A novel approach to describe a U1 snRNA binding site (original) (raw)
Abstract
RNA duplex formation between U1 snRNA and a splice donor (SD) site can protect pre-mRNA from degradation prior to splicing and initiates formation of the spliceosome. This process was monitored, using sub-genomic HIV-1 expression vectors, by expression analysis of the glycoprotein env, whose formation critically depends on functional SD4. We systematically derived a hydrogen bond model for the complementarity between the free 5′ end of U1 snRNA and 5′ splice sites and numerous mutations following transient transfection of HeLa-T4+ cells with 5′ splice site mutated vectors. The resulting model takes into account number, interdependence and neighborhood relationships of predicted hydrogen bond formation in a region spanning the three most 3′ base pairs of the exon (–3 to –1) and the eight most 5′ base pairs of the intron (+1 to +8). The model is represented by an algorithm classifying U1 snRNA binding sites which can or cannot functionally substitute SD4 with respect to Rev-mediated env expression. In a data set of 5′ splice site mutations of the human ATM gene we found a significant correlation between the algorithmic classification and exon skipping (P = 0.018, χ2-test), showing that the applicability of the proposed model reaches far beyond HIV-1 splicing. However, the algorithmic classification must not be taken as an absolute measure of SD usage as it may be modified by upstream sequence elements. Upstream to SD4 we identified a fragment supporting ASF/SF2 binding. Mutating GAR nucleotide repeats within this site decreased the SD4-dependent Rev-mediated env expression, which could be balanced simply by artificially increasing the complementarity of SD4.
INTRODUCTION
Within eukaryotic cells, the formation of an RNA duplex between the free 5′ end of U1 snRNA and its target sequence in pre-mRNA stimulates the assembly of the spliceosome that catalyzes splicing. Prior to intron removal, binding of U1 snRNP, at least in some instances, protects pre-mRNA against nuclear degradation (1–3). Recently, it has been shown that functional 5′ splice sites from randomized intron sequences can be selected through repetitive rounds of in vitro splicing in HeLa cell nuclear extracts containing U1 snRNA with a deleted 5′ end (4). This finding does not controvert the original observation that base pairing between U1 and the 5′ splice site is necessary for the splicing of mRNA precursors in vivo (5). Rather, protein interactions between U1 snRNP and the 5′ splice site may additionally contribute to splice donor (SD) recognition (6–8). Consistent with this concept is the observation upon compiling sequences of exon–intron borders of mammalian 5′ splice sites (9–12) that metazoan splice sites are only poorly conserved. Nevertheless, a 5′ splice site consensus sequence can be derived from the highest frequency with which distinct nucleotides occur in a given position in such data set, viz. AG/GURAGU (where R = purine, and / indicates the exon–intron border) (13). This sequence is complementary to the nucleotides from positions 3 to 10 of the 5′ end of U1 snRNA. However, the significance of such a consensus sequence remains arguable. Numerous approaches that have been suggested for the prediction of 5′ splice sites and that are exclusively represented by nucleotide frequency-based consensus matrices (11,14–16), fail to be highly reliable. An improvement for prediction of 5′ splice sites has been achieved by considering an influence of neighboring bases (17–21) and the consideration that nucleotides of different site positions are mutually dependent (22).
Beyond the nucleotide sequence of the target, binding of U1 snRNP is facilitated by protein components. These include the U1-specific proteins, U1 C (23, 24) and U1 70K (24,25). The latter is bound to loop I of U1 snRNA (26) by its arginine/serine-like domain (27). Other proteins of the SR protein family (serine/arginine-rich proteins) are also involved in 5′ splice site selection, and in the commitment of different pre-mRNAs to the splicing pathway (28). Characteristic for the SR protein family are their RNA binding domain(s) in the N-terminus conferring substrate specificity, and their arginine/serine-rich (RS) domain at the C-terminus, which has been shown to be interchangeable for some SR proteins without affecting substrate specificity (29).
SR protein-dependent splicing is determined by binding of SR proteins to distinct sequence elements, known as exonic or intronic splicing enhancers (ESEs or ISEs, respectively) and exonic or intronic splicing silencers (ESSs or ISSs, respectively), within the upstream (30) or downstream exon (29) or within the intron (31). Moreover, depending on their position on the pre-mRNA, SR proteins can function as activators or repressors of splicing (32). Based on SELEX protocols, short consensus binding sites of 6–10 nt in length were obtained for more than half of the known human SR proteins (33). However, similarity of any sequence to such a consensus motif does not necessarily imply its functional significance (34). Our currently insufficient understanding of the context dependency of ESEs’ and ESSs’ activities (29,35), in particular in relation to the strength of the neighboring splice site (36), complicates any reliable description of and prediction for cognate splice sites and for the binding of U1 snRNP and U2AF to the 5′ and 3′ splice sites, respectively.
In a recent mutational analysis of the HIV-1 5′ splice site SD4 we have shown that a continuous stretch of at least 14 hydrogen bonds mediated envelope glycoprotein (env) mRNA stabilization resulting in unimpaired env expression (3). Mismatches to the U1 snRNA sequence did not decrease env expression, if flanked by a sufficient number of hydrogen bonds.
As an extension of this previous work, we here propose a hydrogen bond model describing the interaction of U1 snRNA and its binding sites. We suggest hydrogen bonding patterns to describe U1 snRNA binding more accurately than the mere similarity to the consensus sequence. In particular, we take into account that for a G:U base pair different hydrogen bonding patterns are possible (37). We limited our study to GT containing U1 snRNA binding sites, since it has been estimated that only ∼3.7% of annotated splice sites do not contain the canonical dinucleotides GT and AG for donor and acceptor sites (38), respectively. We examined our hydrogen bond model of splice site description with 5′ splice site mutations of the human ATM gene containing 66 exons, and we found its applicability confirmed with respect to exon skipping, beyond its authentic context of HIV-1.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides were synthesized and purified as previously described (39). The oligonucleotides are as follows: 1020 (no. 15): 5′-CATCCTTAAGCTTCTCTATCAAAGGGGTGGAAATTACATGTAATGCA. For the following primers, which are identical to 1020 except for the underlined sequence, only these nucleotides are shown: 1226 (no. 16), ATTGTGGGTCA; 1227 (no. 17), TTGGTAAATGT; 1228 (no. 18), TTGGTAGGTTT; 1264 (4G), GCAGTAGGTAG; 1329 (cs-314), CAGGTGTCATG; 1304 (3G7C), GCAGTGAGTCG; 1305 (4G7C), GCAGTAGGTCG; 1332 (-1G3U 7G8U), GCGGTTAGTGT; 1348 (SD2), AAGGTGAAGGG; 884 (-1G3U7U), 5′-CATCAAGCTTCTCTATCAAAGCGGTTAGTTGTACATGTAATGCA. For the following primers, which are identical to 884 except for the underlined sequence, only these nucleotides are shown: 544 (4A), GCAGTAAGTAG; 900 (4U), GCAGTATGTAG; 1184 (U1 5G), 5′-CGAAGATCTCATACGTACCTGGCAGGGGAGAT. For the following primers, which are identical to 1184 except for the underlined sequence, only these nucleotides are shown: 1185 (U1 5A), ATACATACCTG; 1204 (U1 6C), ATACTCACCTG; 1207 (U1 9T10G11C), ATACTTACTGC; 1222 (U1 3T4T10C11C), ATTTTTACCCC; 1230 (U1 5C10C11C), ATACCTACCCC; 1095, 5′-GGGCCTAGGAATTCTCTCTTGAGCTCGCAGTAAGTAGCTTAAGCTCTCCGAAGACAGTGGCAATGAGAGT; 1098, 5′-AATTCAGGAAGAAG CGGAGACAGCGACGAAGAGCT; 1099, 5′-GTCCTTCTTCGCCTCTGTCGCTGCTTC. For detection of double-spliced and skipped RNA the primers 1544 (sense), 5′-CTTGAAAGCGAAAGTAAAGC, and 1542 (antisense), 5′-CACCTTCTTCTTCTATTCCTT, were used. For the detection of the human growth hormone (hGH) mRNA primers 1225 (sense), 5′-CAACAGAAATCCAACCTAGAGCTGCT, and 1224 (antisense), 5′-TCTTCCAGCCTCCCATCAGCGTTTGG, were used.
Recombinant plasmids
The SV40 early env expression vector SV E/X tat– rev– carrying the EcoRI–XhoI fragment of pNLA1 (40) and the 5′ splice site mutations were constructed as previously described (3). SVcrev was constructed by recloning the EcoRI–XhoI fragment from pUHcrev (41) into pSVT7.
For the construction of plasmid pUCBΔU1 the BglII–PstI fragment (nucleotides 440–726) of the parent plasmid pUCBU1 (kindly provided by A. Weiner) was substituted for a linker (1132/1133) containing a unique BglII, PstI and XhoI site. For the mutations, the BglII–XhoI fragment (nucleotides 440–464) of pUCBΔU1 was substituted for a PCR-amplified fragment with a 5′ PCR primer carrying the BglII site and the desired mutation, e.g. 1204 (U1 6C), and the 3′ PCR primer 1131.
Plasmids pBSU1 and pBSU2b used for U1 snRNA and U2 snRNA detection contain U1 snRNA or U2 snRNA sequences in the antisense orientation. Plasmid pDS-H6F1-SF2ΔRS was used for purification of recombinant SF2 protein with a deletion of the RS domain.
pSV-1-env SD2, pSV-1-env –1G3U, pSV-1-env 4G7C and pSV-1-env 4C8U were constructed by substituting the 5′ splice site no. 4 containing SacI–NheI fragment of pSV-1-env with the respective SacI–NheI fragments from the SV E/X tat– rev– constructs including the respective splice site mutation (3).
All PCR-amplified sequences were confirmed by DNA sequencing. The sequences of all constructs are available on request.
Cell culture and transfection
Under our experimental conditions we could not observe any influence of the co-expression of the Env-receptor on the levels of Env protein expression. In order to be able to microscopically observe the formation of syncytia in the course of the transfection experiment, we transfected (3) HeLa-T4+ cells (42) with FuGENE™ 6 (Roche Molecular Biochemicals), rather than using HeLa cells. The transfection efficiency was biochemically monitored by cotransfection of pGL3-control (Promega). The medium was changed after 24 h, and cells were harvested and prepared 48 h after transfection. For RT–PCR cells were transfected with 1 µg of the respective pSV-1-env plasmid and 1 µg of pXGH5 (43). Transfection was performed by using 4 µl of FuGENE™ 6 and total RNA was isolated after 30 h.
Western blot analysis
Cells were scraped from six-well plates into the medium, sedimented at 12 000 g for 14 s, washed twice in PBS and suspended in 200 µl of SDS–polyacrylamide gel electrophoresis sample buffer (44). An aliquot of the PBS washing step was analyzed for luciferase activity (Luciferase Assay System; Promega). Protein concentrations were determined by the Bradford Protein Assay (Bio-Rad). All samples were adjusted to equal levels of luciferase activity and similar protein concentrations by adding extracts of mock-transfected cells. Samples were subjected to electrophoresis on SDS–7% polyacrylamide gel and transferred to a PVDF membrane (Immobilon™ P, pore size 0.45 µm; Millipore) by electroblotting with 70 V in transfer buffer (200 mM glycine, 25 mM Tris, 20% methanol) for 1 h. Blots were blocked in PBS with 10% bovine serum albumin (BSA), 10% Tween®-20, for 20 h. Protein detection was performed in PBS, 1% BSA, 1% Tween®-20, with a monoclonal mouse-anti-gp120 antibody (87-133/026, 1:5000, kindly provided by Dade Behring) for 1 h, washed four times, incubated with a sheep-anti-mouse antibody conjugated with horse-radish peroxidase (NA 931, Amersham, 1:1000), washed four times, rinsed with water and visualized by a chemiluminescence detection system (ECL™-system and ECL™ hyperfilm, Amersham; Super Signal® ultra, Pierce).
Pull-down assay of U1 and U2 snRNP
Biotinylated in vitro transcripts of AflIII-linearized SV E/X tat– rev– plasmids carrying mutations of the tat/rev 5′ splice site were prepared using T7-MEGAshortscript™ (Ambion), Biotin-16-uridin-5′-triphosphat (Roche Molecular Biochemicals) and diluted [32P]UTP as tracer for concentration determination. Complex formation was performed in a 200 µl reaction volume containing 70 µl of HeLa nuclear extract (Cell Culture Center, Belgium), 30 µl buffer D (20 mM HEPES–KOH pH 7.9, 100 mM KCl, 20% glycerol, 0.2 mM EDTA, 0.5 mM DTT), 20 µl of 10 mM ATP, 20 µl of 50 mM creatine phosphate, 20 µl of 25 mM MgCl2 and 1 µg of biotinylated transcripts by incubation at 30°C for 20 min. The Dynabeads® M-280 Streptavidin (Dynal) were preblocked in NET2 buffer (50 mM Tris–HCl, 0.05% Nonidet P-40, 0.5 mM DTT) containing 0.5 mg/ml tRNA (Escherichia coli MRE 600; Roche Molecular Biochemicals) and 0.2 mg/ml BSA, incubated with the reaction volume at 4°C for 1 h, washed four times in 1 ml of washing buffer (0.05% Nonidet P-40, 500 mM KCl, 20 mM HEPES–KOH pH 7.6, 1 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA) over a total of 2 h. The complexed RNAs were eluted by incubating the beads in 300 µl of denaturation buffer (20mM Tris–HCl pH 7.5, 4 M urea, 0.5% SDS, 10 mM EDTA, 0.3 M NaCl) at 37°C for 5 min. After phenol–chloroform extraction and ethanol precipitation, samples were resuspended in formamide loading buffer and incubated at 95°C for 30 s, followed by fractionation on an 8% denaturing polyacrylamide gel. The RNA was blotted onto a positively charged nylon membrane (Roche Molecular Biochemicals) as previously described (3). The complexed snRNAs were identified by probing the membrane with [32P]UTP-labeled antisense transcripts of U1 snRNA and U2 snRNA. Detection of U2 snRNA served as a control for the pull-down assay.
Isolation of RNA
Isolation of total RNA was performed using a modified guanidinium isothiocyanate protocol (45). Cells were washed twice with 2 ml of PBS and cell lysis was performed with 500 µl of buffer D [7.6 µl of 2-mercaptoethanol, 50 µl of 3 M sodium acetate (pH 4)]; 500 µl of phenol and 100 µl of a chloroform–isoamyl alcohol mixture (24:1) were added and mixed for 15 s. After incubation on ice for 15 min, phases were separated by centrifugation (10 600 g, 4°C, 20 min, Eppendorf centrifuge). RNA was precipitated in 1 vol of isopropanol overnight. After centrifugation (10 600 g, 4°C, 20 min, Eppendorf centrifuge) RNA was washed twice with 70% ethanol and dissolved in 10 µl of DMDC-ddH2O.
RT–PCR assay
Prior to RT, 4 µl of RNA samples were subjected to DNAse I digestion using 10 U DNAse I (Roche Molecular Biochemicals) with 50 mM Tris pH 7.5 and 10 mM MgCl2 in a total volume of 10 µl at room temperature for 1 h 45 min. After DNAse I inactivation at 95°C for 10 min, 4.5 µl of the DNAse-digested RNA samples were reverse transcribed with 200 U SuperScript III RNAse H– Reverse Transcriptase (Invitrogen) according to the manufacturer’s protocol using 0.375 mM oligo(dT)15 (Roche Molecular Biochemicals) as primer. As a negative control for the remaining plasmid DNA contamination of each sample, a second assay was performed as described above but replacing reverse transcriptase with ddH2O. PCR was carried out with 1.25 U AmpliTaq (Applied Biosystems) in a total volume of 50 µl according to the manufacturer’s protocol in a Robocycler Gradient 96 Temperature Cycler (Stratagene). All primers were used at a final concentration of 0.2 µM. Double-spliced and skipped RNA was detected with the primer pair 1544/1542 and hGH mRNA with primer pair 1225/1224. Prior to PCR the cDNA reaction mixture was denatured at 94°C for 3 min. To determine the linear PCR-amplification range allowing a semi-quantitative estimation of the relative abundance of pSV-1-env and hGH mRNA, a preliminary PCR test series was carried out using the same cDNA sample but varying the PCR cycle numbers between 15 and 30 [94°C, 0.5 min; 52°C (pSV-1-env) and 56°C (hGH), respectively, 1 min; 72°C, 1 min]. The reactions were completed with a final elongation step of premature amplified products at 72°C for 10 min. Accordingly to the obtained results, PCR analysis was performed with 26 cycles for pSV-1-env as well as hGH PCR amplification.
PCR products were separated on 6% non-denaturating polyacrylamide gels, stained with ethidium bromide (10 min) and visualized with the Lumi-Imager F1 (Roche Molecular Biochemicals).
Gel mobility shift assay
[32P]UTP-labeled in vitro transcripts (30 000 c.p.m) made from AflII-linearized plasmids with (SV GAR Δvpu env) and without (SV ΔGAR Δvpu env) the GAR motifs (trinucleotide sequence with R = A or G) were incubated in a 10 µl reaction volume containing 5 µl of 2× band-shift buffer (20 mM HEPES–KOH pH 7.6, 200 mM KCl, 4 mM MgCl2, 1 mM EDTA, 2 mM DTT, 20% glycerol, 100 ng/µl tRNA), 0.1 µl of RNasin (Promega) and 1 µl of SF2ΔRS (0–1000 ng) diluted in SF2 buffer (0.5 M GuHCl, 20 mM HEPES–KOH pH > 7.9, 42 mM (NH4)2SO4, 0.5 mM DTT, 0.2 mM EDTA, 15% glycerol, 1 mM PMSF) were incubated at room temperature for 15 min and analyzed on a native 5% polyacrylamide gel. Gels were subjected to autoradiography.
RESULTS
A single mismatch within SD4 abolishes U1 snRNP binding in HeLa nuclear extracts
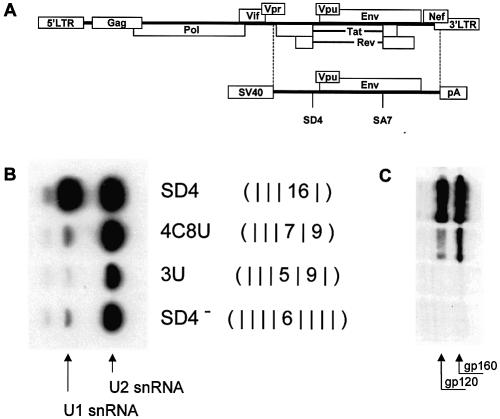
Recently, we have shown that inherently unstable HIV-1 env pre-mRNA can be protected from degradation by RNA duplex formation between the HIV-1 5′ splice site SD4 and U1 snRNA (Fig. 1A and C) (3). To determine what degree of sequence complementarity is necessary for the 5′ splice site to bind U1 snRNP in vitro, we compared U1 snRNP binding to SD4 (GCAGTAAGTAG) and three mutant 5′ splice sites, 4C8U, 3U and SD4–. We used a pull-down assay with biotinylated in vitro transcripts comprising upstream sequences of SD4, the different splice-site mutations and a short portion of the intron sequence. Following incubation of these transcripts with HeLa nuclear extracts, total RNA was extracted from the formed complexes and analyzed by northern blotting for the presence of U1 snRNA (Fig. 1B). Only with the authentic 5′ splice site, SD4 complexes were formed containing significant amounts of U1 snRNA, whereas the mutant SD4 transcripts—even if they carried just one single mismatch—failed to bind appreciable amounts of U1 snRNP (Fig. 1B). This result confirmed our previous observation that, in the case of a discontinuous stretch of hybridization potential, less than 15 hydrogen bonds were insufficient for RNA duplex formation between the HIV-1 5′ splice site SD4 and U1 snRNA. Sensitivity differences between the in vitro and in vivo assays (Fig. 1B and C) could have caused the slightly different result for mutation 4C8U with its intermediate glycoprotein expression. As an additional interesting result, this experiment showed that U2 snRNP binding occurred even in the absence of U1 snRNP binding.
Figure 1.
Mutated SD4 transcripts essentially fail to bind U1 snRNA. (A) Schematic diagram of the HIV-1NL genome. Depicted below is the env expression vector SV E/X tat– rev–. The subgenomic region delimited by the dashed lines was cloned into the SV40 early expression vector pSVT7 (see Materials and Methods). The open-reading frames for tat and rev are translationally silent due to mutations of their translational start codons. (B) Pull-down assay of U1 and U2 snRNP with HIV-1 env transcripts containing different mutant SD4 RNAs. Biotinylated in vitro transcripts carrying mutations of the tat/rev 5′ splice site were incubated with HeLa nuclear extracts. RNA was extracted from the formed complexes with streptavidine beads. Bound U1- and U2-snRNAs were detected by [32P]UTP-labeled antisense transcripts. U2 snRNA served as a loading control for the assay. (C) Immunoblot analysis of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells transfected with SVcrev, pGL3-control and env expression vectors carrying mutations of the tat/rev 5′ splice site SD4. For each plasmid, transfection using FuGENE™ 6 was done with 1 µg of the env expression vector, 1 µg of SVcrev and 1 µg of pGL3-control. The hydrogen bonding patterns (middle) are given as sums of hydrogen bonds within each continuous stretch of complementary nucleotides according to the algorithmic calculation. A mismatch is indicated by a vertical line.
A G:U base pair contributes less to the U1 snRNA-dependent env expression than an A:U base pair
In order to derive a hydrogen bonding model of general significance from the study of interactions between SDs and U1 snRNA, we strived to study alternatives to SD4.
Based on our preliminary hydrogen bonding model derived by Kammler et al. (3), we looked for potential, yet unidentified, U1 snRNA binding sites within the HIV-1 env transcript that would eventually function to stabilize the pre-mRNA. To this end, we scrutinized the env gene between the translational start codon and the splice acceptor SA7 to predict binding sites. For all 11 nt long sequences containing a GT dinucleotide, we counted the number of hydrogen bonds (_N_H) in a continuous or discontinuous stretch of neighboring base pairs. An A:U base pair counted for two and a G:C base pair for three hydrogen bonds. From our previous western blot analyses we classified hydrogen bonding patterns as ‘high complementarity’ (HC) if they ensured env expression and ‘low complementarity’ (LC) otherwise. A hydrogen bonding pattern qualified as HC, if _N_H ≥ 14 for a continuous stretch and _N_H ≥ 15 for a discontinuous stretch, with the additional constraint that for a discontinuous stretch 5′ to the mismatch _N_H ≥ 6 and 3′ to the mismatch _N_H ≥ 5. Beyond the Watson–Crick A:U base pairs analyzed in our previous study, we assumed G:U wobbles to form two hydrogen bonds. Tentatively applying the so defined algorithm, we found four potential U1 snRNA binding sites (designated nos 15–18) within this part of the HIV-1NL genome which so far had not been identified as functional 5′ splice sites. Of note, all four potential U1 snRNA binding sites contained at least one G:U base pair. To examine whether these putative U1 snRNA binding sites enabled 5′ splice site-dependent Rev-mediated env expression, we substituted SD4 with each of them in our env expression vector. Unexpectedly, only one, no. 18, mediated normal env expression; the other three (nos 15–17) showed very little, if any, env expression (Fig. 2).
Figure 2.
Contributions of G:U base pairs to U1 snRNA binding. Immunoblot analysis (left) of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells transfected with 1 µg of env expression vector, 1 µg of SVcrev and 1 µg of pGL3-control. The different env expression vectors carried predicted U1 snRNA binding sites in the position of the tat/rev 5′ splice site SD4. A cellular protein that cross-reacts with the anti-gp120 mAb is marked by an asterisk. Sequences of the predicted U1 snRNA binding sites and their hydrogen bonding patterns are depicted on the right. The three columns indicate U1 snRNA binding site hydrogen bonding patterns on the assumption that different numbers of hydrogen bonds are possible for a G-U base pair (underlined within the sequence): two [G:U (2)], one [G:U (1)] or no [G:U(0)] hydrogen bond(s) are scored per pair. A mismatch is indicated by a vertical line.
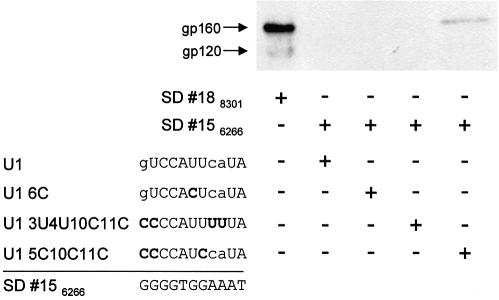
To examine whether env expression mediated by the predicted binding sites 15–17 could be restored by increasing their complementarity to U1 snRNA, we repeated the transfection experiment with the env expression vector carrying site 15 and expression vectors for wild-type U1 snRNA and three different site-mutated U1 snRNAs, normally acting as suppressors. In this case, they served as probes that might increase complementarity to the predicted binding site 15. As already seen in Figure 2, the putative U1 snRNA binding site 18 supported env expression with wild-type U1 snRNA, whereas site 15 did not (Fig. 3). Likewise, site 15 did not support detectable env expression following cotransfection with the mutated U1 snRNAs 6C or 3U4U10C11C, whereas with the site-mutated U1 snRNA 5C10C11C some env expression was detected (Fig. 3). The assumption of a G:U wobble always contributing two hydrogen bonds to the duplex formation could not adequately describe these experimental results.
Figure 3.
Predicted U1 binding site 18 supports env expression with wild-type U1 snRNA, whereas 15 does so only with site-mutated U1 snRNA 5C10C11C. Immunoblot analysis (top) of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells transfected with 1 µg of SVcrev, 1 µg of pGL3-control and 1 µg of env expression vector carrying the predicted U1 snRNA binding site 15 or 18 in the position of the tat/rev 5′ splice site SD4. An expression plasmid coding for wild-type U1 snRNA (U1) or one of three site-mutated U1 snRNAs was cotransfected. Nucleotides of the U1 snRNAs that hypothetically might base pair with the corresponding nucleotide of the predicted binding site SD 156266, i.e. A-U, G-C and G-U, are given in upper case letters, mismatches in lower case letters (left, designations and sequences). Mutated nucleotides of the U1 snRNAs are in bold letters. + and – represent the transfection scheme (right).
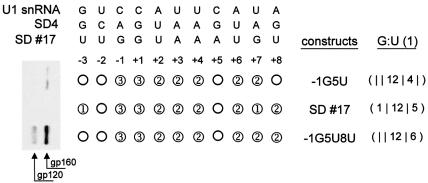
This consideration led us to explore other possibilities. In the duplex r(GGCGUGCC)2, the G:U wobble forms only one hydrogen bond instead of the expected two for the canonical wobble pair (37). Assuming this one-hydrogen-bond model for G:U base pairing in our context, only sites 17 and 18 would have been expected to successfully substitute for SD4 (Fig. 2, column ‘G:U (1)’). However, predicted U1 binding site 17 had been found not to support env expression (Fig. 2), and was thus studied further. In addition to site 17, two site-directed mutants, –1G5U and –1G5U8U, were generated and tested. –1G5U and –1G5U8U contained a continuous stretch of 12 hydrogen bonds identical to site 17 in positions –1 to +5, but had different base pairs in positions +7 and +8. Even if site 17 could form only one hydrogen bond at the G:U wobble in position +7, this assumption would result in a higher total number of hydrogen bonds than site –1G5U. env expression, however, was extremely low upon transfection of HeLa-T4+ cells with the expression vector carrying site 17 (Fig. 4). In particular, it was lower than with –1G5U. From these results, we cautiously concluded that a G:U base pair in position +7, flanked on either side by no more than two hydrogen bonds, did barely contribute to the stability of the RNA duplex. Similar results were obtained analyzing mutants with a G:U base pair in position –2 (data not shown), at the other pre-terminal position of RNA duplex formation.
Figure 4.
A G:U base pair in position +7 does not contribute to U1 snRNA binding without sufficient flanking base pairs. Immunoblot analysis (left) of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells transfected with 1 µg of SVcrev, 1 µg of pGL3-control and 1 µg of of env expression vector carrying alternatives of the tat/rev 5′ splice site SD4, i.e. the predicted U1 binding site 17 (top) or its site-mutated variants, –1G5U or –1G5U8U (right). In the scheme (center), hydrogen bonds that are predicted to form with U1 snRNA are shown as circles (1, one hydrogen bond; 2, two hydrogen bonds; 3, three hydrogen bonds; mismatch, empty circle) assuming one hydrogen bond for G:U wobbles. The right column [G:U (1)] shows the number of hydrogen bonds on either side of the unpaired nucleotide in position +5. A mismatch is indicated by a vertical line.
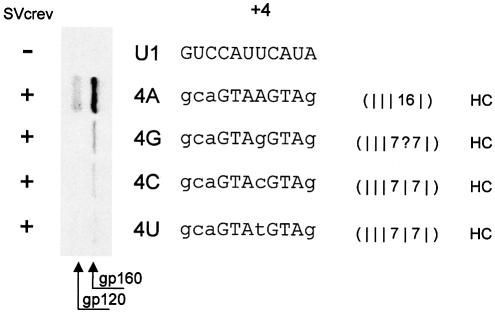
Next, we looked at the behavior of internal G:U wobbles as regards the applicability of the one-hydrogen-bond hypothesis. We examined the contribution of a G:U wobble in position +4 of SD4 to the 5′ splice site-dependent Rev-mediated env expression. SD4 has an A in this position pairing with the U of U1 snRNA. By mutating this position we were able to analyze the effect of a G:U wobble that was flanked by seven hydrogen bonds on either side on RNA duplex formation (Fig. 5). Any hydrogen bond formation within this mutated position +4 would have converted the discontinuous stretch into a continuous one with _N_H > 14 hydrogen bonds. Substitution of the A in +4 with any of the three other possible nucleotides, however, did not lead to a comparable env expression. Therefore, we concluded that a single G:U wobble in +4 does not contribute to RNA duplex formation even if it is flanked by seven Watson–Crick hydrogen bonds on both sides.
Figure 5.
A G:U wobble, like any mismatch, in the center of the complementary RNA sequences of the 5′ splice site SD4 and U1 snRNA abolishes duplex formation. Immunoblot analysis (left) of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells that were transfected with 1 µg of SVcrev, 1 µg of pGL3-control and 0.2 or 1 µg, respectively, of the env expression vector carrying the tat/rev 5′ splice site SD4 (4A) or its counterpart with any possible mutation site-directed to position +4. Within the mutated SD4 sequence of 4G, 4C and 4U (center), wobbles and mismatches to U1 (top) are shown in lower case. env was only expressed with SD4, very little with 4G. Of note, in the transfection experiment 4A only one fifth of the reference construct SV E/X tat– rev– (4A) was used as compared with the experiments 4G, 4C and 4U.
We would not have expected these results on thermodynamic grounds employing two algorithms, Dynalign for finding secondary structures common to two RNA sequences (46) and HyTher, a nearest-neighbor model that predicts hybridization thermodynamics of a given duplex (47). Both algorithms predicted only little difference for the duplex free energies with an A:U base pair or a G:U wobble [Δ_G_° 0.3 and 0.35 kcal/mol (Dynalign and HyTher), respectively] in contrast to a greater difference for the C mismatch [Δ_G_° 4.1 and 4.05 kcal/mol (Dynalign and HyTher), respectively].
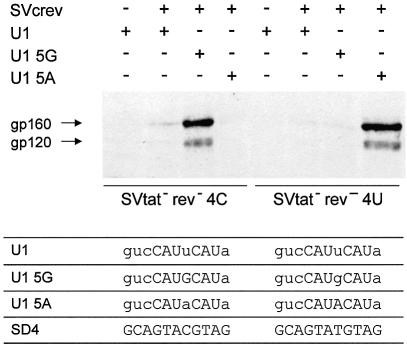
To confirm that in this sequence the G:U wobble at +4 does not contribute to RNA duplex formation, whereas base substitutions that pair by hydrogen bonds do so, cotransfections with site-mutated U1 snRNAs were performed. As shown in Figure 6, env expression of the SD4 mutant 4C was restored only in the presence of the matching site-mutated U1 snRNA, pUCBU1 5G, while the same U1 snRNA failed to restore env expression of the SD4 mutant 4U. env expression of SD4 mutant 4U, on the other hand, was specifically restored in the presence of the matching site-mutated U1 snRNA, pUCBU1 5A, not by pUCBU1 5G.
Figure 6.
Base substitutions in U1 snRNA that reinstate hydrogen bonds to the SD4 mutations 4C and 4U (Fig. 5) restore efficient env expression. Immunoblot analysis (center) of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells that were transfected with 1 µg of SVcrev, 1 µg of pGL3-control, 1 µg of env expression vectors SV E/X tat– rev– 4C or SV E/X tat– rev– 4U and an expression plasmid coding for wild-type U1 snRNA (U1) or site-mutated U1 snRNA (U1 5G and U1 5A) as indicated (+/–) by the scheme (top). The sequences of the free 5′ end of the U1 snRNAs and the mutant splice sites are given at the bottom of the figure. Mismatches within the RNA duplexes are given in lower case letters of the respective U1 snRNA sequence.
This experiment convincingly showed that in this sequence a G:U wobble in +4 does not contribute to RNA duplex formation supporting 5′ splice site-dependent Rev-mediated env expression. However, it did not rule out the possibility that a G:U base pair in position +4 could contribute to the RNA duplex stability if it was flanked by a G:U base pair in position +3. Consequently, the algorithm did not count any hydrogen bonds for a single G:U base pair in position +4.
Different hydrogen bonding patterns are possible for terminal base pairs
Next we set out to study the contribution of a G:U wobble at position +3 to env expression and its dependency on the presence of flanking hydrogen bonds. We used two additional SD4 mutants containing a G:U wobble at position +3. In a continuous stretch of six complementary nucleotides, from –3 to +3, a G:U wobble at +3, terminal to the RNA duplex, did not enable env expression (Fig. 7, see cs –314 versus cs –315) and behaved like cs –313 that has a mismatch t at +3. However, if this G in +3 was flanked on both sides by complementary Watson–Crick base pairs, i.e. was no longer terminal, then the G:U wobble contributed to the env expression, albeit to a lesser extent than an A:U base pair (Fig. 7, see 3G7C and cs –315).
Figure 7.
A terminal G:U wobble does not contribute to RNA duplex formation and env expression. Immunoblot analysis (left) of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells transfected with 1 µg of SVcrev, 1 µg of pGL3-control and 1 µg of env expression vectors carrying mutations in the tat/rev 5′ splice site SD4. The sequences of the splice sites are depicted right of the plasmid designations. The nomenclature refers to the contiguous stretch (cs) of hydrogen bonds starting at the indicated position of the cs within the 5′ splice site and to the number of hydrogen bonds given by the superscript (e.g. cs –314), or to mutated nucleotides of SD4 (e.g. 3U indicates that position +3 of SD4 is mutated from A to T). The hydrogen bonding patterns are given on the right of the figure for different possible weights attributed to single hydrogen bonds. First column, every G:U base pair contributes one hydrogen bond; second column, same as the first column, but terminal G:U base pairs contribute no hydrogen bonds; third column, same as the second column, but a G:C base pair in position –3 contributes two hydrogen bonds. Note that detectability of env expression is consistent with the algorithm classifying the predicted hydrogen bonding pattern as HC or LC with the given threshold values. Calculated free energy of the RNA duplexes using (1)Dynalign (http://rna.chem.rochester.edu/dynalign.html) (46) and (2)HyTher (http://ozone2.chem.wayne.edu/Hyther/hytherm1main.html) (47) is shown utmost right.
To account for these experimental results, we assumed for our algorithm that any G:U wobble terminal to an RNA duplex does not contribute hydrogen bonds to the RNA duplex, in general accordance with the thermodynamic evidence that lack of stacking interactions at helix termini reduces the energy contribution of terminal base pairs. As a quantitative approach to this problem, we calculated the free energies of these mutant duplexes by Dynalign and HyTher. Unfortunately, both programs failed to correlate the free energy of these RNA duplexes with the efficiency of the 5′ splice site-dependent Rev-mediated env expression (Fig. 7, columns ΔG°).
A remaining inconsistency in describing our experimental results by the suggested hydrogen bond model becomes apparent in the mutant sites cs –313, cs –314 and 3G7C. All three are predicted to form a continuous stretch of 13 hydrogen bonds. Assuming a terminal base pair—as here a G:C at position –3—to contribute one hydrogen bond less, assigns 12 hydrogen bonds for cs –313 and cs –314, but leaves 3G7C unchanged with 13 bonds. Calculated this way, a threshold of _N_H ≥ 13 for a continuous stretch of hydrogen bonds was found to correctly describe env expression in all our experiments.
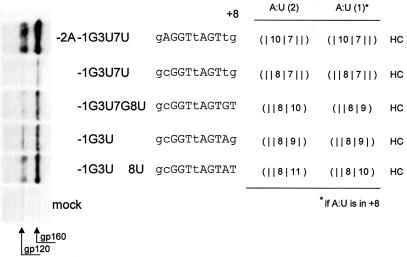
We also examined whether the modification proposed for the calculation of Watson–Crick hydrogen bonds at the 5′ terminus of the 11 nt long sequence would also hold for its 3′ end, i.e. position +8. To examine this question, we compared a further set of SD4 splice sites mutated at the 3′ end (Fig. 8). We purposely chose mutants with suboptimal env expression to avoid saturation effects. Without exception, the classification as HC for the hydrogen bond patterns of these mutants was consistent with env expression.
Figure 8.
A terminal A:U at position +8 contributes only one hydrogen bond. Immunoblot analysis (left) of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells transfected with 1 µg of SVcrev, 1 µg of pGL3-control and 1 µg of env expression vectors carrying mutations in the tat/rev 5′ splice site SD4. The sequences of the splice sites are depicted right of the plasmid designations. Mismatches within the RNA duplexes are given in lower case letters of the respective splice site sequence. The hydrogen bonding patterns are given on the right of the figure for different possible weights attributed to single hydrogen bonds. The scoring rules of Figure 7, columns 1–3, have already been incorporated here. In addition, in the first column every A:U base pair contributes two hydrogen bonds, the second column is the same as the first, but an A:U base pair in position +8 contributes one hydrogen bond. Note that detectability of env expression is consistent with the algorithm classifying the predicted hydrogen bonding pattern as HC.
Exon recognition of HIV-1 exons with SD4 as the downstream 5′ splice site
Classification of hydrogen bonding patterns in terms of HC or LC was derived from experiments with a single intron construct and an _env_-mRNA degradation assay. In order to examine the contribution of U1 snRNA binding to exon recognition and splicing, we supplemented the transcription unit with HIV-1 exon 1 and the 5′ portion of intron 1, thus obtaining a 2-intron HIV-1 sub-genomic env expression construct (as outlined in Fig. 9A).
Figure 9.
5′ Splice sites graded as HC succeed in normal splicing, whereas those classified as LC show impaired exon recognition. (A) Schematic diagram of the pSV-1-env construct. pSV-1-env contains the HIV-1 genome of pNL4-3 but lacks the U3 region of the 5′ LTR, the non-coding region of the 3′ LTR and the region downstream of the gag start codon nearly onto the first coding exon of Rev. The start codons for expression of gag and rev are mutated. The positions of the primers used for RT–PCR are indicated by arrows. GAR, GAR repeats; SV40, SV40 early promotor; p(A), SV40 polyadenylation signal. (B) RT–PCR assay. HeLa-T4+ cells were transfected with 1 µg of pSV-1-env carrying substitutions of SD4 and cotransfected with 1 µg of pXGH5 expressing the hGH mRNA as an internal control. Size marker (M), pSV-1-env SD2 (lane 1), pSV-1-env –1G3U (lane 2), pSV-1-env 4G7C (lane 3) or pSV-1-env 4C8U (lane 4). (C) Determination of the linear amplification range for pSV-1-env and hGH RT–PCR assays. For pSV-1-env RT–PCR, relative amounts over number of cycles for the strongest (1–5 and 4–7) and of the shortest PCR product (1–7) were assessed. (D) Immunoblot analysis of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells transfected with 1 µg of SVcrev, 1 µg of pGL3-control and 1 µg of env expression vectors carrying substitutions of the tat/rev 5′ splice site SD4. SV E/X tat– rev– SD2 (lane 1), SV E/X tat– rev– –1G3U (lane 2), SV E/X tat– rev– 4G7C (lane 3), SV E/X tat– rev– 4C8U (lane 4).
Moreover, we substituted SD4 with four different 5′ splice sites. Two of them [SD2 (80.08), –1G3U (79.34)] revealed high Shapiro–Senapathy scores (11) of ∼80, and the other two [4G7C (65.66), 4C8U (64.84)] had low scores of ∼65. Each pair contained one site graded as HC (–1G3U, 4C8U) according to our hydrogen bond model. These sites were verified as functional 5′ splice sites by detection of an exclusively double-spliced RNA/cDNA pattern (Fig. 9B). Correspondingly, the western blot confirmed env expression (Fig. 9D). The remaining two 5′ splice sites were both graded as LC, showed a single-spliced RNA/cDNA pattern indicative of exon skipping, and the pertinent immunoblots failed to detect Env glycoprotein (Fig. 9B and D). Notably, in this example the high Shapiro–Senapathy score 5′ splice site SD2 (authentic HIV-1 5′ slice site), graded as LC by our hydrogen bond model, led to exon skipping, whereas the low Shapiro–Senapathy score 5′ splice site 4C8U, graded as HC by our hydrogen bond model, showed no exon skipping (Fig. 9B, see lanes 1 and 4).
In contrast to the single-intron env expression construct, where lack of U1 snRNA binding led to complete RNA degradation, in the two-intron construct the RNA degradation was incomplete, possibly due to additional U1 snRNA binding at SD1. If both 5′ splice sites were used, the amount of RNA exceeded the amount found in the case of exon skipping, indicating a possibly additive effect of two U1 snRNA binding sites protecting RNA against degradation.
Analysis of human splice site mutations
A concern was the widespread applicability of our hydrogen bond model since all experimental work was done on HIV-1 SD recognition. To test whether the classification in terms of HC and LC obtained from our hydrogen bond model could be extended to predict exon skipping occurring in human genetic diseases, we chose a prototype human gene known to harbor a high number of exons and splice mutations. The ATM gene (ataxia telangiectasia mutated, MIM 208900, Louis Bar syndrome) includes 66 exons and more than 300 different mutations are documented (ATM Mutation Database, http://www.benaroyaresearch.org/bri_investigators/atm.htm). From all available, published and unpublished mutations and polymorphisms, we selected those involving the nucleotides –3 to +8 of 5′ splice sites, except for the canonical GT dinucleotide in positions +1 and +2. It is well known that point mutations at the 5′ splice site do not necessarily cause alterations of the transcript. Therefore, we further selected only those changes at positions –3 to –1 and +3 to +8 within ATM SD sites where RNA/cDNA studies had been done and pertinent information was available. Given these limitations, a total of 21 sequence changes within 19 different ATM 5′ splice sites were collected (Table 1). Seventeen among these 21 changes in ATM 5′ splice sites led to exon skipping and thus represented mutations. The hydrogen bond patterns of the mutated SDs consistently revealed a lower complementarity than the corresponding patterns of their physiological counterparts (Table 1). Similarly, the Shapiro–Senapathy 5′ splice site score (11) was found to be decreased in all these mutations compared with that of the corresponding wild-type 5′ splice site. Three of the remaining four sequence changes that did not lead to an alteration of the transcript and thus qualified as polymorphisms, had an identical hydrogen bond pattern and also a comparable Shapiro–Senapathy score as their corresponding wild type. Calculations showed that the correlation between exon skipping and the change in classification from HC to LC was significant even in our relatively small data set of 21 sequence alterations in 5′ splice sites of the human ATM gene (Spearman ρ = 0.54, P = 0.018, χ2-test). Thus, our hydrogen bond model proved to be applicable in the characterization of human SD changes.
Table 1. Synopsis of 5′ splice sites of the human ATM gene and mutations therein.
| Change | Transcript alteration | U1 complementary wild type | H-bond wild type | Threshold wild type | U1 complementary mutant | H-bond mutant | Threshold mutant | S&S wild-type 9mer | S&S mutant 9mer | Reference | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IVS4+6a>g | No change | aAGGTAgtaAa | (| 12 | 2 | ) | LC | aAGGTAgtgAa | (| 12 | 2 | ) | LC | 70.65 | 71.63 | a | |||||||
| IVS7+5g>a | ES(7) | tAGGTAtGTtT | (| 12 | 5 | 1) | HC | tAGGTAtaTtT | (| 12 | 2 | 1) | LC | 84.69 | 71.86 | Unpublishedb | |||||||
| 662G>T | ES(8) | gAGGTAAtcta | (| 14 | ) | HC | gAtGTAAtcta | (| 2 | 9 | ) | LC | 78.25 | 65.47 | a | ||||||||
| IVS9+3a>t | ES(9) | aAGGTAtaaAg | (| 12 | 2 | ) | LC | aAGGTttaaAg | (| 10 | 2 | ) | LC | 70.32 | 61.52 | a | |||||||
| IVS14+3del at | ES(14) | atGGTAtGTta | (| | 10 | 5 | ) | HC | atGGTgttatc | (| | 8 | ) | LC | 80.25 | 60.05 | a | ||||||
| 2250G>A | ES(16) | aAGGTAgGaga | (| 12 | 3 | ) | HC | aAaGTAgGaga | (| 2 | 7 | 3 | ) | LC | 83.70 | 71.31 | 55 | ||||||
| 2376G>A | ES(17) | aAGGTAAGatT | (| 17 | 1) | HC | aAaGTAAGatT | (| 2 | 12 | 1) | LC | 93.79 | 81.40 | a | ||||||||
| IVS21+3a>t | ES(21) and ES(21and22) | atcGTAAGaAa | (| | 12 | 2 | ) | LC | atcGTtAGaAa | (| | 5 | 5 | 2 | ) | LC | 72.13 | 63.34 | 55 | ||||
| IVS21+3ins t | ES(21) and ES(21 and 22) | atcGTAAGaAa | (| | 12 | 2 | ) | LC | atcGTtAagAa | (| | 5 | 2 | 2 | ) | LC | 72.13 | 51.48 | a | ||||
| IVS21+5g>a | ES(21) and ES(21 and 22) | atcGTAAGaAa | (| | 12 | 2 | ) | LC | atcGTAAaaAa | (| | 9 | 2 | ) | LC | 72.13 | 59.30 | Unpublishedb | |||||
| 3284G>C | ES(24) | tAGGTAAtggg | (| 14 | ) | HC | tAcGTAAtggg | (| 2 | 9 | ) | LC | 77.97 | 64.51 | a | ||||||||
| 3576G>A | ES(26) | aAGGTAtaTAT | (| 12 | 5) | HC | aAaGTAtaTAT | (| 2 | 7 | 5) | LC | 75.61 | 63.22 | 55 | ||||||||
| IVS28+5g>t | No change | CAGGTAtGgcT | (14 | 3 | 1) | HC | CAGGTAttgcT | (14 | | 1) | HC | 85.05 | 72.00 | a | |||||||||
| 6095G>A | ES(43) | tAGGTAAaTtg | (| 14 | 2 | ) | HC | tAaGTAAaTtg | (| 2 | 9 | 2 | ) | LC | 82.50 | 70.11 | 55 | ||||||
| 7788G>A | ES(54) | gAGGTAttTgg | (| 12 | 2 | ) | LC | gAaGTAttTgg | (| 2 | 7 | 2 | ) | LC | 72.86 | 60.47 | a | ||||||
| 7926A>C | ES(54 and 55) or ES(55) | gAaGTAtGTtT | (| 2 | 7 | 5 | 1) | LC | gcaGTAtGTtT | (| | 7 | 5 | 1) | LC | 73.52 | 65.11 | a | |||||
| 8150A>G | ES(57) | aAGGTGAGccT | (| 16 | 1) | HC | agGGTGAGccT | (| | 14 | 1) | HC | 91.98 | 83.51 | a | ||||||||
| 8266A>T | No change | aAGGTAAcTAT | (| 14 | 5) | HC | tAGGTAAcTAT | (| 14 | 5) | HC | 85.97 | 82.21 | a | |||||||||
| IVS59+5_IVS59+8del gtga | ES(59) | atGGTGAGTga | (| | 16 | ) | HC | atGGTgAcacc | (| | 8 | 2 | ) | LC | 89.03 | 70.62 | 56 | ||||||
| IVS62+8a>c | No change | aAGGTAAGTga | (| 19 | ) | HC | aAGGTAAGTgc | (| 19 | ) | HC | 99.09 | 99.09 | a | |||||||||
| 8850G>T | ES(63) | gAGGTAAagta | (| 14 | ) | HC | gAtGTAAagta | (| 2 | 9 | ) | LC | 79.39 | 66.62 | 57 |
Interestingly, 14 among the regarded 19 physiological ATM 5′ splice sites were classified as HC at the threshold levels defined above. In contrast, four among the remaining five physiological 5′ splice sites that were classified as LC showed a continuous stretch of 12 hydrogen bonds, which is just below the threshold defined from HIV-1. In these cases, splicing of the human ATM transcript may be mediated by an ESE.
Dependency of U1 snRNA binding to SD4 on GAR nucleotide motifs and complementarity of SD4 to U1 snRNA
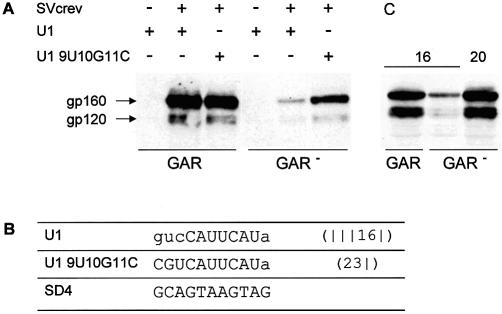
To examine a possible dependence between the presence of ESEs and sequence complementarity of SD4 to U1 snRNA, we analyzed RNA duplex formation of HIV-1 transcripts with and without the GAR motif upstream of SD4, which has been shown to influence U1 snRNA binding (3). As shown here and previously, substituting four single Gs of the GAR motif with Cs (GAR–) led to a reduction of env expression, which was estimated to be 4-fold (Fig. 10A, lanes 2 and 5, and C, lanes 1 and 2). Interestingly, cotransfection of a site-mutated U1 snRNA complementary to nucleotides –3 to +7 of SD4 (U19U10G11C) partially restored env expression in the absence of the upstream GAR repeats (Fig. 10A, lanes 5 and 6). To confirm that an increased number of complementary base pairs could compensate for the lack of the GAR repeats, we analyzed env expression of the construct SV E/X tat– rev– GAR– cs –120 that had two more complementary bases than splice site SD4, but inactivated GAR repeats. As shown in Figure 10C, env expression was fully restored, indicating that increasing the complementarity of the RNA duplex sufficiently facilitated U1 snRNA binding, dispensing with the necessity of GAR support.
Figure 10.
Increased complementarity between SD4 and U1 snRNA compensates for a lack of GAR-mediated enhancing. Immunoblot analysis of Env glycoproteins (gp120 and its precursor gp160) expressed in HeLa-T4+ cells transfected with 1 µg of SVcrev, 1 µg of pGL3-control and 1 µg of env expression vectors SV E/X tat– rev– GAR or SV E/X tat– rev– GAR–, and expression plasmid coding for wild-type U1 snRNA (U1) or a 9U/10G/11C site-mutated U1 snRNA (U1 9U/10G/11C) as indicated (A and C). The numbers above the immunoblot analysis shown in (C) indicate the numbers of hydrogen bonds in the continuous stretch of the 5′ splice sites analyzed (16 for SD4, 20 for mutant cs –220). The sequences of the free 5′ end of the U1 snRNA and the 5′ splice site SD4 are given at the bottom of the figure (B). Mismatches within the RNA duplexes are given in lower case letters of the respective U1 snRNA sequence.
DISCUSSION
Based on our mutational analysis of the HIV-1 5′ splice site SD4, initiated in Kammler et al. (3) and extended in this paper, we described U1 snRNA binding sites with a hydrogen bond model. An algorithm, available at http://www.splicefinder.net/, systematically classified U1 snRNA binding sites as HC or LC according to their capability to functionally replace SD4 in the 5′ splice site-dependent Rev-mediated env expression.
The 5′ splice site consensus sequence only includes nucleotides in positions –2 to +6 (13), taking into account the concept proposed by Shapiro and Senapathy to calculate 8mer 5′ splice site scores (11). However, Hibbert and coworkers (48) and we were able to identify 5′ splice sites, which were functionally dependent on complementary nucleotides in positions +7 or +8 or both (3) (Fig. 4). Moreover, a preference for a purine base at position +7 was also recently found using a strategy to select functional 5′ splice site sequences in vitro from a pool of pre-mRNAs containing randomized 5′ splice sites after three rounds of selection (4). Consistent with these findings, here we demonstrated that in an in vivo RNA splicing assay, complementary nucleotides in positions +7 or +8 contributed to exon recognition. Therefore, we included these two positions into calculations by our algorithm, although statistically the contribution of nucleotides in these positions to splicing is a rare phenomenon, as is apparent from nucleotide frequency distributions.
Recently, it has been shown by NMR spectroscopy that only one hydrogen bond is formed by G:U wobbles, depending on their adjacent base pairs (37). Therefore, we analyzed the contribution of G:U wobbles to the stability of the RNA duplex in all possible positions of the 5′ splice site. In all cases analyzed, a G:U consistently contributed less to the U1 snRNA-dependent Rev-mediated env expression than an A:U base pair in the same position. Since an A:U pair contributed a maximum of two hydrogen bonds, we permitted a maximum of one hydrogen bond for a G:U wobble. However, if a G:U was not stabilized by a sufficient number of adjacent hydrogen bonds, it had to be treated as a mismatch (Fig. 7, see mutants cs –314 versus 3G7C). In this experiment, a point mutation A3G completely suppressed env expression (Fig. 7, see mutants cs –315 versus cs –314), which is unexpected from the point of view of the frequency-based Shapiro–Senapathy concept assigning nearly equal weight to A and G in position +3. This is in marked contrast to X-linked retinitis pigmentosa where the IVS4+3a>g point mutation in the retinitis pigmentosa GTPase regulator (RPGR) gene leads to skipping of exon 4 and disease (49), and our algorithm correctly predicts exon skipping (CAGGTAtagtg, graded as HC with 14 hydrogen bonds, versus CAGGTgtagtg graded as LC with 12 hydrogen bonds). Our experimental results also parallel statistical findings by Burge and Karlin, who observed a dramatic preference for A over G at position +3 in introns carrying a mismatch in position +5 (21).
The amount of env expression as a measure for U1 snRNA binding is described correctly by our hydrogen bond model, when we assume that the terminal base pairs of the 5′ splice site, i.e. –3 and +8, contribute one hydrogen bond less to the stability of the duplex than is given by Watson–Crick pairing. The stability of a base pair in position –3, for instance, might be reduced due to structural hindrance caused by a decreased flexibility of the U1 snRNA in this position next to stem I (50), which is hidden in a doughnut-shaped central element (51).
The accessibility of the U1 snRNA binding site might also be influenced by local intra-strand RNA binding at or around the splice site as has been shown for the HIV-1 splice site A3 (52). To investigate whether secondary structures could have influenced U1 snRNA binding at SD4, we substituted SD4 with the stem–loop structure carrying SD1 (53) in another set of mutant vectors. However, even an extended stem did not lead to a decrease in the 5′ splice site-dependent Rev-mediated env expression (data not shown). Therefore, we consider it unlikely that the mutational analysis presented in this paper was additionally influenced by RNA secondary structure.
Our examination of 5′ splice sites within the human ATM gene has only exemplary character to show the applicability in principle of our algorithm to data other than the HIV-1 genome. This gene includes a total of 64 canonical SDs. Examination of these 64 sequences using our algorithm yielded 51 correct classifications as HC (sensitivity 80%) when we set a threshold of _S_con = 13 hydrogen bonds as derived from our studies with HIV-1 SD4. Lowering the threshold in one of the parameters by one hydrogen bond to _S_con = 12 resulted in 60 correct SD classifications (sensitivity 94%). In a data set of 17 SD mutations comprising the positions –3 to –1 and +3 to +8 of the human ATM gene leading to exon skipping, the same modification of the threshold decreased the sensitivity to classify a mutation as LC from 16/17 (94%) to 14/17 (82%). This is in accordance with the concept that ESE or ISE sequences can compensate for ‘weak’ (non-consensus) 5′ splice sites, and that ‘strong’ splice sites may eliminate enhancer dependency (54). Experimentally, this relationship between the strength of a 5′ splice site (HC/LC) and a splicing enhancer is confirmed by our findings in HIV-1. The dependency on the upstream sequence for the recruitment of U1 snRNP to the 5′ splice site SD4 was abolished simply by increasing the complementarity between SD4 and the free 5′ end of U1 snRNA. This fragment supported ASF/SF2 binding in an in vitro gel mobility shift assay (data not shown). Thus, a more refined classification of a hydrogen bonding pattern as HC or LC will have to take flanking enhancer sequences into account. Leaving the basic counting rules in our hydrogen bond model unchanged, the effects of, for example, different SR protein binding sites, might be included just by suitably redefining the HC conditions in terms of threshold parameters. Up to now, 11 human SR proteins have been described that may support U1 snRNP binding to 5′ splice sites in a hierarchical way at different levels of complementarity. Relatively, HC would require less stabilization so that most SR proteins could support U1 snRNP binding, which might explain their functional redundancy (reviewed in 33). Non-redundant functions might be attributed to the capability of an SR protein to stabilize U1 snRNP binding even in the absence of complementarity. Basically, the SR proteins’ capacity for RNA duplex stabilization might depend on different interactions of their respective RS domains and U1 70K. Although it has been shown that the RS domains of ASF/SF2 and SC35 are functionally interchangeable (29), biochemical evidence has been provided that U1 70K interacts differently with some SR proteins (25).
Due to the availability of the human genome sequence and knowledge of several thousands of alternative and constitutive real 5′ splice sites, our algorithm could generate large data sets of real 5′ splice sites which have comparable capability to support U1 snRNA binding according to their hydrogen bonding patterns. Using such data sets it should be possible to identify common motifs within the flanking sequences of 5′ splice sites that might act as enhancer or silencer sequences.
Any genuine prediction of functional 5′ splice sites will have to take splice enhancers/silencers into account. Thus, at this stage, the power of our algorithm lies in its value to predict the presence of enhancer/silencer sequences.
Acknowledgments
ACKNOWLEDGEMENTS
We dedicate this work to the memory of Andreas Scheid. We are grateful to Xavier Roca for helpful comments on the manuscript. We thank K. Strebel and M. Martin for providing plasmid pNLA1, M. Gething for pSVT7, A. Weiner for pUCBU1, R. Axel for HeLa-T4+ cells (through the MRC AIDS Directed Program Reagent Project), and Dade Behring for the anti-gp120 mAb (87-133/026). We also thank Maria Thieme for excellent technical assistance and Drs K. Köhrer and S. Scheuring for plasmid sequencing. This work was supported by Deutsche Forschungsgemeinschaft grants to A.S. and H.S. (SCHA 909/1-1 and 909/2-1), by a grant from the Heinz-Ansmann-Stiftung and a short-term fellowship of the Boehringer Ingelheim Fonds to M.F., who also had a research fellowship from the Düsseldorf Entrepreneurs Foundation. S.K. was supported by a Graduierten Fellowship from the University of Düsseldorf.
REFERENCES
- 1.Lu X.B., Heimer,J., Rekosh,D. and Hammarskjold,M.L. (1990) U1 small nuclear RNA plays a direct role in the formation of a rev-regulated human immunodeficiency virus env mRNA that remains unspliced. Proc. Natl Acad. Sci. USA, 87, 7598–7602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrett N.L., Li,X. and Carmichael,G.G. (1995) The sequence and context of the 5′ splice site govern the nuclear stability of polyoma virus late RNAs. Nucleic Acids Res., 23, 4812–4817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kammler S., Leurs,C., Freund,M., Krummheuer,J., Seidel,K., Tange,T.O., Lund,M.K., Kjems,J., Scheid,A. and Schaal,H. (2001) The sequence complementarity between HIV-1 5′ splice site SD4 and U1 snRNA determines the steady-state level of an unstable env pre-mRNA. RNA, 7, 421–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lund M. and Kjems,J. (2002) Defining a 5′ splice site by functional selection in the presence and absence of U1 snRNA 5′ end. RNA, 8, 166–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhuang Y. and Weiner,A.M. (1986) A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell, 46, 827–835. [DOI] [PubMed] [Google Scholar]
- 6.Du H. and Rosbash,M. (2001) Yeast U1 snRNP–pre-mRNA complex formation without U1 snRNA-pre-mRNA base pairing. RNA, 7, 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du H. and Rosbash,M. (2002) The U1 snRNP protein U1C recognizes the 5′ splice site in the absence of base pairing. Nature, 419, 86–90. [DOI] [PubMed] [Google Scholar]
- 8.Forch P., Puig,O., Martinez,C., Seraphin,B. and Valcarcel,J. (2002) The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J., 21, 6882–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mount S.M. (1982) A catalogue of splice junction sequences. Nucleic Acids Res., 10, 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Padgett R.A., Grabowski,P.J., Konarska,M.M., Seiler,S. and Sharp,P.A. (1986) Splicing of messenger RNA precursors. Annu. Rev. Biochem., 55, 1119–1150. [DOI] [PubMed] [Google Scholar]
- 11.Shapiro M.B. and Senapathy,P. (1987) RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res., 15, 7155–7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burge C.B., Tuschl,T. and Sharp,P.A. (1999) Splicing of precursors to mRNAs by the spliceosomes. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 525–560. [Google Scholar]
- 13.Moore M.J., Query,C.C. and Sharp,P.A. (1993) Splicing of precursors to mRNA by the spliceosome. In Gesteland,R.F. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 303–357. [Google Scholar]
- 14.Staden R. (1984) Computer methods to locate signals in nucleic acid sequences. Nucleic Acids Res., 12, 505–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Senapathy P., Shapiro,M.B. and Harris,N.L. (1990) Splice junctions, branch point sites and exons: sequence statistics, identification and applications to genome project. Methods Enzymol., 183, 252–278. [DOI] [PubMed] [Google Scholar]
- 16.Stephens R.M. and Schneider,T.D. (1992) Features of spliceosome evolution and function inferred from an analysis of the information at human splice sites. J. Mol. Biol., 228, 1124–1136. [DOI] [PubMed] [Google Scholar]
- 17.Mengeritsky G. and Smith,T.F. (1989) New analytical tool for analysis of splice site sequence determinants. Comput. Appl. Biosci., 5, 97–100. [DOI] [PubMed] [Google Scholar]
- 18.Kel A.E., Ponomarenko,M.P., Likhachev,E.A., Orlov,Y., Ischenko,I.V., Milanesi,L. and Kolchanov,N.A. (1993) SITEVIDEO: a computer system for functional site analysis and recognition. Investigation of the human splice sites. Comput. Appl. Biosci., 9, 617–627. [DOI] [PubMed] [Google Scholar]
- 19.Solovyev V.V., Salamov,A.A. and Lawrence,C.B. (1994) Predicting internal exons by oligonucleotide composition and discriminant analysis of spliceable open reading frames. Nucleic Acids Res., 22, 5156–5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang M.Q. and Marr,T.G. (1993) A weight array method for splicing signal analysis. Comput. Appl. Biosci., 9, 499–509. [DOI] [PubMed] [Google Scholar]
- 21.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 22.Rogozin I.B. and Milanesi,L. (1997) Analysis of donor splice sites in different eukaryotic organisms. J. Mol. Evol., 45, 50–59. [DOI] [PubMed] [Google Scholar]
- 23.Heinrichs V., Bach,M., Winkelmann,G. and Luhrmann,R. (1990) U1-specific protein C needed for efficient complex formation of U1 snRNP with a 5′ splice site. Science, 247, 69–72. [DOI] [PubMed] [Google Scholar]
- 24.Jamison S.F., Pasman,Z., Wang,J., Will,C., Luhrmann,R., Manley,J.L. and Garcia-Blanco,M.A. (1995) U1 snRNP–ASF/SF2 interaction and 5′ splice site recognition: characterization of required elements. Nucleic Acids Res., 23, 3260–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohtz J.D., Jamison,S.F., Will,C.L., Zuo,P., Luhrmann,R., Garcia-Blanco,M.A. and Manley,J.L. (1994) Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature, 368, 119–124. [DOI] [PubMed] [Google Scholar]
- 26.Surowy C.S., van Santen,V.L., Scheib-Wixted,S.M. and Spritz,R.A. (1989) Direct, sequence-specific binding of the human U1-70K ribonucleoprotein antigen protein to loop I of U1 small nuclear RNA. Mol. Cell. Biol., 9, 4179–4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao W. and Garcia-Blanco,M.A. (1998) A serine/arginine-rich domain in the human U1 70k protein is necessary and sufficient for ASF/SF2 binding. J. Biol. Chem., 273, 20629–20635. [DOI] [PubMed] [Google Scholar]
- 28.Fu X.D. (1993) Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature, 365, 82–85. [DOI] [PubMed] [Google Scholar]
- 29.Mayeda A., Screaton,G.R., Chandler,S.D., Fu,X.D. and Krainer,A.R. (1999) Substrate specificities of SR proteins in constitutive splicing are determined by their RNA recognition motifs and composite pre-mRNA exonic elements. Mol. Cell. Biol., 19, 1853–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourgeois C.F., Popielarz,M., Hildwein,G. and Stevenin,J. (1999) Identification of a bidirectional splicing enhancer: differential involvement of SR proteins in 5′ or 3′ splice site activation. Mol. Cell. Biol., 19, 7347–7356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tange T.O., Damgaard,C.K., Guth,S., Valcarcel,J. and Kjems,J. (2001) The hnRNP A1 protein regulates HIV-1 tat splicing via a novel intron silencer element. EMBO J., 20, 5748–5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanopka A., Muhlemann,O. and Akusjarvi,G. (1996) Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature, 381, 535–538. [DOI] [PubMed] [Google Scholar]
- 33.Tacke R. and Manley,J.L. (1999) Determinants of SR protein specificity. Curr. Opin. Cell Biol., 11, 358–362. [DOI] [PubMed] [Google Scholar]
- 34.Liu H.X., Chew,S.L., Cartegni,L., Zhang,M.Q. and Krainer,A.R. (2000) Exonic splicing enhancer motif recognized by human SC35 under splicing conditions. Mol. Cell. Biol., 20, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Staffa A. and Cochrane,A. (1995) Identification of positive and negative splicing regulatory elements within the terminal tat-rev exon of human immunodeficiency virus type 1. Mol. Cell. Biol., 15, 4597–4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Si Z.H., Rauch,D. and Stoltzfus,C.M. (1998) The exon splicing silencer in human immunodeficiency virus type 1 Tat exon 3 is bipartite and acts early in spliceosome assembly. Mol. Cell. Biol., 18, 5404–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen X., McDowell,J.A., Kierzek,R., Krugh,T.R. and Turner,D.H. (2000) Nuclear magnetic resonance spectroscopy and molecular modeling reveal that different hydrogen bonding patterns are possible for G.U pairs: one hydrogen bond for each G.U pair in r(GGCGUGCC)(2) and two for each G.U pair in r(GAGUGCUC)(2). Biochemistry, 39, 8970–8982. [PubMed] [Google Scholar]
- 38.Burset M., Seledtsov,I.A. and Solovyev,V.V. (2000) Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res., 28, 4364–4375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schaal H., Klein,M., Gehrmann,P., Adams,O. and Scheid,A. (1995) Requirement of N-terminal amino acid residues of gp41 for human immunodeficiency virus type 1-mediated cell fusion. J. Virol., 69, 3308–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strebel K., Daugherty,D., Clouse,K., Cohen,D., Folks,T. and Martin,M.A. (1987) The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature, 328, 728–730. [DOI] [PubMed] [Google Scholar]
- 41.Schaal H., Pfeiffer,P., Klein,M., Gehrmann,P. and Scheid,A. (1993) Use of DNA end joining activity of a Xenopus laevis egg extract for construction of deletions and expression vectors for HIV-1 Tat and Rev proteins. Gene, 124, 275–280. [DOI] [PubMed] [Google Scholar]
- 42.Maddon P.J., Dalgleish,A.G., McDougal,J.S., Clapham,P.R., Weiss,R.A. and Axel,R. (1986) The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell, 47, 333–348. [DOI] [PubMed] [Google Scholar]
- 43.Selden R.F., Howie,K.B., Rowe,M.E., Goodman,H.M. and Moore,D.D. (1986) Human growth hormone as a reporter gene in regulation studies employing transient gene expression. Mol. Cell. Biol., 6, 3173–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 45.Chomczynski P. and Sacchi,N. (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 46.Mathews D.H. and Turner,D.H. (2002) Dynalign: an algorithm for finding the secondary structure common to two RNA sequences. J. Mol. Biol., 317, 191–203. [DOI] [PubMed] [Google Scholar]
- 47.Peyret N., Seneviratne,P.A., Allawi,H.T. and SantaLucia,J.,Jr (1999) Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A.A,C.C,G.G and T.T mismatches. Biochemistry, 38, 3468–3477. [DOI] [PubMed] [Google Scholar]
- 48.Hibbert C.S., Gontarek,R.R. and Beemon,K.L. (1999) The role of overlapping U1 and U11 5′ splice site sequences in a negative regulator of splicing. RNA, 5, 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buraczynska M., Wu,W., Fujita,R., Buraczynska,K., Phelps,E., Andreasson,S., Bennett,J., Birch,D.G., Fishman,G.A., Hoffman,D.R. et al. (1997) Spectrum of mutations in the RPGR gene that are identified in 20% of families with X-linked retinitis pigmentosa. Am. J. Hum. Genet., 61, 1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Branlant C., Krol,A., Ebel,J.P., Gallinaro,H., Lazar,E. and Jacob,M. (1981) The conformation of chicken, rat and human U1A RNAs in solution. Nucleic Acids Res., 9, 841–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stark H., Dube,P., Luhrmann,R. and Kastner,B. (2001) Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature, 409, 539–542. [DOI] [PubMed] [Google Scholar]
- 52.Jacquenet S., Ropers,D., Bilodeau,P.S., Damier,L., Mougin,A., Stoltzfus,C.M. and Branlant,C. (2001) Conserved stem-loop structures in the HIV-1 RNA region containing the A3 3′ splice site and its cis-regulatory element: possible involvement in RNA splicing. Nucleic Acids Res., 29, 464–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berkhout B. (1996) Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol., 54, 1–34. [DOI] [PubMed] [Google Scholar]
- 54.Graveley B.R. (2000) Sorting out the complexity of SR protein functions. RNA, 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandoval N., Platzer,M., Rosenthal,A., Dork,T., Bendix,R., Skawran,B., Stuhrmann,M., Wegner,R.D., Sperling,K., Banin,S. et al. (1999) Characterization of ATM gene mutations in 66 ataxia telangiectasia families. Hum. Mol. Genet., 8, 69–79. [DOI] [PubMed] [Google Scholar]
- 56.Buzin C.H., Gatti,R.A., Nguyen,V.Q., Wen,C.Y., Mitui,M., Sanal,O., Chen,J.S., Nozari,G., Mengos,A., Li,X. et al. (2003) Comprehensive scanning of the ATM gene with DOVAM-S. Hum. Mutat., 21, 123–131. [DOI] [PubMed] [Google Scholar]
- 57.Saviozzi S., Saluto,A., Piane,M., Prudente,S., Migone,N., DeMarchi,M., Brusco,A. and Chessa,L. (2003) Six novel ATM mutations in Italian patients with classical ataxia-telangiectasia. Hum. Mutat., 21, 450. [DOI] [PubMed] [Google Scholar]
- 58.Beaudet A.L. and Tsui,L.C. (1993) A suggested nomenclature for designating mutations. Hum. Mutat., 2, 245–248. [DOI] [PubMed] [Google Scholar]
- 59.Platzer M., Rotman,G., Bauer,D., Uziel,T., Savitsky,K., Bar-Shira,A., Gilad,S., Shiloh,Y. and Rosenthal,A. (1997) Ataxia-telangiectasia locus: sequence analysis of 184 kb of human genomic DNA containing the entire ATM gene. Genome Res., 7, 592–605. [DOI] [PubMed] [Google Scholar]