Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing (original) (raw)
Abstract
Massively parallel DNA sequencing technologies have greatly increased our ability to generate large amounts of sequencing data at a rapid pace. Several methods have been developed to enrich for genomic regions of interest for targeted sequencing. We have compared three of these methods: Molecular Inversion Probes (MIP), Solution Hybrid Selection (SHS), and Microarray-based Genomic Selection (MGS). Using HapMap DNA samples, we compared each of these methods with respect to their ability to capture an identical set of exons and evolutionarily conserved regions associated with 528 genes (2.61 Mb). For sequence analysis, we developed and used a novel Bayesian genotype-assigning algorithm, Most Probable Genotype (MPG). All three capture methods were effective, but sensitivities (percentage of targeted bases associated with high-quality genotypes) varied for an equivalent amount of pass-filtered sequence: for example, 70% (MIP), 84% (SHS), and 91% (MGS) for 400 Mb. In contrast, all methods yielded similar accuracies of >99.84% when compared to Infinium 1M SNP BeadChip-derived genotypes and >99.998% when compared to 30-fold coverage whole-genome shotgun sequencing data. We also observed a low false-positive rate with all three methods; of the heterozygous positions identified by each of the capture methods, >99.57% agreed with 1M SNP BeadChip, and >98.840% agreed with the whole-genome shotgun data. In addition, we successfully piloted the genomic enrichment of a set of 12 pooled samples via the MGS method using molecular bar codes. We find that these three genomic enrichment methods are highly accurate and practical, with sensitivities comparable to that of 30-fold coverage whole-genome shotgun data.
The ability to identify genetic alterations underlying disease and phenotypic variation is a major goal of the ongoing genomics revolution. Large-scale medical sequencing holds the promise of elucidating the genetic architecture of virtually all diseases, including the relative role of rare and common genetic variants. Such information should inform our understanding of the pathways involved in disease pathogenesis. Additionally, the ability to apply variant discovery to other organisms with annotated genomes is also of great value. So-called next-generation DNA sequencing technologies that involve massively parallel clonal ensemble sequencing are creating new opportunities for comprehensive genomic interrogation, bypassing some of the limitations associated with high-throughput electrophoresis-based sequencing methods (Margulies et al. 2005; Shendure et al. 2005; Bentley et al. 2008). Although advances in these new technologies are being made at a rapid pace, the cost of sequencing a whole human genome and the associated costs and technical hurdles of data storage and analysis remain high for most large projects. As a result, methods are needed for efficient comprehensive sequencing of targeted genomic regions.
A number of genomic enrichment (or targeted capture) methods have been developed and used with varying levels of success (for reviews, see Garber 2008; Summerer 2009; Turner et al. 2009b; Mamanova et al. 2010). Not surprisingly, each method has both common and unique issues related to the required input DNA, specificity and coverage, sensitivity and accuracy, scalability and potential for automation, and cost-effectiveness. Although multiple publications describe the individual methods (Albert et al. 2007; Hodges et al. 2007; Okou et al. 2007; Porreca et al. 2007; Craig et al. 2008; Krishnakumar et al. 2008; Bau et al. 2009; Gnirke et al. 2009; Herman et al. 2009; Hodges et al. 2009; Li et al. 2009; Summerer et al. 2009; Tewhey et al. 2009), numerous variables are inherent to each experiment, including the nature of the targeted genomic region(s), sequencing platform, analysis software, and performance metrics; these variables make objective comparisons of the different methods challenging to perform.
Here, we report the testing, optimizing, and rigorous comparing of three genomic enrichment methods: Molecular Inversion Probes (MIP) (Porreca et al. 2007), Solution Hybrid Selection (SHS; Agilent) (Gnirke et al. 2009), and Microarray-based Genomic Selection (MGS; Roche-NimbleGen) (Albert et al. 2007; Okou et al. 2007). All three methods were tested for their ability to capture the same 2.61 Mb of noncontiguous DNA sequence from an overlapping set of two HapMap DNA samples. We also report, for the first time, a novel Bayesian genotype-assigning algorithm, Most Probable Genotype (MPG), which was used to analyze the sequence data from all three capture methods. Furthermore, we introduce a pre-capture bar-coding strategy to allow pooling of samples for capture and sequencing. In contrast to previous reviews of genomic enrichment methods, we have directly compared these methods by evaluating genotype sensitivity and accuracy of variant detection, providing insights about overall performance and relative costs.
Results
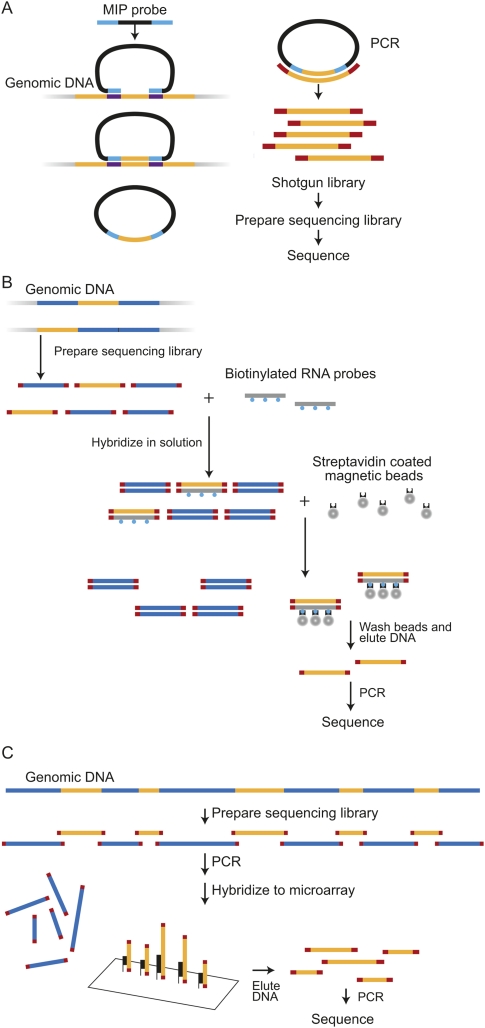
Both of the solution-based methods—MIP (Fig. 1A) and SHS (Fig. 1B)—are highly multiplexed capture methods that can enrich for a large number of genomic regions. We have incorporated published improvements to the original MIP-based method (Li et al. 2009; Turner et al. 2009a), including improvements to facilitate automated and effective probe design; accuracy of the MIP-based method has also been improved by segregating overlapping probes to two separate probe sets. The 384K MGS array (Fig. 1C) also allows for highly multiplexed capture of genomic regions in a single hybridization but uses a solid-phase DNA-oligonucleotide hybridization step; we adapted the original MGS-based protocol (Albert et al. 2007) to increase capture efficiency and to allow for sample pooling to reduce costs. There were a few considerations specific to several of the methods that led us to modify or adapt these methods for the purposes of this study. These are described below, before the overall results of the study are described.
Figure 1.
Three genomic enrichment methods. (A) (MIP) Molecular Inversion Probe: 70 base probes are prepared and hybridized to genomic DNA. Capture occurs by filling in sequence between the probe-targeting arms with polymerase and then sealing the circle with ligase. Total genomic DNA is removed with nucleases. The remaining closed circles undergo shotgun library and sequencing library preparation, followed by sequencing. (B) (SHS) Solution Hybrid Selection: A sequencing library is prepared from genomic DNA. This library is hybridized to biotinylated RNA probes in solution and recovered with streptavidin beads. Eluted products are amplified prior to cluster generation and sequencing. (C) (MGS) Microarray-based Genomic Selection: A sequencing library is prepared from genomic DNA and hybridized to a capture array. Eluted products are amplified prior to cluster generation and sequencing.
Molecular Inversion Probes (MIPs)
In the process of implementing an MIP-based genomic enrichment method, we found that our sequence data included the probe arm sequences that were designed to hybridize to DNA flanking the region of interest (ROI) (data not shown). This occurred when probes were required to overlap each other in order to capture a long region. Because these arm sequences of the MIP probes were sequenced and aligned, they could falsely increase the proportion of the reference allele and yield false-negative results. In addition to the probe arm sequences, some MIP backbone sequence (nonhuman) was also present and aligned to the human reference sequence with several mismatches, which generated significant numbers of false-positive variants.
One approach for avoiding such artifacts involves end-sequencing the captured DNA fragments, thus avoiding shotgun library preparation and the inclusion of probe backbone sequence. The increasing capabilities of sequencers to generate longer read lengths make this approach more feasible, which may improve alignment rates and provide better coverage and higher sensitivities (Turner et al. 2009a). We tested this strategy by designing probes to target ROI fragments of about 105 bases with 76-base end-sequences; however, only 55% of the ROI bases could be covered by this design, which we concluded was unacceptable.
We avoided these artifacts by dividing the capture MIPs into two probe sets. Although the two probe sets overlapped each other, the probes within each set were nonoverlapping. Within a nonoverlapping set, the targeting arm and probe backbone sequences are distinguishable from the captured DNA and thus can be excluded following alignment. For the entire capture-to-sequencing process, each of the two probe sets was treated independently. After sequencing and alignment, the targeting arm and backbone bases were computationally removed, and the data were combined. As this strategy required the separate sequencing of the DNA captured from each probe set, we needed two lanes, which generated twice as much sequence data for the MIP method compared to the other two; we used this full amount of sequence (∼1.1 Gb) in our comparison analyses throughout this study. Future application of this strategy could include indexing of each probe set to allow merging of the individually captured DNA from each set, eliminating the need for two separate sequencing lanes (and the extra sequence) for one sample.
Solution Hybrid Selection (SHS)
Following the initial description of SHS (Gnirke et al. 2009), Agilent Technologies released a commercial version (SureSelect). Due to improvements communicated by the vendor, we chose to implement SHS using these kits and performed the captures as described by the manufacturer.
MGS and pooled bar-coded samples
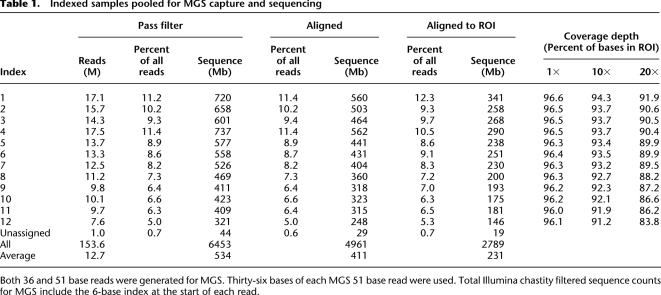
Considering the high cost of MGS (Roche NimbleGen) arrays and the increasing single-lane sequencing capacity of next-generation sequencing platforms, we developed a novel strategy that enables the efficient capture of a pool of 12 individually bar-coded Illumina paired-end (PE) libraries. In contrast to most methods used for bar-coding samples for pool sequencing, this strategy is optimized for use in targeted hybrid capture, followed by sequencing. Specifically, we made a 6-base indexed paired-end library for each of 12 genomic DNA samples (including the same two HapMap samples as were used in the MIP and SHS methods) and simultaneously performed MGS of all 12 samples with one array. The index served as a bar code and provided individual-specific sequence data for the array-enriched targets after sequencing. Reads were assigned to each bar code if the index sequence had no more than one mismatch. Of the reads that passed the quality filter (Illumina GERALD chastity filter), we were able to assign ∼99% to a unique sample using this indexing strategy. A narrow distribution of sequencing depth across samples, or sample uniformity, is critical when sequencing bar-coded pools to avoid over-representation of some samples at the expense of others. Poor sample uniformity would require additional sequencing to provide adequate depth of coverage of the samples with lower representation. Perfect sample uniformity would yield 8.3% of filtered reads originating from each of the 12 samples. The difference between the bar-coded samples with the highest (11.4%) and lowest (5.0%) number of filtered reads was slightly over twofold (Table 1). We conclude that this level of sample uniformity is acceptable for many potential applications of this target enrichment method.
Table 1.
Indexed samples pooled for MGS capture and sequencing
Another important aspect of sequencing pooled samples is to have each ROI base covered to a similar level across all samples in the pool. This allows for consistent interrogation of the same ROI bases for all samples and facilitates follow-up attempts to re-sequence only the smaller subset of missed regions. We found that samples were fairly uniform with respect to well-covered and poorly covered bases. Approximately 78% of the ROI bases had good coverage (≥20-fold redundancy in all 12 samples), 20% had variable coverage (one or more of the indexed samples had one- to 19-fold coverage), and 2% had no coverage in any sample. Bases with no coverage were mainly those for which probes could not be designed.
Input DNA requirements
Sequencing projects often involve analyzing samples with limited DNA resources. We performed MIP capture with 1 μg of input DNA, the lowest amount of the three methods. The SHS and MGS protocols required 3 μg and 4 μg of DNA, respectively. Of note, the modified MGS protocol described here required 4 μg of starting DNA to generate a library suitable for capture and subsequent sequencing on the Illumina GAII platform. Although this reflects an 80% reduction in the amount of input DNA recommended by the Roche/NimbleGen MGS protocol (4 vs. 20 μg), we believe that an even smaller amount of DNA may work as well. We started by shearing 4 μg of input DNA to account for the variability and inaccuracies of measured DNA concentration values; the subsequent library preparation steps (end repair and adapter ligation) proceeded with only 2 μg of sheared DNA. Thus, to further reduce input DNA requirement, more attention could be placed on re-quantifying DNA with more reliable, but more time-consuming, methods.
Comparison of methods
Our regions of interest comprised 2.61 Mb of noncontiguous human genome sequence consisting of exons and conserved elements associated with 528 genes (see Methods). Successful probe designs were achieved for 95.9% (2.506/2.612 Mb) of the ROI bases for the MIP, 92.6% (2.419/2.612 Mb) for the SHS, and 93.8% (2.450/2.612 Mb) for the MGS methods. Since sequencing projects aim to interrogate the entirety of the ROI, we assessed the performance metrics based on the entire 2.61 Mb of ROI rather than only on the regions for which suitable probes could be designed. Furthermore, analysis of the ROI across all three methods allowed us to make more appropriate comparisons for this study.
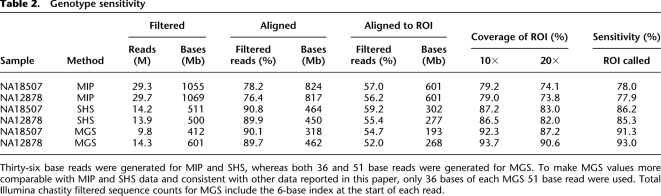
We performed targeted enrichment and sequencing on multiple samples for each of the methods. Although the majority of the samples from the MIP and SHS methods were distinct from those of the indexed samples pooled for the MGS capture, there were two HapMap samples in common across all three methods (NA18507 and NA12878). The amount of sequence generated for each of the indexed samples was comparable to that for the single sample experiments (Table 2). The data presented here compare only these two samples unless otherwise noted (i.e., “extended sample sets” refers to all samples).
Table 2.
Genotype sensitivity
Fraction of sequences that align to ROI
The first metric examined was the fraction of the captured DNA that aligned to the ROI. This metric reflects the ability of the method to enrich for appropriate targets and greatly influences the cost (as more sequencing is required for a lower aligned fraction). Approximately 10 to 14 million filtered reads were obtained for each of the two samples captured with either the SHS or MGS methods; in contrast, about 30 million filtered reads were obtained with the MIP method due to the need to generate two nonoverlapping captures (see above) (Table 2). We used ELAND (Illumina, Inc.) to align the sequence reads to the reference human genome and then calculated depth of coverage for our ROI. The sequencing was performed at different times during the project, and the capability of the sequencer evolved during this time period. Consequently, the amount of generated sequence varied for a given number of reads among the methods. To equalize the comparisons, we only used a maximum of 36 bases of each sequence read when calculating the various metrics in this analysis. Furthermore, the total filtered sequence data count for MGS included the 6-base index at the beginning of each sequence read, accounting for the extra nontarget sequence. Although the proportion of filtered reads that aligned uniquely to the genome varied (76%–90%), the amount that aligned uniquely to the ROI was more consistent (52%–59%) for all methods and samples.
A smaller fraction of MIP-generated reads aligned to the genome as compared to SHS- or MGS-generated reads. Much of this difference was due to the MIP probe-backbone sequence, which is still in the library at this stage; it can only be computationally removed once the alignment is completed, and the captured and designed/backbone bases can be differentiated. As the backbone sequence is not human, it added many mismatches to a given alignment and prevented these reads from aligning to the genome. Even though a smaller fraction of MIP-generated reads aligned to the genome, a higher fraction of the genome-aligned reads (compared to the other two methods) overlapped the ROI, resulting in a total fraction of ROI-aligning reads that was similar to that seen with the other methods.
Depth and uniformity of coverage
Important metrics of the sequence generated by a capture method include the number of sequence reads that overlap each base across the ROI and the uniformity of that coverage depth. This is because accurate genotype assignments depend on a minimum coverage depth (typically 10- to 20-fold) at each base. Uneven depth of coverage across ROI, or poor uniformity, creates the need for more sequencing to provide sufficient data for poorly represented regions.
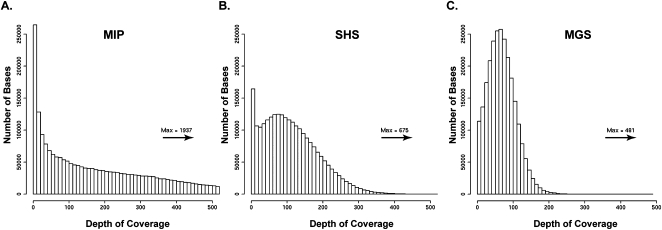
The proportion of ROI bases with adequate depth of coverage was consistent across samples for a given method but varied among the methods (ranging from ∼79%–94% with ≥10-fold and ∼74%–91% with ≥20-fold depth of coverage) (Table 2). A greater proportion of the ROI bases had at least 10-fold coverage depth for MGS, especially compared to the MIP method. This was likely due to differences in the uniformity of coverage for the different methods (Fig. 2). The distribution for MIP-derived sequence depth of coverage was spread over a wide range (one- to 1937-fold, median at 157-fold); this was less the case for SHS (one- to 675-fold, median at 101-fold) and MGS (one- to 481-fold, median at 66-fold). In general, as the depth of coverage distribution widens, bases with excessively high read depth account for a large share of the sequence reads, necessitating more sequence data to ensure that less efficiently captured regions reach the threshold for a high-quality genotype assignment. Due to its more uniform capture, MGS achieved high-quality genotype assignments at more ROI bases. The uniformity for SHS was similar to that of MGS, and its 10-fold and 20-fold coverage depth statistics were almost as high as for MGS. The uniformity for MIP was very broad, resulting in fewer high-quality genotype assignments.
Figure 2.
Depth of coverage distribution. Distributions of depth of coverage at each ROI position. Scales have been standardized for comparison purposes, and maximum coverage depth values are indicated above the arrow. (A) (MIP) Molecular Inversion Probe. (B) (SHS) Solution Hybrid Selection. (C) (MGS) Microarray-based Genomic Selection.
Genotype sensitivity
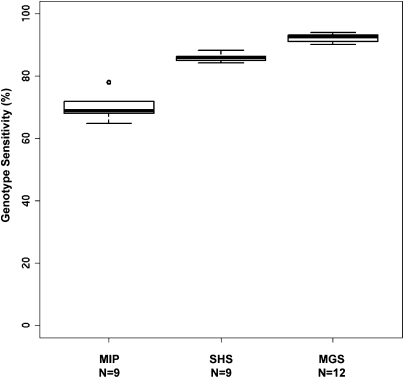
We used a new Bayesian genotype-assigning algorithm, Most Probable Genotype (MPG), to determine genotypes. Specifically, we only accepted genotype assignments with an MPG prediction score of 10 or greater, which we have empirically found to yield a balance between sensitivity and accuracy (see Methods and Supplemental Methods). An MPG score of 10 corresponded roughly with 10× to 20× depth of coverage. We defined genotype sensitivity as the percentage of ROI bases with MPG assignment scores of ≥10. The MIP method had the lowest genotype sensitivity at ∼78% even with twice the amount of sequence data (Table 2). The other two methods were similar to each other, with MGS slightly more sensitive (∼93%) than SHS (∼86%). This high genotype sensitivity was consistently achieved, even when examining an extended set of samples (Fig. 3). We have applied each of these methods to other projects with multiple samples and have observed a similarly low variance in genotype sensitivity among samples within each project (data not shown). These results are especially encouraging since the probes could be designed to capture only 93%–96% of the ROI bases for each of the latter two methods. For example, the MGS probe design covered 94.0% of the ROI bases, and this method had genotype sensitivity of up to 93.0% overall, which suggests that almost all of the designed target bases yielded high-quality genotypes. Similarly, the SHS probe design covered 92.6% of the ROI bases and had a genotype sensitivity of 86.2%, assigning a high-quality genotype to the vast majority of the targeted bases. Thus, we can conclude that genotype sensitivity was good and consistent for all three methods, but that MGS had the highest genotype sensitivity.
Figure 3.
Genotype sensitivity across multiple samples. Boxplots showing distribution of genotype call sensitivities across multiple samples (extended sample set) for each capture method. (N) Number of samples.
Bases with no genotype assignments
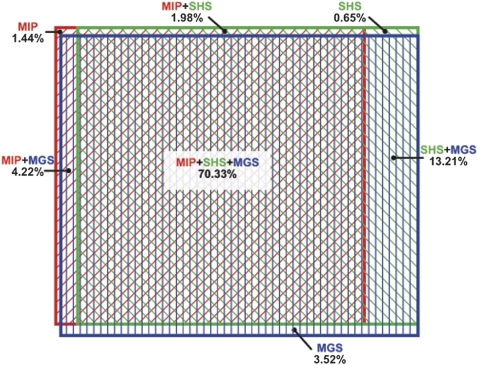
We also determined the characteristics of the ROI bases with no genotype assignments. Such information may prove useful for choosing a genomic enrichment method (or combination of strategies) for a specific project. We analyzed the genotype assignments for all three methods at each ROI base and determined the number of positions in NA18507 covered by all, two, or one method. There was considerable overlap among the three methods. Specifically, the same 70.33% of the ROI bases were assigned a genotype by all three methods, an additional 25.02% was assigned by at least one method, and 4.65% were not assigned by any method (Fig. 4). Overall, SHS and MGS had the greatest overlap, sharing genotype assignments for ∼83% of the bases of the ROI. We would expect by random chance that ∼0.2% of our ROI would be unassigned in all three methods, which is much less than the observed value (4.65%). Similarly, ∼61% of the ROI is expected by chance to have genotype assignments in all three methods, which is also less than the observed value (70.33%). This suggests that similarities among the three methods contribute to the observed overlap. One similarity is the avoidance of repetitive sequence during probe design. Our ROI contains ∼60 kb that overlaps with the Segmental Duplication Track from the UCSC Genome Browser (Bailey et al. 2001). Of these bases, we were unable to assign genotypes for ∼19 kb with SHS, ∼22 kb with MIP, and ∼31 kb with MGS. The high percentage of bases without genotype assignments in these regions confirms that all methods are avoiding repeats as part of probe design, with MGS being slightly more stringent.
Figure 4.
ROI regions with genotype assignments. The Venn diagram of overlapping genotype coverage is area proportional. Colored rectangles identify the proportion of genotype assignments in the ROI for each method: (red) MIP; (green) SHS; (blue) MGS. Note that the greatest overlap is among all three methods, and the second greatest is between SHS and MGS. The numbers sum to 95.35% because 4.65% of the ROI was not assigned a genotype in any method.
Unassigned genotypes specific to each method may have resulted from limitations of probe design, low depth of coverage (due to capture and/or sequencing issues), or a combination of these problems. We analyzed the GC content of the bases that were targeted by the capture probes, but where no genotypes were assigned. The GC content of unassigned SHS bases was 67%, MIP was 61%, and MGS was 58%, compared to 50.5% for the overall ROI. Thus, for all methods, genotypes were not assigned for a similar set of targeted bases, and these bases were slightly biased toward a high GC content.
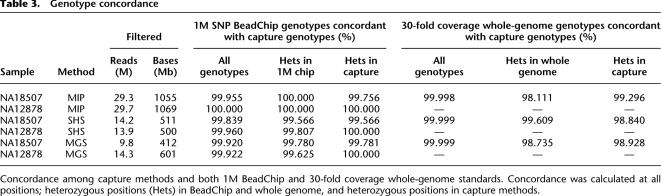
Genotype concordance
One of the most critical metrics we assessed was accuracy of the assigned genotypes. Since there is no “gold standard” data set of known genotypes at every genomic position for a sample that we sequenced, we evaluated accuracy by measuring concordance of our genotype assignments with two different data sets. The first comparison data set was genotypes derived from analysis with the Infinium 1M SNP BeadChip (1M chip). The second was genotypes derived by aligning the 30-fold coverage whole-genome shotgun (WG) sequence data generated from HapMap sample NA18507 (Bentley et al. 2008). We used ELAND and called genotypes using MPG at the same thresholds as those used with our capture data. In each case, we compared genotype assignments at ROI bases where the assessed quality of the data from the capture method and the comparison data set was high.
For the comparison with the 1M chip data, we compared genotypes for the two samples: NA18507 and NA12878. Each of the three capture methods achieved a genotype concordance of >99.84% (Table 3). Similar concordances were seen at heterozygous sites, which are notably more challenging to assign than homozygous sites; specifically, the concordance rate at positions found to be heterozygous with the 1M chip was >99.57% for all methods (Table 3). Thus genotype assignments from targeted capture sequence data are highly accurate, even at variant positions.
Table 3.
Genotype concordance
Since only about 2600 genotyped bases from the 1M chip data were represented in our ROI, we also compared the genotypes from the various capture methods to those derived from the WG NA18507 data, which had genotype assignments for 67.3% (1.76/2.61 Mb) of the ROI bases. These WG data provided a larger basis for comparison, including bases not known to be polymorphic. All three methods yielded genotypes with >99.998% concordance with the WG genotypes. When considering only the heterozygous sites in the WG data, the concordance was lower than it was for all sites: 98.111% for MIP, 98.735% for MGS, and 99.609% for SHS.
To assess the false-positive rate, we compared bases found to be heterozygous in the three capture methods with 1M chip and WG data (Table 3). Depending on the method, 99.57%–99.78% of the heterozygous genotypes derived from capture data agreed with the 1M chip genotypes for NA18507 (one to two discordant positions in each method), while 100% concordant genotypes were seen for NA12878 over the 400–500 positions compared. Further comparison with the more comprehensive WG data for NA18507 showed slightly lower concordance: 98.840% for SHS, 98.928% for MGS, and 99.296% for MIP over the approximately 1000 positions compared. There were six to 12 discordant positions in each method when compared to WG data. Thus, the false-positive rate was low (<1.2%) for all three methods. We conclude that all three capture methods, when coupled with adequate sequence depth of coverage, yield highly accurate genotype calls.
Alignment artifacts affecting genotype assignments
The data presented here were generated using standard ELAND alignments. One significant limitation of the versions of ELAND we used (v1.3.4 and v1.4.0) is that it is a gapless aligner; it does not consider insertions or deletions. Thus, not only are deletion/insertion variants (DIVs) not detected, but errors can be also introduced at these sites. When a sequence read spans a DIV, every base after the start of the DIV can appear as a mismatch. When this occurs within the first 32 bases of a sequence read, it will fail to align due to the high number of mismatches. However, when this occurs beyond the first 32 bases, ELAND will align the read and note the position(s) of the mismatch(es). These mismatches then appear to the genotype-assigning software as closely clustered nonreference variants (in reality, false-positives), not as DIVs.
To detect DIVs, we used a gapped aligner, cross_match (http://www.phrap.org), to align those reads not aligned by ELAND. We then assigned genotypes using both the ELAND and cross_match alignments with MPG. We observed small DIVs (up to 4 bp) using data generated by all three capture methods; a subset of these were 100% validated by Sanger-based sequencing (MIP: 33/33, SHS: 37/37, and MGS: 25/25). It is unclear what was limiting the length of DIV detection. Because cross_match requires aligned sequence on each side of a gap, the short read lengths were likely a factor and could be mitigated by using sequence reads longer than 36 bases. Although we are still refining these methods, we show here that all three of these capture methods can be used to detect small DIVs.
Sequencing requirements
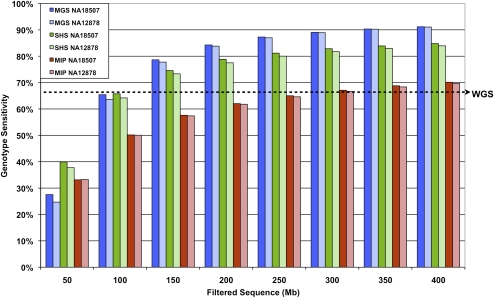
A critical measure of the effectiveness of a genomic enrichment method is the amount of sequencing required to achieve the desired endpoints. Such considerations include the proportion of genotypes accurately assigned in the ROI, the limits of this sensitivity, and the associated costs. We normalized the data to assess the effects of varying the amount of generated sequence data for NA18507 and NA12878 from each of the methods (Fig. 5). For all methods, we observed an initial sharp rise in sensitivity with increasing amounts of filtered sequence data. This was followed by a plateau, during which very little gain was achieved compared to the amount of additional data. The MIP method reached ∼70% coverage with 400 Mb of filtered sequence. For the same amount of sequence, the SHS method attained higher sensitivities of 84%–85%, while the MGS method performed even better with sensitivities of ∼91%. However, if the desire is to pool samples for capture with the MGS method, one must take into account the twofold inter-sample variability in the amount of sequence obtained for individual samples in the pool (Table 1). Thus, we expect the sensitivities of the extended set of 12 samples in a pool will not all be 91%. For example, assuming a normal distribution of the number of sequence reads assigned to each indexed sample, with variance equal to the observed data, we estimate that 11/12 individuals within the pool would have sensitivities ranging from 88% to 93% (when aiming for a per-sample average of 400 Mb of filtered sequence data). To put this filtered sequence requirement into context, our recent experience with the Illumina platform indicates that ∼85% of the generated sequence reads pass the quality filter. Assuming this rate holds true for other experiments, 400 Mb of filtered sequence is roughly equivalent to 470 Mb of raw sequence.
Figure 5.
Genotype sensitivity with increasing sequence data. Percentage of genotypes assigned in the ROI (same for all methods) with increasing filtered sequence data for NA18507 and NA12878. Sequence counts are based on 36 bases per read for MIP and SHS. To account for the 6-base index bar code, 42 bases were used in the sequence count calculations for MGS. The dashed arrow indicates genotype sensitivity level (67.3%) of 30-fold coverage whole-genome shotgun (WG) data for the ROI analyzed in this study.
The determination of adequacy of sequencing will depend on each project and how much one is willing to spend to incrementally improve the sensitivity. However, generating 470 Mb of raw sequence is, as of this writing, achievable with one lane of a single-end 36-base run on the Illumina GAII instrument; thus, the sensitivities compared here are appropriate benchmarks for each method. To evaluate this further, the WG NA18507 data provided genotype coverage for 67.3% of the bases in our ROI. This level of sensitivity was met by all methods with relatively little sequence data (MIP: ∼300 Mb, SHS: ∼110 Mb, and MGS: ∼100 Mb), suggesting a substantial improvement over that provided by early whole-genome sequencing that provided 30-fold average coverage depth.
Discussion
The ever-increasing throughput of DNA sequencing instruments makes feasible the interrogation of the entire genome of individual human research subjects. However, whole-genome shotgun sequencing is not the most appropriate option for all projects. The ability to enrich for genomic regions of interest (e.g., coding exons or regions found by genetic association studies to harbor disease-causing variants) in humans or any other organism at low cost and high efficiency is needed for many experiments. The majority of available genomic enrichment strategies involve liquid- or solid-phase capture, dependent on the hybridization of synthetic probes to complementary target genomic sequences. We evaluated three of these methods with respect to their effectiveness and operational attributes.
An important metric for these methods is the genotype sensitivity represented by the percentage of ROI bases covered by high-quality genotype assignments. All three methods were highly sensitive, with MGS being the most sensitive, followed closely by SHS and then MIP. The sensitivity across the extended sample sets was highly reproducible for SHS and MGS, with the latter having slightly more variation due to differing amounts of sequence data obtained for each sample (within the pool of indexed samples). We then investigated some of the factors that may have accounted for this variability in sensitivity.
Initially, we compared factors at the early steps of the process such as the breadth of coverage of the designed probes across the ROI and the fraction of the quality filtered reads that aligned to the ROI. Despite the fact that the probe-design parameters were different among the three methods, there were no significant differences in the amounts of the ROI covered by each design method. Although MIP had slightly higher design coverage, its overall genotype sensitivity was lower than the other methods. Additionally, the fraction of reads aligning to the ROI was similar among the three methods. Thus, neither of these two factors was a major determinant of the differences in genotype sensitivity.
The fraction of reads aligning to the ROI does not take into account the evenness of enrichment across the targeted regions. We next examined uniformity in the depth of coverage for each method and found this parameter to be the major contributing factor for the variation in genotype assignment sensitivity. The MGS method yielded the most uniform depth of coverage, followed by SHS and then MIP (which was significantly less uniform than the other methods). As uniformity decreases, certain positions have a much higher depth of coverage than is necessary for accurate genotype assignment, while other positions lack coverage and are thus more prone to have absent or incorrect assignments.
Although all methods used different approaches, they captured and assigned genotypes to largely the same positions. This suggests that all three methods have difficulty designing for or enriching for the same regions, or that the loss of these regions occurred during a downstream process common to all the methods (i.e., during sequencing). The implication of these results is that the potential improvement in genotype sensitivity by using two capture methods may not be sufficient to justify the increased cost of implementing multiple methodologies.
A logical follow-up question is whether one can define the characteristics of genomic regions for which genotype assignments are not made. We found that regions lacking genotype assignments were a result of probe design limitations in some instances, while others were due to the capture method and/or sequencing limitations. All probes were designed to avoid repetitive sequence, as these regions are challenging to capture and the resulting reads are difficult to uniquely align even if captured. The ROI bases with poor depth of coverage (this excludes ROI bases not in the probe design) tended to have a higher GC content than those that were sufficiently covered. Of the bases with poor depth of coverage by MGS and SHS methodologies, SHS had a slightly higher percentage of GC bases, suggesting that high GC segments may more adversely affect SHS. Although GC extremes are known to be problematic for polymerases and in hybridization reactions, the Illumina GA also sequences these regions less efficiently (Bentley et al. 2008). We cannot distinguish at which step GC content is preventing sequence recovery, but the effect reduces the sensitivities of these methods and suggests that high-GC regions are problematic for all such methods.
Targeted sequencing is an invaluable tool for variant discovery, and as such, we consider the accuracy of the genotype assignments to be a critical metric. All three of the genomic enrichment methods generated data that agreed well with known genotypes: >99.84% and >99.998% when compared to 1M chip and WG genotypes, respectively. Additionally, of those heterozygous positions identified by these methods, >99.57% and >98.840% agreed with the 1M chip and WG data, respectively, indicating low false-positive rates in all cases.
In addition to substitutions, we also detected small insertion/deletion variants (up to 4 bases) in the captured DNA from each of the three methods, suggesting that such variants are amenable to both polymerase and hybridization-based genomic enrichment. When gapped alignments are available, our genotype-assigning algorithm, MPG, can apply Bayes Theorem to determine insertion/deletion. We used Sanger sequencing to validate insertions/deletions and confirmed all those tested.
One of the most important variables in a genomic enrichment experiment is the amount of sequence needed to achieve a given genotype sensitivity level. By assigning genotypes with varying subsets of random reads, we have shown that for all three methods, there is a point at which additional sequence offers diminishing increases in genotype sensitivity. MGS reached this plateau earlier than the other two methods, presumably due to a more uniform depth of coverage. The amount of sequence required for a given project depends on many variables, but this analysis should be useful in finding the ideal balance between cost and sensitivity. Our data suggest that SHS and MGS provide sensitivity levels in the range of 85%–93% for a 2.61-Mb ROI, with sequence data obtained from the equivalent of one lane of a single-end 36-base run on an Illumina GAII. We used 36-base reads for comparison of the methods in this study as that was the standard read length for the Illumina platform at the time the data was generated. With the longer reads achievable with current technology, this amount of sequence will be in excess of that required for coverage of a 2.61 Mb ROI. More data would be available, for coverage and genotype assignment. However, alignment artifacts resulting from undetected insertion/deletions may increase false-positive rates.
We also analyzed the published WG reads of HapMap individual NA18507 (Bentley et al. 2008), aligned these reads to the genome, and assigned genotypes using the same algorithm as for the enriched samples. Interestingly, the genotype sensitivity of this early whole-genome sequence for our regions of interest was only 67.3%. Although efficiency and data analyses of high-throughput sequencing have improved considerably since the WG sequence was generated, this offers a compelling reason to use genomic enrichment instead of sequencing with whole-genome shotgun approaches. Even as sequencing costs decrease, the amount of whole-genome sequence required to cover a specific region at the sensitivities we have observed may continue to make genome enrichment the preferred strategy for sequencing a subset of the genome.
The total cost (reagents, time, and effort) of a genomic enrichment and sequencing protocol and the number of samples involved must be considered when designing specific experiments. While this study did not include formal cost analysis, some general and relative conclusions can be drawn. Our development and application of a pooled capture strategy for MGS has reduced the cost of not only the genomic enrichment step (array cost and processing), but also the preparation of libraries and sequencing requirements. Methods are available for indexing DNA libraries for pooled sequencing, but methods are not well established for indexing libraries for pooling prior to both enrichment and sequencing. The ability to pool samples in the early stages of library preparation (i.e., immediately after indexed adapter ligation) condenses the majority of the effort for library construction by over a factor of 10. Sequencing of a pool of bar-coded samples is also more cost-efficient than individual sample sequencing as the amount of sequence obtained per lane increases; sequencing only one individual in one lane will provide excessive data for a 2-Mb to 3-Mb ROI. The extra sequencing required for the 6-base index in each read should not be a deterrent, as the indexed MGS samples had the highest sensitivity even when having a lower amount of quality filtered sequence than the other two methods. Although the MIP capture reagents are inexpensive compared to the other methods, it requires the longest processing time, and the separation of MIP probes into two nonoverlapping sets doubles the costs of library preparation and sequencing. At the time of writing, the cost of sequencing is still the significant portion of the total cost of an enrichment-sequencing experiment. SHS was the fastest method, and it did not require specialized equipment. Additionally, the SHS protocol should be amenable to robotic manipulation, theoretically offering a significant increase in throughput. Although we have applied our indexing protocol only in the context of MGS, we believe it can be easily adapted to the solution-based methods. The rapid pace at which sequencing technologies are increasing throughput makes indexing or bar-coding DNA libraries highly relevant for optimizing efficiencies for both targeted enrichment and sequencing.
In this study, we directly compared three methods for genomic enrichment in a rigorous manner. We found MGS to be the most sensitive, with the introduction of sample pooling prior to the capture process reducing its cost further. Single arrays can be ordered, but reagent price scaling, additional equipment cost, and array handling make this method more appropriate for medium to large projects. SHS approaches the sensitivity of MGS and requires no specialized equipment. Reagent price scaling is based on a minimum sample size of 10, making SHS appropriate for both medium and large projects and less so for small projects. Furthermore, the simplicity of SHS benefits large and very large projects. For MIP, reagent cost is low, and it may therefore be appropriate for small projects or for medium projects when sequencing costs are negligible, but decreased sensitivity due to uneven depth of coverage needs to be considered. We conclude that there is no one best method as each has inherent strengths and weaknesses. Researchers have the option to choose among several effective methods and select the one most suitable for their project goals and budgets.
As whole-genome sequencing costs continue to fall, it would appear that the lifetime of genome enrichment methods might be limited. While this may be true in the very long term, these methods will continue to be useful for an extended period of time. Even as whole-genome sequencing becomes less costly, the burden of storing and analyzing large data sets remains high. Thus, one will likely be able to perform targeted sequencing on many more samples with a given amount of resources, allowing greater experimental breadth and power. By focusing on the desired regions, targeted sequencing methods can more effectively achieve higher sensitivity with much less sequence and will remain a valuable strategy for detailed interrogation of genomic regions.
Methods
Targeted genomic regions
We targeted 2.61 Mb of noncontiguous sequence, comprised of the exons and flanking regions from 528 genes. These genes were selected because they were associated with type 2 diabetes (T2D) or related quantitative traits that were either identified from several meta-analyses of genome-wide association studies for T2D or related quantitative traits (318 genes), or were included in the NHGRI ClinSeq Study, a pilot large-scale sequencing effort in a clinical research setting, of cardiovascular disease associated loci (279 genes) (Biesecker et al. 2009). Some genes were in both sets, giving a list of 528 unique genes. More specifically, the gene regions targeted were exons from the UCSC Known Genes and RefSeq Genes tracks (hg18), 2-kb regions centered on the transcriptional start site, and conserved regions according to the UCSC phastConsElements28wayPlacMammal track that occurred within 10 kb upstream of the TSS or in the 3′-UTR.
DNA samples
DNA or cell lines from HapMap individuals were obtained from Coriell Cell Repositories. Finnish DNA samples were collected as part of the Finland–United States Investigation of NIDDM Genetics Study (Valle et al. 1998). ClinSeq DNA samples were collected and consented as described in Biesecker et al. (2009). The collection and use of these samples were approved by the appropriate IRB and/or ethics committees.
Method 1: MIP
Primer design
MIP probes were designed using PrimerTile (Chines et al. 2005), a program for automated large-scale design and in silico testing of PCR primer pairs. This program achieves high target coverage by tiling primer pairs across a given region. This software was adapted for MIP capture by optimizing various design parameters. We used the following parameters to design 14,545 probes, which covered 95.9% of the ROI bases: (1) Probe targeting arms were designed to capture regions between 80 and 360 bases long. (2) GC content of probe targeting arms was between 30% and 60% (most between 35% and 55%). (3) Probe targeting arms did not overlap known dbSNP130 positions. (4) Probe targeting arms did not include Nt.AlwI or Nb.BsrDI restriction sites. We performed multiple iterations of probe design to optimize coverage. Redundant targeting pairs were removed and the remaining targeting arm pairs were oriented into a 100-mer probe as described in Porreca et al. (2007). The probes were separated into two nonoverlapping sets and ordered separately as either Probe Library Synthesis (Agilent Technologies) or OligoMix (LC Sciences). Subsequent captures and sequencing were performed separately for each tube of probes. Sequence data for each tube were filtered to only include positions that were captured in that set, and combined for genotype calling.
Probe preparation
The MIP probes were prepared as described in Porreca et al. (2007) with the following modifications. Ten fmol of synthesized probe library was amplified in a 1-mL PCR reaction (split to 20 tubes) using 10 μL of Amplitaq Gold (Applied Biosystems), 100 μM each primer, and 200 μM each dNTP. Cycling conditions were 5 min at 95°C, 20 cycles of 30 sec at 95°C, 1 min at 54°C, 2 min at 72°C, and finally, 5 min at 72°C.
The PCR products were purified by phenol/chloroform extraction and ethanol precipitation with 1/10th vol of 7.5 M ammonium acetate. The precipitate was resuspended in 85 μL of Elution Buffer (QIAGEN). The purified product was digested with restriction enzymes as described in Porreca et al. (2007), but with a 2-h incubation period. The digested product was again purified by phenol/chloroform extraction and ethanol precipitation. The 70-bp digested product was then size-selected on a 6% denaturing acrylamide gel. Processed, size-selected probes were finally quantitated by running an aliquot against known amounts of similarly sized DNA fragments on a denaturing acrylamide gel.
Capture
Methods were adapted from Porreca et al. (2007), including improvements from Li et al. (2009) and Turner et al. (2009a). For each tube of probes, 500 ng of genomic DNA was combined with capture probes (2.9 pM each probe) in 13 μL of 1× Ampligase buffer (Epicentre Biotechnologies). The reactions were incubated for 4 min at 20°C, 5 min at 95°C, and 36 h at 60°C. One microliter of extension and ligation mix was added (8.75 μM dNTPs, 2 U/μL Taq Stoffel fragment [Applied Biosystems, Inc.], and 5 U/μL Ampligase in 1× Ampligase buffer). The reaction was allowed to proceed for 48 h. Linear genomic DNA was degraded with ExoI (40 U) and ExoIII (200 U) (NEB) for 1 h at 37°C.
Shotgun library preparation
We amplified using normal PCR (instead of rolling circle amplification) with PCR primers CP-2-FA and CP-2-RA (Porreca et al. 2007). The PCR was carried out as follows: 4 μL of capture template, 1× Herculase Reaction Buffer, 0.25 mM each dNTP, 10 μM each primer, and 1 μL of Herculase II Fusion DNA Polymerase (Agilent Technologies, Stratagene Products). Reactions were split into two separate tubes and incubated for 2 min at 98°C, 18 cycles of 20 sec at 98°C, 20 sec at 58°C, and 30 sec at 72°C. Samples were then end-repaired using the End-It End Repair Kit (Epicentre Biotechnologies). Finally, samples were concatenated overnight at 16°C using 2000 U of T4 DNA/Ligase (New England Biolabs), 18% PEG-3350 (Sigma-Aldrich) in 1× T4 ligase buffer. After confirmation of efficient concatenation, samples were prepared for shotgun sequencing on the Illumina GA-2 platform with the Single-End Library Preparation kit (Illumina), using sonication (Covaris) to fragment the samples. Samples were purified using AMPure beads (Beckman Coulter Genomics).
Method 2: SHS
Probe design
Probes were designed using the eArray website and ordered as part of a SureSelect Target Enrichment System kit (Agilent Technologies, Inc.). Default settings were used (120-bp probes, with 60-bp overlap), except that we increased the allowable overlap with the RepeatMasked regions from 20 to 60 bases. The initial targets were “padded” as described below for MGS. The design resulted in 46,869 baits, which covered 92.6% of the ROI bases.
Library preparation and capture
Library preparation and capture were performed as described in the SureSelect Target Enrichment System Protocol, version 1.2 (Agilent Technologies, Inc., April 2009) with several minor modifications. Briefly, 3 μg of sample was sheared with sonication (Covaris), end-repaired, and ligated to Single-End sequencing adapters (Illumina) as described in the SureSelect protocol. The sample was gel-purified and size-selected to 200–300 bp from a 10-cm 4% NuSieve (Lonza) agarose gel (13 V, 17 h) and subsequently amplified. The amplified library was purified on MinElute columns (QIAGEN) and eluted in 10 μL of Buffer EB instead of 50 μL. Five hundred nanograms of this library was mixed with the SureSelect hybridization buffer and denatured for 5 min at 95°C. The biotinylated RNA baits (SureSelect kit) were added, and the reaction was incubated for 24 h at 65°C. The hybridized DNA–bait duplexes were captured on Dynabeads M-280 streptavidin beads (Life Technologies), washed several times with SureSelect wash buffers, and eluted. Following a desalting purification, the sample was amplified using Herculase II Fusion DNA polymerase (Agilent Technologies, Stratagene Products; 18 cycles), purified using AMPure beads (Beckman Coulter Genomics), and quantified, and was then ready for cluster generation.
Method 3: MGS
MGS array design
Overlapping 58- to 104-mer probes were designed (Roche NimbleGen Inc.; probe design version 1.0_2008_06_04) to tile across each region of interest (ROI) segment. Within each tiled region, each successive probe is spaced every 5–11 bases along the genome. To potentially increase the efficiency of hybrid capture, we enlarged small segments to be at least 200 bp in length and merged segments within 200 bp of each other, leading to an ROI-plus target of 3.21 Mb. The final completed array included 3.01 Mb of the ROI-plus target, of which 2.450 Mb covered 93.8% of the 2.61 Mb of ROI bases targeted for the 528 genes.
DNA samples and DNA oligonucleotides
DNA oligonucleotides for adapters, PCR or blocking in hybridization reactions, were purchased from Integrated DNA Technologies.
Bar-coding indexes
We designed six-base indexes to tag DNA from different source individuals. Index base combinations were selected such that each was different from the others at three or more of the five bases that are allowed to vary (Supplemental Table 1). The sixth and last (3′) base is an invariant “T” to allow ligation to “A”-tailed genomic fragments. This strategy was adopted to allow for an incorrect genotype call at one of the index bases, while maintaining a high probability of assigning the read to the correct source individual. Furthermore, we selected index combinations to have balanced representation of each of the four nucleotide types in the first few base positions of the read. This was done to allow Illumina's automated calibration process to calculate appropriate matrix and phasing parameters.
Adapter preparation
Adapters were prepared as described by Craig et al. (2008) but with minor modifications. Briefly, each lyophilized indexed adapter oligonucleotide was resuspended to 100 μM in 10 mM Tris-HCL (pH 8.0). A 50-μL volume mixture consisting of 7.5 μL of each adapter oligonucleotide, 5 μL of 10× annealing buffer (100 mM Tris at pH 8.0, 0.5 M NaCl) and 30 μL of sterile water, was incubated in a thermocycler under the following conditions: 95°C for 5 min, then a 1°C stepdown per minute to cool to a final temperature of 25°C. The resulting 15 μM adapter mix was used as stock in the adapter ligation reaction for Illumina library construction.
Illumina library construction
Paired-end (PE) libraries for sequencing on a GA II sequencer were constructed according to a protocol adapted from the Genome Analyzer Paired End libraries generation kit (Illumina). Slight modifications of the manufacturer's protocol were incorporated to accommodate an indexing strategy. Briefly, 12 PE libraries were made for six HapMap (two CEU, two YRI, and two JPT) and six Finnish individuals. For each library, 4 μg of genomic DNA from one individual was sheared for 65 sec on a Covaris S2 with the following parameters: duty cycle of 5, intensity of 4, and 200 cycles per burst. The resulting fragments (100–700 bp; median ∼250 bp) were quantified on the Model 2100 Bioanalyzer (Agilent Technologies) by comparing with a similarly sheared DNA of known concentration (heretoforth referred to as the “DNA quantification standard”). This normalized DNA concentration was used to calculate 2 μg of each sheared DNA sample for end repair, A-tailing, and adapter ligation as per the PE library protocol, but with an extension to the adapter sequence as described by Craig et al. (2008). A 6-base index was added to one end of each PE adapter oligonucleotide (5′Phos-XXXXXXGATCGGAAGAGCGGTTCAGCAGGAATGCCGAG-3′ and 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTXXXXX*T-3′; X denotes index/complement base and * denotes phosphorothioate bond) such that the annealed adapter complex included a 6-bp index bar code (Supplemental Table 1) on each end of the genomic fragment. After purifying the ligated products with the QIAquick PCR purification kit (QIAGEN) and quantifying with the 2100 Bioanalyzer, equal proportions equivalent to ∼0.5 μg of each sample were pooled (giving a total of 6 μg of DNA) and run for 110 min in two lanes of a 2.0% TAE agarose minigel at 70 V. Library fragments corresponding to ∼250–350 bp were extracted, purified with the QIAGEN gel extraction kit, and quantified with the Nanodrop 1000 spectrophotometer (Nanodrop Products). The library pool was enriched for adapter-modified products with a 12-cycle amplification performed according to the Illumina PE protocol (and with universal Illumina PCR primer PE 1.0 and 2.0), but in six 50-μL replicate tubes and 35 ng of input template per tube. The amplified library pool was purified with 0.8 volumes of Ampure XP Beads according to the manufacturer's instructions (Beckman Coulter Genomics) and then quantified relative to the DNA quantification standard on the 2100 Bioanalyzer.
Hybrid capture and post-capture processing
Hybrid capture of the library pool on the MGS array was performed as per the Sequence Capture protocol (Roche NimbleGen Inc.), but with minor modifications. Briefly, a 4.8-μL solution containing 100 μg of Cot-1 DNA, 4 μg of amplified library pool DNA, and 100× molar excess of each universal PCR primer PE 1.0 and 2.0 (used as blocking oligonucleotides to reduce nonspecific hybridization resulting from annealing of only the PCR primer ends from noncomplementary target sequences) was added to 8 μL of 2× SC hybridization buffer and 3.2 μL of SC hybridization buffer Component A. The resulting 16-μL mixture was heated for 10 min at 95°C, equilibrated to 42°C, and loaded onto a custom 385K array (Roche NimbleGen Inc.). Following a 68-h hybridization period at 42°C with mixing in a MAUI Hybridization System chamber, the array was washed and the captured DNA was eluted with 125 mM NaOH, purified with a MinElute column (QIAGEN), and eluted in 25 μL of EB buffer. The captured DNA was PCR-amplified in five 50-μL replicate tubes with 3 μL of DNA template per tube. Amplification was performed as described in the PE sequencing protocol (Illumina Inc.), but for only 10 cycles. The amplified product was purified with 0.8 volumes of Ampure XP beads according to the manufacturer's instructions, re-suspended in a final volume of 30 μL of sterile water, and prepared for sequencing on the GA II sequencer.
Sequencing
Sequencing was performed as described by the manufacturer's protocols (Illumina Inc.). Samples were quantified for cluster generation using quantitative-PCR to improve cluster number targeting. These samples were sequenced using Version 2 cluster kits and Version 3 sequencing kits.
Alignment of sequence reads and genotype calling
Reads were aligned to the hg18 human reference genome using ELAND as part of the standard Pipeline Analysis v1.3.4 and v1.4.0 (Illumina Inc.). These versions of ELAND use a maximum of 32 bases of each read for alignment, and this maximum 32-base alignment was used for all three methods. Reads that failed the manufacturer's chastity filter were omitted. Sequence alignments and quality scores were passed to the MPG (Most Probable Genotype) program to call genotypes for every reference position at which there are aligned sequence reads. To be included, a read base must have a minimum _phred_-style base quality score of 20. The MPG algorithm is built on a Bayesian model that simulates sampling from one or two chromosomes with sequencing error and then calculates the likelihood of each possible genotype given the observed sequence data. Genotype predictions need an MPG score of at least 10 to be considered accurate.
See Supplemental Methods for further details.
Acknowledgments
This study was supported by the Intramural Research Program of the National Human Genome Research Institute. We express our gratitude to Darryl Leja for his assistance with figure preparation. We thank Agilent Technologies for providing SureSelect probes and Illumina Inc. for genotyping data from the Infinium Human 1Mv1.0 BeadChip. We also thank Emily Turner and Jay Shendure at the University of Washington and Sheila Fisher and Chad Nussbaum at the Broad Institute for ongoing discussions relating to MIP and SHS, respectively. We also thank several individuals for their help with MGS: Kari Kubalanza for technical assistance with the pilot study, Narisu Narisu for help with interim analyses, and Mike Erdos for ongoing discussions.
Footnotes
References
- Albert TJ, Molla MN, Muzny DM, Nazareth L, Wheeler D, Song X, Richmond TA, Middle CM, Rodesch MJ, Packard CJ, et al. 2007. Direct selection of human genomic loci by microarray hybridization. Nat Methods 4: 903–905 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE 2001. Segmental duplications: Organization and impact within the current human genome project assembly. Genome Res 11: 1005–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bau S, Schracke N, Kranzle M, Wu H, Stahler PF, Hoheisel JD, Beier M, Summerer D 2009. Targeted next-generation sequencing by specific capture of multiple genomic loci using low-volume microfluidic DNA arrays. Anal Bioanal Chem 393: 171–175 [DOI] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesecker LG, Mullikin JC, Facio FM, Turner C, Cherukuri PF, Blakesley RW, Bouffard GG, Chines PS, Cruz P, Hansen NF, et al. 2009. The ClinSeq Project: Piloting large-scale genome sequencing for research in genomic medicine. Genome Res 19: 1665–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chines PS, Swift AJ, Bonnycastle LL, Erdos MR, Mullikin JC, Collins FC 2005. PrimerTile: Designing overlapping PCR primers for resequencing. In American Society of Human Genetics Annual Meeting, p. A1257 Salt Lake City, UT [Google Scholar]
- Craig DW, Pearson JV, Szelinger S, Sekar A, Redman M, Corneveaux JJ, Pawlowski TL, Laub T, Nunn G, Stephan DA, et al. 2008. Identification of genetic variants using bar-coded multiplexed sequencing. Nat Methods 5: 887–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K 2008. Fixing the front end. Nat Biotechnol 26: 1101–1104 [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, et al. 2009. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol 27: 182–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman DS, Hovingh GK, Iartchouk O, Rehm HL, Kucherlapati R, Seidman JG, Seidman CE 2009. Filter-based hybridization capture of subgenomes enables resequencing and copy-number detection. Nat Methods 6: 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges E, Xuan Z, Balija V, Kramer M, Molla MN, Smith SW, Middle CM, Rodesch MJ, Albert TJ, Hannon GJ, et al. 2007. Genome-wide in situ exon capture for selective resequencing. Nat Genet 39: 1522–1527 [DOI] [PubMed] [Google Scholar]
- Hodges E, Rooks M, Xuan Z, Bhattacharjee A, Benjamin Gordon D, Brizuela L, Richard McCombie W, Hannon GJ 2009. Hybrid selection of discrete genomic intervals on custom-designed microarrays for massively parallel sequencing. Nat Protoc 4: 960–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar S, Zheng J, Wilhelmy J, Faham M, Mindrinos M, Davis R 2008. A comprehensive assay for targeted multiplex amplification of human DNA sequences. Proc Natl Acad Sci 105: 9296–9301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Gao Y, Aach J, Zhang K, Kryukov GV, Xie B, Ahlford A, Yoon JK, Rosenbaum AM, Zaranek AW, et al. 2009. Multiplex padlock targeted sequencing reveals human hypermutable CpG variations. Genome Res 19: 1606–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ 2010. Target-enrichment strategies for next-generation sequencing. Nat Methods 7: 111–118 [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okou DT, Steinberg KM, Middle C, Cutler DJ, Albert TJ, Zwick ME 2007. Microarray-based genomic selection for high-throughput resequencing. Nat Methods 4: 907–909 [DOI] [PubMed] [Google Scholar]
- Porreca GJ, Zhang K, Li JB, Xie B, Austin D, Vassallo SL, LeProust EM, Peck BJ, Emig CJ, Dahl F, et al. 2007. Multiplex amplification of large sets of human exons. Nat Methods 4: 931–936 [DOI] [PubMed] [Google Scholar]
- Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM 2005. Accurate multiplex polony sequencing of an evolved bacterial genome. Science 309: 1728–1732 [DOI] [PubMed] [Google Scholar]
- Summerer D 2009. Enabling technologies of genomic-scale sequence enrichment for targeted high-throughput sequencing. Genomics 94: 363–368 [DOI] [PubMed] [Google Scholar]
- Summerer D, Wu H, Haase B, Cheng Y, Schracke N, Stahler CF, Chee MS, Stahler PF, Beier M 2009. Microarray-based multicycle-enrichment of genomic subsets for targeted next-generation sequencing. Genome Res 19: 1616–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tewhey R, Warner JB, Nakano M, Libby B, Medkova M, David PH, Kotsopoulos SK, Samuels ML, Hutchison JB, Larson JW, et al. 2009. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat Biotechnol 27: 1025–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner EH, Lee C, Ng SB, Nickerson DA, Shendure J 2009a. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat Methods 6: 315–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner EH, Ng SB, Nickerson DA, Shendure J 2009b. Methods for genomic partitioning. Annu Rev Genomics Hum Genet 10: 263–284 [DOI] [PubMed] [Google Scholar]
- Valle T, Tuomilehto J, Bergman RN, Ghosh S, Hauser ER, Eriksson J, Nylund SJ, Kohtamaki K, Toivanen L, Vidgren G, et al. 1998. Mapping genes for NIDDM. Design of the Finland-United States Investigation of NIDDM Genetics (FUSION) Study. Diabetes Care 21: 949–958 [DOI] [PubMed] [Google Scholar]