Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology (original) (raw)
Abstract
We previously demonstrated high-frequency, targeted DNA addition mediated by the homology-directed DNA repair pathway. This method uses a zinc-finger nuclease (ZFN) to create a site-specific double-strand break (DSB) that facilitates copying of genetic information into the chromosome from an exogenous donor molecule. Such donors typically contain two ∼750 bp regions of chromosomal sequence required for homology-directed DNA repair. Here, we demonstrate that easily-generated linear donors with extremely short (50 bp) homology regions drive transgene integration into 5–10% of chromosomes. Moreover, we measure the overhangs produced by ZFN cleavage and find that oligonucleotide donors with single-stranded 5′ overhangs complementary to those made by ZFNs are efficiently ligated in vivo to the DSB. Greater than 10% of all chromosomes directly incorporate this exogenous DNA via a process that is dependent upon and guided by complementary 5′ overhangs on the donor DNA. Finally, we extend this non-homologous end-joining (NHEJ)-based technique by directly inserting donor DNA comprising recombinase sites into large deletions created by the simultaneous action of two separate ZFN pairs. Up to 50% of deletions contained a donor insertion. Targeted DNA addition via NHEJ complements our homology-directed targeted integration approaches, adding versatility to the manipulation of mammalian genomes.
INTRODUCTION
The insertion of exogenous genetic information into the genome of target cells is broadly used in basic and applied biology. Gene insertion is conventionally achieved via virus-mediated or spontaneous integration of transfected DNA followed by a selection for cells carrying the new DNA. In the context of cell-based medicine, lack of control over the transgene integration site can result in adverse events due to insertional mutagenesis (1). In industrial use, uncontrolled transgene integration gives unwanted phenotypic heterogeneity due to the varying permissivity of integration sites for transgene expression (position-effect variation; 2). In both situations, it would be advantageous to target DNA insertion to a specific, desirable site in the genome.
Targeted gene addition is typically performed by transfection of a selectable marker gene flanked by a substantial amount of DNA homologous to the target locus. Spontaneous DSBs are formed at the target locus, likely from stalled DNA replication forks. While normally repaired inerrantly by homology-directed repair (HDR) templated by the sister chromosome, HDR can instead use the homologous donor DNA to heal the break. When additional DNA sequence is inserted between the two regions of homology in the donor plasmid, the cellular DNA repair machinery unwittingly copies this genetic information into the chromosome (3,4). As this homology-based targeting relies on the capture of very rare DSBs within the region of donor homology, extensive homology to the target locus is needed to obtain targeted integration at a useful frequency. Six to seven kilobases of DNA homologous to the chromosomal target are commonly used in donor construction, although more extensive homology increases targeting efficiency (5). Despite these enormous stretches of homology, the frequency of successful gene targeting is typically in the range of 10−5 to 10−6 prior to selection (5). After selective pressure is applied, resistant clones are screened to identify the minority that contain the intended targeted insertion (6).
Creation of a targeted DSB can dramatically increase the frequency of homologous recombination in mitotically dividing cells (7). The custom engineering of site-specific nucleases has therefore accelerated targeted integration technology. One type of designer nuclease is based on zinc-finger DNA binding motifs, spaced at 3-bp intervals across DNA (8). Zinc fingers that recognize a wide variety of DNA sequences can be joined together to create DNA binding proteins that recognize a 9–18 bp sequence (8–10). Zinc-finger nucleases (ZFNs) are fusions between zinc-finger DNA binding domains and the nuclease domain of the type IIs restriction enzyme FokI (11). When two such ZFN fusions bind at adjacent sites on the chromosome, the nuclease domains interact to create a DSB in the DNA (11–13). The non-homologous end-joining (NHEJ) pathway can directly ligate the broken ends together, often with a gain or loss of several base pairs (14). Alternately, the cell can perform the highly faithful HDR described above.
Exploiting HDR of ZFN-induced DSBs, we previously demonstrated targeted integration of several kilobase transgenes at multiple endogenous loci at frequencies of 5–15% without the need for any selective pressure (15–17). In contrast to the essentially random locations of spontaneous DSBs, the ZFNs specify the position of the DSB in these experiments. Since the large amount of homologous DNA present in conventional targeted-integration donors is necessary primarily to expand the region where a spontaneous DSB can be captured, use of a ZFN to make a site-specific DSB allowed the amount of donor homology to be reduced to ∼1.5 kb. While an improvement, the ZFN-promoted targeted integration process is still hampered by the need for construction of a donor plasmid or virus via conventional recombinant DNA techniques. Additionally, although the majority of targeted gene addition likely happens within the first few days after transfection, experimental output can be assayed only after the 3–4 weeks required for loss of donor–plasmid gene expression. More preferable would be an easily created donor molecule with a short half-life post-transfection. We therefore evaluated whether PCR-generated donors with very limited target site homology could be readily used for targeted gene addition. Use of such donors for manipulation of the yeast Saccharomyces cerevisiae has greatly facilitated molecular genetics in this organism (18).
In addition to improving homology-directed gene targeting, we were interested in providing similar gene addition capability to cell types lacking efficient homology-based DNA repair. Such an approach might prove particularly useful for gene addition in primary, non-dividing cells which preferentially use the NHEJ DNA repair pathway. Gene addition via NHEJ would also be useful for unsequenced genomes (e.g. Cricetulus griseus, CHO cells) as donor construction without a genome sequence requires arduous preliminary cloning and sequencing. Several previous investigators have found that non-specific DNA can be captured at the site of NHEJ-mediated DSB repair (19). Repetitive element and mitochondrial DNA fragments have been observed to integrate at the site of DSBs in S. cerevisiae (20,21). Similarly, exogenous DNA has been found in the output of the NHEJ pathway in mammalian cells (22–24). These observations prompted us to investigate whether the information present in the single-stranded overhangs created by ZFN cleavage could be used to perform targeted DNA integration using the NHEJ DNA repair machinery.
We demonstrate here ZFN-driven targeted integration of genetic information into five loci in mammalian cells using the homology-directed DNA repair pathway. HDR donors with as little as 100 bp of chromosomal homology (50 bp per arm) generally yielded targeted integration frequencies similar to plasmid donors with 15-fold more homology. We determined the types of 5′ overhangs left by ZFNs using high-throughput DNA sequencing of molecules cleaved in vitro and used this knowledge to drive DNA integration via the complementary NHEJ DNA repair pathway. This direct insertion of exogenous DNA into the chromosome was demonstrated at four loci in two cell types at a frequency similar to HDR-based integration (up to 10%). Capture of DNA by NHEJ worked well at both a single DSB and at a deletion created by two DSBs. As the linear donors in both experiments were made by PCR amplification or chemical synthesis, both types allow for rapid experimentation and ascertainment of targeted integration.
METHODS
Donor plasmids and oligonucleotides
Oligonucleotides used to create synthetic linear donors for AAVS1 are _AAVS1_-100F: 5′-c*c*t gtg tcc ccg agc tgg gac cac cTT ATA TTC CCA GGG CCG GTT AAT GTg gct ctg gtt ctg ggt act ttt atc tgt ccc ctc cac ccc aca gtg ggg c-3′ and _AAVS1_-100R: 5′-a*a*t ctg cct aac agg agg tgg ggg tTA GAC CCA ATA TCA GGA GAC TAG GAa gga gga ggc cta agg atg ggg ctt ttc tgt cac caa tcc tgt ccc tag t-3′ where regions of alternating case indicate the termini of analogous oligonucleotides with 75 and 50 bp of AAVS1 homology. Donors were generated by PCR of the AAVS1 donor plasmid described below. These oligonucleotides contain 2 bp of phosphorothioate linkages at their 5′-termini indicated by asterisks. Oligonucleotides used to create synthetic linear donors for _IL2R_γ are GC-50F: 5′-g*t*g tgg atg ggc aga aac gct aca cgt ttc gtg ttc gga gcc gct tta ac-3′ and GC-50R: 5′-t*g*g att ggg tgg ctc cat tca ctc caa tgc tga gca ctt cca cag agt gg-3′. Donors were generated by PCR of the _IL2R_γ donor plasmid (25).
Double-stranded oligonucleotides for direct insertion into the chromosome were annealed in 50 mM NaCl, 10 mM Tris pH 7.5 and 1 mM EDTA at a final concentration of 40 or 500 uM each (Figures 4 and 5, respectively). Correct annealing was verified by non-denaturing polyacrylamide gel electrophoresis. Oligonucleotides are as follows: AAVS1 F, (5′-g*c*c agc tta ggt gag aat tcg gcg gat ccc gaa gct tgc taa ctc agc c-3′); AAVS1 R, (5′-t*g*g cgg ctg agt tag caa gct tcg gga tcc gcc gaa ttc tca cct aag c-3′). These oligonucleotides, identical versions lacking the first four bases, and versions with the first four bases changed to 5′-ctgg-3′ and 5′-ccag-3′, respectively, were used as donors in Figure 4B. These oligonucleotides and versions with the first four bases changed to 5′-aaga-3′ and 5′-tctt-3′, respectively, were used as donors in Figure 4C. Oligonucleotides for insertion into the POU5F1 deletion were loxP F, (5′-t*t*t ggg aat tca taa ctt cgt ata gca tac att ata cga agt tat gga tcc-3′) and loxP R (5′-t*g*c agg atc cat aac ttc gta taa tgt atg cta tac gaa gtt atg aat tc-3′); for BAK, the first 5 bp of the loxP F oligo was replaced by 5′-cagc-3′ in combination with the loxP R oligo with its first 4 bp changed to 5′-ccca-3′. All oligonucleotides were 5′ phosphorylated and contain phosphorothioate linkages between the 5′-terminal two bases unless otherwise noted.
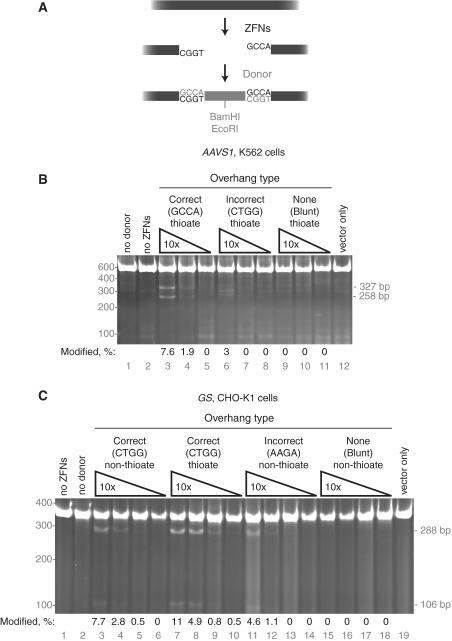
Figure 4.
Targeted DNA integration via non-homologous end joining. (A) Diagram of ZFN cleavage at AAVS1 resulting in 4-bp 5′ overhangs followed by in vivo ligation of a complimentary-overhang donor. The donor contains both BamHI and EcoRI restriction enzyme sites. (B) NHEJ capture at the AAVS1 locus in K562 cells. Three 10-fold dilutions (40–0.4 µM) of donor DNA with the indicated overhang types were co-transfected with the AAVS1 ZFNs. The PCR amplicons were cut with EcoRI, producing 327 and 258 bp products from amplicons with insertion of the oligonucleotide. All donors in this experiment contain terminal phosphorothioate residues. (C) NHEJ capture at the GS locus in CHO-K1 cells. Four 10-fold dilutions of donor DNAs (40–0.04 µM) with the indicated overhang types and phosphorothioate usage were co-transfected with the GS ZFNs. The PCR amplicons were cut with BamHI, producing 288 and 106 bp products from amplicons with insertion of the oligonucleotide. For (B and C), the percentage of modified chromosomes is shown below each lane in black text; the position of the molecular weight markers used is shown in grey text on the left of the gel.
Figure 5.
Targeted DNA integration at deletions via non-homologous end joining. (A) Diagram of dual ZFN cleavage at POU5F1 resulting in 5-bp 5′ overhangs from the left ZFN pair and 4-bp 5′ overhangs from the right ZFN pair followed by in vivo ligation of a complimentary-overhang donor. The donor contains both BamHI and EcoRI restriction enzyme sites. (B) NHEJ capture at a POU5F1 deletion in K562 cells and at BAK in CHO-K1 cells. Left (L) and right (R) ZFN pairs were transfected individually and in combination (LR), with (+) and without donor co-transfection. The sizes of significant PCR products are shown on the right side of the gels. As deletions are heterogeneous, their expected sizes are indicated with tildes. Due to the relatively small deletion made in POU5F1, amplification of the wild-type locus is seen (1956 bp). The deletion quantitiation shown below the gel is from an independent analysis of cell pools described in the main text; the deletion frequency in lane 6 was not measured. (C) Restriction enzyme digestion confirms targeted integration into the POU5F1 and BAK deletions. PCR reactions from lanes 3, 6, 7, 11, 14 and 15 in (B) were divided into thirds, one-third was left uncut (blank), one third was digested with BamHI (B), and one-third was digested with EcoRI (E). The sizes of BamHI and EcoRI digestion products are shown on the right side of the gel, POU5F1 in the left column, BAK in the right. The amount of digested DNA was determined by densitometry.
The plasmid donor for the AAVS1 locus was made by PCR with HAL-F: 5′-tgc ttt ctc tga cca gca tt-3′ and HAL-R: 5′-cca ctg tgg ggt gga ggg ga-3′ for the left homology region and with HAR-F: 5′-tag gga cag gat tgg tga ca-3′ and HAR-R: 5′-ccc tta gag cag agc cag ga-3′. This plasmid has 1641 bp of AAVS1 homology and a 12-bp sequence (5′-ggc aag ctt tac-3′) containing a HindIII site between the regions of homology. A donor transgene was inserted into the HindIII site to make the donor plasmid (17). AAVS1 donors with variable-length homology arms were constructed by cloning of fusion PCR products bounded by the following oligonucleotides: For 750 bp donors, 5′-ctt tct ctg acc agc att ctc tcc-3′ and 5′-ccc tta gag cag agc cag gaa cc-3′; for 500 bp donors, 5′-ggt tcc ctt ttc ctt ctc ctt ctg g-3′ and 5′-acg ggg ctg gct act ggc c-3′; for 250 bp donors, 5′-ctc ccc tac ccc cct tac ctc tc-3′ and 5′-aac cgg gca ggt cac gca tc-3′; for 100 bp donors, 5′-gat cct gtg tcc ccg agc tgg-3′ and 5′-gaa tct gcc taa cag gag gtg gg-3′ and combinations of the above.
The plasmid donor for the GS locus was made by PCR with GJC 185F (5′-tta ctg tcc aga gac agg ag-3′) and GJC 184R (5′-cag gaa tgg gct tgg ggt c-3′) for the left homology region with and GJC 182F (5′-aat ggt gca ggc tgc cat a-3′) and GJC 183R (5′-ttc ttc tcc tgg ccg aca gt-3′) for the right homology region. This plasmid has 1605 bp of GS flanking homology and a 17 bp sequence containing a SalI site (5′-atc gat gtc gac ccg gg-3′) between the homology regions.
The plasmid donors for the CCR5 locus contains a left homology region bounded by 5′-aat tgt tgt caa agc ttc at-3′ and 5′-atg agg atg acc agc atg tt-3′ with a right arm bounded by 5′-aaa ctg caa aag gct gaa ga-3′ and 5′-aaa tca cac atg aaa agt gt-3′. This plasmid has 1878 bp of CCR5 flanking homology and a 52 bp sequence containing two XbaI sites (5′-(t)cta gat cag tga gta tgc cct gat ggc gtc tgg act gga tgc ctc gtc tag a-3′) between the homology regions.
ZFN design, production and assay
ZFNs were designed as described previously (IL2Rγ, AAVS1, POUF5, GS; 17,25,26) or were provided by Sigma-Aldrich (BAK, Rosa). Obligate heterodimer nucleases were constructed as described and are referred to as HiFi nucleases (27). ZFN cleavage in vivo was assayed as described (28).
Cell growth and transfection
CHO-K1 and K562 cells were obtained from the American Type Culture Collection and grown and transfected via nucleofection as described (25,28). For donor capture by NHEJ, one million K562 cells, 3 µg 2A-linked AAVS1 ZFNs, and 2000, 200, 20 or 0 nM donor were transfected in 100 µl. At the GS locus, one million CHO-K1 cells, 3 µg 2A-linked GS ZFNs, and 2000, 200, 20, 2 or 0 nM donor were transfected in 100 µl. For donor capture by NHEJ at the site of a deletion, one million K562 cells, 2 µg each POU5F1 ZFN, and 40, 4 or 0 µM donor were transfected in 100 µl.
Alteration of donor topology, methylation and terminal homology
To compare linear donors versus circular, supercoiled donors, the donor plasmids were cut with ScaI. ScaI cleaves the donor plasmids once, in the β-lactamase gene. Per 200 000 cells, 1 µg of each donor was used in this experiment.
Topologically relaxed and alternately methylated donors for the GS, AAVS1 and CCR5 loci were prepared in parallel to obtain combinations of supercoiled, relaxed, Dam/Dcm-methylated, CpG-methylated and unmethylated DNA. Supercoiled Dam/Dcm-methylated donors were obtained through transformation and culture of Top 10 cells (Invitrogen). Supercoiled, unmethylated donors were obtained by substitution of Top10 cells with the dam−dcm− Escherichia coli strain ER2925 (New England Biolabs). Relaxed donors were produced by a 12-h 37°C incubation of 60 µg of supercoiled donor with 5 U of Topoisomerase I (New England Biolabs) in 1 × Buffer 4 and 100 µg/ml BSA in a 30 µl reaction volume. CpG-methylated supercoiled and relaxed donors were produced by a 12-h 37°C incubation of 50 µg of unmethylated donor with four units of CpG methyltransferase (New England Biolabs) in 1 × TE, 50 mM NaCl, 1 mM DTT and 160 µM SAM in a 30 µl reaction volume. Supercoiling and relaxation were verified by TAE agarose gel electrophoresis. CpG methylation was assayed by the acquisition of insensitivity to NotI.
To ensure ectopic recombination was not responsible for a signal mimicking targeted integration, we made and tested linear, PCR-derived donors terminating in dideoxycytidine. Using the AAVS1 donor plasmid as a template, donors were made with 250 bp or 100 bp of chromosomal homology by PCR with Deep Vent polymerase (New England Biolabs). (DeepVent polymerase lacks terminal transferase activity and therefore produces blunt-ended PCR products.) For 250 bp donors, the oligonucleotides used for PCR were 5′-c*t*c ccc tac ccc cct tac ctc tc-3′ and 5′-a*a*c cgg gca ggt cac gca tc-3′; for 100 bp donors, 5′-g*a*t cct gtg tcc ccg agc tgg-3′ and 5′-g*a*a tct gcc taa cag gag gtg gg-3′, where the asterisks indicate a phosphorothioate linkage. Twenty picomoles (∼7 µg) of these two donor PCR products were tailed with terminal transferase (New England Biolabs) and dideoxycytidine (USB/Affymetrix). Care was taken during oligonucleotide design to ensure that addition of cytidine would maintain homology with the AAVS1 locus (i.e. for both donors the next base in AAVS1 is naturally a C). Per 200 000 cells, 1 µg of the 250 bp plasmid donor, 0.9 µg of the 100 bp plasmid donor, 142 ng of the 250 bp linear donor and 56 ng of the 100 bp linear donor was used in this experiment.
Analysis of donor capture by NHEJ and HDR
All primers used for analysis of HDR-based gene addition are outside the homology regions of plasmid and linear donors.
For analysis of HDR-based gene addition at AAVS1, PCR reactions contained 100 ng genomic DNA, 1 × Accuprime Buffer II and 1 U Accuprime Taq DNA Polymerase High Fidelity (Invitrogen), 4 µCi 32P-dGTP, and 50 uM each of primers HDRF4 (5′-cgg aac tct gcc ctc taa cg-3′) and HDRR5 (5′-ctg gga tac ccc gaa gag tg-3′). PCR reactions were carried out for 23 cycles of amplification; quantitation of 21- and 25-cycle amplicons gave comparable data. The annealing temperature was 62°C and the extension time 3:00.
For analysis of HDR-based gene addition at IL2Rg, PCR reactions contained 50 ng genomic DNA, 1 × Accuprime Buffer II and 1 U Accuprime Taq DNA Polymerase High Fidelity (Invitrogen), 4 µCi 32P-dGTP, and 50 uM each of primers F4 (5′-cca cag ctg gac tgt gag tga cta gg-3′) and R4 (5′-gtg att ctg tgt tct ctg tgc ctg-3′). PCR reactions were carried out for 23 cycles of amplification; quantitation of 21- and 25-cycle amplicons gave comparable data. The annealing temperature was 62°C and the extension time 3:30.
For analysis of HDR-based gene addition at GS, PCR reactions contained 100 ng genomic DNA, 1 × Accuprime Buffer II and 1 U Accuprime Taq DNA Polymerase High Fidelity (Invitrogen), and 50 uM each of primers GJC 180F (5′-agc ttc ctc ccc ata agt tc-3′) and GJC 179R (5′-ggc ggt ctt caa agt aac ct-3′). PCR reactions were carried out for 30 cycles of amplification. The annealing temperature was 60°C and the extension time 1:30. Insertion of the 17 bp sequence (5′-atc gat gtc gac ccg gg-3′) into GS was assayed by digestion with 10 U of SalI for 2 h.
For analysis of HDR-based gene addition at CCR5, PCR reactions contained 100 ng genomic DNA, 1 × Accuprime Buffer II and 1 U Accuprime Taq DNA Polymerase High Fidelity (Invitrogen), and 50 uM each of primers CCR5F (5′-ctg cct cat aag gtt gcc cta ag-3′) and CCR5R (5′-cca gca ata gat gat cca act caa att cc-3′). PCR reactions were carried out for 30 cycles of amplification. The annealing temperature was 60°C and the extension time 1:30. Insertion of the XbaI-containing sequence was assayed by digestion with 10 U of XbaI for 2 h.
Southern blot analysis at AAVS1 was done with XmnI-digested chromosomal DNA probed with a radiolabelled 474 bp BamHI fragment of the left AAVS1 homology arm.
For analysis of donor capture via NHEJ, the AAVS1 locus was PCR-amplified with AAVS1 CEL-I F2 (5′-ccc ctt acc tct cta gtc tgt gc-3′) and AAVS1 CEL-I R1 (5′-ctc agg ttc tgg gag agg gta g-3′). The GS locus was PCR-amplified with GS F5928 (5′-ggg tgg ccc gtt tca tct-3′) and GS R6272 (5′-cgt gac aac ttt ccc ata tca ca-3′). POU5F1 was amplified using Group3F (5′-gat aga acg aga ttc cgt ctt ggt gg-3′) and Group4R (5′-gca gag ctt tga tgt cct ggg act-3′). BAK was amplified using GJC 24F (5′-cat ctc aca tct gga cca cag ccg-3′) and GJC 163R (5′-ctg cgg gca aat aga tca c-3′). All PCR reactions done for analysis of donor capture by NHEJ-contained 100 ng genomic DNA, 1 × Accuprime Buffer II, and 1 U Accuprime Taq DNA Polymerase High Fidelity (Invitrogen), and 50 uM each of the appropriate primer. PCR reactions were carried out for 30 cycles of amplification. The annealing temperature was 60°C and the extension time 0:30.
Quantitation of all gels was performed by densitometry with Imagequant 5.1 software. Care was taken during photography and autoradiography to ensure that no portion of the image was saturated. Longer exposure gel photographs are displayed in the figures to show sometimes low-abundance bands.
Sequencing of donor insertions
Samples containing transgene insertion into AAVS1 were PCR-amplified with the HDRF4 and HDRR5 primers. The insert band was gel purified and nested PCR was performed with the CEL-I F2 and CEL-I R1 primers. Insert-containing bands were excised, cloned, and sequenced.
Samples from Figure 4C, lanes 3 and 7 were PCR amplified with GJC 172F (5′-atc cgc atg gga gat cat ct-3′) and GJC 171R (5′-gcc ttg gtg cta aag ttg gt-3′), electrophoresed on a 10% polyacrylamide gel, and donor-specific bands excised and purified. A second round of PCR was performed and the resulting fragments cloned and sequenced. Sequence reads that were wild-type or consistent with NHEJ events not resulting in donor insertion were not considered further.
The donor-specific bands from Figure 5B, lanes 7 and 14 were excised from the gel, purified and re-amplified with Group3F and Group4R (POU5F1) or GJC 24F and GJC 163R (BAK), cloned and sequenced. Sequence reads of deletions that did not contain inserts were not considered further.
Determination of ZFN-generated overhangs
Oligos containing ZFN target sites for the AAVS1 (5′-tgt ccc ctc cAC CCC ACA GTG Ggg cca cTA GGG ACA GGA Ttg gtg aca ga-3′), GS (5′-gac cCC AAG CCC ATT CCT GGG Aac tgg aAT GGT GCA GGC Tgc cat acc aa-3′) and IL2Rγ (5′-gtt tcg tgt tCG GAG CCG CTT Taa ccc ACT CTG TGG AAG tgc tca gca tt-3′) ZFN pairs were annealed to their reverse complements in 50 mM NaCl, 10 mM Tris pH 7.5 and 1 mM EDTA. Capital letters denote the ZFN binding sites, while lowercase letters denote flanking and spacer sequence. The double-stranded products were then cloned into the EcoRV site of the pBluescript II KS−.
ZFNs were synthesized in vitro by means of a T7-coupled transcription/translation kit using rabbit reticulocyte lysate (Promega). For the 2A-linked ZFNs AAVS1 (SBS 15 556 and 15 590) and GS (SBS 9372 and 9075), 30 ng of plasmid were used; for the unlinked IL2Rγ ZFNs (SBS 7263 and 7264), 20 ng of each plasmid were used. Transcription and translation reactions (60 µl) were supplemented with 500 µM ZnCl2 and incubated for 1.5 h at 30°C. ZFN-containing lysates were used for DNA cleavage within 30 min. With the exception of those targeting IL2Rγ, all ZFNs used were of the HiFi variety (27).
Cleavage reactions (35 µl) contained 2.5 µg of target plasmid, 28.5 µl of reticulocyte lysate, 10 mM EGTA and 1 × Restriction Buffer 2 (New England Biolabs) and were incubated at 37°C. Control experiments with HindIII in ZFN-free lysate, and Hind III in 1 × NEB Buffer 2 were also conducted. Plasmid linearization required the presence of the correct ZFN pair (data not shown). Reactions with AAVS1, IL2Rγ and HindIII were terminated after 2 min and the GS reaction after 5 min by addition of 10 mM Tris/1 mM EDTA to 200 µl, followed by phenol extraction and ethanol precipitation. Linearized plasmids were gel purified by agarose gel electrophoresis and incubated for 30 min at 37°C with 0.05 U Klenow DNA polymerase (New England Biolabs) in 1 × Buffer 2, plus 50 µM dNTPs. Klenow polymerase was inactivated by incubation at 75°C for 20 min, followed by addition of 20 U of T4 DNA Ligase (New England Biolabs) and ATP to 1 mM.
Ligation reactions were amplified with 30 cycles of PCR using target-specific primers containing standard Illumina sequencing regions. PCR products were purified with the QIAquick Gel Extraction Kit, then re-purified with a GeneJET PCR Purification Kit (Fermentas), and eluted in 0.1 × elution buffer. Samples were mixed together at an equimolar ratio and submitted for 34 bp read length Illumina DNA sequencing (Elim Biopharmaceuticals). Sequencing reads with a quality score of at least 30 were binned using a custom Python script. A quality score cutoff of 2 was used for AAVS1 reads due to a template-specific sequencing anomaly that reduced quality scores without an actual adverse effect on sequence interpretability. Wild-type target sequences (5–15% of the total) were discarded and the top 10 bins for each target were analyzed manually. Percentages given in the text were calculated using the relevant bin as the numerator and the entire collection of reads as the denominator. The percentages shown do not sum to 100% as the unanalyzed sequences (∼1500 bins with 0.2–0.0001% each) were not analyzed. For HindIII in buffer 2, 573 490 sequence reads were analyzed; for HindIII in reticulocyte lysate, 3 473 683; for IL2Rγ, 1 985 413; for GS, 2 389 486; AAVS1, 3 111 505.
RESULTS
We assembled a ZFN pair which cleaves ∼33% of chromosomes in intron 1 of the AAVS1 gene (PPP1R12C) and a plasmid donor molecule containing two ∼750-bp regions of AAVS1 sequence flanking a transgene (Figure 1A; 29). To explore the requirements of the targeted integration reaction, analogous donors were PCR-amplified using oligonucleotides containing 5′ extensions of 50, 75 or 100 bp of AAVS1 sequence from both sides of the DSB created by the AAVS1 ZFNs (Figure 1B). The 5′-terminal two phosphates in the oligonucleotides were derived from phosphorthioamidite nucleotides to make the resulting PCR product resistant to cellular exonucleases.
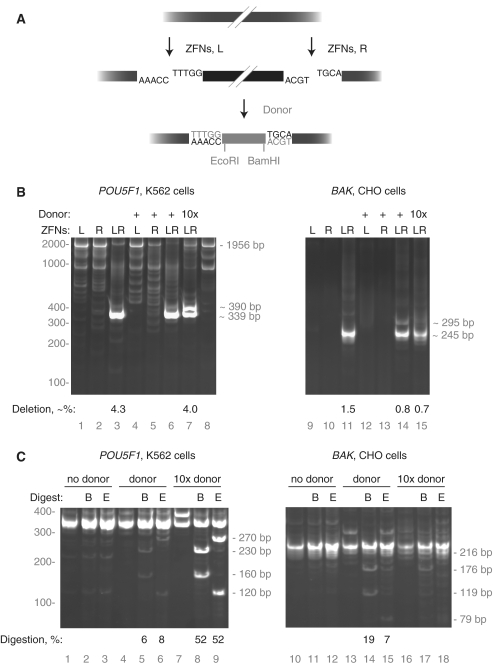
Figure 1.
Homology-based targeted integration using short-homology synthetic donors. (A) Diagram of ZFNs binding to the AAVS1 locus. (B) Schematic of short-homology synthetic donor creation via PCR and comparison with plasmid donor. (C) Targeted integration at AAVS1 with plasmid and short-homology synthetic donors assayed by PCR. Homology region length is given in base pairs for linear donors and kilobase pairs for plasmid donors. DNA type is either plasmid (Pl) or linear phosphorothioate (S). Co-transfection of 7 µg of a linear donor is stoichiometrically equal to 50 µg of this plasmid donor. As PCR preferentially amplifies shorter molecules, the assay slightly under-represents the true frequency of targeted integration. (D) Targeted integration at AAVS1 with plasmid and short-homology synthetic donors assayed by Southern blot. Genomic DNAs used as a PCR template in (B) were analyzed directly by probing for the 5′ region of AAVS1 homology present in the donor plasmid. A map of the locus below the blot shows the position of the homology arms (white box), the probe (grey region with in the white box), the XmnI sites used for digestion. The XmnI fragment from wild-type chromosome is 5336 bp; from an integrated chromosome, 6881 bp. (E) Targeted integration using short-homology synthetic donors at IL2Rγ. Obligate FokI heterodimer ZFNs are referred to as HiFi ZFNs. The DNA type descriptor refers to either plasmid (Pl) or whether the ends of linear donors are normal (O) or phosphorothioate DNA (S). Co-transfection of 5 µg of a linear donor is approximately stoichiometrically equal to 50 µg of this plasmid donor. Homology region length is given in base pairs for linear donors and kilobase pairs for plasmid donors. For (C, D and F) the percentage of modified chromosomes is shown below each lane in black text.
The donor molecules were transfected into the human erythroleukemia cell line K562 with and without co-transfection of the AAVS1 ZFNs. Targeted integration of the donor transgene was monitored by PCR and Southern blot. Integration produced a 3050 bp PCR product that was dependent on the presence of both the donor and the AAVS1 ZFNs (Figure 1C); only the 1953 bp wild-type amplicon was seen in the absence of ZFNs. Surprisingly, the efficiency of integration with short-homology synthetic donors was similar to the plasmid donor even though the plasmid donor has a ∼15-fold longer region of AAVS1 homology (Figure 1C, lanes 7, 8–12). These cell populations were expanded and targeted integration confirmed by Southern blot. Between 6% and 10% of chromosomes contained a correctly targeted transgene insertion in these samples (Figure 1D, lanes 6–10).
To demonstrate the general applicability of these results, we tested analogous reagents at the _IL2R_γ locus (25). Insertion of the donor into the _IL2R_γ locus will produce a 3326 bp product upon PCR whereas the wild-type amplicon is only 1738 bp. Similar to targeted integration at the AAVS1 locus, integration at _IL2R_γ worked about as well with synthetic donor molecules with 50 bp of _IL2R_γ homology at each end as with a conventional plasmid donor with a total of 1500 bp of _IL2R_γ homology (Figure 1E, lanes 5 versus 6, and 8 versus 9). Targeted integration worked both with and without the use of modified FokI domains (27; Figure 1E, lanes 5–7 versus 8–10). Nuclease-resistant phosphorothioate DNA yielded insertion frequencies slightly higher than donors with a conventional DNA backbone (Figure 1E, lane 6 versus 7 and lane 9 versus lane 10).
To ensure that gene addition using short-homology synthetic donor molecules proceeded via the homology-directed DNA repair pathway, the PCR amplicon from the AAVS1 transgene insertion using 50 bp homology regions was cloned and sequenced. Fifty-eight of 60 clones were consistent with transgene insertion via a homology-based process; i.e. showed a perfectly-specific introduction of the new sequence into an otherwise wild-type locus. In one instance, the transgene:AAVS1 junction had mutations consistent with insertion via NHEJ; in the remaining clone, HDR seems to have been used at one end of the molecule and NHEJ at the other. Together, these data demonstrate that linear donor molecules with as little as 50 bp of chromosomal homology are sufficient to drive efficient and targeted transgene insertion via HDR.
The unexpected integration efficiency of short-homology donors prompted us to examine the specific activities of different donor molecules. These two donor types have differing topology, methylation, homology lengths and homology at donor termini. We performed a systematic series of experiments at non-saturating donor concentrations designed to reveal the effect of each of these four differences on HDR.
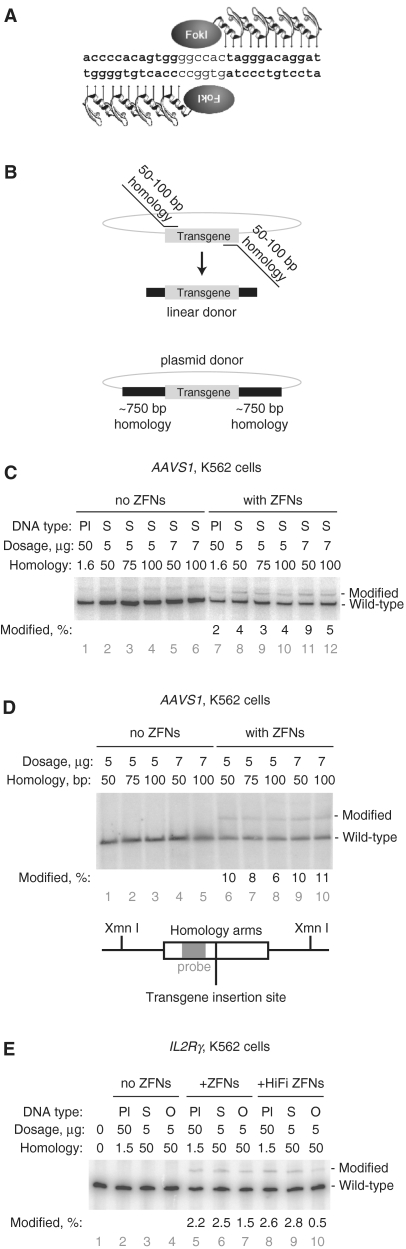
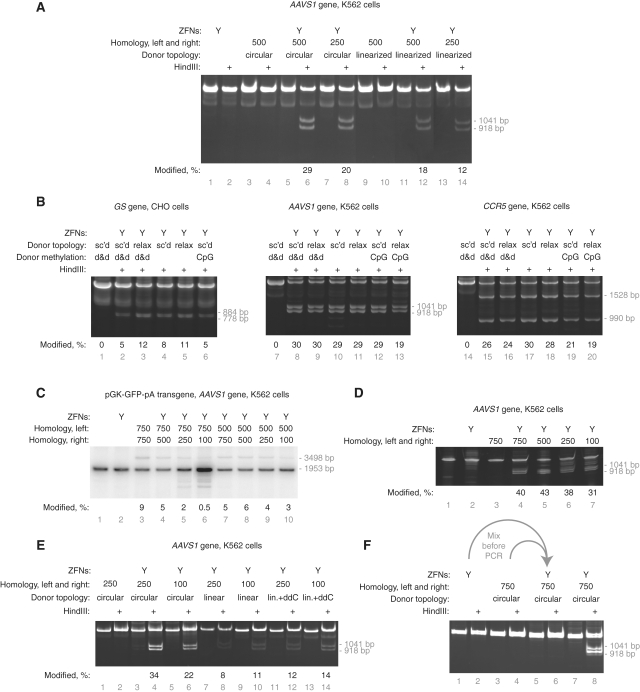
To examine the influence of donor topology on HDR, we measured the integration of a HindIII restriction enzyme site into AAVS1 from supercoiled plasmid donors and from identical donors linearized outside the region of chromosomal homology. We found that circular, supercoiled donors resulted in ∼60% more targeted integration than their linear counterparts. (Figure 2A, compare lanes 6 and 8 with lanes 12 and 14). We then tested a similar AAVS1 donor plasmid and analogous donors for the CCR5 and GS loci, all of which had been relaxed with topoisomerase I (Figure 2B). Donor supercoiling had only a minor affect on targeted integration.
Figure 2.
The effect of donor topology, methylation and terminal sequence homology on targeted integration of a small, HindIII site-containing patch. (A) K562 cells were transfected with supercoiled or linearized (ScaI) plasmid donors containing either 500 or 250 bp of flanking AAVS1 homology. Targeted integration was assayed by HindIII digestion of PCR-amplified chromosomes into 1041 and 918 bp products. (B) Targeted integration with topologically relaxed and differently methylated donors. Donor type is indicated as follows: supercoiled, sc’d; relaxed with topoisomerase I, relax; dam and dcm methylated, d&d; CpG methylated, CpG. Targeted integration was assayed by SalI (GS), HindIII (AAVS1) or XbaI (CCR5) digestion of PCR-amplified chromosomes as appropriate. (C) Insertion of a pGK-GFP-pA transgene into the AAVS1 locus. The length of the homology arms was varied as indicated and targeted integration measured by PCR. PCR amplification of the wild-type locus produces a 1953 bp product; integration of the transgene results in a 3498 bp product. (D) Insertion of a small HindIII-containing patch into the AAVS1 locus in K562 cells. The length of the homology arms was varied as indicated and targeted integration measured by PCR and HindIII digestion. Targeted integration produces 1041 and 918 bp HindIII digestion products. (E) Targeted integration into AAVS1 using donors with and without terminal sequence homology and extendibility. Targeted integration was performed with equimolar amounts of donors containing 250 or 100 bp of flanking homology. Circular donors are supercoiled plasmids, linear donors are PCR products. Linear donors tailed with dideoxycytidine are indicated as lin.+ddC. HindIII site integration was assayed by digestion as above. (F) The targeted integration signal is generated in the cell not during PCR. The indicated samples were mixed after DNA preparation but before PCR. HindIII site integration was assayed as above. The percentage of modified chromosomes is shown below each lane in black text.
PCR-derived donors lack DNA methylation. In contrast, plasmid donors share the methylation pattern of the bacterial chromosome. We prepared plasmid donors from bacteria with and without active Dam and Dcm methyltransferases. Donor plasmids from dam− dcm− bacteria were then treated with CpG methyltransferase and the specific activity of all three donor methylforms assayed (Figure 2B). While CpG methylation consistently modestly reduced donor-specific activity, the human and hamster recombination machinery was indifferent to bacterial methylation.
Next, we performed several experiments to assay the importance of target homology length in HDR. We varied homology arm length in the context of plasmid donors and assayed targeted integration of both a transgene and a restriction site at three loci in four cell types (Figure 2B and C; Supplementary Figure S1a–d). In brief, donor homology length is relatively unimportant when HDR copies a small (17 bp) insert into the chromosome yet becomes more important when transgene-size (∼1.5 kb) segments are copied (Figure 2B and C).
Plasmid and PCR-derived donors are also different in that PCR-derived donors retain chromosomal homology at the donor termini; plasmid donors necessarily disrupt this homology at the junction of the plasmid backbone and the homology region. This disruption might reduce the specific activity of plasmid donors. To test this hypothesis, we compared the ability of plasmid donors and PCR-derived donors with identical regions of homology to drive targeted integration. Similar to the results in Figure 2A, supercoiled donors resulted in ∼2-fold more targeted integration of a HindIII site (Figure 2E, compare lanes 4 and 6 with 8, 10, 12 and 14). We conclude that terminal sequence homology is not important for targeted integration of this short HindIII-containing patch.
In a process which is the inverse of HDR, linear donor molecules can be extended at their termini using the homology with chromosomal DNA. Such extended donors can integrate into the genome in a non-homologous manner but generate a false-positive signal in targeted integration assays if the extended regions contain either the primer-binding or restriction enzyme sites used for PCR and Southern blot analysis, respectively (30). To control for this possibility, we added a dideoxycytidine residue to the 3′-ends of the linear donors in the previous experiment. This modification conserves chromosomal homology yet renders the donor incapable of extension by cellular polymerases. Relative to linear donors without a 3′-dideoxycytidine, we observed a slight increase in targeted integration signal, suggesting that this type of ectopic recombination does not confound our assays (Figure 2E, compare lanes 8 and 10 with lanes 12 and 14). Furthermore, extension by Taq polymerase of ZFN-cleaved chromosomal DNA when annealed with a donor molecule could also produce a false-positive targeted-integration signal. We eliminated this possibility by demonstrating that genomic DNA from cells transfected with only the ZFN plasmid, when mixed prior to PCR with genomic DNA from cells transfected with only donor DNA, does not produce any targeted-integration-like signal (Figure 2F).
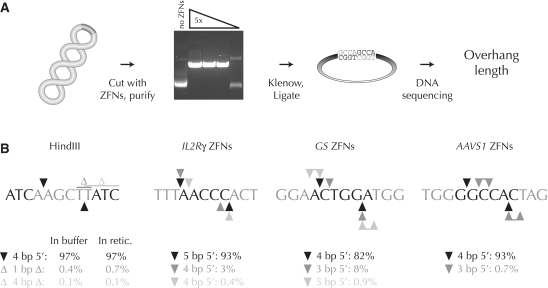
Having demonstrated HDR-based gene addition using donors with minimal target homology, we then asked whether the non-homologous end joining DNA repair pathway was similarly amenable to the deliberate insertion of exogenous DNA. ZFNs produce 5′ overhangs which could be exploited to capture DNA with complementary 5′ extensions. Successful donor capture at ZFN cleavage sites would therefore require knowledge of the exact types of overhangs produced by ZFN cleavage. Previous work demonstrated that ZFNs spaced 6 bp apart leave mainly 4 bp 5′ overhangs (31). As ZFNs with different designs have been developed since this report, we devised a simple assay to measure ZFN cleavage overhangs. In brief, a ZFN-cleaved target plasmid is purified, treated with Klenow polymerase to create blunt-ended fragments, the fragments ligated in cis, and the ligated region sequenced (Figure 3A). This procedure yields short duplications between the ZFN binding sites from which the identity of the overhangs can be deduced. The use of high-throughput DNA sequencing allows the full spectrum of cleavage products to be revealed. We validated this strategy by measuring the 4 bp 5′ overhangs generated by the well-characterized HindIII restriction enzyme, then used the assay to determine the overhangs created by the IL2Rγ, GS and AAVS1 ZFNs (Figure 3B). For IL2Rγ where the ZFN monomers are 5 bp apart, 5 bp 5′ overhangs comprised 93% of all overhang types. Secondary and tertiary classes of 4 bp overhangs were seen due to 1 bp shifts in the top and bottom strand nicking sites. Analogous results were obtained for GS and AAVS1: these 6-bp spaced ZFNs produced predominantly 4 bp overhangs with secondary products generated from 1 bp shifts in the FokI nuclease cleavage. Importantly, cleavage in reticulocyte lysate had no effect on the types of overhangs generated (Figure 3B).
Figure 3.
Analysis of the overhang types created by ZFNs. (A) Scheme to determine ZFN overhangs. A supercoiled plasmid with a ZFN cleavage site is cut by a titration of in vitro transcribed and translated ZFNs. ZFN-linearized plasmids are purified by gel electrophoresis, 5′ overhangs filled in with Klenow polymerase (grey nucleotides), and the resulting blunt ends ligated. The mixture is subjected to high-throughput DNA sequencing. (B) Overhang types generated by a control restriction enzyme (HindIII) and three of the ZFN pairs used in this work. For clarity, only one DNA strand is shown. Enzyme binding sites are shown in grey; only the flanking three nucleotides are shown for ZFN binding sites. Primary cleavage sites, black triangles; secondary and tertiary cleavage sites, dark and light grey triangles, respectively; deletions, Δ. Microhomology within the target site can prevent unambiguous deduction of the overhang type. In such situations the possible overhangs are shown as joined triangles. Either of the two indicated thymidine residues may have been deleted after HindIII digestion.
Armed with this knowledge, we synthesized two 49 bp 5′ phosphorylated oligonucleotides designed to have 4 bp 5′ overhangs complementary to those produced by the AAVS1 ZFNs when annealed (Figure 4A). The double-stranded oligonucleotide contains two phosphorothioate nucleotide residues at each 5′-end and a site for the EcoRI restriction enzyme. Control donor oligonucleotides were created that have either no 5′ overhangs or 4 bp 5′ overhangs predicted not to base-pair with those created by the AAVS1 ZFNs. These double-stranded DNA donors were co-transfected with the AAVS1 ZFNs. Two days post-transfection, the AAVS1 locus was amplified by PCR and donor insertion into the AAVS1 site assayed by EcoRI digestion (Figure 4B). Successful insertion will produce 327 and 258 bp EcoRI fragments; if insertion were to occur in the opposite orientation, 308 and 277 bp bands would result. More than 7% of PCR products produced the expected EcoRI fragments in a donor concentration-, overhang- and ZFN-dependent manner (Figure 4B, lane 3). As measured by the CEL-I mutation detection assay (27), 28 ± 5% of chromosomes were cleaved by the ZFNs in this experiment; the efficiency of donor capture was therefore as high as 27% (7.6/28, Supplementary Figure S2a). The donor that could not correctly base pair with the AAVS1 overhangs was inserted into the chromosome in the opposite orientation at a lower frequency (3%), (Figure 4B, lane 6). The donor without 5′ overhangs was not detectably integrated (Figure 4B, lane 9).
To demonstrate that NHEJ-capture of a linear donor was neither locus nor cell-type specific, we extended this same technique to the glutamine synthase (GS) gene in Chinese hamster ovary cells (CHO cells; C. griseus). In this experiment, donors analogous to those described above were co-transfected with ZFNs that cleave the GS gene (26). Donor insertion was measured two days post-transfection by BamHI digestion of the PCR product into 288 and 106 bp fragments. Eleven percent of chromosomes contained an insertion of the donor DNA (Figure 4C, lane 7). As at least 24 ± 3% of GS loci were ZFN-cleaved in this experiment, the efficiency of donor capture was as high as 46% (11/24, Supplementary Figure S2b). When a non-phosphorothioate donor was used, 8% of chromosomes accepted a donor insertion and insertion became more sensitive to low donor concentration (Figure 4C, lanes 3–6). Similar to the results obtained at the AAVS1 locus, synthetic donor insertion at GS took place at lower frequency with non-complementary-overhang donors and was abolished when blunt-ended donors were used. (Figure 4C, lanes 11–18). At the GS locus, the frequency of donor integration via NHEJ was comparable to HDR-mediated targeted integration using a conventional plasmid donor (compare Figure 4C, lane 7 and Figure 2B, lane 2).
To confirm the results of our PCR-based donor insertion assay, we isolated CHO cell clones bearing insertions of donor sequence. One hundred and thirty-five clones were screened by BamHI digestion to find 11 clones (8%) with bona fide donor insertion as confirmed by DNA sequencing.
Insertion of donors with incorrectly base-paired ends requires the inexact joining mode of NHEJ. The lower-but-appreciable frequency of inexact end joining suggested that some donors might not have been faithfully inserted even when perfectly complementary overhangs were provided. To determine the fidelity of donor insertion, a pool of donor-dependent PCR products was cloned and sequenced. Fifty-five percent of insertions contained perfectly ligated junctions when phosphorothioate donors were used; this frequency dropped to only 9% with use of standard DNA donors (Table 1). Exonuclease digestion of the donor and chromosomal ends at the break resulted in imperfect insertion in the remainder of events.
Table 1.
Fidelity of donor capture by NHEJ at GS in CHO cells
| Normal donor | Phosphorothioate donor | |
|---|---|---|
| Total sequence reads | 56 | 32 |
| Perfect insertions | 5 | 17 |
| Deletion of donor only | 33 | 8 |
| Deletion of chromosome only | 0 | 2 |
| Deletion of donor and chromosome | 18 | 5 |
| Perfect, as percentage of total | 9 | 55 |
| Estimate of cells with perfect insertion (%) | 0.7 | 6 |
Co-transfection of two separate ZFN pairs results in the creation of two DSBs and occasionally, loss of the intervening DNA to create a deletion via microhomology-mediated end joining (MMEJ; 26,32). To see if donors could be captured at the site of deletions, we created donors compatible with the outer two overhangs generated by two ZFN pairs targeted to the POU5F1 locus in K562 cells and the BAK locus in CHO-K1 cells (Figure 5A; 17). ZFN pairs were transfected individually and in combination with the second pair, both with and without inclusion of a donor oligonucleotide containing a loxP site (Supplementary Figure S2c and d). Deletion formation was assayed by PCR amplification of the POU5F1 and BAK loci. Only when both ZFN pairs were co-transfected did deletion-specific PCR products appear (Figure 5B). For POU5F1, deletion of ∼1617 bp resulted in formation of a ∼339 bp deletion-specific band (e.g. lane 3). For BAK, deletion of ∼5833 resulted in formation of ∼245 deletion-specific band (e.g. lane 11). When a donor was co-transfected along with both ZFN pairs, a new band appeared with a size corresponding to donor insertion at the deletion (Figure 5B, lanes 6 and 7, 390 bp; lanes 14 and 15, 295 bp). The efficiency of insertion into POU5F1 increased proportionally when the donor concentration was raised 10-fold to 50 µM; in contrast, insertion into BAK was reduced when the donor concentration was increased to 50 µM (compare lanes 6, 7, 14 and 15).
The donor used in these experiments contains both BamHI and EcoRI restriction enzyme sites. When the deletion PCR products from Figure 5B were incubated with either BamHI or EcoRI, the donor-dependent bands were digested. For both POU5F1 and BAK, the sizes of the digestion products exactly matched the sizes expected from donor insertion (for POU5F1, 230 and 160 bp BamHI products and 270 and 120 bp EcoRI products, Figure 5C, lanes 5 and 8, 6 and 9; for BAK, 176 and 119 bp BamHI products and 216 and 79 bp EcoRI products, lanes 14 and 17, 15 and 18). Quantitation of the digests in Figure 5C indicated that 52% of POU5F1 deletions and 7–19% of BAK deletions acquired a donor insertion.
Asymmetry of the BamHI and EcoRI sites within the donor allows digestion to report the orientation specificity of donor insertion. Insertion in the reverse orientation will yield an approximate reversal of the digestion products (similar to Figure 4B, lane 6). A detectable but very minor fraction of POU5F1 and BAK insertions are in the incorrect orientation (seen most clearly in Figure 5C, lanes 9 and 15).
To confirm these results and to determine the deletion and deletion plus insertion frequency, cells from all lanes in Figure 5C containing a deletion-specific PCR product were diluted, grown for 2 weeks, and 96 or more 10-cell pools (>960 cells) assayed by PCR as above. Approximately 4% of K562 cells treated with POU5F1 ZFNs and 1% of CHO-K1 cells treated with BAK ZFNs contained either a deletion or a deletion and a donor insertion. The deletion frequency did not increase when donor was present. These data are shown under their respective lanes in Figure 5B and C.
The overall fidelity of donor insertion at deletions was determined by cloning and sequencing donor insertion events. Similar to the 55% perfect insertion frequency found at GS, 42% of POU5F1 donors and 69% of BAK donors were faithfully inserted (Table 2). A major failure mode for correct insertion into BAK resulted in disruption of the EcoRI site (data not shown). Consistent with this, EcoRI treatment did not completely digest the donor-dependent band for BAK (Figure 5C, lane 15).
Table 2.
Fidelity of donor capture at deletions by NHEJ
| POU5F1 | BAK | |
|---|---|---|
| Total sequence reads | 33 | 32 |
| Perfect insertions | 14 | 22 |
| Deletion of donor only | 6 | 4 |
| Deletion of chromosome only | 4 | 2 |
| Deletion of donor and chromosome | 9 | 4 |
| Perfect, as percentage of total | 42 | 69 |
| Estimate of cells with perfect insertion (%) | ∼0.9 | ∼0.1 |
Together, the data show that the NHEJ machinery in mammalian cells is generally capable of capturing exogenous linear donor DNA at targeted DSBs and that this reaction is strongly promoted by the presence of complementary single-strand donor overhangs.
DNA can also integrate into DSBs via non-homology-dependent mechanisms. DSBs are spontaneously generated in the cell due to errors in DNA metabolism and can also created by inappropriate ZFN action. We searched for off-target integration events at AAVS1 by inspection of the 10 most-likely off-target sites predicted ab initio from the known specificity of the AAVS1 ZFNs (17). A PCR primer specific to each of the ten loci was paired with a PCR primer in either the transgene donor or in the oligonucleotide donor. Pools of cells treated with ZFNs and donor molecules were assayed for the junction between the donor and each off-target site (Supplementary Figure S3a and b). No such junctions were observed.
We also assayed off-target ZFN activity by attempting to force deliberate misintegration of a transgene donor (Supplementary Figure S3c). When a GFP-containing AAVS1 donor was co-transfected with the AAVS1 ZFNs, 2.7% of cells became GFP-positive. In contrast, even in the presence of cleavage at ∼33% of AAVS1 loci (in addition to potential cleavage at AAVS1 ZFN off-target sites), only 0.2% of cells became GFP-positive when a donor with IL2Rγ homology was co-transfected. In contrast, both donors integrate readily at sites of non-specific DNA cleavage by etoposide. We infer that the large majority of transgene integration with matched ZFNs and donors is in fact targeted integration.
DISCUSSION
Our work expands upon earlier targeted gene addition experiments in which the cellular DNA repair machinery copies a transgene into the chromosome at the site of a DSB. Use of site-specific nucleases enabled active induction of this process and a reduction in target homology to ∼750 bp or less on each side of the transgene (7,15,33). We show here that linear donor molecules with as little as 50 bp of homology on both ends can efficiently hijack the HDR process and result in transgene insertion. In parallel, we show efficient and directed integration of synthetic donor molecules based on a completely different DNA repair pathway, NHEJ. In these experiments, donor insertion is targeted by the information contained in the short 5′ overhangs generated by ZFN cleavage rather than any longer donor homology. These two techniques use very different mechanisms for DNA integration but similarly improve our capacity to manipulate the mammalian genome.
Linear, synthetic donors for use in HDR-mediated targeted gene addition can be made easily and rapidly; in contrast, conventional cloning consumes at least a week to produce a donor plasmid. Short-homology synthetic donors should have a much reduced level and duration of background transgene expression, allowing prompt evaluation of experimental results. Compared to circular plasmid donors, linear donors with short homology regions are likely to have less spurious transcription and/or a shorter post-transfection half-life.
It is formally possible that linear donors are themselves extended using the AAVS1 locus as a template followed by integration elsewhere in the genome. Extension of exogenous DNA has been observed in mammalian cells and in plants and might be expected to be more common when linear fragments with terminal chromosomal homology are present (30,34,35). We tested this hypothesis by preventing donor extension via addition of dideoxycytidine to the donor 3′ ends (Figure 2C). This modification actually had a modest positive effect on targeted integration, ruling out this type of donor misintegration.
It is somewhat surprising that such short regions of homology can be readily used by the HDR machinery, as Rad51 is thought to require ∼100 bp for efficient homology searching (36,37). Extension of the donor by the chromosome would also serve to increase donor homology length, potentially improving its suitability as a donor in a subsequent, separate round of conventional HDR. Therefore, our experiment preventing donor extension also indicates that the HDR we observed employed the intended amount of chromosomal homology.
The experiments in Figure 2 were performed with approximately 10-fold lower concentrations of donor DNA than those in Figure 1 to allow differences in donor specific activity to be seen. This fact likely accounts for the less efficient usage of the linear AAVS1 donor seen in Figure 2A and C compared to Figure 1C and D. While linear donors are intrinsically less efficient that plasmid donors, addition of linear donors to saturating levels can readily compensate for their lower specific activity.
We found that HDR-based copying of a short region of DNA was insensitive to donor homology arm length whereas copying of a longer transgene was diminished with shorter homology arms (Supplementary Figure S1). One explanation for this observation is that short Rad51 filaments may be less stable than those created with longer stretches of homology and that the importance of Rad51 filament stability is proportional to the length of the region copied.
The Rad51-independent single-strand annealing (SSA) pathway may have been used for transgene insertion (37,38). SSA-based gene addition is likely relatively inefficient with donors containing phosphorothioate 5′-ends as their nuclease resistance should prevent efficient generation of the 3′-ends required for SSA (39). Our data therefore suggest that use of donors with as little as 50 bp of flanking homology is supported by classical HDR.
Use of short, synthetic oligonucleotide donors for insertion by NHEJ relies on accurate prediction of the overhangs produced by ZFN cleavage. We measured the overhangs produced during in vitro DNA cleavage with the IL2Rγ, GS and AAVS1 ZFNs. Despite use of a very different ZFP-FokI linker, the main 4 bp 5′ overhang class made by 6-bp spaced ZFNs was similar to that found previously (31). In contrast, ZFNs spaced 5-bp apart produced 5 bp 5′ overhangs. The production of 2 bp 5′ overhangs by 6-bp spaced ZFNs has been asserted (40); we find no evidence consistent with 2 bp 5′ overhangs.
At both AAVS1 and GS we found approximately one-third of measurable NHEJ events contained a donor insertion (Figures 4B and C). Faithful NHEJ capture of donor molecules at DSBs competes kinetically with both accurate and error-prone NHEJ. Accurate repair of the ZFN-induced DSB simply precludes donor insertion. In error-prone NHEJ, any resection of the chromosome makes accurate donor insertion unlikely. Exonuclease activity on donor molecules will also prevent faithful insertion. Consistent with this, we found that use of phosphorothioate-containing donors markedly improved the fidelity of donor insertion by NHEJ capture (Table 1). In contrast, degradation of linear donors intended for use in HDR should not be detrimental to faithful gene addition unless degradation proceeds below the minimum length usable by HDR.
NHEJ often heals DSBs correctly; resection and subsequent microhomology-mediated ligation are additional, separate events. As the non-complementary ends joined to form deletions are necessarily repaired via MMEJ, it was possible that a donor with perfect complementarity to the outside ends of both DSBs would obviate the need for resection and MMEJ, increasing the efficiency of deletion formation. Despite successful use of such donors, we did not observe a meaningful change in deletion frequency (Figure 5B).
Our NHEJ-capture of linear donors occurred at a similar frequency to previous reports of exogenous DNA integration at DSBs, but is otherwise different from these experiments (22,23,41). Specifically, our technique uses the sequence information contained in the 5′ overhang to add DNA to the break and inserts DNA without loss of chromosomal sequence. In contrast, previous experiments involving co-transfection of blunt-ended or single-stranded fragments required both target and donor resection to reveal microhomology needed for fragment joining (23,41). While informative as to the plasticity of the DNA repair machinery, the near impossibility of chromosomal and donor sequence conservation reduces the utility of this previous approach for directed DNA addition. Furthermore, NHEJ and MMEJ are distinct DNA repair pathways with different cofactor requirements (42). Exogenous single-stranded oligonucleotides have been used to repair DSBs in yeast via SSA but this homology-based repair is also fundamentally different than the NHEJ-repair used here (43,44).
Significant effort has gone into the development of recombinases and resolvases with engineered specificities (45–49). The oligonucleotide donors inserted into POU5F1 and BAK deletions contain loxP sites. ZFN-mediated integration of a recombinase site followed by use of a wild-type recombinase could achieve targeted transgene integration without HDR or custom recombinases, albeit via a less elegant two-step process. Transgene recombination into such sites could be used to replace the deleted regions with variants of the original gene, allowing study of isolated haplotypes. Indeed, any transgene too large to be efficiently cloned in bacteria or integrated via HDR might be better integrated via a recombinase-mediated process. For example, a yeast artificial chromosome donor functionalized with a loxP site could be site-specifically integrated after transfection (50).
Unlike HDR-mediated gene addition, donor capture by NHEJ results in the direct incorporation of foreign DNA into the chromosome. Experiments using phosphorothioate donors therefore result in the chromosomal insertion of chemically abnormal DNA. Another potential use for the NHEJ capture technique is the creation of cells with a variety of non-native DNA bases and backbones. In particular, insertion of DNA with methylated cytosines might serve to establish an area of persistent transcriptional quiescence.
NHEJ normally operates on DNA ends bound by Ku, the protein which binds to DNA ends and helps align overhanging bases to promote ligation (51). It is unknown whether the donors involved in NHEJ capture are Ku-bound, or if Ku binding is even likely given the very high donor concentration required for efficient insertion and the limited cellular pool of Ku. NHEJ can be Ku independent when high-GC content overhangs are present (52). All overhangs tested here contain at least two G or C residues. Additional work will be needed to determine if potential saturation of endogenous Ku pools is consequential for NHEJ capture when mostly A- and T-containing overhangs are present.
The flexibility of ZFN design and the speed of linear donor creation we describe here will accelerate targeted transgene integration into mammalian genomes, both via homology-directed and non-homologous DNA repair. Finally, both use of HDR donors with short homology regions and the directed capture of exogenous DNA should prove extensible to DSBs created by other nucleases (such as meganucleases) which leave defined overhangs amenable to rational donor design.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Sangamo BioSciences.
Conflict of interest statement. All authors are current or former full-time employees of Sangamo Biosciences. Sangamo Biosciences has filed a patent application on the basis of the data in this article.
Supplementary Material
[Supplementary Data]
ACKNOWLEDGEMENTS
We thank Ed Rebar, Dave Paschon, Lei Zhang, Sarah Hinkley, George Katibah, Gladys Dulay, Anna Vincent; Rainier Amora for ZFN construction and cloning; and Sandra Cristea for quantitation of the POU5F1 and BAK deletion frequency. We thank the anonymous reviewers of this paper for their helpful criticism.
REFERENCES
- 1.Hacein-Bey-Abina S, von Kalle C, Schmidt M, Le Deist F, Wulffraat N, McIntyre E, Radford I, Villeval JL, Fraser CC, Cavazzana-Calvo M, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 2.Kwaks TH, Otte AP. Employing epigenetics to augment the expression of therapeutic proteins in mammalian cells. Trends Biotechnol. 2006;24:137–142. doi: 10.1016/j.tibtech.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Thomas KR, Folger KR, Capecchi MR. High frequency targeting of genes to specific sites in the mammalian genome. Cell. 1986;44:419–428. doi: 10.1016/0092-8674(86)90463-0. [DOI] [PubMed] [Google Scholar]
- 4.Koller BH, Hagemann LJ, Doetschman T, Hagaman JR, Huang S, Williams PJ, First NL, Maeda N, Smithies O. Germ-line transmission of a planned alteration made in a hypoxanthine phosphoribosyltransferase gene by homologous recombination in embryonic stem cells. Proc. Natl Acad. Sci. USA. 1989;86:8927–8931. doi: 10.1073/pnas.86.22.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng C, Capecchi MR. Reexamination of gene targeting frequency as a function of the extent of homology between the targeting vector and the target locus. Mol. Cell Biol. 1992;12:3365–3371. doi: 10.1128/mcb.12.8.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sedivy JM, Al J. Gene Targeting. Oxford: Oxford University Press; 1992. [Google Scholar]
- 7.Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc. Natl Acad. Sci. USA. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 9.Choo Y, Sanchez-Garcia I, Klug A. In vivo repression by a site-specific DNA-binding protein designed against an oncogenic sequence. Nature. 1994;372:642–645. doi: 10.1038/372642a0. [DOI] [PubMed] [Google Scholar]
- 10.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Annu. Rev. Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 11.Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl Acad. Sci. USA. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith J, Berg JM, Chandrasegaran S. A detailed study of the substrate specificity of a chimeric restriction enzyme. Nucleic Acids Res. 1999;27:674–681. doi: 10.1093/nar/27.2.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weterings E, van Gent DC. The mechanism of non-homologous end-joining: a synopsis of synapsis. DNA Repair. 2004;3:1425–1435. doi: 10.1016/j.dnarep.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lombardo A, Genovese P, Beausejour CM, Colleoni S, Lee YL, Kim KA, Ando D, Urnov FD, Galli C, Gregory PD, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 17.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haviv-Chesner A, Kobayashi Y, Gabriel A, Kupiec M. Capture of linear fragments at a double-strand break in yeast. Nucleic Acids Res. 2007;35:5192–5202. doi: 10.1093/nar/gkm521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore JK, Haber JE. Capture of retrotransposon DNA at the sites of chromosomal double-strand breaks. Nature. 1996;383:644–646. doi: 10.1038/383644a0. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 22.Lin Y, Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Y, Waldman AS. Promiscuous patching of broken chromosomes in mammalian cells with extrachromosomal DNA. Nucleic Acids Res. 2001;29:3975–3981. doi: 10.1093/nar/29.19.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malphettes L, Freyvert Y, Chang J, Liu P-Q, Chan E, Miller JC, Zhou Z, Nguyen T, Tsai C, Snowden AW, et al. Highly-efficient deletion of FUT8 in CHO cell lines using zinc-finger nucleases yields cells which produce completely nonfucosylated antibodies. Biotechnol. Bioeng. 2010 doi: 10.1002/bit.22751. doi: 10.1002/bit.22751 [Epub ahead of print 8 April 2010] [DOI] [PubMed] [Google Scholar]
- 25.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 26.Liu PQ, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, et al. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol. Bioeng. 2009;106:97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- 27.Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat. Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 28.Cost GJ, Freyvert Y, Vafiadis A, Santiago Y, Miller JC, Rebar E, Collingwood TN, Snowden A, Gregory PD. BAK and BAX deletion using zinc-finger nucleases yields apoptosis-resistant CHO cells. Biotechnol. Bioeng. 2009;105:330–340. doi: 10.1002/bit.22541. [DOI] [PubMed] [Google Scholar]
- 29.DeKelver RC, Choi VM, Moehle EA, Paschon DE, Hockemeyer D, Meijsing SH, Sancak Y, Cui X, Steine EJ, Miller JC, et al. Functional genomics, proteomics, and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010 doi: 10.1101/gr.106773.110. doi: 10.1101/gr.106773.110 [Epub ahead of print 27 may 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanin M, Volrath S, Bogucki A, Briker M, Ward E, Paszkowski J. Gene targeting in Arabidopsis. Plant J. 2001;28:671–677. doi: 10.1046/j.1365-313x.2001.01183.x. [DOI] [PubMed] [Google Scholar]
- 31.Smith J, Bibikova M, Whitby FG, Reddy AR, Chandrasegaran S, Carroll D. Requirements for double-strand cleavage by chimeric restriction enzymes with zinc finger DNA-recognition domains. Nucleic Acids Res. 2000;28:3361–3369. doi: 10.1093/nar/28.17.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2009;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richardson C, Jasin M. Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol. Cell Biol. 2000;20:9068–9075. doi: 10.1128/mcb.20.23.9068-9075.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aratani Y, Okazaki R, Koyama H. End extension repair of introduced targeting vectors mediated by homologous recombination in mammalian cells. Nucleic Acids Res. 1992;20:4795–4801. doi: 10.1093/nar/20.18.4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Merrihew RV, Marburger K, Pennington SL, Roth DB, Wilson JH. High-frequency illegitimate integration of transfected DNA at preintegrated target sites in a mammalian genome. Mol. Cell Biol. 1996;16:10–18. doi: 10.1128/mcb.16.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldman AS, Liskay RM. Dependence of intrachromosomal recombination in mammalian cells on uninterrupted homology. Mol. Cell Biol. 1988;8:5350–5357. doi: 10.1128/mcb.8.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haber JE. In vivo biochemistry: physical monitoring of recombination induced by site-specific endonucleases. Bioessays. 1995;17:609–620. doi: 10.1002/bies.950170707. [DOI] [PubMed] [Google Scholar]
- 39.Eckstein F. Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 2000;10:117–121. doi: 10.1089/oli.1.2000.10.117. [DOI] [PubMed] [Google Scholar]
- 40.Zeevi V, Tovkach A, Tzfira T. Increasing cloning possibilities using artificial zinc finger nucleases. Proc. Natl Acad. Sci. USA. 2008;105:12785–12790. doi: 10.1073/pnas.0803618105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Radecke S, Radecke F, Cathomen T, Schwarz K. Zinc-finger nuclease-induced gene repair with oligodeoxynucleotides: wanted and unwanted target locus modifications. Mol. Ther. 2010;18:743–753. doi: 10.1038/mt.2009.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Storici F, Durham CL, Gordenin DA, Resnick MA. Chromosomal site-specific double-strand breaks are efficiently targeted for repair by oligonucleotides in yeast. Proc. Natl Acad. Sci. USA. 2003;100:14994–14999. doi: 10.1073/pnas.2036296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storici F, Snipe JR, Chan GK, Gordenin DA, Resnick MA. Conservative repair of a chromosomal double-strand break by single-strand DNA through two steps of annealing. Mol. Cell Biol. 2006;26:7645–7657. doi: 10.1128/MCB.00672-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ivics Z, Katzer A, Stuwe EE, Fiedler D, Knespel S, Izsvak Z. Targeted Sleeping Beauty transposition in human cells. Mol. Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 46.Feng X, Bednarz AL. and Colloms, S.D. Precise targeted integration by a chimaeric transposase zinc-finger fusion protein. Nucleic Acids Res. 38:1204–1216. doi: 10.1093/nar/gkp1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gersbach CA, Gaj T, Gordley RM. and Barbas, C.F. III. Directed evolution of recombinase specificity by split gene reassembly. Nucleic Acids Res. doi: 10.1093/nar/gkq125. doi: 10.1093/nar/gkq125 [Epub ahead of print 1 March 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gordley RM, Smith JD, Graslund T, Barbas CF., III Evolution of programmable zinc finger-recombinases with activity in human cells. J. Mol. Biol. 2007;367:802–813. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 49.Gordley RM, Gersbach CA, Barbas CF., III Synthesis of programmable integrases. Proc. Natl Acad. Sci. USA. 2009;106:5053–5058. doi: 10.1073/pnas.0812502106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marschall P, Malik N, Larin Z. Transfer of YACs up to 2.3 Mb intact into human cells with polyethylenimine. Gene Ther. 1999;6:1634–1637. doi: 10.1038/sj.gt.3300975. [DOI] [PubMed] [Google Scholar]
- 51.Walker JR, Corpina RA, Goldberg J. Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair. Nature. 2001;412:607–614. doi: 10.1038/35088000. [DOI] [PubMed] [Google Scholar]
- 52.Sandoval A, Labhart P. High G/C content of cohesive overhangs renders DNA end joining Ku-independent. DNA Repair. 2004;3:13–21. doi: 10.1016/j.dnarep.2003.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]