastritis. The human-subject protocol was approved by the Institutional Review Board (IRB) at the Venezuelan Institute of Scientific Research and at the University of Puerto Rico (0809-051), which allows available specimens and cultures to be used without patient identifiers. H. pylori V225 pure culture (01-225) has been available since 2001 in the clinical isolate repository of the microbial ecology laboratory at the University of Puerto Rico. Genomic DNA was extracted, using a DNeasy tissue kit (Qiagen, Chatsworth, CA), from strain V225 samples originating from gastric corpus biopsy specimens previously homogenized in 200 μl of 0.9% saline solution with approximately 0.1 ml of 0.5-mm glass beads in 1.5-ml tubes and mixed at high speed for 20 s in a bead beater (29). The quality and quantity of genomic DNA were assessed using a bioanalyzer (Agilent Technologies, Santa Clara, CA). An aliquot of DNA (10 μg) was used for Roche/454 pyrosequencing, as described below.
DNA sequencing and annotation.
Initially, two rounds of Roche/454 GS-20 sequencing and one round of genome sequencer flexible system (GS-FLX) sequencing were performed on H. pylori strain V225 (01-225) DNA, resulting in over 800,000 reads (average read lengths of 222 bp for the GS-FLX run and 110 bp for the GS-20 runs). During the assembly, we observed that the DNA sample was a 4:1 mixture of two distinct strains (designated V225d and V225b, respectively) of H. pylori. The two strains were clonally purified, and two rounds of sequencing using Roche/454 GS-FLX were performed on the V225d clone. Shotgun and paired-end libraries were prepared following the manufacturer's instructions (48). A round of shotgun sequencing generated 62-fold coverage (a total of 422,314 reads). The second round of paired-end sequencing generated 31,448 paired reads. The sequence reads were assembled to obtain a draft genome assembly consisting of 29 contigs in 4 scaffolds. Both replicon sequences were completed using a combination of in silico repeat resolution and targeted Sanger sequencing. Automated nucleic acid and protein sequence annotation was accomplished using the PATRIC pipeline (74). The annotation protocol containing the full list of applications and parameters is available online at http://patric.vbi.vt.edu/about/standard_procedures.php.
Sequencing of the V225-RE cag PAI was performed using targeted Sanger sequencing, with primers designed from the V225d cag PAI as a template (see Table SA1 in the supplemental material). Primers were designed using primer-BLAST (http://www.ncbi.nlm.nih.gov) to amplify an 800- to 1,000-bp region of the V225d sequence, with a 50- to 200-bp overlap between adjacent amplicons. Sequencing was performed by the Virginia Bioinformatics Institute (Blacksburg, VA) Core Laboratory Facility. Contig assembly was executed using the SeqMan program (Lasergene, Madison, WI).
Genome alignment.
Multiple alignments of whole genomes and the cag pathogenicity island (cag PAI) were prepared for seven complete H. pylori genomes (strains V225d, Shi470, P12, G27, HPAG1, J99, and 26695) by using MAUVE 2.2.0 (17).
Phylogenetic analyses.
A phylogenetic tree was prepared from the analysis of 1,931 phylotypes from the _m_ulti_l_ocus _s_equence _t_yping (MLST) database (35, 47). Specifically, in order to compare the consistencies of protein phylogenetic trees with maximum-likelihood trees that contain a larger number of sequences, we used sequences from 7 housekeeping genes, atpA (627 bp), efp (410 bp), mutY (420 bp), ppa (398 bp), trpC (456 bp), ureI (585 bp), and yphC (510 bp) (35, 47), in a concatenated string of 3,406 bp. The multilocus sequences (MLSs) were from H. pylori strains from hosts from Africa, Europe, Asia, and the Americas. The data set was partitioned into the seven genes, and each gene was partitioned into one fragment containing the third-codon positions and another containing the first- and second-codon positions. The maximum-likelihood program RAxML (77) was used, with its GTRGAMMA model. For a more robust phylogenetic analysis, the protein sequences of H. pylori V225d were collected along with those from the complete or incomplete genome projects for 10 other H. pylori strains, eight additional Helicobacter species, and two organisms from other genera of the Helicobacteraceae. The proteins were sorted into 3,432 families by using OrthoMCL (44), and the 603 families that were represented once and only once in each of the 10 complete Helicobacter genomes were processed further. The families were subjected to “decimation,” removal of the 10% with the most divergent phylogenies, as follows. For each family, proteins were aligned using MUSCLE (20), masked using Gblocks (80) in its default mode except using the −b5 = h setting, which allows a position to contain gaps if they occur in less than half of the sequences. Trees were prepared for 50 bootstrap samples by using RAxML, with its PROTGAMMAWAG model, as previously shown (18, 67, 89, 91). Taxon bipartitions occurring in ≥75% of the bootstrap trees were collected and compared pairwise with the bipartition set for each other family. The 10% of families whose bipartition set most frequently conflicted with the others (which may include cases of horizontal gene transfer) were eliminated, and masked alignments for the remaining 543 families were concatenated, with the maximum-likelihood tree built using RAxML as described above. Bootstrap support values were obtained using the quick mode of RAxML. Rooting was based on trees prepared similarly for all of the genomes, which also included the genome from Campylobacter jejuni RM1221 as an outgroup.
For V225d and the other six available complete H. pylori genomes, the conformity of proteins with their orthologs was measured. The proteins were sorted into families by using OrthoMCL (44), and the 1,231 families with representation in all seven genomes were retained. Within each family, all pairwise Smith-Waterman alignments were performed, and for each protein, the conformity score was defined as the average score of alignment to the best-scoring representative from each of the other six genomes, normalized to the self-alignment score. In cases in which a genome had multiple representations in a family, only the protein with the highest conformity score was retained.
Classification of CagA EPIYA units.
The 1,118 unique CagA sequences available at GenBank on 4 April 2009 were collected, and CagA and CagA2 from H. pylori V225d were added to the collection, after artificial correction of the frameshift mutation of the latter. A seed alignment of the 11 complete CagA sequences from nine H. pylori genomes was prepared by manual adjustment of the MUSCLE (20) alignment. The remaining sequences were incorporated into the alignment sequentially in descending order of length, using the profile mode of MUSCLE, with periodic pauses to adjust the alignment manually. A portion consisting of the phosphotyrosine (PY) region and a small number of flanking positions (corresponding to N884 to L1000 of H. pylori 26695 CagA) was extracted from the alignment, and all sequences not reaching both flanks were removed. The remaining 834 PY regions were divided into overlapping segments consisting of one EPIYA motif continued in both directions up to but not including the neighboring EPIYA motif. The unique segments were aligned without gaps according to their EPIYA motifs, and a pairwise scoring system was implemented. Successive positions in either direction from the center of the aligned EPIYA motif were weighted less by a factor of 1.07, and for each position in which the pair had identical amino acids, the weight was added to the score. The matrix of similarity scores was processed by the Markov cluster algorithm (84), with an inflation factor of 1.2, which sorted the unit segments into four clusters. Segment sequences in each cluster were aligned. Within each cluster, the right portions of the segments were heterogeneous, but the left portions of the segments were substantially homogeneous, corresponding to the previously described A, B, C, and D EPIYA unit types (32). By comparing in a cluster the segments from the right flank of the PY region to the PY-internal segments, the highly conserved position corresponding to S999 of H. pylori 26695 CagA was clearly identified as the endpoint of the shared sequence. The corresponding position could be identified in all four clusters and was used to precisely identify PY region endpoints and to divide each PY region into abutting EPIYA units. The B cluster was readily divided into Bc (usually neighboring C units) and Bd (usually neighboring D units) subclusters. A small number of miscategorized units were moved to their proper cluster. Each EPIYA unit was typed according to its cluster or subcluster, as well as whether it represented fusion with another unit or with a PY-flanking sequence and whether it had insertions or deletions of >2 codons, and each PY region was typed as the concatenation of its unit types. Sequence logos were generated with WebLogo (http://weblogo.berkeley.edu/) after removal of positions in unit alignments missing in >50% of the sequences and removal of redundant identical copies of the trimmed units.
For the CagA phylogenetic tree, the alignment of the 110 complete protein sequences was split into the left and right flanks of the PY region (itself excluded due to its poor ability to be aligned) and converted to the corresponding nucleotide alignments. Maximum-likelihood analysis was performed as described above for the MLST data.
Analyses of VacA.
A total of 88 full-length vacA gene sequences and their corresponding protein sequences were downloaded from GenBank on 4 April 2009. The sequences were aligned using MUSCLE, and phylogenetic trees were constructed as described above. To identify genotype variants of H. pylori VacA based on the 5′ terminus (s region) and the midregion (m region), we used electronic PCR (71) to perform in silico PCR analysis using vacA diagnostic primers (78). Up to 2 mismatches and 2 gaps were allowed during primer annealing.
Bacterial strains used for coculture experiments and experimental infection.
All H. pylori strains were cultured under microaerobic conditions (N2, 85%; O2, 5%; CO2, 10%) at 37°C as previously described (86). Two H. pylori strains were used as controls for determining IL-8 secretion and the hummingbird phenotype on the human gastric adenocarcinoma cell line AGS, the potent proinflammatory cag PAI-positive H. pylori strain ATCC 43504 (39) and the weakly proinflammatory cag PAI-negative H. pylori strain B38 (81).
The V225-RE strain was obtained after reisolation from the stomach of an experimentally infected C57BL/6J mouse 33 weeks postinfection. Briefly, 7-week-old C57BL/6J female mice (Charles River Laboratories, L'Arbresle, France) were infected by oral gavage with 200 μl of 108 CFU/ml of H. pylori V225d suspension every other day for a total of three doses. Mice were housed at five per microisolator cage on ventilated shelves on a 12-h day/12-h night cycle, with constant humidity and temperature control and with food and water ad libitum. Control groups were given peptone trypsin broth alone (100 μl). At 33 weeks postinfection, mice were sacrificed by cervical dislocation, and each stomach was isolated. H. pylori culture was performed on the aseptically collected antrum and body part of the stomach, and cultures were homogenized, plated onto selective medium, and incubated under microaerobic conditions at 37°C for 3 to 5 days as previously described (42). Helicobacter growth was confirmed morphologically by phase-contrast microscopy and Gram staining and biochemically by urease, catalase, and oxidase reactions. All animal experiments were performed in accordance with institutional guidelines as determined by the Central Animal Facility Committee of the University Victor Segalen Bordeaux 2, in conformity with the French Ministry of Agriculture Guidelines for Animal Care. All animal protocols were approved by the Institutional Animal Care and Use Committee and met or exceeded the requirement of the Office of Laboratory Animal Welfare at the National Institutes of Health and the Animal Welfare Act.
Cell lines and culture conditions.
AGS cells (ATCC CRL-1739; ATCC, Manassas, VA) were maintained in Ham's F-12K culture medium (Invitrogen, Cergy-Pontoise, France) supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen) and 50 μg/ml of vancomycin (Sigma, Saint-Quentin-Fallavier, France) at 37°C in a 5% CO2 humidified atmosphere. Cells were seeded onto culture plates or glass coverslips 24 h before addition of bacteria to the culture medium.
Coculture experiments.
All coculture experiments were performed at a multiplicity of infection (MOI) of 100 bacteria/cell. Cells were seeded on glass coverslips at a density of 5 × 104 cells/well in 24-well plates 24 h before addition of bacteria. The H. pylori strains grown on agar plates were collected in phosphate-buffered saline (PBS) (Invitrogen), and the optical density (600 nm) was adjusted to 1.0, corresponding to a concentration of 2 × 108 bacteria/ml. The volume corresponding to an MOI of 100 was added to each cell culture well for a further 24 h of incubation at 37°C in a 5% CO2 humidified atmosphere. Coculture supernatants were collected and deep-frozen after centrifugation, and cells were processed for immunofluorescence staining.
Similar experiments were performed with semipermeable Transwell culture inserts (0.2 μm pore size, Anopore; Nunc, Naperville, IL) fitted into culture wells containing epithelial cells. In some experiments, H. pylori was heat killed by a 20-min incubation of part of the bacterial suspension at 60°C as previously described (86, 88). The absence of bacterial viability of the heated bacterial suspension was verified after 72 h of culture on blood agar under microaerobic conditions; in parallel, the unheated part of the bacterial suspension was used as a positive control to determine bacterial growth.
Cytokine measurement.
IL-8 levels in culture supernatants were determined by an enzyme-linked immunosorbent assay performed using a D8000C kit (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions and an ETIMax-3000 reader (DiaSorin, Saluggia, Italy), as described previously (86). In these assays, the lower and upper limits of detection were 5 and 4,500 pg/ml, respectively.
Antibodies.
Monoclonal antibodies generated against human vinculin (hvin1; Sigma) were used at a 1:400 final dilution for immunofluorescent staining.
Immunofluorescence and image analysis.
Cells were seeded on glass coverslips at a density of 5 × 104 cells/well in 24-well plates 24 h before the addition of bacteria. After 24 h of coculture with bacteria, cells were fixed with 3% paraformaldehyde prepared in cytoskeletal buffer and processed as described previously (86, 87). Cell cultures grown on glass coverslips were washed two times with PBS to remove cellular debris/bacteria and then fixed with 3% paraformaldehyde prepared in cytoskeletal buffer and processed as described previously (86, 87). Coverslips were washed in water and mounted on microscope slides with Fluoromount (Clinisciences SA, Montrouge, France). Cells were analyzed by fluorescence imaging using a Nikon fluorescence microscope (Nikon France S.A.S., Champigny-sur-Marne, France) equipped with NIS-Elements BR (basic research) acquisition software and a 63× (numerical aperture, 1.4) oil immersion objective. Triple-color imaging with Hoechst 33342 compound, phalloidin-Alexa Fluor 568, and Alexa Fluor 488-labeled secondary antibodies (Molecular Probes, Eugene, OR) was performed using selective optical filters. Fluorescent images were processed with Adobe Photoshop 7.0.
Statistical analysis.
Each experiment was performed in triplicate. Quantification values represent the means of the triplicate values ± standard deviations (SD) from one representative experiment out of three. Significance was determined using Student's t test.
Nucleotide sequence accession numbers.
All random shotgun and paired-end sequencing data from this study are available in the NCBI Sequence Read Archive under accession number SRA008809. The entire chromosomal and plasmid sequences of H. pylori V225d have been deposited in GenBank under accession numbers CP001582 and CP001583, respectively. The plasmid sequence of H. pylori V225-RE has been deposited in GenBank under accession number GU370068.
RESULTS
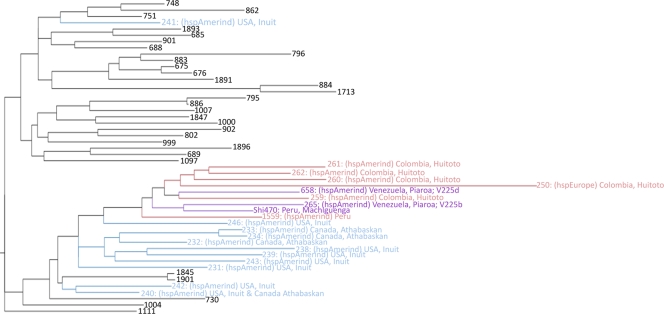
In the initial phase of sequencing, we found that the original V225 strain from the gastric biopsy specimen of a Piaroa Amerindian was a 4:1 mixture of two closely related strains. These two strains were purified from the mixture and renamed V225b and V225d; V225d was selected for further sequence characterization, since it represented the major component of the original sample and was also found to be more distant than V225b from the other available Amerindian genome, Shi470 (Fig. 1).
FIG. 1.
Multilocus sequence typing (MLST) scheme of the Helicobacter pylori hspAmerind cluster. A maximum-likelihood phylogenetic tree was prepared for the 1,939 unique 7-gene concatenations (types, identified numerically) available from the MLST database and from H. pylori genome projects (GTRGAMMA, unpartitioned) and midpoint rooted, which effectively roots with the hpAfrica2 population. Only the hspAmerind portion of the tree is shown, with arctic hspAmerind types shown in blue, equatorial hspAmerind types in red, and three types of special interest in purple.
Genome anatomy of H. pylori strain V225d.
The genome of H. pylori V225d comprises two replicons: a 1,588,278-bp circular chromosome and a 7,326-bp circular plasmid, pHPV225d (see Fig. SA1 in the supplemental material). The average G+C content is 38.97% for the chromosome and 32.88% for the plasmid. Based on sequence coverage, we calculate 6.2 plasmid copies per chromosome. The plasmid encodes 11 coding sequences (CDS). The chromosome has properties similar to those of other complete H. pylori genomes (see Table SA2 in the supplemental material), containing a total of 1,544 CDS and 40 RNA features. Of the 1,544 predicted open reading frames (ORFs), more than 68% had BLASTP hits to the cluster of orthologous groups (COG) database with an E value of <1e−4. The COG classifications of V225d and other complete genomes of H. pylori are provided in Table SA2 in the supplemental material.
Genome alignment with other H. pylori strains.
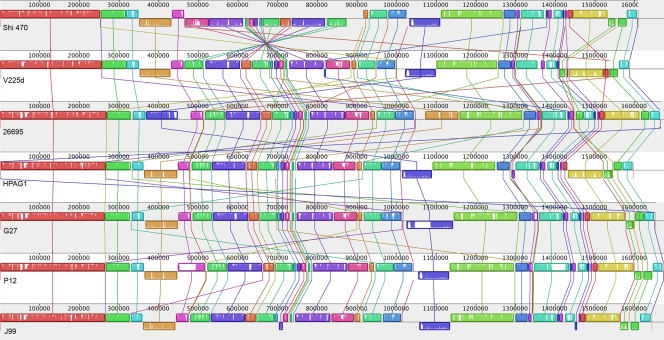
Chromosomal sequences for the seven complete H. pylori genomes were aligned. MAUVE alignment returned 11,660 matches constituting 52 locally colinear blocks spanning 1.5 Mb of homologous sequence common to all strains (Fig. 2). The majority of rearrangements in H. pylori genomes are inversions and translocations. Chief among them is an inversion in Shi470 of an ∼400-kb segment including the cag pathogenicity island. Additionally, an ∼100-kb segment (around coordinate 1.5 Mb of V225d) was rearranged, apparently independently, in V225d and 26695.
FIG. 2.
Alignment of seven complete Helicobacter pylori genomes. Each genome is represented horizontally, with homologous segments presented as colored rectangles. Each region is oriented relative to the corresponding region from H. pylori V225d. Segments aligned between genomes are joined by vertical lines. Average sequence similarities within a segment are proportional to the heights of the interior colored bars. Large sections of white within blocks and gaps between blocks indicate strain-specific sequences. Colored rectangles below the median line indicate inversions. Numbers above the rectangles measure the genome lengths in base pairs.
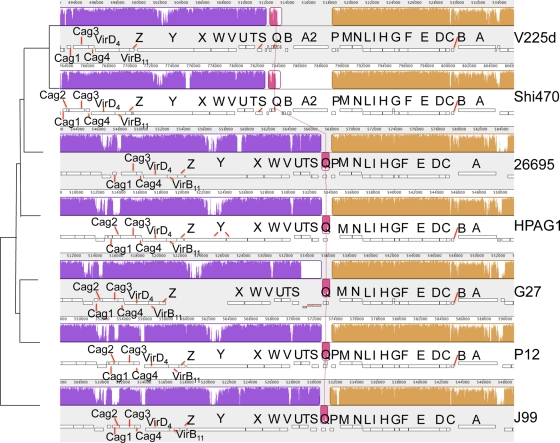
On a finer scale, the cag pathogenicity island of V225d was compared to those of the other six complete H. pylori genomes by use of MAUVE (Fig. 3). Rearrangements specific to the two Amerindian strains are a small inversion of 750 bp and an adjacent duplication of a 4.5-kb segment containing the genes cagA and cagB; we designate these duplicates _cagA_2 and _cagB_2, respectively. In both Amerindian genomes, _cagA_2 is shorter than the parent gene cagA and contains several deletions and insertions that may have rendered it nonfunctional, especially for V225d _cagA_2, which has a single frameshift mutation.
FIG. 3.
Cytotoxin-associated gene (cag) pathogenicity islands in seven complete Helicobacter pylori genomes. The topology of the phylogeny represented in Fig. 4 is indicated on the left side. The single letters represent CagA (A) through CagZ (Z). Genes encoding non-Cag proteins are not labeled. Numbers above the rectangles measure the genome lengths in base pairs. See the legend for Fig. 2 for descriptions of other features.
Phylogenetic analysis of Helicobacteraceae genomes.
The multilocus sequence typing (MLST) scheme for H. pylori, employing a segment from each of seven housekeeping genes, allows comparison to >1,900 genotypes, over half of which have been assigned to a specific H. pylori population and/or subpopulation (23). The seven gene segments from V225d and other available genomes were included with those from the MLST database, and a maximum-likelihood tree was prepared. This allowed assignment of all of the H. pylori genomes studied here to populations and delineation of the relationships among the hspAmerind genotypes. V225d was closest to the genotypes found among the Huitoto people of Colombia, while V225b was closest to Shi470, isolated from the Machiguenga people of Peru, and these two clades were sisters (Fig. 1).
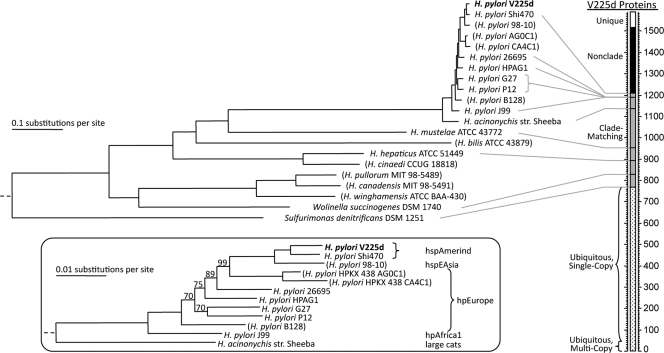
The availability of many more genes allows more-robust phylogenetic analysis of genomic data. V225d was placed among other Helicobacteraceae genomes by use of a maximum-likelihood phylogenetic analysis that employed over 500 protein families (Fig. 4). While Sulfurimonas denitrificans was outside the Helicobacter clade, the other available non-Helicobacter genome Wolinella succinogenes was within the Helicobacter clade, specifically with the H. canadensis/H. pullorum/H. winghamensis subgroup, indicating that the taxonomy of this subgroup needs revision. The H. pylori strains, together with H. acinonychis, were tightly clustered and distant from other Helicobacter strains. The close relationship between H. pylori and H. acinonychis, found in large cats, has been noted before and has been attributed to infection of a cat by ingestion of an early human (22, 50, 84). A comparison of the available H. pylori strain sequences revealed a pattern of human migration: the hpAfrica1 genotype is basal, passing through hpEurope genotypes to a tight clade with the hspEAsia genotype as a sister to the two hspAmerind genotypes. The common sequences among V225d and other completely sequenced Helicobacter genomes are represented in Table SA3 in the supplemental material. Specifically, V225d proteins were classified based on orthology group memberships established by considering only the other 11 fully sequenced Helicobacteraceae genomes. The first level of classification comprised the following categories: unique, clade matching (membership perfectly matching a clade represented in Fig. 4), or non-clade matching. The clade-matching proteins were subdivided according to which V225d-containing clade they matched or into the ubiquitous category (found in all taxa).
FIG. 4.
Phylogenetic analysis of Helicobacteraceae genomes. (Left) Maximum-likelihood tree based on a concatenation of masked alignments for 543 protein families. Incomplete genomes are indicated in parentheses. Bootstrap support values for the concatenated alignment were 100% for all nodes except those indicated in the inset (an elongated view of the H. acinonychis/H. pylori clade, denoting genotypes). Rooting was based on a similar analysis that included Campylobacter jejuni RM1221 (omitted, but represented by dashed line at left) as an outgroup. (Right) Stacked bar categorizing the 1,589 V225d proteins according to membership in families with proteins from the other 11 complete genomes. The proteins were sorted into four main categories: (i) unique proteins found only in V225d (white segment), (ii) non-clade-matching V225d proteins in families whose membership breaks the tree topology (black segment), (iii) clade-matching V225d proteins whose family membership matches an exclusive clade of the tree (gray segment), and (iv) ubiquitous proteins found in all 12 complete genomes (dotted segment, marking the fractions that are strictly single copy or multicopy, with at least one case of duplication). Within the clade-matching segment, proteins are stacked according to the size of the V225d-containing clade, with the smallest clade at the top; lines from the 11 other complete genomes indicate the numbers of V225d proteins shared by that taxon and all closer relatives of V225d. For example, the line from H. hepaticus shows that 718 V225d proteins are shared with all other complete Helicobacter genomes.
Characterization of the core H. pylori genome.
Ortholog group (OG) analysis was performed for all complete Helicobacteraceae genomes (n = 6), resulting in 765 OGs common to all of the genomes (containing a total of 908 V225d proteins) (Fig. 4). We identified 1,231 core ortholog families (comprising 1,428 V225d proteins) that are represented in all seven complete H. pylori genomes by using OrthoMCL, which employs the Markov clustering algorithm (21, 84). These results are consistent with prior studies of different genomes and methods that reported core sets ranging from 1,091 to 1,281 genes (30, 31, 50, 70). We compared this set of core genes with lists of H. pylori genes shown to be essential for microaerobic growth on rich medium (33 genes) (13) or for gastric colonization in gerbils (47 genes) (38) or mice (23 genes) (6). All of these essential genes were present in our core gene set, except that the RNase P protein gene was missing from our set because it had not been annotated in H. pylori G27. However, upon reexamination, it was found to be intact in the G27 genome (data not shown). An in silico metabolic-reconstruction study (82) predicted 128 genes essential for growth in nutrient-rich medium, all of which were present in the core OG set. A microarray survey identified 1,150 H. pylori genes present in all 56 strains tested (30), all but 77 of which were present in the core OG set. These studies provide independent partial validations of our methodology.
We identified 112 strain-specific genes in V225d, most with no known function and not closely related to any genes/proteins in the nonredundant databases (BLASTN E value of ≤1e−10). However, most of the strain-specific genes matched the genes of partially sequenced genomes of H. pylori strains HPKX_438_AG0C1 and 98-10 and H. acinonychis strain Sheeba. Thus, only two V225d genes, encoding FtsK/SpoIIIE and DjlA, have no known orthologs in the Helicobacter genus.
We identified 110 proteins unique to V225d, 60 singletons and 50 from 16 multimember OGs specific to V225d (see Table SA4 in the supplemental material). Of these, 90 proteins have no known function. We examined the remaining proteins to remove proteins that were smaller than 40 amino acids (aa) or that had no significant hits (E value of ≤1e−10) to the nonredundant protein database and rechecked their uniqueness. Most notably, the remaining 19 singleton proteins are outer membrane proteins, including a truncated HomB homolog, an FtsK/SpoIIIE family protein, and two type III restriction enzyme R proteins (see Table SA4 in the supplemental material). We identified 26 OGs specific to the Amerindian strains of V225d and Shi470; all but one (exodeoxyribonuclease 7 large-subunit protein) have no known function.
Divergent genes in H. pylori V225d.
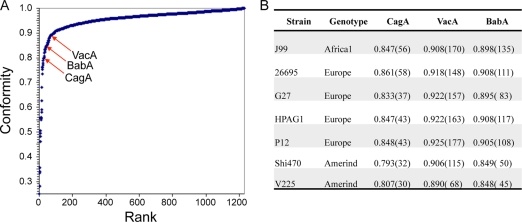
The V225d representative may not be uniformly divergent from other H. pylori strains in all of its OGs. Conformity scores for V225d were obtained for each of 1,231 proteins from the core OGs, by averaging the Smith-Waterman scores for pairwise alignment to the orthologs from the other six genomes and normalizing the data (Fig. 5 A). Interestingly, among the least conforming V225d proteins were three well-known interaction proteins, CagA, VacA, and the adhesin BabA; others are listed in Table SA5 in the supplemental material. According to the corresponding 1,231 conformity scores for the other six genomes, these three important interaction proteins have even lower conformity scores in the two hspAmerind strains than in the other tested strains (Fig. 5B).
FIG. 5.
Low conformity of key interactive proteins of Helicobacter pylori strain V225d. (A) Conformity scores (average normalized scores of alignment to orthologs) for 1,231 proteins of V225d. (B) Conformity scores (and ranks among 1,231 families, shown in parentheses) for key interactive proteins of complete H. pylori genomes.
Unusual CagA.
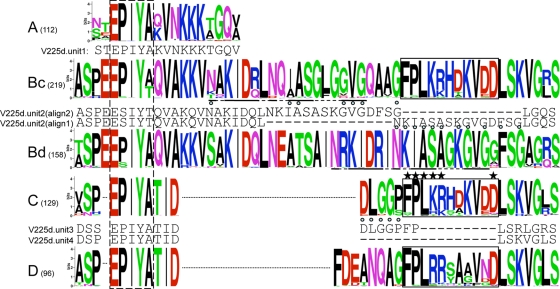
CagA is a multifunctional effector protein injected into host cells by the cag type IV secretion system. The phosphotyrosine (PY) region near the C terminus of CagA has variable numbers of units, each containing the phosphorylation site motif EPIYA. These units have been grouped into four classes, A, B, C, and D (32, 33); among these, the classes C (associated with hpEurope and hpAfrica1 genotypes) and D (associated with hspEAsia genotypes) are phosphorylated at substantially higher frequencies than are classes A and B (associated with hspEAsia and hpEurope genotypes) (33). A higher multiplicity of C or D class units is associated with increased gastric cancer rates (4, 36). A portion of the C unit outside its EPIYA sequence, termed the CRPIA (conserved repeat responsible for phosphorylation-independent activity) motif, has phosphorylation-independent effects (26, 54). We freshly analyzed 839 available complete PY region sequences, including those from the two known _cagA_2 alleles, and we found that 834 out of 839 were unique, including those from the two known _cagA_2 alleles. As in previous studies, our analysis initially identified four EPIYA unit classes, although upon refining the boundaries of the units, the B unit could be divided into two classes differing in their C-terminal portions: Bc, which usually is followed by a C unit, and Bd, which usually is followed by a D unit. Sequence logos for the unit classes are shown in Fig. 6. Interesting sequence relationships among the unit classes were observed. The Bc unit has a CRPIA motif virtually identical to that originally described for the C unit (79), and the D unit has a very similar sequence in the same location, which probably also has a CRPIA function. The Bc and Bd units share a motif that is displaced in our alignment (underlined in Fig. 6). Unit classification was refined by adding a notation when a unit had undergone an insertion, deletion, or fusion. We also noted that some PY regions included various extents of duplication from either the left or the right flank of the PY region. The most striking is represented by GenBank accession no. BAC10448, which has a nearly perfect 165-amino-acid sequence duplicating an ABdD cluster with portions of both its left and right flanking sequences.
FIG. 6.
CagA EPIYA units. All unique sequences (numbers in parentheses) for each of the five EPIYA unit classes were aligned, and a sequence logo was produced. Some logos are split (dotted lines) to maximize cross-class alignment. An additional sequence relationship between Bc and Bd is underlined. The dashed box marks the EPIYA segment, and the solid boxes mark the active portion of the CRPIA domain (starred positions reduced NF-κB activation by >40% upon Ala replacement [79]). CagA usually contains combinations of either Bc and C units or Bd and D units. V225d CagA (and five additional South American examples) has a B unit (V225.2) with a deletion; assignment to the Bd type (alignment 1) is preferred (11 distinguishing matches; blue circles), and V225d may therefore unusually mix Bd with C, but an alternative placement of the deletion (alignment 2) shows similarity to Bc. The last two EPIYA units of V225d (V225.3 and V225.4) have also undergone deletions, but V225.3 is readily assigned to the C type, while V225.4 has lost all positions that can distinguish between the C and D classes. Additional CagA sequences that mix EPIYA units include one from an Alaskan hspAmerind strain, with a fused Bd/Bc unit followed by a C unit, and two from Japan that mix Bc with D. Amino acid types are color coded, and the total height of each column represents its information content.
By classifying each unit, all 839 PY regions could be subtyped. This analysis yielded 55 types, although only 6 of these occurred more than seven times. Most (72%) PY regions were of either the ABdD or the ABcC type, and the next most common types were ABcCC, ABcCCC, ABc, and ABdBdD (see Table SA6 in the supplemental material). C units were never found together with D units. Only one sequence (GenBank accession no. BAD13935) mixes Bc with a D unit, and the seven sequences that appear to mix Bd with C are described below.
The PY region of V225d CagA was originally typed ABd−C−C−, signifying that all of the last three EPIYA units bear deletions. In fact, the terminal unit has such an extensive deletion that it cannot be distinguished as C or D; we designated it C because there is no precedent for C/D mixing. According to its type, V225d CagA is one of a very small group (7 [0.9%] of 839 sequences) that appear to mix Bd with C units; five others from equatorial South America, including Shi470 CagA, mix Bd with C (three of these were ABd−C−C−, one was ABd−C, and one was ABd−+C [bearing a deletion and an insertion in Bd]), and an Alaskan CagA has a unit that fuses Bd and Bc followed by a C unit. These analyses raise the question of whether the V225d CagA sequence might reflect recombination. From an alignment of the 109 available full-length CagA (and CagA2) sequences, the left and right flanks of the PY region were used for separate phylogenetic analyses. Trees for both flanks produced three clusters; one associated with D class EPIYA units, another associated with C class units, and the third consisted of the CagA and CagA2 proteins of the only two hspAmerind genotype strains in the analysis (Fig. 6). Only a few genomes allow assignment of these CagA groups to H. pylori populations, the C cluster to the hpEurope and hspWAfrica populations and the D cluster to the hspEAsia population. These trees indicate recombination between C- and D-type flanks or PY regions, but the distinctness of the Amerindian CagA, consistently in both PY region flanks, suggests that the unusual mixture of Bd− and C EPIYA units may be a long-term association. Ancient recombination between Bd and C EPIYA units may not have occurred, because the gene may be under positive selection, the distinguishing segments are short, and an alternative alignment of the Bd unit is a reasonable match to the Bc consensus (Fig. 6).
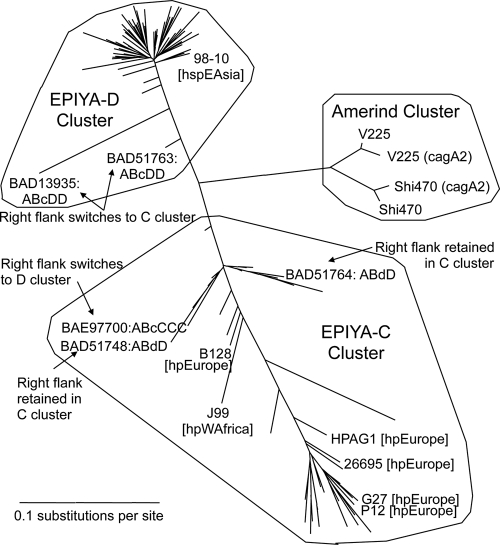
A second copy of cagA (_cagA_2) is currently known only from V225d and another hspAmerind genome, Shi470. Both of these _cagA_2 products differ from the cagA product by a DESL peptide unit repeat near the N terminus (17 copies in V225d and 8 in Shi470). Associated with this repeat in V225d (but not Shi470) CagA2 is an insertion of one additional nucleotide that breaks the reading frame, indicating that V225d _cagA_2 may be a pseudogene or subject to phase variation. The two CagA2 PY regions also are highly unusual, typed as C−C− (V225d) and CC− (Shi470), and have deletions in the left flank of the PY region. An interesting aspect of the CagA phylogenetic tree (Fig. 7) is that both CagA2 sequences, at both flanks of the PY region, match their within-genome CagA partners better than they match each other. This suggests gene conversion events (66) that have homogenized the cagA and _cagA_2 sequences within each genome, subsequent to an ancestral duplication event that produced _cagA_2.
FIG. 7.
Phylogeny of the flanks of the CagA phosphotyrosine (PY) region. All 109 full-length CagA sequences were aligned, and the regions on the left and right flanks of the PY region were extracted for separate phylogenetic analysis and converted to the corresponding gene alignments. The unrooted tree for the left flank is shown here; the tree for the right flank is highly similar, with the exceptions noted. Three main clusters appear: a C cluster, whose members all (except those marked) contain Bc or C EPIYA units and have the right flank in the same cluster; a D cluster, as described above but with Bd or D EPIYA units; and an hspAmerind cluster, consisting of CagA and CagA2 from Shi470 and V225d. The indicated exceptions in the C and D clusters suggest from one to three recombination events. Both left and right flanks in the hspAmerind cluster form an isolated cluster approximately equidistant from the C and D clusters. Note that the CagA2 sequences are less similar to each other than to their within-strain CagA sequences.
Induction of cag PAI-dependent phenotypes in AGS cells by strain V225d.
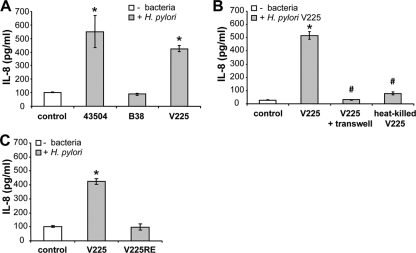
The unusual features of the hspAmerind cag pathogenicity island (cag PAI) raise the possibility that this PAI is no longer functional. We tested for PAI function, assaying host cells for secretion of the cytokine IL-8 and for the hummingbird phenotype. V225d strongly induced IL-8 secretion in human gastric adenocarcinoma AGS cells at a level similar to that for the cag PAI-positive strain 43504 (Fig. 8 A). As anticipated, the H. pylori cag PAI-negative strain B38 did not induce IL-8 secretion. These results provide evidence that V225d possesses a functional cag PAI, despite its atypical cagA. Stimulation of IL-8 secretion was blocked when bacterial adherence to AGS cells was inhibited by separate coculture across Transwell inserts. Moreover, live bacteria were necessary to stimulate IL-8 secretion, as heat-killed bacteria did not do so (Fig. 8B).
FIG. 8.
Helicobacter pylori V225d induces interleukin-8 secretion in gastric epithelial cells, a process that requires bacterial adherence. (A) Quantitation of IL-8 secretion by AGS cells under basal conditions (control) and after 24 h of coculture with H. pylori V225, 43504, or B38. (B) Cocultures were performed in the absence or in the presence of Transwell inserts (0.22 μm pore size), preventing cell-bacterium contact, and with live or heat-killed bacteria. (C) Cocultures were performed as described for panel A, but V225 and V225-RE were compared. *, P < 0.05 compared to control; #, P < 0.05 compared to the live-H. pylori V225-induced response without Transwell inserts. Error bars indicate SD from triplicates in one representative experiment.
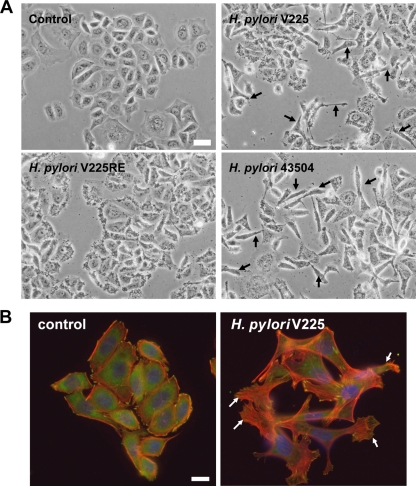
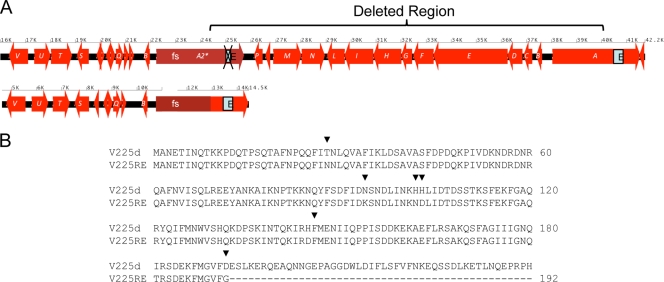
AGS cells in culture displayed a typical epithelial cell morphology, with polygon-shaped adherent cellular clusters (Fig. 9). H. pylori V225d induced the hummingbird phenotype, marked by the elongation of AGS cells and the disruption of cell/cell junctions (Fig. 9). V225d showed an activity similar to that of the positive control, the cag PAI-positive strain 43504, both stimulating IL-8 secretion and inducing the hummingbird phenotype. On the contrary, the H. pylori V225-RE strain, which had been isolated from a mouse 33 weeks after inoculation of V225d, had lost the ability to induce the hummingbird phenotype (Fig. 9B) and to stimulate IL-8 secretion (Fig. 8C). Based on these functional differences in the phenotypes of V225d and V225-RE in AGS cells, we sequenced the cag PAI of V225-RE and performed comparative genomic analyses with that of V225d. Our data demonstrate a 15-kb-segment deletion in the cag PAI region that can be explained by homologous recombination between the cagA and _cagA_2 genes of V225d (Fig. 10). This deleted region contained 13 cag PAI ORFs, including cagP, cagM, cagN, cagL, cagI, cagH, cagG, cagF, cagE, cagD, cagC, and cagB, many of which are essential for the type IV secretion system that injects the CagA protein into epithelial cells. The deletion also effectively eliminated cagA, since its upstream portion was replaced with that from _cagA_2, including the frameshift mutation that severely truncates the CagA protein.
FIG. 9.
Helicobacter pylori strain V225d induces the formation of the “hummingbird phenotype” in AGS cells. AGS cells were cultured under basal conditions (control) or for 24 h with H. pylori strain 43504, V225, or V225-RE. (A) Phase-contrast microscopy imaging of AGS cells. Bar, 20 μm. (B) Fluorescence microscopy imaging of the actin cytoskeleton and focal adhesions of AGS cells. Shown are merged images of the fluorescent staining of F-actin (with Alexa Fluor 568-labeled phalloidin, red), vinculin (with primary and Alexa Fluor 488-labeled secondary antibodies, green), and DNA (with Hoechst 33342 compound, blue). Control cells are organized in clusters of polygon-shaped cells, with focal adhesions at the cell periphery, few actin stress fibers, and no lamellipodia. Arrows indicate cells after coculture that exhibit cytoskeleton remodeling, with dissociation of cell clusters, loss of the polygon shape, cellular elongation with lamellipodia and stress fiber formation, and an increase in focal-adhesion number and size, defining the hummingbird phenotype. Bar, 10 μm.
FIG. 10.
Helicobacter pylori strain V225-RE contains a large deletion in the cag pathogenicity island. (A) A 15-kb region affecting 13 cag genes, between cagA and the pseudogene _cagA_2, is missing in the strain V225-RE, reisolated from experimentally infected mice. The single letters represent cagA (A) through cagX (X). Genes encoding non-Cag proteins are not labeled. A boxed E represents the EPIYA region. (B) Alignment of V225d cagA and predicted coding sequence of the V225-RE _cagA_2-cagA fusion, subject to the frameshift (fs) mutation in _cagA_2, which results in the formation of a stop codon at amino acid 193.
VacA analysis.
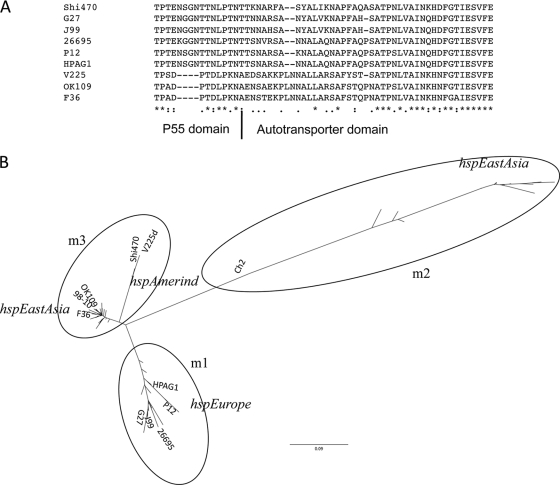
The vacuolating protein VacA is another important H. pylori interaction factor. In addition to the single distinctive VacA ortholog found in all H. pylori genomes, a variable number of longer VacA-like paralogs are found in each genome (46, 59), but their function is not understood. Multiple sequence alignment of H. pylori VacA orthologs from the seven complete genomes shows a V225d-specific 4-aa deletion in the p55 domain, followed by a region of marked dissimilarity (Fig. 11 A). We collected 88 complete VacA protein sequences from GenBank to analyze whether this deletion was specific to V225d. Two East Asian strains (F36 and OK109) also share similar changes in their putative VacA products (Fig. 11A; also see Fig. SA2 in the supplemental material). We also collected nucleotide sequences for these 88 vacA genes and performed in silico PCR using primers for the s and m regions. A maximum-likelihood tree of VacA proteins reveals three distinct clusters (see Figure SA3 in the supplemental material). The m1 and m2 clusters consist of all VacA proteins whose genes were amplified by m1 and m2 primers, respectively. We found that the m regions of V225d and 42 other vacA sequences clustered in a new category (termed m3) that would not be amplifiable by these classical primers. A maximum-likelihood tree of the isolated m region revealed a similar topology, with the exception of Shi470 and V225d, which are on a separate branch within the new m3 cluster (Fig. 11B).
FIG. 11.
Comparison of VacA sequences. (A) Multiple sequence alignment of VacA proteins. Sequences from seven complete genomes and two VacA sequences with changes similar to that for V225d are indicated. The multiple sequence alignment of all 88 complete VacA sequences is shown in Fig. SA2 in the supplemental material. Stars, identical residues; dots, conserved substitutions; semicolons, semiconserved substitutions. (B) Phylogenetic tree of the m regions of VacA proteins from 88 strains. The VacA proteins of the seven complete genomes are labeled. The m1 and m2 clusters contain VacA proteins whose genes can be amplified by m1 and m2 genotyping primers, while the newly designated m3 cluster contains VacA proteins whose gene sequences fail to amplify with either m1 or m2 primers.
DISCUSSION
In this study, we compared the genome sequence of an H. pylori strain isolated from a Venezuelan Piaroa Amerindian with six available H. pylori whole-genome sequences and other H. pylori sequences, and to relate genomic differences to strain-specific variation in induction of inflammatory responses, we also performed functional studies. Sequencing Amerindian strains is important due to the bias of available genomes toward strains that are predominantly from people of European ancestry. The genomes are quite similar in size, number of genes and proteins, and structural organization of the chromosome. By phylogenetic analysis, V225d is most proximal to the Peruvian strain Shi470, in the hspAmerind cluster, which shows that they form a clade that is related to the hspEAsia cluster, confirming prior reports that H. pylori traces the Asian ancestry of New World natives (24). Indeed, our tree robustly (based on 543 protein families) supports the prehistoric human migration out of Africa, through Europe, to Asia, and on to the New World, as described using independent methods (1-5).
Using pairwise Smith-Waterman alignment within each H. pylori core OG, we identified the least conforming (most divergent) proteins of V225d, which included the key interaction proteins CagA, VacA, and BabA. These proteins were more divergent for the Amerindian genomes than they were for the other genomes studied (Fig. 5), yet these genomes were not particularly divergent in the species tree (Fig. 4). This difference suggests that there has been a greater impact of host interaction on the hspAmerind lineage than on other H. pylori lineages. The finding of BabA as highly divergent is consistent with the highly skewed distribution of its ligand, blood group O antigen, in Amerindians (2) and suggests that parallel, yet to be identified host polymorphism skewing relevant to both CagA and VacA interactions exists in Amerindians.
It has been claimed that Amerindian strains either do not carry the cag pathogenicity island or carry only a vestigial, incomplete form (30). We now show that CagA of V225d is distinctive in many ways from its hpEurope and hspEAsia counterparts, suggesting altered activities that will be important to address experimentally. Its phosphotyrosine region contains four EPIYA units, one of the short A class and three that are usually longer but that contain deletions. As part of this variation, the cagA product contains no intact CRPIA domain, to which phosphorylation-independent activities have been ascribed (69, 79). The deletions may also reduce the high frequency of tyrosine phosphorylation that is usually associated with units of the C class. hspAmerind CagA appears long isolated from hpEurope and hspEAsia CagA, likely subject to positive selection, but further evolutionary studies are needed.
There is great allelic diversity particularly near the 5′ terminus of vacA (the s region) and in the midregion of the H. pylori gene (the m region) (16). Subtypes of vacA alleles (combinations of s and m allelic variants) have a geographically defined distribution (85). V225d vacA is of the s1a type and clusters with East Asian vacA. The phylogenetic tree of the m region of the VacA protein shows that the Amerindian strains V225d and Shi470 form a distinct subgroup along with the other East Asian strain. However, the distinctive 4-aa deletion in the oligomerization region of the p55 domain, followed by a region of marked dissimilarity in the autotransporter domain, in V225d may have the same origin as those in the two East Asian strains. One hypothesis (85) is that H. pylori strains have been under selective pressure to coevolve into either a high-VacA-level (CagA+) or a low-VacA-level (CagA−) phenotype, with few intermediate forms. The variation in both of these loci in the Amerindian strains provides further support that their selection may be linked. Another plausible explanation is that this mosaic form in V225d might have arisen via homologous recombination among vacA alleles from East Asian strains.
Functionally, in terms of inducing inflammatory responses, V225d resembled the H. pylori cag PAI-positive strain 43504 and had a greater IL-8-stimulating ability than the cag PAI-negative strain B38, providing evidence that V225d, albeit atypical, carries a functional cag PAI capable of inducing inflammatory responses in human gastric epithelial cells. That the IL-8-stimulating ability was suppressed when bacterial adherence to AGS cells was blocked or after heat inactivation of the bacterium indicates that V225d-induced IL-8 secretion requires bacterial adherence and an active mechanism (cag PAI-encoded type IV secretion system). Consistent with prior studies (65, 76), the passage of V225d through a mouse stomach for 33 weeks and reisolation as V225-RE resulted in an impaired ability of the bacterium to stimulate IL-8 secretion and the hummingbird phenotype. However, the genomic basis for cag inactivation during murine passage had not been thoroughly defined. Now, sequencing of the cag PAI from strain V225-RE and comparative genomic analyses with V225d revealed a 15-kb deletion. This missing region contains 13 genes, including cagM, cagL, cagI, cagG, and cagE, reported to play critical roles in both the operation of the type IV secretion system (63) and the activation of the transcription factor NF-κB, involved in mediating IL-8 secretion (62), as well as cagF and cagH, involved only with the type IV secretion system (63). This work demonstrates that the missing cag PAI segment in V225-RE contains genes critical to the operation of the type IV secretion pathway as well as the stimulation of IL-8 secretion and inflammation.
In conclusion, this study presents an analysis of the genome sequence of the Amerindian H. pylori strain V225d. A comprehensive reanalysis of the CagA PY region revealed the prevalences and geographical distributions of different EPIYA types in H. pylori. Our robust multiprotein phylogenetic tree does not show a great divergence of the hspAmerind genomes from an hspEAsia pangenome, consistent with MLST data. However, the single-gene trees for cagA and vacA show strong divergence from both hspEAsia and hpEurope counterparts. The exaggerated evolution of these genes that has occurred over the ∼15,000 years since the arrival of Amerindian ancestors to the Americas makes them less suitable for deducing evolutionary relationships but highlights the need to assess the physiological activities of the Amerindian alleles, as they may contribute to the effects of hspAmerind H. pylori on gastric inflammation and human health. It remains unknown whether the atypical nature of CagA observed in H. pylori strains from Africa and Latin America (Amerindian strains) contributes to the low incidence of _H. pylori_-associated gastric cancer reported in related locales.
Supplementary Material
[Supplemental material]
Acknowledgments
This project was funded by Virginia Bioinformatics Institute (VBI) support to B.W.S., an exploratory VBI grant to J.B.-R., UPR FIPI grant 880314 to M.G.D.-B., and NIH grant R01 GM 63270 to M.J.B.
Footnotes
▿
Published ahead of print on 16 April 2010.
REFERENCES
- 1.Amieva, M. R., R. Vogelmann, A. Covacci, L. S. Tompkins, W. J. Nelson, and S. Falkow. 2003. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300**:**1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aspholm-Hurtig, M., G. Dailide, M. Lahmann, A. Kalia, D. Ilver, N. Roche, S. Vikstrom, R. Sjostrom, S. Linden, A. Backstrom, C. Lundberg, A. Arnqvist, J. Mahdavi, U. J. Nilsson, B. Velapatino, R. H. Gilman, M. Gerhard, T. Alarcon, M. Lopez-Brea, T. Nakazawa, J. G. Fox, P. Correa, M. G. Dominguez-Bello, G. I. Perez-Perez, M. J. Blaser, S. Normark, I. Carlstedt, S. Oscarson, S. Teneberg, D. E. Berg, and T. Boren. 2004. Functional adaptation of BabA, the H. pylori ABO blood group antigen binding adhesin. Science 305**:**519-522. [DOI] [PubMed] [Google Scholar]
- 3.Atherton, J. C., and M. J. Blaser. 2009. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J. Clin. Invest. 119**:**2475-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azuma, T., S. Yamazaki, A. Yamakawa, M. Ohtani, A. Muramatsu, H. Suto, Y. Ito, M. Dojo, Y. Yamazaki, M. Kuriyama, Y. Keida, H. Higashi, and M. Hatakeyama. 2004. Association between diversity in the Src homology 2 domain-containing tyrosine phosphatase binding site of Helicobacter pylori CagA protein and gastric atrophy and cancer. J. Infect. Dis. 189**:**820-827. [DOI] [PubMed] [Google Scholar]
- 5.Bagnoli, F., L. Buti, L. Tompkins, A. Covacci, and M. R. Amieva. 2005. Helicobacter pylori CagA induces a transition from polarized to invasive phenotypes in MDCK cells. Proc. Natl. Acad. Sci. U. S. A. 102**:**16339-16344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baldwin, D. N., B. Shepherd, P. Kraemer, M. K. Hall, L. K. Sycuro, D. M. Pinto-Santini, and N. R. Salama. 2007. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect. Immun. 75**:**1005-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Invest. 113**:**321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blaser, M. J., and S. Falkow. 2009. What are the consequences of the disappearing human microbiota? Nat. Rev. Microbiol. 7**:**887-894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boncristiano, M., S. R. Paccani, S. Barone, C. Ulivieri, L. Patrussi, D. Ilver, A. Amedei, M. M. D'Elios, J. L. Telford, and C. T. Baldari. 2003. The Helicobacter pylori vacuolating toxin inhibits T cell activation by two independent mechanisms. J. Exp. Med. 198**:**1887-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourzac, K. M., and K. Guillemin. 2005. _Helicobacter pylori_-host cell interactions mediated by type IV secretion. Cell. Microbiol. 7**:**911-919. [DOI] [PubMed] [Google Scholar]
- 11.Brandt, S., T. Kwok, R. Hartig, W. Konig, and S. Backert. 2005. NF-kappaB activation and potentiation of proinflammatory responses by the Helicobacter pylori CagA protein. Proc. Natl. Acad. Sci. U. S. A. 102**:**9300-9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Censini, S., C. Lange, Z. Xiang, J. E. Crabtree, P. Ghiara, M. Borodovsky, R. Rappuoli, and A. Covacci. 1996. Cag, a pathogenicity island of Helicobacter pylori, encodes type I-specific and disease-associated virulence factors. Proc. Natl. Acad. Sci. U. S. A. 93**:**14648-14653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183**:**1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, Y., and M. J. Blaser. 2007. Inverse associations of Helicobacter pylori with asthma and allergy. Arch. Intern. Med. 167**:**821-827. [DOI] [PubMed] [Google Scholar]
- 15.Covacci, A., S. Censini, M. Bugnoli, R. Petracca, D. Burroni, G. Macchia, A. Massone, E. Papini, Z. Xiang, N. Figura, et al. 1993. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc. Natl. Acad. Sci. U. S. A. 90**:**5791-5795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cover, T. L., and S. R. Blanke. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3**:**320-332. [DOI] [PubMed] [Google Scholar]
- 17.Darling, A. C., B. Mau, F. R. Blattner, and N. T. Perna. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14**:**1394-1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degnan, P. H., Y. Yu, N. Sisneros, R. A. Wing, and N. A. Moran. 2009. Hamiltonella defensa, genome evolution of protective bacterial endosymbiont from pathogenic ancestors. Proc. Natl. Acad. Sci. U. S. A. 106**:**9063-9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dominguez-Bello, M. G., M. E. Perez, M. C. Bortolini, F. M. Salzano, L. R. Pericchi, O. Zambrano-Guzman, and B. Linz. 2008. Amerindian Helicobacter pylori strains go extinct, as European strains expand their host range. PLoS One 3**:**e3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5**:**113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Enright, A. J., S. Van Dongen, and C. A. Ouzounis. 2002. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30**:**1575-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eppinger, M., C. Baar, B. Linz, G. Raddatz, C. Lanz, H. Keller, G. Morelli, H. Gressmann, M. Achtman, and S. C. Schuster. 2006. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet. 2**:**e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Falush, D., M. Stephens, and J. K. Pritchard. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164**:**1567-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falush, D., T. Wirth, B. Linz, J. K. Pritchard, M. Stephens, M. Kidd, M. J. Blaser, D. Y. Graham, S. Vacher, G. I. Perez-Perez, Y. Yamaoka, F. Megraud, K. Otto, U. Reichard, E. Katzowitsch, X. Wang, M. Achtman, and S. Suerbaum. 2003. Traces of human migrations in Helicobacter pylori populations. Science 299**:**1582-1585. [DOI] [PubMed] [Google Scholar]
- 25.Figueiredo, C., J. C. Machado, P. Pharoah, R. Seruca, S. Sousa, R. Carvalho, A. F. Capelinha, W. Quint, C. Caldas, L. J. van Doorn, F. Carneiro, and M. Sobrinho-Simoes. 2002. Helicobacter pylori and interleukin 1 genotyping: an opportunity to identify high-risk individuals for gastric carcinoma. J. Natl. Cancer Inst. 94**:**1680-1687. [DOI] [PubMed] [Google Scholar]
- 26.Franco, A. T., D. A. Israel, M. K. Washington, U. Krishna, J. G. Fox, A. B. Rogers, A. S. Neish, L. Collier-Hyams, G. I. Perez-Perez, M. Hatakeyama, R. Whitehead, K. Gaus, D. P. O'Brien, J. Romero-Gallo, and R. M. Peek. 2005. Activation of β-catenin by carcinogenic Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 102**:**10646-10651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galmiche, A., J. Rassow, A. Doye, S. Cagnol, J. C. Chambard, S. Contamin, V. de Thillot, I. Just, V. Ricci, E. Solcia, E. Van Obberghen, and P. Boquet. 2000. The N-terminal 34 kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. EMBO J. 19**:**6361-6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghose, C., G. I. Perez-Perez, M. G. Dominguez-Bello, D. T. Pride, C. M. Bravi, and M. J. Blaser. 2002. East Asian genotypes of Helicobacter pylori strains in Amerindians provide evidence for its ancient human carriage. Proc. Natl. Acad. Sci. U. S. A. 99**:**15107-15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghose, C., G. I. Perez-Perez, L. J. van Doorn, M. G. Dominguez-Bello, and M. J. Blaser. 2005. High frequency of gastric colonization with multiple Helicobacter pylori strains in Venezuelan subjects. J. Clin. Microbiol. 43**:**2635-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gressmann, H., B. Linz, R. Ghai, K. P. Pleissner, R. Schlapbach, Y. Yamaoka, C. Kraft, S. Suerbaum, T. F. Meyer, and M. Achtman. 2005. Gain and loss of multiple genes during the evolution of Helicobacter pylori. PLoS Genet. 1**:**e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han, Y. H., W. Z. Liu, Y. Z. Shi, L. Q. Lu, S. Xiao, Q. H. Zhang, and G. P. Zhao. 2007. Comparative genomics profiling of clinical isolates of Helicobacter pylori in Chinese populations using DNA microarray. J. Microbiol. 45**:**21-28. [PubMed] [Google Scholar]
- 32.Hatakeyama, M. 2004. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer 4**:**688-694. [DOI] [PubMed] [Google Scholar]
- 33.Higashi, H., R. Tsutsumi, A. Fujita, S. Yamazaki, M. Asaka, T. Azuma, and M. Hatakeyama. 2002. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc. Natl. Acad. Sci. U. S. A. 99**:**14428-14433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Islami, F., and F. Kamangar. 2008. Helicobacter pylori and esophageal cancer risk: a meta-analysis. Cancer Prev. Res. 1**:**329-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jolley, K. A., M. S. Chan, and M. C. Maiden. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5**:**86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, K. R., Y. M. Joo, S. Jang, Y.-J. Yoo, H. S. Lee, I.-S. Chung, C. H. Olsen, J. M. Whitmire, D. S. Merrell, and J.-H. Cha. 2009. Polymorphism in the CagA EPIYA motif impacts development of gastric cancer. J. Clin. Microbiol. 47**:**959-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamada, T., J. Hata, H. Kusunoki, M. Ito, S. Tanaka, Y. Kawamura, K. Chayama, and K. Haruma. 2005. Eradication of Helicobacter pylori increases the incidence of hyperlipidaemia and obesity in peptic ulcer patients. Dig. Liver Dis. 37**:**39-43. [DOI] [PubMed] [Google Scholar]
- 38.Kavermann, H., B. P. Burns, K. Angermuller, S. Odenbreit, W. Fischer, K. Melchers, and R. Haas. 2003. Identification and characterization of Helicobacter pylori genes essential for gastric colonization. J. Exp. Med. 197**:**813-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Keates, S., A. C. Keates, M. Warny, R. M. Peek, Jr., P. G. Murray, and C. P. Kelly. 1999. Differential activation of mitogen-activated protein kinases in AGS gastric epithelial cells by cag+ and cag− Helicobacter pylori. J. Immunol. 163**:**5552-5559. [PubMed] [Google Scholar]
- 40.Kidd, M., J. A. Louw, and I. N. Marks. 1999. Helicobacter pylori in Africa: observations on an ‘enigma within an enigma.’ J. Gastroenterol. Hepatol. 14**:**851-858. [DOI] [PubMed] [Google Scholar]
- 41.Kimura, M., S. Goto, A. Wada, K. Yahiro, T. Niidome, T. Hatakeyama, H. Aoyagi, T. Hirayama, and T. Kondo. 1999. Vacuolating cytotoxin purified from Helicobacter pylori causes mitochondrial damage in human gastric cells. Microb. Pathog. 26**:**45-52. [DOI] [PubMed] [Google Scholar]
- 42.Lee, A., J. O'Rourke, M. C. De Ungria, B. Robertson, G. Daskalopoulos, and M. F. Dixon. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112**:**1386-1397. [DOI] [PubMed] [Google Scholar]
- 43.Leunk, R. D., P. T. Johnson, B. C. David, W. G. Kraft, and D. R. Morgan. 1988. Cytotoxic activity in broth-culture filtrates of Campylobacter pylori. J. Med. Microbiol. 26**:**93-99. [DOI] [PubMed] [Google Scholar]
- 44.Li, L., C. J. Stoeckert, Jr., and D. S. Roos. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13**:**2178-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linz, B., F. Balloux, Y. Moodley, A. Manica, H. Liu, P. Roumagnac, D. Falush, C. Stamer, F. Prugnolle, S. W. van der Merwe, Y. Yamaoka, D. Y. Graham, E. Perez-Trallero, T. Wadstrom, S. Suerbaum, and M. Achtman. 2007. An African origin for the intimate association between humans and Helicobacter pylori. Nature 445**:**915-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Löwer, M., C. Weydig, D. Metzler, A. Reuter, A. Starzinski-Powitz, S. Wessler, and G. Schneider. 2008. Prediction of extracellular proteases of the human pathogen Helicobacter pylori reveals proteolytic activity of the Hp1018/19 protein HtrA. PLoS One 3**:**e3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U. S. A. 95**:**3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437**:**376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marini, E., A. L. Maldonado-Contreras, S. Cabras, G. Hidalgo, R. Buffa, A. Marin, G. Floris, W. Racugno, L. R. Pericchi, M. E. Castellanos, M. Groschl, M. J. Blaser, and M. G. Dominguez-Bello. 2007. Helicobacter pylori and intestinal parasites are not detrimental to the nutritional status of Amerindians. Am. J. Trop. Med. Hyg. 76**:**534-540. [PubMed] [Google Scholar]
- 50.McClain, M. S., C. L. Shaffer, D. A. Israel, R. M. Peek, Jr., and T. L. Cover. 2009. Genome sequence analysis of Helicobacter pylori strains associated with gastric ulceration and gastric cancer. BMC Genomics 10**:**3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moodley, Y., B. Linz, Y. Yamaoka, H. M. Windsor, S. Breurec, J. Y. Wu, A. Maady, S. Bernhoft, J. M. Thiberge, S. Phuanukoonnon, G. Jobb, P. Siba, D. Y. Graham, B. J. Marshall, and M. Achtman. 2009. The peopling of the Pacific from a bacterial perspective. Science 323**:**527-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moss, S. F., E. M. Sordillo, A. M. Abdalla, V. Makarov, Z. Hanzely, G. I. Perez-Perez, M. J. Blaser, and P. R. Holt. 2001. Increased gastric epithelial cell apoptosis associated with colonization with cagA+ Helicobacter pylori strains. Cancer Res. 61**:**1406-1411. [PubMed] [Google Scholar]
- 53.MSAS. 2000. Anuario de epidemiología y estadística vital, 1999. Ministerio de Sanidad y Asistencia Social, Caracas, Venezuela.
- 54.Murata-Kamiya, N., Y. Kurashima, Y. Teishikata, Y. Yamahashi, Y. Saito, H. Higashi, H. Aburatani, T. Akiyama, R. M. Peek, Jr., T. Azuma, and M. Hatakeyama. 2007. Helicobacter pylori CagA interacts with E-cadherin and deregulates the [beta]-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene 26**:**4617-4626. [DOI] [PubMed] [Google Scholar]
- 55.Nakayama, M., M. Kimura, A. Wada, K. Yahiro, K. Ogushi, T. Niidome, A. Fujikawa, D. Shirasaka, N. Aoyama, H. Kurazono, M. Noda, J. Moss, and T. Hirayama. 2004. Helicobacter pylori VacA activates the p38/activating transcription factor 2-mediated signal pathway in AZ-521 cells. J. Biol. Chem. 279**:**7024-7028. [DOI] [PubMed] [Google Scholar]
- 56.Nomura, A., G. N. Stemmermann, P.-H. Chyou, G. I. Perez-Perez, and M. J. Blaser. 1994. Helicobacter pylori infection and the risk for duodenal and gastric ulceration. Ann. Intern. Med. 120**:**977-981. [DOI] [PubMed] [Google Scholar]
- 57.Nomura, A. M. Y., G. I. Perez-Perez, J. Lee, G. Stemmermann, and M. J. Blaser. 2002. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am. J. Epidemiol. 155**:**1054-1059. [DOI] [PubMed] [Google Scholar]
- 58.Reference deleted.
- 59.Oh, J. D., H. Kling-Backhed, M. Giannakis, J. Xu, R. S. Fulton, L. A. Fulton, H. S. Cordum, C. Wang, G. Elliott, J. Edwards, E. R. Mardis, L. G. Engstrand, and J. I. Gordon. 2006. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. U. S. A. 103**:**9999-10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohnishi, N., H. Yuasa, S. Tanaka, H. Sawa, M. Miura, A. Matsui, H. Higashi, M. Musashi, K. Iwabuchi, M. Suzuki, G. Yamada, T. Azuma, and M. Hatakeyama. 2008. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc. Natl. Acad. Sci. U. S. A. 105**:**1003-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parsonnet, J., G. D. Friedman, N. Orentreich, and H. Vogelman. 1997. Risk for gastric cancer in people with CagA positive or CagA negative Helicobacter pylori infection. Gut 40**:**297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peek, R. M., Jr. 2005. Orchestration of aberrant epithelial signaling by Helicobacter pylori CagA. Sci. STKE 2005**:**pe14. [DOI] [PubMed] [Google Scholar]
- 63.Peek, R. M., Jr., M. J. Blaser, D. J. Mays, M. H. Forsyth, T. L. Cover, S. Y. Song, U. Krishna, and J. A. Pietenpol. 1999. Helicobacter pylori strain-specific genotypes and modulation of the gastric epithelial cell cycle. Cancer Res. 59**:**6124-6131. [PubMed] [Google Scholar]
- 64.Permin, H., and L. P. Andersen. 2005. Inflammation, immunity, and vaccines for Helicobacter infection. Helicobacter 10(Suppl. 1)**:**21-25. [DOI] [PubMed] [Google Scholar]
- 65.Philpott, D. J., D. Belaid, P. Troubadour, J.-M. Thiberge, J. Tankovic, A. Labigne, and R. L. Ferrero. 2002. Reduced activation of inflammatory responses in host cells by mouse-adapted Helicobacter pylori isolates. Cell. Microbiol. 4**:**285-296. [DOI] [PubMed] [Google Scholar]
- 66.Pride, D. T., and M. J. Blaser. 2002. Concerted evolution between duplicated genetic elements in Helicobacter pylori. J. Mol. Biol. 316**:**629-642. [DOI] [PubMed] [Google Scholar]
- 67.Reyes-Prieto, A., and D. Bhattacharya. 2007. Phylogeny of nuclear-encoded plastid-targeted proteins supports an early divergence of glaucophytes within Plantae. Mol. Biol. Evol. 24**:**2358-2361. [DOI] [PubMed] [Google Scholar]
- 68.Rothenbacher, D., M. J. Blaser, G. Bode, and H. Brenner. 2000. Inverse relationship between gastric colonization of Helicobacter pylori and diarrheal illnesses in children: results of a population-based cross-sectional study. J. Infect. Dis. 182**:**1446-1449. [DOI] [PubMed] [Google Scholar]
- 69.Saadat, I., H. Higashi, C. Obuse, M. Umeda, N. Murata-Kamiya, Y. Saito, H. Lu, N. Ohnishi, T. Azuma, A. Suzuki, S. Ohno, and M. Hatakeyama. 2007. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature 447**:**330-333. [DOI] [PubMed] [Google Scholar]
- 70.Salama, N., K. Guillemin, T. K. McDaniel, G. Sherlock, L. Tompkins, and S. Falkow. 2000. A whole-genome microarray reveals genetic diversity among Helicobacter pylori strains. Proc. Natl. Acad. Sci. U. S. A. 97**:**14668-14673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schuler, G. D. 1997. Sequence mapping by electronic PCR. Genome Res. 7**:**541-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 96**:**14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Selbach, M., S. Moese, C. R. Hauck, T. F. Meyer, and S. Backert. 2002. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J. Biol. Chem. 277**:**6775-6778. [DOI] [PubMed] [Google Scholar]
- 74.Snyder, E. E., N. Kampanya, J. Lu, E. K. Nordberg, H. R. Karur, M. Shukla, J. Soneja, Y. Tian, T. Xue, H. Yoo, F. Zhang, C. Dharmanolla, N. V. Dongre, J. J. Gillespie, J. Hamelius, M. Hance, K. I. Huntington, D. Jukneliene, J. Koziski, L. Mackasmiel, S. P. Mane, V. Nguyen, A. Purkayastha, J. Shallom, G. Yu, Y. Guo, J. Gabbard, D. Hix, A. F. Azad, S. C. Baker, S. M. Boyle, Y. Khudyakov, X. J. Meng, C. Rupprecht, J. Vinje, O. R. Crasta, M. J. Czar, A. Dickerman, J. D. Eckart, R. Kenyon, R. Will, J. C. Setubal, and B. W. Sobral. 2007. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 35**:**D401-D406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reference deleted.
- 76.Sozzi, M., M. Crosatti, S.-K. Kim, J. Romero, and M. J. Blaser. 2001. Heterogeneity of Helicobacter pylori cag genotypes in experimentally infected mice. FEMS Microbiol. Lett. 203**:**109-114. [DOI] [PubMed] [Google Scholar]
- 77.Stamatakis, A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22**:**2688-2690. [DOI] [PubMed] [Google Scholar]
- 78.Strobel, S., S. Bereswill, P. Balig, P. Allgaier, H.-G. Sonntag, and M. Kist. 1998. Identification and analysis of a new vacA genotype variant of Helicobacter pylori in different patient groups in Germany. J. Clin. Microbiol. 36**:**1285-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suzuki, M., H. Mimuro, K. Kiga, M. Fukumatsu, N. Ishijima, H. Morikawa, S. Nagai, S. Koyasu, R. H. Gilman, D. Kersulyte, D. E. Berg, and C. Sasakawa. 2009. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe 5**:**23-34. [DOI] [PubMed] [Google Scholar]
- 80.Talavera, G., and J. Castresana. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56**:**564-577. [DOI] [PubMed] [Google Scholar]
- 81.Thiberge, J. M., P. Lehours, C. Bouchier, L. Ma, S. Creno, C. Medigue, D. Vallenet, C. Ecobichon, C. Bourseaux-Eude, and A. Labigne. 2006. Sequence of the first Helicobacter pylori strains involved in low grade gastric mucosa associated tissue (MALT) lymphoma. Helicobacter 11**:**02.01. [Google Scholar]
- 82.Thiele, I., T. D. Vo, N. D. Price, and B. O. Palsson. 2005. Expanded metabolic reconstruction of Helicobacter pylori (iIT341 GSM/GPR): an in silico genome-scale characterization of single- and double-deletion mutants. J. Bacteriol. 187**:**5818-5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tummuru, M. K., T. L. Cover, and M. J. Blaser. 1993. Cloning and expression of a high-molecular-mass major antigen of Helicobacter pylori: evidence of linkage to cytotoxin production. Infect. Immun. 61**:**1799-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Van Dongen, S. 2000. Graph clustering by flow simulation. Ph.D. thesis. University of Utrecht, Utrecht, Netherlands.
- 85.Van Doorn, L. J., C. Figueiredo, F. Megraud, S. Pena, P. Midolo, D. M. Queiroz, F. Carneiro, B. Vanderborght, M. D. Pegado, R. Sanna, W. De Boer, P. M. Schneeberger, P. Correa, E. K. Ng, J. Atherton, M. J. Blaser, and W. G. Quint. 1999. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology 116**:**823-830. [DOI] [PubMed] [Google Scholar]
- 86.Varon, C., A. Duriez, P. Lehours, A. Menard, S. Laye, F. Zerbib, F. Megraud, and D. Laharie. 2009. Study of Helicobacter pullorum proinflammatory properties on human epithelial cells in vitro. Gut 58**:**629-635. [DOI] [PubMed] [Google Scholar]
- 87.Varon, C., F. Tatin, V. Moreau, E. Van Obberghen-Schilling, S. Fernandez-Sauze, E. Reuzeau, I. Kramer, and E. Genot. 2006. Transforming growth factor beta induces rosettes of podosomes in primary aortic endothelial cells. Mol. Cell. Biol. 26**:**3582-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Viala, J., C. Chaput, I. G. Boneca, A. Cardona, S. E. Girardin, A. P. Moran, R. Athman, S. Memet, M. R. Huerre, A. J. Coyle, P. S. DiStefano, P. J. Sansonetti, A. Labigne, J. Bertin, D. J. Philpott, and R. L. Ferrero. 2004. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 5**:**1166-1174. [DOI] [PubMed] [Google Scholar]
- 89.Wattam, A. R., K. P. Williams, E. E. Snyder, N. F. Almeida, Jr., M. Shukla, A. W. Dickerman, O. R. Crasta, R. Kenyon, J. Lu, J. M. Shallom, H. Yoo, T. A. Ficht, R. M. Tsolis, C. Munk, R. Tapia, C. S. Han, J. C. Detter, D. Bruce, T. S. Brettin, B. W. Sobral, S. M. Boyle, and J. C. Setubal. 2009. Analysis of ten Brucella genomes reveals evidence for horizontal gene transfer despite a preferred intracellular lifestyle. J. Bacteriol. 191**:**3569-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Willhite, D. C., T. L. Cover, and S. R. Blanke. 2003. Cellular vacuolation and mitochondrial cytochrome c release are independent outcomes of Helicobacter pylori vacuolating cytotoxin activity that are each dependent on membrane channel formation. J. Biol. Chem. 278**:**48204-48209. [DOI] [PubMed] [Google Scholar]
- 91.Williams, K. P., J. J. Gillespie, B. W. S. Sobral, E. K. Nordberg, E. E. Snyder, J. M. Shallom, and A. W. Dickerman. 2010. Phylogeny of gammaproteobacteria. J. Bacteriol. 192**:**2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wirth, T., X. Wang, B. Linz, R. P. Novick, J. K. Lum, M. Blaser, G. Morelli, D. Falush, and M. Achtman. 2004. Distinguishing human ethnic groups by means of sequences from Helicobacter pylori: lessons from Ladakh. Proc. Natl. Acad. Sci. U. S. A. 101**:**4746-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]