miR-328 Functions as an RNA Decoy to Modulate hnRNP E2 Regulation of mRNA Translation in Leukemic Blasts (original) (raw)
. Author manuscript; available in PMC: 2010 Aug 20.
SUMMARY
MicroRNAs and heterogeneous ribonucleoproteins (hnRNPs) are posttranscriptional gene regulators that bind mRNA in a sequence-specific manner. Here, we report that loss of miR-328 occurs in blast crisis chronic myelogenous leukemia (CML-BC) in a BCR/ABL dose- and kinase-dependent manner through the MAPK-hnRNP E2 pathway. Restoration of miR-328 expression rescues differentiation and impairs survival of leukemic blasts by simultaneously interacting with the translational regulator poly(rC)-binding protein hnRNP E2 and with the mRNA encoding the survival factor PIM1, respectively. The interaction with hnRNP E2 is independent of the microRNA’s seed sequence and it leads to release of CEBPA mRNA from hnRNP E2-mediated translational inhibition. Altogether, these data reveal the dual ability of a microRNA to control cell fate both through base pairing with mRNA targets and through a decoy activity that interferes with the function of regulatory proteins.
INTRODUCTION
Dysfunctional posttranscriptional gene regulation by sequence-specific RNA-binding proteins (RBPs) plays a critical role in the pathogenesis and evolution of human diseases (Carpenter et al., 2006; Glisovic et al., 2008; Keene, 2007), including the blastic phase of chronic myelogenous leukemia (Melo and Barnes, 2007; Perrotti and Neviani, 2007). In this disease stage, increased activity of the BCR/ABL oncoprotein alters processing, export, and/or translation of mRNAs encoding tumor suppressors, oncoproteins, and critical regulators of differentiation by aberrantly modulating the expression and function of RBPs with sequence-specific mRNA-binding activity (Eiring et al., 2008; Perrotti and Neviani, 2007). Differentiation arrest in myeloid blast crisis chronic myelogenous leukemia (CML-BC) depends on the BCR/ABL-MAPK-induced activity of hnRNP E2 (Chang et al., 2007; Perrotti et al., 2002), a poly(rC)-binding protein that controls translation of specific mRNAs (Ostareck-Lederer and Ostareck, 2004). In CML-BC, but not chronic phase (CML-CP) CD34+ bone marrow (BM) progenitors, hnRNP E2 is highly expressed and, upon interaction with the C-rich element in the 5′ untranslated region (UTR) of CEBPA mRNA, suppresses translation (Chang et al., 2007; Perrotti et al., 2002) of this master regulator of myeloid differentiation (Tenen, 2003).
hnRNP E2 RNA-binding activity does not require posttranslational modifications, and its expression is transiently induced by cytokines (e.g., IL-3) in myeloid precursors (Chang et al., 2007). Nonetheless, the early stages of myelopoiesis are characterized by cytokine-dependent proliferation and C/EBPα-mediated differentiation, suggesting that the hnRNP E2:CEBPA interaction might be prevented by a hnRNP E2-interacting protein or RNA, or by another RBP interacting with the CEBPA 5′UTR. Because small noncoding RNAs are regulators of critical cell functions including normal and leukemic hematopoiesis (Chen et al., 2004; Garzon et al., 2006), we hypothesized that differentiation arrest of myeloid CML-BC blasts results from impaired expression of a microRNA (miRNA) that, in addition to exerting its gene silencing activity through canonical binding to mRNA regulatory regions (Bartel, 2009; Friedman et al., 2009), also strongly competes with CEBPA mRNA for binding to hnRNP E2. Here, we report the existence of miRNA-dependent posttranslational control of biological processes through both base pairing with complementary mRNAs and sequence-dependent interference with the mRNA-regulatory function of RNA-binding proteins (decoy activity).
RESULTS
BCR/ABL-Dependent Downregulation of miR-328 Expression
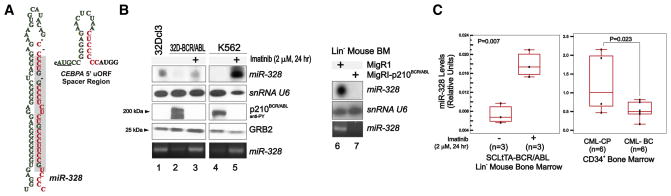
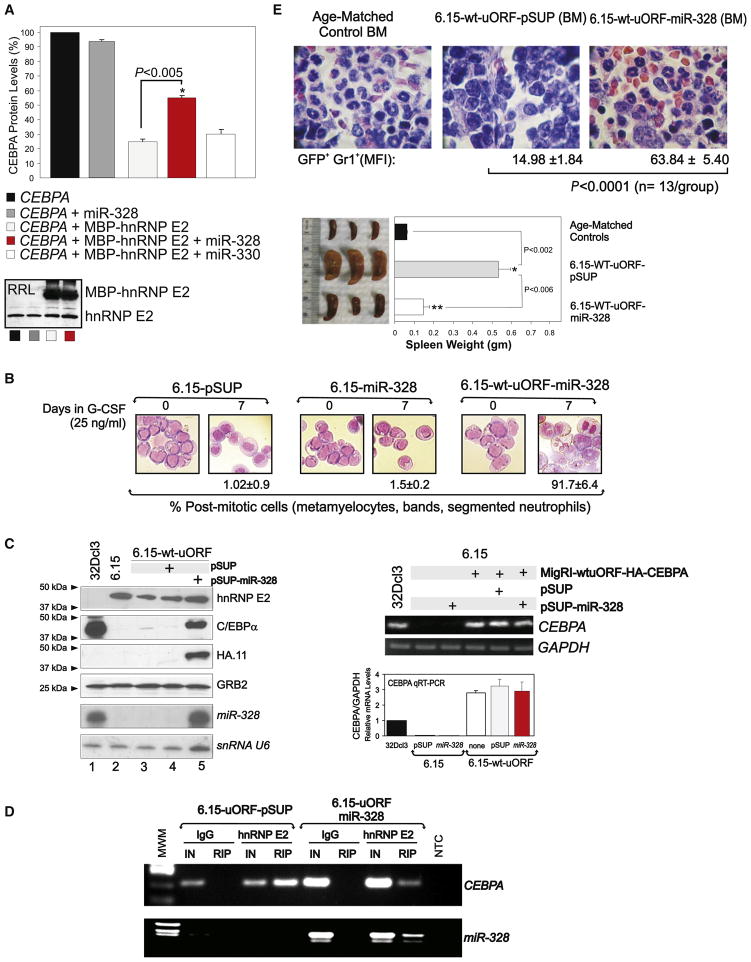
Comparative miRNA arrays revealed that a discrete number of miRNAs were more than 3-fold modulated in a BCR/ABL dose-and kinase-dependent manner in cell line models and patient-derived myeloid CML-BCCD34+ versus CML-CPCD34+ BM progenitors (not shown). Among the miRNAs downregulated in CML-BCCD34+, we focused on miR-328 because its mature form harbors a C-rich element that resembles the negative regulatory hnRNP E2-binding site contained in the CEBPA intercistronic mRNA region (Figure 1A).
Figure 1. miR-328 Is Downregulated in CML-BC.
(A) hnRNP E2 consensus binding sites (red) in the precursor and mature (gray box) miR-328 and in the CEBPA uORF/spacer region.
(B) Left: Northern blot and RT-PCR show miR-328 levels and western blot shows BCR/ABL levels in 32Dcl3 and untreated or imatinib-treated 32D-BCR/ABL cells; right: northern blot and RT-PCR show miR-328 levels in vector- and BCR/ABL-infected lineage-negative (Lin−) mouse BM cells.
(C) qRT-PCR shows miR-328 levels in untreated (n = 3) and imatinib-treated (n = 3) Lin− BM cells from leukemic SCLtTA-BCR/ABL mice (left) and in CD34+ BM progenitors from CML-CP (n = 6) and CML-BC (n = 6) patients (right). snRNA U6 and GRB2 levels were used as loading controls (mean ± standard error of the mean [SEM]).
Northern blot and qRT-PCR analyses confirmed that miR-328 downregulation is markedly induced by BCR/ABL expression in 32Dcl3 myeloid precursors (32D-BCR/ABL) and primary lineage-negative (Lin−) mouse BM cells (Figure 1B). Reduction of miR-328 expression depends on BCR/ABL kinase activity as imatinib treatment (2 μM; 24 hr) significantly increased miR-328 expression in 32D-BCR/ABL and K562 cells (Figure 1B), as well as in Lin− BM cells from leukemic SCLtTA-BCR/ABL mice (n = 3/group) (Figure 1C). As expected, imatinib did not rescue miR-328 levels in imatinib-resistant BCR/ABL(T315I) cells (not shown). Finally, miR-328 expression was also significantly reduced in myeloid CML-BCCD34+ (n = 6) compared to CML-CPCD34+ (n = 6) BM patient cells (Figure 1C), suggesting its possible involvement in blastic transformation.
miR-328 Competes with CEBPA mRNA for Binding to hnRNP E2
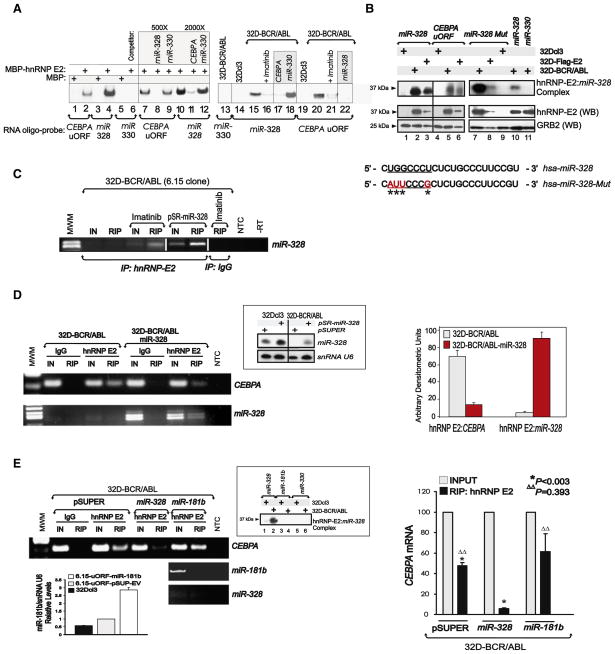
To determine whether the C-rich element present in mature miR-328 is a bona fide hnRNP E2-binding site, RNA electrophoretic mobility shift assays (REMSA), UV crosslinking, and RNA immunoprecipitation (RIP) assays were performed using maltose-binding protein (MBP)-tagged hnRNP E2 (MBP-hnRNP E2), 32D-BCR/ABL cytoplasmic cell lysates expressing high levels of hnRNP E2 (Perrotti et al., 2002), and/or lysates of 32Dcl3 and 32D-BCR/ABL cells expressing a Flag-tagged hnRNP E2. REMSA (Figure 2A and Figure S1A available online) and UV crosslinking (Figure 2B) revealed that a 32P-labeled miR-328 oligoribonucleotide efficiently formed a complex with recombinant, endogenous, and ectopic hnRNP E2 proteins but not with MBP alone or lysates of 32Dcl3 cells. Accordingly, the mobility of hnRNP E2:miR-328 complexes was identical to that formed using the uORF/spacer region of CEBPA mRNA (CEBPA uORF) oligoribonucleotide (Figures 2A and 2B), and miR-328 binding to hnRNP E2 was four times stronger than that of CEBPA uORF (Figure 2A). By contrast, the BCR/ABL-regulated miR-330, which is not C-rich, neither interacted with hnRNP E2 nor disturbed hnRNP E2:miR-328 complex formation (Figures 2A and 2B). miR-328 and CEBPA uORF binding to endogenous hnRNP E2 was also impaired by imatinib (Figure 2A) that, reportedly, inhibits hnRNP E2 expression in BCR/ABL+ cells (Chang et al., 2007).
Figure 2. miR-328 Competes with CEBPA mRNA for Binding to hnRNP E2.
(A) REMSA shows binding of miR-328, CEBPA uORF, and miR-330 RNA probes to MBP-tagged hnRNP E2 (lanes 2, 4, 6, 7–12), MBP (lanes 1, 3, 5), and cytoplasmic lysates of 32Dcl3 (lanes 14, 19) and untreated or imatinib-treated 32D-BCR/ABL cells (lanes 13, 15–18, 20–22). Cold competitor RNAs were used as indicated (lanes 8–9, 11–12, 17–18, 22).
(B) Top: UV crosslinking shows binding of miR-328 (lanes 1–3 and 10), CEBPA uORF (lanes 4–6), the seed sequence-mutated miR-328 (miR-328-Mut) (lanes 7–9), and miR-330 (lane 11) to hnRNP E2 in 32Dcl3, 32D-BCR/ABL, and 32D-Flag-E2 cell lysates (lanes 1–6). Bottom: Western Blots show levels of hnRNP E2 and GRB2 in the lysates used in UV crosslinking. Sequence of the wild-type and seed sequence-mutated miR-328 with substituted nucleotides indicated by asterisks.
(C) RNA Immunoprecipitation (RIP) assay for miR-328 performed on anti-hnRNP E2 immunoprecipitates (IPs) from lysates of untreated (lane 3), imatinib-treated (lane 5), and pSR-miR-328-transduced (lane 7) 32D-BCR/ABL (6.15 clone) cells. RIP with a nonrelated IgG served as controls (lane 8). IN: input RNA; MWM: molecular weight marker (lane 1); NTC: nontemplate control;-RT: no reverse transcribed PCR reaction. RIP was also observed with ectopically expressed Flag-hnRNP E2 proteins (see Figure S1).
(D) Left: RIP assays for CEBPA mRNA (top) and miR-328 (bottom) performed on anti-hnRNP E2 (lanes 5 and 9) and nonrelated IgG (lanes 3 and 7) IPs from parental (lanes 2–5) and miR-328-expressing (lanes 6–9) 32D-BCR/ABL cells. IN: RNA input; MWM: molecular weight marker (lane 1); NTC: nontemplate PCR control. hnRNP E2 RIP assays for a nonrelated mRNA (i.e., SET) are reported in Figure S1. Inset top right: Northern blot shows levels of ectopic miR-328 in vector- (lanes 1 and 3) and miR-328-transduced (lanes 2 and 4) 32Dcl3 and 32D-BCR/ABL cells. snRNA U6 was analyzed for normalization. Right: Densitometric analyses of the levels of CEBPA mRNA and miR-328 associated to hnRNP E2 evaluated by RIP assays (n = 4) performed with parental (light bars) and miR-328-expressing (dark bars) 32D-BCR/ABL cells (mean ± SEM).
(E) Left: RIP assays for CEBPA mRNA (top) on anti-hnRNP E2 IPs from vector (pSuper)- (lane 5), miR-328- (lane 7), and miR-181b-transduced (lane 9) 32D-BCR/ABL cells; RIP assays for miR-181b (middle) and miR-328 (bottom) on anti-hnRNP E2 IPs from miR-181b-expressing 32D-BCR/ABL cells (lanes 8 and 9). Inset top right: UV crosslinking with miR-328, miR-181b, and miR-330 32P-labeled oligoribonucleotides and cytoplasmic lysates of 32Dcl3 (lanes 1, 3, and 5) and 32D-BCR/ABL (lanes 2, 4, and 6) cells. Binding of hnRNP E2 to miR-328 is shown in lane 2. Inset bottom left: Graph shows qRT-PCR analysis of miR-181b expression in 32Dcl3 (black), vector- (gray), and pSUP-miR-181b-transduced (white) 32D-BCR/ABL cells (mean ± SEM). Right: Densitometric analysis of the RIP’ed CEBPA mRNA associated to hnRNP E2 (n = 3), expressed as percentage of the input RNA (IN), in vector-, miR-328-, and miR-181b-transduced 32D-BCR/ABL cells (mean ± SEM). Controls for endogenous and ectopic hnRNP E2 protein and miR-328 expression levels and the specificity of the RIP protocol are shown in Figure S1.
To determine whether hnRNP E2:miR-328 interaction requires integrity of the miRNA seed sequence (Figure 2B, underlined nucleotides) that, as known, is essential for canonical miRNA: mRNA target interaction, UV crosslinking assays were performed using 32D-BCR/ABL and 32D-Flag-E2 lysates, as well as a miR-328 oligoribonucleotide harboring a mutated seed sequence that retains the wild-type C-rich character (miR-328-Mut) (Figure 2B). As shown (Figure 2B), miR-328-Mut binds both endogenous and ectopic hnRNP E2 at higher affinity than wild-type miR-328, suggesting that the C-rich character rather than the seed sequence is important for hnRNP E2 interaction. Furthermore, the hnRNP E2:miR-328 interaction was also demonstrated in living cells by RIP assay. In fact, endogenous miR-328 was found associated with endogenous hnRNP E2 in anti-hnRNP E2 but not anti-IgG immunoprecipitates (IP) from imatinib-treated 32D-BCR/ABL cells (Figure 2C). As expected, no binding was detected in untreated IL-3-cultured 32D-BCR/ABL cells (Figure 2C). Ectopic expression of miR-328 at physiological levels (Figure 2D and Figure S1B) also resulted in complex formation with both ectopic (Figure S1C) and endogenous (Figures 2C and 2D and Figure S1D) hnRNP E2. Note that Flag-hnRNP E2 did not significantly alter total hnRNP E2 levels (Figure S1A) and that miR-328 binding to hnRNP E2 impaired (~80% inhibition) hnRNP E2:CEBPA association (Figures 2D and 2E and Figure S1D). Furthermore, qRT-PCR analyses performed on anti-hnRNP E2 RIPs revealed that 4.26% ± 0.56% (n = 3) of the total miR-328 was associated to hnRNP E2 (not shown). As expected, hnRNP E2 did not interact with either a BCR/ABL-induced mRNA (i.e., SET) or the endogenous C-rich miR-223 (Figure S1D). Accordingly, ectopic miR-181b did neither interact with hnRNP E2 nor significantly alter levels of the hnRNP E2:CEBPA complex (Figure 2E). Thus, miR-328 specifically competes with CEBPA mRNA for binding to hnRNP E2.
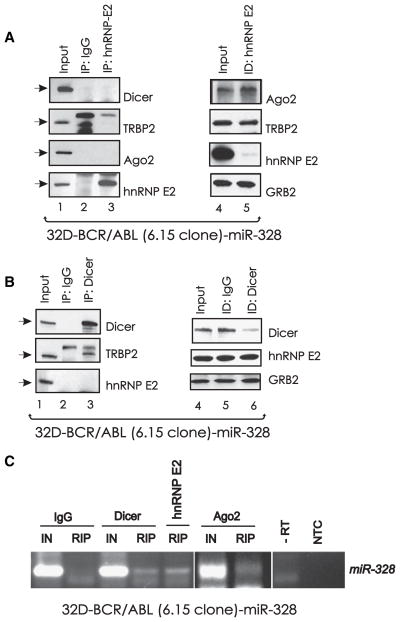
hnRNP E2:miR-328 Interaction Does Not Associate with the RISC Loading Complex
To assess whether the hnRNP E2:miR-328 interaction occurs in the presence of major components of the RISC loading complex (Chendrimada et al., 2005), we evaluated whether hnRNP E2 is in complex with Dicer, Ago2, or TRBP2. Anti-hnRNP E2 and anti-Dicer (Figures 3A and 3B) coimmunoprecipitation (coIP) experiments clearly show that hnRNP E2 did not interact with Dicer, TRBP2, or Ago2. Likewise, hnRNP E2 or Dicer immunodepletion did not alter levels of RISC components (Ago2 and TRBP2) or hnRNP E2, respectively (Figures 3A and 3B). As expected, TRBP2 was readily detectable in anti-Dicer IPs (Figure 3B). Thus, hnRNP E2:miR-328 interaction does not appear to require association with RISC loading components. However, RIP assays revealed miR-328 in complex with both Dicer and Ago2 (Figure 3C), suggesting that miR-328 also functions as a silencer of mRNA targets in a canonical RISC-dependent manner and that, most likely, its binding to hnRNP E2 is not mutually exclusive. Indeed, the presence of readily detectable hnRNP E2:miR-328 complex in RIP assays on Dicer-immunodepleted lysates (Figure 3C, lane 5) suggests that miR-328 might interact with hnRNP E2 in a RISC-independent manner.
Figure 3. hnRNP E2:miR-328 Interaction Does Not Associate with the RISC Loading Complex.
Western blots show levels of Dicer, TRBP2, Ago2, and/or hnRNP E2 in cytoplasmic lysates (input; lanes 1 and 4), in anti-hnRNP E2 (A) and anti-Dicer (B) IPs (lane 3), in IPs with an isotype-matched IgG (lane 2), and in hnRNP E2-and Dicer-immunodepleted (ID) lysates (lane 5) of 32D-BCR/ABL-miR-328cells. (C) RIP assay for miR-328 on anti-Dicer (lane 4), anti-hnRNP E2 (lane 5), and anti-Ago2 (lane 7) IPs from 32D-BCR/ABL-miR-328 cell lysates. Note that the anti-hnRNP E2 RIP for miR-328 was performed on Dicer-immunodepleted lysates. RIP with a nonrelated IgG served as a negative control.
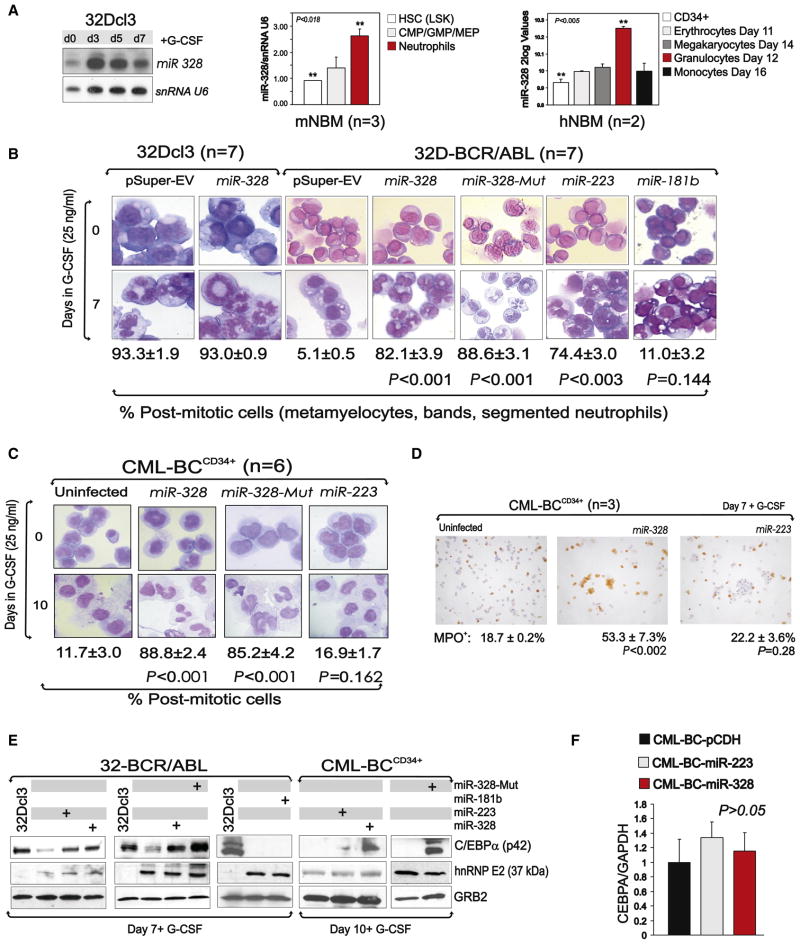
miR-328 Rescues C/EBPα-Driven Granulocytic Differentiation of CML-BCCD34+ Progenitors
As loss of miR-328 may influence the phenotype of CML-BC myeloid blasts, we evaluated proliferation, clonogenic potential, and G-CSF-driven differentiation in miR-328-transduced GFP+ BCR/ABL+ cell lines, BCR/ABL+ Lin− mouse BM, and/or CML-BCCD34+ cells. As controls, we assessed the effects of ectopic miR-328-Mut, miR-181b, and the myeloid differentiation-related (Chen et al., 2004) miR-223 in BCR/ABL+ cells. Of note, ectopic miR-328 or miR-223 levels in BCR/ABL+ cells were similar to those in nontransformed cells (Figure 2D and Figure S2A), thus excluding off-target effects due to overexpression.
Although endogenous miR-328 (Figure 1) and, to a lower extent, miR-223 (Figure S2B) were downregulated in 32D-BCR/ABL and K562 cells, their ectopic expression did not have a significant effect on IL-3-dependent and/or -independent growth (Figure S2B). Likewise, ectopic miR-328 did not accelerate the kinetics of 32Dcl3 neutrophil maturation (Figure 4B), consistent with the barely detectable levels of hnRNP E2 in 32Dcl3 cells (Perrotti et al., 2002) and the increased expression of endogenous miR-328 in 32Dcl3, human CD34+ (n = 2) and mouse Lin−/Sca+/Kit+ (n = 3) BM (NBM) progenitors undergoing granulocytic differentiation (Figure 4A). Conversely, miR-328 levels were not significantly different in normal CD34+ BM cells before and after differentiation toward other hematopoietic lineages (Figure 4A). As expected (Fazi et al., 2005), miR-223 enhanced 32Dcl3 differentiation (not shown).
Figure 4. miR-328 Rescues Granulocytic Differentiation through Restoration of C/EBPα Expression.
(A) miR-328 levels in (left) 32Dcl3 cells undergoing G-CSF-induced differentiation; (middle) Lin−/Sca+/Kit+ HSC, CMP/GMP/MEP committed progenitors and mature neutrophil BM subpopulations from wild-type C57BL/6 mice (mean ± SEM); and (right) CD34+ human BM cells undifferentiated (white) and induced to differentiate for the indicated time toward the erythroid (light gray), megakaryocytic (dark gray), granulocytic (red), or monocytic (black) lineages (mean ± SEM).
(B) Wright-Giemsa-stained cytospins of G-CSF-treated (0–7 days) pSuper-, miR-328-, miR-328-Mut-, miR-223-, and/or miR-181b-infected 32Dcl3 and/or 32D-BCR/ABL cells (mean ± SEM). Levels of miR-223 in BCR/ABL+ cell lines and primary cells and effect of ectopic miR-223 on cell proliferation are reported in Figure S2.
(C) Wright-Giemsa-stained cytospins of primary G-CSF-treated (0–10 days) uninfected and miR-328-, miR-328-Mut-, and miR-223-infected CML-BCCD34+ BM progenitors (mean ± SEM). For levels of ectopic miR-328 and miR-223 expression in primary CML-BC cells, see Figure S2.
(D) Myeloperoxidase (MPO) immunostaining of G-CSF-treated uninfected and miR-328- and miR-223-transduced CML-BCCD34+ BM cells. Data are representative of three independent experiments (mean ± SEM).
(E) Western blot shows C/EBPα, hnRNP E2, and GRB2 levels in G-CSF-treated 32Dcl3 and empty vector-, miR-328-, miR-223-, miR-328-Mut-, and/or miR-181b-infected 32D-BCR/ABL and CML-BCCD34+ BM cells (right).
(F) qRT-PCR shows levels of CEBPA in empty vector-, miR-223-, or miR-328-infected CML-BC cells (mean ± SEM). qRT-PCR showing CEBPA mRNA levels in empty vector-, miR-223-, or miR-328-transduced 32D-BCR/ABL cells (mean ± SEM) is reported in Figure S2.
In agreement with the potential role of miR-328 as an antagonist of hnRNP E2 differentiation inhibitory activity, forced expression of miR-328 at physiological levels (Figure 2D and Figure S2A) efficiently rescued granulocytic differentiation of newly established (15 days after BCR/ABL infection) 32D-BCR/ABL cells (Figure 4B). In fact, the majority (82.1% ± 3.9%) of miR-328-expressing 32D-BCR/ABL cells were postmitotic metamyelo-cytes, bands, and segmented neutrophils after 7 days of G-CSF-supplemented culture (Figure 4B). As expected, G-CSF-treated vector- and miR-181b-transduced BCR/ABL+ cells remained blasts (5.1% ± 0.5% and 11.0% ± 3.2% postmitotic cells) (Figure 4B). By contrast, miR-328-Mut expression efficiently induced 32D-BCR/ABL differentiation (88.6% ± 3.1% postmitotic cells) (Figure 4B).
Although terminal differentiation was also a characteristic of miR-223-overexpressing 32D-BCR/ABL cells, only ectopic miR-328 but not miR-223 expression (Figure S2C) restored G-CSF-driven maturation of GFP+ CD34+ BM progenitors from myeloid CML-BC patients (n = 6) (Figure 4C). In fact, both miR-328- and miR-328-Mut-expressing CML-BCCD34+ BM cultures became bands and segmented neutrophils (88.8% ± 2.4% and 85.2% ± 4.2% postmitotic cells) after 10 days in rhG-CSF (25 ng/ml). By contrast, morphology of miR-223-expressing CML-BCCD34+ (n = 6) progenitors remained similar to untransduced cells (n = 6), appearing arrested at the myeloblast stage after 7–10 days in G-CSF-containing medium (16.9% ± 1.7% postmitotic cells) (Figure 4C) with unchanged levels of the granulocyte/macrophage markers CD11b or CD14 (not shown).
Consistent with the essential role of C/EBPα in neutrophil maturation of BCR/ABL+ blasts (Ferrari-Amorotti et al., 2006; Perrotti et al., 2002; Wagner et al., 2006), C/EBPα expression was readily detectable in miR-223- and miR-328-transduced 32D-BCR/ABL myeloid precursors, and in miR-328- but not miR-223-expressing CML-BCCD34+ BM progenitors and miR-181b-expressing 32D-BCR/ABL cells (Figure 4E). This was dependent neither on hnRNP E2 downregulation (Figure 4E) nor on increased CEBPA mRNA levels (Figure 4F and Figure S2C). In agreement with its ability to bind hnRNP E2, miR-328-Mut also restored C/EBPα expression (Figure 4E), indicating that the differentiation-promoting effects of miR-328 do not result from the seed sequence-dependent silencing of miR-328 mRNA targets. Furthermore, myeloperoxidase (MPO), a marker of granulocyte/macrophage commitment and a direct transcriptional target of C/EBPα (Rosmarin et al., 1989; Wang et al., 2001), was also significantly increased in G-CSF-cultured miR-328-(18.7% ± 0.2% [uninfected] versus 53.3% ± 7.3% [miR-328]; p < 0.004) but not miR-223- (18.7% ± 0.2% [uninfected] versus 22.2% ± 3.6% [miR-223]; p = 0.28) expressing CML-BCCD34+ cells (n = 3) (Figure 4D). Finally, the dissimilar response of miR-223-transduced 32D-BCR/ABL versus CML-BCCD34+ progenitors to G-CSF might depend on differences in the mechanism(s) controlling expression of NFI-A (Figure S2D), a miR-223-negative regulator (Fazi et al., 2005).
miR-328 Restores CEBPA mRNA Translation Both In Vitro and In Vivo
As hnRNP E2:miR-328 binding in vitro is more efficient than that of CEBPA uORF (Figure 2), and miR-328 expression antagonizes hnRNP E2:CEBPA interaction (Figures 2D and 2E) most likely by competing for binding to hnRNP E2, it is plausible that miR-328 releases CEBPA from the translation inhibitory effects of hnRNP E2. Thus, we assessed the effect of miR-328 on CEBPA translation in rabbit reticulocyte lysate and in an in vivo mouse model of myeloid CML-BC. In the latter, BCR/ABL+ cell differentiation is driven solely by ectopic C/EBPα expression, which is under the control of its uORF/spacer mRNA element (Chang et al., 2007). As reported (Perrotti et al., 2002), translation of CEBPA mRNA was markedly impaired (~80% inhibition) in in vitro translation reactions programmed with a CEBPA construct containing the uORF/spacer intercistronic region (pcDNA3-WT-uORF-C/EBPα) and the recombinant fusion protein MBP-hnRNP E2 (Figure 5A) but not when performed in the absence of exogenous hnRNP E2 (Figure 5A). Addition of 1000-fold excess of mature miR-328 but not miR-330 resulted in an almost 100% increase of newly synthesized 35S-C/EBPα protein (CEBPA+hnRNP E2 versus CEBPA+hnRNP E2+miR-328; p < 0.005) (Figure 5A). Note that addition of miR-328 in the absence of MBP-hnRNP E2 did not significantly affect CEBPA mRNA translation (Figure 5A). Moreover, the large amount of MBP-hnRNP E2 (Figure 5A) might justify the incomplete rescue of CEBPA translation.
Figure 5. In Vitro and In Vivo Interference of miR-328 with hnRNP E2 Translation Inhibition of C/EBPα Expression.
(A) Levels of newly synthesized 35S-C/EBPα protein in RRL translation reactions programmed with CEBPA mRNA (derived from pcDNA3-WT-uORF-C/EBPα) (black), CEBPA mRNA and mature miR-328 RNA oligonucleotides (dark gray), CEBPA mRNA and recombinant MBP-hnRNP E2 protein either alone (light gray) or in the presence of mature miR-328 (red), or miR-330 (white; negative control) RNA oligonucleotides. Data are expressed as percentage of the mean ± SEM and are representative of three different experiments performed in duplicate. Inset: Western blot shows levels of both endogenous RRL hnRNP E2 and recombinant MBP-hnRNP E2.
(B) Wright-Giemsa-stained cytospins of G-CSF-treated (0–7 days) 6.15-pSUP, 6.15-miR-328, and 6.15-WT-uORF-miR-328 cells (mean ± SEM).
(C) Left: Levels of hnRNP E2, endogenous and HA-tagged C/EBPα, and GRB2 proteins and miR-328 and snRNA U6 in parental 32Dcl3, 6.15-pSUP-transduced, and miR-328-transduced 6.15-WT-uORF cells; right: RT-PCR and qRT-PCR show levels of CEBPA mRNA in 32Dcl3, 6.15, 6.15-miR-328, and 6.15-WT-uORF-HA-CEBPA cells either uninfected or infected with pSUP or miR-328 constructs. GAPDH levels were measured for normalization (mean ± SEM).
(D) RIP assays for CEBPA mRNA (top) and miR-328 (bottom) on anti-hnRNP E2 (lanes 5 and 9) and nonrelated IgG (lanes 3 and 7) IPs from 6.15-uORF-pSUP (lanes 2–5) and 6.15-uORF-miR-328 cells (lanes 6–9). IN: input RNA.
(E) Top: H&E-staining of BM shows maturation of BCR/ABL+ cells in mice injected with p-SUPERIOR vector- (middle) and miR-328-transduced (right) 6.15-WT-uORF cells. Age-matched mice (left) served as a control. FACS analysis shows mean fluorescence intensity (MFI; mean ± SEM) of differentiated GFP+Gr1+BCR/ABL+ cells at 3 weeks post-transplant from BM of 3 mice/group. Bottom: Visual analysis and weight of spleens from the same groups of mice (mean ± SEM).
To assess whether forced miR-328 expression rescues neutrophilic maturation of differentiation-arrested BCR/ABL+ blasts through restoration of CEBPA mRNA translation, we used the aberrant 32D-BCR/ABL long-term cultured 6.15 cell clone that exhibits extremely high levels of BCR/ABL and hnRNP E2 but is unable to undergo G-CSF-driven differentiation due to transcriptional suppression of CEBPα expression (Figure 5C). Indeed, 6.15 cells completely rely on translation of ectopic CEBPA mRNA for differentiation. Thus, parental and 6.15 cells expressing a GFP-WT-uORF/spacer-C/EBPα (6.15-WT-uORF), which contains the hnRNP E2 translation inhibitory element, were retrovirally transduced with the pSUPERIOR-retro-puro-miR-328 (6.15-WT-uORF-miR-328) or with the empty vector (6.15-WT-uORF-pSUP). Differentiation assays confirmed that the ability of miR-328 to induce neutrophil maturation is dependent on and mediated by the presence of CEBPA mRNA, as expression of miR-328 in parental 6.15 cells failed to rescue differentiation, whereas 91.7% ± 6.4% of 6.15-WT-uORF-miR-328 cells were postmitotic after 7 days of culture in G-CSF (Figure 5B). Furthermore, forced miR-328 expression neither decreased hnRNP E2 protein nor increased CEBPA mRNA levels (Figure 5C), suggesting that the restoration of C/EBPα protein expression in 6.15-WT-uORF-miR-328 cells (Figure 5C) results from the ability of miR-328 to interfere with hnRNP E2 translation inhibitory activity. Accordingly, anti-hnRNP E2 RIP assays performed with 6.15-WT-uORF-pSUP and 6.15-WT-uORF-miR-328 lysates revealed that miR-328 expression and, therefore, formation of the hnRNP E2:miR-328 complex (Figure 5D) markedly decreased levels of the hnRNP E2-bound CEBPA mRNA (Figure 5D).
To determine whether miR-328 influences the CML-BC-like disease process induced by transplantation of BCR/ABL-expressing cells, SCID mice (n = 13 per group) were intravenously injected with 6.15-WT-uORF-miR-328 or 6.15-WT-uORF-pSUP cells (5 × 105 GFP+-puromycin-selected cells/mouse), and engraftment was assessed 1 week later by nested RT-PCR-mediated BCR/ABL detection in peripheral blood (not shown). After 3 weeks, three mice/group were sacrificed for visual and histopathologic examination of hematopoietic organs and for flow cytometric quantification of GFP+/GR1+ differentiated BM cells. Consistent with the almost complete hnRNP E2-dependent translational inhibition of C/EBPα expression in 6.15-WT-uORF cells (Chang et al., 2007), hematoxylin/eosin-stained sections of BM (Figure 5E), spleen, and liver (not shown) from 6.15-WT-uORF-pSUP-injected mice showed splenomegaly and massive infiltration of myeloid blasts with a low degree of differentiation. A few myeloid cells undergoing terminal neutrophil differentiation were occasionally observed in BM from 6.15-WT-uORF-pSUP-injected mice (mean fluorescence intensity [MFI]: 14.98 ± 1.84 GFP+/GR1+ BCR/ABL+ cells). By contrast, spleens from 6.15-WT-uORF-miR-328-injected mice appeared normal in weight or slightly hyperplastic, and histopathologic analysis of BM (Figure 5E), spleen, and liver (not shown) showed marked infiltration by mature neutrophils and myeloid precursors at postmitotic stages of differentiation (MFI: 63.84 ± 5.40 GFP+/GR1+ BCR/ABL+ cells) when compared to age-matched controls (Figure 5E), suggesting that miR-328 also negatively regulates survival pathways in CML-BC although its major effect appears to be on differentiation. In fact, although no significant difference in survival time was noted, the remaining 6.15-WT-uORF-pSUP-injected mice died of a CML-BC-like leukemia, whereas 6.15-WT-uORF-miR-328-injected animals succumbed from an aggressive CML-CP-like myeloproliferative disorder (not shown). Altogether, these in vitro and in vivo results indicate that rescue of granulocytic maturation of differentiation-arrested BCR/ABL+ cells by miR-328 is likely due to its direct binding to hnRNP E2 that, in turn, prevents translational inhibition of CEBPA mRNA.
A BCR/ABL-MAPK-hnRNP E2 Pathway Suppresses miR-328 Transcription through Inhibition of C/EBPα
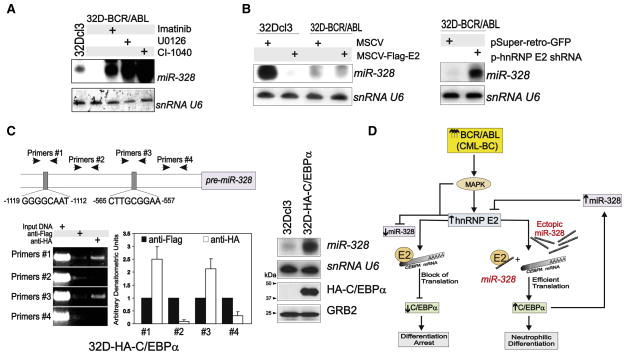
We recently reported that high levels of BCR/ABL expression/kinase activity, as observed in CML-BC (Jamieson et al., 2004; Schultheis et al., 2005), impair C/EBPα expression through the MAPK(ERK1/2)-dependent regulation of hnRNP E2 expression/activity (Chang et al., 2007). To determine whether BCR/ABL uses the same signaling pathway to suppress miR-328 expression in CML-BC, miR-328 levels were evaluated in G-CSF-cultured (24–48 hr) parental and newly established 32D-BCR/ABL cells treated with imatinib (2 μM) or MEK1 inhibitors U0126 (25 μM) and CI-1040 (10 μM) and after overexpression (MSCV-Flag-E2) or shRNA-mediated downregulation (pSR-hnRNP E2 shRNA) of hnRNP E2. Inhibition of BCR/ABL or MEK1 kinases strongly enhanced miR-328 expression (Figure 6A), suggesting that BCR/ABL-mediated suppression of miR-328 requires MAP-K(ERK) activity. Ectopic hnRNP E2 impaired miR-328 expression in 32Dcl3 cells, with no noticeable effect in 32D-BCR/ABL cells (Figure 6B) that already express high levels of hnRNP E2 (Perrotti et al., 2002). By contrast, shRNA-mediated downregulation of hnRNP E2 efficiently rescued miR-328 expression (Figure 6B). Thus, the BCR/ABL-MAPK-induced hnRNP E2 may directly regulate miR-328 nuclear export, processing, and/or stability or indirectly influence miR-328 transcription. Of interest, modulation of hnRNP E2 levels did not alter miR-223 expression in 32D-BCR/ABL cells, whereas hnRNP E2 overexpression inhibited miR-223 in 32Dcl3 cells (Figure S2E), consistent with the notion that hnRNP E2 reduces C/EBPα expression (Perrotti et al., 2002), thereby averting C/EBPα-dependent miR-223 trans-activation (Fazi et al., 2005).
Figure 6. Pathways Regulating miR-328 Expression.
miR-328 levels in 32Dcl3 and/or 32D-BCR/ABL cells (A) treated with imatinib or the MAPK inhibitors U0126 and CI-1040 or (B) expressing a Flag-hnRNP E2 (left) or a shRNA-targeting hnRNP E2 (right).
(C) Top: Representation of the C/EBPα-binding sites within the miR-328 promoter; bottom: Chromatin immunoprecipitation (ChIP) with anti-HA antibody shows binding of HA-C/EBPα to miR-328 promoter sequences in 32D-HA-C/EBPα cells (left: ChIP blot; middle: densitometric analysis). Anti-Flag immunoprecipitates served as negative controls. Bars indicate the mean ± SEM from three independent experiments; northern and western blots (right) show levels of miR-328 and HA-C/EBPα, respectively, in parental and 32D-HA-C/EBPα cells. U6 snRNA and GRB2 protein levels were used as controls.
(D) Model of the molecular network regulating miR-328 expression in CML-BC and miR-328 decoy activity in BCR/ABL+ myeloid cell differentiation by direct interference with hnRNP E2 translation inhibition of C/EBPα expression.
Transcription Element Search System-mediated (http://www.cbil.upenn.edu/cgi-bin/tess) sequence analysis revealed four putative C/EBPα-binding sites scattered within 1500 bp upstream of the mouse pre-miR-328 (Figure 6C). Thus, chromatin immunoprecipitation (ChIP) assays were performed using nuclear extracts from GFP-sorted HA-C/EBPα-expressing 32Dcl3 cells (32D-HA-C/EBPα) and four sets of primers, each encompassing one of the potential C/EBPα-binding sites. In vivo physical interaction between HA-C/EBPα and the miR-328 promoter region was detected in ChIP assays performed on anti-HA but not anti-Flag (negative control) immunoprecipitates with primer sets #1 and #3 containing the human/mouse-conserved C/EBPα-binding sites located at nucleotides −1119 to −1112 and −565 to −557, respectively (Figure 6C). Accordingly, ectopic C/EBPα expression markedly induced miR-328 levels in myeloid precursors (Figure 6C), altogether suggesting that a BCR/ABL-MAPK-hnRNP E2 pathway downregulates miR-328 expression through inhibition of C/EBPα, thus impeding enhancement of miR-328 transcription (Figure 6D).
miR-328 Impairs CML-BCCD34+ Clonogenic Potential and Canonically Suppresses PIM1 Expression
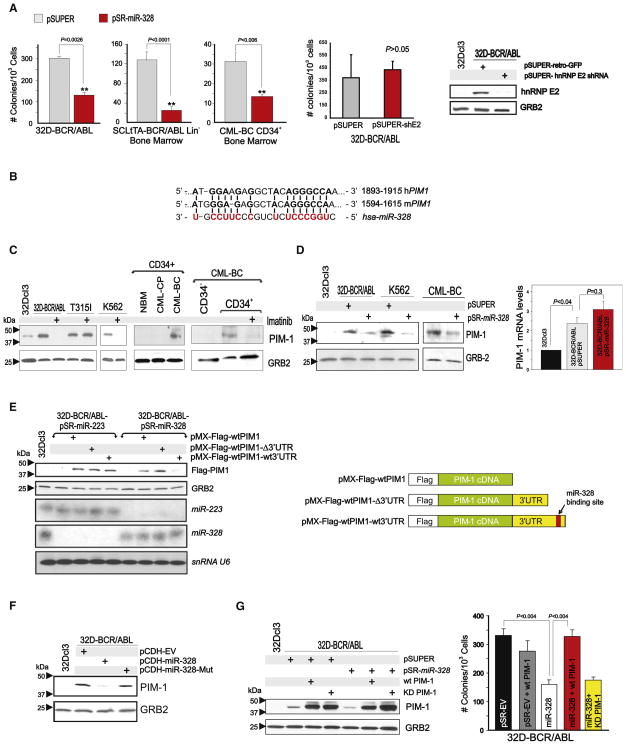
Ectopic miR-328 expression reduced colony formation in 32D-BCR/ABL, Lin− SCLtTA-BCR/ABL (n = 5), and CML-BCCD34+ (n = 2) BM cells by 75%, 83%, and 75%, respectively (Figure 7A). The effect of miR-328 on clonogenicity of BCR/ABL+ cells is independent from hnRNP E2:miR-328 interaction, as shRNA-mediated hnRNP E2 downregulation did not affect BCR/ABL-driven colony formation (Figure 7A). Thus, miRanda, PicTar, and TargetScan bioinformatics algorithms were utilized to identify miR-328 targets regulating CML-BC progenitor cell survival. Among the predicted miR-328 human/mouse mRNA targets, PIM1 (Figure 7B) kinase is important for survival of BCR/ABL+ cell lines (Nieborowska-Skorska et al., 2002). PIM1 protein was strongly upregulated in a BCR/ABL kinase-dependent manner in cell lines and CD34+ CML-BC (n = 2) versus CML-CP and NBM BM cells (Figure 7C).
Figure 7. miR-328 Impairs Survival through Targeting of PIM1 Kinase mRNA.
(A) IL-3-independent or -dependent methylcellulose colony formation (mean ± SEM from triplicates of three independent experiments) of vector- (gray bars) and miR-328-transduced (red bars) 32D-BCR/ABL cells (IL-3-independent), leukemic Lin− SCLtTA-BCR/ABL (n = 5) (IL-3-dependent) cells, CML-BCCD34+ (n = 2) (IL-3 dependent) BM progenitors, and vector- (pSUPER) or hnRNP E2 shRNA (pSUPER-shE2)-infected 32D-BCR/ABL cells (IL-3-independent). Inset: Western blot shows hnRNP E2 levels upon shRNA knockdown.
(B) miR-328-binding site (red) within the 3′UTR of mouse and human PIM1 mRNA.
(C) PIM1 protein levels in 32Dcl3, untreated or imatinib-treated 32D-BCR/ABL (wild-type and T315I) and K562 cells (left); in CD34+ BM cells from healthy donors (NBM), CML-CP, and CML-BC patients (middle); and in the CD34−, untreated and imatinib-treated CD34+ BM fractions from a CML-BC patient.
(D) Left: Effect of miR-328 expression on PIM1 protein levels in 32D-BCR/ABL, K562, and CML-BCCD34+ BM cells. 32Dcl3 cells were used as a negative control; right: PIM1 mRNA expression by qRT-PCR in 32Dcl3, vector-transduced, and miR-328-transduced 32D-BCR/ABL. Representative of triplicates from three independent experiments (mean ± SEM).
(E) Left: Levels of ectopic Flag-PIM1 proteins from constructs lacking (pMX-Flag-WTPIM1) and harboring the wild-type (pMX-Flag-WTPIM1-WT3′UTR) or 196 base pair end-terminal-deleted (pMX-Flag-WTPIM1-Δ3′UTR) PIM1 3′UTR in miR-223- or miR-328-expressing 32D-BCR/ABL. Northern blot shows levels of miR-223, miR-328, and snRNA U6. Right: Schematic representation of the Flag-PIM1 constructs.
(F) Effect of seed sequence-mutated (miR-328-Mut) on endogenous PIM1 expression in parental 32Dcl3 and empty vector (pCDH)-, miR-328-, and miR-328-Mut-infected 32D-BCR/ABL cells.
(G) Left: Endogenous and ectopic (wild-type [WT PIM-1] and kinase-deficient [KD PIM-1]) PIM1 protein levels in 32Dcl3, pSUPER- and pSR-miR-328-infected 32D-BCR/ABL cells; right: graph shows rescue of IL-3-independent clonogenic activity of miR-328-expressing (white) 32D-BCR/ABL cells to normal levels (black) by ectopic wild-type (red) but not kinase-deficient (yellow) PIM1 construct lacking the 3′UTR. Effect of PIM1 forced expression on vector-transduced clonogenicity (gray). Bars represent the mean ± SEM of colony numbers from three independent experiments.
Ectopic miR-328 expression in 32D-BCR/ABL, K562, and CML-BCCD34+ cells decreased PIM1 protein without significantly affecting its mRNA levels (Figure 7D), suggesting that miR-328 might impair mRNA translation upon interaction with the PIM1 3′UTR. To formally demonstrate that miR-328 silences PIM1 expression through its interaction with the miR-328-binding site, we cloned the wild-type (pMX-Flag-WTPIM1-WT3′UTR) and miR-328-binding site-deleted (pMX-Flag-WTPIM1-Δ3′UTR) PIM1 3′UTR (Figure 7E) into a pMX-Flag-WTPIM1 plasmid and transduced these constructs into 32D-BCR/ABL-miR-328 or, as a negative control, 32D-BCR/ABL-miR-223 cells. As expected, ectopic Flag-PIM1 expression was lower in pMX-Flag-WTPIM1-WT3′UTR-transduced 32D-BCR/ABL-miR-328 cells but not in cells transduced with pMX-Flag-WTPIM1-Δ3′UTR or with PIM1 cDNA only (pMX-Flag-WTPIM1) (Figure 7E). Accordingly, Flag-PIM1 expression derived from pMX-Flag-WTPIM1-WT3′UTR was barely detectable in miR-328- but not in miR-223-transduced cells, in which its expression was similar to that of Flag-WTPIM1 and Flag-WTPIM1-Δ3′UTR (Figure 7E), indicating that decreased ectopic PIM1 levels are not due to loss of other miRNA-binding sites within the 196 bp-deleted 3′UTR. Expression of the seed sequence-mutated mir-328 (miR-328-Mut) impaired the ability of miR-328 to canonically suppress PIM1 expression (Figure 7F). Thus, miR-328 specifically silences PIM1 expression through interaction with the PIM1 3′UTR. In agreement with the importance of PIM1 for survival of BCR/ABL+ cells, expression of a wild-type PIM1 cDNA lacking the 3′UTR (WT PIM1) but not of a kinase-deficient (KD PIM1) PIM1 cDNA into 32D-BCR/ABL-miR-328 cells (Figure 7G) completely restored IL-3-independent colony formation (Figure 7G), suggesting that miR-328-dependent inhibition of BCR/ABL-driven clonogenic potential results from direct PIM1 downregulation. Note that WT PIM1 alone did not affect 32D-BCR/ABL-pSR-EV clonogenic potential (Figure 7G).
DISCUSSION
Altered miRNA expression has been tightly associated with cancer development and progression (Friedman et al., 2009; Garzon et al., 2006). Among the miRNAs differentially expressed in CML, we focused on miR-328 and provided a series of evidence highlighting two important concepts. The first represents a paradigm shift to the notion that miRNAs act primarily as negative posttranscriptional regulators of gene expression and proposes for miRNAs a function termed decoy activity. The second identifies miR-328 as a molecular relay, the loss of which is important for the differentiation arrest of progressing CML-BC blasts.
miR-328 Decoy Activity
As miRNAs base pair with mRNA 3′UTRs in a sequence-specific manner (Bartel, 2009), it is conceivable that miRNAs could interfere with the activity of RNA-binding proteins (e.g., hnRNPs), either indirectly by pairing with RBP-binding sites contained in specific mRNAs (George and Tenenbaum, 2006) or directly through binding the RBP itself and impeding RBP:mRNA interaction. Herein we demonstrated that miRNAs can act as direct inhibitors of RBP activity. In fact, miR-328 specifically interacts in a seed sequence-independent manner and, most likely, through its C-rich clusters, with the translational inhibitor poly (rC)-binding protein hnRNP E2. This, in turn, prevents and/or displaces CEBPA mRNA binding to hnRNP E2 and rescues CEBPA mRNA translation both in vitro and in vivo. In support of the notion that miR-328 and, most likely, other miRNAs may act as “decoy” molecules for RBPs, which upon binding could control synthesis, processing, export, stability, and/or translation of specific mRNA subsets, a proteomics-based study in epithelial A431 cells reported that forced miR-328 expression not only decreased levels of different genes but also upregulated a subset of proteins (Wang et al., 2008). Interestingly, 37% of these upregulated proteins have mRNAs with complex 5′UTRs (e.g., uORF or multiple ATGs) containing C-rich elements representing potential hnRNP E2-binding sites. Thus, it is reasonable to speculate that upregulation of some of these proteins might result from interference with hnRNP E2 activity. Furthermore, there is evidence that miRNAs, other components of the RISC complex (Parker and Sheth, 2007) and RBPs (e.g., hnRNP E2) are present in processing bodies (P bodies) (Fujimura et al., 2008), dynamic subcellular structures where mRNAs are complexed with RBPs and/or miRNAs for translational suppression or decay (Parker and Sheth, 2007). In this scenario, miR-328 may compete with CEBPA mRNA for binding to hnRNP E2 that, in turn, releases CEBPA and allows its loading onto poly-somes for translation. It is also possible that hnRNP E2 not only prevents C/EBPα-dependent induction of pri-miR-328 transcription but also directly promotes miR-328 decay. Accordingly, an inverse correlation exists between hnRNP E2 and miR-328 expression in CML-BC, and hnRNP E2 more efficiently binds to miR-328 than to CEBPA. However, as hnRNP E2 continuously shuttles between nucleus and cytoplasm as well as in and out of P bodies (Fujimura et al., 2008; Makeyev and Liebhaber, 2002), knowledge of the subcellular location where initial binding of hnRNP E2 to miR-328 or CEBPA occurs remains elusive, although a plausible assumption is the cytoplasm as their association was detected using cytoplasmic extracts.
MicroRNA interaction with sequence-specific RBPs is not unprecedented; however, none of the reported mechanisms match the case of hnRNP E2 and miR-328. For example, hnRNP A1, another RBP upregulated in CML-BC (Perrotti and Neviani, 2007), binds the primary miR-17-92 transcript to allow processing of pre-miR-18a (Guil and Caceres, 2007). Interestingly, expression of the miR-17-92 cluster is upregulated in CML and is important for proliferation and reduced susceptibility to apoptosis of K562 cells (Venturini et al., 2007). However hnRNP E2:miR-328 interaction does not seem to affect miR-328 biogenesis, as no accumulation of primary or precursor miR-328 was detected in BCR/ABL+ cells (not shown). It was also shown that miR-369-3 interacts with the AU-rich region (ARE) of TNF-α mRNA, which recruits the AGO2-FXR1 RBP complex to the ARE element itself and upregulates or represses translation under serum-starved or proliferating conditions, respectively (Vasudevan et al., 2007). However, it is unlikely that hnRNP E2 is recruited to CEBPA mRNA by miR-328, as no miR-328 target sites are present in the 5′ and 3′UTRs of human and mouse CEBPA mRNA. Because our data indicate that miR-328 binds hnRNP E2 in a seed sequence-independent manner and strongly prevents its interaction with the C-rich intercistronic element of CEBPA mRNA, it is safe to conclude that hnRNP E2 binding to miR-328 and CEBPA mRNA are mutually exclusive. Interestingly, as binding to hnRNP E2 is mediated by the C-rich nucleotide clusters present in miR-328, it is possible that miR-328 also alters the function of other hnRNPs (PCBP1 and hnRNP K) that, like hnRNP E2, bind C-rich elements within the 5′ and 3′UTRs (Ostareck-Lederer and Ostareck, 2004). Indeed, preliminary data indicate that miR-328 interacts with hnRNP K, albeit with a lower affinity than the interaction with hnRNP E2 (not shown). Because hnRNP K expression/activity is also enhanced in CML-BCCD34+ cells in a BCR/ABL-MAPK-dependent manner (Notari et al., 2006), and both hnRNP K and hnRNP E2 are essential for IRES-mediated induction of MYC mRNA translation (Evans et al., 2003), we can speculate that loss of miR-328 in CML might favor disease progression by also promoting hnRNP K/E2-dependent MYC translation activation. Seemingly, we can also speculate that the mRNA stabilizing and destabilizing activity of the AU-rich element (ARE) RNA-binding proteins AUF1 (hnRNP D) and HuR (Lal et al., 2004), respectively, can be influenced by the putative decoy activity of miRNAs bearing the ARE element in their nucleotide sequence (e.g., miR-95; uucaacggguAUUUAuugagca). Indeed, TargetScan-based analysis revealed that several mature miRNAs contain the AUUUA element in their sequence. Thus, it is conceivable that these miRNAs can antagonize the ability of AUF1 and HuR to regulate expression of mRNAs encoding cytokine receptors (e.g., GM-CSF, TNF-α, and interleukins), oncogenes (e.g., c-Myc and FOS), tumor suppressors (e.g., p53), and cell-cycle regulators (e.g., p21 and cyclin D1) (Hinman and Lou, 2008; Khabar, 2005; Lal et al., 2004). Likewise, the function of the CGG-repeat binding FMRP RBP, which is the cause of the Fragile X mental retardation syndrome and normally regulates translation of mRNAs controlling synaptic function (Brown et al., 2001; Darnell et al., 2001; Zalfa et al., 2003), might be influenced by the altered expression of CGG-rich miRNAs (e.g., miR-572 and miR-638). Thus, it is likely that the decoy activity of miRNAs is not limited to miR-328 but can be extended to other miRNAs containing nucleotide sequences resembling the consensus RNA-binding sites for RBPs that are involved in different normal cell functions and in neoplastic as well as non-cancer-related diseases.
miR-328: A Molecular Relay in CML Disease Progression
Even though a few miRNAs are aberrantly regulated in CML (Agirre et al., 2008; Bueno et al., 2008; Venturini et al., 2007), evidence of their involvement in disease progression is still lacking. The failure of myeloid CML-BC progenitors to undergo maturation depends on increased BCR/ABL activity which, in addition to enhancing survival, proliferation, genomic instability, and self-renewal (Melo and Barnes, 2007), allows the hnRNP E2 inhibitory effect on C/EBPα that, per se, is sufficient to reinstate differentiation of Ph(+) blasts (Ferrari-Amorotti et al., 2006; Perrotti et al., 2002). We demonstrated that loss of miR-328 occurs in CML-BCCD34+ but not CML-CPCD34+ myeloid progenitors, and that forced miR-328 expression at physiological levels rescues C/EBPα-driven granulocytic maturation and impairs survival of CML-BC blasts. However, the proapoptotic effect of miR-328 does not seem to depend on its decoy activity and, therefore, on C/EBPα-induced differentiation and growth arrest (Keeshan et al., 2003), as shRNA-mediated hnRNP E2 downregulation does not influence BCR/ABL-driven clonogenic potential. Rather, we showed that impaired colony formation is the consequence of miR-328 canonical activity that targets PIM1 mRNA, thus repressing PIM1 expression and survival-promoting activity (Hoover et al., 2001; Nieborowska-Skorska et al., 2002). Indeed, forced expression of a wild-type, but not a kinase-deficient, PIM1 cDNA lacking the 3′UTR into miR-328-expressing cells fully rescued BCR/ABL clonogenicity. However, the main in vivo effect of miR-328 seems to be on differentiation rather than survival, as forced miR-328 expression did not delay leukemogenesis but reverted the blast crisis-like phenotype to a disease that resembles a myeloproliferative-like disorder, although the absence of marked splenomegaly may suggest that a portion of miR-328+/BCR/ABL+ cells underwent apoptosis. Although miR-223 was also described as a positive regulator of neutrophil maturation in APL cells (Fazi et al., 2005), our data in primary leukemic samples and work in miR-223 knockout animals (Johnnidis et al., 2008) argue against a general role for miR-223 as an inducer of myeloid differentiation.
Mechanistically, we showed that BCR/ABL uses the same MAPK(ERK1/2)-hnRNP E2 signaling pathway (see model in Figure 6D) to suppress C/EBPα (Chang et al., 2007) and miR-328 expression. Notably, constitutive MAPK activation by BCR/ABL occurs in CML-BC but not CML-CP (Notari et al., 2006). Moreover, similar to the positive feedback loop described for miR-223 and C/EBPα (Fazi et al., 2005), we showed that C/EBPα also interacts with the miR-328 promoter, thus enhancing its transcription.
In conclusion, the discovery of dual activities for miR-328 that profoundly affect myeloid cell differentiation and survival not only add a new layer to the complexity of mechanisms regulating the phenotype of CML-BC progenitors but, more importantly, highlight the ability of miRNAs to alter mRNA metabolism by acting also as molecular decoys for RNA-binding proteins.
EXPERIMENTAL PROCEDURES
Additional details on all the methods are available online in the Extended Experimental Procedures.
Clonogenic and Viability Analysis
Methylcellulose clonogenic assays were carried out by plating 103 32Dcl3 and derivative cell lines, 104 CML-BCCD34+, or 104 Lin− SCLtTA-BCR/ABL BM progenitors in 0.9% MethoCult (Stem Cell Technologies) in the presence or absence of rIL-3 (100 ng/ml). Colonies (>100 μm) from cell lines and primary cells were scored 7 and 15 days later, respectively. Human and mouse cell lines and primary cell source and culture conditions as well as plasmids and retro/lentiviral vectors are reported in the Extended Experimental Procedures.
In Vitro and In Vivo Differentiation Assays
In vitro granulocytic differentiation was induced for 7–10 days with 25 ng/ml rG-CSF. Morphologic differentiation was assessed by Wright/Giemsa staining. For in vivo differentiation, 10-week-old ICR-SCID mice (n = 13 per group) were intravenously (i.v.) injected (5 × 105 cells/mouse) with pSUPERIOR.retro.puro-or pSUP-miR-328-transduced 6.15-WT-uORF-CEBPA(GFP+) cells.
RNA Extraction, Northern Blot, and Real-Time PCR
Total RNA was used in northern blot, RT-PCR, and/or qRT-PCR for the analysis of miRNA and mRNA expression. U6 snRNA and GAPDH levels were analyzed for normalization of miRNA and mRNA PCRs, respectively.
REMSA, UV Crosslinking, and RNA Immunoprecipitation
Recombinant MBP-hnRNP E2 (Chang et al., 2007) and 32Dcl3 or 32D-BCR/ABL cytoplasmic extracts were used in REMSA and UV crosslinking as described (Perrotti et al., 2002). RIP and miR-328 RT-PCR were performed as described (Keene et al., 2006; Wang et al., 2008).
In Vitro Translation Assay
In vitro translation assays using the transcription/translation-coupled rabbit reticulocyte lysate system (Promega) were performed with pcDNA3-WT-uORF-C/EBPα in the presence or absence of 1 μg recombinant MBP-hnRNP E2 (Perrotti et al., 2002) either with or without 1000× mature miR-328 or miR-330 oligoribonucleotides.
Western Blotting, Coimmunoprecipitation, and ChIP Assays
For western blot, 1 × 107 cells were lysed (0°C; 30 min) in 50–100 μl RIPA buffer, clarified, and subjected to SDS-PAGE. For C/EBPα detection, 106 cells were directly lysed in 20 μl Laemmli buffer and denatured prior to SDS-PAGE and transfer to nitrocellulose. For coIP, cells were lysed on ice with immunoprecipitation buffer and 1.0 mg of protein was used in coimmunoprecipitation assays. Chromatin immunoprecipitation (ChIP) assays were performed using an EZ-Chip kit (Millipore).
Statistics
Data were analyzed as follows: (1) two-tailed paired Student’s t test for assays with identical cell lines, untreated and imatinib-treated SCLtTA-BCR/ABL cells, RIP assay densitometric and qRT-PCR, and in vitro translation assays; (2) two-tailed independent Student’s t test for clonogenic assays with unpaired miRNA-infected Lin− BM cells; and (3) the Mann-Whitney rank sums test for assays with unpaired CML patient samples. A p value of less than 0.05 was considered statistically significant.
Supplementary Material
Supplementary Text
Supplementary Video
Acknowledgments
This work was supported in part by grants from the National Cancer Institute CA095512 (D.P.), CA16058 (OSU-CCC), NIH, Bethesda, MD, USA; the US Army, CML Research Program, W81XWH-07-1-0270 (D.P.); the American-Italian Cancer Foundation (P.N.); Fonds de la Recherche en Sante du Quebec (D.C.R.); and AGGRS from The OSU Graduate School (A.M.E.). D.P. is a Scholar of The Leukemia and Lymphoma Society. G.A.C. is a Fellow of the M.D. Anderson Research Trust and Scholar of the University of Texas System Regents. We thank J.-S. Chang, M. Odeh, and A. Martin for technical assistance and S. Lee for editorial assistance; T. Skorski (Temple University, Philadelphia, PA, USA) for providing PIM1 cDNAs; F. Racke (OSU, Columbus OH, USA) for histopathology analysis; B. Calabretta (TJU, Philadelphia, PA, USA), R. Arlinghaus (MDACC, Houston, TX, USA), and K. Huebner (OSU) for critical reading of the manuscript.
Footnotes
Supplemental Information includes Extended Experimental Procedures and two figures and can be found with this article online at doi:10.1016/j.cell. 2010.01.007.
References
- Agirre X, Jimenez-Velasco A, San Jose-Eneriz E, Garate L, Bandres E, Cordeu L, Aparicio O, Saez B, Navarro G, Vilas-Zornoza A, et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol Cancer Res. 2008;6:1830–1840. doi: 10.1158/1541-7786.MCR-08-0167. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, et al. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Bueno MJ, Perez de Castro I, Gomez de Cedron M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernandez-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Carpenter B, MacKay C, Alnabulsi A, MacKay M, Telfer C, Melvin WT, Murray GI. The roles of heterogeneous nuclear ribonucleoproteins in tumour development and progression. Biochim Biophys Acta. 2006;1765:85–100. doi: 10.1016/j.bbcan.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Chang JS, Santhanam R, Trotta R, Neviani P, Eiring AM, Briercheck E, Ronchetti M, Roy DC, Calabretta B, Caligiuri MA, Perrotti D. High levels of the BCR/ABL oncoprotein are required for the MAPK-hnRNP E2 dependent suppression of C/EBPalpha-driven myeloid differentiation. Blood. 2007;110:994–1003. doi: 10.1182/blood-2007-03-078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Eiring AM, Neviani P, Santhanam R, Oaks JJ, Chang JS, Notari M, Willis W, Gambacorti-Passerini C, Volinia S, Marcucci G, et al. Identification of novel posttranscriptional targets of the BCR/ABL oncoprotein by ribonomics: requirement of E2F3 for BCR/ABL leukemogenesis. Blood. 2008;111:816–828. doi: 10.1182/blood-2007-05-090472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, Willis AE. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
- Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Ferrari-Amorotti G, Keeshan K, Zattoni M, Guerzoni C, Iotti G, Cattelani S, Donato NJ, Calabretta B. Leukemogenesis induced by wild-type and STI571-resistant BCR/ABL is potently suppressed by C/EBPalpha. Blood. 2006;108:1353–1362. doi: 10.1182/blood-2006-01-011833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura K, Kano F, Murata M. Identification of PCBP2, a facilitator of IRES-mediated translation, as a novel constituent of stress granules and processing bodies. RNA. 2008;14:425–431. doi: 10.1261/rna.780708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- George AD, Tenenbaum SA. MicroRNA modulation of RNA-binding protein regulatory elements. RNA Biol. 2006;3:57–59. doi: 10.4161/rna.3.2.3250. [DOI] [PubMed] [Google Scholar]
- Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat Struct Mol Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–3181. doi: 10.1007/s00018-008-8252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoover RR, Gerlach MJ, Koh EY, Daley GQ. Cooperative and redundant effects of STAT5 and Ras signaling in BCR/ABL transformed hematopoietic cells. Oncogene. 2001;20:5826–5835. doi: 10.1038/sj.onc.1204549. [DOI] [PubMed] [Google Scholar]
- Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by micro-RNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. 2006;1:302–307. doi: 10.1038/nprot.2006.47. [DOI] [PubMed] [Google Scholar]
- Keeshan K, Santilli G, Corradini F, Perrotti D, Calabretta B. Transcription activation function of C/EBPalpha is required for induction of granulocytic differentiation. Blood. 2003;102:1267–1275. doi: 10.1182/blood-2003-02-0477. [DOI] [PubMed] [Google Scholar]
- Khabar KS. The AU-rich transcriptome: more than interferons and cytokines, and its role in disease. J Interferon Cytokine Res. 2005;25:1–10. doi: 10.1089/jir.2005.25.1. [DOI] [PubMed] [Google Scholar]
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J. 2004;23:3092–3102. doi: 10.1038/sj.emboj.7600305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev AV, Liebhaber SA. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo JV, Barnes DJ. Chronic myeloid leukaemia as a model of disease evolution in human cancer. Nat Rev Cancer. 2007;7:441–453. doi: 10.1038/nrc2147. [DOI] [PubMed] [Google Scholar]
- Nieborowska-Skorska M, Hoser G, Kossev P, Wasik MA, Skorski T. Complementary functions of the antiapoptotic protein A1 and serine/threonine kinase pim-1 in the BCR/ABL-mediated leukemogenesis. Blood. 2002;99:4531–4539. doi: 10.1182/blood.v99.12.4531. [DOI] [PubMed] [Google Scholar]
- Notari M, Neviani P, Santhanam R, Blaser BW, Chang JS, Galietta A, Willis AE, Roy DC, Caligiuri MA, Marcucci G, Perrotti D. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood. 2006;107:2507–2516. doi: 10.1182/blood-2005-09-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostareck-Lederer A, Ostareck DH. Control of mRNA translation and stability in haematopoietic cells: the function of hnRNPs K and E1/E2. Biol Cell. 2004;96:407–411. doi: 10.1016/j.biolcel.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K, Iervolino A, Condorelli F, Gambacorti-Passerini C, Caligiuri MA, Calabretta B. BCR-ABL suppresses C/EBPalpha expression through inhibitory action of hnRNP E2. Nat Genet. 2002;30:48–58. doi: 10.1038/ng791. [DOI] [PubMed] [Google Scholar]
- Perrotti D, Neviani P. From mRNA metabolism to cancer therapy: chronic myelogenous leukemia shows the way. Clin Cancer Res. 2007;13:1638–1642. doi: 10.1158/1078-0432.CCR-06-2320. [DOI] [PubMed] [Google Scholar]
- Rosmarin AG, Weil SC, Rosner GL, Griffin JD, Arnaout MA, Tenen DG. Differential expression of CD11b/CD18 (Mo1) and myeloperoxidase genes during myeloid differentiation. Blood. 1989;73:131–136. [PubMed] [Google Scholar]
- Schultheis B, Szydlo R, Mahon FX, Apperley JF, Melo JV. Analysis of total phosphotyrosine levels in CD34+ cells from CML patients to predict the response to imatinib mesylate treatment. Blood. 2005;105:4893–4894. doi: 10.1182/blood-2005-01-0210. [DOI] [PubMed] [Google Scholar]
- Tenen DG. Disruption of differentiation in human cancer: AML shows the way. Nat Rev Cancer. 2003;3:89–101. doi: 10.1038/nrc989. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Venturini L, Battmer K, Castoldi M, Schultheis B, Hochhaus A, Muckenthaler MU, Ganser A, Eder M, Scherr M. Expression of the miR-17-92 polycistron in chronic myeloid leukemia (CML) CD34+ cells. Blood. 2007;109:4399–4405. doi: 10.1182/blood-2006-09-045104. [DOI] [PubMed] [Google Scholar]
- Wagner K, Zhang P, Rosenbauer F, Drescher B, Kobayashi S, Radomska HS, Kutok JL, Gilliland DG, Krauter J, Tenen DG. Absence of the transcription factor CCAAT enhancer binding protein alpha results in loss of myeloid identity in bcr/abl-induced malignancy. Proc Natl Acad Sci USA. 2006;103:6338–6343. doi: 10.1073/pnas.0508143103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CH, Lee DY, Deng Z, Jeyapalan Z, Lee SC, Kahai S, Lu WY, Zhang Y, Yang BB. MicroRNA miR-328 regulates zonation morphogenesis by targeting CD44 expression. PLoS ONE. 2008;3:e2420. doi: 10.1371/journal.pone.0002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Wang X, Ward AC, Touw IP, Friedman AD. C/EBPalpha and G-CSF receptor signals cooperate to induce the myeloperoxidase and neutrophil elastase genes. Leukemia. 2001;15:779–786. doi: 10.1038/sj.leu.2402094. [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Supplementary Video