Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation (original) (raw)
Abstract
Cardiac valve formation is crucial for embryonic and adult heart function. Valve malformations constitute the most common congenital cardiac defect, but little is known about the molecular mechanisms regulating valve formation and homeostasis. Here, we show that endocardial Notch1 and myocardial Bmp2 signal integration establish a valve-forming field between 2 chamber developmental domains. Patterning occurs through the activation of endocardial epithelial-to-mesenchymal transition (EMT) exclusively in prospective valve territories. Mice with constitutive endocardial Notch1 activity ectopically express Hey1 and Heyl. They also display an activated mesenchymal gene program in ventricles and a partial (noninvasive) EMT in vitro that becomes invasive upon BMP2 treatment. Snail1, TGF-β2, or Notch1 inhibition reduces BMP2-induced ventricular transformation and invasion, whereas BMP2 treatment inhibits endothelial Gsk3β, stabilizing Snail1 and promoting invasiveness. Integration of Notch and Bmp2 signals is consistent with Notch1 signaling being attenuated after myocardial Bmp2 deletion. Notch1 activation in myocardium extends Hey1 expression to nonchamber myocardium, represses Bmp2, and impairs EMT. In contrast, Notch deletion abrogates endocardial Hey gene transcription and extends Bmp2 expression to the ventricular endocardium. This embryonic Notch1-Bmp2-Snail1 relationship may be relevant in adult valve disease, in which decreased NOTCH signaling causes valve mesenchyme cell formation, fibrosis, and calcification.
Introduction
Congenital cardiac valvuloseptal defects are the most frequently diagnosed developmental malformations during the first year of human life (1), and several valvular defects manifest in adult disease (2). Evidence from animal models has linked many genes and signaling pathways to cardiac valve development (3); however, mutations of the human homologs seldom correlate with human valve defects (4). Moreover, the similar phenotypes of targeted mutant mice suggest they affect functionally interconnected signaling pathways (5).
Cardiac valve formation (CVF) in mice begins at approximately E9.5. In the developing atrioventricular canal (AVC) and outflow tract (OFT), myocardial signals induce adjacent endocardial cells (ENCs) to undergo epithelial-to-mesenchymal transition (EMT) (5). These cells invade the underlying matrix (cardiac jelly) to generate the heart valve primordia (endocardial cushions). Cardiac EMT begins with ENC activation, in which ENCs swell due to the enlargement of the Golgi and rough endoplasmic reticulum. Transformation follows, in which ENCs lose cell:cell contacts, acquire the ability to move into the plane of the monolayer, polarize their Golgi, and form migratory appendages. EMT is completed once the transformed ENCs invade the cardiac jelly to form the valvuloseptal mesenchyme (6). ENC transformation is dependent on the initiation of EMT-specific transcriptional machinery, the physical detachment of epithelial cells, and the activation and stabilization of a migratory and invasive mesenchymal phenotype.
Defining the primary signal or signals inducing cardiac EMT has been problematic due to the difficulties of extrapolating results from chick and mouse (5) and because the exact extent of spatiotemporal interaction between myocardium and endocardium cannot be fully reproduced in vitro. Data from collagen gel assays with chicken tissue suggested TGF-β2 or TGF-β3 as the main myocardial EMT inducers (7); however, _Tgfb2_- or _Tgfb3_-deficient mice show no endocardial EMT defect, suggesting induction of mammalian EMT by other signals (8). Indeed, in mice, Bmp2 is critical for EMT induction in vitro (9) and in vivo (10) and Bmp2 alone can specify a field of cardiac progenitors as a heart valve–inducing region (11).
Within the endocardium, EMT is regulated by Notch (12), an ancient local cell signaling system in which both ligands (Delta and Jagged) and receptors (Notch) are membrane bound (13). Ligand-receptor binding initiates 2 consecutive receptor cleavage steps, the last one involving γ-secretase (13), to generate the biologically active Notch intracellular domain (NICD). In the nucleus, NICD binds the RBPJK/CSL transcription factor and regulates target gene expression and thus cell fate specification (13). Notch1 activity delineates AVC and OFT endocardium in the mouse E9.5 heart (14), and the AVC of RBPJk- and _Notch1_-targeted mutants almost completely lacks mesenchymal cushion cells (12). Although ENCs from E9.5 _RBPJk_-deficient AVC show some features of activated premigratory endocardium, they remain in close association, maintain adherens junctions, and fail to invade the cardiac jelly. This correlates with reduced transcription of the cadherin repressor Snail1, which is normally expressed in AVC and OFT endocardium and mesenchyme (12). Loss of Snail1 expression thus appears to block EMT by preventing downregulation of ENC cell adhesion. RBPJk and Notch1 mutant AVC explants cultured on collagen gels fail to undergo EMT, a finding confirmed by Notch inhibition in WT mouse AVC explants and zebrafish embryos (12). Moreover, deletion of the Notch target Hey2 produces AV valve defects (15), and double Hey1;Hey2 or Hey1;Heyl mutants additionally show severe cardiac EMT defects (16, 17).
Recent data have suggested that N1ICD overexpression in zebrafish AVC endocardium inhibits EMT (18), while on the other hand, Mesp1-Cre driven N1ICD activation in the mouse cardiac lineage impairs AVC myocardial differentiation without affecting EMT (19). These apparently inconsistent data indicate that the precise role of Notch in the regulation of endocardial EMT, its downstream molecular mechanism, and relationship with other activators of this process (e.g., TGF-βs and Bmps) remain to be defined.
Using a variety of mouse genetic models, in vitro assays, and molecular studies, we show that Notch1 is sufficient to activate a cell-autonomous, promesenchymal gene expression program in ENCs. Constitutive Notch1 activity in endocardium enables ectopic, noninvasive EMT of ventricular ENCs, conferring “valvular” features to an otherwise “nonvalvular,” EMT-refractory ventricular endocardium. BMP2 treatment of these ventricular ENCs drives EMT to completion by promoting Notch- and Snail1-dependent invasion. Ectopic myocardial Notch1 expression and loss-of-function experiments indicate that Notch1 represses Bmp2 in cardiac cells by Hey target activation. Bmp2 inactivation in the myocardium impairs Notch1 activity, suggesting a functional link between these 2 signaling pathways. Snail1 expression induced by Notch1 and Bmp2 and Snail1 protein stabilization mediated by Bmp2 via Gsk3β inhibition suggest that Notch1 and Bmp2 signals converge to promote ENC invasiveness. We propose a model in which the interplay between myocardial Bmp2 and endocardial Notch1 restricts EMT to prospective valve territory. In this setting, the prevalvular endocardium is not a passive tissue, simply responding to inductive myocardial signals, but plays an active role in promoting EMT.
Results
Endocardial Notch1 activation causes heart dysmorphogenesis and ectopic target gene expression.
To investigate whether ectopic Notch1 activation in the heart was sufficient to drive EMT, we activated Notch1 in the endocardium, myocardium, or both tissues. We used the conditional N1ICD transgenic line R26N1ICD, which upon Cre activation expresses the Notch1 intracellular domain (amino acids 1749–2293) and nuclear-localized enhanced GFP (20). R26N1ICD mice (from now on, N1ICD) were crossed with the endothelium/endocardium-specific Tie2-Cre driver line (ref. 21 and Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI42666DS1). At E9.5, Tie2-Cre;N1ICD embryos were smaller than WT littermates and showed pericardial distension and enlarged LV and atrium (Figure 1A). EGFP expression delineated the endocardium, indicating N1ICD expression (Figure 1B). Scanning electron microscopy revealed AVC mesenchymal cells invading the cushion region in E9.5 WT embryos (Figure 1, C and E). Transgenic littermates had relatively fewer mesenchymal cells in AVC cushion tissue (Figure 1, D and F), probably due to a slight developmental delay. H&E-stained sections were consistent with these findings (Supplemental Figure 1, B–E) and revealed poorly formed trabeculae in E9.5 transgenic embryos (Supplemental Figure 1, F and G).
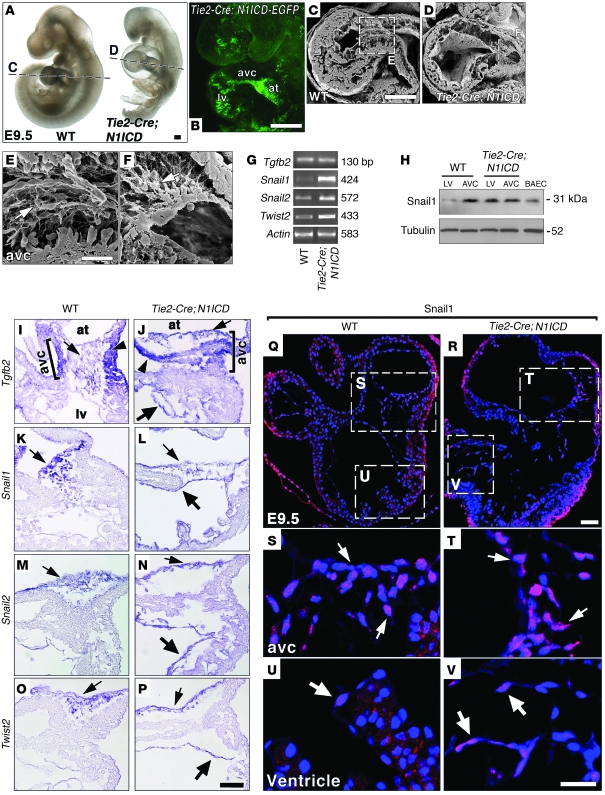
Figure 1. E9.5 Tie2-Cre;N1ICD mice ectopically express mesenchyme genes in chamber endocardium.
(A) WT and Tie2-Cre;N1ICD embryos. Dotted lines indicate C and D section planes. (B) Endocardial EGFP expression in a Tie2-Cre;N1ICD embryo. at, atrium. (C and D) SEM images of longitudinal WT (C) and Tie2-Cre;N1ICD (D) heart sections. (E and F) Details of AVC in WT (E) and Tie2-Cre;N1ICD hearts (F). Arrows, mesenchymal cells. (G) Semiquantitative RT-PCR analysis in hearts. (H) Snail1 Western blot. (I–P) WISH, heart details. Arrowheads, AVC myocardium; thin arrows, AVC endocardium; thick arrows, ventricular endocardium. (I) Tgfb2 expression in WT AVC myocardium and endocardium (bracket). (J) Normal Tgfb2 expression in AVC and ectopic expression in ventricular endocardium of Tie2-Cre;N1ICD embryos. (K and L) WT mice express Snail1 in AVC endocardium and mesenchyme (K); Tie2-Cre;N1ICD embryos show ectopic expression in ventricular endocardium (L). Tie2-Cre;N1ICD hearts also show ectopic ventricular expression of Snail2 (N) and Twist2 (P). (Q–V) Snail1 expression (red) in E9.5 heart. Nuclei are DAPI counterstained (blue). (Q) General view of an E9.5 WT heart. (S) Detail of AVC region. Arrows, nuclear Snail1 in endocardial and mesenchyme cells. (U) Detail of LV region with an ENC weakly expressing Snail1 (arrow). (R) General view of Tie2-Cre;N1ICD heart. (T) Detail of AVC. Arrows, nuclear Snail1 staining in endocardium and mesenchyme. (V) Ectopic Snail1 staining in ventricular endocardium. Scale bars: 100 μm (A and B); 125 μm (C and D); 20 μm (E and F); 10 μm (I–P); 30 μm (Q and R); 20 μm (S–V).
Whole-mount in situ hybridization (WISH) and RT-PCR in Tie2-Cre;N1ICD embryos showed that endocardial expression of N1ICD and its targets Hey1 and Hey2 (22), Heyl (17), Delta-like 4 (23), Nrarp (24), and c-myc (25) was increased and expanded from the AVC to the ventricles (Supplemental Figure 1, H–T), indicating increased Notch activity throughout the endocardium. CD31/PECAM and MLC2v expression, indicative of early endocardial and myocardial differentiation, were normal (Supplemental Figure 2 and data not shown). The expression domains of Anf (26) and Chisel (27) in the AVC were less well defined in Tie2-Cre;N1ICD embryos compared with WT animals (Supplemental Figure 2, A and B), though their expression levels were unaffected (Supplemental Figure 2K and data not shown). Irx5, normally expressed in ventricular endocardium (ref. 28 and Supplemental Figure 2C), was strongly reduced in Tie2-Cre;N1ICD embryos (Supplemental Figure 2, D and K), suggesting loss of chamber identity.
The expression of endothelial identity markers was examined, given that the Tie2-Cre driver is active in the vascular endothelium as well (21). Tie2-Cre;N1ICD embryos ectopically expressed the Dll4 and Efnb2 markers in veins (Supplemental Figure 3, A–D), while expression of the venous marker Ephb4 was reduced (Supplemental Figure 3, E and F), suggesting that the vascular endothelium of transgenic embryos had lost its venous identity. Tie2-Cre;N1ICD embryos died at E11.0, probably as a consequence of defective vascular and hematopoietic development (P. Melgar and J.L. de la Pompa, unpublished observations).
Activation of a mesenchyme gene expression program characteristic of AVC in the ventricular endocardium of Tie2-Cre;N1ICD embryos.
Bmp2 is expressed in AVC myocardium at E9.5 and triggers the EMT of adjacent ENCs (9). Bmp2 expression was slightly weaker at E9.5 and was unaffected in E10.5 Tie2-Cre;N1ICD embryos (Supplemental Figure 2, E, F, and K). Tgf-β2, which acts downstream of Bmp2 (9) is transcribed in AVC myocardium at E9.5 and at lower levels in AVC endocardium and mesenchyme (ref. 8, Figure 1I, and Supplemental Figure 5A). Tgfb2 showed slightly weaker expression in Tie2-Cre;N1ICD AVC and was ectopically expressed in ventricular endocardium (Figure 1J and Supplemental Figure 5B). RT-PCR revealed normal Tgfb2 expression, probably because of ectopic expression in the ventricles (Figure 1G). Snail1, expressed in response to Notch (12) and TGF-β2 (29), was detected in AVC endocardium and mesenchymal cells of WT embryos (Figure 1K and Supplemental Figure 5C). In transgenic embryos, Snail1 transcription was extended to the ventricular endocardium (Figure 1L and Supplemental Figure 5D) and increased (Figure 1G). Ectopic Snail1 expression in ventricles was confirmed by Western blot (Figure 1H). Immunostaining showed Snail1 expression in the nuclei of WT and Tie2-Cre;N1ICD AVC endocardial and mesenchyme cells (Figure 1, Q–T). Transgenic embryos displayed ectopic nuclear Snail1 expression in ventricular ENCs (Figure 1, U and V). Sox9, a Tgf-β2 target in mesenchymal cells implicated in endocardial cushion cell differentiation (30), was transcribed in AVC endocardium and mesenchyme (Supplemental Figure 4A) and its expression was increased and expanded to the ventricles in transgenic embryos (Supplemental Figure 4, B and I).
Affymetrix microarray analysis to identify additional genes differentially expressed in Tie2-Cre;N1ICD E9.5 hearts uncovered 516 genes, 66 of which were downregulated and 450 of which were upregulated (Supplemental Excel file). Many of the upregulated genes were associated with EMT initiation, mesenchymal phenotype maintenance, cell migration, invasion, adhesion, and collagen deposition (Supplemental Table 1). These included Snail2, Twist2, Bmp6, and the Bmp receptor genes Alk3 and Alk6 (Supplemental Table 1), all of which are essential for EMT in different cellular contexts (31–33). Snail2, like Snail1, is required for Notch-induced EMT (31), and its expression was expanded in Tie2-Cre;N1ICD embryos from AVC endocardium and mesenchyme to the ventricular endocardium (Figure 1, M and N, and Supplemental Figure 5, E and F) and increased (Figure 1G). Twist1 is required downstream of Bmp2 during AVC EMT (10), but Twist1 expression was not altered in the microarray (Supplemental Excel file) or by WISH or RT-PCR (Supplemental Figure 2, G, H, and K), suggesting that Bmp2-dependent signaling in Tie2-Cre;N1ICD embryos was unaltered. In contrast, Twist2 expression was expanded from AVC to ventricles (Figure 1, O and P, and Supplemental Figure 5, G and H) and increased (Figure 1G), but the late EMT marker Has2 (34) was unaltered (Supplemental Figure 2, I–K). Similarly, Bmp6, normally expressed in AVC endocardium (Supplemental Figure 4E) and required for cushion morphogenesis (33), was ectopically expressed in the ventricle of transgenic embryos (Supplemental Figure 4, F and I). Expression of the chemokine receptor gene Cxcr4, involved in cell migration and metastasis (35), was increased in transgenic embryos (Supplemental Table 1 and Supplemental Figure 4I) and expanded from AVC endocardium to ventricular endocardium (Supplemental Figure 4, G and H). A further 12 genes involved in EMT, cell migration, and invasion were significantly increased in transgenic embryos (Supplemental Table 1), and upregulation of the Notch pathway genes detected by WISH and RT-PCR (Supplemental Figure 1, H–T) was also confirmed. Thus, Tie2-Cre;N1ICD embryos show the expansion of mesenchymal gene expression from AVC to ventricles. Moreover, expression of the chamber marker Hand2 was strongly reduced (Supplemental Table 1), confirming the loss of ventricular chamber identity in Tie2-Cre;N1ICD embryos.
Tie2-Cre;N1ICD ventricular explants undergo Tgf-β2 and Snail1-dependent EMT.
Culture of AVC explants on a type I collagen gel lattice is an excellent functional assay to evaluate EMT (36). To quantify differences between WT and transgenic explants, we determined the “transformation index” (TI) in 2 and 3 dimensions. 2D TI is the fraction of ENCs able to detach and migrate on the gel surface without invading the collagen matrix. 3D TI is the fraction able to invade the gel. After 72 hours, E9.5 WT AVC explants produced a halo of mesenchymal cells around the myocardium (Supplemental Figure 6A), with mesenchymal cells invading the collagen depths (Supplemental Figure 6A and Supplemental Video 1). E9.5 Tie2-Cre;N1ICD explants also produced invasive mesenchymal cells (Supplemental Figure 6B and Supplemental Video 1), but while 3D TI was similar for both genotypes (3D = 0.1), 2D TI was 3-fold higher in transgenic explants (0.35 vs. 0.15; Supplemental Figure 6C). Expression of EMT-inducing molecules in WT and transgenic ENCs was similar. Similar to the situation in vivo, Hey1 was ectopically expressed and upregulated in transgenic ENCs, together with Hey2, Heyl, and the mesenchymal marker Vimentin (Supplemental Figure 6D).
To determine whether ectopic Notch1 activity conferred transforming ability on ventricular tissue, we performed explant cultures with the distal tip of the LV. WT explants generated an endocardial monolayer surrounding the myocardium (Figure 2A), and ENCs grew as a coherent epithelium on the collagen surface (Figure 2A and Supplemental Video 2). In contrast, in Tie2-Cre;N1ICD explants, ENCs migrated over the gel and had a mesenchymal phenotype revealed by α-SMA staining (Figure 2B). These transformed cells did not invade the collagen matrix (Figure 2B and Supplemental Video 2), but their migratory ability was reflected in a sharply increased 2D TI (0.9 vs. 0.1; Figure 2C). Unlike WT cells, ENCs of transgenic explants expressed the mesenchymal markers Snail1, Snail2, Tgfb2, Vimentin, Periostin (37), and very low levels of Has2 (ref. 34 and Figure 2D) and Collagen. Twist1 was unaffected, consistent with the in vivo data (Figure 2D).
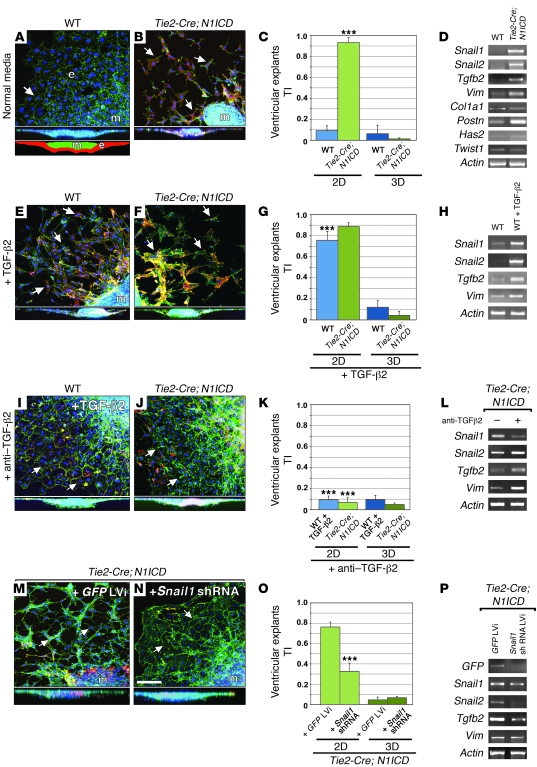
Figure 2. Tie2-Cre;N1ICD ventricular explants undergo Tgf-β2– and Snail1-mediated ectopic EMT.
(A, E, and I) Details of WT explants. (B, F, J, M, and N) Details of Tie2-Cre;N1ICD explants. Full lateral views of explants are shown below panels A–N. Schematic of a full lateral view of explant is shown at the bottom of panel A. e, endocardium (red); m, myocardium (green). All explants were stained with phalloidin-FITC (green), anti–α-SMA–Cy3 (red), and DAPI (blue). Arrows mark ENCs. (C, G, K, and O) Quantitative analysis of 2D and 3D TI. (D, H, L, and P) RT-PCR of explant endocardium. (A) WT. Arrow, ENCs growing as a monolayer. The lateral section shows ENC outgrowth on the collagen surface. (B) Tie2-Cre;N1ICD. Arrows, scattered ENCs that have undergone partial EMT. (C) 2D TI is increased in Tie2-Cre;N1ICD explants (P = 3.7 × 10–4). (D) Snail1, Snail2, Tgfb2, Vimentin, and Periostin expression is upregulated; Has2 is slightly increased and Twist1 appears unaffected. (E, F, and G) ENCs scatter without invading the collagen in TGF-β2–treated WT explants, and 2D TI is increased with respect to untreated WT ones (C; P = 1.8 × 10–7) and Tie2-Cre;N1ICD explants. (H) Increased Snail1, Snail2, and Tgfb2 expression. (I and K) ENCs of WT explants cultured with TGF-β2 and anti–TGF-β2 antibody grow as a monolayer and show reduced 2D TI with respect to TGF-β2–treated WT (G; P = 8.1 × 10–6). (J and K) Anti–TGF-β2 reduces ENC migration in Tie2-Cre;N1ICD explants, reflected in a reduced 2D TI (P = 2.3 × 10–5) and attenuated Snail1 expression (L). (M–O) Lentiviral-mediated shRNA Snail1 downregulation in transgenic explants reduces ENC migration with respect to GFP-transduced control explants (P = 3.3 × 10–8). LVi, lentivirus. (P) Expression of Snail2 and Tgfb2 is reduced. Scale bar: 50 μm. Results are expressed as mean + SD. ***P < 0.001.
The transformed ventricular ENCs of Tie2-Cre;N1ICD explants showed increased expression of Tgfb2 (Figure 2D), a key EMT inducer in AVC cells (8). Addition of TGF-β2 to WT ventricular explants mimicked the results with Tie2-Cre;N1ICD explants, inducing surface migration of ENCs but not invasion (Figure 2E). TGF-β2–treated WT ventricular explants had a 2D TI similar to untreated transgenic explants (0.75 vs. 0.9; Figure 2G) and expressed Snail1, Snail2, and Vimentin (Figure 2H). TGF-β2 had no additive effect on Tie2-Cre;N1ICD explants (Figure 2, F and G). A neutralizing anti–TGF-β2 antibody reverted the phenotype of Tie2-Cre;N1ICD and TGF-β2–treated WT ventricular explants to the endocardial monolayer seen in untreated WT explants (Figure 2, I–K), accompanied by reduced mesenchymal marker expression (Figure 2L).
During AVC EMT, Snail1, and Snail2 are key downstream effectors of Notch (12, 31) and TGF-β2 (38). To test the role of the Snail genes in the acquisition of the mesenchymal phenotype by ventricular ENCs, we transduced ventricular explant cells with Snail1 shRNAS. Two Snail1 shRNAs reduced the 2D TI of Tie2-Cre;N1ICD ventricular explants effectively (Figure 2, M–O). RT-PCR confirmed reduced Snail1 expression, though the reduction was greater for Snail2 (Figure 2P), consistent with the reported requirement of Snail1 for Snail2 expression (39). Tgfb2 expression was also reduced by Snail1 shRNAs (Figure 2P). This is consistent with the dose-dependent activation by Snail1 of a TGF-β2 reporter (data not shown), suggesting a functional link between Snail1 and Tgf-β2.
Ectopic endocardial N1ICD expression was examined in a stage-dependent manner using the tamoxifen-inducible line Cdh5(PAC)-CreERT2 (40). Recombination was induced from E9.5 to E10.5 and at E11.5, Cdh5(PAC)-CreERT2;N1ICD embryos showed hemorrhage in the heart and trunk region (Supplemental Figure 7, A and B) and increased N1ICD and Snail1 cardiac expression (Supplemental Figure 7C). Importantly, ENCs in ventricular trabeculae showed a fibroblastic, star-like morphology, contrasting with the flat ENCs in WT trabeculae (Supplemental Figure 7, D–I). Cdh5(PAC)-CreERT2;N1ICD ENCs transformed and migrated in ventricular explant assays (Supplemental Figure 7, J–L) but like Tie2-Cre;N1ICD ENCs were not invasive.
BMP2 confers invasivity on ventricular ENCs.
Functional studies have shown that Bmp2 is a critical inducer of AVC EMT upstream of Tgf-β2 (9, 10). Bmp2 expression was unaffected in Tie2-Cre;N1ICD embryos (Supplemental Figure 2, E, F, and K), suggesting that Notch acts either downstream or independently of Bmp2 during cardiac EMT. BMP2 treatment of WT and Tie2-Cre;N1ICD ventricular explants produced invasive mesenchymal cells, significantly increasing 2D and 3D TI in WT explants and 3D TI in transgenic explants (Figure 3, A, B, D, and E, and Supplemental Video 3). Thus, BMP2 confers invasive ability to transformed ventricular ENCs. Expression of Snail1, Has2, and Periostin (but not Twist1) was upregulated in WT ventricular cells treated with BMP2 (Figure 3F), indicating that BMP2-treated WT ventricular explants acquire full mesenchymal features.
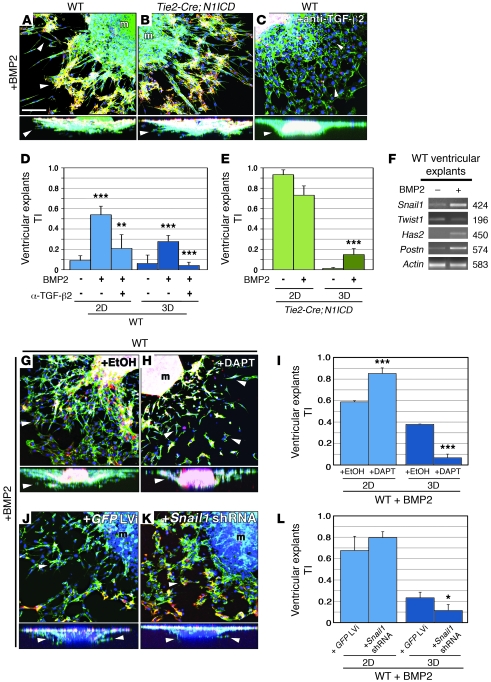
Figure 3. BMP2 induces ventricular explants to undergo Tgf-β2–, Notch-, and Snail1-mediated invasive EMT.
All explants were triple stained as in Figure 2. (A–C, G, H, J, and K) Details of explants. Full lateral views are shown below. Arrowheads mark invading ENCs. (A and D) BMP2 treatment of WT ventricular explants induces invasive EMT, increasing 2D and 3D TI (P = 2 × 10–4 and 2.7 × 10–4). (B and E) BMP2 treatment increases the 3D TI of Tie2-Cre;N1ICD explants (P = 9 × 10–4) at the expense of reduced 2D TI. (C and D) Anti–TGF-β2 reduces the 2D and 3D TI of BMP2-treated WT explants (P = 4.1 × 10–3 and 1.8 × 10–7). (F) BMP2-treated WT ventricular explants upregulate Snail1, Has2, and Periostin, but Twist1 appears unaffected. (G–I) Inhibition of Notch with DAPT reduces invasive capacity (3D TI) of BMP2-treated WT ventricular explants (P = 1.02 × 10–8), while increasing 2D TI (P = 1.3 × 10–4). (J–L) Lentiviral-mediated shRNA Snail1 downregulation reduces the invasive capacity of BMP2-treated WT explants. (P = 3.4 × 10–2). m, myocardium. Scale bar: 50 μm. Results are expressed as mean + SD. **P < 0.01; ***P < 0.001.
Anti–TGF-β2 antibody inhibited the effect of BMP2 on WT explants (Figure 3, C and D), indicating that, as in AVC EMT, TGF-β2 acts downstream of Bmp2 during ventricular EMT. Although Bmp6 was ectopically expressed in Tie2-Cre;N1ICD ventricular endocardium (Supplemental Table 1 and Supplemental Figure 4, E, F, and I), BMP6 did not induce EMT in WT ventricular explants (not shown).
To examine the requirement for Notch in BMP2-induced ventricular EMT, we cultured WT ventricular explants with BMP2 plus the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) (41). DAPT reduced invasion after 72 hours, increasing 2D TI while reducing 3D TI (Figure 3, G–I). Thus, inhibition of Notch in BMP2-treated WT ventricular explants led to accumulation of scattered cells that did not invade. BMP2-induced invasion and 3D TI were similarly reduced in WT ventricular explants transduced with Snail1-shRNA (Figure 3, J–L). Thus, Tgf-β2, Notch, and Snail1 participate in the molecular mechanism underlying BMP2-induced invasive EMT in ventricular explants.
Myocardial-specific Notch1 activation leads to Bmp2 and Tgfb2 repression and ectopic Snail1 expression.
Studies in chick and mouse have shown that Hey transcription factors repress Bmp2 expression in chamber myocardium, restricting it to AVC and OFT territory (42, 43). To determine whether myocardially expressed N1ICD impaired endocardial EMT via Hey-mediated Bmp2 repression, we used the Nkx2.5-Cre driver line, which is active in both endocardium and myocardium from E7.5 (44). Supplemental Figure 8A shows Cre reporter activity in these tissues at E9.5. N1ICD-GFP was expressed throughout the heart of Nkx2.5-Cre;N1ICD embryos (Supplemental Figure 8B). At E9.5, Nkx2.5-Cre;N1ICD embryos showed pericardial distension and heart dilation, with poorly defined boundaries between chamber and nonchamber tissue, an AVC region devoid of transformed cells, and a thin ventricular myocardium with underdeveloped trabeculae (Supplemental Figure 8, C–G). Hey1 was expressed throughout the myocardium and endocardium, including the AVC (Figure 4, A and B). The expanded Hey1 domain was reflected in increased transcription by RT-PCR (Figure 4P). Myocardial Hey2 and Heyl expression was unaltered (not shown), but was increased overall (Figure 4P and data not shown) due to stronger endocardial expression.
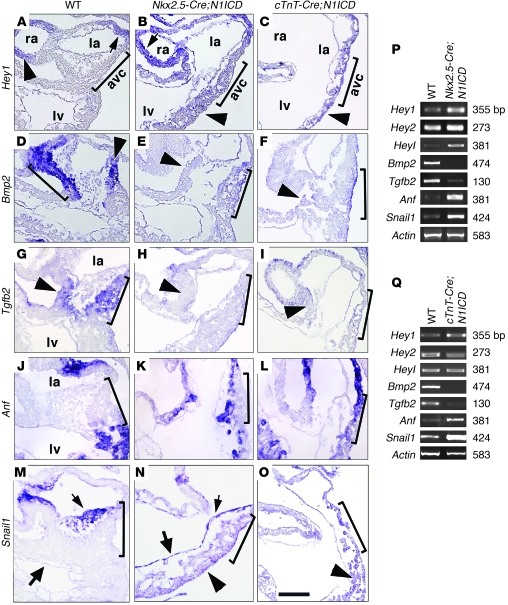
Figure 4. Myocardial Notch1 activation leads to loss of AVC identity.
(A–O) WISH of E9.5 hearts, showing detail of AVC and LV. Brackets mark the AVC. Arrowheads, myocardium; arrows, endocardium. la, left atrium; ra, right atrium. (A–C) Hey1. Expression is restricted to atrial myocardium and endocardium in WT hearts, extends throughout myocardium and endocardium of Nkx2.5-Cre;N1ICD hearts (B) and throughout the myocardium of cTnT-Cre;N1ICD hearts (C). (D–F) Bmp2 expression in WT AVC is markedly reduced in transgenic hearts. (G–I) Tgfb2. Expression in WT AVC is reduced in transgenic hearts. (J–L) Anf is restricted to WT atrium and ventricle myocardium, and is extended to AVC myocardium in transgenic hearts. (M–O) Snail1 is expressed in WT AVC endocardium and mesenchyme (M) and extends throughout the myocardium and endocardium of Nkx2.5-Cre;N1ICD hearts (N) and throughout the myocardium of cTnT-Cre;N1ICD hearts (O). Scale bar: 30 μm. (P and Q) RT-PCR of E9.5 WT and Nkx2.5-Cre;N1ICD hearts (P) and cTnT-Cre;N1ICD hearts (Q).
The ectopic Hey1 expression was accompanied by reduced Bmp2 in Nkx2.5-Cre;N1ICD AVC myocardium (Figure 4, D, E, and P). Tgfb2 transcription in AVC myocardium (Figure 4G) was strongly reduced in transgenic embryos (Figure 4, H and P). Expression of the myocardial chamber marker Anf was expanded to the AVC (Figure 4, J, K, and P), suggesting that AVC identity was lost in myocardial cells. Snail1 was exclusively expressed in AVC and OFT endocardium and mesenchyme of WT embryos (Figure 4M), while Snail1 transcript and protein were ectopically expressed throughout the endocardium and myocardium of Nkx2.5-Cre;N1ICD embryos (Figure 4N and data not shown).
To substantiate these findings, Notch1 was activated exclusively in the myocardium by breeding into the cTnT-Cre line, which is active in every heart segment from E7.5 onwards (45). Supplemental Figure 8H shows myocardial Cre reporter activity at E9.5. cTnT-Cre;N1ICD embryos expressed N1ICD-GFP throughout the myocardium (Supplemental Figure 8I). E10.5 transgenic embryos were smaller than WT and showed pericardial distension, poorly defined cardiac regions, and a lack of mesenchymal cells in the AVC (Supplemental Figure 8, J–M). Trabeculae were apparent (Supplemental Figure 8, L and N) but slightly delayed compared with WT embryos (Supplemental Figure 1, B and F). Hey1 was expressed throughout the myocardium of cTnT-Cre;N1ICD embryos, including the AVC (Figure 4C and data not shown), and was increased (Figure 4Q). Bmp2 and Tgfb2 expression were reduced (Figure 4, F, I, and Q), and as in Nkx2.5-Cre;N1ICD embryos, Anf expression was extended to AVC myocardium (Figure 4, L and Q), suggesting loss of AVC identity. Likewise, Snail1 was abnormally expressed in the myocardium (Figure 4O) and increased (Figure 4Q).
The aberrant Hey1 and Bmp2 expression in the myocardium of Nkx2.5-Cre;N1ICD and cTnT-Cre;N1ICD embryos supports the idea that N1ICD-driven ectopic Hey1 expression represses Bmp2 in AVC myocardium. Hey and Bmp2 expression were examined in Notch loss of function (LOF) (Supplemental Figure 9). The expression of Hey1 and Hey2 was detected in chamber myocardium (Supplemental Figure 9, A and C). Hey1 was also found in atrial endocardium (Supplemental Figure 9A), Hey2 throughout the endocardium (Supplemental Figure 9C), and Heyl in AVC endocardium (Supplemental Figure 9E). In RBPJk mutants, myocardial Hey1 and Hey2 expression were unaffected (Supplemental Figure 9, B and D), whereas endocardial Hey gene expression was reduced (Supplemental Figure 9, B, D, F, and I). In RBPJk mutants (Supplemental Figure 9, G and H), Bmp2 expression was not affected in the myocardium but was upregulated in the endocardium (Supplemental Figure 9, H and I). These data suggest that Hey2 acts downstream of Notch in repressing Bmp2 expression in the endocardium.
Ectopic myocardial Notch1 activity impairs EMT in a Bmp2-dependent fashion.
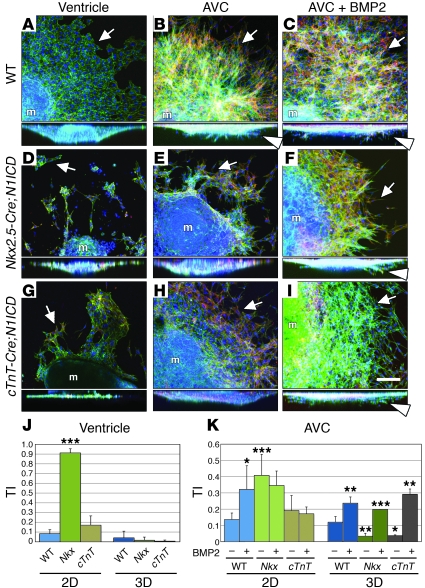
To study the effect of deregulated Bmp2 and Snail1 expression on EMT, we performed cardiac explant experiments with E9.5 Nkx2.5-Cre;N1ICD and cTnT-Cre;N1ICD embryos. As expected, WT ventricular endocardium grew as a monolayer (Figure 5A) with a few scattered ENCs at the explant edge and almost no invasion (Figure 5J). WT AVC explants showed ENC outgrowth and several invading mesenchymal cells (Figure 5B), with 2D and 3D TI between 0.1 and 0.15 (Figure 5K). Addition of BMP2 significantly increased surface migration and invasion (Figure 5C and Supplemental Video 3), reflected in 2D and 3D TI values of greater than 0.3 and greater than 0.2 (Figure 5K).
Figure 5. BMP2 treatment rescues EMT in Notch1-expressing myocardium.
Heart explants triple stained as in Figure 2. (A–I) Details of explants. Full lateral views are shown below. (A) WT ventricular explants show an endocardial monolayer. (B and C) WT AVC explants ± BMP2 undergo EMT. (D) In Nkx2.5-Cre;N1ICD ventricular explants, ENCs migrate across the collagen surface, but are not invasive. (E) Nkx2.5-Cre;N1ICD AVC explants produce migratory mesenchymal cells that show reduced invasion, which is rescued by BMP2 (F). (G) In cTnT-Cre;N1ICD ventricular explants, most ENCs grow as a monolayer and are noninvasive. (H) Mesenchymal cells in cTnT-Cre;N1ICD AVC explants migrate but show reduced invasion, which is rescued by BMP2 (I). Arrows, ENCs; arrowheads, invasive mesenchymal cells. (J) TI analysis of ventricular explants. Most ENCs in Nkx2.5-Cre;N1ICD explants (Nkx) migrate over the gel surface (P = 2.2 × 10–7 for 2D TI versus WT explants), whereas few cells in cTnT-Cre;N1ICD explants (abbreviated cTnT) migrate in 2D. 3D TI is very low for both genotypes. (K) TI analysis of AVC explants. BMP2 treatment of WT explants significantly increases 2D and 3D TI (P = 3.7 × 10–2 and 6.1 × 10–3). Nkx2.5-Cre;N1ICD explants (Nkx) show increased 2D TI (P = 3.5 × 10–4) but a markedly reduced 3D TI compared with WT (P = 5.7 × 10–3). BMP2 treatment increases Nkx2.5-Cre;N1ICD 3D TI (P = 6.7 × 10–4) and slightly reduced 2D TI. BMP2 treatment sharply increases the 3D TI of cTnT-Cre;N1ICD explants (cTnT; P = 1.9 × 10–3). m, myocardium. Scale bar: 50 μm. Results are expressed as mean + SD. *P < 0.005; **P < 0.01; ***P < 0.001.
Ventricular explants of Nkx2.5-Cre;N1ICD embryos generated scattered, noninvasive ENCs (Figure 5D) with a 2D TI (0.9) significantly higher than that of WT explants (Figure 5J). Nkx2.5-Cre;N1ICD AVC explants (Figure 5E and Supplemental Video 4) showed relatively high surface migration (2D TI = 0.4; Figure 5K) but severely reduced invasion (3D TI < 0.1; Figure 5K). Addition of BMP2 restored the WT phenotype (3D TI = 0.2; Figure 5, F and K, and Supplemental Video 4).
cTnT-Cre;N1ICD ventricular explants behaved similarly to WT ventricular explants, with no endocardial transformation (Figure 5, G and J), consistent with the lack of ectopically activated endocardial Notch1 in this model. cTnT-Cre;N1ICD AVC explants (Figure 5H) transformed very poorly (3D TI < 0.1; Figure 5K), but addition of BMP2 restored invasiveness (3D TI = 0.3; Figure 5, I and K).
These data indicate that Notch1 activation in endocardium endows chamber ENCs with transforming ability. Notch1 activation in myocardium leads to Bmp2 and Tgfb2 downregulation by ectopic Hey1 expression.
Bmp2 promotes Snail1 expression and stability, and its inactivation impairs Notch1 signaling in AVC.
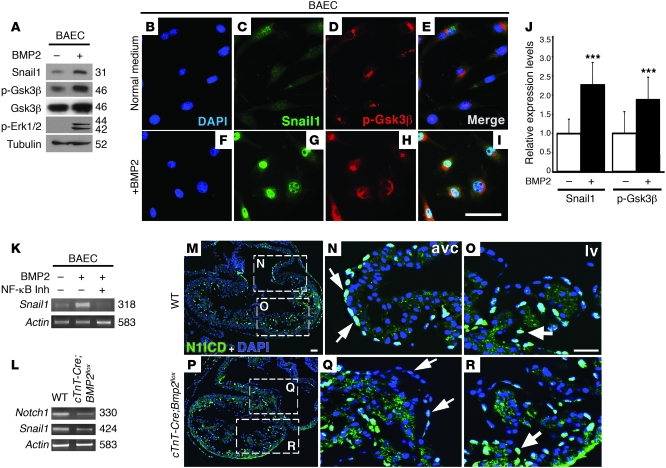
During EMT, SNAIL1 is regulated by 2 sequential GSK3β-mediated phosphorylation events. The first SNAIL1 phosphorylation event induces nuclear export; the second, proteasomal degradation (46). In endothelial cells, Bmp2 induces ERK1/2 phosphorylation (activation), which itself phosphorylates (inactivates) GSK3β (47). We tested the putative link among Bmp2 signaling, Gsk3β inhibition, and Snail1 stabilization in ENCs. Immunoblotting of bovine aortic endothelial cells (BAECs) incubated with the proteasome inhibitor MG132 (46) detected a 31-kDa Snail1 band, while the levels of p-Gsk3β and p-Erk1/2 were low (Figure 6A). Addition of BMP2 markedly increased Snail1 expression as well as the levels of p-Gsk3β and p-Erk1/2, while the total amount of Gsk3β was unaffected (Figure 6A). Immunofluorescence of unstimulated BAECs detected weak nuclear Snail1 expression (Figure 6, B and C) and faint, predominantly perinuclear staining of p-Gsk3β (Figure 6, D and E). In BMP2-treated BAECs, Snail1 nuclear staining was increased (120% increase) and perinuclear p-Gsk3β staining was also stronger (80%; Figure 6, G–J).
Figure 6. BMP2 inhibits Gsk3β, leading to Snail1 expression and nuclear stabilization.
Deletion of myocardial Bmp2 reduces Notch1 signaling in AVC. (A) Immunoblot of BAECs cultured ± BMP2. Weak Snail1 expression is detected in the absence of BMP2. BMP2 induces increased Snail1 levels and increases phosphorylation of Gsk3β and Erk1/2, suggesting Gsk3β inhibition. (B–I) Immunostaining of BAECs cultured ± BMP2. (B and F) DAPI-counterstained nuclei. (C and G) Snail1 expression; nuclear staining in BMP2-treated cells is stronger and more punctate. (D and H) p-Gsk3β; predominant perinuclear p-Gsk3β staining is stronger after BMP2 treatment. (E and I) Merged showing DAPI, Snail1, and p-Gsk3β staining. (J) Quantification of Snail1 and p-Gsk3β protein. Expression is increased upon BMP2 treatment. (K) RT-PCR analysis of Snail1 expression in BAECs cultured ± BMP2 and ± NF-κB inhibitor. Snail1 expression is reduced in the presence of BMP2 and NF-κB inhibitor. (L) RT-PCR of E10.5 WT and cTnT-Cre;Bmp2flox hearts. Notch1 and Snail1 expression is reduced in mutant hearts. (M–R) N1ICD immunostaining (green). Nuclei are DAPI counterstained (blue). (M–O) E10 WT heart. (P–R) cTnT-Cre;Bmp2flox heart. (M and P) General views. (N and Q) AVC detail. Arrows mark N1ICD-positive nuclei in WT AVC endocardium (N) and weakly positive nuclei in the corresponding region of the mutant (Q). (O and R) Detail of LV. Arrows mark N1ICD-positive nuclei in endocardium at the base of trabeculae in WT (O) and mutant embryos (R). Scale bars: 20 μm. Results are expressed as mean + SD. ***P < 0.001.
In epithelial cells, Gsk3β has also been shown to regulate Snail1 at the transcriptional level, together with Gsk3α, via NF-κB inhibition (48). We tested to determine whether this mechanism contributes to the effect of BMP2 in BAECs. Figure 6K shows that adding BMP2 to BAECs markedly increased Snail1 mRNA while BMP2 plus an NF-κB inhibitor markedly reduced it. We tested the effect of NF-κB inhibition on the BMP2-driven EMT of ventricular explants. WT ventricular explants cultured with BMP2 underwent invasive EMT (Supplemental Figure 10, A and C), which was notably reduced when an NF-κB inhibitor was added to the culture medium (Supplemental Figure 10, B and C). This effect was also reflected by the marked reduction of Snail1 mRNA upon addition of the NF-κB inhibitor (Supplemental Figure 10D). Together these observations suggest that BMP2 treatment of endothelial cells augments Snail1 mRNA in an NF-κB–dependent manner and stabilizes Snail1 protein by Gsk3β phosphorylation/ inactivation.
Our data show that Bmp2 and Notch1 cooperate to induce invasive EMT in ventricular explants. This process is Notch dependent, and the noninvasive phenotype of AVC explants ectopically expressing Notch1 in the myocardium is rescued by exogenous BMP2. To explore the Bmp2-Notch1 relationship in vivo, we crossed a conditional Bmp2 mutant (49) with the myocardial-specific cTnT-Cre driver line. E10.5 cTnT-Cre;Bmp2flox embryos lacked defined AVC and EC tissue (Supplemental Figure 11, A–F), similar to the phenotype obtained with the Nkx2.5-Cre driver (10, 11). Cardiac expression of Notch1 (and Snail1) was markedly reduced (Figure 6L), which, given the exclusive endocardial Notch1 expression at this stage, suggests that myocardial Bmp2 deletion impairs endocardial Notch1 expression. In E10 WT embryos, N1ICD is localized in most ENCs in the AVC and to ENCs at the base of trabeculae in the ventricles (Figure 6, M–O, and ref. 14). In cTnT-Cre;Bmp2flox mutants, N1ICD staining was strongly reduced in AVC endocardium (Figure 6, P and Q) but ventricular staining was unaffected (Figure 6, P and R). These data suggest that Bmp2 deletion in the myocardium specifically impairs Notch1 expression and activity in AVC endocardium.
Discussion
We previously showed that Notch is a key signal during CVF that via Snail1 activation, promotes the EMT of ENCs to form the valve primordium (12). Notch/RBPJK directly regulate Snail1 transcription (12, 50), and Notch inactivation leads to a downregulation of Snail1 expression in AVC endocardium (12). This earlier report did not, however, provide information about the role of Notch targets in EMT and neither did it define the domain-specific (chamber vs. nonchamber) and tissue-specific (endocardium vs. myocardium) patterning of Notch function or determine the extent of Notch interaction with myocardial signals that trigger EMT (e.g., Tgf-β2 and Bmp2). The main goal of the present report was to shed light on the integration of EMT-driving signals and the establishment of valve developmental domains in the embryonic heart.
Transgenic mice constitutively expressing N1ICD throughout the endocardium, myocardium, or both have allowed us to establish Notch as a key endocardial signal that acts in concert with myocardial Bmp2 to promote a valve-forming field in the developing heart. We propose that Snail1 is a critical point of convergence between Notch1 and Bmp2 signaling based on the following experimental evidence: (a) ectopic N1ICD expression in either endocardium or myocardium leads to Snail1 upregulation in these tissues; (b) the noninvasive EMT of Tie2-CRE;N1ICD ENCs in ventricular explants is mediated by Snail1, and the complete EMT driven by BMP2 is also Snail1-dependent; (c) targeted Bmp2 inactivation in the myocardium leads to reduced Snail1 expression in AVC endocardium (ref. 10 and this report); and (d) BMP2 stimulation increases Snail1 expression in BAECs.
An important issue raised by these findings is how these signals are restricted to specific tissues and their activities coordinated. Two reports have examined the effect of ectopic Notch1 activation on cardiovascular development. In the first study, Notch1 was activated in cardiogenic mesoderm using the Mesp1-Cre driver line, demonstrating that transgenic mice show defective AVC myocardial differentiation and ventricular maturation (19). Similarly to WT embryos, the AVC of transgenic embryos contained mesenchymal cells, but also showed ectopic myocardial masses of uncertain origin. Since N1ICD activation occurred in both endocardium and myocardium, these authors did not study the specific effect of Notch1 activation in either of these layers (19). The second study used the Tie2-Cre driver line to express N1ICD in the endocardium and vascular endothelium (51). These authors reported defective trabeculation, thinner myocardium, and delayed endocardial cushion formation, but the cellular and molecular bases of these defects were not examined.
Our results provide the first demonstration of the expansion of the characteristic AVC-restricted transcription of Notch target genes (Hey1, Heyl, Dll4, c-myc, and Nrarp) to the ventricular endocardium after N1ICD overexpression in this tissue (Tie2-Cre;N1ICD). In light of earlier observations that Hey1 and Dll4 are downregulated in RBPJk and Notch1 mutants (12), we suggest that Notch target gene expression is regulated by a positive feedback loop and that Notch signals in the endocardium by lateral induction, with AVC ENCs behaving as a developmental field. We find a set of genes involved in EMT, cell migration, metastasis, and invasion to be upregulated and ectopically expressed in the ventricular endocardium of Tie2-Cre;N1ICD embryos (Supplemental Table 1). These data indicate that WT Notch1 activity in AVC ENCs promotes EMT by driving, in a cell-autonomous manner, a mesenchyme gene program that can be experimentally extended to the ventricles by endocardial NICD overexpression. This notion is consistent with the downregulation of the endocardial chamber marker Irx5, suggesting that ventricular ENCs of Tie2-Cre;N1ICD embryos have lost their chamber identity.
AVC explants of Tie2-Cre;N1ICD embryos undergo EMT, with ENCs invading the collagen matrix (3D EMT) and showing increased migratory capacity on the gel surface (2D EMT) with respect to WT AVC. In contrast to the characteristic EMT-refractory ventricular endocardium of WT mice, ENCs of Tie2-Cre;N1ICD ventricular explants (or those from Cdh5[PAC]-CreERT2;N1ICD mice) generate mesenchymal cells expressing Snail1, Snail2, and Tgfb2 that lose contact and scatter over the gel surface, but are unable to invade the gel matrix. Addition of TGF-β2 can mimic this effect in WT ventricular endocardium, whereas lentiviral- or antibody-mediated inhibition of Snail1 and Tgf-β2 inhibits transformation of Tie2-Cre;N1ICD explants, indicating that these molecules lie downstream of Notch in EMT. Thus, Notch alone is able to promote noninvasive EMT in a Snail1 and Tgf-β2–dependent manner, but cannot trigger a fully invasive EMT. WT ventricular ENCs become fully invasive after BMP2 addition and inhibition of Notch, Tgf-β2, or Snail1 severely attenuates BMP2-induced invasion, indicating that Notch and Bmp2 are critical signals acting in concert during EMT.
Constitutive Notch1 activation in the myocardium (Nkx2.5-Cre;N1ICD or cTnT-Cre;N1ICD) caused cardiac dysmorphogenesis. The expansion of myocardial chamber markers Anf and Chisel to the AVC and the Hey1 upregulation and concomitant Bmp2 downregulation in AVC myocardium suggested that myocardial cells have lost their AVC identity. Thus, ectopic myocardial Notch activity — via the activation of Hey1 — prevents Bmp2 expression in chamber myocardium, supporting the idea that Hey proteins regulate Bmp2 expression and thus AVC patterning (42, 43). Loss of Bmp2 expression is consistent with the poor ENC invasion observed in AVC explants from cTnT-Cre;N1ICD embryos. Remarkably, Snail1 expression is upregulated in both the endocardium and myocardium of Nkx2.5-Cre;N1ICD embryos, whereas Bmp2 is downregulated in the AVC (presumably via ectopic Hey1 activity), in agreement with the very poor transforming ability of AVC explants from these embryos. The rescue of invasiveness by BMP2 indicates that this molecule is critical for the acquisition of invasiveness by ENCs and that Notch promotes the transformed mesenchymal phenotype in AVC or OFT endocardium. Thus, Notch is likely to cooperate with Bmp2 to promote full deployment of invasive properties by transformed ENCs.
What is the role of Notch, Bmp2, and Hey in cardiac patterning? Supplemental Figure 12 summarizes the data from studies of Notch (this report and ref. 19) and Hey1 and Hey2 (42, 43) gain of function (GOF) and LOF, in terms of their effect on the Bmp2 expression domain and cardiac patterning. In the myocardium, Bmp2 expression is confined to the AVC by Hey1 and Hey2, which are expressed in the chambers (ref. 42 and 43, and Supplemental Figure 12A). In the endocardium, Notch1-dependent Hey1, Hey2, and Heyl are differentially expressed in the AVC and chambers and repress Bmp2 (Supplemental Figure 9 and Supplemental Figure 12A). Ectopic N1ICD expression in endocardium expands AVC patterning to ventricular endocardium but does not affect Hey1, Hey2, or Bmp2 myocardial expression or patterning (Supplemental Figure 12B). Ectopic N1ICD expression in the myocardium leads to expansion of Hey1 and loss of Bmp2 expression, and chamber markers expand to the AVC (Figure 4, K and L, and Supplemental Figure 12C). In both N1ICD GOF models, Bmp2 is repressed in the endocardium, similarly to the WT situation. Systemic abrogation of Notch signaling leads to downregulation of Hey genes and ectopic endocardial Bmp2 expression, indicating that Notch represses Bmp2 in the endocardium via Hey proteins (Supplemental Figure 9 and Supplemental Figure 12D) and suggesting that Notch might normally prevent ENCs from acquiring primitive cardiomyocyte features, such as those of the AVC. Notch inactivation impairs endocardial patterning of the AVC and chambers (refs. 12 and 52, and Supplemental Figure 12D), but does not affect myocardial patterning or the myocardial expression domains of Hey1, Hey2, or Bmp2 (Supplemental Figure 9 and Supplemental Figure 12D).
Our findings also shed light on how Notch1 and Bmp2 signals are integrated to promote EMT. Our data establish that Bmp2 triggers full EMT in WT and Tie2-Cre;N1ICD ventricular explants and that cell invasion in WT explants can be markedly reduced by inhibiting Tgf-β2, Notch, or Snail1. This supports the idea that these molecules are required for Bmp2-driven invasive EMT and indicate that Bmp2 and Notch1 signaling converge during AVC (and OFT) differentiation and the establishment of EMT competence in ENCs. At least 2 lines of evidence suggest that Bmp2 acts upstream of endocardial Notch activity during cardiac EMT: (a) impaired Notch1-Snail1–dependent signaling in AVC endocardium of Bmp2 myocardial mutants (cTnT-Cre; BMP2flox) indicates that Bmp2 is required for Notch activity in AVC territory; (b) Notch deficiency, while blocking transformation in vivo and in vitro by loss of Snail1 expression (12), does not affect Bmp2 expression in AVC myocardium (Supplemental Figure 9H). Moreover, ectopic Notch activity in endocardium upregulates and expands Snail1-dependent expression of Twist2, but not that of Bmp2-dependent Twist1 (10), suggesting that Bmp2 and Notch1 elicit overlapping but different mesenchyme gene expression programs in the AVC endocardium.
The concerted activities of Notch1 and Bmp2 in valve formation are reflected in the activation of Snail1 expression in chamber endocardium in response to ectopic Notch1 activity and in the reduced Snail1 expression after myocardial Bmp2 deletion. Moreover, inhibition of Gsk3β in BMP2-stimulated BAECs suggests that whereas Notch1 promotes Snail1 expression, Bmp2 stimulates both Snail1 expression and protein stabilization by inhibiting Gsk3β activity. The interplay between Notch and Bmp2 during the acquisition of the invasive phenotype by AVC ENCs involves Notch1- and Bmp2-induced Snail1 expression and Bmp2-mediated Snail1 nuclear accumulation, leading to sustained expression of mesenchymal genes. This provides what we believe is a novel mechanism for the integration of Notch1 and Bmp2 activity in EMT. Our results, summarized in Figure 7, emphasize the importance of endocardium-myocardium interplay during CVF and suggest that endocardial Notch1 converges with myocardial Bmp2 to promote and regulate the extent of EMT in prospective valve territory in both the AVC and OFT.
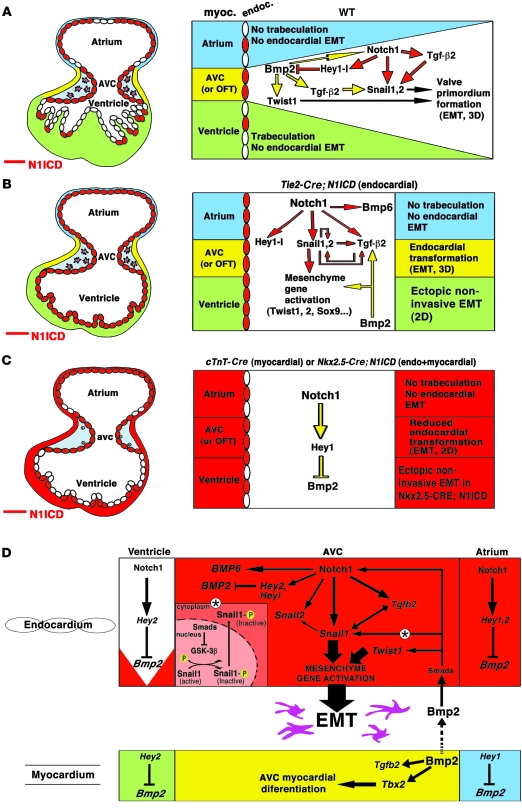
Figure 7. A model for concerted Notch1 and Bmp2 activities in CVF.
Left, schematic representations of the E9.5 heart. Right, schematics showing the different cardiac regions and developmental processes occurring within them. Green, ventricular myocardium; yellow, AVC myocardium; blue, atrial myocardium. ENCs expressing N1ICD are labeled red. Pink, invasive mesenchyme cells. Yellow arrows, myocardial signals; red arrows, endocardial signals. (A) WT embryo. AVC myocardial Bmp2 is required for Tgfb2, Notch1, Snail1, Snail2, and Twist1 expression. Endocardial Notch1 is required for Tgfb2 expression, activates Snail1 and Snail2, and represses Bmp2 in endocardium via Hey proteins. Bmp2 and Notch1 signals converge in AVC endocardium to promote complete EMT. (B) Tie2-Cre;N1ICD embryo. Ectopic N1ICD expression in endocardium (left) activates mesenchymal genes, promotes noninvasive EMT in ventricles but not in atria, and leads to loss of chamber identity. (C) cTnT-Cre;N1ICD and Nkx2.5-Cre;N1ICD embryos. Ectopic N1ICD expression in myocardium leads to ectopic Hey1 expression and Bmp2 repression. Myocardial AVC identity is lost, and EMT is severely affected. (D) Molecular pathways downstream of Notch during cardiac EMT. LOF and GOF data (this report and refs. 19, 42, and 43) indicate that Notch represses Bmp2 via Hey target activation. Endocardial Notch1 activates a mesenchyme gene program. The double-headed arrow linking Tgfb2 and Snail1 indicates the interdependence of both genes. Myocardial Bmp2 converges with endocardial Notch1 signaling to promote mesenchyme gene activation and EMT in the AVC. Convergence of Notch1 and Bmp2 is reflected in Notch activation of Snail1 expression and Bmp2-mediated Snail1 nuclear stabilization, via Gsk3β inhibition (*).
The results presented in this study open new avenues of research into the origin of cardiac valves and septal defects, which are the most common types of cardiac congenital abnormality and a major cause of adult morbidity and mortality (1, 53). Moreover, alterations of any component of the embryonic Notch1-Bmp2-Snail1 axis described here might be relevant in calcific aortic valve stenosis (4) or atherosclerosis (54), as these diseases share features such as EMT, fibrosis, and calcification. This analysis will be instrumental in the dissection of the cellular and molecular components of cardiac valve disease, the prevalent condition among congenital heart anomalies.
Methods
Mouse strains.
Mouse strains were R26N1ICD (20), Bmp2flox (49), RBPJk (55), Tie2-Cre (21), Nkx2.5-Cre (44), cTnT-Cre (45), and Cdh5(PAC)-CreERT2 (40). For simplicity, we use Tie2-Cre;N1ICD to refer to Tie2-Cre/+;R26N1ICD/+ mice, Nkx2.5-Cre;N1ICD for Nkx2.5-Cre/+;R26N1ICD/+ mice, cTnT-Cre;N1ICD for cTnT-Cre/+;R26N1ICD/+ mice, Cdh5(PAC)-CreERT2;N1ICD for Cdh5(PAC)-CreERT2/+;R26N1ICD/+ mice and cTnT-Cre;Bmp2flox for cTnT-Cre/+;Bmp2flox/flox mice. R26N1ICD mice were provided by D.A. Melton (Harvard University, Cambridge, Massachusetts, USA), and Nkx2.5-Cre mice were provided by R.P. Harvey (Victor Chang Cardiac Research Institute, Darlinghurst, Australia). cTnT-Cre driver lines were provided by S. Evans (University of California San Diego, La Jolla, California, USA). Bmp2 conditional lines were provided by V. Rosen (Harvard University School of Dental Medicine, Boston, Massachusetts, USA). Details are provided in Supplemental Data and Supplemental Table 2.
All experimental procedures and protocols were approved by the Ethics Committee and Animal Welfare Committee of the CNIC and conformed to European Union guidelines for the care and use of laboratory animals.
AVC and LV explants.
A solution (1.5 mg/ml) of rat-tail collagen type I (BD Biosciences) was dispensed into 4-well microculture dishes and allowed to solidify inside a 37°C, 5% CO2 incubator. Collagen gels were washed several times with DMEM containing 8% FBS, 0.1% insulin-transferrin-seleniun (ITS, GIBCO; Invitrogen), and antibiotics, and drained. AVCs were harvested in sterile PBS from E9.5 or E11.5 embryos. LV were carefully dissected, avoiding contamination from AVC tissue. Explants were placed with the endocardium face down and allowed to attach (16 hours, 37°C, 5% CO2). Medium (100 μl/well in AVC explants or 50 μl/well in LV) was added and explants cultured for up to 3 days (AVC) or 4 days (LV). Explants were fixed and stained with phalloidin-FITC (1:100; Sigma-Aldrich) to reveal the actin cytoskeleton and α-SMA-Cyan3 (1:100; Sigma-Aldrich) to detect mesenchymal cells as described (34). Explants were mounted on excavated slides in Vectashield medium containing DAPI. For explant treatments, the medium was supplemented with human TGF-β2 (30 ng/ml; Peprotech), human BMP2 (200 ng/ml; R&D Biosciences), anti-human TGF-β2 antibody (1 μg/ml; R&D Biosciences), or NF-κB SN50 inhibitor (18 μM; Calbiochem). For treatments with DAPT (γ-secretase inhibitor IX; 565770; Calbiochem), collagen gels were conditioned in explant media containing 50 μM DAPT or vehicle (ethanol). Medium was replaced every 24 hours.
Lentivirus production and Snail shRNA interference.
Pseudotyped lentivectors were produced according to ref. 56. Subconfluent HEK293T cells cultured in 10-cm plates with DMEM-Glutamax (Invitrogen), 10% FBS, and antibiotics were transiently cotransfected using Lipofectamine Plus (Invitrogen) with the following plasmids: 5 μg LV-eGFP control vector or lentiviral Snail-shRNA vectors TRCN0000096620 and TRCN000009622 (mission shRNA library; Sigma-Aldrich), 5 μg packaging plasmid pCMVdR8.74, and 2 μg vesicular stomatitis virus G envelope protein plasmid pMD2G (plasmid 12259; Addgene). Supernatants were collected after 2 days and added together with polybrene (10 μg/ml) to ventricular explants on collagen gels. Snail mAb was provided by I. Virtanen (University of Helsinki, Helsinki, Finland).
Histology, lacZ staining, and in situ hybridization.
See Supplemental Data.
Immunohistochemistry.
See Supplemental Data.
Confocal imaging.
See Supplemental Data.
Statistics.
The TI of AVC and ventricular explants is the ratio of the number of scattered cells (2D migration) or the number of cells invading the collagen gel (3D migration) to the total number of cells (2D cells + 3D cells + nonmigrating ENCs) as follows: 2D TI = 2D/(2D + 3D + nonmigrating ENCs); 3D TI = 3D/(2D + 3D + nonmigrating ENCs).
Cell numbers were determined by counting DAPI-stained nuclei in the Z stack, distinguishing between 2D migrating cells, 3D migrating cells, and nonmigrating ENCs. At least 15 explants were assayed per genotype and condition.
Results are expressed as mean + SD. An unpaired 2-tailed Student’s t test was performed to assess differences among groups. P < 0.05 was considered significant.
RNA isolation and semiquantitative RT-PCR.
See Supplemental Data. For primers and conditions, see Supplemental Table 3.
Microarray hybridization and scanning.
See Supplemental Data.
Western blot.
See Supplemental Data.
Supplementary Material
Supplemental data
Supplemental video 1
Supplemental video 2
Supplemental video 3
Supplemental video 4
Supplemental Excel file 5
Acknowledgments
We thank D.A. Melton for the R26N1ICD mice; R.P. Harvey for the Nkx2.5-Cre mice; S. Evans for the cTnT-Cre driver lines; V. Rosen for the Bmp2 conditional lines; I. Virtanen for the Snail mAb; A. García-Herreros, F.J. Díaz-Benjumea and D. MacGrogan for helpful discussions; M. Manzanares for critical reading of the manuscript; and S. Bartlett for English editing. L. Luna-Zurita is supported by a fellowship linked to grant LSHM-CT-2005-018630 (HeartRepair, European Union FP6) and the Spanish Society of Cardiology. The CNIC is supported by the Spanish Ministry of Science and Innovation and the Pro-CNIC Foundation. This work was funded by grants SAF2007-62445 (Spanish Ministry of Science and Innovation), P-2006/BIO-194 (Regional Government of Madrid), RD06/0014/0038 and RD06/0010/1013 (Spanish Ministry of Health), and LSHM-CT-2005-018630 (HeartRepair, European Union FP6) to J.L. de la Pompa.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(10):3493–3507. doi:10.1172/JCI42666.
Joaquim Grego-Bessa’s present address is: Sloan-Kettering Institute, New York, New York, USA.
References
- 1.Loffredo CA. Epidemiology of cardiovascular malformations: prevalence and risk factors. Am J Med Genet. 2000;97(4):319–325. doi: 10.1002/1096-8628(200024)97:4<319::AID-AJMG1283>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 2.Siu SC, Silversides CK. Bicuspid aortic valve disease. J Am Coll Cardiol. 2010;55(25):2789–2800. doi: 10.1016/j.jacc.2009.12.068. [DOI] [PubMed] [Google Scholar]
- 3.Joziasse IC, et al. Genes in congenital heart disease: atrioventricular valve formation. Basic Res Cardiol. 2008;103(3):216–227. doi: 10.1007/s00395-008-0713-4. [DOI] [PubMed] [Google Scholar]
- 4.Garg V, et al. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437(7056):270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 5.Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. doi: 10.1016/S0074-7696(05)43005-3. [DOI] [PubMed] [Google Scholar]
- 6.Markwald RR, Fitzharris TP, Manasek FJ. Structural development of endocardial cushions. Am J Anat. 1977;148(1):85–119. doi: 10.1002/aja.1001480108. [DOI] [PubMed] [Google Scholar]
- 7.Boyer AS, Ayerinskas II, Vincent EB, McKinney LA, Weeks DL, Runyan RB. TGFbeta2 and TGFbeta3 have separate and sequential activities during epithelial-mesenchymal cell transformation in the embryonic heart. Dev Biol. 1999;208(2):530–545. doi: 10.1006/dbio.1999.9211. [DOI] [PubMed] [Google Scholar]
- 8.Camenisch TD, et al. Temporal and distinct TGFbeta ligand requirements during mouse and avian endocardial cushion morphogenesis. Dev Biol. 2002;248(1):170–181. doi: 10.1006/dbio.2002.0731. [DOI] [PubMed] [Google Scholar]
- 9.Sugi Y, Yamamura H, Okagawa H, Markwald RR. Bone morphogenetic protein-2 can mediate myocardial regulation of atrioventricular cushion mesenchymal cell formation in mice. Dev Biol. 2004;269(2):505–518. doi: 10.1016/j.ydbio.2004.01.045. [DOI] [PubMed] [Google Scholar]
- 10.Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132(24):5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- 11.Rivera-Feliciano J, Tabin CJ. Bmp2 instructs cardiac progenitors to form the heart-valve-inducing field. Dev Biol. 2006;295(2):580–588. doi: 10.1016/j.ydbio.2006.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Timmerman LA, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Monte G, Grego-Bessa J, Gonzalez-Rajal A, Bolos V, De La Pompa JL. Monitoring Notch1 activity in development: Evidence for a feedback regulatory loop. Dev Dyn. 2007;236(9):2594–2614. doi: 10.1002/dvdy.21246. [DOI] [PubMed] [Google Scholar]
- 15.Kokubo H, et al. Targeted disruption of hesr2 results in atrioventricular valve anomalies that lead to heart dysfunction. Circ Res. 2004;95(5):540–547. doi: 10.1161/01.RES.0000141136.85194.f0. [DOI] [PubMed] [Google Scholar]
- 16.Kokubo H, Miyagawa-Tomita S, Nakazawa M, Saga Y, Johnson RL. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev Biol. 2005;278(2):301–309. doi: 10.1016/j.ydbio.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 17.Fischer A, et al. Combined loss of Hey1 and HeyL causes congenital heart defects because of impaired epithelial to mesenchymal transition. Circ Res. 2007;100(6):856–863. doi: 10.1161/01.RES.0000260913.95642.3b. [DOI] [PubMed] [Google Scholar]
- 18.Beis D, et al. Genetic and cellular analyses of zebrafish atrioventricular cushion and valve development. Development. 2005;132(18):4193–4204. doi: 10.1242/dev.01970. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe Y, et al. Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development. 2006;133(9):1625–1634. doi: 10.1242/dev.02344. [DOI] [PubMed] [Google Scholar]
- 20.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci U S A. 2003;100(25):14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230(2):230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 22.Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol. 1999;216(1):72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- 23.Krebs LT, et al. Notch signaling is essential for vascular morphogenesis in mice. Genes Dev. 2000;14(11):1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 24.Lamar E, et al. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 2001;15(15):1885–1899. doi: 10.1101/gad.908101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palomero T, et al. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103(48):18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeller R, Bloch KD, Williams BS, Arceci RJ, Seidman CE. Localized expression of the atrial natriuretic factor gene during cardiac embryogenesis. Genes Dev. 1987;1(7):693–698. doi: 10.1101/gad.1.7.693. [DOI] [PubMed] [Google Scholar]
- 27.Palmer S, et al. The small muscle-specific protein Csl modifies cell shape and promotes myocyte fusion in an insulin-like growth factor 1-dependent manner. J Cell Biol. 2001;153(5):985–998. doi: 10.1083/jcb.153.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christoffels VM, Keijser AG, Houweling AC, Clout DE, Moorman AF. Patterning the embryonic heart: identification of five mouse Iroquois homeobox genes in the developing heart. Dev Biol. 2000;224(2):263–274. doi: 10.1006/dbio.2000.9801. [DOI] [PubMed] [Google Scholar]
- 29.Jamora C, et al. A signaling pathway involving TGF-beta2 and snail in hair follicle morphogenesis. PLoS Biol. 2005;3(1):e11. doi: 10.1371/journal.pbio.0030011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akiyama H, et al. Essential role of Sox9 in the pathway that controls formation of cardiac valves and septa. Proc Natl Acad Sci U S A. 2004;101(17):6502–6507. doi: 10.1073/pnas.0401711101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niessen K, Fu Y, Chang L, Hoodless PA, McFadden D, Karsan A. Slug is a direct Notch target required for initiation of cardiac cushion cellularization. . J Cell Biol. 2008;182(2):315–325. doi: 10.1083/jcb.200710067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ansieau S, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14(1):79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 33.Kim RY, Robertson EJ, Solloway MJ. Bmp6 and Bmp7 are required for cushion formation and septation in the developing mouse heart. Dev Biol. 2001;235(2):449–466. doi: 10.1006/dbio.2001.0284. [DOI] [PubMed] [Google Scholar]
- 34.Camenisch TD, et al. Disruption of hyaluronan synthase-2 abrogates normal cardiac morphogenesis and hyaluronan-mediated transformation of epithelium to mesenchyme. J Clin Invest. 2000;106(3):349–360. doi: 10.1172/JCI10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, et al. Roles of Chemokine Receptor 4 (CXCR4) and Chemokine Ligand 12 (CXCL12) in Metastasis of Hepatocellular Carcinoma Cells. Cell Mol Immunol. 2008;5(5):373–378. doi: 10.1038/cmi.2008.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Runyan RB, Markwald RR. Invasion of mesenchyme into three-dimensional collagen gels: a regional and temporal analysis of interaction in embryonic heart tissue. Dev Biol. 1983;95(1):108–114. doi: 10.1016/0012-1606(83)90010-6. [DOI] [PubMed] [Google Scholar]
- 37.Kruzynska-Frejtag A, Machnicki M, Rogers R, Markwald RR, Conway SJ. Periostin (an osteoblast-specific factor) is expressed within the embryonic mouse heart during valve formation. Mech Dev. 2001;103(1–2):183–188. doi: 10.1016/S0925-4773(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 38.Romano LA, Runyan RB. Slug is an essential target of TGFbeta2 signaling in the developing chicken heart. Dev Biol. 2000;223(1):91–102. doi: 10.1006/dbio.2000.9750. [DOI] [PubMed] [Google Scholar]
- 39.Aybar MJ, Nieto MA, Mayor R. Snail precedes slug in the genetic cascade required for the specification and migration of the Xenopus neural crest. Development. 2003;130(3):483–494. doi: 10.1242/dev.00238. [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, et al. Ephrin-B2 controls VEGF-induced angiogenesis and lymphangiogenesis. Nature. 2010;465(7297):483–486. doi: 10.1038/nature09002. [DOI] [PubMed] [Google Scholar]
- 41.Dovey HF, et al. Functional gamma-secretase inhibitors reduce beta-amyloid peptide levels in brain. . J Neurochem. 2001;76(1):173–181. doi: 10.1046/j.1471-4159.2001.00012.x. [DOI] [PubMed] [Google Scholar]
- 42.Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133(21):4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kokubo H, Tomita-Miyagawa S, Hamada Y, Saga Y. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development. 2007;134(4):747–755. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- 44.Stanley EG, et al. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3’UTR-ires-Cre allele of the homeobox gene Nkx2-5. Int J Dev Biol. 2002;46(4):431–439. [PubMed] [Google Scholar]
- 45.Jiao K, et al. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17(19):2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou BP, et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat Cell Biol. 2004;6(10):931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 47.de Jesus Perez VA, et al. Bone morphogenetic protein 2 induces pulmonary angiogenesis via Wnt-beta-catenin and Wnt-RhoA-Rac1 pathways. J Cell Biol. 2009;184(1):83–99. doi: 10.1083/jcb.200806049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bachelder RE, Yoon SO, Franci C, de Herreros AG, Mercurio AM. Glycogen synthase kinase-3 is an endogenous inhibitor of Snail transcription: implications for the epithelial-mesenchymal transition. . J Cell Biol. 2005;168(1):29–33. doi: 10.1083/jcb.200409067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsuji K, et al. BMP2 activity, although dispensable for bone formation, is required for the initiation of fracture healing. Nat Genet. 2006;38(12):1424–1429. doi: 10.1038/ng1916. [DOI] [PubMed] [Google Scholar]
- 50.Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci U S A. 2008;105(17):6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Venkatesh DA, Park KS, Harrington A, Miceli-Libby L, Yoon JK, Liaw L. Cardiovascular and hematopoietic defects associated with Notch1 activation in embryonic Tie2-expressing populations. Circ Res. 2008;103(4):423–431. doi: 10.1161/CIRCRESAHA.108.177808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grego-Bessa J, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12(3):415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol. 2002;39(12):1890–1900. doi: 10.1016/s0735-1097(02)01886-7. [DOI] [PubMed] [Google Scholar]
- 54.Ii M, et al. Notch signaling regulates endothelial progenitor cell activity during recovery from arterial injury in hypercholesterolemic mice. Circulation. 2010;121(9):1104–1112. doi: 10.1161/CIRCULATIONAHA.105.553917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oka C, et al. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 1995;121(10):3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 56.Salmon P, Trono D. Production and titration of lentiviral vectors. Curr Protoc Neurosci. 2006;Chapter 4:Unit 4.21. doi: 10.1002/0471142301.ns0421s37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data
Supplemental video 1
Supplemental video 2
Supplemental video 3
Supplemental video 4
Supplemental Excel file 5