A mouse model for the human pathogen Salmonella Typhi (original) (raw)
. Author manuscript; available in PMC: 2011 Apr 21.
Published in final edited form as: Cell Host Microbe. 2010 Oct 21;8(4):369–376. doi: 10.1016/j.chom.2010.09.003
SUMMARY
Salmonella enterica serovar Typhi (S. Typhi) is the cause of typhoid fever, a life-threatening disease of humans. The lack of an animal model due to S. typhi's strict human host specificity has been a significant obstacle in the understanding of its pathogenesis and the development of a safe and effective vaccine against typhoid fever. We report here the development of a mouse model for S. Typhi infection. We showed that immunodeficient Rag2 -/- γc -/- mice engrafted with human fetal liver hematopoietic stem and progenitor cells were able to support S. Typhi replication and persistent infection. A S. Typhi strain carrying a mutation in a gene required for its virulence in humans was not able to replicate in these humanized mice. In contrast, another mutant strain unable to produce the recently identified typhoid toxin, exhibited increased replication suggesting a potential role for this toxin in the establishment of persistent infection. Furthermore, infected animals mounted a human innate and adaptive immune response to S. Typhi resulting in the production of cytokines and pathogen-specific antibodies. These results therefore indicate that this animal model can be used to study S. Typhi pathogenesis and to evaluate potential vaccine candidates against typhoid fever.
Keywords: bacterial pathogenesis, innate immunity, stem cells, humanized mouse, typhoid fever
INTRODUCTION
Typhoid fever remains a global health problem, resulting in more than 200,000 annual deaths, mostly children in developing countries (Pang et al., 1998) (Crump and Mintz, 2010 ). Unlike other Salmonella enterica serovars such as S. Typhimurium or S. Enteritidis, which are associated with gastroenteritis (i. e. “food poisoning”) and can infect a variety of hosts, S. Typhi is an exclusive human pathogen (Parry et al., 2002). Furthermore, S. Typhi can cause life-long infections in humans, most often by colonizing the gall bladder. The molecular bases for its host adaptation and ability to cause persistent infection are not known. However, it is believed that a combination of genome degradation and acquisition of new genetic information has conferred on S. Typhi its unique pathogenic properties 4 (Sabbagh et al., 2010). Although much is known about the pathogenic mechanisms of S. enterica in general, and some serovars in particular, remarkably little is known about the unique pathogenic features of S. Typhi. There are currently no effective vaccines against typhoid fever, and no vaccines that can be used in young children. The isolation of multi drug resistant S. Typhi has raised the worrisome possibility of the reemergence of untreatable typhoid fever (Mirza et al., 1996). Since S. Typhi is restricted to humans, there is no suitable animal model (other than higher primates) to study S. Typhi pathogenesis and to test potential vaccines. To study typhoid fever pathogenesis, investigators have made use of S. Typhimurium, which in mice carrying a mutation in nramp1 produces a disease that resembles typhoid fever (O'Brien et al., 1980). Furthermore, S. Typhimurium infection of nramp1 +/+ mice, which are resistant to infection, has been used as a model of persistent infection (Monack et al., 2004). Although these models have been useful to the understanding of systemic infections by S. enterica in general, they have been of limited value to the study of pathogenic mechanisms specific to S. Typhi. Since S. Typhi is in essence a pathogen of the reticuloendothelial system (Parry et al., 2002) (House et al., 2001) it is possible that determinants of host specificity and restriction may reside within the reticuloendothelial system since this is the most variable compartment across different animal species (Flajnik and Kasahara, 2010) (Barreiro and Quintana-Murci, 2010). Therefore we sought to investigate the ability of a mouse with a humanized immune system to support infection by S. Typhi. We found that immunodeficient Rag2 -/- γc -/- mice engrafted with human fetal liver hematopoietic stem and progenitor cells support S. Typhi replication and persistent infection. Infected animals mounted a human innate and adaptive immune response to S. Typhi resulting in the production of cytokines and pathogen-specific antibodies. These results therefore indicate that this animal model can be used to study S. Typhi pathogenesis and to evaluate potential vaccine candidates against typhoid fever.
RESULTS AND DISCUSSION
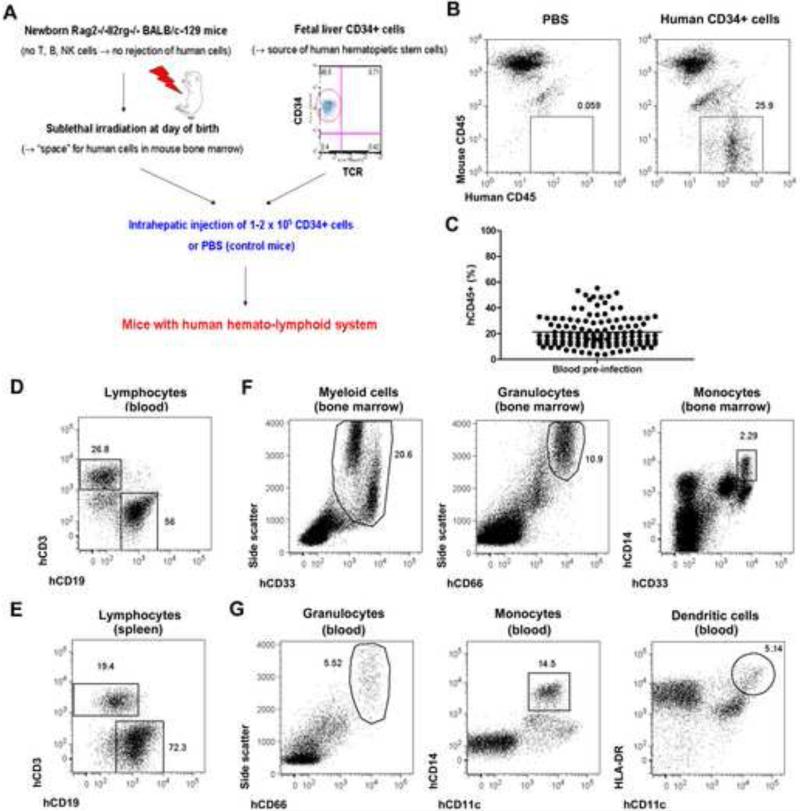
Immunodeficient mice engrafted with human hematopoietic stem and progenitor cells have been used to study human diseases including immune responses to microbial pathogens (Shultz et al., 2007) (Legrand et al., 2006) (Manz, 2007a) 15. We therefore engrafted fetal liver CD34+ human hematopoietic stem cells into the livers of Rag2 -/- γc -/- mice 16. Previous studies have shown that these animals support reconstitution of a functional human immune system 16 17 (Baenziger et al., 2006) (Kuruvilla et al., 2007.) (Kwant-Mitchell et al., 2009 ) (Yu et al., 2008). As controls we used conditioned newborn Rag2 -/- γc -/- mice injected with PBS only (Fig. 1a and 1b). Average engraftment with human CD45+ hematopoietic cells was 21.3% (range: 3.7-55.4%) in the animals used in this study (Fig. 1c). Engrafted mice developed human lymphocytes (Fig. 1d and 1e) as well as human myeloid cells (Fig. 1f and 1g).
Figure 1. Reconstitution of a human immune system in immunodeficient mice.
(A) Diagram depicting the generation of mice with a human hemato-lymphoid system. (B) Representative flow cytometric analysis of blood cells from mice injected with PBS or with human CD34+ cells. Numbers next to boxed areas indicate the percentages of human hematopoietic (hCD45+) cells. (C) Frequency of human hematopoietic (hCD45+) cells in blood in mice engrafted with human CD34+ cells (n = 125) determined by flow cytometry. Horizontal bar indicates mean frequency. (D-G) Flow cytometric analysis of human immune cell populations in engrafted mice. Representative examples of human T (hCD3+) and B cells (hCD19+) in blood (D) and spleens (E) are shown. Human myeloid (hCD33+ Side Scatterhi), granulocytes (hCD66+ Side Scatterhi), monocytes (hCD14hi), and dendritic cells (hCD11chi HLA-DRhi) in bone marrow (F) and blood (G) are also shown. All plots are gated on hCD45+ mCD45- cells. Numbers next to boxed/circled areas indicate percentages of cells. Mice were analyzed 8-14 weeks post-engraftment.
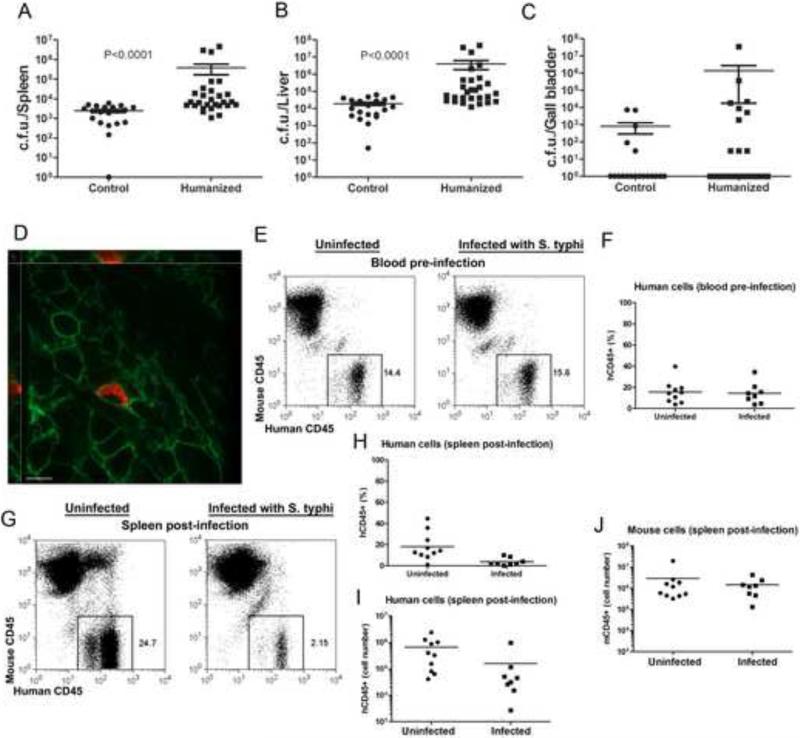
We infected groups of engrafted and control animals with an intraperitoneal dose of wild-type S. Typhi, which in pilot experiments conducted in non-engrafted animals, did not result in significant numbers of bacteria recovered from systemic tissues 4 weeks after infection. Neither the control nor the engrafted animals showed any symptoms as a consequence of the infection. However, large numbers of S. Typhi colony forming units (CFU) were recovered from liver and spleen of engrafted animals at 4 weeks after infection (Fig. 2a and 2b). In contrast, in control animals, bacteria were recovered in much lower numbers (Fig. 2a and 2b). Furthermore, the engrafted group had a higher proportion of animals with larger number of S. Typhi CFU in the gall bladder when compared to the non-engrafted controls, although this difference did not reach statistical significance (Fig. 2c). This observation may be of interest since the gall bladder is the preferred site of S. Typhi residency in persistently infected humans (Sinnott and Teall, 1987). We observed mouse-to-mouse variation in the levels of S. Typhi CFU in humanized mice not only between different experiments but also within a given experiment. We hypothesize that this difference may have been due to different levels of engraftment, differences in the lineages of the human cell populations generated in the engrafted animals, differences in the genetic make up of the human cell donors, or a combination of all these factors. Similar observations have been made in previous studies using humanized mice (Brehm et al., 2010).
Figure 2. Humanized mice can support S. Typhi replication.
(A-C) CFU recovered from S. Typhi-infected control (left) and humanized (right) mouse spleen (A), liver (B), and gall bladder (C). Control and humanized mice were infected intraperitoneally with 104 S. Typhi and CFU of the organs of the infected animals were examined 4 weeks after infection. Data in (A) and (B) represent three pooled independent experiments (p < 0.0001 for the difference between the number of CFU recovered from humanized and control animals). Data in C represent two pooled independent experiments (n=19 for control and n=25 for humanized mice) in which 5 control mice and 10 humanized mice showed S. typhi colonization in the gall bladder. Each symbol represents the actual number of CFU per organ of a single mouse. The mean ± s. e. m. are shown. (D) Immunofluorescence staining of intracellular S. Typhi in humanized mouse spleen. S. Typhi, red; Human CD45+ cells, green (scale bar = 10 μm). (E-J) Flow cytometric analysis of blood cells from mice before (E, F) and splenocytes 4 weeks after (G-J) infection with 104 S. Typhi. Control mice were left uninfected. Cells were stained with antibodies specific for mouse or human CD45+. The frequency (F-H) and absolute number (I) of human hematopoietic (hCD45+) cells and the absolute number of mouse hematopoietic (mCD45+) cells (J) are shown. The p values of the difference between frequency/number of the indicated cells in infected and uninfected animals are 0.722 (F), 0.004 (H), 0.011 (I), and 0.896 (J).
The mouse Nramp1 protein is known to be very important for resistance to S. Typhimurium infection in mice, and nramp1 -/- mice are up to ~1,000-fold more susceptible to bacterial infection (Iacus et al., 1998). However, we observed no significant differences between the levels of S. Typhi CFU recovered from engrafted Rag2 -/- γc -/- nramp1 -/- or Rag2 -/- γc -/- nramp1 +/+ mice (Fig. S1). These results indicate that the Nramp1 status of the mice, which was variable due to the mixed BALB/c x 129 genetic background, does not influence the outcome of S. Typhi infection of humanized mice, and they are consistent with a previous observation indicating that Nramp1 is not involved in the restriction of S. Typhi replication in mice (O'Brien, 1982). Although these results are consistent with extensive epidemiological data that has found no association between nramp1 alleles and typhoid fever susceptibility in humans (Dunstan et al., 2001), it is also possible that mouse cells may not contribute to S. Typhi replication and hence only the genetic make up of the engrafted human cells may be relevant for bacterial replication. Indeed, human CD45+ splenocytes containing large numbers of S. Typhi were detected in the spleens of infected animals, indicating that human cells are capable of supporting bacterial replication (Fig. 2d). In contrast, we did not observe S. Typhi within mouse CD45+ cells (data not shown). We occasionally observed isolated bacteria in areas of the spleen not stained by either antibody, which may represent extracellular bacteria or non-dividing bacteria contained within mouse CD45- cells. We also observed a specific depletion of human CD45+ cells 4 weeks after infection in the spleens of infected animals (Fig. 2e-2i), suggesting that bacterial replication may lead to cell death. In contrast, we observed no depletion of mouse CD45+ cells (Fig. 2j). Taken together, these results suggest that human cells may provide a replicative niche for S. Typhi although we cannot rule out that human cells only indirectly contribute to bacterial replication.
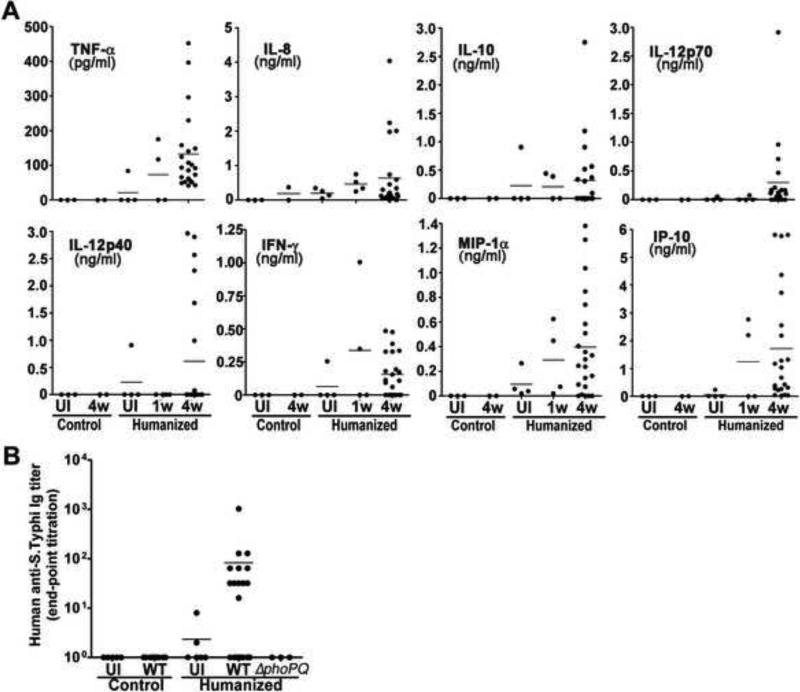
Innate and adaptive immune responses are essential to control S. Typhi infection (Raffatellu et al., 2008) (Sztein, 2007). Therefore, we tested whether S. Typhi infection resulted in a human innate immune response in the engrafted animals by measuring the levels of several human cytokines in peripheral blood 1 and 4 weeks after infection. Only a few infected animals showed levels of human cytokines above those of control uninfected animals 1 week after infection (Fig. 3a). In contrast, 4 weeks after infection, a large proportion of the infected animals showed increased levels of the human cytokines TNFα, IL-8, IL-10, IL-12, IFNγ, MIP-1α, and IP-10 (Fig. 3a). These results indicate that engrafted animals were able to mount a human innate immune response to S. Typhi. We also tested whether S. Typhi infection could elicit an acquired human immune response by measuring the levels of S. Typhi-specific antibody responses in the sera of engrafted animals 4 weeks after infection. Significant levels of human antibodies to S. Typhi were detected in ~25% of infected animals but not in the uninfected controls (Fig. 3b). These results indicate that this model could be useful for pre-clinical testing of S. Typhi vaccine candidates.
Figure 3. S. Typhi stimulates a human innate and acquired immune response in humanized mice.
(A) Human cytokine and chemokine concentrations in the sera of control and humanized animals. Sera were collected from the uninfected or infected animals 1 wk (1w) or 4 wks (4w) after infection. (B) Human immunoglobulin (IgG, M, and A) levels against S. Typhi in the sera of control and humanized animals that were left uninfected or infected for 4 wks with either wild type (WT) or Δ_phoP_Δ_phoQ_ mutant (Δ_phoPQ_) S. Typhi.
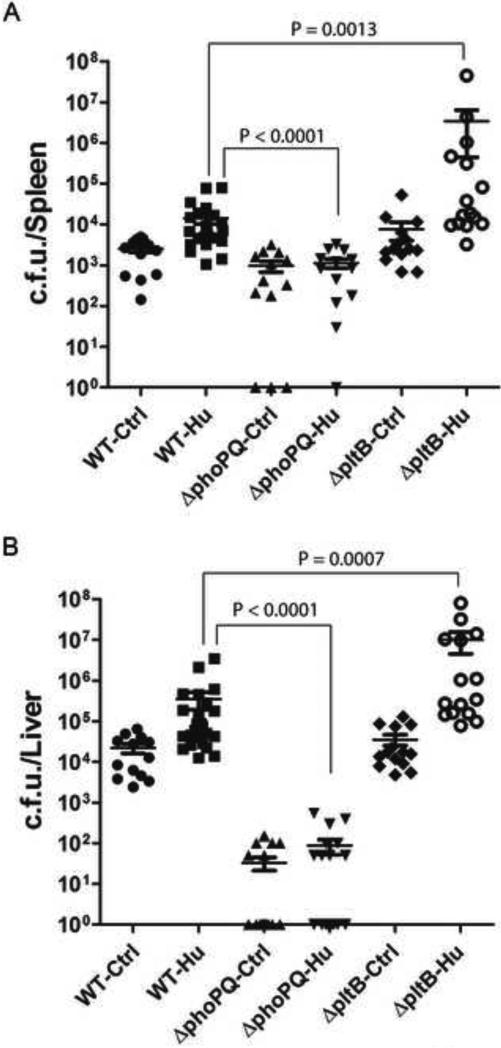
A useful animal model should be able to resolve differences between strains of different virulence. Only a limited number of S. Typhi mutant strains have been tested in humans for their virulence properties (Galen et al., 2009 ). One of them is the S. Typhi Δ_phoP_Δ_phoQ_ mutant, which has been shown to be avirulent in human volunteer trials, and is being considered as a potential vaccine candidate (Hohmann et al., 1996). PhoP/PhoQ is a two component regulatory system that controls the expression of several genes that are critical for virulence (Kato and Groisman, 2008). We therefore compared the levels of CFU recovered from tissues of engrafted animals infected with either wild type or an isogeneic Δ_phoP_ Δ_phoQ_ mutant 4 weeks after infection. We found that the levels of wild type S. Typhi were significantly higher than those of the Δ_phoP_ Δ_phoQ_ mutant, and contrary to wild type, the mutant strain was either not detected in tissues or was recovered in much lower numbers (Fig. 4a and 4b). Consistent with its poor replication, the Δ_phoP_ Δ_phoQ_ mutant did not stimulate a human immune response in the engrafted animals (Fig. 3b). Typhoid toxin is a recently identified potential virulence factor of S. Typhi (Haghjoo and Galan, 2004; Spano et al., 2008). Exclusively produced by S. Typhi and S. Paratyphi, this toxin is composed of one “B” subunit known as PltB, and two active subunits, known as CdtB and PltA, which have nuclease and the ADP-ribosyl transferase activities. Although it is known that this toxin is exclusively produced by intracellularly located bacteria (Haghjoo and Galan, 2004), the role of typhoid toxin in S. Typhi pathogenesis and infection is unknown. We therefore tested the virulence of a S. typhi Δ_pltB_ mutant strain. PltB is essential for toxin delivery so a strain lacking this subunit is unable to intoxicate cultured epithelial cells (Spano et al., 2008). Surprisingly, we found that this mutant strain exhibited increased ability to replicate in the humanized mouse model. Significantly higher CFU of this mutant were detected in the liver and spleen of humanized mice. These results suggest the possibility that typhoid toxin may contribute to the establishment of persistent infection perhaps by slowing down S. Typhi replication in infected tissues. Additional studies will be required to understand the role of typhoid toxin in S. Typhi infection. Taken together, these results indicate that this model can be used to evaluate the in-vivo replication of S. Typhi mutant strains and could therefore be useful in the pre-clinical evaluation of virulence-attenuated vaccine candidates.
Figure 4. Replication of S. Typhi Δ_phoP_ Δ_phoQ_ or Δ_pltB_ (typhoid toxin-defective) mutant strains in humanized mice.
Total CFU in the spleen (A) and liver (B) of control (Ctrl) and humanized (Hu) animals 4 weeks after intraperitoneal administration of 104 CFU of wild-type S. Typhi (WT), or its isogenic mutant strains Δ_phoP_ Δ_phoQ_ or a Δ_pltB_ (typhoid toxin defective). Data represent two pooled independent experiments. Each dot represents the number of CFU per organ of a single mouse. The mean ± s. e. m. of the CFU for each group of mice are shown. The p values of the difference between the CFU obtained from humanized animals infected with wild type or the Δ_phoP_ Δ_phoQ_ mutant, and the difference between the CFU obtained from animals infected with wild type or the Δ_pltB_ mutant strains are also shown.
We have shown here that an immunodeficient mouse engrafted with human hematopoietic stem and progenitor cells can support the replication of S. Typhi and can serve as a model to study this human-specific pathogen. Humanizing the immune system of the mouse effectively relieved the potent host-specificity barrier that blocks S. Typhi replication in this species, and resulted in persistent infection. It is not yet clear how the presence of human cells render mice permissive for S. typhi replication. Human cells may simply provide a permissive niche for bacterial replication. Indeed, we have observed replicating S. Typhi within human cells in the engrafted animals. However, it is also possible that the human cells help S. Typhi to overcome some (yet unknown) “bottle neck” that prevents the progression of S. Typhi infection. Our results have shown that S. Typhi infection in humanized mice is very different from that of S. Typhimurium in conventional mice. This is highlighted by the observation that the Nramp1 protein, which plays a dominant role in conferring resistance to S. Typhimurium, did not influence the susceptibility of humanized mice to S. Typhi. Infection of humanized mice did not lead to acute lethal infection (i. e. murine typhoid), although it led to persistent infection, a very poorly understood aspect of S. Typhi pathogenesis. It is not clear why humanized animals did not develop symptoms of systemic infection despite the presence of large number of bacteria in various tissues. However, it is likely that the inability of the human cytokines to effectively act on mouse tissues (Libert et al., 1990; Manz, 2007b; Schoenhaut et al., 1992) may prevent the development of systemic pathology. Future improvements on this mouse model may allow the modeling of systemic pathology and may increase the consistency of the engrafted animals to support infection and immune responses. The detailed study of S. typhi infection in the humanized mouse model described here may lead to a better understanding of several aspects of human S. Typhi pathogenesis. One of the most significant hurdles for the development of S. Typhi vaccines has been the absence of a convenient animal model that can be used to test virulence-attenuated mutants and evaluate their vaccine potential. It is known that immunization or even virulence studies in conventional mice often do not translate well into humans. The model described here may therefore greatly facilitate the development and pre-clinical testing of novel anti typhoid fever therapeutic and prevention strategies.
EXPERIMENTAL PROCEDURES
Engraftment of Mice with Human Hematopoietic CD34+ Stem and Progenitor Cells
Rag2-/- γc-/- mice were provided by Regeneron Pharmaceuticals. These mice were derived from a V17 embryonic stem cell line (BALB/c × 129 heterozygote) targeted for the Rag2 and Il2rg genes. Human fetal liver samples were obtained under approval from the Yale University Human Investigation Committee from Albert Einstein Medical College New York. CD34+ cells were purified by density gradient centrifugation and immunomagnetic selection using CD34 microbeads (Miltenyi Biotec). Newborn pups were irradiated and engrafted with human CD34+ cells (1-2 ×105 cells in PBS) as previously described (Traggiai et al., 2004). Control mice were injected with PBS only. Engraftment with human hematopoietic cells was determined 8-14 weeks post-transplantation by retro-orbital bleeding and flow cytometry as described below. Mice used for experiments had blood engraftment levels of ≥15% hCD45+ cells. Mice were maintained under specific pathogen-free conditions and received prophylactic antibiotics (Sulfatrim) in the drinking water to prevent opportunistic infections until two weeks prior to S. Typhi infection. All animal work was approved by the Yale University Institutional Animal Care and Use Committee (IACUC) and conducted in accordance with IACUC regulations.
Flow Cytometry
Cell suspensions were prepared from bone marrow, spleen and blood of mice 10-14 weeks post-transplantation. Lysis of RBC was performed using ACK lysis buffer (Lonza). Samples were then stained with fluorochrome-labeled monoclonal antibodies (mAbs) against mouse and human cell surface antigens. The following mAbs were used: (1) Anti-human: CD3 (UCHT1), CD11c (B-ly6), CD14 (MoP9), CD19 (HIB19), CD33 (WM53), CD45 (HI30 and 2D1), CD66 (B1.1). (2) Anti-mouse: CD45 (30-F11). CD45 (30-F11) mAb was from eBioscience. All other mAbs were from BD Biosciences. Samples were analyzed on a FACSCalibur or LSRII flow cytometer (BD Biosciences).
Bacterial Strains, Media, and Growth Conditions
The wild-type Salmonella enterica serovar Typhi strain ISP2825 and its isogenic Δ_pltB_ mutant have been described previously (Galán and Curtiss III, 1991; Spano et al., 2008). A mutant strain carrying an in-frame deletion in _phoP pho_Q in the chromosome was constructed by standard recombinant DNA and allelic exchange procedures as previously described (Kaniga et al., 1994). S. Typhi strains were grown overnight at 37°C in 2 ml LB broth. For mouse experiments, overnight cultures of bacteria were freshly inoculated at a dilution of 1:50 in LB broth containing 0.3M NaCl and grown at 37°C to an OD600 of <0.8. Indicated numbers of bacteria were prepared in buffered saline with gelatin (BSG) (8.5 g of NaCl, 0.3 g of KH2PO4, 0.6 g of Na2HPO4, 0.1 g gelatin in 1000 ml distilled water, pH 7.0) for mouse experiments.
Bacterial Infections and CFU Assay
All animal experiments were conducted according to protocols approved by Yale University's Institutional Animal Care and Use Committee. Groups of control and humanized 13-16 week-old mice were infected intraperitoneally with either 104 or 105 of wild-type S. Typhi or the isogenic Δ_phoP_Δ_phoQ_ or Δ_pltB_ mutant strains in 100 μl of BSG. Infected animals were sacrificed and organs were harvested at 5, 11, 21, 28 days post infection to perform in vivo CFU assays. Spleens, livers, and gall bladders were aseptically removed and mechanically homogenized in 3 ml (for spleens and gall bladders) or 5 ml (for livers) of sterile PBS containing 0.05% sodium deoxycholate. Bacterial loads were determined by plating 10-fold serial dilutions of homogenates on LB agar plates and incubated overnight at 37°C. Colonies were counted and the number of total CFU recovered was calculated.
Immunofluorescence of Spleen Sections
Control and humanized 12-14 week-old mice were infected intraperitoneally with 106 S. Typhi in 100 μl BSG. Five days after infection, spleens were aseptically removed, fixed in 3.7% formaldehyde overnight, paraffin-embeded, and cut 5 μm thick. Tissue sections were pretreated at 95°C for 20 min in 10 mM citrate buffer (pH 6.0) to retrieve the epitope and followed by 30 min incubation at RT with the primary antibodies. Monoclonal mouse anti-human CD45, leukocyte common antigen (LCA) clones 2B11+PD7/26 (Dako; 1:100 dilution) and monoclonal rat anti-mouse CD45, LCA clone 30-F11 (BD Pharmingen; 1;40 dilution) antibodies were used to detect human and mouse CD45+ cells on spleen sections. Polyclonal rabbit anti-S. Typhi lipopolysaccharide (Difco Laboratories; 1:1000 dilution) antibody was used to detect intracellular bacteria. The tissue sections were then incubated for 20 min at RT with secondary anti-mouse, anti-rat, or anti-rabbit antibodies conjugated with FITC (BioCare Medical) or DyLight549 (Biocare Medical). Slides were mounted using a mounting reagent (Vector Laboratories, H1200).
Peripheral Blood Leukocyte (PBL) Preparation, Splenocyte Preparation, Immunostaining, and FACS Analysis
Peripheral blood of uninfected and infected humanized mice was collected into heparinized tubes. In order to remove RBCs, ~100 μl heparinized blood was incubated with 1 ml ACK lysis buffer (Lonza) for 5 min on ice, washed with 2 ml PBS, and centrifuged to collect PBLs. This RBC-lysis step was repeated once more and PBLs were immediately processed for an immunostaining as described below. Spleens of uninfected and infected humanized mice were aseptically harvested, mechanically homogenized, and filtered using a nylon mesh to obtain single cell suspensions. RBCs were lysed by incubating them with 1 ml ACK lysis buffer. After washing, cells were immediately incubated for 30 min on ice with 100 μl of an antibody cocktail containing 1) anti-mouse: CD45 (30-F11; eBioscience) and 2) anti-human CD45 (HI30 and 2D1; BD Biosciences), CD3 (UCHT1; BD Biosciences), and CD19 (HIB19; BD Biosciences). The cells were then washed with 2 ml FACS buffer (0.16% BSA/0.1% NaN3/PBS), resuspended in 100 μl FACS fix buffer (1% FCS/1% Formaldehyde/PBS), kept at 4 °C overnight, and used for FACS analyses. Total splenocyte numbers were determined by cell counting using a hemocytometer.
Human Cytokine/Chemokine Measurement
Human cytokine/chemokine concentration in the sera of control and humanized animals were measured using a human cytokines/chemokines/growth factors multiplex kit (Millipore) according to the manufacturer's protocol. The following human cytokines were assayed: Eotaxin, G-CSF, GM-CSF, IFN-α2, IFN-γ, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-17, IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IP-10, MCP-1, MIP-1α, MIP-1β, TNF-α, and TNF-β. Only human cytokines that showed a significant elevation in serum after S. Typhi infection are shown in Fig. 3. The other cytokines were either not detected or detected in a mall subset of infected animals.
Measurement of Human S. Typhi-Specific Antibody Responses
Human S. Typhi-specific antibody levels in the sera of infected humanized mice were examined using a direct ELISA. ELISA plates were coated with a S. Typhi total cell lysate prepared as follows. S. Typhi was cultured in 50 ml of LB broth, heat-killed at 70°C for 20 min, centrifuged, and resuspended in 50 mM Tris buffer, pH 8.0 containing 1% Triton X-100, and a protease inhibitor cocktail (Roche). The bacterial cells were then sonicated and unbroken cells removed by centrifugation. Ninety six-well plates (Maxisorp™; Nunc) were coated with 30 μg bacterial cell lysates in 100 μl plate coating buffer (0.5 M carbonate-bicarbonate buffer, pH 9.6) (per well) and incubated overnight at 4 °C. Wells were washed with PBS containing 0.05% Tween-20 and blocked with PBS containing 1% BSA for 1 hr at 37°C. Two-fold serial dilution of the sera contained in 50 μl of PBS/0.05% Tween-20/0.5% BSA was added into each well and incubated for 2 hrs at 37°C. After washing, bound antibodies were detected with HRP-conjugated anti-human immunoglobulin (IgM, IgG, and IgA; Sourthern Biotech) at a 1:4000 dilution in PBS/0.05% Tween-20/0.5% BSA. Wells were then incubated with a HRP substrate Tetramethylbenzidine (TMB; Sigma) for 10-30 min and the reaction was stopped by addition of 100 μl of 1M H3PO4. Relative serum antibody levels were determined by standard end-point ELISA titration.
nramp1 Genotyping
Tail biopsies from sacrificed animals were incubated in 500 μl of DNA digestion buffer (50 mM Tris-HCl, pH8.0, 100 mM EDTA, pH8.0, 100 mM NaCl, 1% SDS, and 0.5 mg/ml protease K) overnight at 53°C with gentle shaking. The following day, supernatants were taken after microcentrifugation at maximum speed for 10 min. One volume of isopropanol was mixed with the supernatants to precipitate DNA. DNA pellet was obtained by microcentrifugation, resuspended in 50 μl of TE buffer, and used for _nramp_1 PCR amplification. A 514-bp fragment of the _nramp_1 gene was amplified by PCR using primers 5′-AAGTGACATCTCGCCATAGGTGCC-3′ (forward) and 5′-TTCTCTCACCATAGTTATCCAAG AAG-3′ (reverse). The purified PCR product was then sequenced using the primer 5′-CCCCCATCTATGTTATCACCC-3′.
Statistics
Statistical analysis was conducted with the two-tailed Mann-Whitney test.
Supplementary Material
01
ACKNOWLEDGEMENTS
This work was supported by NIAID Grants AI079022 and AI070949 to J. E. G. and by a grant from the Bill & Melinda Gates Foundation through the Grand Challenges in Global Health Initiative to R. A. F. who is an investigator of the Howard Hughes Medical Institute. The authors would like to thank Regeneron Pharmaceuticals for providing Rag2-/- γc-/- mice. We thank Stefania Spano for useful suggestions, A.M. Franco for isolation of CD34+ cells, P. Ranney for mouse breeding, R. Webber for mouse breeding and engraftment, J. Alderman for organizational support, and members of the Galán laboratory for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental information includes two figures.
REFERENCES
- Baenziger S, Tussiwand R, Schlaepfer E, Mazzucchelli L, Heikenwalder M, Kurrer M, Behnke S, Frey J, Oxenius A, Joller H, et al. Disseminated and sustained HIV infection in CD34+ cord blood cell-transplanted Rag2-/-gamma c-/- mice. Proc Natl Acad Sci U S A. 2006;103:15951–15956. doi: 10.1073/pnas.0604493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro L, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11:17–30. doi: 10.1038/nrg2698. [DOI] [PubMed] [Google Scholar]
- Brehm M, Cuthbert A, Yang C, Miller D, DiIorio P, Laning J, Burzenski L, Gott B, Foreman O, Kavirayani A, et al. Parameters for establishing humanized mouse models to study human immunity: analysis of human hematopoietic stem cell engraftment in three immunodeficient strains of mice bearing the IL2rgamma(null) mutation. Clin Immunol. 2010;135:84–98. doi: 10.1016/j.clim.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump J, Mintz E. Global trends in typhoid and paratyphoid Fever. Clin Infect Dis. 2010;50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan S, Ho V, Duc C, Lanh M, Phuong C, Luxemburger C, Wain J, Dudbridge F, Peacock C, House D, et al. Typhoid fever and genetic polymorphisms at the natural resistance-associated macrophage protein 1. J Infect Dis. 2001;183:1156–1160. doi: 10.1086/319289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik M, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galen J, Pasetti M, Tennant S, Ruiz-Olvera P, Sztein M, Levine M. Salmonella enterica serovar Typhi live vector vaccines finally come of age. Immunol Cell Biol. 2009;87:400–412. doi: 10.1038/icb.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghjoo E, Galan JE. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc Natl Acad Sci U S A. 2004;101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann EL, Oletta CA, Killeen KP, Miller SI. phoP/phoQ-deleted Salmonella typhi (Ty800) is a safe and immunogenic single-dose typhoid fever vaccine in volunteers. Journal of Infectious Diseases. 1996;173:1408–1414. doi: 10.1093/infdis/173.6.1408. [DOI] [PubMed] [Google Scholar]
- House D, Bishop A, Parry C, Dougan G, Wain J. Typhoid fever: pathogenesis and disease. Curr Opin Infect Dis. 2001;14:573–578. doi: 10.1097/00001432-200110000-00011. [DOI] [PubMed] [Google Scholar]
- Iacus S, Skamene E, Schurr E, Gros P. Infection genomics: Nramp1 as a major determinant of natural resistance to intracellular infections. Annu Rev Med. 1998;49:275–287. doi: 10.1146/annurev.med.49.1.275. [DOI] [PubMed] [Google Scholar]
- Kaniga K, Bossio JC, Galán JE. The Salmonella typhimurium invasion genes invF and invG encode homologues to the PulD and AraC family of proteins. Mol Microbiol. 1994;13:555–568. doi: 10.1111/j.1365-2958.1994.tb00450.x. [DOI] [PubMed] [Google Scholar]
- Kato A, Groisman E. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv Exp Med Biol. 2008;631:7–21. doi: 10.1007/978-0-387-78885-2_2. 7-21. [DOI] [PubMed] [Google Scholar]
- Kuruvilla JG, Troyer R, Devi S, Akkina R. Dengue virus infection and immune response in humanized RAG2–/–{gamma}c–/– (RAG-hu) mice. Virology. 2007;369:143–152. doi: 10.1016/j.virol.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Kwant-Mitchell A, Ashkar A, Rosenthal K. Mucosal innate and adaptive immune responses against herpes simplex virus type 2 in a humanized mouse model. J Virol. 2009;83:10664–10676. doi: 10.1128/JVI.02584-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legrand N, Weijer K, Spits H. Experimental models to study development and function of the human immune system in vivo. J Immunol. 2006;176:2053–2058. doi: 10.4049/jimmunol.176.4.2053. J Immunol 176, 2053-2058. [DOI] [PubMed] [Google Scholar]
- Libert C, Brouckaert P, Shaw A, Fiers W. Induction of interleukin 6 by human and murine recombinant interleukin 1 in mice. Eur J Immunol. 1990;20:691–694. doi: 10.1002/eji.1830200333. [DOI] [PubMed] [Google Scholar]
- Manz M. Human-hemato-lymphoid-system mice: opportunities and challenges. Immunity. 2007a;26:537–541. doi: 10.1016/j.immuni.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Manz M. Human-hemato-lymphoid-system mice: opportunities and challenges. Immunity. 2007b;26:537–541. doi: 10.1016/j.immuni.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Mirza S, Beeching N, Hart C. Multi-drug resistant typhoid: a global problem. J Med Microbiol. 1996;44:317–319. doi: 10.1099/00222615-44-5-317. [DOI] [PubMed] [Google Scholar]
- Monack D, Bouley D, Falkow S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med. 2004;199:231–241. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien A. Innate resistance of mice to Salmonella typhi infection. Infect Immun. 1982;38:948–952. doi: 10.1128/iai.38.3.948-952.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien AD, Rosenstreich DL, Taylor BA. Control of natural resistance to Salmonella typhimurium and Leishmania donovani in mice by closely linked but distinct genetic loci. Nature. 1980;287:440–442. doi: 10.1038/287440a0. [DOI] [PubMed] [Google Scholar]
- Pang T, Levine MM, Ivanoff B, Wain J, Finlay BB. Typhoid fever--important issues still remain. Trends Microbiol. 1998;6:131–133. doi: 10.1016/s0966-842x(98)01236-0. [DOI] [PubMed] [Google Scholar]
- Parry C, Hien TT, Dougan G, White N, Farrar J. Typhoid fever. N Engl J Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- Raffatellu M, Wilson R, Winter S, Bäumler A. Clinical pathogenesis of typhoid fever. J Infect Dev Ctries. 2008;2:260–266. doi: 10.3855/jidc.219. [DOI] [PubMed] [Google Scholar]
- Sabbagh S, Forest C, Lepage C, Leclerc J, Daigle F. So similar, yet so different: uncovering distinctive features in the genomes of Salmonella enterica serovars Typhimurium and Typhi. FEMS Microbiol Lett. 2010 Jan 20; doi: 10.1111/j.1574-6968.2010.01904.x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Schoenhaut D, Chua A, Wolitzky A, Quinn P, Dwyer C, McComas W, Familletti P, Gately M, Gubler U. Cloning and expression of murine IL-12. J Immunol. 1992;148:3433–3440. [PubMed] [Google Scholar]
- Shultz L, Ishikawa F, Greiner D. Humanized mice in translational biomedical research. Nat Rev Immunol. 2007;7:118–130. doi: 10.1038/nri2017. [DOI] [PubMed] [Google Scholar]
- Sinnott C, Teall A. Persistent gallbladder carriage of Salmonella typhi. The Lancet. 1987;329:976. doi: 10.1016/s0140-6736(87)90319-9. [DOI] [PubMed] [Google Scholar]
- Spano S, Ugalde JE, Galan JE. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe. 2008;3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Sztein M. Cell-mediated immunity and antibody responses elicited by attenuated Salmonella enterica Serovar Typhi strains used as live oral vaccines in humans. Clin Infect Dis. 2007;45:S15–19. doi: 10.1086/518140. [DOI] [PubMed] [Google Scholar]
- Yu CI, Gallegos M, Marches F, Zurawski G, Ramilo O, Garcia-Sastre A, Banchereau J, Palucka A. Broad influenza-specific CD8+ T-cell responses in humanized mice vaccinated with influenza virus vaccines. Blood. 2008;112:3671–3678. doi: 10.1182/blood-2008-05-157016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01