RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex (original) (raw)
. Author manuscript; available in PMC: 2010 Nov 25.
SUMMARY
Compelling evidence indicates that the CRISPR-Cas system protects prokaryotes from viruses and other potential genome invaders. This adaptive prokaryotic immune system arises from the clustered regularly interspaced short palindromic repeats (CRISPRs) found in prokaryotic genomes, which harbor short invader-derived sequences, and the CRISPR-associated (Cas) protein-coding genes. Here we have identified a CRISPR-Cas effector complex that is comprised of small invader-targeting RNAs from the CRISPR loci (termed prokaryotic silencing (psi)RNAs) and the RAMP module (or Cmr) Cas proteins. The psiRNA-Cmr protein complexes cleave complementary target RNAs at a fixed distance from the 3' end of the integral psiRNAs. In Pyrococcus furiosus, psiRNAs occur in two size forms that share a common 5' sequence tag but have distinct 3' ends that direct cleavage of a given target RNA at two distinct sites. Our results indicate that prokaryotes possess a unique RNA silencing system that functions by homology-dependent cleavage of invader RNAs.
INTRODUCTION
RNAs that arise from the clustered regularly interspaced short palindromic repeats (CRISPRs) found in prokaryotic genomes are hypothesized to guide proteins encoded by CRISPR-associated (cas) genes to silence potential genome invaders in prokaryotes (Makarova et al., 2006). CRISPRs consist of multiple copies of a short repeat sequence (typically 25 – 40 nucleotides) separated by similarly-sized variable sequences that are derived from invaders such as viruses and conjugative plasmids (Godde and Bickerton, 2006; Lillestol et al., 2006; Makarova et al., 2006; Mojica et al., 2005; Pourcel et al., 2005; Sorek et al., 2008; Tyson and Banfield, 2008). CRISPR loci are found in nearly all sequenced archaeal genomes and approximately half of bacterial genomes (Godde and Bickerton, 2006; Haft et al., 2005; Makarova et al., 2006). cas genes are strictly found in the genomes of prokaryotes that possess CRISPRs, frequently in operons in close proximity to the CRISPR loci (Haft et al., 2005; Jansen et al., 2002; Makarova et al., 2006). Over 40 cas genes have been described, a subset of which is found in any given organism (Haft et al., 2005; Jansen et al., 2002; Makarova et al., 2006). The proteins encoded by the cas genes include predicted RNA binding proteins, endo- and exo-nucleases, helicases, and polymerases (Haft et al., 2005; Jansen et al., 2002; Makarova et al., 2006). Recent studies have demonstrated that CRISPRs and cas genes function in invader defense in prokaryotes. Exposure of microorganisms that possess the CRISPR-Cas system to a virus results in the appearance of new virus-derived sequences at the leader-proximal end of CRISPR loci in the genomes of surviving individuals (Barrangou et al., 2007; Deveau et al., 2008). Moreover, the acquisition or loss of invader-specific CRISPR elements or of Cas protein genes has been directly correlated with virus and plasmid resistance or sensitivity, respectively (Barrangou et al., 2007; Brouns et al., 2008; Deveau et al., 2008). This rapidly evolving immune system influences the ecology of natural microbial populations (Andersson and Banfield, 2008; Heidelberg et al., 2009; Tyson and Banfield, 2008).
RNAs from the CRISPR loci are hypothesized to guide the CRISPR-Cas defense response based on their potential to base pair with invading nucleic acids. Available data indicate that entire CRISPR loci are transcribed from the leader region, producing primary transcripts containing the full set of CRISPR repeats and embedded invader-derived (or guide) sequences (Hale et al., 2008; Jansen et al., 2002; Lillestol et al., 2006; Lillestol et al., 2009; Tang et al., 2002; Tang et al., 2005). These large precursor RNAs are processed (or diced) into shorter (∼60–70 nucleotide) intermediate RNAs that contain individual invader-targeting sequences (∼25–40 nucleotides) by Cas endonucleases that cleave within the repeats (Brouns et al., 2008; Carte et al., 2008). However, the ultimate products of the CRISPR loci appear to be smaller RNAs (Brouns et al., 2008; Hale et al., 2008; Lillestol et al., 2009). In Pyrococcus furiosus, the most abundant CRISPR RNAs are two species of ∼45 nucleotides and ∼39 nucleotides (Hale et al., 2008). These small, abundant products of the CRISPR loci are thought to be the prokaryotic silencing (psi)RNAs of the CRISPR-Cas RNA silencing pathway (Brouns et al., 2008; Hale et al., 2008; Makarova et al., 2006).
Intriguingly, the protein-mediated functions of the CRISPR-Cas system are apparently carried out by distinct sets of Cas proteins in different organisms (Haft et al., 2005). Six “core” CRISPR-associated genes (cas1 - cas 6) are found in many and diverse organisms, however, most organisms have only a subset of these 6 genes and only cas1 is present in nearly all organisms that appear to possess the system (Haft et al., 2005; Makarova et al., 2006). Furthermore, the core cas genes in a given organism are complemented by one or more sets of additional cas genes: the cse, csy, csn, csd, cst, csh, csa, csm and cmr genes (Haft et al., 2005). These sets are comprised of 2 to 6 CRISPR-associated genes that co-segregate, and are mostly designated for a prototypical organism (e.g. the cse or Cas subtype Escherichia coli genes) (Haft et al., 2005). (The cmr (Cas module RAMP) gene set is named for its 4 RAMP (repeat-associated mysterious proteins; see below) gene members.) E. coli K12, for example, has 3 core cas genes and the full set of 5 cse genes (which includes the E. coli subtype member of the core Cas5 gene family, cas5e) (Brouns et al., 2008). Phylogenetic analyses suggest that the cas genes are distributed by lateral gene transfer (Haft et al., 2005; Jansen et al., 2002; Makarova et al., 2002). The functional consequences of the differences in the complement of Cas proteins found among organisms are not yet known.
Functional classes have been predicted for many of the Cas proteins based on sequence, but very few of the proteins have been characterized. Only one of the core Cas proteins, Cas6, has a clearly established function which is to process precursor CRISPR RNAs to release individual invader-targeting RNAs (Carte et al., 2008). Cas1 was recently shown to be a DNA-specific endonuclease with properties consistent with a role in processing invader DNA into fragments that become incorporated into CRISPR loci (Wiedenheft et al., 2009). The five E. coli subtype Cas proteins (Cse1–4 and Cas5e (Haft et al., 2005)) have been shown to form a complex that processes precursor CRISPR RNAs in E. coli (which lacks Cas6) (Brouns et al., 2008). Many of the Cas proteins are members of the large superfamily of RAMP proteins, which have features of RNA binding proteins (Haft et al., 2005; Makarova et al., 2002; Makarova et al., 2006). At least a few of the RAMPs (including for example Cas6) have been found to possess previously unpredicted nuclease activity (Beloglazova et al., 2008; Brouns et al., 2008; Carte et al., 2008). The Cas proteins are expected to function in various aspects of maintenance of CRISPR gene loci (including addition of new invader-derived elements in response to infection) as well as psiRNA biogenesis and psiRNA-mediated resistance to invaders.
While there is very strong evidence that CRISPR RNAs and Cas proteins function to silence potential invaders in prokaryotes (Barrangou et al., 2007; Brouns et al., 2008; Deveau et al., 2008), the effector complexes and silencing mechanisms of the CRISPR-Cas pathway remain unknown. Recent studies in Staphylococcus species and E. coli (Brouns et al., 2008; Marraffini and Sontheimer, 2008) indicate that the CRISPR-Cas systems present in those organisms (comprised of the Csm or Cse proteins and several core Cas proteins, respectively) target invader DNA rather than RNA, but the effectors and mechanisms of silencing in these organisms remain unknown. The results presented here demonstrate that the Cmr or RAMP module proteins function with mature psiRNAs to cleave target RNAs. These findings define psiRNA-guided RNA cleavage as a mechanism for the function of the CRISPR-Cas system in organisms that possess the RAMP module of Cas proteins.
RESULTS
Isolation of a complex containing mature psiRNAs and a subset of Cas proteins
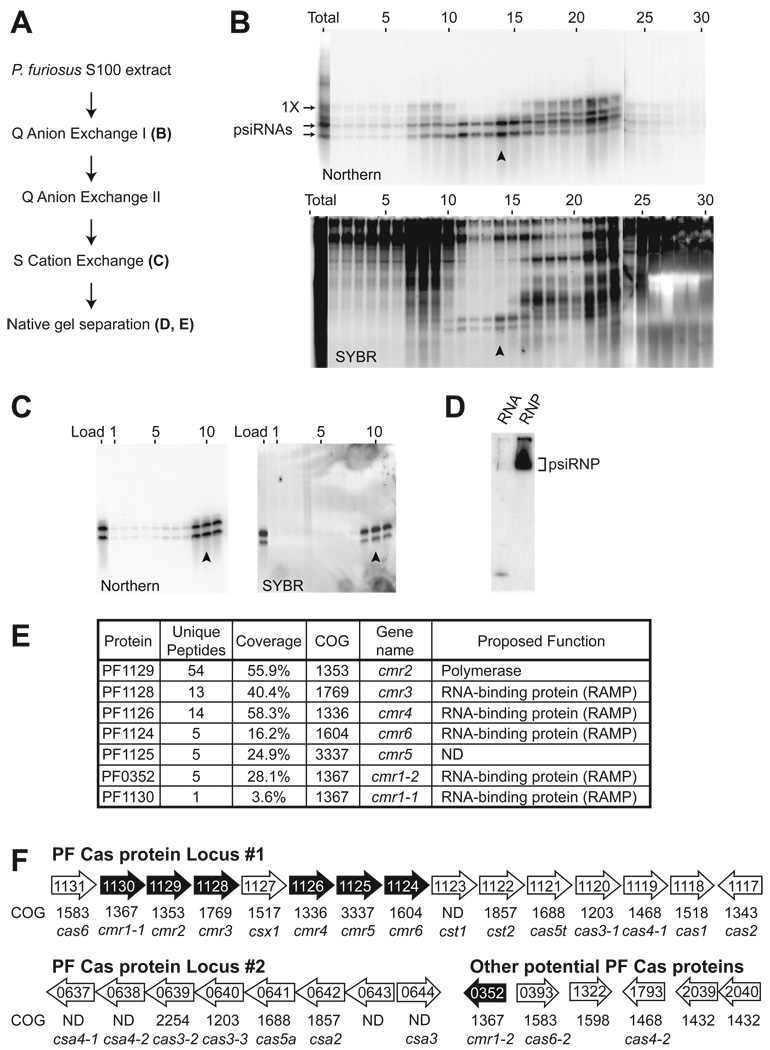
PsiRNAs are hypothesized to guide Cas proteins to effect invader silencing in prokaryotes (Brouns et al., 2008; Hale et al., 2008; Makarova et al., 2006). P. furiosus is a hyperthermophilic archaeon whose genome encodes 200 potential psiRNAs (organized in seven CRISPR loci) and at least 29 potential Cas proteins (largely found in 2 gene clusters), including members of all 6 core Cas protein families and 3 sets of additional Cas proteins: the Cmr, Cst and Csa proteins (see Figure 1F). In P. furiosus, most psiRNAs are processed into 2 species of ∼45 nucleotides and ∼39 nucleotides (Hale et al., 2008). To gain insight into the functional components of the CRISPR-Cas invader defense pathway, we isolated complexes containing the mature psiRNA species from P. furiosus cellular extract on the basis of psiRNA fractionation profiles (Figure 1). The doublet of psiRNAs, detectable both by Northern blotting of an individual psiRNA and total RNA staining (SYBR), was purified away from larger CRISPR-derived RNAs (including the 1X intermediate; Hale, 2008) as well as other cellular RNAs (Figure 1C).
Figure 1. Identification of a ribonucleoprotein complex containing psiRNAs and Cas proteins.
A) psiRNP purification scheme. Letters indicate the location of corresponding data within Figure 1. B) psiRNA (top panel) and total RNA (bottom panel) profiles across the initial Q-sepharose anion exchange fractions and an unfractionated sample (total). Northern analysis (top panel) was performed for P. furiosus psiRNA 7.01. The positions of the mature psiRNAs and 1X intermediate RNA (Hale et al., 2008) are indicated. The lower panel shows all RNAs detected by SYBR Gold staining. The peak fraction is indicated by an arrow in each panel. C) psiRNA (Northern analysis of psiRNA 7.01, left panel) and total RNA (SYBR staining, right panel) profiles across the S-sepharose cation exchange fractions and starting material (load). The peak fraction is indicated by an arrow in each panel. D) Native gel Northern analysis of the psiRNP. The peak S-sepharose fraction (arrow, C) was fractionated by native gel electrophoresis and analyzed by Northern blotting for psiRNA 7.01. RNA extracted from the same fraction was co-analyzed. The position of the psiRNP is indicated. E) Cas proteins identified by tandem mass spectrometry. The isolated psiRNP (D) was subject to in-gel trypsin digestion and tandem mass spectrometry. Sequence coverage and the number of unique peptides for Cas proteins identified with 99% confidence are shown. P. furiosus cas gene names are as given (Haft et al., 2005) and proposed functions are as predicted (Haft et al., 2005; Makarova et al., 2006). F) Genome organization of predicted P. furiosus cas genes. Operon organization and COG assignments were adapted from NCBI database. Core cas genes (cas) and Cas module-RAMP (cmr), Cas subtype Apern (csa) and Cas subtype Tneap (cst) genes are indicated. Proteins identified by mass spectrometry are indicated in black.
To determine whether the psiRNAs are components of RNA-protein complexes in the purified fraction (Figure 1C), we performed native gel northern analysis. The mobility of the psiRNAs on native gel electrophoresis was reduced in the purified fraction relative to a sample from which proteins were extracted (Figure 1D), indicating the presence of psiRNA-protein complexes in the purified fraction. We gel purified the psiRNA-containing complex from the native gel and analyzed the sample by mass spectrometry. The sample contained a mixture of proteins that included seven Cas proteins identified with 99% confidence: Cmr1-1, Cmr1–2, Cmr2, Cmr3, Cmr4, Cmr5, and Cmr6 (Figure 1E).
The identities of the non-Cas proteins found in the sample are listed in Supplemental Table 1. Analysis of a native gel-purified psiRNP obtained by an alternate chromatography scheme revealed a similar Cas protein profile (Cmr2, Cmr3, Cmr4, and Cmr6), but few common non-Cas proteins (Supplemental Table 1). The five common co-purifying non-Cas proteins are denoted in Supplemental Table 1 None of these proteins has any known link to the CRISPR-Cas system.
Remarkably, the seven Cas proteins associated with the complex are all encoded by the tightly linked RAMP module or cmr genes (Haft et al., 2005). Moreover, the identified proteins comprise the complete set of Cmr proteins (Haft et al., 2005). (The independently defined “polymerase cassette” is closely related to the RAMP module (Makarova et al., 2006).) There are 6 cmr genes: cmr2 encodes a predicted polymerase with HD nuclease domains, and cmr1, cmr3, cmr4, and cmr6 encode repeat-associated mysterious proteins (RAMPs) (Haft et al., 2005; Makarova et al., 2002). The P. furiosus genome contains two cmr1 genes and a single representative of each cmr2 – cmr6, and all seven corresponding proteins were found in the purified psiRNP complex (Figure 1E). The organization of the genes encoding the seven identified proteins is shown in Figure 1F. Six of the seven identified Cas proteins are encoded in a nearly contiguous region of one of the two major cas gene loci in P. furiosus. This locus is located directly adjacent to CRISPR locus 7, and also encodes core Cas proteins Cas1 - Cas4, Cas5t and Cas6. The striking correlation between the evolutionary co-segregation and physical association of the 6 Cmr proteins strongly supports the co-function of the proteins. Our findings indicate that the two mature psiRNA species are components of complexes containing the RAMP module or Cmr proteins in P. furiosus.
psiRNAs possess a 5’ psiRNA-tag sequence
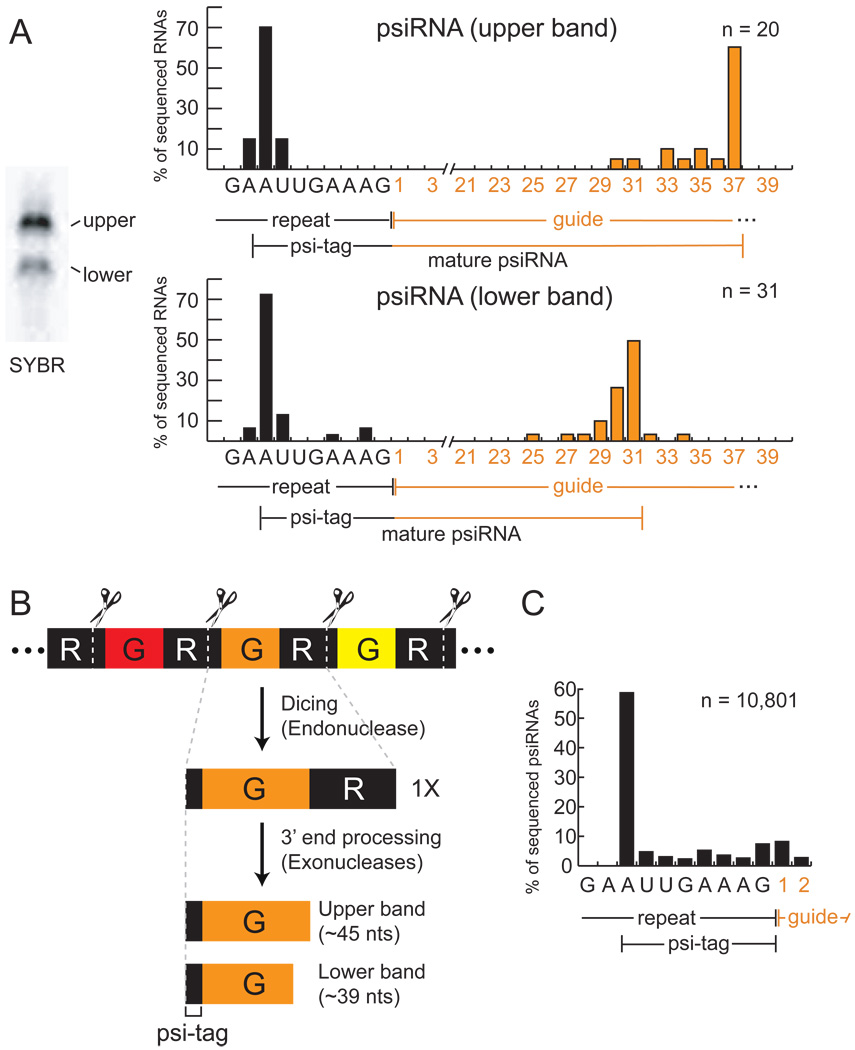
In order to better understand the nature of the two psiRNA species that are components of the purified complexes, each of the two RNA bands present in the final chromatography sample (Figure 2A) was extracted and cloned. We obtained sequences of 51 RNAs (20 from the upper band and 31 from the lower band) that included psiRNAs from all seven P. furiosus CRISPR loci (Supplemental Table 2). Six RNAs with the same guide sequence were represented in both the upper and lower bands, consistent with Northern analysis that has shown that most psiRNAs exist in both size forms (Hale et al., 2008).
Figure 2. psiRNA species in the RNP contain a common 5’ sequence element and distinct 3’ termini.
A) Sequence analysis of RNAs associated with the complex. RNA species present in the S-sepharose fraction (visualized by SYBR Gold staining) are shown in left panel. RNAs in the upper and lower bands were isolated, cloned, and sequenced. Graphs show the percentage of sequenced RNAs with 5’ ends located at specific positions within the repeat sequence (black), and with indicated numbers of guide sequence nucleotides downstream of the repeat sequence (orange). The average guide sequence is 37 nucleotides in P. furiosus. A consensus for each psiRNA species is diagrammed under each graph. The 8-nucleotide repeat sequence found at the 5’ end of the majority of the psiRNAs is indicated as the psi-tag. B) Model for biogenesis of the two psiRNA species in P. furiosus. CRISPR locus transcripts containing alternating repeat (R, black segments) and guide (G, colored segments) elements are cleaved at a specific site within the repeat by the Cas6 endoribonuclease (Carte et al., 2008), ultimately producing 1X intermediate RNAs that contain a full invader-targeting sequence flanked on both sides by segments of the repeat. The mature RNAs retain the 5’ end repeat sequence (psi-tag). Uncharacterized 3’ end processing of the 1X intermediate by endo- and/or exo-nucleases forms the two major mature psiRNAs: a 45-nucleotide species that contains the 8-nucleotide psi-tag and a full guide sequence, and a 39-nucleotide species that contains a shorter 31-nucleotide guide sequence. C) Deep sequencing of small RNAs from P. furiosus confirms the presence of the psi-tag. The 5’ ends of the sequenced psiRNAs are graphed as in A. The number of total clones analyzed (n) is indicated in the graphs of panels A and C.
The cloned psiRNAs consisted primarily of an individual guide (invader-targeting or “spacer”) sequence, however, all of the clones retained a portion of the common repeat sequence at the 5’ end. Indeed, the majority (∼70%) of the RNAs in both bands contained an identical 5’ end consisting of an 8-nucleotide segment of the repeat sequence (Figure 2A). The difference between the two psiRNA size forms was found at the 3’ ends. Downstream of the repeat sequence, the majority of the clones from the top band contained 37 nucleotides of guide sequence (the full length of a typical guide element in P. furiosus) (Figure 2A, top panel). The 3’ ends of most of the clones from the bottom band were located within the guide sequence. The majority of these RNAs contained 31 nucleotides of guide sequence downstream of the repeat sequence (Figure 2A, bottom panel).
The psiRNAs are processed from long CRISPR locus transcripts (Brouns et al., 2008; Hale et al., 2008; Lillestol et al., 2006; Lillestol et al., 2009; Tang et al., 2002; Tang et al., 2005) (Figure 2B). In P. furiosus, the Cas6 endoribonuclease cleaves CRISPR RNAs at a site within the repeat element located 8 nucleotides upstream of the guide sequence, generating the precise 5’ end observed in the two psiRNA species found in the complex (Figure 2B; (Carte et al., 2008)). Our results indicate that the 5’ end generated by the Cas6 endoribonuclease is maintained in the mature psiRNAs, but that the RNAs undergo further processing at the 3’ end to generate psiRNAs that contain either ∼37 or ∼31 nucleotides of guide sequence (Figure 2B). The mechanism that defines the two distinct 3’ end boundaries is not known. The larger ∼45-nucleotide mature psiRNA species is generally more abundant than the smaller ∼39-nucleotide species ((Hale et al., 2008), Figure 1 and Figure 2A).
The short repeat sequence that remains at the 5’ end of mature psiRNAs in P. furiosus provides a common identifying sequence tag for the psiRNAs that could function in recognition of the RNAs by the proteins in the CRISPR-Cas pathway. In order to more rigorously delineate the potentially important psiRNA-tag or “psi-tag”, we purified small RNAs from P. furiosus, performed deep sequencing and obtained the sequences of the 5’ ends of more than 10,000 CRISPR-derived RNAs (from loci 1–7). The 5’ ends of the majority of the RNAs mapped 8 nucleotides upstream of the guide sequence (Figure 2C), verifying the presence of a discrete psi-tag on small CRISPR-derived RNAs in P. furiosus.
The sequences of CRISPR repeats (from which psi-tags are derived) are generally conserved within groups of organisms, but can vary widely (Godde and Bickerton, 2006; Kunin et al., 2007). Thus, while the sequence of the psi-tag found on most P. furiosus psiRNAs (AUUGAAAG) can be found in the repeat sequence of numerous organisms, psi-tags of distinct sequence and length would be expected in others. We found evidence to support this prediction in the psiRNAs from P. furiosus CRISPR locus 8, which contains a single nucleotide deletion in the psi-tag region of the repeat. The majority (60%) of the 640 sequenced RNAs that mapped to CRISPR locus 8 possessed a 7-nucleotide AUUGAAG psi-tag. In E. coli, CRISPR transcripts are cleaved by a different endoribonuclease (Cse3 of the Cse complex), which nonetheless appears to generate RNAs with an 8-nucleotide AUAAACCG repeat sequence at the 5’ end (Brouns et al., 2008). An 8-nucleotide ACGAGAAC repeat sequence is also present at the 5’ termini of CRISPR RNAs in S. epidermidis (Marraffini and Sontheimer, 2008), suggesting that the psi-tag is a general feature of the psiRNAs. Interestingly, the distinct CRISPR repeat sequences found in various genomes are accompanied by distinct subsets of Cas proteins (Kunin et al., 2007), which may reflect coupling of specific series of Cas proteins with the psi-tagged RNAs that they recognize.
Homology-dependent cleavage of a target RNA
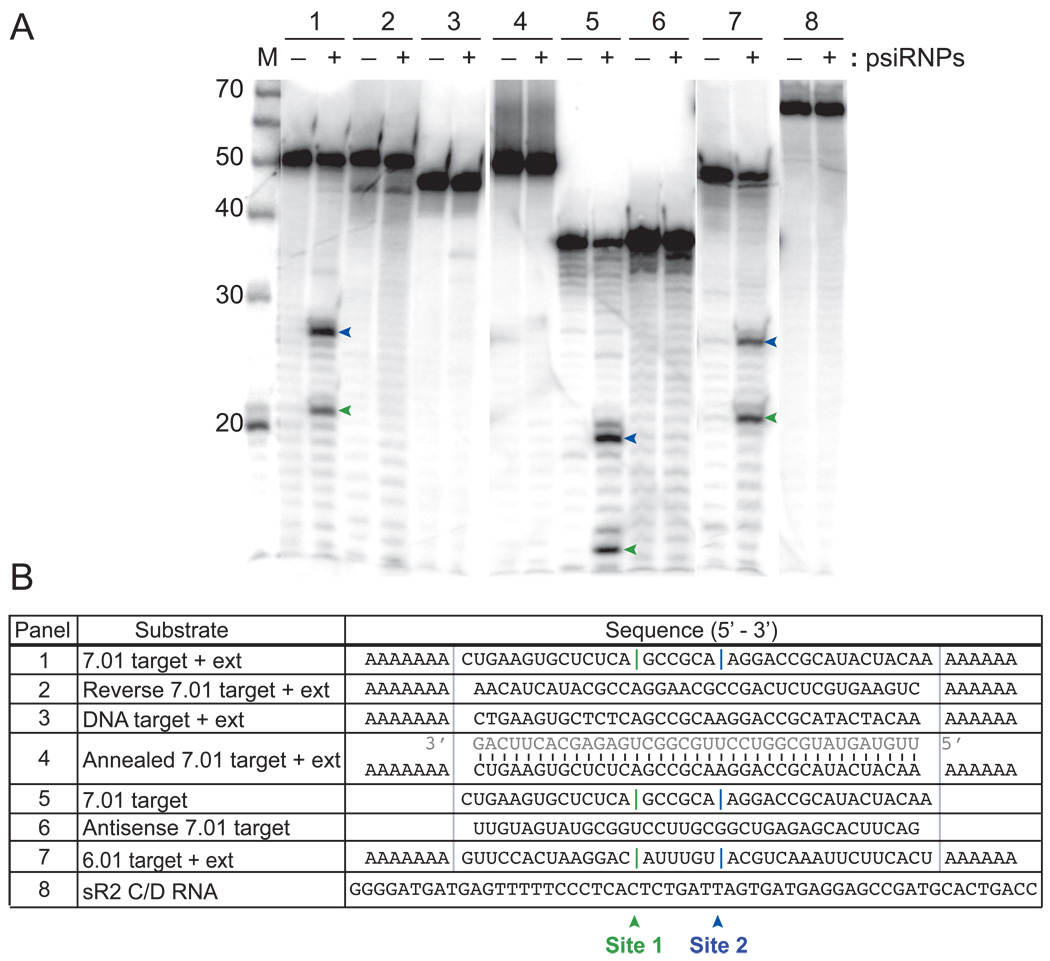
One hypothesis for the mechanism by which CRISPR RNAs and Cas proteins mediate genome defense is psiRNA-guided cleavage of invader nucleic acids (Makarova et al., 2006). Therefore, we tested the ability of the isolated psiRNP complexes to recognize and cleave a labeled RNA and DNA target complementary to endogenous P. furiosus psiRNA 7.01 (first psiRNA encoded in CRISPR locus 7, which Northern analysis indicated is present in the native complexes, see Figure 1). The 5’ end-labeled 7.01 target RNA was cleaved at two sites (site 1 indicated with green vertical line and site 2 indicated with blue vertical line, Figure 3B, panel 1) yielding 5’ end-labeled products of 27 and 21 nucleotides (indicated with corresponding green and blue arrowheads, Figure 3A, panel 1). The single-stranded DNA 7.01 target sequence was not cleaved (Figure 3, panel 3).
Figure 3. Specific cleavage of complementary target RNAs.
The indicated 5’ end-labeled substrates were incubated in the presence (+) or absence (−) of the native psiRNPs (Figure 1C). Products were resolved by denaturing gel electrophoresis. The primary cleavage products are indicated by green and blue arrows (upper panel), and the corresponding sites of cleavage are indicated with green (site 1) and blue (site 2) vertical lines in the substrate sequences shown in the lower panel. The sizes of RNA markers (M) are indicated. “Target” substrates (panels 1, 3, 4, 5, 7) contain regions of perfect complementarity to the guide sequence of the indicated P. furiosus psiRNA. Grey bars demarcate the guide sequences in the lower panel. “+ ext” substrates (panels 1, 2, 3, 4, 7) contain 5’ and 3’ polyA extensions. In panel 4, a synthetic psiRNA (sequence shown in grey) was pre-annealed to the 7.01 target RNA + ext. Panel 2 shows a reverse target sequence substrate and panel 6 shows an antisense (AS) target substrate. Panel 3 shows a DNA substrate; all other substrates are RNA. Panel 8 shows unrelated RNA sR2.
Further characterization of the cleavage activity revealed that the psiRNP complexes cleave the target RNA on the 5’ side of the phosphodiester bond. The 3’ end generated by the complex is not a substrate for polyadenylation (supplemental Figure 1A), indicating the presence of a 3’ phosphate (or 2’, 3’ cyclic phosphate) end. In addition, cleavage activity is lost in the presence of 0.1 mM EDTA indicating that the enzyme depends on divalent cations (supplemental Figure 1B). Activity was restored by the addition of 1 mM Mg2+, Mn2+, Ca2+, Zn2+, Ni2+ or Fe2+ with no detectable change in cleavage sites with any of the metals, but was not supported by Co2+or Cu2+ (supplemental Figure 1B). Cleavage of the target RNA did not require sequences extending beyond the 37-nucleotide region of complementarity with the psiRNA, and occurred at the same two sites in the target RNA lacking sequence extensions (Figure 3, panel 5). No activity was observed toward RNAs that lacked homology with known P. furiosus psiRNAs, including the reverse 7.01 target sequence, antisense 7.01 target sequence, and a box C/D RNA (Figure 3, panels 2, 6, and 8). Pre-annealing a synthetic psiRNA 7.01 to the 7.01 target RNA (to form a double-stranded RNA target) blocked cleavage by the psiRNPs (Figure 3, panel 4). Finally, we tested a target for endogenous P. furiosus psiRNA 6.01 and observed cleavage that generates 2 products of the same sizes observed for the 7.01 target RNA (Figure 3, panel 7).
These results demonstrate the presence of cleavage activity in P. furiosus that is specific for single-stranded RNAs that are complementary to psiRNAs. The activity is associated with a purified fraction that contains 2 mature psiRNA species and 7 RAMP module (Cmr) proteins.
Cleavage of the target RNA occurs a fixed distance from the 3’ end of the psiRNA
To investigate the mechanism of psiRNA-directed RNA cleavage, we analyzed the results of cleavage assays with a series of truncations of the 7.01 target RNA (Figure 4A). We found that the target RNA truncations analyzed did not affect the locations of the two cleavage sites. The full-length 7.01 target RNA is cleaved at sites 1 and 2 to generate 14- and 20-nucleotide 5’ end-labeled products, respectively (Figure 3 and Figure 4A). The 3’ end-truncated target RNAs were cleaved at the same two sites to yield the same two 5’ end-labeled cleavage products (except where truncation eliminated cleavage site 2, Δ20–37, Figure 4A). On the other hand, in the case of the 5’ end-truncated target RNAs, cleavage at the same sites would be expected to generate shorter 5’ end-labeled cleavage products. The 14-nucleotide product that results from cleavage of the Δ1–6 target RNA at site 2 was observed (Figure 4A), but cleavage at site 1 could not be assessed because the size of the product is below that which could be detected in the experiment. If the twelve- and eighteen-nucleotide 5’ end-truncated target RNAs were cleaved at the same two sites, the products would also be outside the range of detection, however, interestingly, very little cleavage of these RNAs was observed (Figure 4, Δ1–18 and Δ1–12, compare substrate band +/- complex).
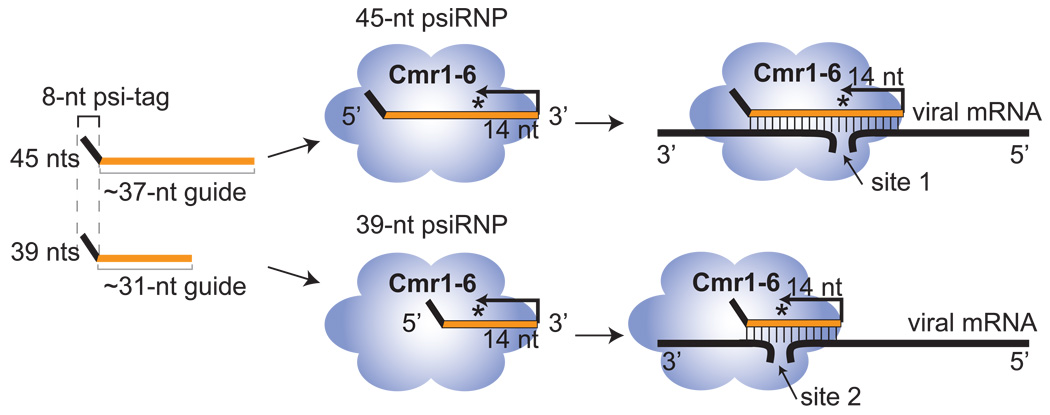
Figure 4. Cleavages occurs 14 nucleotides from the 3’ ends of the psiRNAs.
A)The indicated 5’ end-labeled (*) substrates were incubated in the presence (+) or absence (−) of the psiRNP (Figure 1C). The substrates were full-length 7.01 target RNA (F.L.), and the indicated truncations as diagramed in the lower panel. As in Figure 3, the locations of observed cleavages at sites 1 (green) and 2 (blue) are indicated on the full-length target and truncated RNAs (lower panel) and the corresponding cleavage products are indicated with green and blue arrows in the upper panels. Question mark in the lower panel indicates cleavage that could not be assessed. B) Model for cleavage at two sites directed by two psiRNAs. The 45-nucleotide psiRNA species guides cleavage at site 1 and the 39-nucleotide psiRNA guides cleavage at site 2 on each of the substrate RNAs as indicated. In both cases, cleavage occurs 14 nucleotides from the 3’ end of the psiRNA. Observed products are shown in green (site 1) and blue (site 2) and correspond to products in Figure 3 and Figure 4A.
Strikingly, the difference in the sizes of the two cleavage products observed with the various substrates is the same as the difference in the sizes of the two endogenous psiRNA species (6 nucleotides in both cases, Figure 3). This size difference as well as the specific product sizes suggest that the two cleavages occur a fixed distance (14 nucleotides) from the 3’ ends of the two psiRNAs. Figure 4B illustrates the proposed mechanism by which the 45- and 39-nucleotide psiRNAs guide cleavage at target sites 1 and 2, respectively, for each of the target RNAs analyzed here. For example, using the full-length 7.01 target RNA we observed 20- and 14-nucleotide cleavage products (Figure 3, panel 5) suggesting cleavage of the bound target RNA 14 nucleotides from the 3’ end of the 39- and 45-nucleotide psiRNAs, respectively (Figure 4B, F.L.). In addition, a 7-nucleotide extension at the 5’ end of the target RNA resulted in a pair of 5’ end-labeled products 27 and 21 nucleotides in length (Figure 3, panel 1), consistent with cleavage of the substrate 14 nucleotides from the ends of the two psiRNAs (Figure 4B, F.L.+ext). The anchor for this counting mechanism is the 3’ end of the psiRNA. While reductions in the extent of duplex formation between the 5’ end of the psiRNA and the cleavage site (3’ truncations to within 6 nucleotides of the cleavage site) did not have an observable effect on cleavage efficiency, truncations that reduced duplex formation between the 3’ end of the psiRNA and the cleavage site had a strong negative impact, suggesting that basepairing of the last 14 nucleotides of the psiRNA with the target is critical for cleavage activity.
The results of these studies indicate that both of the mature psiRNA species are active in guiding target RNA cleavage by a mechanism that depends upon the distance from the 3’ end of the psiRNA.
Analysis of reconstituted Cmr-psiRNA complexes
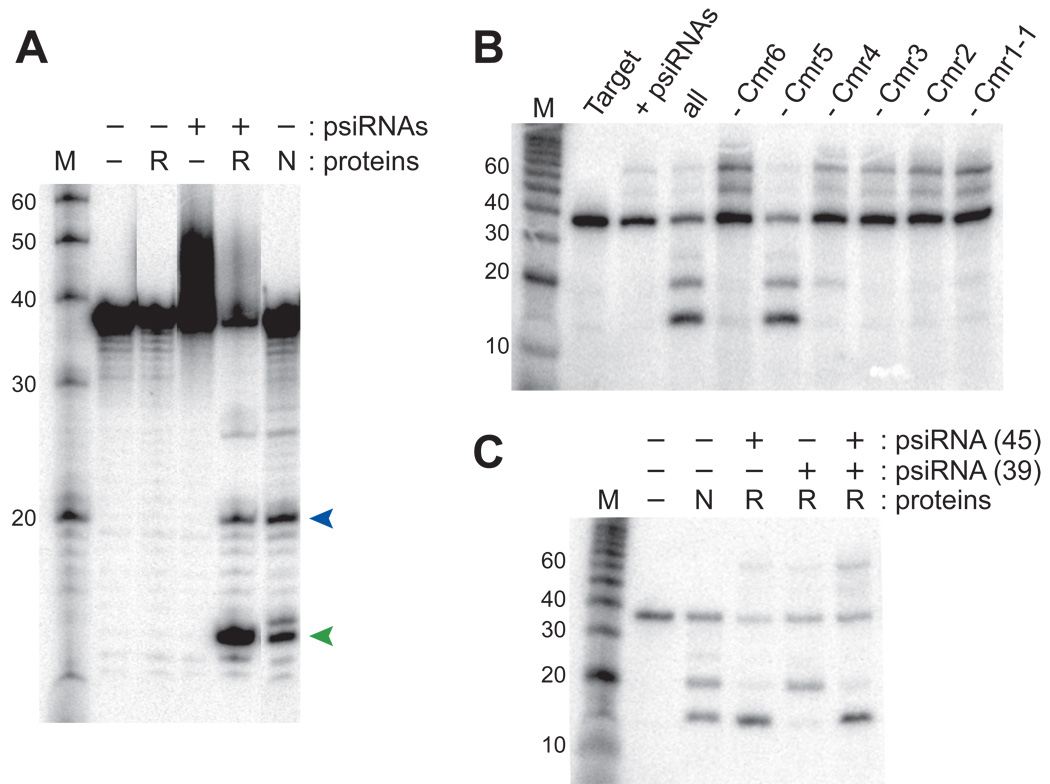
Identification of the Cmr proteins in the purified psiRNP complex (Figure 1) along with the evolutionary evidence for their co-function with the CRISPRs (Haft et al., 2005; Jansen et al., 2002; Makarova et al., 2002) strongly suggests that the Cmr proteins and psiRNAs function as a complex to cleave target RNAs (Figure 3). In order to determine whether the Cmr proteins and psiRNAs are sufficient for function (independent of other co-purifying P. furiosus components), we tested the ability of purified recombinant Cmr proteins and synthetic psiRNAs to cleave target RNAs (Figure 5). A reconstituted set of six P. furiosus Cmr proteins (Cmr1-1, Cmr2 – Cmr6) and two mature psiRNA species (45- and 39-nucleotide psiRNA 7.01, found in the native complex based on Northern analysis (Figure 1) and activity of the native complex against the 7.01 target (Figure 3)) cleaved the target RNA at 2 sites generating the same size products as those observed with the isolated native complex (Figure 5A). While both P. furiosus isoforms of the Cmr1 protein are present in the isolated complexes (Figure 1), we found that only one of the two proteins (Cmr1-1) was required for a functional reconstituted complex (Figure 5A), suggesting that the isoforms may perform redundant functions. No activity was observed in the absence of the psiRNAs or in the absence of the Cmr proteins (Figure 5A), indicating that both are necessary. These results demonstrate that the RAMP module Cas proteins and psiRNAs function together to cleave complementary target RNAs.
Figure 5. Target RNA cleavage requires five Cmr proteins and a single psiRNA species.
A) 5' end-labeled 7.01 target RNA was incubated in the absence of added psiRNAs or proteins (−), in the presence of synthetic psiRNAs (+) or purified recombinant P. furiosus Cmr proteins (R) or in the presence of purified native psiRNPs (N) as indicated. The synthetic psiRNAs were 45- and 39-nucleotide forms of psiRNA 7.01. The six added recombinant Cmr proteins were P. furiosus Cmr1-1, Cmr2, Cmr3, Cmr4, Cmr5 and Cmr6. Products were resolved by denaturing gel electrophoresis. The products corresponding to cleavage at site 1 and site 2 (see Figure 3) are indicated by green and blue arrows, respectively. The sizes of RNA markers (M) are indicated. B) The 7.01 target RNA (Target) was incubated with the synthetic 7.01 psiRNAs (both 45-and 39-nucleotide species) in the absence (+ psiRNAs) and presence of the purified recombinant P. furiosus Cmr proteins (all), and also with combinations of proteins lacking individual Cmr proteins as indicated (e.g. - Cmr6). C) Cleavage activity of the recombinant psiRNP (R) reconstituted with either the individual 7.01 psiRNA species (45- or 39-nt) or both. Cleavage by the native psiRNP (N) is included for comparison.
In order to determine whether all of the six Cmr proteins are essential for psiRNA-guided RNA cleavage, we assayed cleavage activity in the absence of each of the individual proteins (Figure 5B). Omission of Cmr5 did not observably affect the activity of the complex (Figure 5B). However, cleavage was significantly reduced in the absence of any one of the other proteins (Figure 5B), indicating that 5 of the 6 RAMP module proteins are required for activity of the psiRNA-Cmr protein complex.
Finally, we had reconstituted the same cleavage activity profile observed for the native complexes using both psiRNA species (45- and 39-nucleotides) (e.g. Figure 5A). Our model for the mechanism of cleavage predicts that each of the psiRNAs guides a distinct cleavage: the 45-nucleotide psiRNA at site 1, and the 39-nucleotide psiRNA at site 2 (see Figure 4B). To determine whether both psiRNAs are required for activity, and whether each guides the distinct cleavage that is predicted by the model, we tested the activity of complexes reconstituted with a single psiRNA. As predicted, we found that the 45-nucleotide psiRNA guided cleavage at site 1 producing a 14-nucleotide 5’ end-labeled product, and the 39-nucleotide psiRNA guided cleavage at site 2 producing a 20-nucleotide 5’ end-labeled product (Figure 5C). Based on our truncation analysis (Figure 4, Δ20–37), the larger product of the cleavage guided by the 39-nucleotide psiRNA could act as a substrate for cleavage guided by the 45-nucleotide psiRNA, and consistent with this, we often obtain more of the smaller cleavage product in cleavage assays using both the native complex and the reconstituted complex containing both psiRNAs (e.g. Figure 5A). The results of these experiments demonstrate that each of the psiRNA species is competent to form functional psiRNPs and guides cleavage 14 nucleotides from its 3’ end.
DISCUSSION
The findings presented here reveal the mechanism of action of an RNA-protein complex implicated in a novel RNA silencing pathway that functions in invader defense in prokaryotes. Previous work had shown that both invader-specific sequences within CRISPRs and Cas protein genes are important in virus and plasmid resistance in prokaryotes (Barrangou et al., 2007; Brouns et al., 2008; Deveau et al., 2008; Marraffini and Sontheimer, 2008). The results presented here establish how small RNAs from CRISPRs and the RAMP module Cas proteins function together to destroy RNAs recognized by the CRISPR RNAs. The major findings and models established in this work are summarized in Figure 6.
Figure 6. Model for the function of psiRNA-Cmr protein complexes in silencing molecular invaders.
Based on the results of this study, a psiRNA with a conserved 5’ sequence element derived from the CRISPR repeat (psi-tag) and a region of invader-targeting sequence assembles with six Cas module-RAMP proteins (Cmr1-6). The assembled psiRNP interacts with an invader RNA (e.g., viral mRNA) through base pairing between the psiRNA and invader RNA, positioning the region of the RNA-RNA duplex 14 nucleotides from the 3’ end of the psiRNA in proximity to the active site (star) of the enzyme. In P. furiosus, there are two prominent size forms of each psiRNA with different 3’ ends that guide cleavage of viral mRNAs at two distinct sites. There are also two Cmr1 proteins in P. furiosus that are both found in purified preparations and likely function redundantly.
Our findings indicate that the RAMP module of the CRISPR-Cas system silences invaders by psiRNA-guided cleavage of invader RNAs (Figure 6). Specifically, the results indicate that psiRNAs present in complexes with the Cmr proteins recognize and bind an invader RNA such as a viral mRNA (via the psiRNA guide sequence co-opted from the invader by another branch of the CRISPR-Cas system), and that the complex then cleaves the invader RNA, destroying the message and presumably blocking the viral life cycle. The psiRNA-Cmr complexes cleave complementary RNAs (Figure 3 and Figure 5). Five of the six Cmr proteins are required for target RNA cleavage (Figure 5) and the component of the complex that provides catalytic activity remains to be determined. Cmr2 contains a predicted nuclease domain (Makarova et al., 2002; Makarova et al., 2006), however the other four essential proteins (Cmr1, 3, 4 and 6) belong to the RAMP superfamily, members of which have been found to be ribonucleases (Beloglazova et al., 2008; Brouns et al., 2008; Carte et al., 2008). It will be important in future work to identify the catalytic component(s) of the psiRNA-Cmr protein complex. Our data indicate that the Cmr ribonuclease generates products with 3’ phosphate (or 2’, 3’ cyclic phosphate) and 5’ hydroxy termini and requires divalent metal ions for activity (Supplemental Figure 1).
Our results also establish a simple model for the mechanism of cleavage site selection by the psiRNA-Cmr effector complex - a 14-nucleotide ruler anchored by the 3’ end of the psiRNA (Figure 6). We found that P. furiosus psiRNAs occur in two lengths that share a 5’ psi-tag (derived from the CRISPR repeat) and contain either ∼37 or ∼31 nucleotides of guide sequence (Figure 1 and Figure 2). Both psiRNA species are associated with the Cmr effector complex (Figure 1) and each guides cleavage at a distinct site (Figure 5C). Analysis of the cleavage products of both psiRNAs and of a series of substrate RNAs (Figure 3, Figure 4 and Figure 5) indicates that the complex cleaves based on a 14-nucleotide counting mechanism anchored by the 3’ end of the psiRNA. The results suggest that the 3’ end of the psiRNA places the bound target RNA relative to the enzyme active site (Figure 6).
The activity of the psiRNA-Cmr protein complex (RNA-guided RNA cleavage) bears an interesting resemblance to that of Argonaute 2 (a.k.a. Slicer) (Liu et al., 2004), an enzyme with an analogous function in the eukaryotic RNAi pathway, however there is little similarity between the enzymes. There is no significant sequence homology between the Cmr proteins and Argonaute 2 (or between any of the Cas proteins and known components of the eukaryotic RNAi pathway). Both the psiRNA-Cmr complex and Argonaute 2 employ a ruler mechanism for cleavage site selection; however, in the case of Argonaute 2, the site of cleavage is located ∼10–11 nucleotides from the 5’ end of the siRNA (Elbashir et al., 2001a; Elbashir et al., 2001b). The activity of both enzymes requires divalent metal ions (Supplemental Figure 1 and (Schwarz et al., 2004)), however for the psiRNA-Cmr RNP, it is not yet clear whether the metal is involved in cleavage catalysis or is required for some other essential aspect of the functionality of this multi-component complex. Finally, Argonaute 2 cleaves target RNAs on the 3’ side of the phosphodiester bond, leaving 3’ OH and 5’ phosphate termini (Martinez and Tuschl, 2004). Interestingly, eukaryotes also exploit small RNA-guided gene silencing pathways to combat viruses and other mobile genetic elements that they encounter (Ghildiyal and Zamore, 2009; Malone and Hannon, 2009).
Figure 6 also illustrates the Cmr-psiRNA effector complex model that arises from the findings presented here. Both size classes of psiRNAs and all seven Cmr proteins are found in complexes in active, purified fractions (Figure 1), however accurate RNA-guided cleavage activity can be reconstituted with either psiRNA species and with a single Cmr1 isoform (Figure 5). We hypothesize that each psiRNA associates with a single set of six Cmr proteins, and that Cmr1-1 and Cmr1–2 function redundantly in P. furiosus. Five unrelated proteins that co-purified with the complexes (Supplemental Table 1) are not essential for reconstitution of cleavage activity in vitro (Figure 5) and are not included in our model, but could play a role in function in vivo. Recognition of the psiRNAs by the Cmr proteins and psiRNA-Cmr complex assembly likely depend upon conserved features of the RNAs that could include 5’ and 3’ end groups and folded structure as well as the psi-tag. Our data reveal that the psiRNA-Cmr complex can utilize psiRNAs of different sizes to cleave a target RNA at distinct sites (Figure 5C). Thus, the two size forms of psiRNAs present in P. furiosus may provide more certain and efficient target destruction.
Our data indicate that the function of the RAMP module of Cas proteins is psiRNA-guided destruction of invading target RNA. The widespread occurrence of the cmr genes in diverse archaea (including Sulfolobus and Archaeoglobus species) and bacteria (including Bacillus and Myxococcus species) indicates that invader RNA cleavage is a mechanism utilized by many prokaryotes for viral defense (Haft et al., 2005; Jansen et al., 2002; Makarova et al., 2006). However, not all prokaryotes with the CRISPR-Cas system possess the RAMP module (Cmr) proteins. In these numerous other organisms, it is possible that a different set of Cas proteins mediates psiRNA-guided RNA cleavage or that Cas proteins effect invader resistance by another mechanism. Indeed, very recent work indicates that the CRISPR-Cas system targets invader DNA in a strain of Staphylococcus epidermidis and perhaps E. coli (Brouns et al., 2008; Lillestol et al., 2006; Makarova et al., 2006), which possess the Mtube (Csm) and Ecoli (Cse) subtype Cas protein modules, respectively (Haft et al., 2005; Jansen et al., 2002; Makarova et al., 2006). The prokaryotes include evolutionarily distant and very diverse organisms. Diversity in the core components of the eukaryotic RNAi machinery has led to a tremendous variety of observed RNA-mediated gene silencing pathways that can act at post-transcriptional or transcriptional levels (Chapman and Carrington, 2007; Farazi et al., 2008; Hutvagner and Simard, 2008; Zaratiegui et al., 2007). The diversity of Cas proteins found in CRISPR-containing prokaryotes may reflect significantly different mechanisms of CRISPR element integration, CRISPR RNA biogenesis, and invader silencing.
EXPERIMENTAL PROCEDURES
Chromatography
P. furiosus S100 extract was prepared from approximately 4 grams of cells. Cells were resuspended in 20 mL of 50 mM Tris (pH 7.0), 100 U RNase-free DNase (Promega), and 0.5 mM phenylmethanesulphonyl fluoride (PMSF) at room temperature by stirring. The resulting whole cell extract was subject to ultracentrifugation at 100,000 × g for 1.5 hours using an SW 41 Ti rotor (Beckman). The resulting S100 extract was loaded onto a 5 mL Q-sepharose Fast Flow (GE) pre-packed column. Proteins were eluted using a 0–1 M NaCl gradient. Fractions were analyzed by Northern analysis by isolating RNA from 100 ul of each fraction using Trizol LS (Invitrogen, following manufacturer’s instructions). The RNAs were separated on 15% TBE-urea gels (Criterion, Bio-Rad), blotted and analyzed for the presence of a single guide sequence as described previously (Hale et al., 2008). Peak fractions containing the psiRNA doublet were further separated on a second 5 mL Q-sepharose column, eluted with 220–430 mM NaCl. Fractions were analyzed as described above. Peak fractions were pooled, diluted in 50 mM sodium phosphate buffer, pH 7.0, and loaded onto a 5 mL S-sepharose column (GE). Bound proteins were eluted with a gradient of 0–1 M NaCl. Native gel northern analysis was performed as described previously (Hale et al., 2008). The secondary data shown in Supplemental Table 1 was obtained from S100 extract fractionated on a DEAE column as previously described (Hale et al., 2008) followed by a hydroxyapatite column eluted with a gradient of 5–500 mM sodium phosphate buffer, pH 6.5, and further purified by native gel electrophoresis.
Protein assignment by tandem mass spectrometry
In-gel and in-solution tryptic digests were performed as previously described (Lim et al., 2008; Wells et al., 2002). Desalted tryptic peptides were analyzed by nLC-MS/MS on a linear ion-trap (LTQ, ThermoFisher) as previously described (Lim et al., 2008). Acquired data was searched against a P. furiosus-specific database (forward and inverted) using the TurboSEQUEST algorithm (ThermoFisher). Data was collated and filtered to obtain a 1% false discovery rate at the protein level using the ProteoIQ software package (BioInquire) that is based on the PROVALT algorithm (Weatherly et al., 2005).
Cloning and sequencing of psiRNAs from the purified complexes
RNAs from S-column fractions (isolated as described above for Northern analysis) were treated with 1 U calf intestinal alkaline phosphatase (Promega) for 1 hour at 37°C, followed by extraction with phenol:chloroform:isoamyl alcohol (PCI; pH 5.2, Fisher) and ethanol precipitation. The resulting RNAs were separated by 15% polyacrylamide, TBE-urea gels (Criterion, Bio-Rad), visualized by SYBR Gold staining (Invitrogen) and the visible bands were excised. RNAs were passively eluted overnight in 0.5 M ammonium acetate, 0.1% SDS, 0.5 mM EDTA, followed by ethanol precipitation. A 5’-phosphorylated, 3’ capped oligonucleotide (5’-pCTCGAGATCTGGATCCGGG-ddC3’; IDT) was ligated with T4 RNA ligase to the 3’ end of the RNAs. The ligated RNAs were PCI extracted, ethanol precipitated, gel purified, and subject to reverse transcription using Superscript III (Invitrogen) RT (as described by the manufacturer), followed by gel purification. The gel-purified cDNAs were polyA-tailed for 15 minutes at 37°C using terminal deoxynucleotide transferase (Roche) using manufacturer’s recommendations. PCR was performed to amplify the cDNA libraries using the following primers: 5’-CCCGGATCCAGATCTCGAG-3’, 5’-GCGAATTCTGCAG(T)30-3’). cDNAs were cloned into the TOPO pCRII (Invitrogen) cloning vector and transformed into TOP10 cells. White and light-blue colonies were chosen for plasmid DNA preparation, and sequencing using the M13 Reverse and T7 promoter sequencing primers was performed by the University of Georgia Sequencing and Synthesis Facility.
Small RNA deep sequencing
Small RNA libraries were prepared using the Illumina small RNA Sample preparation kit as described by the manufacturer (Illumina). Briefly, total RNA was isolated from P. furiosus and fractionated on a 15% polyacrylamide/urea gel, and small RNAs 18–65 nt in length were excised from the gel. 5' and 3' adapters were sequentially ligated to the small RNAs and the ligation products were gel-purified between each step. The RNAs were then reverse-transcribed and PCR-amplified for 16 cycles. The library was purified with a Qiagen QuickPrep column and quantitated using an Agilent Bioanalyzer and a nanodrop. The sample was diluted to a concentration of 2 pM and subjected to 42 cycles of sequencing on the Illumina Genome Analyzer II.
Small RNA Analysis
Sequence data was extracted from the images generated by the Illumina Genome Analyzer II using the software applications Firecrest and Bustard. The adapter sequences were then trimmed from the small RNA reads, which were then mapped to the P. furiosus genome using btbatchblast. Only reads that mapped perfectly to the genome over their entire length were used for further analysis. The location and number of reads that initiate within the CRISPR repeats were determined using a perlscript. As the maximal read length of the sequences was 42 nt, it was not possible to be certain that the 3’ end of a read represented the actual 3’ end of the small RNA. Therefore, the deep sequencing data was only used to determine the 5’ ends.
Nuclease assays
To detect target RNA cleavage, 2 µL of the peak S-column fractions (Figure 1C) or 500 nM each of recombinant proteins was incubated with 0.05 pmoles of 32P-5’ end-labeled synthetic target RNAs (Figure 3, Figure 4 and Figure 5) and 0.5 pmoles of each unlabeled psiRNA (Figure 5) for 1 hour at 70°C in 20 mM HEPES pH 7.0, 250 mM KCl, 1.5 mM MgCl2, 1 mM ATP, 10 mM DTT, in the presence of 1 unit of SUPERase-In ribonuclease inhibitor (Applied Biosystems). For assays with recombinant proteins, the psiRNAs were first incubated with the proteins for 30 minutes at 70°C prior to the addition of target RNA. Reaction products were isolated by treatment with 800 ng of proteinase K for 30 minutes at room temperature, followed by PCI extraction and ethanol precipitation. The resulting RNAs were separated on 15% polyacrylamide, TBE 7M urea gels and visualized by phosphorimaging. 5’ end-labeled RNA size standards (Decade Markers, Applied Biosystems) were used to determine the sizes of the observed products. Annealed RNAs were prepared by mixing equimolar amounts of RNAs in 30 mM HEPES pH 7.4, 100 mM potassium acetate, 2 mM magnesium acetate and incubating for 1 minute at 95°C, followed by 1 hour at 37°C. Annealing was confirmed by non-denaturing 8% PAGE.
For analysis of the chemical ends of the cleavage products, cleavage reactions were performed using 5’-end labeled target as described above. The resulting RNA products were isolated by PCI extraction and ethanol precipitation, and subject to polyadenylation by incubation with 5 U E. coli polyA polymerase (NEB) for 15 minutes at 37°C as described by the manufacturer. The reaction was stopped by PCI extraction, followed by ethanol precipitation. The resulting products were analyzed on 15% polyacrylamide, TBE 7M Urea gels as described above.
In order to determine the divalent metal requirements of the purified complex, cleavage reactions were performed for 1 hour at 70°C in 50 mM HEPES pH 7.0, 250 mM KCl, 1 mM ATP, 10 mM DTT, 0.1 mM EDTA, and 1 mM metal (if applicable) in the presence of 1 unit of SUPERase-In ribonuclease inhibitor (Applied Biosystems). Certified metal reference solutions (Spex CertiPrep except calcium obtained from Fisher Scientific) were added to 1 mM final concentration. The resulting products were isolated and analyzed as described above.
Expression and purification of recombinant proteins
The genes encoding P. furiosus Cmr1-1 (PF1130), Cmr2 (PF1129), Cmr3 (PF1128), Cmr4 (PF1126), Cmr5 (PF1125) and Cmr6 (PF1124) were amplified by PCR from genomic DNA or existing constructs and cloned into a modified version of pET24d (PF1124, PF1125 and PF1126) or pET200D (PF1128, PF1129 and PF1130). The recombinant proteins were expressed in E. coli BL21-RIPL cells (DE3, Stratagene). The cells (400 mL cultures) were grown to a OD600 of 0.7, and expression of the proteins was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG) overnight at room temperature. The cells were pelleted, resuspended in 20 mM sodium phosphate buffer (pH 7.6), 500 mM NaCl and 0.1 mM phenylmethylsulfonyl fluoride (PMSF), and disrupted by sonication. The sonicated sample was centrifuged at 4,500 rpm for 15 min at 4°C. The supernatant was heated at 75–78°C for 20 min, centrifuged at 4,500 rpm for 20 min at 4°C, and filtered (0.8 µm pore size Millex filter unit, Millipore). The recombinant histidine-tagged proteins were purified by batch purification using 50 µl Ni–NTA agarose beads (Qiagen) equilibrated with resuspension buffer. Following 3 washes (resuspension buffer), the bound proteins were eluted with resuspension buffer containing 500 mM imidazole. The protein samples were dialyzed at room temperature against 40 mM HEPES (pH 7.0) and 500 mM KCl prior to performing activity assays.
Synthetic psiRNAs
The 45- and 39-nucleotide psiRNAs were chemically synthesized (Integrated DNA Technologies). The sequence of the 45-nucleotide psiRNA 7.01 is: AUUGAAAGUUGUAGUAUGCGGUCCUUGCGGCUGAGAGCACUUCAG. The sequence of the 39-nucleotide psiRNA 7.01 is: AUUGAAAGUUGUAGUAUGCGGUCCUU GCGGCUGAGAGCA.
Supplementary Material
01. SUPPLEMENTAL DATA.
Supplemental data include two tables and one figure with legend. Sequences of the psiRNAs are available in the Gene Expression Omnibus.
ACKNOWLEDGMENTS
We extend special thanks to Tim Davies (Director, University of Georgia Bioexpression & Fermentation Facility) for P. furiosus cells, the University of Connecticut Health Center Translational Genomics Core Facility for use of the Illumina Genome Analyzer, Frank Sugar (University of Georgia Bioexpression Facility) for expert advice on chromatography, Mike Adams and the Southeast Collaboratory for Structural Genomics (University of Georgia) for constructs, Lindsay Jones, Joshua Elmore and Sonali Majumdar (Terns Lab, University of Georgia) for generation of protein expression constructs, and Claiborne Glover (University of Georgia) for critical reading. L.W. is a Georgia Cancer Coalition Distinguished Scientist. This work was supported by National Institutes of Health grants RO1GM54682 (M.T. and R.T.) and R01GM062516 (B.R.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Caryn R. Hale, Department of Biochemistry and Molecular Biology, and Genetics, University of Georgia, Athens, GA 30602, USA
Peng Zhao, Department of Biochemistry and Molecular Biology, and Genetics, University of Georgia, Athens, GA 30602, USA.
Sara Olson, Department of Genetics and Developmental Biology, University of Connecticut Stem Cell Institute, University of Connecticut Health Center, 263 armington Avenue, Farmington, CT 06030-3301, USA.
Michael O. Duff, Department of Genetics and Developmental Biology, University of Connecticut Stem Cell Institute, University of Connecticut Health Center, 263 armington Avenue, Farmington, CT 06030-3301, USA
Brenton R. Graveley, Department of Genetics and Developmental Biology, University of Connecticut Stem Cell Institute, University of Connecticut Health Center, 263 armington Avenue, Farmington, CT 06030-3301, USA
Lance Wells, Department of Biochemistry and Molecular Biology, and Genetics, University of Georgia, Athens, GA 30602, USA.
Rebecca M. Terns, Department of Biochemistry and Molecular Biology, and Genetics, University of Georgia, Athens, GA 30602, USA
Michael P. Terns, Department of Biochemistry and Molecular Biology, and Genetics, University of Georgia, Athens, GA 30602, USA
REFERENCES
- Andersson AF, Banfield JF. Virus population dynamics and acquired virus resistance in natural microbial communities. Science. 2008;320:1047–1050. doi: 10.1126/science.1157358. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Beloglazova N, Brown G, Zimmerman MD, Proudfoot M, Makarova KS, Kudritska M, Kochinyan S, Wang S, Chruszcz M, Minor W, et al. A Novel Family of Sequence-specific Endoribonucleases Associated with the Clustered Regularly Interspaced Short Palindromic Repeats. J Biol Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. Embo J. 2001b;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–729. doi: 10.1007/s00239-005-0223-z. [DOI] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale C, Kleppe K, Terns RM, Terns MP. Prokaryotic silencing (psi)RNAs in Pyrococcus furiosus. RNA. 2008;14:2572–2579. doi: 10.1261/rna.1246808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg JF, Nelson WC, Schoenfeld T, Bhaya D. Germ warfare in a microbial mat community: CRISPRs provide insights into the co-evolution of host and viral genomes. PLoS ONE. 2009;4:e4169. doi: 10.1371/journal.pone.0004169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61. doi: 10.1186/gb-2007-8-4-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillestol RK, Redder P, Garrett RA, Brugger K. A putative viral defence mechanism in archaeal cells. Archaea. 2006;2:59–72. doi: 10.1155/2006/542818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillestol RK, Shah SA, Brugger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009 doi: 10.1111/j.1365-2958.2009.06641.x. [DOI] [PubMed] [Google Scholar]
- Lim JM, Sherling D, Teo CF, Hausman DB, Lin D, Wells L. Defining the regulated secreted proteome of rodent adipocytes upon the induction of insulin resistance. J Proteome Res. 2008;7:1251–1263. doi: 10.1021/pr7006945. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Grishin NV, Shabalina SA, Wolf YI, Koonin EV. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol Direct. 2006;1:7. doi: 10.1186/1745-6150-1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez J, Tuschl T. RISC is a 5’ phosphomonoester-producing RNA endonuclease. Genes Dev. 2004;18:975–980. doi: 10.1101/gad.1187904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Sorek R, Kunin V, Hugenholtz P. CRISPR--a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- Tang TH, Bachellerie JP, Rozhdestvensky T, Bortolin ML, Huber H, Drungowski M, Elge T, Brosius J, Huttenhofer A. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc Natl Acad Sci U S A. 2002;99:7536–7541. doi: 10.1073/pnas.112047299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang TH, Polacek N, Zywicki M, Huber H, Brugger K, Garrett R, Bachellerie JP, Huttenhofer A. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol Microbiol. 2005;55:469–481. doi: 10.1111/j.1365-2958.2004.04428.x. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–207. doi: 10.1111/j.1462-2920.2007.01444.x. [DOI] [PubMed] [Google Scholar]
- Weatherly DB, Atwood JA, 3rd, Minning TA, Cavola C, Tarleton RL, Orlando R. A Heuristic method for assigning a false-discovery rate for protein identifications from Mascot database search results. Mol Cell Proteomics. 2005;4:762–772. doi: 10.1074/mcp.M400215-MCP200. [DOI] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Cole RN, Cronshaw JM, Matunis MJ, Hart GW. Mapping sites of O-GlcNAc modification using affinity tags for serine and threonine post-translational modifications. Mol Cell Proteomics. 2002;1:791–804. doi: 10.1074/mcp.m200048-mcp200. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Zhou K, Jinek M, Coyle SM, Ma W, Doudna JA. Structural basis for DNase activity of a conserved protein implicated in CRISPR-mediated genome defense. Structure. 2009;17:904–912. doi: 10.1016/j.str.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01. SUPPLEMENTAL DATA.
Supplemental data include two tables and one figure with legend. Sequences of the psiRNAs are available in the Gene Expression Omnibus.