HIV-1 Reactivation Induced by the Periodontal Pathogens Fusobacterium nucleatum and Porphyromonas gingivalis Involves Toll-Like Receptor 4 and 9 Activation in Monocytes/Macrophages (original) (raw)
Abstract
Although oral coinfections (e.g., periodontal disease) are highly prevalent in human immunodeficiency virus type 1-positive (HIV-1+) patients and appear to positively correlate with viral load levels, the potential for oral bacteria to induce HIV-1 reactivation in latently infected cells has received little attention. We showed that HIV-1 long terminal repeat (LTR) promoter activation can be induced by periodontopathogens in monocytes/macrophages; nevertheless, the mechanisms involved in this response remain undetermined. Since Toll-like receptor 2 (TLR2), TLR4, and TLR9 activation have been involved in HIV-1 recrudescence, we sought to determine the role of these TLRs in HIV-1 reactivation induced by the periodontal pathogens Fusobacterium nucleatum and Porphyromonas gingivalis using BF24 monocytes/macrophages stably transfected with the HIV-1 promoter driving chloramphenicol acetyltransferase (CAT) expression and THP89GFP cells, a model of HIV-1 latency. We demonstrated that TLR9 activation by F. nucleatum and TLR2 activation by both bacteria appear to be involved in HIV-1 reactivation; however, TLR4 activation had no effect. Moreover, the autocrine activity of tumor necrosis factor alpha (TNF-α) but not interleukin-1β (IL-1β) produced in response to bacteria could impact viral reactivation. The transcription factors NF-κB and Sp1 appear to be positively regulating HIV-1 reactivation induced by these oral pathogens. These results suggest that oral Gram-negative bacteria (F. nucleatum and P. gingivalis) associated with oral and systemic chronic inflammatory disorders enhance HIV-1 reactivation in monocytes/macrophages through TLR2 and TLR9 activation in a mechanism that appears to be transcriptionally regulated. Increased bacterial growth and emergence of these bacteria or their products accompanying chronic oral inflammatory diseases could be risk modifiers for viral replication, systemic immune activation, and AIDS progression in HIV-1+ patients.
AIDS is caused by the retrovirus human immunodeficiency virus type 1 (HIV-1), which targets immune cells, such as CD4+ T cells, monocytes, macrophages, and dendritic cells (67). After a few weeks of HIV-1 infection, a massive CD4+ T-cell depletion occurs mainly at mucosal surfaces, and viral reservoirs (i.e., latently infected cells) are formed (11, 29). Although CD4+ T cells are the most stable and well-characterized viral reservoir, monocytes/macrophages have received particular attention because they, unlike CD4+ T cells that are depleted by apoptosis during AIDS, exhibit a resistance to virus-mediated apoptotic death. This promotes a long-term persistence of the HIV infection and a potential contribution to development of the immunodeficiency symptoms. In addition to being a critical viral reservoir, infected monocytic cells may also transfer HIV to uninfected T cells (73). Although the mechanisms associated with AIDS progression are not fully understood, it has been shown that active production of HIV-1 proteins (i.e., Tat, Nef, and Vpr) during viral replication and bacterial translocation of coinfecting non-HIV-1 pathogens or their products, such as lipopolysaccharide (LPS) and DNA from mucosal surfaces, appear to be critical factors associated with chronic immune activation in HIV-1+ patients and AIDS progression, even in the presence of highly active antiretroviral therapy (HAART) (10, 41, 69).
The cellular and molecular mechanisms by which opportunistic commensal and pathogenic microorganisms could enhance HIV reactivation in latently infected cells remain somewhat unclear; however, growing evidence suggests a role for Toll-like receptors (TLRs) and various signaling molecules and transcription factors involved in TLR activation. To date, HIV reactivation associated with TLR2, TLR4, and TLR9 has been shown with different cell types (22, 23). Nevertheless, Nordone et al. in a recent study found that TLR4 stimulation did not induce HIV production, by using an ultrapurified LPS from Escherichia coli (57). In addition to TLR agonists, soluble factors and proinflammatory cytokines (e.g., tumor necrosis factor alpha [TNF-α]) produced in response to bacterial challenge could also be involved in HIV-1 reactivation acting in an autocrine manner (6, 32, 46). Normally both TLRs and cytokine receptors activate transcription factors downstream (e.g., NF-κB, AP-1, Sp1, etc.) that not only will turn on proinflammatory cytokine/chemokine genes but also could activate the HIV-1 promoter, which also has consensus binding sequences for them (42, 60). Particularly, NF-κB is considered a positive regulator of proviral reactivation in latently infected cells; however, its upregulation alone may not be sufficient to activate the HIV promoter (57, 75). On the other hand, the role of Sp1 remains controversial since both positive and negative regulation of the HIV-1 promoter by this transcription factor has been reported mainly in T cells (40, 51, 68, 76). Whether or not Sp1 is involved in TLR-induced HIV-1 reactivation in monocyte and macrophage cells remains unclear.
Similarly to the gut, the oral cavity is colonized by a high number and variety of microorganisms that upon immunosuppression caused by HIV-1 infection will grow and cause disease. In fact, a high prevalence of oral bacterial (e.g., HIV-1 gingivitis and periodontitis), fungal (e.g., pseudomembranous candidiasis), and viral (e.g., oral hairy leukoplakia and Kaposi's sarcoma) coinfections in HIV-1+ patients has been described widely (18, 55). Evidence also suggests that periodontal disease, oral candidiasis, and oral hairy leukoplakia positively correlate with higher HIV viral loads (1, 2, 15, 28, 59), although the potential for oral pathogens to stimulate latently infected cells and induce HIV-1 reactivation has received little attention (27). We have recently shown using gene reporter assays that periodontal pathogens have the capacity to differentially enhance HIV-1 promoter activation in T cells, monocytes, macrophages, and dendritic cells in vitro (35). In addition, a recent study showed that the bacterial metabolic product butyric acid, which can be produced by various oral Gram-negative bacteria, including Porphyromonas gingivalis, has the ability to induce HIV-1 replication in latently infected T cells, monocytes, and macrophages through histone deacetylase inhibition. Furthermore, the same group reported that stimulation with isolated LPS or fimbria (TLR2 agonists) from Porphyromonas gingivalis did not induce HIV-1 reactivation (36). Thus, the potential of TLR activation by engagement of oral pathogens to induce HIV-1 reactivation remains undetermined. Herein we demonstrate that oral Gram-negative bacteria (Fusobacterium nucleatum and P. gingivalis) associated with oral (i.e., periodontal disease) and systemic (i.e., atherosclerosis and diabetes) chronic inflammatory disorders (26, 52) have the ability to induce HIV-1 reactivation in monocytes/macrophages through TLR2 and TLR9 activation. Importantly, in spite of a reduced TNF-α production by virally infected cells (i.e., THP89GFP) in response to bacteria, TNF-α appears to play a critical role in this response. Lastly, HIV-1 reactivation appears to be positively regulated by NF-κB and Sp1 transcription factors. These results suggest that a fine balance of signaling pathways and transcription factors activated by these TLRs along with other receptors (e.g., cytokine receptors) could be critical in defining either HIV-1 reactivation or maintenance of latency in HIV-1-chronically infected monocytes/macrophages exposed to these periodontal pathogens.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study were Porphyromonas gingivalis (ATCC 33277) grown in anaerobic broth (Becton Dickinson, Sparks, MD), and Fusobacterium nucleatum (ATCC 25586) grown in Trypticase soy broth supplemented with yeast extract (TSBYE). Bacterial cultures were grown at 37°C under anaerobic conditions (80% N2, 10% H2, and 10% CO2) as described previously (31, 39, 44). Bacterial extracts were obtained starting with 5 colonies grown on CDC anaerobic 5% sheep blood agar plates (Remel, Lenexa, KS), which were further placed into 25 ml of corresponding broth and incubated for 24 h. Then, bacterial cultures were transferred into 500 ml of corresponding broth and incubated under the same conditions for 24 h. The bacterial suspension was washed three times with sterile phosphate-buffered saline (PBS) and centrifuged at 10,000 × g for 20 min at 4°C. The pellet was reconstituted in 15 ml of PBS with complete EDTA-free protease inhibitor cocktail (Roche, Mannheim, Germany), followed by sonication of the bacterial pellet using an ultrasonic disrupter (Branson Sonifier model 450; Branson, Danbury, CT). Disruption of bacteria was confirmed by light microscopy. The crude extract after sonication was centrifuged at 13,000 × g for 10 min at 4°C, and supernatants were evaluated for protein concentration by a bicinchoninic acid (BCA) assay (Pierce, Rockford, IL) and used to challenge BF24 and THP89GFP cells in concentrations between 1 and 10 μg/ml of protein.
Cell cultures.
BF24 cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH, from Barbara K. Felber and George N. Paviakis (25). The BF24 cell line is a subclone of the monocytic leukemia cell line THP-1 that was stably transfected with the HIV-1 long terminal repeat (LTR) promoter driving the chloramphenicol acetyltransferase (CAT) reporter gene expression. THP89GFP cells were a generous gift of David Levy (New York University). These cells were infected with an HIV-1 virus that expresses the enhanced green fluorescent protein (EGFP) under the control of the HIV-1 promoter without removing any viral sequences. Fluorescence and virus production are tightly coupled in THP89GFP cells (49). Parental THP-1 cells were used as a negative control for analyses of EGFP expression by flow cytometry. All cell types were cultured in RPMI 1640 with l-glutamine and 10% fetal bovine serum (FBS) and maintained in a 5% CO2 atmosphere at 37°C. In addition, 100 U/ml penicillin and 100 μg/ml streptomycin were used for THP89GFP cultures.
DNA isolation from oral bacteria.
Bacterial DNA was isolated using the Wizard Genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's instructions. Briefly, 600 μl of bacteria (109 cells/ml) resuspended in 50 mM EDTA were centrifuged at 1,000 × g for 2 min at room temperature. Further, 600 μl of lysis buffer was added into the pellets, and cells were incubated at 80°C for 5 min. Then, RNA and protein were removed by incubation with 3 μl RNase followed by treatment with 200 μl of protein precipitation solution. After centrifugation at 10,000 × g for 3 min, DNA-containing supernatants were transferred and mixed with isopropanol to precipitate the bacterial DNA. Finally, DNA-containing pellets were obtained by centrifugation at 10,000 × g for 2 min and washed once with 70% ethanol. Bacterial DNA was resuspended in 50 μl of endotoxin-free water, and the DNA concentration was obtained using the spectrophotometer and software Nanodrop (ND-1000) (Thermo Scientific, Wilmington, DE).
Stimulation of cells with bacterial extracts and TLR agonists.
Monocytes/macrophages were seeded in 24-well plates at a cell density of 2.5 × 105 cells/well in 1 ml of RPMI 1640 medium supplemented with 2% FBS and incubated 16 h either with media alone or in the presence of different concentrations of each bacterial extract (1 to 10 μg/ml) obtained as described above and purified lipopolysaccharide (LPS) (1 to 100 ng/ml) from E. coli 0111:B4 (TLR4 agonist) and P. gingivalis (TLR2 agonist) or CpG ODN2006 (TLR9 agonist) (InvivoGen, San Diego, CA). The bacterial extract concentrations used to stimulate the cells are representative of a cell/bacterium ratio range of 1:10 to 1:100. Unstimulated cells were used as a negative control. For neutralization assays, cells were preincubated 1 h with the monoclonal antibody mouse IgG2a anti-human TLR2, mouse IgG1 anti-human TNF-α, and anti-human interleukin 1β (IL-1β) with their correspondent isotype controls (eBioscience and BD Pharmingen, San Diego, CA), or inhibitory oligodeoxynucleotide (ODN) TTAGGG (InvivoGen, San Diego, CA) before challenge with bacterial extracts or TLR agonists. Chloroquine (Sigma, St. Louis, MO), as well as enzymatic pretreatment of bacterial DNA from F. nucleatum with 5 U DNase or RNase for 1 h at 37°C followed by inactivation at 65°C for 15 min, was also used to inhibit TLR9 activation as previously shown (24, 53).
HIV-1/CAT ELISA.
HIV-1 LTR CAT activation was measured by a sandwich enzyme-linked immunosorbent assay (ELISA) using a commercially available kit (Roche, Mannheim, Germany). Briefly, BF24 cells were harvested after several treatments and washed with 1× PBS at 2,600 × g for 15 min. Then, pellets were resuspended in 220 μl lysis buffer provided in the kit for 30 min at room temperature (RT). Further, 210 μl of lysed cells were placed in 96-well plates precoated with an anti-chloramphenicol acetyltransferase (anti-CAT) for 2 h at 37°C. After washing, a primary antibody anti-CAT conjugated with digoxigenin (anti-CAT-DIG) was added for 1 h, followed by a secondary antibody anti-digoxigenin conjugated with peroxidase (anti-DIG-POD). Finally, the reaction was revealed using 200 μl/well of ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate for 40 min at RT. The absorbance was measured using a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA) at 405 nm.
Detection of p24 by ELISA.
Supernatants from THP89GFP cells were harvested after different treatments and evaluated for the presence of HIV-1 p24 protein by ELISA using a commercially available kit (Advanced BioScience Laboratories, Inc., Kensington, MD). The manufacturer's instructions were followed, and absorbance was measured using a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA) at 450 nm.
Fluorescence microscopy.
THP89GFP cells were photographed in culture through a Nikon Eclipse TS100 inverted microscope using ×100 magnification with a QICAM Fast1394 device camera (QImaging, Surrey, BC, Canada). To detect EGFP fluorescence, the 495-nm green fluorescent protein filter set was used.
Flow cytometry analysis.
Flow cytometry analysis was performed with a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA). After different treatments, THP89GFP cells were harvested, washed with 1× PBS at 1,300 × g for 5 min, and fixed with 2% paraformaldehyde solution for 20 min before fluorescence-activated cell sorter (FACS) analysis.
Fluorometry.
After different treatments, THP89GFP cells were harvested and washed in 1× PBS at 1,300 × g for 5 min. Further, pellets were resuspended in 150 μl of lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Nonidet P-40) for 20 min on ice. Then, cell lysates were centrifuged at 10,000 × g for 5 min at 4°C, and 100 μl of supernatants was added into Optilux 96-well clear-bottom plates (BD Falcon, Franklyn Lakes, NJ) for fluorescence analysis using the microplate fluorescence reader FLx800 (Bio-Tek Instruments, Inc., Winooski, VT). The wavelengths used for excitation and emission were 485 nm and 528 nm, respectively.
Analysis of cytokine production.
Supernatants from cell cultures were harvested after different treatments and centrifuged at 2,600 × g, 4°C for 10 min. Further supernatants were evaluated for the presence of TNF-α, IL-1β, IL-6 (eBioscience, San Diego, CA), and IL-8 (R&D Systems, Minneapolis, MN) using commercially available ELISA kits and following the manufacturer's instructions. The reaction was revealed by adding substrate solution at RT, and absorbance was measured using a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA) at 450 nm.
Transcription factor analysis.
To determine the role of transcription factors in HIV-1 reactivation induced by bacteria, cells were challenged with 10 μg/ml of bacterial extracts in the presence of different concentrations of the specific NF-κB transcription factor inhibitor BAY 11-7082 (Calbiochem, San Diego, CA) dissolved in dimethyl sulfoxide (DMSO). In addition, THP89GFP cells transfected with either Sp1 small interfering RNA (siRNA) or a nonsilencing (NS) siRNA (Qiagen Sciences, Germantown, MD) were used to determine the role of Sp1 in bacterium-induced HIV-1 reactivation. Briefly, 1 × 105 cells/well were seeded in 6-well plates with 1.6 ml RPMI 1640 supplemented with 10% FBS overnight. Then, cells were transfected with 2 μM siRNA using DharmaFECT 2 (Dharmacon, Chicago, IL). Sp1 silencing was confirmed by Western blotting using the monoclonal antibodies mouse IgG1 anti-Sp1 and anti-β-actin (Santa Cruz Biotechnology, Santa Cruz, CA). THP89GFP cells were exposed to bacterial extracts 48 h after transfection, and HIV-1/EGFP levels in cell lysates as well as p24 levels in supernatants were determined 48 h later by fluorometry and ELISA, respectively, as described previously.
Statistical analysis.
The significance of any difference between the means for the experimental groups was determined by Student's t test. The means were considered significantly different if P was ≤0.01.
RESULTS
Periodontal pathogens F. nucleatum and P. gingivalis induce HIV-1 promoter activation in monocytes/macrophages.
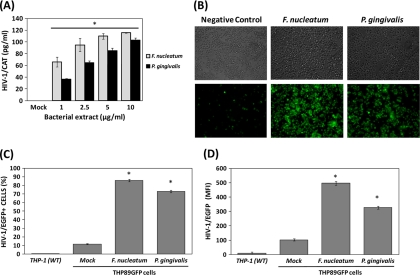
The abilities of several opportunistic and pathogenic microorganisms (i.e., Mycobacterium tuberculosis, Streptococcus pneumoniae, and Neisseria gonorrhoeae) commonly found during HIV-1 infections to directly induce HIV-1 reactivation in latently infected cells have previously been shown (7, 12, 16); however, the potential impact of oral microorganisms associated with highly prevalent oral chronic infectious diseases in HIV-1+ patients in HIV-1 exacerbation remains undetermined. Herein, we evaluated the ability of two pathogens associated with oral and systemic chronic inflammatory disorders to induce HIV-1 promoter activation in monocytes/macrophages. Both extracts from P. gingivalis and F. nucleatum induced HIV-1 LTR CAT promoter activation in a dose-dependent manner in BF24 cells (Fig. 1 A). Consistently with these results, HIV-1 reactivation in THP89GFP cells examined by fluorescence microscopy was remarkably upregulated by oral Gram-negative bacteria compared with unstimulated cells (Fig. 1B). Quantitative analyses by FACS showed that about 80 to 90% of the total cell population exhibited HIV-1/EGFP promoter activation compared with 10% of unstimulated cells after treatment with bacterial extracts (Fig. 1C), which correlated with the HIV-1 promoter activation determined as mean fluorescence intensity by FACS (Fig. 1D).
FIG. 1.
Effect of F. nucleatum and P. gingivalis extracts on HIV-1 promoter activation in monocytes/macrophages. BF24 and THP89GFP cells (2.5 × 105/ml) were either exposed or not (mock) to bacterium extracts. Parental THP-1 (WT) cells were used as a negative control for FACS. (A) HIV-1 CAT promoter activation in BF24 cells challenged with several extract concentrations of each bacterium for 16 h was determined by ELISA. (B) Induction of HIV-1/EGFP in THP89GFP monocytes/macrophages incubated with 10 μg/ml bacterial extracts for 24 h was visualized by light microscopy and fluorescence microscopy using ×100 magnification. Percentage of EGFP-positive cells (C) and mean fluorescence intensity (MFI) of HIV-1/EGFP cells (D) described in the legend to panel B were quantified by flow cytometry as described in Materials and Methods. The data are representative of two independent experiments with triplicate determinations (n = 6). Data are expressed as means ± standard deviations. *, P < 0.01 compared to the results for the control (mock), as determined by Student's t test.
Effect of TLR2, TLR4, and TLR9 activation on HIV-1 promoter activation in monocytes/macrophages.
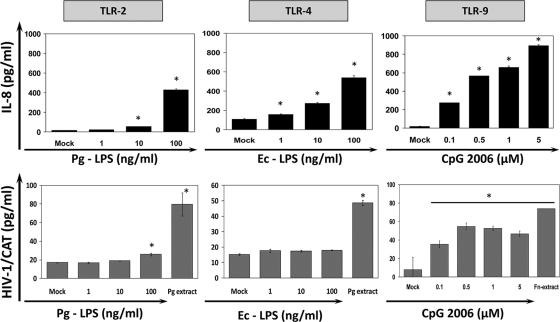
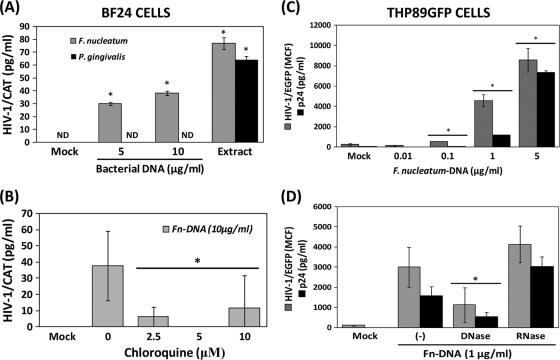
As mentioned earlier, an important mechanism by which mammalian cells recognize bacteria is mediated by evolutionary, highly conserved receptors named pattern recognition receptors (PRRs), including Toll-like receptors (TLRs) and nucleotide oligomerization domain (Nod)-like receptors that promote downstream activation of transcription factors (i.e., NF-κB, Sp1, CREB, and C/EBP), which generally regulate the production of several proinflammatory mediators (5). Given that the HIV-1 promoter harbors binding sequences for these transcription factors, it has been suggested that TLR activation could be involved in HIV-1 reactivation from latently infected cells (60, 63). Attempting to determine the role of TLRs in HIV-1 promoter activation, BF24 cells were first tested for their ability to respond to purified TLR2, TLR4, and TLR9 agonists, and HIV-1 LTR CAT was further determined. Although all TLR agonists enhanced IL-8 production (Fig. 2), only CpG 2006 (TLR9) and LPS from P. gingivalis (TLR2) at higher concentrations (100 ng/ml), but not E. coli (TLR4), induced HIV-1 promoter activation (Fig. 2). Based on these results and the natural ability of bacterial DNA to activate TLR9, the HIV-1 promoter activity was further evaluated in cells exposed to isolated DNA from F. nucleatum and P. gingivalis. The bacterial DNA concentrations obtained were 343 μg/ml for P. gingivalis and 1,006 μg/ml for F. nucleatum with purity (_A_260/_A_280 ratio) between 1.95 and 2.12. The higher bacterial extract concentrations (10 μg/ml) had DNA levels between 2 and 10 μg/ml, a range of DNA concentrations that was used to challenge BF24 cells. Only DNA from F. nucleatum, but not P. gingivalis, activated the HIV-1 promoter; however, consistently with the previous results, the whole bacterial extracts from both periodontal pathogens increased HIV-1 LTR activity (Fig. 3 A). Preincubation of BF24 cells with the TLR9 inhibitor chloroquine before challenge with DNA from F. nucleatum reduced the HIV-1 LTR activation (Fig. 3B). A similar effect was observed when BF24 cells were preincubated with the inhibitory CpG TTAGGG (data not shown).
FIG. 2.
Effect of Toll-like receptor agonists on HIV-1 CAT promoter activation in BF24 monocytes/macrophages. Cells were incubated with either 10 μg/ml bacterial extracts or different concentrations of LPS from P. gingivalis (TLR2 ligand), LPS from E. coli (TLR4 ligand), or CpG ODN2006 (TLR9 ligand). IL-8 production in culture supernatants was analyzed by ELISA, and HIV-1 promoter activation was measured by determination of CAT levels in cell lysates as described in Materials and Methods. The data are representative of three independent experiments with triplicate determinations (n = 9). Data are expressed as means ± standard deviations. *, P < 0.01 compared to the results for the control (mock), as determined by Student's t test.
FIG. 3.
TLR9 activation by DNA from F. nucleatum is involved in HIV-1 reactivation in monocytes andmacrophages. (A) BF24 cells were incubated with either purified DNA or whole bacterial extracts from F. nucleatum or P. gingivalis for 16 h, and HIV-1 CAT promoter activation in cell lysates was determined by ELISA. ND denotes “nondetected” (values below the level of detection). (B) HIV-1 CAT promoter activity induced by 10 μg/ml DNA from F. nucleatum in BF24 cells preincubated (or not) for 1 h with the TLR-9 inhibitor chloroquine. (C) Induction of HIV-1/EGFP promoter activity in cell lysates (gray bars) and p24 levels in supernatants (black bars) by several concentrations of DNA from F. nucleatum for 24 h was determined in THP89GFP cells by fluorometry and ELISA, respectively, as described in Materials and Methods. MCF, mean cumulative fluorescence. (D) Effect of enzymatic pretreatment of DNA from F. nucleatum with DNase or RNase in HIV-1 reactivation after 24 h of incubation is shown. The data are representative of three independent experiments with triplicate determinations (n = 9). Data are expressed as means ± standard deviations. *, P < 0.01 compared to the results for the control (mock), as determined by Student's t test.
Similarly, when THP89GFP cells were stimulated with DNA from F. nucleatum, HIV-1/EGFP activity and viral replication were upregulated in a dose-dependent manner determined by fluorometry and ELISA (Fig. 3C). Both HIV-1/EGFP promoter activity and viral replication determined by p24 levels in supernatants were reduced when DNA from F. nucleatum was pretreated with DNase but not RNase (Fig. 3D).
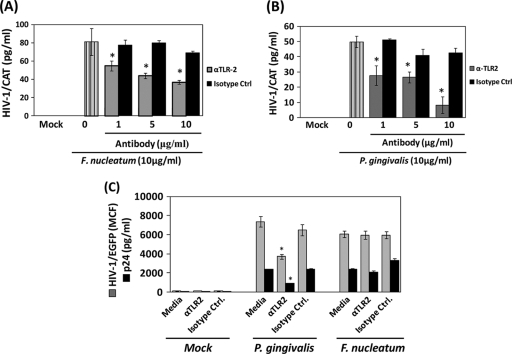
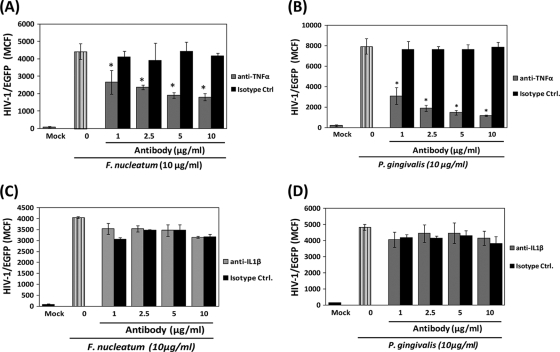
To better rule out the potential role for TLR2 activation by P. gingivalis and F. nucleatum in HIV-1 promoter activity, we determined the HIV-1 promoter activity induced by the whole extract of these periodontal pathogens in monocytes/macrophages preincubated with monoclonal anti-TLR2 or its correspondent isotype control. Neutralization of TLR2 reduced by 50% the HIV-1 promoter activity induced by the F. nucleatum extract in BF24 cells (Fig. 4 A), while the response induced by the P. gingivalis extract was nearly completely abrogated (approximately 90%), compared with cells preincubated with the correspondent isotype control (Fig. 4B). Furthermore, neutralization of TLR2 in THP89GFP cells reduced the HIV-1/EGFP and p24 levels induced by P. gingivalis (Fig. 4C); however, TLR2 blocking was less efficient for reducing bacterium-induced HIV-1 reactivation in virally infected THP89GFP cells compared with the effect of blocking TLR2 previously observed with BF24 cells. In contrast, the presence of anti-TLR2 in THP89GFP cells challenged with F. nucleatum did not affect HIV-1/EGFP levels or p24 production compared with cells incubated with the isotype control (Fig. 4C).
FIG. 4.
Blocking of TLR2 inhibits bacterium-induced HIV-1 reactivation. Cells (2.5 × 105/ml) were preincubated for 1 h with different concentrations of either a monoclonal anti-TLR2 neutralizing antibody or its respective isotype control before challenge with 10 μg/ml of bacterial extracts. Effect of TLR2 neutralization in HIV-1 CAT promoter activation induced by F. nucleatum (A) and P. gingivalis (B) in BF24 cells exposed to bacteria for 16 h is shown. α, anti-. (C) Effects of TLR2 neutralization in HIV-1/EGFP promoter activation (gray bars) in cell lysates and p24 levels (black bars) in supernatants of THP89GFP cells exposed to bacteria for 48 h are shown. The data are representative of two independent experiments with triplicate determinations (n = 6). Data are expressed as means ± standard deviations. *, P < 0.01 compared to the results for the cells challenged with bacteria in the presence of anti-TLR2 versus the isotype control, as determined by Student's t test.
TNF-α produced in response to F. nucleatum and P. gingivalis is involved in HIV-1 promoter activation.
Normally, the activation of TLRs in monocytes/macrophages by bacteria leads to the production of proinflammatory cytokines (e.g., TNF-α, IL-1β, and IL-6). Importantly, these proinflammatory cytokines have been suggested as an important mechanism of HIV-1 reactivation in latently infected cells (20, 61). Hence, to test whether production of these cytokines in response to F. nucleatum and P. gingivalis was playing a role in HIV-1 promoter activation, we first determined the levels of TNF-α, IL-1β, and IL-6 produced by either THP89GFP cells or their parental THP-1 cells in response to different concentrations of bacterial extracts. Although detectable levels of all proinflammatory cytokines were observed with supernatants from THP-1 parental cells, only TNF-α and IL-1β, not IL-6, were detected in supernatants from virally infected THP89GFP cells exposed to bacterial extracts. Of note, the levels of TNF-α and IL-1β produced by THP89GFP cells in response to bacterial extracts were significantly lower (5- to 15-fold) compared with the cytokine levels produced by THP-1 parental cells (data not shown). Further, in order to determine the potential for TNF-α and IL-1β produced in response to bacterial extracts to enhance HIV-1/EGFP promoter activity in THP89GFP cells, neutralization assays using monoclonal antibodies against these cytokines or correspondent isotype controls were performed. Neutralization of TNF-α reduced by about 40 to 50% the HIV-1/EGFP promoter activation induced by F. nucleatum and by nearly 90% the response induced by P. gingivalis compared with the effect of its correspondent isotype control (Fig. 5 A and B). In contrast, neutralization of IL-1β did not induce changes in bacterium-induced HIV-1/EGFP promoter activation (Fig. 5C and D).
FIG. 5.
TNF-α, but not IL-1β, produced by THP89GFP cells in response to F. nucleatum and P. gingivalis is involved in HIV-1/EGFP promoter reactivation. THP89GFP cells (2.5 × 105/ml) were incubated 24 h with 10 μg/ml of bacterial extracts either in the presence of a specific neutralizing antibody (gray bars) or its matched isotype control (black bars). Effects of TNF-α (A and B) and IL-1β neutralization (C and D) in HIV-1/EGFP activity induced by F. nucleatum and P. gingivalis extracts are shown. The data are representative of two independent experiments with triplicate determinations (n = 6). Data are expressed as means ± standard deviations. *, P < 0.01 compared to the results for the cells challenged with bacteria in the presence of neutralizing antibody versus the isotype control, as determined by Student's t test.
Transcription factors NF-κB and Sp1 are positive regulators of periodontal pathogen-induced HIV-1 reactivation in monocytes/macrophages.
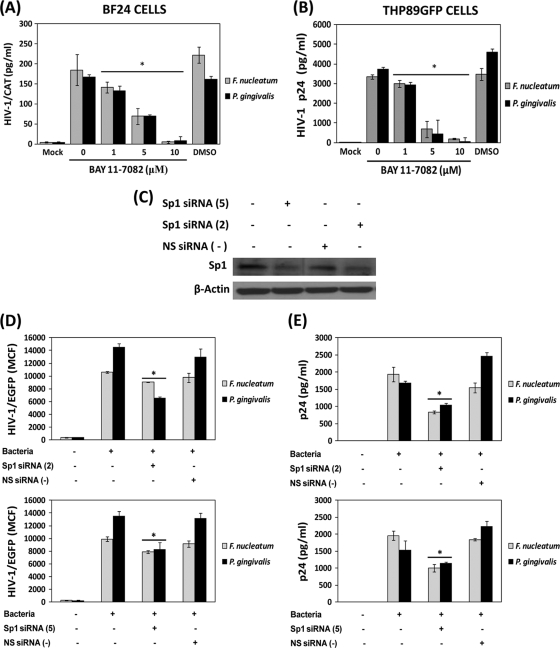
Growing evidence suggests that upon TLR activation several signaling pathways and transcription factors are activated, including NF-κB and Sp1, among others (42, 58). Since both NF-κB and Sp1 transcription factors have also been shown to be important regulators of HIV-1 reactivation (38, 40), we evaluated the role of these transcription factors in HIV-1 reactivation induced by F. nucleatum and P. gingivalis. Monocytes/macrophages were challenged with periodontal pathogen extracts, either in the presence or absence of the specific NF-κB inhibitor BAY 11-7082. The presence of BAY 11-7082 completely abrogated the HIV-1 LTR CAT activity induced by F. nucleatum and P. gingivalis in BF24 monocytes/macrophages; however, this response was not affected by the solvent DMSO (Fig. 6 A). Consistently, viral replication determined by p24 levels in supernatants of THP89GFP cells exposed to bacterial extracts was substantially blocked by the presence of the NF-κB inhibitor (Fig. 6B). Furthermore, to determine the role of Sp1 transcription factor in bacterium-induced HIV-1 reactivation, Sp1 protein expression was silenced by transient transfection of THP89GFP cells with specific Sp1 siRNAs. The transfection of THP89GFP cells with two different Sp1 siRNAs significantly reduced the protein levels of Sp1 compared with cells transfected with nonsilencing siRNA and nontransfected cells as determined by Western blotting (Fig. 6C). Both HIV-1/EGFP promoter activation and viral replication induced by F. nucleatum and P. gingivalis were significantly reduced in THP89GFP monocytes/macrophages transfected with Sp1 siRNAs but not nonsilencing siRNA (Fig. 6D and E).
FIG. 6.
HIV-1 reactivation induced by F. nucleatum and P. gingivalis is positively regulated by NF-κB and Sp1. BF24 or THP89GFP monocytes/macrophages were challenged with 10 μg/ml of F. nucleatum or P. gingivalis extracts either in the presence or absence of different concentrations of the specific NF-κB inhibitor BAY 11-7082 dissolved in DMSO. (A) HIV-1 CAT promoter activity in BF24 cell lysates was determined by ELISA. (B) p24 levels were determined in THP89GFP cell supernatants as described in Materials and Methods. (C) Western blot analyses of the total cell extract from THP89GFP cells transfected individually with two different Sp1siRNAs (Sp1siRNA 5 and 2) or with an NS siRNA as described in Materials and Methods. Detection of β-actin was used as a loading control. HIV-1/EGFP promoter activation (D) and viral replication (p24 levels) (E) induced by F. nucleatum (gray bars) and P. gingivalis (black bars) in THP89GFP cells differentially transfected are shown. The data are representative of two independent experiments with triplicate determinations (n = 6). Data are expressed as means ± standard deviations. *, P < 0.01 compared to the results for the cells transfected with Sp1 siRNA or NS siRNA versus nontransfected cells, as determined by Student's t test.
DISCUSSION
Translocation of bacterial products (e.g., LPS and DNA) into the systemic circulation through impaired mucosal surfaces has been associated with HIV-1 exacerbation, chronic immune activation, and AIDS progression (10, 41). Similarly to the gut, the oral cavity is colonized by a wide variety and number of microorganisms, including commensal, opportunistic, and pathogenic species (about 1 × 108 to 1 × 109 bacteria/mg of plaque) (66). Growing evidence suggests that periodontal pathogens, including F. nucleatum and P. gingivalis, or their products can not only cause local inflammation at the oral cavity but also appear to emerge systemically and enhance inflammatory responses associated with conditions such as atherosclerosis, preterm birth, and diabetes (13, 21, 37, 47, 52). A high prevalence of oral opportunistic infections, including severe forms of periodontal disease in HIV-1+ patients, has been demonstrated widely (18, 48). Nevertheless, the potential for oral pathogens to enhance HIV-1 exacerbation has received little attention. Supporting our previous findings herein, we show that HIV-1 promoter activity was increased significantly when BF24 and THP89GFP monocytes/macrophages were exposed to bacterial extracts from periodontal pathogens F. nucleatum and P. gingivalis (35).
Oral bacteria, including F. nucleatum and P. gingivalis, can activate TLR2, TLR4, and TLR9, inducing the production of cytokines and chemokines (45, 50, 56, 71, 72), and the same group of TLRs has been involved in HIV-1 reactivation from latently infected cells (22, 23). Initial treatment of BF24 monocytes/macrophages with purified agonists for these TLRs showed that only TLR9 (CpG2006) but not TLR2 (P. gingivalis LPS) and TLR4 (E. coli and other LPS) engagement was able to induce HIV-1 LTR activation. These findings led us further to evaluate the HIV-1 promoter activity induced by isolated DNA from F. nucleatum and P. gingivalis as a natural TLR9 ligand. Interestingly, only DNA from F. nucleatum triggered HIV-1 promoter activation in a dose-dependent manner. It has been shown that differences in the abilities of DNA from different bacterial strains to activate TLR9 could be related to the frequency of CpG motifs in the bacterial genome (19). In effect, differences in cytokine production by monocytic cells and gingival fibroblasts in response to DNA from P. gingivalis and Actinobacillus actinomycetemcomitans have been associated with the frequency of CpG motifs in their genomes (56). To the best of our knowledge there is not published evidence addressing the effect of TLR9 activation by DNA from F. nucleatum in monocytic cells; however, its increased ability to trigger HIV-1 promoter activation in BF24 cells, with respect to the effect induced by DNA from P. gingivalis, suggests that there could be important variations in the CpG motif frequency in its genome. Since the HIV-1 promoter response associated with TLR9 activation by bacterial DNA did not completely recapitulate the levels of HIV-1 promoter activity observed with the whole extract of F. nucleatum, there could be other microbial structures involved in this response or synergism between bacterial DNA, and other TLR ligands, as well as host cytokines, would be potentially contributing to the magnitude of response. A recent study demonstrated that systemic levels of bacterial DNA in HIV-1+ patients correlated with chronic immune activation and AIDS progression (41). F. nucleatum is found not only in oral mucosa but also at the urogenital level, at which it has been related to vaginosis and undesirable pregnancy outcomes (33, 34). Of note, HIV-1 promoter activity triggered by DNA from F. nucleatum in monocytes/macrophages correlated with viral replication. Thus, overgrowth of F. nucleatum at mucosal surfaces as well as systemic translocation of this microorganism could increase the risk of HIV-1 viral exacerbation, disease progression, and perhaps viral transmission.
Early studies suggested that LPS from Gram-negative bacteria was a strong inducer of HIV-1 reactivation (22, 62). Nevertheless, it was recently shown by Nordone et al. that LPS from E. coli that activates TLR-4 appeared not to be involved in HIV-1 reactivation (57). The differences in the effect of LPS in HIV-1 promoter activation were attributed to contamination of LPS with TLR2 agonists, as well as the use of different cell lines. In our study, stimulation of BF24 cells with LPS from different enteric and oral bacterial strains was not sufficient to recapitulate the HIV-1 promoter activity induced by the whole extracts of F. nucleatum and P. gingivalis (data not shown). These results indicate that TLR4 activation itself could not be sufficient to trigger HIV-1 promoter activation in these latently infected monocytic cells. It does not allow us to rule out the potential for TLR4 activation by LPS as a potential modulator of HIV-1 promoter activation induced by other TLRs or cytokine receptors. Indeed, synergism between LPS and granulocytic macrophage and monocytic colony stimulator factor (GM-CSF) for inducing HIV-1 reactivation was recently reported (58).
On the other hand, TLR2 activation by LPS from P. gingivalis did not increase substantially HIV-1 promoter activity; however, previous reports have shown that stimulation with other TLR2 agonists (e.g., Pam3CSK4) or whole bacteria (e.g., M. tuberculosis) induces HIV-1 reactivation in different cell types, including monocytic cells (4, 57). The possibility of TLR2 interacting with different bacterial structures (i.e., peptidoglycan, lipoteichoic acid, etc.) and forming heterodimers with TLR1 and TLR6 presents the likelihood of different cellular responses, depending upon the combination of this group of alternatives. It has been shown that LPS from P. gingivalis is recognized by the heterodimer TLR2/TLR1, and fimbriae from the same microorganism interact with both TLR2/TLR1 and TLR2/TLR6 heterodimers in order to induce TNF-α production (30). Thus, it is possible that TLR2/TLR6 activation by fimbriae could be triggering HIV-1 promoter activation in our system; however, Imai et al. showed that recombinant fimbriae from P. gingivalis were not sufficient to induce HIV-1 reactivation in latently infected monocytes and T cells (36). To confirm the lack of a role for TLR2 activation by periodontopathogens in HIV-1 promoter activation, neutralization assays using a monoclonal anti-TLR2 were performed. Interestingly, HIV-1 promoter activity was partially reduced by anti-TLR2 in cells challenged with F. nucleatum, and it was virtually abrogated in cells stimulated with P. gingivalis. These results suggest that other structural components rather than LPS or fimbriae from P. gingivalis or F. nucleatum with affinity for TLR2 could be involved in HIV-1 promoter activation. Nevertheless, since a cross talk between TLRs, Nod receptors, and cytokine receptors has been suggested to be critical for the final cellular response (70, 77), it is also likely that individual TLR2 and TLR4 activation by isolated structural components of periodontal pathogens may not be sufficient to induce HIV-1 promoter activation, but these TLRs could synergize with other cellular receptors in order to trigger this response.
Several in vitro and in vivo studies have shown that proinflammatory cytokines (e.g., TNF-α, IL-1β, IL-6, etc.) could be playing a role in HIV-1 reactivation in latently infected cells, including monocytes/macrophages. Moreover, increased TNF-α levels in serum from HIV-1+ patients have been associated with HIV-1 exacerbation and disease progression (32, 74). In agreement with these observations we showed that even though there was a substantial reduction in proinflammatory cytokine production in HIV-1-infected cells (i.e., THP89GFP cells), with regard to noninfected THP-1 cells, autocrine activity of TNF-α, but not IL-1β, produced in response to both periodontal pathogens appears to play a critical role in HIV-1 reactivation. Importantly, the impaired cytokine production by THP89GFP cells compared with the parental cell line, as well as the reduced effect of anti-TLR2 in bacterium-induced HIV-1 reactivation, with regard to BF24 cells, supports recent findings in which HIV-1 infection altered the expression and activation of TLRs, with a consequent reduction in cytokine expression (54).
Finally, the presence of consensus binding sequences for TLRs and cytokine-activated transcription factors, such as NF-κB, AP-1, and Sp1 among others, in the HIV-1 LTR promoter has been shown, which suggests that inflammatory responses against pathogens triggered by TLR engagement will enhance HIV-1 exacerbation (14, 43, 63). Using the specific chemical inhibitor BAY 11-7082, we showed that NF-κB appears to be critically necessary for _F. nucleatum_- and _P. gingivalis_-induced HIV-1 reactivation. These results add more weight to the role of NF-κB as a positive regulator of HIV-1 reactivation that has particularly been related to heterodimers p50/p65 in monocytic cells (38). Of note, in spite of an increase in IL-8 production upon TLR2 or TLR4 activation by purified agonists, an event that involves NF-κB activation, there were no changes in the HIV-1 promoter response in monocytes/macrophages. These results suggest that activation of certain transcription factors (e.g., NF-κB, AP-1, etc.) which also have the ability to activate the HIV-1 promoter may not be sufficient to trigger HIV-1 reactivation as previously suggested (57). Importantly, it has been demonstrated recently that the kinetic of NF-κB activation is critical in controlling the magnitude of TLR-mediated cytokine production (8), an event that could also play a role in TLR-induced HIV-1 promoter activation. Likewise, silencing of the Sp1 transcription factor significantly reduced HIV-1 reactivation induced by F. nucleatum and P. gingivalis in monocytes/macrophages. The role of Sp1 in HIV-1 recrudescence is not fully understood; although studies suggest that Sp1 and Sp4 members of the Sp1 multigene family are transcriptional activators of HIV-1 LTR, Sp3 appears to be a transcriptional repressor (51). In contrast to our findings, recent evidence showed that Sp1 could also maintain HIV-1 latency in T cells through recruitment of histone deacetylase-1 to the HIV-1 promoter (40). Thus, the ability of cellular transcription factors to positively or negatively regulate HIV-1 LTR transcription could be determined by the cell type that is latently infected as well as the magnitude and time of activation of particular transcription factors, which could be different depending upon the stimuli. Besides, since some HIV-1 proteins appear to interact with host cell transcription factors modulating their activity (3, 17, 64), it is likely that the regulatory function of transcription factors to either enhance or inhibit viral reactivation in HIV-1 latently infected cells might be altered with regard to uninfected cells.
An additional observation of these studies is that both periodontal pathogens induced HIV-1 reactivation in a Tat-independent manner (i.e., absence of Tat in BF24 cell model). The transcriptional transactivator Tat is a critical HIV-1 protein that promotes a successful elongation during transcription of HIV-1 proviral forms (9). These findings support the idea of a mechanism for viral promoter activation, irrespective of the presence of Tat that has been proposed by others (65), in which a sustained NF-κB activation would be critically necessary to enhance HIV-1 promoter activity. Hence, studies to determine the effect of viral proteins, such as Tat, in HIV-1 promoter activity induced by oral bacteria are warranted.
Overall, we demonstrated for the first time that bacteria associated with oral and systemic chronic inflammatory disorders enhance HIV-1 reactivation in monocytes/macrophages through TLR2 and TLR9 activation in a mechanism that appears to be transcriptionally regulated and may not require the viral transactivator Tat. Because HIV-1+ patients are treated with a combination of antiretrovirals (HAART), whether or not antiretroviral drugs ameliorate HIV-1 reactivation in latently infected cells induced by periodontal pathogens in monocytes/macrophages remains to be determined. Finally, even though these are in vitro studies and more basic and additional clinical studies are necessary, we hypothesize that the appropriate and efficient control of oral infections, including HIV-1-associated periodontal disease in HIV-1+ patients, would be expected to contribute to minimizing the likelihood of viral recrudescence, HAART failure, and AIDS progression.
Acknowledgments
We thank Sreenatha Kirakodu and Yelena Alimova from COHR College of Dentistry, as well as Greg Bauman and Jennifer Strange from the FACS facility, Department of Microbiology, Immunology, and Molecular Genetics, for their valuable technical support.
This study was supported by USPHS grant P20 RR020145 from the National Institute for Research Resources and funds from the University of Kentucky, College of Dentistry.
Footnotes
▿
Published ahead of print on 7 July 2010.
REFERENCES
- 1.Alpagot, T., J. Remien, M. Bhattacharyya, K. Konopka, W. Lundergan, and N. Duzgunes. 2007. Longitudinal evaluation of prostaglandin E2 (PGE2) and periodontal status in HIV+n patients. Arch. Oral Biol. 52**:**1102-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alpagot, T., V. Suzara, and M. Bhattacharyya. 2006. The associations between gingival crevice fluid matrix metalloproteinase-9, tissue inhibitor of metalloproteinase-1 and periodontitis in human immunodeficiency virus-positive patients. J. Periodontal Res. 41**:**491-497. [DOI] [PubMed] [Google Scholar]
- 3.Amini, S., M. Saunders, K. Kelley, K. Khalili, and B. E. Sawaya. 2004. Interplay between HIV-1 Vpr and Sp1 modulates p21(WAF1) gene expression in human astrocytes. J. Biol. Chem. 279**:**46046-46056. [DOI] [PubMed] [Google Scholar]
- 4.Báfica, A., C. A. Scanga, M. L. Schito, S. Hieny, and A. Sher. 2003. Cutting edge: in vivo induction of integrated HIV-1 expression by mycobacteria is critically dependent on Toll-like receptor 2. J. Immunol. 171**:**1123-1127. [DOI] [PubMed] [Google Scholar]
- 5.Beutler, B. A. 2009. TLRs and innate immunity. Blood 113**:**1399-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, P., B. Mantelli, F. Delfanti, M. Cota, G. Vallanti, C. de Filippi, M. Mengozzi, E. Vicenzi, A. Lazzarin, and G. Poli. 2001. Tumor necrosis factor-alpha drives HIV-1 replication in U937 cell clones and upregulates CXCR4. Cytokine 13**:**55-59. [DOI] [PubMed] [Google Scholar]
- 7.Blanchard, A., L. Montagnier, and M. L. Gougeon. 1997. Influence of microbial infections on the progression of HIV disease. Trends Microbiol. 5**:**326-331. [DOI] [PubMed] [Google Scholar]
- 8.Bode, K. A., F. Schmitz, L. Vargas, K. Heeg, and A. H. Dalpke. 2009. Kinetic of RelA activation controls magnitude of TLR-mediated IL-12p40 induction. J. Immunol. 182**:**2176-2184. [DOI] [PubMed] [Google Scholar]
- 9.Brady, J., and F. Kashanchi. 2005. Tat gets the “green” light on transcription initiation. Retrovirology 2**:**69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenchley, J. M., D. A. Price, T. W. Schacker, T. E. Asher, G. Silvestri, S. Rao, Z. Kazzaz, E. Bornstein, O. Lambotte, D. Altmann, B. R. Blazar, B. Rodriguez, L. Teixeira-Johnson, A. Landay, J. N. Martin, F. M. Hecht, L. J. Picker, M. M. Lederman, S. G. Deeks, and D. C. Douek. 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12**:**1365-1371. [DOI] [PubMed] [Google Scholar]
- 11.Brenchley, J. M., T. W. Schacker, L. E. Ruff, D. A. Price, J. H. Taylor, G. J. Beilman, P. L. Nguyen, A. Khoruts, M. Larson, A. T. Haase, and D. C. Douek. 2004. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. J. Exp. Med. 200**:**749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bush, C. E., R. M. Donovan, N. P. Markowitz, P. Kvale, and L. D. Saravolatz. 1996. A study of HIV RNA viral load in AIDS patients with bacterial pneumonia. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13**:**23-26. [DOI] [PubMed] [Google Scholar]
- 13.Cahill, R. J., S. Tan, G. Dougan, P. O'Gaora, D. Pickard, N. Kennea, M. H. Sullivan, R. G. Feldman, and A. D. Edwards. 2005. Universal DNA primers amplify bacterial DNA from human fetal membranes and link Fusobacterium nucleatum with prolonged preterm membrane rupture. Mol. Hum. Reprod. 11**:**761-766. [DOI] [PubMed] [Google Scholar]
- 14.Carmody, R. J., and Y. H. Chen. 2007. Nuclear factor-kappaB: activation and regulation during Toll-like receptor signaling. Cell. Mol. Immunol. 4**:**31-41. [PubMed] [Google Scholar]
- 15.Chattopadhyay, A., D. J. Caplan, G. D. Slade, D. C. Shugars, H. C. Tien, and L. L. Patton. 2005. Risk indicators for oral candidiasis and oral hairy leukoplakia in HIV-infected adults. Community Dent. Oral Epidemiol. 33**:**35-44. [DOI] [PubMed] [Google Scholar]
- 16.Chen, A., I. C. Boulton, J. Pongoski, A. Cochrane, and S. D. Gray-Owen. 2003. Induction of HIV-1 long terminal repeat-mediated transcription by Neisseria gonorrhoeae. AIDS 17**:**625-628. [DOI] [PubMed] [Google Scholar]
- 17.Chun, R. F., O. J. Semmes, C. Neuveut, and K. T. Jeang. 1998. Modulation of Sp1 phosphorylation by human immunodeficiency virus type 1 Tat. J. Virol. 72**:**2615-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coogan, M. M., J. Greenspan, and S. J. Challacombe. 2005. Oral lesions in infection with human immunodeficiency virus. Bull. World Health Organ. 83**:**700-706. [PMC free article] [PubMed] [Google Scholar]
- 19.Dalpke, A., J. Frank, M. Peter, and K. Heeg. 2006. Activation of Toll-like receptor 9 by DNA from different bacterial species. Infect. Immun. 74**:**940-946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devadas, K., N. J. Hardegen, L. M. Wahl, I. K. Hewlett, K. A. Clouse, K. M. Yamada, and S. Dhawan. 2004. Mechanisms for macrophage-mediated HIV-1 induction. J. Immunol. 173**:**6735-6744. [DOI] [PubMed] [Google Scholar]
- 21.Ebersole, J. L., M. J. Novak, B. S. Michalowicz, J. S. Hodges, M. J. Steffen, J. E. Ferguson, A. Diangelis, W. Buchanan, D. A. Mitchell, and P. N. Papapanou. 2009. Systemic immune responses in pregnancy and periodontitis: relationship to pregnancy outcomes in the Obstetrics and Periodontal Therapy (OPT) Study. J. Periodontol. 80**:**953-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Equils, O., E. Faure, L. Thomas, Y. Bulut, S. Trushin, and M. Arditi. 2001. Bacterial lipopolysaccharide activates HIV long terminal repeat through Toll-like receptor 4. J. Immunol. 166**:**2342-2347. [DOI] [PubMed] [Google Scholar]
- 23.Equils, O., Z. Madak, C. Liu, K. S. Michelsen, Y. Bulut, and D. Lu. 2004. Rac1 and Toll-IL-1 receptor domain-containing adapter protein mediate Toll-like receptor 4 induction of HIV-long terminal repeat. J. Immunol. 172**:**7642-7646. [DOI] [PubMed] [Google Scholar]
- 24.Ewaschuk, J. B., J. L. Backer, T. A. Churchill, F. Obermeier, D. O. Krause, and K. L. Madsen. 2007. Surface expression of Toll-like receptor 9 is upregulated on intestinal epithelial cells in response to pathogenic bacterial DNA. Infect. Immun. 75**:**2572-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Felber, B. K., and G. N. Pavlakis. 1988. A quantitative bioassay for HIV-1 based on trans-activation. Science 239**:**184-187. [DOI] [PubMed] [Google Scholar]
- 26.Gibson, F. C., III, T. Ukai, and C. A. Genco. 2008. Engagement of specific innate immune signaling pathways during Porphyromonas gingivalis induced chronic inflammation and atherosclerosis. Front. Biosci. 13**:**2041-2059. [DOI] [PubMed] [Google Scholar]
- 27.González, O. A., J. L. Ebersole, and C. B. Huang. 2009. Oral infectious diseases: a potential risk factor for HIV virus recrudescence? Oral Dis. 15**:**313-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenspan, D., E. Komaroff, M. Redford, J. A. Phelan, M. Navazesh, M. E. Alves, H. Kamrath, R. Mulligan, C. E. Barr, and J. S. Greenspan. 2000. Oral mucosal lesions and HIV viral load in the Women's Interagency HIV Study (WIHS). J. Acquir. Immune Defic. Syndr. 25**:**44-50. [DOI] [PubMed] [Google Scholar]
- 29.Guadalupe, M., E. Reay, S. Sankaran, T. Prindiville, J. Flamm, A. McNeil, and S. Dandekar. 2003. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J. Virol. 77**:**11708-11717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hajishengallis, G., R. I. Tapping, E. Harokopakis, S. Nishiyama, P. Ratti, R. E. Schifferle, E. A. Lyle, M. Triantafilou, K. Triantafilou, and F. Yoshimura. 2006. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell. Microbiol. 8**:**1557-1570. [DOI] [PubMed] [Google Scholar]
- 31.Han, Y. W., R. W. Redline, M. Li, L. Yin, G. B. Hill, and T. S. McCormick. 2004. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect. Immun. 72**:**2272-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbein, G., A. Varin, A. Larbi, C. Fortin, U. Mahlknecht, T. Fulop, and B. B. Aggarwal. 2008. Nef and TNFalpha are coplayers that favor HIV-1 replication in monocytic cells and primary macrophages. Curr. HIV Res. 6**:**117-129. [DOI] [PubMed] [Google Scholar]
- 33.Hill, G. B. 1998. Preterm birth: associations with genital and possibly oral microflora. Ann. Periodontol. 3**:**222-232. [DOI] [PubMed] [Google Scholar]
- 34.Holst, E., A. R. Goffeng, and B. Andersch. 1994. Bacterial vaginosis and vaginal microorganisms in idiopathic premature labor and association with pregnancy outcome. J. Clin. Microbiol. 32**:**176-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, C. B., K. A. Emerson, O. A. Gonzalez, and J. L. Ebersole. 2009. Oral bacteria induce a differential activation of human immunodeficiency virus-1 promoter in T cells, macrophages and dendritic cells. Oral Microbiol. Immunol. 24**:**401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Imai, K., K. Ochiai, and T. Okamoto. 2009. Reactivation of latent HIV-1 infection by the periodontopathic bacterium Porphyromonas gingivalis involves histone modification. J. Immunol. 182**:**3688-3695. [DOI] [PubMed] [Google Scholar]
- 37.Ishihara, K., A. Nabuchi, R. Ito, K. Miyachi, H. K. Kuramitsu, and K. Okuda. 2004. Correlation between detection rates of periodontopathic bacterial DNA in stenotic artery plaque and in dental plaque samples. J. Clin. Microbiol. 42**:**1313-1315. (Erratum, **42:**5437.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacqué, J. M., B. Fernandez, F. Arenzana-Seisdedos, D. Thomas, F. Baleux, J. L. Virelizier, and F. Bachelerie. 1996. Permanent occupancy of the human immunodeficiency virus type 1 enhancer by NF-kappa B is needed for persistent viral replication in monocytes. J. Virol. 70**:**2930-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji, S., J. Hyun, E. Park, B. L. Lee, K. K. Kim, and Y. Choi. 2007. Susceptibility of various oral bacteria to antimicrobial peptides and to phagocytosis by neutrophils. J. Periodontal Res. 42**:**410-419. [DOI] [PubMed] [Google Scholar]
- 40.Jiang, G., A. Espeseth, D. J. Hazuda, and D. M. Margolis. 2007. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J. Virol. 81**:**10914-10923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang, W., M. M. Lederman, P. Hunt, S. F. Sieg, K. Haley, B. Rodriguez, A. Landay, J. Martin, E. Sinclair, A. I. Asher, S. G. Deeks, D. C. Douek, and J. M. Brenchley. 2009. Plasma levels of bacterial DNA correlate with immune activation and the magnitude of immune restoration in persons with antiretroviral-treated HIV infection. J. Infect. Dis. 199**:**1177-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kawai, T., and S. Akira. 2007. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 13**:**460-469. [DOI] [PubMed] [Google Scholar]
- 43.Kawai, T., and S. Akira. 2006. TLR signaling. Cell Death Differ. 13**:**816-825. [DOI] [PubMed] [Google Scholar]
- 44.Kesavalu, L., S. C. Holt, and J. L. Ebersole. 2003. In vitro environmental regulation of Porphyromonas gingivalis growth and virulence. Oral Microbiol. Immunol. 18**:**226-233. [DOI] [PubMed] [Google Scholar]
- 45.Kikkert, R., M. L. Laine, L. A. Aarden, and A. J. van Winkelhoff. 2007. Activation of toll-like receptors 2 and 4 by gram-negative periodontal bacteria. Oral Microbiol. Immunol. 22**:**145-151. [DOI] [PubMed] [Google Scholar]
- 46.Kitaura, H., N. Ohara, K. Kobayashi, and T. Yamada. 2001. TNF-alpha-mediated multiplication of human immunodeficiency virus in chronically infected monocytoid cells by mycobacterial infection. APMIS 109**:**533-540. [DOI] [PubMed] [Google Scholar]
- 47.Kozarov, E. V., B. R. Dorn, C. E. Shelburne, W. A. Dunn, Jr., and A. Progulske-Fox. 2005. Human atherosclerotic plaque contains viable invasive Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Arterioscler. Thromb. Vasc. Biol. 25**:**e17-18. [DOI] [PubMed] [Google Scholar]
- 48.Kroidl, A., A. Schaeben, M. Oette, M. Wettstein, A. Herfordt, and D. Haussinger. 2005. Prevalence of oral lesions and periodontal diseases in HIV-infected patients on antiretroviral therapy. Eur. J. Med. Res. 10**:**448-453. [PubMed] [Google Scholar]
- 49.Kutsch, O., E. N. Benveniste, G. M. Shaw, and D. N. Levy. 2002. Direct and quantitative single-cell analysis of human immunodeficiency virus type 1 reactivation from latency. J. Virol. 76**:**8776-8786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu, H., R. W. Redline, and Y. W. Han. 2007. Fusobacterium nucleatum induces fetal death in mice via stimulation of TLR4-mediated placental inflammatory response. J. Immunol. 179**:**2501-2508. [DOI] [PubMed] [Google Scholar]
- 51.Majello, B., P. De Luca, G. Hagen, G. Suske, and L. Lania. 1994. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. 22**:**4914-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Makiura, N., M. Ojima, Y. Kou, N. Furuta, N. Okahashi, S. Shizukuishi, and A. Amano. 2008. Relationship of Porphyromonas gingivalis with glycemic level in patients with type 2 diabetes following periodontal treatment. Oral Microbiol. Immunol. 23**:**348-351. [DOI] [PubMed] [Google Scholar]
- 53.Miyazato, A., K. Nakamura, N. Yamamoto, H. M. Mora-Montes, M. Tanaka, Y. Abe, D. Tanno, K. Inden, X. Gang, K. Ishii, K. Takeda, S. Akira, S. Saijo, Y. Iwakura, Y. Adachi, N. Ohno, K. Mitsutake, N. A. Gow, M. Kaku, and K. Kawakami. 2009. Toll-like receptor 9-dependent activation of myeloid dendritic cells by deoxynucleic acids from Candida albicans. Infect. Immun. 77**:**3056-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicol, M. Q., J. M. Mathys, A. Pereira, K. Ollington, M. H. Ieong, and P. R. Skolnik. 2008. Human immunodeficiency virus infection alters tumor necrosis factor alpha production via Toll-like receptor-dependent pathways in alveolar macrophages and U1 cells. J. Virol. 82**:**7790-7798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nokta, M. 2008. Oral manifestations associated with HIV infection. Curr. HIV/AIDS Rep. 5**:**5-12. [DOI] [PubMed] [Google Scholar]
- 56.Nonnenmacher, C., A. Dalpke, S. Zimmermann, L. Flores-De-Jacoby, R. Mutters, and K. Heeg. 2003. DNA from periodontopathogenic bacteria is immunostimulatory for mouse and human immune cells. Infect. Immun. 71**:**850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nordone, S. K., G. A. Ignacio, L. Su, G. D. Sempowski, D. T. Golenbock, L. Li, and G. A. Dean. 2007. Failure of TLR4-driven NF-kappa B activation to stimulate virus replication in models of HIV type 1 activation. AIDS Res. Hum. Retroviruses 23**:**1387-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Osiecki, K., L. Xie, J. H. Zheng, R. Squires, M. Pettoello-Mantovani, and H. Goldstein. 2005. Identification of granulocyte-macrophage colony-stimulating factor and lipopolysaccharide-induced signal transduction pathways that synergize to stimulate HIV type 1 production by monocytes from HIV type 1 transgenic mice. AIDS Res. Hum. Retroviruses 21**:**125-139. [DOI] [PubMed] [Google Scholar]
- 59.Patton, L. L. 2000. Sensitivity, specificity, and positive predictive value of oral opportunistic infections in adults with HIV/AIDS as markers of immune suppression and viral burden. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 90**:**182-188. [DOI] [PubMed] [Google Scholar]
- 60.Pereira, L. A., K. Bentley, A. Peeters, M. J. Churchill, and N. J. Deacon. 2000. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 28**:**663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poli, G., P. Bressler, A. Kinter, E. Duh, W. C. Timmer, A. Rabson, J. S. Justement, S. Stanley, and A. S. Fauci. 1990. Interleukin 6 induces human immunodeficiency virus expression in infected monocytic cells alone and in synergy with tumor necrosis factor alpha by transcriptional and post-transcriptional mechanisms. J. Exp. Med. 172**:**151-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pomerantz, R. J., M. B. Feinberg, D. Trono, and D. Baltimore. 1990. Lipopolysaccharide is a potent monocyte/macrophage-specific stimulator of human immunodeficiency virus type 1 expression. J. Exp. Med. 172**:**253-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rohr, O., C. Marban, D. Aunis, and E. Schaeffer. 2003. Regulation of HIV-1 gene transcription: from lymphocytes to microglial cells. J. Leukoc. Biol. 74**:**736-749. [DOI] [PubMed] [Google Scholar]
- 64.Rossi, A., R. Mukerjee, P. Ferrante, K. Khalili, S. Amini, and B. E. Sawaya. 2006. Human immunodeficiency virus type 1 Tat prevents dephosphorylation of Sp1 by TCF-4 in astrocytes. J. Gen. Virol. 87**:**1613-1623. [DOI] [PubMed] [Google Scholar]
- 65.Sgarbanti, M., A. L. Remoli, G. Marsili, B. Ridolfi, A. Borsetti, E. Perrotti, R. Orsatti, R. Ilari, L. Sernicola, E. Stellacci, B. Ensoli, and A. Battistini. 2008. IRF-1 is required for full NF-kappaB transcriptional activity at the human immunodeficiency virus type 1 long terminal repeat enhancer. J. Virol. 82**:**3632-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Socransky, S. S., and A. D. Haffajee. 2005. Periodontal microbial ecology. Periodontol. 2000 38**:**135-187. [DOI] [PubMed] [Google Scholar]
- 67.Stevenson, M. 2003. HIV-1 pathogenesis. Nat. Med. 9**:**853-860. [DOI] [PubMed] [Google Scholar]
- 68.Stojanova, A., C. Caro, R. J. Jarjour, S. K. Oster, L. Z. Penn, and R. J. Germinario. 2004. Repression of the human immunodeficiency virus type-1 long terminal repeat by the c-Myc oncoprotein. J. Cell. Biochem. 92**:**400-413. [DOI] [PubMed] [Google Scholar]
- 69.Swingler, S., A. Mann, J. Jacque, B. Brichacek, V. G. Sasseville, K. Williams, A. A. Lackner, E. N. Janoff, R. Wang, D. Fisher, and M. Stevenson. 1999. HIV-1 Nef mediates lymphocyte chemotaxis and activation by infected macrophages. Nat. Med. 5**:**997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tada, H., S. Aiba, K. Shibata, T. Ohteki, and H. Takada. 2005. Synergistic effect of Nod1 and Nod2 agonists with Toll-like receptor agonists on human dendritic cells to generate interleukin-12 and T helper type 1 cells. Infect. Immun. 73**:**7967-7976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takeshita, A., K. Imai, and S. Hanazawa. 1999. CpG motifs in Porphyromonas gingivalis DNA stimulate interleukin-6 expression in human gingival fibroblasts. Infect. Immun. 67**:**4340-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tietze, K., A. Dalpke, S. Morath, R. Mutters, K. Heeg, and C. Nonnenmacher. 2006. Differences in innate immune responses upon stimulation with gram-positive and gram-negative bacteria. J. Periodontal Res. 41**:**447-454. [DOI] [PubMed] [Google Scholar]
- 73.Wahl, S. M., T. Greenwell-Wild, G. Peng, G. Ma, J. M. Orenstein, and N. Vazquez. 2003. Viral and host cofactors facilitate HIV-1 replication in macrophages. J. Leukoc. Biol. 74**:**726-735. [DOI] [PubMed] [Google Scholar]
- 74.Wig, N., P. Anupama, S. Singh, R. Handa, P. Aggarwal, S. N. Dwivedi, B. L. Jailkhani, and J. P. Wali. 2005. Tumor necrosis factor-alpha levels in patients with HIV with wasting in South Asia. AIDS Patient Care STDS 19**:**212-215. [DOI] [PubMed] [Google Scholar]
- 75.Williams, S. A., H. Kwon, L. F. Chen, and W. C. Greene. 2007. Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1. J. Virol. 81**:**6043-6056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yedavalli, V. S., M. Benkirane, and K. T. Jeang. 2003. Tat and trans-activation-responsive (TAR) RNA-independent induction of HIV-1 long terminal repeat by human and murine cyclin T1 requires Sp1. J. Biol. Chem. 278**:**6404-6410. [DOI] [PubMed] [Google Scholar]
- 77.Zhong, B., P. Tien, and H. B. Shu. 2006. Innate immune responses: crosstalk of signaling and regulation of gene transcription. Virology 352**:**14-21. [DOI] [PubMed] [Google Scholar]