Jarid2/Jumonji Coordinates Control of PRC2 Enzymatic Activity and Target Gene Occupancy in Pluripotent Cells (original) (raw)
. Author manuscript; available in PMC: 2010 Jul 29.
SUMMARY
Polycomb Repressive Complex 2 (PRC2) regulates key developmental genes in embryonic stem(ES) cells and during development. Here we show that Jarid2/Jumonji, a protein enriched in pluripotent cells and a founding member of the Jumonji C (JmjC) domain protein family, is a PRC2 subunit in ES cells. Genome-wide ChIP-seq analyses of Jarid2, Ezh2, and Suz12 binding reveal that Jarid2 and PRC2 occupy the same genomic regions. We further show that Jarid2 promotes PRC2 recruitment to the target genes while inhibiting PRC2 histone methyltransferase activity, suggesting that it acts as a “molecular rheostat” that finely calibrates PRC2 functions at developmental genes. Using Xenopus laevis as a model we demonstrate that Jarid2 knockdown impairs the induction of gastrulation genes in blastula embryos and results in failure of differentiation. Our findings illuminate a mechanism of histone methylation regulation in pluripotent cells and during early cell-fate transitions.
INTRODUCTION
Histone methylation by the PRC2 complex regulates developmental gene expression patterns in multicellular organisms (Schuettengruber et al., 2007; Simon and Kingston, 2009). PRC2 contains three core subunits: Ezh2, Suz12 and Eed, all of which are essential for trimethylation of histone H3 lysine 27 (H3K27me3), a mark that has been correlated with the silent state of target genes (Schuettengruber et al., 2007; Simon and Kingston, 2009).
In ES cells PRC2 represses developmental genes involved in cellular differentiation and organismal development (Boyer et al., 2006; Lee et al., 2006). Deletion of any of the PRC2 core components in mice results in gastrulation defects and early embryonic lethality (Faust et al., 1998; O'Carroll et al., 2001; Pasini et al., 2004). Nevertheless, mouse ES cells lacking Eed, Suz12 or Ezh2 can be derived from the respective homozygous knockout blastocysts and propagated in vitro (Morin-Kensicki et al., 2001; Pasini et al., 2007; Shen et al., 2008). However, loss of PRC2 function leads to defects in ES cell differentiation (Chamberlain et al., 2008; Pasini et al., 2007; Shen et al., 2008), emphasizing the essential role of PRC2 in executing differentiation programs during early development.
Despite detailed molecular studies of the PRC2 components, some outstanding questions remain largely unanswered: What molecular mechanisms control PRC2 recruitment to the target genes? What is the role of PRC2 in transitions from pluripotent to restricted developmental fates? We used a combination of biochemical, genomic and embryological approaches to provide the first evidence that Jarid2/Jumonji (hereafter referred to as Jarid2), a JmjC-domain protein enriched in pluripotent cells, coordinates control of PRC2 occupancy and enzymatic activity at target genes in ES cells and early embryos.
RESULTS
Jarid2 Associates with the PRC2 Complex in Mouse ES Cells
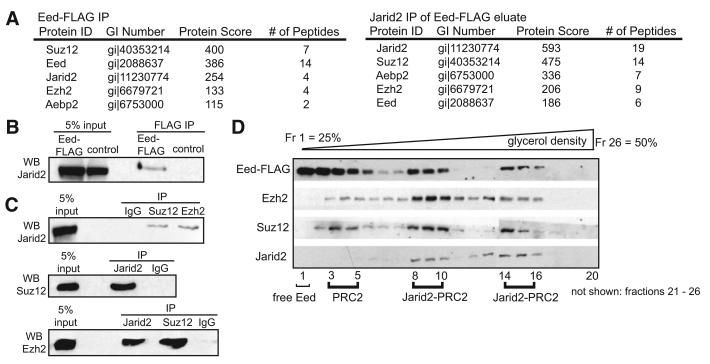
To screen for novel PRC2 partners we immunopurified and identified Eed-associated proteins using clonal mouse ES transgenic lines stably-expressing FLAG epitope-tagged Eed, as diagrammed in Figure S1A, available online. In addition to previously characterized PRC2 components—Eed, Suz12, Ezh2 and Aepb2—mass spectrometry analysis identified Jarid2 in Eed-FLAG immunoprecipitates, but not control extracts (Figure 1A, left panel; all identified peptides are listed in Table S1). Anti-Jarid2 immunoblot analysis of Eed-FLAG eluates confirmed association between Jarid2 and Eed (Figure 1B). To address whether Jarid2 interacts with the intact PRC2 complex, we subjected the Eed-FLAG eluate to another round of immunoaffinity purification with anti-Jarid2 IgG or control IgG (Figure S1B). Mass spectrometry analysis after this two-step purification identified all core PRC2 subunits in addition to Jarid2, indicating that Jarid2 interacts with the intact PRC2 complex (Figure 1A, right panel; peptides listed in Table S1).
Figure 1. Isolation of the PRC2 Complex from ES Cells Identified Jarid2 as a Novel Component.
(A) Jarid2 associates with PRC2 in mouse ES cells. Left panel: proteins specifically identified in Eed-FLAG purification. Right panel: proteins specifically identified in Eed-FLAG/Jarid2 double purification. Protein identification scores (Mascot) and numbers of tryptic peptides identified are shown. See Figure S1 for purification schematics.
(B) Confirmation of Jarid2-Eed association. FLAG immunoprecipitates from wt and Eed-FLAG ES cells were analyzed by anti-Jarid2 immunoblotting.
(C) Association between endogenous Jarid2 and PRC2. Endogenous Suz12, Ezh2, and Jarid2 proteins were immunoprecipitated from ES cell nuclear extracts and analyzed by immunoblotting with indicated antibodies.
(D) Jarid2 cosediments with PRC2. Eed-FLAG eluates were separated on a 25%–50% glycerol density gradient and fractions analyzed by immunoblotting with indicated antibodies.
Next, we showed that endogenous Suz12 and Ezh2 immunoprecipitated endogenous Jarid2 from mouse ES cell nuclear extracts and conversely, Jarid2 immunoprecipitated Suz12 and Ezh2 (Figure 1C). Furthermore, Jarid2 co-sedimented with PRC2 in two high-density peaks in the glycerol-gradient sedimentation analysis of the Eed-FLAG eluates (Figure 1D). Eed, Suz12 and Ezh2 co-sedimented in fractions 3-5 in the absence of Jarid2, suggesting that Jarid2 is not required for the assembly of core PRC2 complex in mouse ES cells, consistent with previous reports that the three core subunits—Eed, Ezh2, and Suz12—form a stable complex (Cao and Zhang, 2004; Martin et al., 2006). Nevertheless, the majority of PRC2 in ES cells appears to be bound to Jarid2 (Figure 1D). Sedimentation analyses of nuclear extracts from Jarid2 shRNA-expressing cells (in which Jarid2 is downregulated to 30%–40% of wild-type levels; described in detail below), revealed that the remaining Jarid2 co-sediments with PRC2 in a single peak (Figure S1C). These data suggest that the formation and/or stability of the largest complex is sensitive to Jarid2 levels.
Numerous studies demonstrated that Jarid2 expression is under the control of ES transcriptional circuitry, including transcription factors Nanog, Oct4, Sox2, Klf4 and Tcf3 (Boyer et al., 2005; Cole et al., 2008; Kim et al., 2008; Loh et al., 2006; Zhou et al., 2007). Jarid2 mRNA is among the transcripts most highly enriched in undifferentiated mouse and human ES cells and human oocytes (Assou et al., 2009; Sun et al., 2008; Zhou et al., 2007). Interestingly, due to conserved amino acid changes that preclude cofactor binding (Figure S2A), Jarid2 lacks histone demethylase activity characteristic of other JmjC domain proteins (Klose et al., 2006; Lan et al., 2008; Shirato et al., 2009). We reasoned that Jarid2 represents an attractive candidate regulator of the PRC2 function.
A Short Motif Conserved in Jarid Proteins Is Required for Interaction of Jarid2 and Jarid1a with Suz12
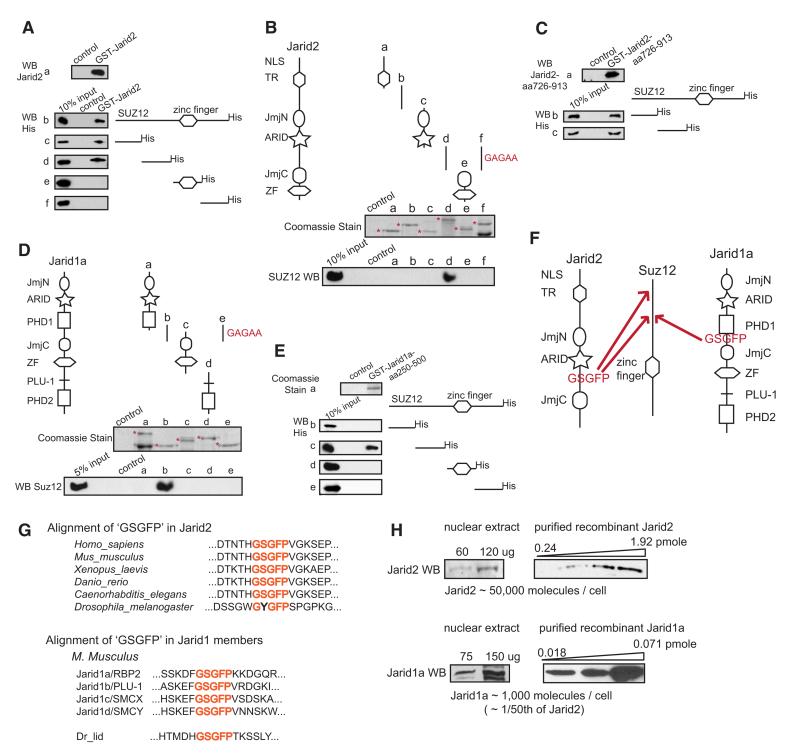
The mass spectrometry analysis of Jarid2 associated proteins showed Suz12 as the most enriched PRC2 member (Figure 1A, right panel), suggesting a direct interaction. In agreement, purified recombinant GST-Jarid2 recovered recombinant Suz12 in a pull-down assay (Figure 2A, a and b). Two N-terminal regions of Suz12 bound GST-Jarid2 (Figure 2A, c–f), and a single Jarid2 region corresponding to amino acids 726-913 (fragment d) was sufficient for binding to both Suz12 N-terminal fragments (Figures 2B and 2C). In contrast, we failed to detect interaction of Jarid2 with recombinant Ezh2, Eed, Aebp2 or RbAp48 (Figures S2B and S2C).
Figure 2. Jarid2 and Jarid1a Directly Bind Suz12 via a Conserved Amino Acid Motif.
(A) Recombinant Jarid2 associates with two N-terminal regions of Suz12. Full-length recombinant GST-Jarid2 protein (a) was used as a bait to pull full-length Suz12-His (b), or Suz12-His protein fragments (c–f). Bound proteins were visualized by anti-His immunoblotting. In all experiments, ‘control’ represents glutathione beads incubated with bacterial extracts not expressing bait proteins.
(B) Jarid2 region corresponding to aa 726–913 binds Suz12 in a “GSGFP” motif dependent manner. Recombinant GST-Jarid2 protein fragments were used to pull recombinant full-length Suz12. Purified GST-fusion proteins were visualized by Coomasie staining (a-e, top panel, respective fusion proteins are marked with asterisks), and Suz12 binding was assayed by anti-Suz12 immunoblotting (a–e, bottom panel). The “GSGFP” motif embedded in the fragment d (aa 726–913 of mouse Jarid2), was substituted with ‘GAGAA’ sequence, and the mutated GST-fusion protein assayed for Suz12 binding (f).
(C) aa 726-913 fragment of Jarid2 is sufficient for binding to both N-terminal regions of Suz12. GST-Jarid2 aa 726-913 fusion protein (a) was used as a bait to pull two N-terminal Suz12 fragments (aa 1-185 and aa 185-370 of mouse Suz12). Bound proteins were visualized by anti-His immunoblotting (b and c).
(D) Jarid1a region corresponding to aa 250-500 binds Suz12 in a “GSGFP” motif dependent manner. Recombinant GST-Jarid1a protein fragments were used to pull full-length Suz12. Purified GST-fusion proteins were visualized by Coomasie staining (a–d, top panel, respective fusion proteins are marked with asterisks), and Suz12 binding was assayed by anti-Suz12 immunoblotting (a–d, bottom panel). The GSGFP motif embedded in fragment b (aa 250–500 of mouse Jarid1a), was substituted with GAGAA sequence, and the mutated GST-fusion protein assayed for Suz12 binding (e).
(E) aa 250-500 fragment of Jarid1a binds a single Suz12 N-terminal region, corresponding to aa 185-370. GST-Jarid1a (shown in a) was used to pull Suz12-His fragments (b-e); bound proteins were visualized by anti-His immunoblotting.
(F) Schematic diagram summarizing binding results presented in (A)–(E).
(G) Conservation of the ‘GSGFP’ motif among Jarid proteins. Top panel: a sequence alignment of the “GSGFP”-containing regions in Jarid2 proteins from indicated species. Bottom panel: a sequence alignment of the “GSGFP”-containing regions in the four M. musculus Jarid1 family members, and in D. melanogaster Jarid1 homolog, Lid. Motif is highlighted in red.
(H) Jarid2 and Jarid1a protein levels in mouse ES cells. Immunoblot signals of endogenous Jarid2 or Jarid1a from a defined amount of ES nuclear extract were compared to those of a serial dilution of purified, recombinant Jarid2 or Jarid1a protein fragments of a known concentration. Left and right panels represent same exposure of the same blot. Estimated number of molecules per cell nucleus is shown at the bottom. Calculations can be found in Supplemental Experimental Procedures.
Jarid2 is most closely related to the four Jarid1 family proteins, which also contain JmjC domain and ARID (AT-rich interaction domain) (Kortschak et al., 2000). Jarid1 proteins are active H3K4me3 demethylases (Lan et al., 2008), and Jarid1a/Rbp2 was shown to associate with PRC2 (Pasini et al., 2008). Although we failed to identify Jarid1a peptides in mass spectrometry analysis of Eed-FLAG eluates (Table S1), we detected Jarid1a-PRC2 association by coimmunoprecipitation and immunoblotting (Figure S2D), and hypothesized that investigating the mode of Jarid1 association with PRC2 may shed light on the molecular recognition of PRC2 by Jarid proteins.
First, we tested whether Jarid1a, like Jarid2, binds Suz12. Recombinant GST-fusion protein corresponding to amino acids 250-500 of human Jarid1a (fragment b) bound the second, but not the first, N-terminal region of Suz12 (Figures 2D and 2E). Thus, both Jarid2 and Jarid1a interact with Suz12 amino acid region 185-370, but Jarid2 recognizes additional residues within the first N-terminal fragment.
Jarid2 and Jarid1a regions responsible for Suz12 binding do not overlap with any discernible structural domains (Figure 2F) and display low similarity, with the exception of a highly homologous short sequence “GSGFP.” We hypothesized that this motif may play a role in Suz12 recognition. Indeed, mutations of GSGFP to GAGAA diminished binding of Jarid2 and Jarid1a fragments to full-length Suz12 (compare d and f in Figure 2B, and b and e in Figure 2D). This motif is conserved in all vertebrate Jarid2 proteins, as well as in C. elegans Jarid2 (Figure 2G), whereas D. melanogaster and other Drosophila species contain a non-conservative substitution within the motif (GYGFP). The GSGFP motif is also conserved in all four Jarid1 family proteins: Jarid1a/RBP2, Jarid1b/PLU-1, Jarid1c/SMCX and Jarid1d/SMCY, as well as in the single Jarid1 homolog in Drosophila, Lid (Figure 2G). The presence of the GSGFP motif in metazoan Jarid proteins suggests that the association with Suz12 may be a common feature of Jarid family members. However, we cannot exclude the possibility that additional molecular interactions control Jarid-PRC2 complex formation in vivo.
Mouse ES Cells Contain High Levels of Jarid2 Protein
The preferential recovery of Jarid2 in the Eed-FLAG purification indicates that Jarid2 is the major Jarid family member associated with PRC2 in ES cells. To estimate the relative molar amounts of Jarid2 and Jarid1a proteins in ES nuclear extracts we compared immunoblot signals of endogenous Jarid2 or Jarid1a to signals from a serial dilution of purified, recombinant protein fragments of known concentrations (Figure 2H). From this analysis we calculated that 1 mg of ES nuclear extract contains 6 pmols of Jarid2 and 0.12 pmole of Jarid1a. Although such measurements are not precise, we further estimated that a single ES cell nucleus contains about 50,000 Jarid2 and about 1000 Jarid1a molecules. Interestingly, downregulation of Jarid2 results in a modest upregulation of Jarid1a protein, but not RNA levels (Figures S2E and S2F), perhaps via stabilization of Jarid1a through PRC2 association.
Jarid2 Occupies PRC2 Targets Genome-Wide
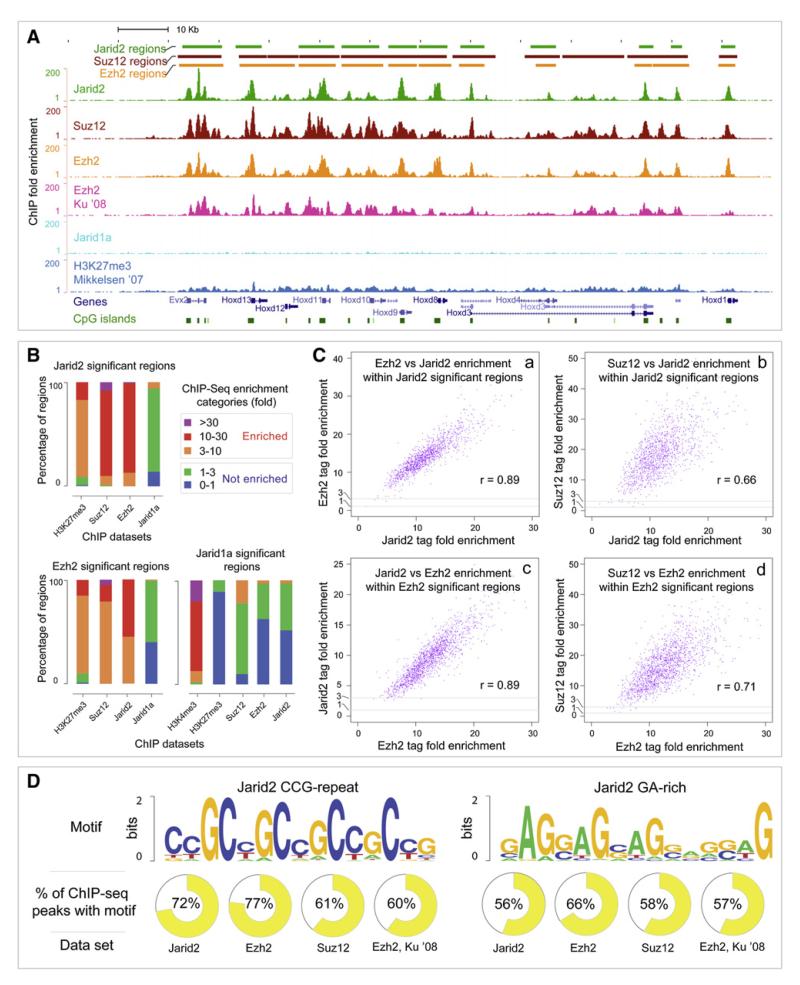
To determine the genome-wide occupancy of Jarid2 in mouse ES cells and to analyze the extent to which it overlaps with PRC2 and Jarid1a binding, we used chromatin immunoprecipitation coupled with massively parallel DNA sequencing (ChIPseq; (Barski et al., 2007; Johnson et al., 2007). Illumina Genome Analyzer was used to generate 13.5, 12.7, 18.7, 11, and 28.5 million mapped sequence reads from Jarid2, Ezh2, Suz12, Jarid1a, and control libraries, respectively. QuEST ChIP-Seq analysis software (Valouev et al., 2008) identified 1337, 1692, 2073 and 1764 “significant regions” enriched within Jarid2, Ezh2, Suz12 and Jarid1a ChIP-seq datasets [at the false discovery rate (FDR) of less than 2.8%]. Significant regions were identified for all proteins with high stringency threshold of having at least one position with 50-fold or higher enrichment.
Comparison of Jarid2, Suz12 and Ezh2 ChIP-Seq enrichment signals revealed a nearly complete overlap of binding patterns (as illustrated by the Hoxd gene cluster and Sox9 gene locus, Figures 3A and S3A). We also observed good overlaps between our binding data and those reported in the literature (Figures 3A and S3A, compare top three panels with the panels displaying Ezh2 data [Ku et al., 2008] and H3K27me3 data [Mikkelsen et al., 2007]). However, we did not detect significant binding of Jarid1a over PRC2 bound loci (Figures 3A and S3A). Next, we analyzed genome-wide co-occupancy of the interrogated proteins. Within Jarid2, Ezh2 and Jarid1a significant regions (defined as having a peak of 50-fold or higher enrichment) we identified relative fold enrichment of Suz12, Ezh2, Jarid2 and Jarid1a (defined as the enrichment of sequence tags relative to control across the entire region; less than 3-fold enrichment falls within experimental variability and was considered as not enriched). 99.2% and 99.8% of Jarid2 significant regions was enriched for Suz12 and Ezh2, respectively, with the majority of regions enriched over 10-fold (Figure 3B). Conversely, 99.7% and 99.7% of Ezh2 significant regions was enriched for Suz12 and Jarid2, respectively. The majority of Ezh2 regions were enriched for Jarid2 more than 10-fold (Figure 3B). Colocalization of Jarid2 with PRC2 was also supported by high correlation (0.66-0.89) of ChIP-seq binding signals among the Jarid2, Ezh2 and Suz12 datasets (Figure 3C). There was also good correlation of Jarid2 and Ezh2 binding with H3K27me3 (Figure 3B).
Figure 3. Jarid2 and PRC2 Occupy Same Genomic Targets in ES Cells.
(A) Genome browser representation of Jarid2, Suz12, Ezh2, Jarid1a and H3K27me3 binding patterns at the Hoxd gene cluster in mouse ES cells. The top three tracks represent calls for significantly enriched regions of Jarid2, Suz12 and Ezh2 ChIP-seq experiments as determined by QuEST software (Valouev et al., 2008). Following are six tracks displaying calculated ChIP-Seq enrichment values across the locus for Jarid2, Suz12, Ezh2 from this study, Ezh2 (Ku et al., 2008), Jarid1a from this study, and H3K27me3 (Mikkelsen et al., 2007) ChIP-seq datasets. Relative positions of genes and CpG islands are shown at the bottom.
(B) Comparisons of coenrichment between ChIP-seq experiments. The three bar graphs represent relative levels of enrichment (defined as the fold enrichment of sequence tags relative to control across the entire region) of Suz12, Ezh2, Jarid2, Jarid1a, H3K27me3 and H3K4me3, as indicated, within Jarid2, Ezh2 and Jarid1a significant regions determined by QuEST. Three enriched categories correspond to ChIP-to-control tag enrichment ranges of 3-10 (orange), 10-30 (red) and over 30 (purple). Non-enriched categories correspond to relative tag enrichment levels of 0-1 (blue) and 1-3 (green).
(C) Genome-wide analysis of Jarid2, Ezh2, and Suz12 co-occupancy. Scatter plots display mutual enrichment between indicated ChIP-Seq datasets relative to the input. Correlation values are shown.
(D) Sequence motifs enriched in Jarid2-PRC2 bound regions. Logos (Crooks et al., 2004) for the two significant motifs identified using MEME software (from 200 bp windows around Jarid2 peaks) are shown. % of ChIP-seq regions containing the motif at 5% FDR is shown.
In sharp contrast, only 1% and 2% of Jarid2 and Ezh2 significant regions, respectively, was enriched for Jarid1a signals, and none had over 10-fold enrichment (Figure 3B). Overall, Jarid1a occupancy showed little correlation with PRC2 binding (0.18-0.37, Figure S4A) or with enrichment for H3K27me3 (Figure 3B). Instead, Jarid1a significant regions overlapped with H3K4me3 (Figure 3B and S3B).
Jarid2-PRC2 Targets Are Enriched for Unique DNA Sequence Motifs
De novo search for motifs overrepresented within the Jarid2, Ezh2 and Suz12 binding peaks identified two significant motifs (Figure 3D) that were enriched at peaks and also throughout the bound regions. The first motif is a tandem repeat of CCG and is present within 61%–72% of Jarid2, Ezh2 and Suz12 regions (5% FDR). A second, GA-rich motif was present within 56%–66% of PRC2 and Jarid2 regions (5% FDR). Both motifs were also significantly enriched within a previously reported Ezh2 dataset from Ku et al. (2008) (60% and 57% of regions contained these motifs, respectively).
Jarid2 and PRC2 Co-occupy Promoters of Developmental Genes in Mouse and Human ES Cells
Jarid2 bound regions typically overlap with transcription start sites (TSS; 69% overlap, Figure S5A) and exhibit a mean and median size of 3.3 kb and 2.7 kb, respectively (Figure S5D). Functional classification of identified targets via GO term analysis (Beissbarth and Speed, 2004) showed highly significant enrichment in genes involved in development, morphogenesis, and transcription (Figure S5B), similarly to what was previously observed for PRC2 (Boyer et al., 2006). A complete list of Jarid2 bound genes is provided in Table S2.
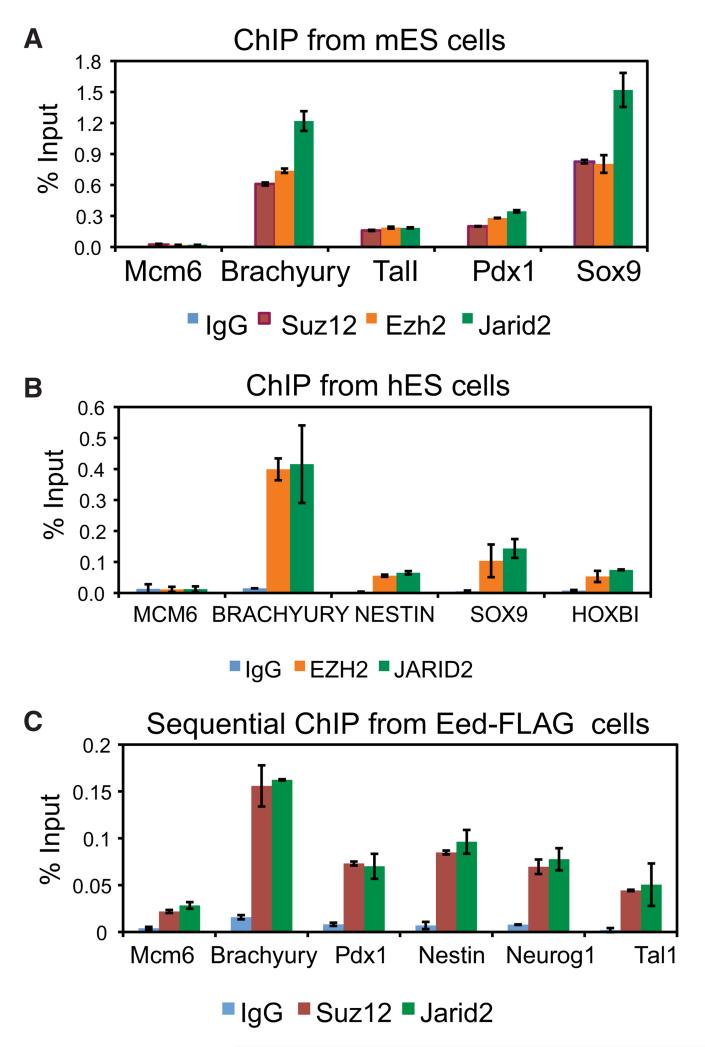
To validate ChIP-seq results, we performed ChIP-qPCR analyses of selected PRC2 target genes and, as a control, a gene not bound by the PRC2 complex (Mcm6) using independently isolated DNA from mouse and human ES cells. Relative Jarid2, Ezh2 and Suz12 occupancy levels were correlated for all tested PRC2 target genes in both mouse and human ES cells, indicating that Jarid2 association with PRC2 targets is conserved between mouse and human (Figures 4A and 4B).
Figure 4. Jarid2 and PRC2 Simultaneously Bind Target Genes.
(A) ChIP-qPCR analyses of Jarid2, Ezh2 and Suz12 binding at selected target genes in mouse ES cells.
(B) ChIP-qPCR analyses of JARID2, EZH2 and SUZ12 binding at selected target genes in human ES cells.
(C) Sequential ChIP of Suz12 and Jarid2 from Eed-FLAG-bound chromatin. y axis shows percent of input recovery. Error bars represent standard deviation calculated from triplicate qPCR reactions. Findings were confirmed by multiple biological replicates.
To demonstrate that Jarid2 and PRC2 can simultaneously bind to the same chromatin regions, we performed sequential ChIP analysis. Chromatin from Eed-FLAG mouse ES cells was first immunoprecipitated with FLAG antibody, followed by a second ChIP step with either Jarid2, Suz12, or non-specific IgG antibody. For all interrogated PRC2 targets, we detected simultaneous Jarid2 and PRC2 binding (Figure 4C). In sum, biochemical association, direct binding between Jarid2 and Suz12 in vitro, and target co-occupancy across the genome strongly suggest Jarid2 is an integral PRC2 subunit in ES cells.
Jarid1a/Rbp2 Occupies Promoters of Genes Involved in RNA Metabolism and Mitochondrial Function
Although we identified 1764 genomic regions enriched for Jarid1a, we failed to detect an overlap with PRC2 binding. Consistently, functional classification of identified Jarid1a targets showed enrichment of genes involved in RNA processing and mitochondrial function, but not in development (Figure S5C). 82% of Jarid1a significant regions overlapped with H3K4me3 (representing 15% of all H3K4me3 significant regions in mouse ES cells). These targets were not bivalently marked, however, as we failed to detect an overlap between Jarid1a and H3K27me3 (Figure 3B). We also noted a quantitative difference in the size of Jarid1a and Jarid2 bound regions (Figure S5D). De novo motif search analysis revealed enrichment for a consensus recognition site of the Ets family transcription factor GABP in 55% of Jarid1a bound regions (Figure S4B). Intriguingly, our observations parallel those made in a previous ChIP-chip study of JARID1A occupancy in human promonocytic U937 cells (Lopez-Bigas et al., 2008). These parallels include: (i) overrepresentation of RNA metabolism and mitochondrial gene targets, (ii) enrichment for Ets family binding sites, and (iii) strong overlap with H3K4me3 in the absence of H3K27me3. Strikingly, 79% (185 out of 232) of JARID1A targets reported by Lopez-Bigas et al. (2008) in U937 cells are found in our Jarid1a dataset, despite differences in cell type, species and antibody used in the two studies. The findings outlined above suggest that a significant subset of Jarid1a targets represents house-keeping genes active across many cell types in humans and mice.
To confirm that our results are not an artifact of antibody cross-reactivity, we performed ChIP-qPCR analyses using five different anti-Jarid1a antibodies. We failed to detect Jarid1a binding to PRC2 targets with any of the antibodies (Figure S6A), but observed Jarid1a enrichment at all interrogated Jarid1a target genes identified by ChIP-seq with all tested antibodies (Figure S6B). Taken altogether, our data strongly argue that Jarid2, not Jarid1a, is the major PRC2 partner at chromatin targets in ES cells. We therefore focused our subsequent analyses on the mechanisms by which Jarid2 regulates PRC2 functions.
Jarid2 Is Important for Recruitment and/or Stabilization of PRC2 at Target Genes
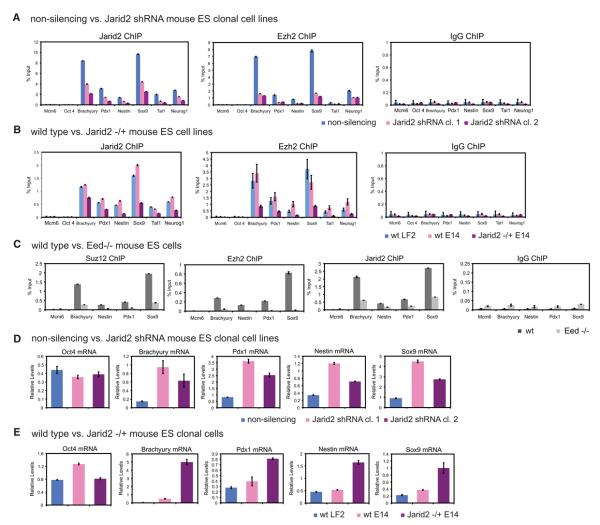
To address whether Jarid2 downregulation affects PRC2 target occupancy we developed stable, clonal mouse ES cell lines expressing shRNA targeting Jarid2 or, as a control, a non-targeting shRNA, in a doxycycline (Dox) inducible manner. Dox treatment of Jarid2 shRNA cells resulted in Jarid2 downregulation to 30%–40% of control levels (Figure S7A), without affecting total Ezh2 and Suz12 protein levels (Figure S7A), Oct4 expression (Figure 5D), or ES cell proliferation (data not shown). ChIP-qPCR analysis showed varied Jarid2 levels at different targets, nevertheless in all cases both Jarid2 and Ezh2 occupancy were significantly reduced upon Jarid2 knockdown (Figure 5A).
Figure 5. Jarid2 Controls PRC2 Target Occupancy in ES Cells.
(A) Knockdown of Jarid2 results in diminished Ezh2 target binding. ChIP-qPCR analyses of Jarid2 and Ezh2 occupancy at selected target genes in clonal mouse ES cell lines expressing shRNA targeting Jarid2 (Jarid2 shRNA cl.1 and 2) versus non-silencing shRNA. y axis shows percent of input recovery.
(B) Ezh2 association with gene targets is decreased in Jarid2 −/+ ES cells. ChIP-qPCR analyses are of wt LF2, wt E14, and E14 Jarid2 −/+ mouse ES cell lines.
(C) Jarid2 and PRC2 target occupancy is mutually dependent. ChIP-qPCR analyses of Suz12, Ezh2 and Jarid2 occupancy at selected target genes in wt and Eed −/− mouse ES cells.
(D) Jarid2 regulates expression of PRC2 target genes. RT-qPCR analysis of mRNA levels in Jarid2 shRNA cl.1 and 2 and non-silencing shRNA mouse ES cell lines. Expression was normalized to Pdha1 mRNA levels.
(E) Expression of PRC2 targets is upregulated in Jarid2 −/+ ES cells. RT-qPCR analysis of mRNA levels (normalized to Pdha1), in wt LF2, wt E14, and E14 Jarid2 −/+ mouse ES cell lines.
Error bars represent standard deviation calculated from triplicate qPCR reactions.
To exclude a possibility that diminished PRC2 binding is an artifact of shRNA off-target effects, we used the heterozygous Jarid2 gene trap mouse ES line (Davisson, 2006) to assay Jarid2 and Ezh2 protein levels and target occupancy. As this line was developed in a different genetic background than the one used throughout this study (E14 versus LF2), we compared wt LF2 cells, wt E14 cells and Jarid2 −/+ E14 (CSA 131) cells. Total levels of Jarid2 protein were higher in LF2 as compared to E14 cells, and were further diminished in Jarid2 −/+ E14 cells (Figure S7B). Nevertheless, Jarid2 target occupancy was comparable between LF2 and E14 wt cells, with reduced binding in Jarid2 −/+ E14 cells (Figure 5B), indicating that even a modest two-fold reduction of Jarid2 levels is sufficient to diminish Ezh2 association with target genes.
Jarid2 and PRC2 Association with Target Genes Is Mutually Dependent
If Jarid2 and PRC2 cooperate in target recognition, then Jarid2 chromatin binding should be PRC2-dependent. To test this prediction, we assayed occupancy of Ezh2, Suz12 and Jarid2 at selected target genes in Eed −/− ES cells. Consistent with Eed serving as a linchpin for the PRC2 complex, Suz12 and Ezh2 binding was diminished in Eed −/− as compared to wt cells (Figure 5C). Jarid2 binding was concomitantly reduced, indicating that Jarid2 and PRC2 target association is mutually dependent (Figure 5C). However, we observed a two-fold down-regulation of Jarid2 protein levels in Eed −/− cells (Figure S7C), which may in part account for the diminished association of Jarid2 with targets.
Jarid2 Knockdown Results in Derepression of PRC2 Target Genes in ES Cells
Loss of PRC2 components in ES cells results in upregulation of PRC2 target genes (Boyer et al., 2006; Pasini et al., 2007). To test whether Jarid2 knockdown leads to a similar effect, we assayed mRNA expression levels of selected PRC2 target genes in ES cell lines expressing Jarid2 or non-silencing shRNAs. Whereas Oct4 mRNA levels were comparable among all shRNA lines, expression of interrogated PRC2 target genes was upregulated upon Jarid2 knockdown (Figure 5D). Similarly, expression of PRC2 target genes was also upregulated in Jarid2 −/+ E14 cells, as compared to wt E14 and LF2 ES lines (Figure 5E).
Jarid2 Negatively Regulates PRC2 Enzymatic Activity
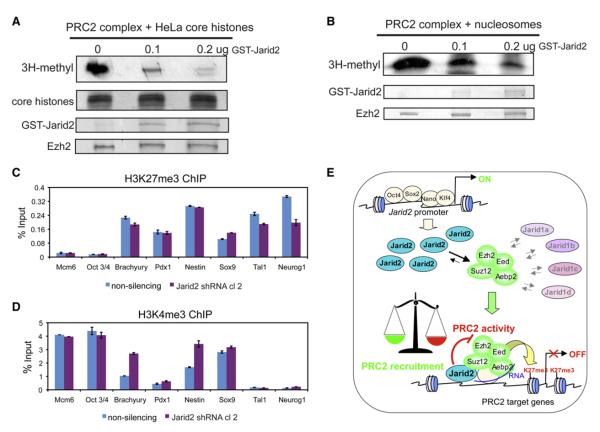
To test whether, in addition to targeting, Jarid2 can also regulate PRC2 enzymatic activity, we performed histone methyltransferase (HMT) assays with purified reconstituted recombinant PRC2 complex either in the absence or in the presence of the recombinant full-length Jarid2, and using native core HeLa histones as a substrate. Addition of sub-stoichiometric amounts of Jarid2 was sufficient to inhibit PRC2 HMT activity in a dose-dependent manner (Figure 6A), while addition of other recombinant chromatin proteins had no effect (Figure S8A). Similarly, Jarid2 inhibited PRC2 HMT activity on nucleosomal substrates (Figure 6B). However, addition of Jarid2 to H3K4 methyltransferase complexes purified via the Ash2 core subunit had no effect on its HMT activity (Figure S8B). Moreover, Jarid2 stimulated HMT activity of recombinant, purified H3K9 methyltransferase G9a (Figure S8C), which was previously shown to bind Jarid2 (Shirato et al., 2009). In sum, these results demonstrate that Jarid2 specifically inhibits PRC2 HMT activity in vitro and suggest that it plays diverse roles in regulation of histone methyltransferases in distinct cellular contexts. As HMT assays were performed in the absence of cofactors other than S-adenosyl-methionine (SAM), Jarid2-mediated inhibition of PRC2 HMT likely occurs through a nonenzymatic mechanism. Interestingly, we found that recombinant Jarid1a also inhibited PRC2 HMT activity in vitro (Figure S8D), indicating that other Jarid family members may regulate PRC2 enzymatic function.
Figure 6. Jarid2 Directly Inhibits PRC2 Histone Methyltransferase Activity.
(A) Jarid2 inhibits PRC2 HMT activity on core histones. Purified reconstituted PRC2 complex was pre-bound to 0, 0.1, and 0.2 ug of purified recombinant Jarid2 and used in HMT assays with native HeLa core histones as substrates and tritium-labeled S-adenosyl-methionine (3H-SAM) as a cofactor. 3H-methyl incorporation was visualized by fluorography (top panel). Histones, Jarid2 and Ezh2 present in each reaction were visualized by Coomasie staining, allowing for direct comparisons of the relative protein levels (bottom panels).
(B) Jarid2 inhibits PRC2 HMT activity on nucleosomes. Reactions were performed as in (A) except that in vitro assembled recombinant nucleosomal templates were used as substrates.
(C) H3K27me3 levels at selected PRC2 target genes are not significantly affected by Jarid2 knockdown. ChIP-qPCR analysis of H3K27me3 levels in mouse ES line expressing Jarid2 shRNA (Jarid2 shRNA cl. 2) versus non-silencing shRNA. We verified that histone recognition by H3K27me3 antibody is PRC2 dependent (Figure S8E).
(D) H3K4me3 levels at selected PRC2 target genes in Jarid2 shRNA ES cells. ChIP-qPCR analysis of H3K4me3 levels in clonal mouse ES line expressing Jarid2 shRNA (Jarid2 shRNA cl. 2) versus non-silencing shRNA.
(E) Model of Jarid2 in modulation of PRC2 recruitment and enzymatic activity. In ES cells, Jarid2 gene is under control of ES regulatory circuitry, resulting in high levels of Jarid2 expression and preferential formation of Jarid2-PRC2 complexes. Jarid2 simultaneously promotes the recruitment and inhibits the enzymatic activity of PRC2 (indicated as a balance). PRC2 targeting is mediated via a combinatorial mechanism involving Jarid2, other proteins and non-coding RNAs.
Next, we studied the effects of Jarid2 downregulation on H3K27me3 levels in vivo. Although a modest reduction of Jarid2 levels achieved in our ES lines is sufficient to reduce Ezh2 target occupancy by 2-6 fold (Figure 5A), we observed little to no effect on H3K27me3 levels at the same gene targets (Figure 6C). For example, the Sox9 gene exhibited the strongest relative downregulation of Ezh2 occupancy, yet a slight upregulation of H3K27me3 levels (Figure 6C), suggesting that the Jarid2-containing PRC2 complex is less active. Interestingly, despite unaffected H3K27me3 levels, expression of PRC2 target genes was upregulated upon Jarid2 knockdown (Figure 5D), and this upregulation correlated with a modest increase in H3K4me3 levels (Figure 6D). Our results demonstrate that Jarid2 negatively regulates PRC2 HMT activity.
Jarid2 Depletion Results in Gastrulation Defects in Xenopus Embryos
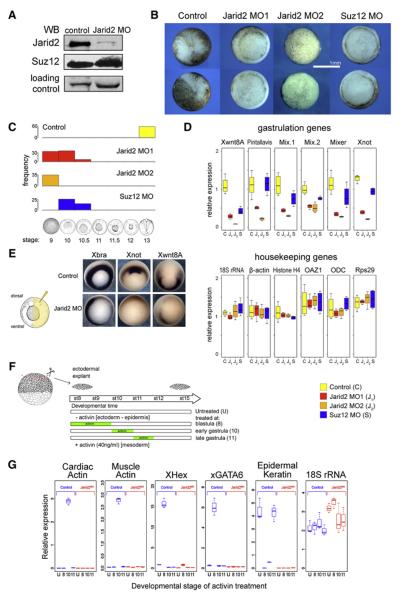
Jarid2 promotes PRC2 recruitment to the target genes while inhibiting PRC2 enzymatic activity, suggesting that it modulates PRC2 function at developmental genes, perhaps to sensitize them for subsequent activation during differentiation (Figure 6E). To address whether Jarid2 is important for gene regulation during early embryogenesis, we downregulated Jarid2 levels in Xenopus laevis embryos by injecting morpholino oligonucleotides targeting the translation start sites of both non-allelic copies of X. laevis Jarid2 (Jarid2 MO1). Jarid2 amino acid sequence and domain composition is highly conserved between frogs and mammals (Figure S9).
Immunoblot analysis of embryonic extracts showed that Jarid2 MO1 injection resulted in the downregulation of Jarid2, without affecting Suz12 protein levels (Figure 7A). Jarid2 MO1 injected embryos exhibited gastrulation arrest, whereas control embryos proceeded to develop normally and were assayed at the neurula stage (Figure 7B; phenotype penetrance in Figure 7C). To ensure that the observed phenotype does not result from off-target effects, we designed two additional translation-blocking MOs, each matching one of the two non-allelic Jarid2 copies (Jarid2 MO2a and Jarid2 MO2b). Injection of either of the MOs resulted in phenocopy of Jarid2 MO1 phenotype. When co-injected together in equimolar amounts (referred to as Jarid2 MO2) at 2-4 fold lower concentration than either Jarid2 MO1, Jarid2 MO2a, or Jarid2 MO2b alone, these two MOs showed a strong synergistic effect resulting in 100% penetrant developmental arrest at the late blastula stage (Figure 7B; quantified in Figure 7C, protein knockdown verified in Figure S10A). Injection of Suz12 MO also resulted in arrest prior to completion of gastrulation (Figure 7B and C), consistent with gastrulation defects reported in Suz12 −/− mice (Pasini et al., 2004). In contrast, injection of MOs targeting H3K4 methyltransferase MLL1 or Jarid1a had no effects on gastrulation (not shown).
Figure 7. Jarid2 Is Required for Induction of Gastrulation Programs in Xenopus Embryos.
(A) Knockdown of Jarid2 protein in Xenopus embryos. Immunoblotting analysis of nuclear extracts from gastrula stage embryos derived from control or Jarid2 MO1-injected two-cell stage embryos.
(B) Downregulation of Jarid2 or Suz12 results in gastrulation arrest. Representative images of control, Jarid2 MO1-, Jarid2 MO2-, and Suz12 MO- treated embryos developed from the same batch of zygotes. Control embryos completed gastrulation and proceeded to neuru-late (imaged at stage 13), whereas Jarid2 MO1, Jarid2 MO2, and Suz12 MO embryos arrested at the late blastula stage or during early gastrulation (stages 9-10.5).
(C) Penetrance of gastrulation defects shown in (B).
(D) Impaired induction of gastrulation genes in Jarid2 MO1, Jarid2 MO2, and Suz12 MO embryos. RT-qPCR analyses of mRNA levels in control (C), Jarid2 MO1-treated (J1), Jarid2 MO2-treated (J2) and Suz12 MO-treated (S) late blastula stage embryos. mRNA levels of selected gastrulation genes (top panel) and housekeeping genes (bottom panel) are shown.
(E) Jarid2 knockdown perturbs germ layer formation. Whole mount in situ detection of Xbra, Xnot, and Wnt8A transcripts in gastrula stage embryos derived from control or embryos asymmetrically injected with Jarid2 MO1 into one blastomere at the two-cell stage.
(F) Schematic diagram of ectodermal explant assay. Ectodermal explants were isolated from the animal poles of early blastula embyos (stage 7) and cultured in the absence (“U”) or in presence of Activin A during: blastula stages 8 and 9 (“8”), early gastrulation between stages 10 and 11 (“10”), or late gastrulation between stages 11 and 12 (“11”). At developmental stage 15, explants were harvested for quantitative RT-qPCR assay.
(G) Impaired mesoderm induction in response to Activin in Jarid2 MO explants. Results of RT-qPCR gene expression analysis of indicated genes relative to the whole sibling embryo at the same stage. Blue and red boxes indicate control and Jarid2 MO treated explants, respectively. In panels D and G the central line of the box and whisker plot represent median value, the hinges approximate quartiles and the whiskers extremal values (minimum and maximum).
Jarid2 Is Required for Activation of Gastrulation Gene Expression Program
In Xenopus embryos, genes necessary for orchestrating gastrulation events are induced during the mid- and late blastula stage. The late blastula stage corresponds to the critical transition period during which cells exit pluripotency and restrict their developmental potential, but have not yet differentiated to form three germ layers. In agreement with a recent report (Akkers et al., 2009), H3K27me3 levels at selected early differentiation genes significantly increased during the transition from mid- to late blastula stage (Figure S10B and C, compare % input recovery scales on y axis in B and C). Importantly, Jarid2 knockdown resulted in further upregulation of H3K27me3, evident particularly at the late blastula stage (Figure 10B and C) and consistent with repression of PRC2 HMT activity by Jarid2.
To assay the effects of Jarid2 downregulation on gene expression, we performed quantitative RT-qPCR analyses from late blastula stage embryos, either untreated or injected with Jarid2 MO1, Jarid2 MO2 or Suz12 MO. Expression of genes involved in germ layer formation was downregulated by all assayed MOs (Figure 7D), although generally Jarid2 MOs displayed a more prominent effect than Suz12 MO. In contrast, analysis of a cohort of non-developmental genes revealed no significant effect on expression (Figure 7D), indicating that MO treatment did not cause a global transcriptional failure. Furthermore, whole-mount RNA in situ hybridization analysis of gastrula embryos asymmetrically injected with Jarid2 MO1 at the two-cell stage (resulting in Jarid2 depletion in half of the embryo) revealed diminished expression of gastrulation genes Brachyury, Wnt8 and Xnot on the MO-injected side of the embryo (Figure 7E). In sum, our observations indicate that Jarid2 function is essential for induction of gastrulation programs at the exit from pluripotency.
Jarid2 Knockdown Results in Failure to Induce Mesoderm in Response to Activin Signaling
Xenopus ectodermal explants isolated from blastula stage embryos can be induced in vitro to form mesoderm by treatment with Activin, a ligand for Nodal signaling, but the competence to induce mesoderm is lost by the end of gastrulation (Kimelman, 2006).
To test whether Jarid2 depletion affects induction of mesodermal genes or, alternatively, results in the delayed or prolonged competence for induction, we isolated ectodermal explants from the early blastula stage control or Jarid2 MO-treated embryos, cultured them either in the absence of Activin or in the presence of Activin pulse at blastula, early gastrula or late gastrula stages, respectively, and analyzed gene expression by RT-qPCR (Figure 7F). Control explants not exposed to Activin established a default epidermal fate, as evidenced by the expression of epidermal Keratin. Activin treatment of control explants at blastula stages resulted in the induction of the mesodermal markers XHex and GATA6 and suppression of the default epidermal fate, whereas treatment at early gastrula stages lead to the induction of cardiac and muscle actin, again accompanied by the suppression of the epidermal fate (Figure 7G). The competence for mesoderm induction was lost by late gastrula stages.
In contrast, Jarid2 depleted embryos failed to induce mesodermal markers in response to Activin treatment at any of the assayed stages, suggesting a differentiation defect rather than altered timing of developmental competence. Remarkably, Jarid2 depleted explants also failed to realize their default developmental potential as evidenced by the absence of epidermal Keratin expression (Figure 7G), indicating that Jarid2 depletion results in differentiation failure.
DISCUSSION
The work presented here provides important insights into the regulation of PRC2 recruitment and activity in ES cells and early embryos, illuminates a novel mechanism of histone methylation regulation by Jarid proteins and suggests that JmjC domain proteins exhibit important, but previously unappreciated functions outside of their activities as histone demethylases.
Jarid2 Function in PRC2 Targeting
Jarid2 contains a DNA-binding ARID domain and was shown to associate with DNA-binding proteins (Kim et al., 2004), suggesting a role for Jarid2-mediated DNA recognition in PRC2 recruitment. Our de novo sequence motif analysis identified a CCG-repeat motif as highly overrepresented among Jarid2-PRC2 targets. Interestingly, the ARID domain of another Jarid family member preferentially binds CCG-rich DNA (Tu et al., 2008). Perhaps the ARID domain of Jarid2 exhibits similar specificity and accounts for the previously reported preferential association of PRC2 with CpG-island rich promoters (Ku et al., 2008).
Alternative to a direct role in DNA binding, Jarid2 may coordinate assembly of the higher order PRC2 complex, competent for proper targeting. Regardless of the exact mechanism, we favor the view that in vertebrate organisms the nature of PRC2 recruitment is combinatorial and tissue-specific, with PRC2 complex subunits and/or additional PRC2-associated protein and RNA cofactors cooperate with Jarid2 in ES cells and early embryos (Figure 6E). For example, a PRC2 subunit Aebp2, YY1 and long non-coding RNAs have all been suggested to contribute to PRC2 targeting in mammalian cells (Kim et al., 2009; Simon and Kingston, 2009).
Jarid2 Coordinates Regulation of PRC2 Recruitment and Enzymatic Activity
Although Jarid2 promotes PRC2 targeting and its binding is tightly correlated with H3K27me3 distribution genome-wide, counter-intuitively, Jarid2 represses PRC2 enzymatic activity. Perhaps such fine calibration of PRC2 functions is necessary to maintain a repressed, yet poised state of developmental genes and to permit subsequent gene activation during differentiation. Indeed, depletion of Jarid2 in early Xenopus embryos results in failure of differentiation and developmental gene induction, concomitant with accumulation of H3K27me3.
A modest downregulation of Jarid2 in ES cells is sufficient to diminish PRC2 target gene occupancy, leading to derepression of PRC2 target genes, despite relatively unaffected H3K27me3 levels. This observation suggests that the Jarid2-PRC2 complex can silence gene expression in part independently of H3K27me3.
Jarid Family Proteins as PRC2 Modulators
Both Jarid2 and Jarid1a directly inhibit PRC2 HMT activity in vitro in the absence of cofactors other than SAM, and in particular in the absence of cofactors required for histone demethylation, implying a nonenzymatic mechanism of inhibition. Intriguingly, Jarid1a, an active H3K4me3 demethylase (Christensen et al., 2007; Klose et al., 2007), binds genomic regions enriched for H3K4me3. Thus, perhaps in certain biological contexts Jarid1a regulates both H3K4 and H3K27 methylation levels, albeit via distinct molecular mechanisms.
The conservation of Suz12 binding motif suggests that association with PRC2 is a common feature of Jarid family proteins. In ES cells, where the Jarid2 gene is under control of the pluripotency circuitry and approximately 50,000 Jarid2 molecules are present in the nucleus of each cell, the complex formation equilibrium is shifted toward Jarid2-PRC2 (Figure 6E). Nevertheless, the observation that Jarid1a and Jarid2-PRC2 exhibit virtually non-overlapping genomic binding patterns indicates that additional mechanisms beyond Suz12 recognition must regulate target specificity of Jarid-PRC2 complexes.
Jarid2 Is a Key Regulator of Vertebrate Development
Jarid2 is important for proper organogenesis of tissues derived from all three germ layers, including brain, heart and liver [reviewed in (Takeuchi et al., 2006)]. Jarid2 gene trap homozygous mice gastrulate normally but show defects in neural plate formation and neural tube closure (Takeuchi et al., 1995), resulting in the cruciform-like structure (hence the name “Jumonji,” Japanese for cruciform). Although these observations underscore a critical role of Jarid2 in development, they differ from our observation that Jarid2 is essential for gastrulation in Xenopus. The discrepancy may be explained in several ways: (i) gene trap embryos partially retain Jarid2 function, (ii) maternal Jarid2 transcripts provide a significant contribution (translation of frog maternal transcripts is blocked by MO injection), (iii) other Jarid proteins complement Jarid2 more efficiently in mice, (iv) Jarid2 is not important for early development in mice. Although at present we cannot distinguish among the aforementioned possibilities, the last scenario is unlikely: regulation of Jarid2 transcription by multiple components of ES transcriptional circuitry in mice and humans indicate that an evolutionary pressure existed in mammals to preserve Jarid2 function in the early embryo. Moreover, Jarid2 downregulation affects the induction of differentiation genes during embryoid body formation from mouse ES cells (J.P. and J.W., unpublished data).
The Role of Jarid2-PRC2 during the Transition from Pluripotent to Restricted Cell Fates
Elegant transplantation experiments demonstrated that cells of early blastula stage Xenopus embryos remain pluripotent (Heasman et al., 1984; Snape et al., 1987). This multilineage differentiation potential becomes gradually restricted in late blastula embryos when cells exit pluripotency, culminating in the first differentiation events during gastrulation (Heasman et al., 1984; Snape et al., 1987).
We demonstrated that H3K27me3 levels at developmental genes in X. laevis embryos increase by an order of magnitude just prior to the onset of gastrulation; this temporal regulation of H3K27me3 was also reported in X. tropicalis (Akkers et al., 2009). Jarid2 depletion results in a further aberrant increase of H3K27me3 levels, concomitant with the failure of developmental gene induction in the late blastula embryos. Thus, Jarid2-mediated modulation of PRC2 activity at the exit from pluripotency appears critical for the subsequent execution of developmental programs.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies used in this study are listed in Table S4.
cDNAs
Primers used for cloning are described in Table S7.
Cell Lines
Please see Supplemental Experimental Procedures.
Protein Extraction and Immunoprecipitation
Nuclear extracts were prepared from ES cells according to the Dignam protocol (Dignam et al., 1983). For FLAG IP, nuclear extracts were incubated with M2 FLAG agarose (Sigma), beads were washed and eluted with 0.25mg/ml 3xFLAG peptides (Sigma) in PBS 0.2% Triton X-100.
Glycerol Gradients
4.5 ml 25%–50% glycerol density gradient was prepared using a Hoefer SG15 gradient maker. Eed-FLAG eluates were laid on top of each gradient and separated by 40,000 RPM centrifugation for 3 hr in a SW50.1 rotor (Beckman); 180 ul fractions were collected.
HPLC/MS/MS Analysis
HPLC/MS/MS analysis was performed with the Agilent 1100 nanoflow liquid chromatography system and a LTQ two-dimensional ion trap mass spectrometer equipped with a nanoelectrospray ionization source. Database search was conducted with Mascot 2.1 and the NCBI nr database. Details can be found in the Supplemental Experimental Procedures.
Chromatin Immunoprecipitation, ChIP-qPCR, and ChIP-seq Library Preparation
ChIP from ES cells and Xenopus embryos was performed according to Boyer et al. (2005) and Blythe et al. (Blythe et al., 2009), respectively, using 100–500 ug of chromatin per IP. ChIP-qPCR signals were calculated as % of input. Primer sequences are included in Table S5. For sequential ChIP, 8 mg of chromatin prepared from Eed-FLAG cells were incubated with M2 agarose, and Eed-FLAG chromatin was eluted with 3XFLAG peptide. The eluted Eed-FLAG chromatin was subsequently immunoprecipitated with Suz12 and Jarid2 antibodies using the standard ChIP protocol. ChIP-seq libraries were prepared according to Illumina protocol and sequenced using Illumina Genome Analyzer; 26–36 base-pair-long reads were obtained.
ChIP-seq Data Analysis
All sequences (including those from Ku et al., 2008 and Mikkelsen et al., 2007) were mapped by ELAND software (Illumina Inc) and analyzed by QuEST 2.4 software (Valouev et al., 2008) using a “histone” or “punctate binding site” setting (bandwidth of 100 bp, region size of 1000 bp) and default stringency (corresponding to 50-fold ChIP to input enrichment for seeding the regions, and 3-fold ChIP enrichment for extending the regions).
ChIP-seq Data Availability
All ChIP-Seq datasets have been deposited into GEO repository (GSE18776).
shRNA Lentivirus Infection and Selection
The Jarid2 shRNAmir construct was purchased from Open Biosystems (Cat #:RMM4431-98858628) and subcloned into the pTRIPZ Tet-on inducible vector. Verified nonsilencing shRNA (RHS4335) were purchased from Open Biosystems. VSVG pseudotyped lentiviruses were produced in 293T cells using standard procedure. Selection was done by 1.5ug/ml puromycin for 2 weeks and clones screened by RFP expression after 4-day doxycyclin (1ug/ml) induction.
Morpholino Design
Translation-blocking morpholinos designed to Xenopus laevis Jarid2 (Jarid2 MO1 5′ TCCTCTTGGGCCTTTCCTTGCTCAT 3′; Jarid2 MO2a 5′ GGCCTTTC CTTGCTCATCCTGTTAT 3′ and Jarid2 MO2b 5′ GGCCTTTCCTTGCTCAT CCTGTTCT 3′) and Suz12 (Suz12 MO 5′ CCATGCGGGATACTACGAGTGA TAA 3′) were obtained from Gene Tools LLC (http://www.gene-tools.com) and resuspended at 1mM.
Additional details can be found in the Supplemental Experimental Procedures.
Supplementary Material
S1
Table S1
Table S3
ACKNOWLEDGMENTS
Authors thank E. Benevolenskaya for Jarid1a antibodies; G. Narlikar for nucleosomes; Y. Zhang and Y. Shi, for the PRC2 and Jarid1a expression constructs; A. Vonica for the Xbra cDNA; and E. Heard (LF2), T. Magnuson (Eed−/−), and B. Panning (E14) for ES cell lines. We thank Z. Weng, P. Lacroute, and Stanford Pathology/Genetics UHT Sequencing Initiative for sequencing ChIP libraries; A. Sun for help with Eed-FLAG line derivation; R. Bajpai and Z. Ma for advice on ES culture and HMT assay, respectively. We thank members of the Wysocka lab for discussions; and E. Duncan, E. Grow, Z. Ma, A. Ring, A. Rada, and A. Roos for comments on the manuscript. This work was supported by the CIRM New Faculty (RN1005791), W.M. Keck Distinguished Young Scholar, and Searle Scholar Awards for J.W., American Cancer Society Postdoctoral Fellowship (PF0816001DDC) for J.C.P., and NIH R01DK082664 for Y.Z.
Footnotes
REFERENCES
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Francoijs KJ, Stunnenberg HG, Veenstra GJ. A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Dev. Cell. 2009;17:425–434. doi: 10.1016/j.devcel.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Cerecedo D, Tondeur S, Pantesco V, Hovatta O, Klein B, Hamamah S, De Vos J. A gene expression signature shared by human mature oocytes and embryonic stem cells. BMC Genomics. 2009;10:10. doi: 10.1186/1471-2164-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Blythe SA, Reid CD, Kessler DS, Klein PS. Chromatin immunoprecipitation in early Xenopus laevis embryos. Dev. Dyn. 2009;238:1422–1432. doi: 10.1002/dvdy.21931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PAC, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 Belongs to a Family of Demethylases, Specific for Tri-and Dimethylated Lysine 4 on Histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davisson M. FIMRe: Federation of International Mouse Resources: global networking of resource centers. Mamm. Genome. 2006;17:363–364. doi: 10.1007/s00335-006-0001-2. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust C, Lawson KA, Schork NJ, Thiel B, Magnuson T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development. 1998;125:4495–4506. doi: 10.1242/dev.125.22.4495. [DOI] [PubMed] [Google Scholar]
- Heasman J, Wylie CC, Hausen P, Smith JC. Fates and states of determination of single vegetal pole blastomeres of X. laevis. Cell. 1984;37:185–194. doi: 10.1016/0092-8674(84)90314-3. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic Acids Res. 2009;37:2940–2950. doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TG, Chen J, Sadoshima J, Lee Y. Jumonji represses atrial natriuretic factor gene expression by inhibiting transcriptional activities of cardiac transcription factors. Mol. Cell. Biol. 2004;24:10151–10160. doi: 10.1128/MCB.24.23.10151-10160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimelman D. Mesoderm induction: from caps to chips. Nat. Rev. Genet. 2006;7:360–372. doi: 10.1038/nrg1837. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Yan Q, Tothova Z, Yamane K, Erdjument-Bromage H, Tempst P, Gilliland DG, Zhang Y, Kaelin WG., Jr. The Retinoblastoma Binding Protein RBP2 Is an H3K4 Demethylase. Cell. 2007;128:889–900. doi: 10.1016/j.cell.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Kortschak RD, Tucker PW, Saint R. ARID proteins come in from the desert. Trends Biochem. Sci. 2000;25:294–299. doi: 10.1016/s0968-0004(00)01597-8. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr. Opin. Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, DeWaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. Genome-wide Analysis of the H3K4 Histone Demethylase RBP2 Reveals a Transcriptional Program Controlling Differentiation. Mol. Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Cao R, Zhang Y. Substrate Preferences of the EZH2 Histone Methyltransferase Complex. J. Biol. Chem. 2006;281:8365–8370. doi: 10.1074/jbc.M513425200. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin-Kensicki EM, Faust C, LaMantia C, Magnuson T. Cell and tissue requirements for the gene eed during mouse gastrulation and organogenesis. Genesis. 2001;31:142–146. doi: 10.1002/gene.10017. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol. Cell. Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y, Takeuchi T. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J. Biol. Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of Polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009 doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Snape A, Wylie CC, Smith JC, Heasman J. Changes in states of commitment of single animal pole blastomeres of Xenopus laevis. Dev. Biol. 1987;119:503–510. doi: 10.1016/0012-1606(87)90053-4. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li H, Liu Y, Mattson MP, Rao MS, Zhan M. Evolutionarily conserved transcriptional co-expression guiding embryonic stem cell differentiation. PLoS ONE. 2008;3:e3406. doi: 10.1371/journal.pone.0003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev. Dyn. 2006;235:2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Yamazaki Y, Katoh-Fukui Y, Tsuchiya R, Kondo S, Motoyama J, Higashinakagawa T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995;9:1211–1222. doi: 10.1101/gad.9.10.1211. [DOI] [PubMed] [Google Scholar]
- Tu S, Teng YC, Yuan C, Wu YT, Chan MY, Cheng AN, Lin PH, Juan LJ, Tsai MD. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat. Struct. Mol. Biol. 2008;15:419–421. doi: 10.1038/nsmb.1400. [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat. Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:16438–16443. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
S1
Table S1
Table S3