DNA Replication Fidelity and Cancer (original) (raw)
. Author manuscript; available in PMC: 2011 Oct 15.
Abstract
Cancer is fueled by mutations and driven by adaptive selection. Normal cells avoid deleterious mutations by replicating their genomes with extraordinary accuracy. Here we review the pathways governing DNA replication fidelity and discuss evidence implicating replication errors (point mutation instability or PIN) in carcinogenesis.
Keywords: DNA polymerase, proofreading, mismatch repair, mutator, cancer
1. Genetic instability and cancer
Tumor development is a multistep process requiring the accumulation of mutations that activate oncogenes or inactivate tumor suppressors [1–3]. To maintain normal cell functions, genetic stability is strictly controlled [4]. Therefore it is argued that defects in pathways governing genetic stability will facilitate tumorigenesis by fueling the reiterative process of mutation, selection and clonal expansion that drives cancer progression (reviewed and debated in [5–13]).
There is considerable evidence that genetic instability plays a role in cancer (reviewed in [3, 13–16]). Cancer cells exhibit high frequencies of chromosomal aberrations [17, 18], and the rates of gene rearrangements and amplifications are increased in many tumor cell lines [19–21]. Moreover, inherited deficiencies in genome maintenance systems contribute to human cancer susceptibility syndromes [22–24]. Defects in genes required for nucleotide excision repair (NER; Xeroderma pigmentosum), double-strand break recognition and repair (Ataxia telangectasia, Nijmegen breakage syndrome), genetic recombination (Bloom, Werner and Rothmund-Thomson syndromes and BRCA1/2) and mismatch repair (MMR; Lynch syndrome) all cause genetic instability and are associated with human cancer syndromes. Thus, it is well established that chromosome instability (CIN) and microsatellite instability (MIN or MSI) predispose to cancer. Recently, sporadic cancers were shown to have elevated frequencies of random nucleotide point mutations, thus implicating point mutation instability (PIN) in oncogenesis [25, 26].
Perhaps the strongest early evidence that increased spontaneous mutation (i.e., mutator phenotype) contributes to human cancer was the discovery that defective mismatch repair (MMR) causes hereditary colon cancer (reviewed in [27–31]). In the early 1990s, colorectal cancer samples from Lynch syndrome pedigrees (also called hereditary nonpolyposis colon cancer or HNPCC) were noted to have microsatellite instability, normal cytogenetics, and were associated with a unique clinical presentation. Two groups simultaneously reported that these families carried mutations in MSH2, the gene encoding one of the primary proteins required for MMR [32, 33]. Shortly thereafter, MLH1, the gene encoding another essential MMR protein, was cloned and found to be mutated in additional Lynch syndrome families [34, 35]. The majority of Lynch syndrome patients inherit a mutation in either MSH2 or MLH1, with a smaller percentage inheriting mutations in PMS2 or MSH6 [36, 37]. The wild-type allele is then lost in tumors through LOH or gene silencing. Patients with inherited MMR deficiency are primarily susceptible to early-onset colorectal cancer, but also have an increased risk for extra-intestinal neoplasms. Inherited MMR defects are only responsible for a small number (1–5%) of colorectal cancer cases; thus, most colorectal cancers with MSI (~15% of all colorectal cancer cases) result from acquired defects in MMR, almost exclusively due to MLH1 promoter hypermethylation [38]. MMR defects and MSI are also detected in non-colonic sporadic tumors, most commonly in endometrial, lung and gastric cancer [38, 39].
MMR-deficient human tumor cell lines display increased spontaneous mutation rates with a preference for frameshifts and base substitution mutations [40–43]. Accordingly, microsatellite instability is a hallmark of MMR loss [31]. Microsatellite instability may be particularly relevant for colorectal cancer as many genes involved in intestinal carcinogenesis (TGF_β_R2, APC, KRAS, BRAF, and others) have repetitive DNA in their coding regions [44]. MSH6 defects are a less common cause of Lynch syndrome and result in predominantly extra-colonic tumors [45–49]. Interestingly, cells defective for MSH6 have elevated rates of base-substitution mutations and lower levels of frameshifts, due to selective inactivation of MutSα [43, 50, 51]. Thus, intestinal carcinogenesis in Lynch syndrome may be directly related to MSI, whereas extra-colonic tumors may result from elevated base substitutions.
2. Determinants of DNA replication fidelity
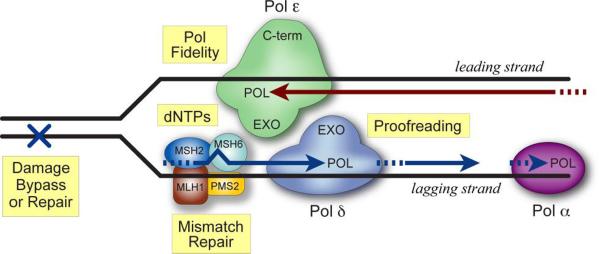
Normal cells replicate their DNA with extraordinary fidelity (~10−10 mutations per base pair per cell division) [52]. This is achieved through the combined actions of polymerase base selectivity, 3′→5′ exonucleolytic proofreading, mismatch correction and DNA damage repair (Fig. 1; reviewed in [53–71]).
Fig. 1.
Determinants of DNA replication fidelity. Schematic of a DNA replication fork with Pol ε and Pol δ on the leading- and lagging-strands, respectively. Major determinants of faithful DNA synthesis are highlighted in yellow. The polymerase domains (POL) of Pols ε and δ discriminate between correct and incorrect dNTPs prior to phosphodiester bond formation. If an error occurs, these are corrected primarily by the intrinsic proofreading exonuclease (EXO) present in each polymerase. Errors that escape proofreading are rectified by mismatch repair (MMR), which acts on both lagging (shown here) and leading (not shown) strands. DNA damage repair and dNTP pool ratios also influence replication fidelity.
Proofreading and MMR both contribute substantially to the overall fidelity of cellular DNA replication and mutation avoidance. Genetic experiments in E. coli [53, 54] and S. cerevisiae [50, 51, 72–76] show that loss of either proofreading or MMR results in a 10- to 1000-fold increase in spontaneous mutation rate. Although studies in mammalian cells are more limited, cell-free fidelity assays [77, 78] and experiments with MMR-deficient [40–43, 79, 80] and proofreading-deficient [81–84] cells also point to these repair pathways as major determinants of replication fidelity in higher eukaryotes. The prevailing model (Figs. 1 and 2) is that spontaneous errors by the replicative lagging- and leading-strand DNA polymerases (Pols δ and ε, respectively [85–87]) trigger proofreading by their intrinsic 3′-exonucleases. Occasional errors escape proofreading, and these are corrected by the MMR machinery. It is estimated that replicative eukaryotic DNA polymerases make errors approximately once every 104 – 105 nucleotides polymerized [58, 59]. Thus, each time a diploid mammalian cell replicates, at least 100,000 and up to 1,000,000 polymerase errors occur.1 The majority of these are base•base mispairs and ±1 slippage events [58, 59], which must be corrected with almost 100% efficiency to achieve a spontaneous mutation rate of ~10−10 per base pair per cell division [52].
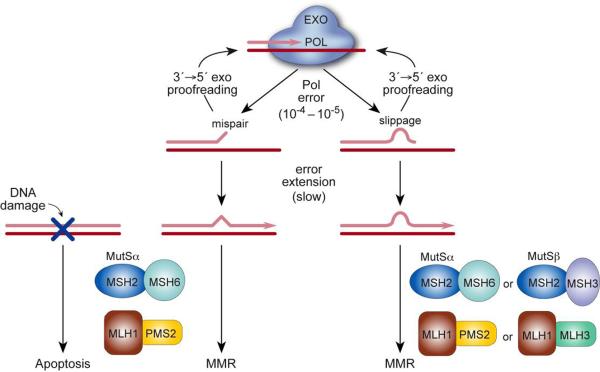
Fig. 2.
Pathways correcting DNA polymerase errors. During DNA synthesis, rare polymerase errors [base•base mispairs (left) or primer•template slippage (right)] impede primer extension and thus trigger transfer of the growing DNA strand from the polymerase active site POL) to the 3′→5′ exonuclease active site (EXO) where the errant bases are excised by proofreading. Errors that escape 3′→5′ exonucleolytic proofreading are corrected, at least in part, by two partially redundant pathways of the MMR system. Studies in yeast indicate that Pol proofreading and MMR act in series along a common pathway [75, 76]. MutSα and MutSβ have overlapping substrate specificities, and MutSα also recognizes DNA damage and signals apoptosis. [Note: mammalian Pms2 (shown here) is equivalent to yeast Pms1.]
Repair of promutagenic DNA damage (both spontaneous and induced; [22, 68–71]) and maintenance of normal dNTP pools [88–90] are also important determinants of replication fidelity. Similar to proofreading and mismatch repair, defects in individual enzymes affecting dNTP pool ratios confer spontaneous mutator phenotypes [88–90]. In contrast, most single-gene defects in DNA damage repair pathways exhibit near-normal spontaneous mutation rates and reveal themselves as “conditional mutators” when cells are challenged with DNA damaging agents [71]. One exception is the repair of 8-oxo-G lesions by the MutM/MutY/MutT “GO” system [54, 91–95]. In E. coli, loss of either MutM or MutT confers a moderate-to-strong mutator phenotype in the absence of exogenous oxidative stress [54, 92, 96]. However, defects in homologous mouse genes have only modest effects on spontaneous mutation rates, presumably due to different interactions of redundant pathways that prevent or repair oxidative DNA damage in mammals [71, 93–95].
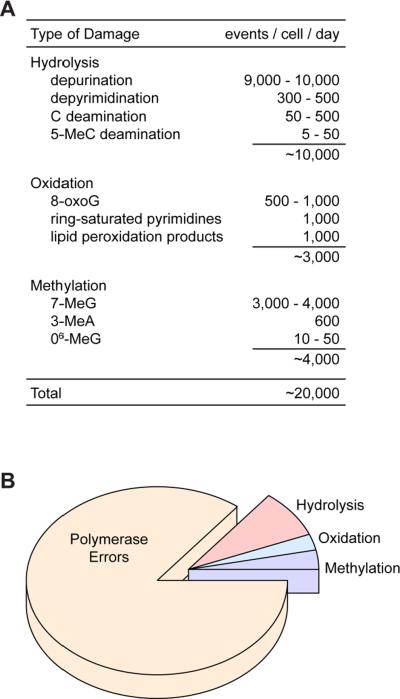
Quantitative estimates of spontaneous DNA degradation in cells suggest that the daily dose of promutagenic damage is substantial [97–99] (Fig. 3A). These lesions result from the intrinsic chemical instability of DNA under physiological conditions and the exposure of DNA to active oxygen and other reactive metabolites and coenzymes that are generated by normal cells [97, 99]. Altogether, it is estimated that ~20,000 potentially mutagenic lesions arise per diploid mammalian cell per day. Most of these lesions are repaired by the base excision repair (BER) pathway [68, 69]. This repair must occur efficiently prior to DNA replication for cells to maintain a low spontaneous mutation rate. The toll of 20,000 spontaneous lesions per cell per day is high, and this is in addition to the 100,000 – 1,000,000 DNA polymerase errors that occur in replicating cells over the same approximate time frame (Fig. 3B).
Fig. 3.
DNA “lesions” arising spontaneously in mammalian cells. (A) Spontaneous chemical decay of deoxyribonucleic acid. Rates are expressed as the number of decay events estimated to occur per mammalian cell per day under physiological conditions ([97, 98]; T. Lindahl, personal communication). (B) Distribution of DNA lesions formed per cell per day in dividing mammaliaan cells. Values for hydrolysis, oxidation and methylation products are from panel A. Polymerase errors (100,000 or more per cell division) are based on measurements of Pol δ and Pol ε error rates in vitro in the absence of proofreading as described in the text and footnote 1. This assumes that actively dividing cells replicate approximately once every 24 hours. Shorter or longer cell cycles will occur in vivo depending on cell type, tissue, stage of embryonic development, and responses to environmental conditions. Panel B shows the distribution when there are 100,000 polymerase errors per day, but this could be as high as 1,000,000 errors per day (see footnote 1).
Thus, there are a number of repair systems that ensure faithful DNA replication. Among these, polymerase proofreading and MMR play primary roles as evidenced by the strong mutator phenotypes conferred by loss of either pathway in the absence of exogenous DNA damage.
2.1. Proofreading polymerases
There are 3 eukaryotic DNA polymerases that have intrinsic 3′→5′ exonucleolytic proofreading activity: Pol δ, Pol ε and Pol γ [60, 100]. Only Pols δ and ε are nuclear, while Pol γ is mitochrondrial. Similar to other proofreading DNA polymerases, Pols δ and ε are comprised of multiple subunits with the catalytic peptide of each enzyme harboring both polymerase (pol) and exonuclease (exo) activities. These proofreading polymerases, together with Pol α:primase, are the primary replicative enzymes functioning at DNA replication forks (reviewed in [87, 101, 102]). Pol α primes both leading- and lagging-strand synthesis and copies relatively short stretches of DNA, while Pols δ and ε are responsible for the bulk of chromosomal DNA synthesis during cell division. In yeast, Pols ε and δ are principal leading- and lagging-strand DNA polymerases, respectively [85–87].
A large literature shows that Pol δ and Pol ε play essential roles in the replication and repair of chromosomal DNA [101–108]. These roles are most readily demonstrated using genetic approaches in yeast. Pol δ contributes to yeast lagging-strand DNA synthesis from replication origins [86, 108] and the maturation of Okazaki fragments [102, 109]. Pol ε participates in leading-strand elongation [85] (perhaps preferentially during late S phase [110]) in addition to its more specialized roles at DNA replication origins [111–114], in S phase checkpoint control [115], and sister chromatid cohesion [116]. Studies in mammalian cells also establish essential roles for Pol δ and Pol ε in chromosomal DNA replication. Both polymerases associate with nascent chromosomal DNA [110, 117, 118], and anti-Pol δ and -Pol ε antibodies inhibit chromosomal replication when introduced into human cells [118, 119]. Interestingly, SV40 DNA replication requires only Pols α and δ and occurs by a Pol ε-independent mechanism [117, 119]. Thus, SV40 replication is not a complete model of S phase-regulated cellular DNA replication [102]. Finally, Pols δ and ε catalyze DNA synthesis in several repair pathways including nucleotide excision repair, MMR, the long-patch pathway of base excision repair, and double-strand break repair [71, 101, 120]. Thus, Pols δ and ε participate in most cellular pathways that require nuclear DNA synthesis, and together they synthesize the bulk of chromosomal DNA during cell division.
Point mutations that selectively inactivate exonucleolytic proofreading activities of Pol δ or Pol ε confer moderate-to-strong mutator phenotypes in yeast [75, 76, 121–128]. For reasons not fully understood, this mutator effect tends to be greater in Pol δ than Pol ε proofreading mutants. Consistent with their leading- and lagging-strand functions, there is good evidence that the proofreading exonucleases of Pol δ and Pol ε correct DNA replication errors on opposite strands [76, 127, 129]. However, it is not clear whether they always correct their own errors. Studies of double mutants show that the proofreading activities of Pol δ and Pol ε synergize to suppress spontaneous mutations and that this synergy reflects a competitive relationship [76, 122, 126]. This suggests that yeast Pol δ and Pol ε can act on the same pool of DNA replication errors and may proofread for each other. Recent studies indicate that Pol δ and Pol ε can also proofread for Pol α [130, 131], suggesting that these and other 3′-exonucleases in the cell [60] could function as extrinsic proofreading enzymes [132, 133]. Evidence for overlapping polymerase functions also comes from the observation that the polymerase domain of Pol ε is dispensable in yeast when it is deleted (albeit with severe growth consequences; [105, 106, 122, 134]), while inactivating point mutations in the intact pol domain of Pol ε are lethal [105]. Thus, it appears that yeast Pols δ, ε, α and perhaps other polymerases interact and can serve partially redundant functions. The roles of Pol δ and Pol ε proofreading and their interactions in mammalian cells are largely unknown.
2.2. Mismatch repair
Eukaryotes have multiple MMR proteins that exhibit distinct substrate preferences (reviewed in [62–67]). In mammals, polymerase errors are recognized and bound by complexes containing the Msh2 protein. Both the Msh6 and Msh3 proteins heterodimerize with Msh2 and dictate substrate preferences. Biochemical studies show that base•base and small insertion/deletion mispairs are primarily targeted by the Msh2/Msh6 heterodimer (MutSα), while larger insertion/deletion loops are bound primarily by a Msh2/Msh3 complex (MutSβ) [135–138]. Similar substrate binding preferences are observed for yeast and mammalian Msh proteins in vitro [138], and genetic studies in yeast confirm that MutSα preferentially repairs base•base mispairs and 1-bp loops while MutSβ prefers larger loops [50, 51, 62]. However, MutSα and MutSβ are redundant with respect to the repair of small insertion/deletion mispairs [50, 51], and MutSβ recognizes a limited set of base•base mispairs in certain sequence contexts [139]. There is also functional overlap between MutSα and MutSβ in human cells [43]. To initiate repair, the MutS complexes interact with a heterodimer of MutL homologues (MutLα comprised of Mlh1/Pms1 in yeast, equivalent to Mlh1/Pms2 in mammals), which contains endonucleolytic activity that nicks the nascent strand both 5′ and 3′ of the mismatch [140, 141]. These nicks provide entry points for enzymes that remove the mismatched sequence, allowing re-synthesis of the DNA [140, 142].
In addition to correcting mispairs, MMR proteins play an important role as sensors of DNA damage that signal cell death ([79, 143]; reviewed in [67, 144–148]). Although initially thought to confer sensitivity to a broad range of DNA damaging agents, it is now apparent that MMR-mediated cytotoxicity occurs primarily in response to Sn1 alkylating agents, 6-thioguanine and cisplatin [149]. There is also evidence that some DNA adducts (e.g., 8-oxo-G and O6-methyl-G) are repaired by MMR [150, 151] and that misincorporated bases opposite some adducts are excised by MMR [152, 153], potentially resulting in a futile repair cycle [154]. These MMR-dependent, damage-response functions are mediated by MutSα complexed with MutLα. Thus, cells with defective Msh2, Msh6 or Mlh1 are resistant to DNA damage-induced apoptosis, whereas Msh3 mutants remain sensitive and indistinguishable from wild type [155–157]. Studies examining mechanisms of MMR-dependent apoptosis show that MMR proteins recognize and bind to DNA lesions and initiate a signal transduction cascade that activates G2/M cell cycle checkpoints and engages the apoptotic machinery [67, 147, 158].
2.3. Cooperativity of proofreading and MMR
Genetic studies in yeast show that Pol proofreading and MMR serve overlapping functions in the repair of DNA replication errors and that they exhibit a synergistic relationship [75, 76, 121, 126, 159]. Proofreading defects in Pol δ or Pol ε are synthetically lethal in haploid cells when combined with loss of pms1, mlh1 or msh2 [75, 126], and defective Pol δ proofreading is synthetically lethal with nullizygous msh6 [160]. In diploids proofreading/MMR double mutants are viable, but grow slowly and exhibit extremely high spontaneous mutation rates [75, 76, 124–126]. This implies that deficiencies in either proofreading or MMR can be partially compensated by the function of the other and that loss of both pathways results in the accumulation of replication errors and compromised growth. These data also indicate that proofreading and MMR act in series along a common pathway (see discussion in [75, 76]). This is supported by evidence that MMR proteins interact with components of the DNA replication apparatus [125, 161–164]. Synergy occurs primarily with the Msh6 pathway of MMR [125], which is consistent with proofreading and Msh6-dependent MMR being the principal pathways that correct base•base mispairs [58–67]. Interestingly, the fidelity of lagging-strand DNA synthesis in yeast is greater than that of leading-strand [165], because MMR preferentially corrects lagging-strand replication errors [153].
Evidence for cooperativity in mammals comes from recent studies of mice with defects in both proofreading and MMR [83, 84]. Loss of either Pol δ or Pol ε proofreading combined with defective MMR results in embryonic lethality [83]. Interestingly, Pol δ and Pol ε proofreading defects confer different phenotypes in embryos. In the absence of MMR, early mouse development requires proofreading by Pol δ but not Pol ε, and this dependency is reversed later in development (see discussion in [83]). These results suggest that Pols δ and ε have unique roles at different stages of mammalian development. Moreover, they indicate that embryogenesis requires faithful DNA replication and that dual loss of proofreading and MMR exceeds an error threshold.
3. Mutator mice
Although mechanisms of DNA replication fidelity have been studied extensively in simple microbes, less is known about the corresponding pathways and their functions in higher eukaryotes. Mammals require faithful DNA replication to avoid deleterious mutations in critical subpopulations of cells (gametes, stem cells, developing embryos) and to sustain essential somatic functions throughout the adult reproductive life span. At the same time, increased mutation (hypermutation) is required for normal functions in some somatic tissues [166]. Thus, the complexities of mammalian biology impose unique constraints on DNA replication fidelity that vary among tissues, cell lineages and genetic loci.
To study the effects of repair deficiencies on tumor predisposition, mice containing knockout and knockin alleles of a number of DNA repair genes have been constructed [167]. These mouse lines are powerful experimental tools that reveal the biological functions of mammalian mutation-avoidance pathways and the genetic interactions that occur among repair genes, oncogenes and tumor suppressors. Furthermore, cells obtained from these animals are used to probe mechanisms of mammalian DNA repair at the cellular level.
Interestingly, deficiencies in individual components of the major DNA damage repair pathways (BER and NER) do not generally manifest spontaneous cancer phenotypes. This contrasts with deficiencies in MMR (reviewed in [168, 169]) or proofreading [81–84] which, by themselves, confer strong spontaneous cancer phenotypes in the absence of DNA damaging agents or other cancer-susceptibility genes. Thus, MMR and Pol proofreading are different from most DNA repair pathways studied in mice in that the simple loss of either pathway alone is sufficient to cause spontaneous cancer with almost 100% penetrance.
3.1. DNA damage repair
A number of laboratories have produced mice with mutations in genes that affect repair of endogenous DNA damage (summarized in [167]). Knockouts of uracil-DNA glycosylase (Ung/Udg), O6-methylguanine-DNA methyltransferase (Mgmt), 3-methyladenine-DNA glycosylase (Aag/Apng), 8-oxoguanine-DNA glycosylase (Ogg1), or NER genes do not result in a significant spontaneous cancer predisposition, although indolent lung tumors are observed in Xpc mutant mice late in life [170]. Mice defective for Mth1 (an 8-oxo-dGTPase) or Myh (an adenine DNA glycosylase that repairs A:8-oxo-G mispairs) have relatively weak spontaneous cancer phenotypes [171–174]. However, when _Myh_-null mice are exposed to oxidative stress, the incidence and numbers of intestinal tumors are dramatically increased [174]. Mgmt-deficient mice as well as mice lacking components of the NER pathway also exhibit conditional cancer-susceptibility phenotypes when exposed to exogenous DNA damaging agents [22, 167, 175–177].
Some repair gene knockouts reveal phenotypes when combined with defects in other cancer-susceptibility genes [167, 178–181] or combined with defects in redundant repair pathways. A good example of redundancy is in the repair of 8-oxo-G lesions. Mice with deficient Ogg1 or Myh alone develop few tumors and live normal lifespans [172–174, 182, 183]. In contrast, the combined loss of Ogg1 and Myh results in a high spontaneous tumor incidence and cancer mortality [173]. Thus, different pathways of repair often have overlapping functions that can influence the biological impact of a single class of DNA lesions [71].
3.2. Mismatch repair
Soon after the discovery of MMR defects in Lynch syndrome, several groups generated and characterized mouse strains with knockout and knockin alleles of the MMR genes (reviewed in [168, 169]. Many aspects of the Lynch syndrome phenotype are recapitulated in mice deficient for Msh2, Mlh1, or Msh6. Homozygous mutant mice display increased tumor susceptibility, consisting primarily of gastrointestinal tract tumors and lymphomas. Mouse embryonic fibroblasts obtained from Msh2 or Mlh1 knockouts have increased frameshift and base-substitution mutation rates and exhibit resistance to DNA alkylating agents much like human MMR-deficient tumor cells [80, 184, 185]. Tumors as well as somatic tissue in these mice show microsatellite instability [186] and increased mutant frequencies as revealed in transgenic reporter systems [187–190].
It is tempting to conclude that cancer results from the mutator phenotype associated with loss of MMR. However, as discussed above, MMR proteins also play an important role as sensors of DNA damage that signal apoptosis [67, 144–148]. This has important implications for tumorigenesis [191]. In addition to acquiring a mutator phenotype, inactivation of MMR reduces the apoptotic response to endogenous and exogenous sources of DNA damage. The combination of a mutator phenotype and defective apoptosis is expected to accelerate the reiterative process of mutation and clonal expansion that drives tumor progression [5]. While a mutator phenotype will affect any rapidly dividing cell, defective apoptosis will have the greatest impact in tissues under genotoxic stress (e.g., gut, skin, lungs and inflammation). Thus, MMR-defective tumors may result from loss of mismatch repair, loss of apoptosis or both, and the relative contribution of these factors likely varies in different tissues. Studies of mice carrying mutations that selectively inactivate the mismatch repair activities of Msh2 or Msh6 but preserve their damage-response functions indicate that the increased mutation rate in MMR-deficient cells is sufficient to drive tumorigenesis [192, 193] and that defective apoptosis can accelerate this tumorigenesis [193].
While the mouse MMR gene knockouts have a phenotype similar to that found in Lynch syndrome patients, there are also notable differences. The Msh2 and Mlh1 knockout mice primarily develop lymphomas (which are not generally found in Lynch syndrome) as well as intestinal adenomas and adenocarcinomas. However, a major difference is that only homozygous null mice (_i.e., Msh2_−/− and _Mlh1_−/−) develop tumors, while the heterozygotes do not. Lynch syndrome patients carry heterozygous mutations of the MMR genes, and with rare exceptions [194], normal somatic tissues from these individuals do not show evidence of microsatellite instability or increased mutation rate. There are a number of factors that could account for these discrepancies: life span, diet, environmental exposures, differences in human and mouse somatic cell biology or DNA repair pathways.
3.3. Polymerase proofreading
The studies of MMR-deficient mice clearly demonstrate that MMR genes function as tumor suppressors and suggest that unrepaired polymerase errors may contribute to tumorigenesis. To more directly examine the role of polymerase errors in cancer, we created mice with inactivating point mutations in the proofreading exonuclease domains of DNA Pols δ and ε and examined the effects of defective proofreading on spontaneous mutation and tumorigenesis [81–84].
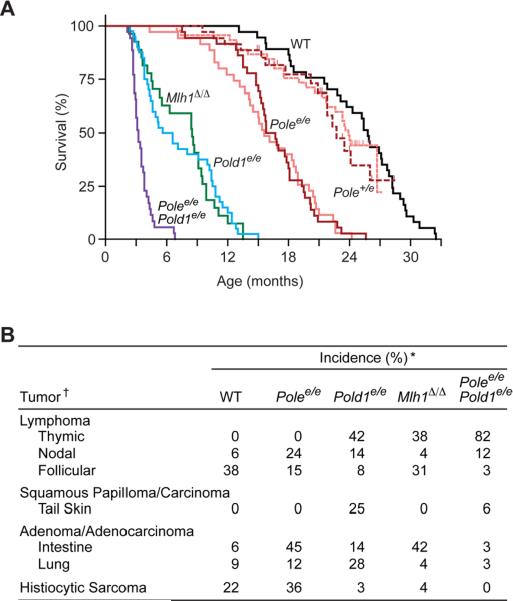
Our initial studies focused on Pol δ [81, 82]. We used a knockin strategy to introduce a single mutation in Pold1, the gene encoding the catalytic subunit of Pol δ. This mutation (designated Pold1e) eliminates a catalytic carboxylate residue (D400A) in the exonuclease domain of Pol δ that is required for proofreading activity. The analogous substitution in yeast Pol δ (D407A) confers a strong spontaneous mutator phenotype [72] characterized by elevated base-substitutions and ±1 frameshifts. We observed that Pold1e inactivates the 3′→5′ exonuclease of mouse Pol δ and causes a recessive mutator and cancer phenotype. Greater than 95% of homozygous Pold1e/e mice spontaneously develop cancer and die by 15 months of age (Fig. 4A). Most of the cancers are epithelial in origin, and one-third of the animals develop tumors in more than one tissue. Interestingly, the prevalent cancers in Pold1e/e mice (thymus, skin & lung tumors, but not intestine) differ from those observed in MMR knockout mice (thymus and intestinal tumors, but not skin; [168, 169]; Fig. 4B). Thus, MMR is rate-limiting for tumorigenesis in intestinal epithelium and both thymic and splenic lymphocytes, while Pol δ proofreading is rate-limiting in lung epithelium, skin epidermis and only thymic lymphocytes. This study shows that mammalian Pol δ proofreading suppresses spontaneous mutation and tumor development and strongly suggests that unrepaired polymerase errors contribute to carcinogenesis.
Fig. 4.
Survival and cancer phenotypes of Pol δ and Pol ε proofreading-deficient mice. (A) Kaplan-Meier survival estimates. Mice were followed for long-term survival and observed daily until moribund or unexpected natural death. dark red, Polee/e (n=36) and Pole+/e (n=35) in C57BL/6 genetic background after removal of the neomycin selection cassette (_Neo_−); light red, Polee/e (n=35) and Pole+/e (n=45) in a mixed C57BL/6:129/Sv genetic background with the neomycin selection cassette still present (Neo+); blue, Pold1e/e (n=40) in C57BL/6 (_Neo_−); purple, Polee/ePold1e/e (n=35) in C57BL/6 (_Neo_−); black, wild-type (WT; n=37) C57BL/6; green, _Mlh1_Δ/Δ (n=27) in C57BL/6. One month = 30.4 days. (B) Spontaneous tumor incidences. Moribund mice were euthanized and necropsied, and tumors were diagnosed by histology. *Incidences among 32 wild-type (WT), 33 Polee/e, 36 Pold1e/e, 26 _Mlh1_Δ/Δ and 34 Polee/ePold1e/e mice. †Tumors with ≥15% incidence in one or more groups. Figure from Albertson et al. [83].
Using a similar allelic replacement strategy, we recently engineered mice with defective Pol ε proofreading [83]. In this case, two amino acid substitutions (D272A and E274A) were introduced into the highly conserved Exo I motif of the Pol ε exonuclease domain. The encoding allele, Polee, is equivalent to yeast pol2–4, which selectively inactivates proofreading while preserving normal polymerase activity [73]. To directly compare the effects of defective Pol ε proofreading, Pol δ proofreading and MMR, mutant congenic mouse strains were generated in the C57BL/6 genetic background. We found that homozygous loss of Pol ε proofreading confers a strong, base-substitution mutator phenotype and high incidence of spontaneous neoplasms. Surprisingly, Pold1e/e and Polee/e mice exhibit very different survivals and cancer susceptibilities (Fig. 4). Consistent with our initial findings, Pold1e/e mice died with thymic lymphomas, lung adenocarcinomas and skin squamous cell carcinomas during the first year of life. In contrast, Polee/e mice developed spontaneous intestinal adenocarcinomas, histiocytic sarcomas and nonthymic lymphomas and survived up to 24 months of age. The combined loss of both Pol δ and Pol ε proofreading (Pold1e/e Polee/e) further accelerates cancer mortality, while proofreading/MMR double mutant mice die in utero [83]. Strong cooperativity between proofreading and MMR was also demonstrated in MMR nullizygotes expressing heterozygous proofreading alleles; these mice rapidly succumb to aggressive metastatic lymphomas [84]. Taken together, these studies distinguish Pol ε and δ functions in vivo and reveal tissue-specific requirements for DNA replication fidelity. Moreover, they demonstrate strong synthetic interactions between proofreading and MMR in mammals.
The tissue-specific cancer phenotypes of Polee/e and Pold1e/e mice suggest that Pol ε and Pol δ proofreading participate in separate pathways of tumor suppression that are contingent on cell type. Several molecular and cellular mechanisms may contribute. Pols ε and δ are involved in DNA damage repair and lesion bypass [71, 101]. Thus, Pol ε may preferentially protect intestinal epithelium from natural carcinogens in the gut, while Pol δ may be required for faithful repair or bypass of DNA damage unique to skin. Cell lineage-specific differences in the expression levels of each polymerase, of MMR or other repair pathways that interact with Pol ε or Pol δ would similarly shape the cancer phenotype. There is also evidence that Pols ε and δ can function independently in the cell [110, 118], perhaps replicating different regions of the genome that encode tissue-specific oncogenes or tumor suppressors. And these cancer genes may have “hotspots” that are preferentially mutated by one polymerase or the other. Finally, tissue specificity may not be a direct consequence of increased spontaneous mutation [8]. Secondary functions of proofreading exonucleases [60, 61, 195–197] and actions of Pol ε in cell cycle control [115] and gene silencing [198] could modify cell turnover and oncogenesis in select tissues. Other models have been proposed to explain the unique cancer spectrum caused by defective MMR [199]. Additional studies are needed to identify mechanisms that govern tissue-specific DNA replication fidelity and somatic disease.
4. Pol δ and ε mutations in human cancer
The studies of Pol δ and ε proofreading-deficient mice underscore the importance of DNA polymerase fidelity in the suppression of spontaneous tumorigenesis. It is reasonable to speculate that polymerase mutator alleles also increase the risk of human cancers [200, 201]. The mouse studies suggest roles for Pol ε mutators in the genesis of intestinal tumors and Pol δ mutators in skin and lung carcinogenesis. Pol ε and δ mutators are also implicated in hematologic malignancies, and aggressive lymphomas or other pediatric neoplasms may result from the combined loss of proofreading and MMR. In addition to proofreading defects, mutations affecting base selectivity by Pols δ and ε also confer mutator phenotypes in yeast [85, 86, 202] and mice [203]. Thus, similar mutations or polymorphisms could be oncogenic in humans.
4.1. Single-nucleotide polymorphisms (SNPs) in Pol δ and Pol ε genes
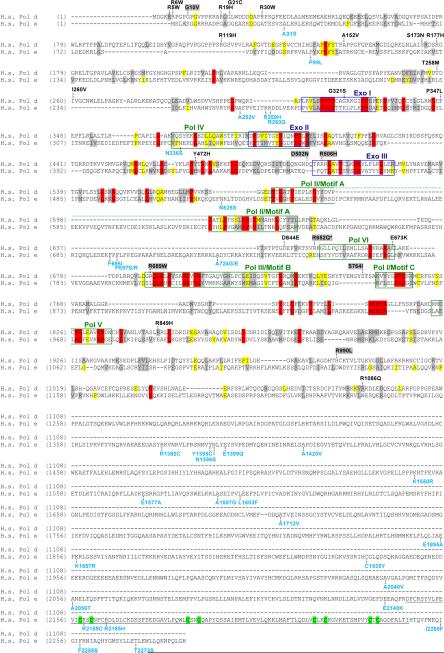
The catalytic subunits of human Pols δ and ε are encoded by POLD1 and POLE. Review of the NCBI dbSNP (www.ncbi.nlm.nih.gov/SNP) and NIEHS geneSNP (www.genome.utah.edu/genesnps) databases currently identifies 18 SNPs in POLD1 and 33 SNPs in POLE that result in amino acid changes (Fig. 5). Several SNPs (POLD1 G321S and E673K; POLE N336S) are located in Pol or Exo motifs, which are conserved between diverse species and contain important residues for polymerase and exonucleolytic proofreading functions [60, 204, 205]. Only four SNPs (POLD1 R849H and R1086Q; POLE F695I and E1577A) have been modeled in yeast, and none were found to be mutators in the presence or absence of MMR [206].
Fig. 5.
Pol δ and Pol ε amino acid changes encoded by human SNPs and found in tumor cells. Alignment of human Pol δ and Pol ε (H.s. Pol d and H.s. Pol e, respectively). To generate this alignment, yeast and human Pol δ and ε sequences were first aligned as described [205], and conserved amino acid residues were highlighted as follows: red, absolutely conserved among the four sequences; yellow, identical in the majority of sequences; gray, similar in majority of sequences. Yeast sequences were then remove from the figure for clarity. Conserved polymerase and exonuclease motifs are indicated by colored frames: blue, exonuclease motifs; green, polymerase motifs [204]. The zinc finger in Pol ε (residues 2146–2246) is underlined with component cysteines highlighted in green. Amino acid changes encoded by SNPs are placed above (Pol δ, black) or below (Pol ε, cyan) the alignment at the relevant positions. Data are from the NCBI dbSNP (www.ncbi.nlm.nih.gov/SNP) and NIEHS geneSNP (www.genome.utah.edu/genesnps) databases. Pol δ changes found in human colorectal cancer cell lines or primary tumors [214, 215] are highlighted in gray above the Pol δ sequence. Asterisk (*) marks the amino acid substitution found in rat Novikoff hepatoma cells (rat R648Q equivalent to human R652Q [216]).
There are relatively few studies examining the cancer risk associated with non-synonymous SNPS in POLD1 or POLE. POLE E2140K has been associated with an increased risk of breast cancer [207], but not cervical cancer [208]; A252V is associated with decreased overall survival in lung cancer patients [209]; and N1396S is not associated with meningioma risk [210]. The POLD1 R119H SNP has been reported in multiple genome-wide SNP studies, and no association was detected with meningioma, bladder or breast cancer risk [210–213]. Surprisingly, most studies looking for associations with increased cancer risk have studied POLD1 or POLE intronic or synonymous coding SNPs that are unlikely to affect polymerase expression or fidelity.
4.2. Pol δ variants in cancer
Several POLD1 mutations have been found in colorectal cancer cell lines and primary tumors [214, 215], and one mutation was identified in a rat hepatoma cell line [216] (Fig. 5). The MMR-deficient human cancer cell lines HCT15 and DLD-1 carry four heterozygous mutations that result in non-synonymous amino acid changes in Pol δ: G10V, R506H, R689W, and S746I [214, 215]. Shcherbakova et al. [206] recently used a yeast system to study phenotypes conferred by R506H and R689W, which are positioned in Pol δ's Exo III and Pol III motifs, respectively. They found that R506H was a mild mutator (2.5-fold over wild-type rates) in an MMR-defective background, while R689W was lethal in haploid cells. Heterozygous R689W did not influence the spontaneous mutation rate in diploid yeast proficient for MMR; MMR-deficient diploids were not examined [206]. Interestingly, haploid or diploid R689W yeast expressing low levels of wild-type Pol δ exhibited extreme rates of spontaneous mutation, and the purified Pol δ R689W enzyme was highly error-prone [206]. The R648Q substitution present in rat hepatoma cells [216] is also located in a conserved polymerase region (equivalent to human R652Q next to Pol VI), and this substitution affects the biochemical properties and fidelity of partially purified Pol δ [216–219].
Two additional POLD1 mutations, D502N and R990L, were identified in primary colorectal cancer samples [214, 215]. Both alleles were also present in surrounding normal colon tissue. Thus, these variants could be low-incidence SNPs, pre-neoplastic mutations in the colonic epithelium, or novel inherited mutations. D502N was introduced into homologous positions of Pol δ in yeast (D506N and E508N) and found not to be a mutator in the presence or absense of MMR (Singh and Preston, unpublished data).
Lastly, a POLD1 frameshift mutation, which results in an early stop codon, was identified in the colon cancer cell lines HCT116 and SW48 [215]. The resultant Pol δ lacks two conserved DNA binding domains in the carboxy terminus and is expressed at decreased levels in mutant cells [215]. Depletion of Pol δ or Pol α is known to increase insertion/deletion events and chromosomal instability in yeast [220–224]. Alleles impacting mouse Pol δ activity also increase spontaneous mutation rates and chomosomal instability and accelerate tumorigenesis [203]. Taken together, these findings suggest that decreased polymerase fidelity or activity may play a role in tumorigenesis in humans. A weakness of many studies examining tumor cell lines, however, is the lack of control DNA from uncultured tumor samples and normal tissues from the patient. Thus, it is unclear whether the observed POLD1 mutations were present in vivo or aquired during establishment of the cell lines.
Interestingly, most heterozygous POLD1 mutations are found in MMR-deficient tumors and cell lines. This may simply be a bias due to the limited number and types of human tumors analyzed for POLD1 mutations. Alternatively, heterozygous mutations in polymerase genes may be more oncogenic when MMR is also defective. In yeast and mammalian cells, heterozygous mutations that affect the fidelity of Pols δ or ε result in mild mutator phenotypes [75, 83, 130, 225]. However, when MMR is inactivated in yeast that are heterozygous for these mutator polymerases, the mutation rate is synergistically elevated [75, 159, 225]. Similarly, mice heterozygous for polymerase proofreading and nullizygous for MMR exhibit extreme mutator phenotypes and rapidly develop aggressive disseminated lymphomas ([84] and unpublished results). Thus, heterozygous mutations in human POLD1 or POLE may have an enhanced tumorigenic effect in a MMR-deficient cell. Moreover, SNPs that mildly decrease polymerase fidelity could have strong mutator effects in cells that have epigenetically silenced MMR.
4.3. Pol ε variants in cancer
Very few studies have examined human tumors for mutations in the Pol ε genes. Zhou et al. surveyed cancer-associated POLE or POLE2 mutations in tumors from breast, colon and brain [226–229]. Analyses of ~160 breast tumor samples revealed no mutations in the highly conserved regions of POLE [226]. However, when the survey was expanded to include the coding region and flanking intronic sequences of POLE2, the gene for Pol ε's 55-kD subunit, they found an intronic 4-bp deletion in breast and colorectal tumors that was not present in control tissues from the same patients [227, 228]. In contrast, no POLE2 mutations were found in a series of brain tumors [229]. The authors propose that this POLE2 site may be a mutational hotspot in some cancers, although the distance of this intronic deletion from the nearest exon suggests it is not pathogenic. Additional work is required to determine if this small POLE intronic deletion affects splicing or expression of POLE2, supporting its role in tumorigenesis.
5. Summary and perspectives
At the beginning of the 20th century, cytogenetic techniques revealed the first chromosomal abnormalities in malignant cells and led to the hypothesis that genetic instability fueled cancer [230, 231]. Since then it has become clear that multiple changes in the genome, including point mutations, insertion/deletion events, large chromosomal rearrangements and epigenetic modifications of gene expression dysregulate cellular pathways on the road to malignant transformation [1–3, 13, 14]. In cultured human cells, the introduction of only three oncogenes is sufficient for malignant transformation, and rodent cells can be transformed with as few as two oncogenes [232]. Kinetic analyses of age-related tumor incidences in humans suggests 4 to 6 events are required in vivo [233]. Recent genome-wide sequence analyses show that, although a small number of mutations may be required for tumor formation, most cancer cells contain significantly more: as many as 50,000 clonal mutations [234–237] and a large, undetermined number of non-clonal mutations that occur randomly throughout the genome of each cancer cell [238, 239].
Loeb et al. proposed the “mutator hypothesis” in 1974 to reconcile the number of mutations needed for carcinogenesis and the low spontaneous mutation rate of normal cells [6]. He argued that a cell must acquire a mutator phenotype, through defects in DNA replication or repair, early in the evolution of a tumor. As described in this review, multiple pathways function to maintain genetic integrity in normal cells. Accordingly, tumors exhibit different types of genetic instability. These can be categorized into three general classes: 1) chromosome instability (CIN) involving gross chromosomal aberrations (translocations, large deletions, gene amplifications, chromosome losses or gains), 2) microsatellite instability (MSI or MIN) involving widespread expansions and contractions of short tandem repeat sequences (“microsatellites”), and 3) point mutation instability (PIN) involving subtle nucleotide sequence changes (base substitutions, small deletions/insertions). Chromosome instabilities in cancer are readily revealed through cytogenetic and molecular analyses and are abundantly documented in the literature. Microsatellite instability is also relatively easy to detect and is observed in cancers associated with loss of mismatch repair as typified by Lynch syndrome. In contrast, nucleotide sequence instabilities are experimentally difficult to detect, and this difficulty has limited the search for PIN mutator phenotypes in tumors [25, 26].
PIN is a potentially important pathway to cancer. Nucleotide sequence changes activate oncogenes and inactivate tumor suppressors, and at least one human cancer syndrome (Lynch syndrome with defective Msh6) is associated with PIN, minimal MSI and no CIN. The recent demonstration of increased spontaneous cancer in proofreading-deficient mice [81–84] provides additional evidence that PIN causes cancer and suggests that other PIN-associated cancers will be discovered in humans as improved methods of detection are developed. Additional studies are needed to screen tumors for DNA polymerase variants that may be error-prone. Because Pols δ and ε are required for cell viability, error-prone variants will likely result from missense mutations and not large insertions, deletions, translocations or LOH that would disrupt vital polymerase functions. Thus, it will be necessary to sequence the coding regions of POLD1, POLE and other essential “fidelity genes” to detect mutator variants. Mutations affecting conserved regions of Pol δ and Pol ε are of interest, particularly loss-of-function alleles that disrupt proofreading, which is not essential for cell viability.
Finally, cooperation of polymerase mutators with reduced MMR may significantly impact cancer risk and tumor progression. As tumor cells evolve and transit through new microenvironments, changing selective pressures will favor high mutation rates, which increase the likelihood of genetic adaptation. However, after adaptation to a new niche or tissue subcompartment in the body, these mutator cells will progressively lose fitness as they accumulate deleterious mutations in essential genes. Thus, mutators may rise and fall in tumor populations during the course of cancer progression [240]. Transient mutator phenotypes are possible via epigenetic silencing of repair genes and the emergence of mutator suppressors.
Acknowledgements
Our sincere thanks to Larry Loeb for his critical insight and creative spirit. We also thank past and present laboratory members for valuable discussions and experimental contributions. Our research was supported by the National Institutes of Health (R01 ES09927, R01 CA098243, R01 CA111582, P20 CA103728, P01 AG01751 and P01 CA77852) and the National Institute of Environmental Health Sciences-sponsored Center for Ecogenetics and Environmental Health at the University of Washington (P30 ES07033). Tina Albertson was supported by a Young Investigators Award from Seattle Children's Hospital and Regional Medical Center and is a St. Baldrick's Foundation Scholar. Alan Herr holds a Hitchings-Elion Fellowship from the Burroughs Wellcome Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest The authors declare that there are no conflicts of interest.
1
Calculated as follows: The mammalian diploid genome has 6 × 109 base pairs, which corresponds to 1.2 × 1010 nucleotides that must be polymerized each time a cell divides. DNA polymerases δ and δ replicate the bulk of genomic DNA and have error rates of 10−4 to 10−5 per nucleotide polymerized (prior to correction by proofreading or MMR). Thus, if the error rate is 10−5, then 1010 nucleotides per genome X 10−5 errors per nucleotide = 100,000 polymerase errors per genome per cell division. If the error rate is 10−4, then 1010 nucleotides per genome X 10−4 errors per nucleotide = 1,000,000 errors per genome per cell division.
References
- [1].Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- [2].Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–99. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- [3].Weinberg RA. The Biology of Cancer. Garland Science; New York: 2007. [Google Scholar]
- [4].Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–7. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- [5].Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–8. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- [6].Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–21. [PubMed] [Google Scholar]
- [7].Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–7. doi: 10.1158/0008-5472.CAN-07-5835. [DOI] [PubMed] [Google Scholar]
- [8].Bodmer W. Genetic instability is not a requirement for tumor development. Cancer Res. 2008;68:3558–60. doi: 10.1158/0008-5472.CAN-07-6544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Prehn RT. Cancers beget mutations versus mutations beget cancers. Cancer Res. 1994;54:5296–300. [PubMed] [Google Scholar]
- [10].Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- [11].Anderson GR, Stoler DL, Brenner BM. Cancer: the evolved consequence of a destabilized genome. Bioessays. 2001;23:1037–46. doi: 10.1002/bies.1149. [DOI] [PubMed] [Google Scholar]
- [12].Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, Velculescu VE, et al. Genetic progression and the waiting time to cancer. PLoS Comput Biol. 2007;3:e225. doi: 10.1371/journal.pcbi.0030225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability--an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11:220–8. doi: 10.1038/nrm2858. [DOI] [PubMed] [Google Scholar]
- [14].Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- [15].Special Issue on Genome Instability. Science. 2002;297:543–69. [Google Scholar]
- [16].Special Issue on Genome Instability. Nat Rev Mol Cell Biol. 2010;11:165–228. [Google Scholar]
- [17].Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–45. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- [18].Mitelman F, Johansson B, Mertens F. Mitelman database of chromosome aberrations and gene fusions in cancer. 2010 http://cgap.nci.nih.gov/Chromosomes/Mitelman.
- [19].Tlsty TD, Margolin BH, Lum K. Differences in the rate of gene amplification in nontumorigenic and tumorigenic cell lines as measured by Luria-Delbruk fluctuation analysis. Proc Natl Acad Sci USA. 1989;86:9441–5. doi: 10.1073/pnas.86.23.9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Phear G, Bhattacharyya NP, Meuth M. Loss of heterozygosity and base substitution at the APRT locus in mismatch repair-proficient and -deficient colorectal carcinoma cell lines. Mol Cell Biol. 1996;16:6516–23. doi: 10.1128/mcb.16.11.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–7. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- [22].Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–74. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- [23].Levitt NC, Hickson ID. Caretaker tumour suppressor genes that defend genome integrity. Trends Mol Med. 2002;8:179–86. doi: 10.1016/s1471-4914(02)02298-0. [DOI] [PubMed] [Google Scholar]
- [24].Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10:756–68. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- [25].Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–42. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Klein CA. Random mutations, selected mutations: A PIN opens the door to new genetic landscapes. Proc Natl Acad Sci USA. 2006;103:18033–4. doi: 10.1073/pnas.0609000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med. 2003;348:919–32. doi: 10.1056/NEJMra012242. [DOI] [PubMed] [Google Scholar]
- [28].de la Chapelle A. Genetic predisposition to colorectal cancer. Nat Rev Cancer. 2004;4:769–80. doi: 10.1038/nrc1453. [DOI] [PubMed] [Google Scholar]
- [29].Abdel-Rahman WM, Peltomaki P. Lynch syndrome and related familial colorectal cancers. Crit Rev Oncog. 2008;14:1–22. doi: 10.1615/critrevoncog.v14.i1.10. discussion 3–31. [DOI] [PubMed] [Google Scholar]
- [30].Lynch HT, Lynch PM, Lanspa SJ, Snyder CL, Lynch JF, Boland CR. Review of the Lynch syndrome: history, molecular genetics, screening, differential diagnosis, and medicolegal ramifications. Clin Genet. 2009;76:1–18. doi: 10.1111/j.1399-0004.2009.01230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–87. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–38. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- [33].Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–25. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- [34].Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–61. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- [35].Papadopoulos N, Nicolaides NC, Wei Y-F, Ruben SM, Carter KC, Rosen CA, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–9. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- [36].Peltomaki P, Vasen H. Mutations associated with HNPCC predisposition -- Update of ICG-HNPCC/INSiGHT mutation database. Dis Markers. 2004;20:269–76. doi: 10.1155/2004/305058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Jasperson KW, Tuohy TM, Neklason DW, Burt RW. Hereditary and familial colon cancer. Gastroenterology. 2010;138:2044–58. doi: 10.1053/j.gastro.2010.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–80. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- [39].Arzimanoglou II, Gilbert F, Barber HR. Microsatellite instability in human solid tumors. Cancer. 1998;82:1808–20. doi: 10.1002/(sici)1097-0142(19980515)82:10<1808::aid-cncr2>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- [40].Parsons R, Li G-M, Longley MJ, Fang W-h, Papadopoulos N, Jen J, et al. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–36. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- [41].Bhattacharyya NP, Skandalis A, Ganesh A, Groden J, Meuth M. Mutator phenotypes in human colorectal carcinoma cell lines. Proc Natl Acad Sci USA. 1994;91:6319–23. doi: 10.1073/pnas.91.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Eshleman JR, Lang EZ, Bowerfind GK, Parsons R, Vogelstein B, Willson JK, et al. Increased mutation rate at the hprt locus accompanies microsatellite instability in colon cancer. Oncogene. 1995;10:33–7. [PubMed] [Google Scholar]
- [43].Umar A, Risinger JI, Glaab WE, Tindall KR, Barrett JC, Kunkel TA. Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics. 1998;148:1637–46. doi: 10.1093/genetics/148.4.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62:2447–54. [PubMed] [Google Scholar]
- [45].Akiyama Y, Sato H, Yamada T, Nagasaki H, Tsuchiya A, Abe R, et al. Germ-line mutation of the hMSH6/GTBP gene in an atypical hereditary nonpolyposis colorectal cancer kindred. Cancer Res. 1997;57:3920–3. [PubMed] [Google Scholar]
- [46].Miyaki M, Konishi M, Tanaka K, Kikuchi-Yanoshita R, Muraoka M, Yasuno M, et al. Germline mutation of MSH6 as the cause of hereditary nonpolyposis colorectal cancer. Nat Genet. 1997;17:271–2. doi: 10.1038/ng1197-271. [DOI] [PubMed] [Google Scholar]
- [47].Kolodner RD, Tytell JD, Schmeits JL, Kane MF, Gupta RD, Weger J, et al. Germ-line msh6 mutations in colorectal cancer families. Cancer Res. 1999;59:5068–74. [PubMed] [Google Scholar]
- [48].Wijnen J, de Leeuw W, Vasen H, van der Klift H, Moller P, Stormorken A, et al. Familial endometrial cancer in female carriers of MSH6 germline mutations. Nat Genet. 1999;23:142–4. doi: 10.1038/13773. [DOI] [PubMed] [Google Scholar]
- [49].Baglietto L, Lindor NM, Dowty JG, White DM, Wagner A, Gomez Garcia EB, et al. Risks of Lynch syndrome cancers for MSH6 mutation carriers. J Natl Cancer Inst. 2010;102:193–201. doi: 10.1093/jnci/djp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–20. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- [51].Johnson RE, Kovvali GK, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2 dependent genomic stability. J Biol Chem. 1996;271:7285–8. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- [52].Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–86. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–5. [PubMed] [Google Scholar]
- [54].Miller JH. Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol. 1996;50:625–43. doi: 10.1146/annurev.micro.50.1.625. [DOI] [PubMed] [Google Scholar]
- [55].Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- [56].Kunkel TA, Bebenek K. DNA replication fidelity. Annu Rev Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- [57].Johnson KA. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim Biophys Acta. 2010;1804:1041–8. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–61. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Shevelev IV, Hübscher U. The 3′–5′ exonucleases. Nat Rev Mol Cell Biol. 2002;3:364–76. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- [61].Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804:1049–63. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- [62].Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- [63].Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- [64].Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–23. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- [65].Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–46. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- [66].Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- [67].Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 2008;129:391–407. doi: 10.1016/j.mad.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Barnes DE, Lindahl T. Repair and genetic consequences of endogenous DNA base damage in mammalian cells. Annu Rev Genet. 2004;38:445–76. doi: 10.1146/annurev.genet.38.072902.092448. [DOI] [PubMed] [Google Scholar]
- [69].Meira LB, Burgis NE, Samson LD. Base excision repair. Adv Exp Med Biol. 2005;570:125–73. doi: 10.1007/1-4020-3764-3_5. [DOI] [PubMed] [Google Scholar]
- [70].Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–70. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- [71].Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed. ASM Press; Washington, D.C.: 2006. [Google Scholar]
- [72].Simon M, Giot L, Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase δ subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10:2163–70. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Morrison A, Bell JB, Kunkel TA, Sugino A. Eukaryotic DNA polymerase amino acid sequence required for 3′→5′ exonuclease activity. Proc Natl Acad Sci USA. 1991;88:9473–7. doi: 10.1073/pnas.88.21.9473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Strand M, Prolla TA, Liskay RM, Petes TD. Destabilization of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature. 1993;365:274–6. doi: 10.1038/365274a0. [DOI] [PubMed] [Google Scholar]
- [75].Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–73. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Morrison A, Sugino A. The 3′→5′ exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–96. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- [77].Roberts JD, Thomas DC, Kunkel TA. Exonucleolytic proofreading of leading and lagging strand DNA replication errors. Proc Natl Acad Sci USA. 1991;88:3465–9. doi: 10.1073/pnas.88.8.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hauser J, Levine AS, Dixon K. Fidelity of DNA synthesis in a mammalian in vitro replication system. Mol Cell Biol. 1988;8:3267–71. doi: 10.1128/mcb.8.8.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Kat A, Thilly WG, Fang W-H, Longley MJ, Li G-M, Modrich P. An alkylation-tolerant, mutator human cell line is deficient in strand-specific mismatch repair. Proc Natl Acad Sci USA. 1993;90:6424–8. doi: 10.1073/pnas.90.14.6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Reitmair AH, Risley R, Bristow RG, Wilson T, Ganesh A, Jang A, et al. Mutator phenotype in Msh2-deficient murine embryonic fibroblasts. Cancer Res. 1997;57:3765–71. [PubMed] [Google Scholar]
- [81].Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, et al. Defective DNA polymerase-δ proofreading causes cancer susceptibility in mice. Nat Med. 2001;7:638–9. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- [82].Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, et al. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc Natl Acad Sci USA. 2002;99:15560–5. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, et al. DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci USA. 2009;106:17101–4. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Treuting PM, Albertson TM, Preston BD. Case series: acute tumor lysis syndrome in mutator mice with disseminated lymphoblastic lymphoma. Toxicol Pathol. 2010;38:476–85. doi: 10.1177/0192623310362249. [DOI] [PubMed] [Google Scholar]
- [85].Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase ε participates in leading-strand DNA replication. Science. 2007;317:127–30. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Nick McElhinny SA, Gordenin DA, Stith CM, Burgers PM, Kunkel TA. Division of labor at the eukaryotic replication fork. Mol Cell. 2008;30:137–44. doi: 10.1016/j.molcel.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–7. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Meuth M. The molecular basis of mutations induced by deoxyribonucleoside triphosphate pool imbalances in mammalian cells. Exp Cell Res. 1989;181:305–16. doi: 10.1016/0014-4827(89)90090-6. [DOI] [PubMed] [Google Scholar]
- [89].Kunz BA, Kohalmi SE, Kunkel TA, Mathews CK, McIntosh EM, Reidy JA. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat Res. 1994;318:1–64. doi: 10.1016/0165-1110(94)90006-x. [DOI] [PubMed] [Google Scholar]
- [90].Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–14. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- [91].Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–5. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Tajiri T, Maki H, Sekiguchi M. Functional cooperation of MutT, MutM and MutY proteins in preventing mutations caused by spontaneous oxidation of guanine nucleotide in Escherichia coli. Mutat Res. 1995;336:257–67. doi: 10.1016/0921-8777(94)00062-b. [DOI] [PubMed] [Google Scholar]
- [93].Sekiguchi M, Tsuzuki T. Oxidative nucleotide damage: consequences and prevention. Oncogene. 2002;21:8895–904. doi: 10.1038/sj.onc.1206023. [DOI] [PubMed] [Google Scholar]
- [94].Tsuzuki T, Nakatsu Y, Nakabeppu Y. Significance of error-avoiding mechanisms for oxidative DNA damage in carcinogenesis. Cancer Sci. 2007;98:465–70. doi: 10.1111/j.1349-7006.2007.00409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Russo MT, De Luca G, Degan P, Bignami M. Different DNA repair strategies to combat the threat from 8-oxoguanine. Mutat Res. 2007;614:69–76. doi: 10.1016/j.mrfmmm.2006.03.007. [DOI] [PubMed] [Google Scholar]
- [96].Cox EC. Bacterial mutator genes and the control of spontaneous mutation. Ann Rev Genet. 1976;10:135–56. doi: 10.1146/annurev.ge.10.120176.001031. [DOI] [PubMed] [Google Scholar]
- [97].Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- [98].Lindahl T, Barnes DE. Repair of endogenous DNA damage. Cold Spring Harb Symp Quant Biol. 2000;65:127–33. doi: 10.1101/sqb.2000.65.127. [DOI] [PubMed] [Google Scholar]
- [99].De Bont R, van Larebeke N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004;19:169–85. doi: 10.1093/mutage/geh025. [DOI] [PubMed] [Google Scholar]
- [100].Hübscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–63. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- [101].Pavlov YI, Shcherbakova PV, Rogozin IB. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol. 2006;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- [102].Burgers PM. Polymerase dynamics at the eukaryotic DNA replication fork. J Biol Chem. 2009;284:4041–5. doi: 10.1074/jbc.R800062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Sitney KC, Budd ME, Campbell JL. DNA polymerase III, a second essential DNA polymerase, is encoded by the S. cerevisiae CDC2 gene. Cell. 1989;56:599–605. doi: 10.1016/0092-8674(89)90582-5. [DOI] [PubMed] [Google Scholar]
- [104].Boulet A, Simon M, Faye G, Bauer GA, Burgers PMJ. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989;8:1849–54. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Dua R, Levy DL, Campbell JL. Analysis of the essential functions of the C-terminal protein/protein interaction domain of Saccharomyces cerevisiae pol epsilon and its unexpected ability to support growth in the absence of the DNA polymerase domain. J Biol Chem. 1999;274:22283–8. doi: 10.1074/jbc.274.32.22283. [DOI] [PubMed] [Google Scholar]
- [106].Feng W, D'Urso G. Schizosaccharomyces pombe cells lacking the amino-terminal catalytic domains of DNA polymerase epsilon are viable but require the DNA damage checkpoint control. Mol Cell Biol. 2001;21:4495–504. doi: 10.1128/MCB.21.14.4495-4504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Waga S, Masuda T, Takisawa H, Sugino A. DNA polymerase ε is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc Natl Acad Sci USA. 2001;98:4978–83. doi: 10.1073/pnas.081088798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Fukui T, Yamauchi K, Muroya T, Akiyama M, Maki H, Sugino A, et al. Distinct roles of DNA polymerases delta and epsilon at the replication fork in Xenopus egg extracts. Genes Cells. 2004;9:179–91. doi: 10.1111/j.1356-9597.2004.00716.x. [DOI] [PubMed] [Google Scholar]
- [109].Kao HI, Bambara RA. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit Rev Biochem Mol Biol. 2003;38:433–52. doi: 10.1080/10409230390259382. [DOI] [PubMed] [Google Scholar]
- [110].Fuss J, Linn S. Human DNA polymerase ε colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J Biol Chem. 2002;277:8658–66. doi: 10.1074/jbc.M110615200. [DOI] [PubMed] [Google Scholar]
- [111].Aparicio OM, Stout AM, Bell SP. Differential assembly of Cdc45p and DNA polymerases at early and late origins of DNA replication. Proc Natl Acad Sci USA. 1999;96:9130–5. doi: 10.1073/pnas.96.16.9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol Cell Biol. 2000;20:2809–17. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–65. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Feng W, Rodriguez-Menocal L, Tolun G, D'Urso G. Schizosacchromyces pombe Dpb2 binds to origin DNA early in S phase and is required for chromosomal DNA replication. Mol Biol Cell. 2003;14:3427–36. doi: 10.1091/mbc.E03-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Navas TA, Zhou Z, Elledge SJ. DNA polymerase ε links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- [116].Edwards S, Li CM, Levy DL, Brown J, Snow PM, Campbell JL. Saccharomyces cerevisiae DNA polymerase epsilon and polymerase sigma interact physically and functionally, suggesting a role for polymerase epsilon in sister chromatid cohesion. Mol Cell Biol. 2003;23:2733–48. doi: 10.1128/MCB.23.8.2733-2748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zlotkin T, Kaufmann G, Jiang Y, Lee MY, Uitto L, Syvaoja J, et al. DNA polymerase epsilon may be dispensable for SV40- but not cellular-DNA replication. EMBO J. 1996;15:2298–305. [PMC free article] [PubMed] [Google Scholar]
- [118].Rytkönen AK, Vaara M, Nethanel T, Kaufmann G, Sormunen R, Läärä E, et al. Distinctive activities of DNA polymerases during human DNA replication. FEBS J. 2006;273:2984–3001. doi: 10.1111/j.1742-4658.2006.05310.x. [DOI] [PubMed] [Google Scholar]
- [119].Pospiech H, Kursula I, Abdel-Aziz W, Malkas L, Uitto L, Kastelli M, et al. A neutralizing antibody against human DNA polymerase epsilon inhibits cellular but not SV40 DNA replication. Nucleic Acids Res. 1999;27:3799–804. doi: 10.1093/nar/27.19.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Hicks WM, Kim M, Haber JE. Increased mutagenesis and unique mutation signature associated with mitotic gene conversion. Science. 2010;329:82–5. doi: 10.1126/science.1191125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3'→5' exonuclease of DNA polymerase delta can substitute for the 5' flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci USA. 2001;98:5122–7. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Ohya T, Kawasaki Y, Hiraga S, Kanbara S, Nakajo K, Nakashima N, et al. The DNA polymerase domain of polε is required for rapid, efficient, and highly accurate chromosomal DNA replication, telomere length maintenance, and normal cell senescence in Saccharomyces cerevisiae. J Biol Chem. 2002;277:28099–108. doi: 10.1074/jbc.M111573200. [DOI] [PubMed] [Google Scholar]
- [123].Tran HT, Keen JD, Kricker M, Resnick MA, Gordenin DA. Hypermutability of homonucleotide runs in mismatch repair and DNA polymerase proofreading yeast mutants. Mol Cell Biol. 1997;17:2859–65. doi: 10.1128/mcb.17.5.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Tran HT, Degtyareva NP, Gordenin DA, Resnick MA. Genetic factors affecting the impact of DNA polymerase delta proofreading activity on mutation avoidance in yeast. Genetics. 1999;152:47–59. doi: 10.1093/genetics/152.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Tran HT, Gordenin DA, Resnick MA. The 3′→5′ exonucleases of DNA polymerases δ and ε and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:2000–7. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Greene CN, Jinks-Robertson S. Spontaneous frameshift mutations in Saccharomyces cerevisiae: accumulation during DNA replication and removal by proofreading and mismatch repair activities. Genetics. 2001;159:65–75. doi: 10.1093/genetics/159.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Karthikeyan R, Vonarx EJ, Straffon AF, Simon M, Faye G, Kunz BA. Evidence from mutational specificity studies that yeast DNA polymerases δ and ε replicate different DNA strands at an intracellular replication fork. J Mol Biol. 2000;299:405–19. doi: 10.1006/jmbi.2000.3744. [DOI] [PubMed] [Google Scholar]
- [128].Babudri N, Pavlov YI, Matmati N, Ludovisi C, Achilli A. Stationary-phase mutations in proofreading exonuclease-deficient strains of the yeast Saccharomyces cerevisiae. Mol Genet Genomics. 2001;265:362–6. doi: 10.1007/s004380000424. [DOI] [PubMed] [Google Scholar]
- [129].Shcherbakova PV, Pavlov YI. 3′→5′ exonucleases of DNA polymerases ε and δ correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142:717–26. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Pavlov YI, Maki S, Maki H, Kunkel TA. Evidence for interplay among yeast replicative DNA polymerases alpha, delta and epsilon from studies of exonuclease and polymerase active site mutations. BMC Biol. 2004;2:11. doi: 10.1186/1741-7007-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Pavlov YI, Frahm C, Nick McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr Biol. 2006;16:202–7. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- [132].Albertson TM, Preston BD. DNA replication fidelity: proofreading in trans. Curr Biol. 2006;16:R209–11. doi: 10.1016/j.cub.2006.02.031. [DOI] [PubMed] [Google Scholar]
- [133].Nick McElhinny SA, Pavlov YI, Kunkel TA. Evidence for extrinsic exonucleolytic proofreading. Cell Cycle. 2006;5:958–62. doi: 10.4161/cc.5.9.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol Cell. 1999;3:679–85. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- [135].Alani E. The Saccharomyces cerevisiae Msh2 and Msh6 proteins form a complex that specifically binds to duplex oligonucleotides containing mismatched DNA base pairs. Mol Cell Biol. 1996;16:5604–15. doi: 10.1128/mcb.16.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Acharya S, Wilson T, Gradia S, Kane MF, Guerrette S, Marsischky GT, et al. hMSH2 forms specific mispair-binding complexes with hMSH3 and hMSH6. Proc Natl Acad Sci USA. 1996;93:13629–34. doi: 10.1073/pnas.93.24.13629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Genschel J, Littman SJ, Drummond JT, Modrich P. Isolation of MutSβ from human cells and comparison of the mismatch repair specificities of MutSβ and MutSα. J Biol Chem. 1998;273:19895–901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- [138].Marsischky GT, Kolodner RD. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2–MSH6 complex and mispaired bases in DNA. J Biol Chem. 1999;274:26668–82. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- [139].Harrington JM, Kolodner RD. Saccharomyces cerevisiae Msh2–Msh3 acts in repair of base-base mispairs. Mol Cell Biol. 2007;27:6546–54. doi: 10.1128/MCB.00855-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLalpha in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- [141].Erdeniz N, Nguyen M, Deschenes SM, Liskay RM. Mutations affecting a putative MutL[alpha] endonuclease motif impact multiple mismatch repair functions. DNA Repair. 2007;6:1463–70. doi: 10.1016/j.dnarep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci USA. 2009;106:8495–500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Branch P, Aquilina G, Bignami M, Karran P. Defective mismatch binding and a mutator phenotype in cells tolerant to DNA damage. Nature. 1993;362:652–4. doi: 10.1038/362652a0. [DOI] [PubMed] [Google Scholar]
- [144].Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis. 2001;22:1931–7. doi: 10.1093/carcin/22.12.1931. [DOI] [PubMed] [Google Scholar]
- [145].Bignami M, Casorelli I, Karran P. Mismatch repair and response to DNA-damaging antitumour therapies. Eur J Cancer. 2003;39:2142–9. doi: 10.1016/s0959-8049(03)00569-0. [DOI] [PubMed] [Google Scholar]
- [146].Stojic L, Brun R, Jiricny J. Mismatch repair and DNA damage signalling. DNA Repair. 2004;3:1091–101. doi: 10.1016/j.dnarep.2004.06.006. [DOI] [PubMed] [Google Scholar]
- [147].O'Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27:682–92. doi: 10.1093/carcin/bgi298. [DOI] [PubMed] [Google Scholar]
- [148].Wang JY, Edelmann W. Mismatch repair proteins as sensors of alkylation DNA damage. Cancer Cell. 2006;9:417–8. doi: 10.1016/j.ccr.2006.05.013. [DOI] [PubMed] [Google Scholar]
- [149].Papouli E, Cejka P, Jiricny J. Dependence of the cytotoxicity of DNA-damaging agents on the mismatch repair status of human cells. Cancer Res. 2004;64:3391–4. doi: 10.1158/0008-5472.CAN-04-0513. [DOI] [PubMed] [Google Scholar]
- [150].Duckett DR, Bronstein SM, Taya Y, Modrich P. hMutSα- and hMutLα-dependent phosphorylation of p53 in response to DNA methylator damage. Proc Natl Acad Sci USA. 1999;96:12384–8. doi: 10.1073/pnas.96.22.12384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Colussi C, Parlanti E, Degan P, Aquilina G, Barnes D, Macpherson P, et al. The mammalian mismatch repair pathway removes DNA 8-oxodGMP incorporated from the oxidized dNTP pool. Curr Biol. 2002;12:912–8. doi: 10.1016/s0960-9822(02)00863-1. [DOI] [PubMed] [Google Scholar]
- [152].Ni TT, Marsischky GT, Kolodner RD. MSH2 and MSH6 are required for removal of adenine misincorporated opposite 8-oxo-guanine in S. cerevisiae. Mol Cell. 1999;4:439–44. doi: 10.1016/s1097-2765(00)80346-9. [DOI] [PubMed] [Google Scholar]
- [153].Pavlov YI, Mian IM, Kunkel TA. Evidence for preferential mismatch repair of lagging strand DNA replication errors in yeast. Curr Biol. 2003;13:744–8. doi: 10.1016/s0960-9822(03)00284-7. [DOI] [PubMed] [Google Scholar]
- [154].York SJ, Modrich P. Mismatch repair-dependent iterative excision at irreparable O6-methylguanine lesions in human nuclear extracts. J Biol Chem. 2006;281:22674–83. doi: 10.1074/jbc.M603667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].de Wind N, Dekker M, Claij N, Jansen L, van Klink Y, Radman M, et al. HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat Genet. 1999;23:359–62. doi: 10.1038/15544. [DOI] [PubMed] [Google Scholar]
- [156].Buermeyer AB, Wilson-Van Patten C, Baker SM, Liskay RM. The human MLH1 cDNA complements DNA mismatch repair defects in Mlh1- deficient mouse embryonic fibroblasts. Cancer Res. 1999;59:538–41. [PubMed] [Google Scholar]
- [157].Zhang H, Richards B, Wilson T, Lloyd M, Cranston A, Thorburn A, et al. Apoptosis induced by overexpression of hMSH2 or hMLH1. Cancer Res. 1999;59:3021–7. [PubMed] [Google Scholar]
- [158].Yoshioka K, Yoshioka Y, Hsieh P. ATR kinase activation mediated by MutSα and MutLα in response to cytotoxic O6-methylguanine adducts. Mol Cell. 2006;22:501–10. doi: 10.1016/j.molcel.2006.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Drotschmann K, Clark AB, Tran HT, Resnick MA, Gordenin DA, Kunkel TA. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc Natl Acad Sci USA. 1999;96:2970–5. doi: 10.1073/pnas.96.6.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Sokolsky T, Alani E. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics. 2000;155:589–99. doi: 10.1093/genetics/155.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Umar A, Buermeyer AB, Simon JA, Thomas DC, Clark AB, Liskay RM, et al. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- [162].Clark AB, Valle F, Drotschmann K, Gary RK, Kunkel TA. Functional interaction of proliferating cell nuclear antigen with MSH2–MSH6 and MSH2–MSH3 complexes. J Biol Chem. 2000;275:36498–501. doi: 10.1074/jbc.C000513200. [DOI] [PubMed] [Google Scholar]
- [163].Amin NS, Nguyen MN, Oh S, Kolodner RD. exo1-Dependent mutator mutations: model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21:5142–55. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Lau PJ, Kolodner RD. Transfer of the MSH2.MSH6 complex from proliferating cell nuclear antigen to mispaired bases in DNA. J Biol Chem. 2003;278:14–7. doi: 10.1074/jbc.C200627200. [DOI] [PubMed] [Google Scholar]
- [165].Pavlov YI, Newlon CS, Kunkel TA. Yeast origins establish a strand bias for replicational mutagenesis. Mol Cell. 2002;10:207–13. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- [166].Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, et al. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- [167].Friedberg EC, Meira LB. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage. DNA Repair (Amst) (Version 7) 2006;5:189–209. doi: 10.1016/j.dnarep.2005.09.009. [DOI] [PubMed] [Google Scholar]
- [168].Wei K, Kucherlapati R, Edelmann W. Mouse models for human DNA mismatch-repair gene defects. Trends Mol Med. 2002;8:346–53. doi: 10.1016/s1471-4914(02)02359-6. [DOI] [PubMed] [Google Scholar]
- [169].Taketo MM, Edelmann W. Mouse models of colon cancer. Gastroenterology. 2009;136:780–98. doi: 10.1053/j.gastro.2008.12.049. [DOI] [PubMed] [Google Scholar]
- [170].Hollander MC, Philburn RT, Patterson AD, Velasco-Miguel S, Friedberg EC, Linnoila RI, et al. Deletion of XPC leads to lung tumors in mice and is associated with early events in human lung carcinogenesis. Proc Natl Acad Sci USA. 2005;102:13200–5. doi: 10.1073/pnas.0503133102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Tsuzuki T, Egashira A, Igarashi H, Iwakuma T, Nakatsuru Y, Tominaga Y, et al. Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase. Proc Natl Acad Sci USA. 2001;98:11456–61. doi: 10.1073/pnas.191086798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Sakumi K, Tominaga Y, Furuichi M, Xu P, Tsuzuki T, Sekiguchi M, et al. Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption. Cancer Res. 2003;63:902–5. [PubMed] [Google Scholar]
- [173].Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, et al. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- [174].Sakamoto K, Tominaga Y, Yamauchi K, Nakatsu Y, Sakumi K, Yoshiyama K, et al. MUTYH-null mice are susceptible to spontaneous and oxidative stress induced intestinal tumorigenesis. Cancer Res. 2007;67:6599–604. doi: 10.1158/0008-5472.CAN-06-4802. [DOI] [PubMed] [Google Scholar]
- [175].Meira LB, Reis AM, Cheo DL, Nahari D, Burns DK, Friedberg EC. Cancer predisposition in mutant mice defective in multiple genetic pathways: uncovering important genetic interactions. Mutat Res. 2001;477:51–8. doi: 10.1016/s0027-5107(01)00097-5. [DOI] [PubMed] [Google Scholar]
- [176].Lin Q, Clark AB, McCulloch SD, Yuan T, Bronson RT, Kunkel TA, et al. Increased susceptibility to UV-induced skin carcinogenesis in polymerase eta-deficient mice. Cancer Res. 2006;66:87–94. doi: 10.1158/0008-5472.CAN-05-1862. [DOI] [PubMed] [Google Scholar]
- [177].Ohkumo T, Kondo Y, Yokoi M, Tsukamoto T, Yamada A, Sugimoto T, et al. UV-B radiation induces epithelial tumors in mice lacking DNA polymerase eta and mesenchymal tumors in mice deficient for DNA polymerase iota. Mol Cell Biol. 2006;26:7696–706. doi: 10.1128/MCB.01076-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Cheo DL, Meira LB, Hammer RE, Burns DK, Doughty AT, Friedberg EC. Synergistic interactions between XPC and p53 mutations in double-mutant mice: neural tube abnormalities and accelerated UV radiation-induced skin cancer. Curr Biol. 1996;6:1691–4. doi: 10.1016/s0960-9822(02)70794-x. [DOI] [PubMed] [Google Scholar]
- [179].Cheo DL, Meira LB, Burns DK, Reis AM, Issac T, Friedberg EC. Ultraviolet B radiation-induced skin cancer in mice defective in the Xpc, Trp53, and Apex (HAP1) genes: genotype-specific effects on cancer predisposition and pathology of tumors. Cancer Res. 2000;60:1580–4. [PubMed] [Google Scholar]
- [180].Meira LB, Cheo DL, Hammer RE, Burns DK, Reis A, Friedberg EC. Genetic interaction between HAP1/REF-1 and p53. Nat Genet. 1997;17:145. doi: 10.1038/ng1097-145. [DOI] [PubMed] [Google Scholar]
- [181].Meira LB, Cheo DL, Reis AM, Claij N, Burns DK, te Riele H, et al. Mice defective in the mismatch repair gene Msh2 show increased predisposition to UVB radiation-induced skin cancer. DNA Repair. 2002;1:929–34. doi: 10.1016/s1568-7864(02)00143-x. [DOI] [PubMed] [Google Scholar]
- [182].Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, et al. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc Natl Acad Sci USA. 1999;96:13300–5. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Minowa O, Arai T, Hirano M, Monden Y, Nakai S, Fukuda M, et al. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc Natl Acad Sci USA. 2000;97:4156–61. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–30. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- [185].Prolla TA, Baker SM, Harris AC, Tsao JL, Yao X, Bronner CE, et al. Tumour susceptibility and spontaneous mutation in mice deficient in Mlh1, Pms1 and Pms2 DNA mismatch repair. Nat Genet. 1998;18:276–9. doi: 10.1038/ng0398-276. [DOI] [PubMed] [Google Scholar]
- [186].Reitmair AH, Redston M, Cai JC, Chuang TC, Bjerknes M, Cheng H, et al. Spontaneous intestinal carcinomas and skin neoplasms in Msh2-deficient mice. Cancer Res. 1996;56:3842–9. [PubMed] [Google Scholar]
- [187].Andrew SE, Reitmair AH, Fox J, Hsiao L, Francis A, McKinnon M, et al. Base transitions dominate the mutational spectrum of a transgenic reporter gene in MSH2 deficient mice. Oncogene. 1997;15:123–9. doi: 10.1038/sj.onc.1201180. [DOI] [PubMed] [Google Scholar]
- [188].Baross-Francis A, Makhani N, Liskay RM, Jirik FR. Elevated mutant frequencies and increased C: G-->T: A transitions in Mlh1−/− versus Pms2−/− murine small intestinal epithelial cells. Oncogene. 2001;20:619–25. doi: 10.1038/sj.onc.1204138. [DOI] [PubMed] [Google Scholar]
- [189].Andrew SE, Xu XS, Baross-Francis A, Narayanan L, Milhausen K, Liskay RM, et al. Mutagenesis in PMS2- and MSH2-deficient mice indicates differential protection from transversions and frameshifts. Carcinogenesis. 2000;21:1291–5. [PubMed] [Google Scholar]
- [190].Hegan DC, Narayanan L, Jirik FR, Edelmann W, Liskay RM, Glazer PM. Differing patterns of genetic instability in mice deficient in the mismatch repair genes Pms2, Mlh1, Msh2, Msh3 and Msh6. Carcinogenesis. 2006;27:2402–8. doi: 10.1093/carcin/bgl079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Fishel R. The selection for mismatch repair defects in hereditary nonpolyposis colorectal cancer: revising the mutator hypothesis. Cancer Res. 2001;61:7369–74. [PubMed] [Google Scholar]
- [192].Yang G, Scherer SJ, Shell SS, Yang K, Kim M, Lipkin M, et al. Dominant effects of an Msh6 missense mutation on DNA repair and cancer susceptibility. Cancer Cell. 2004;6:139–50. doi: 10.1016/j.ccr.2004.06.024. [DOI] [PubMed] [Google Scholar]
- [193].Lin DP, Wang Y, Scherer SJ, Clark AB, Yang K, Avdievich E, et al. An Msh2 point mutation uncouples DNA mismatch repair and apoptosis. Cancer Res. 2004;64:517–22. doi: 10.1158/0008-5472.can-03-2957. [DOI] [PubMed] [Google Scholar]
- [194].Parsons R, Li G-M, Longley M, Modrich P, Liu B, Berk T, et al. Mismatch repair deficiency in phenotypically normal human cells. Science. 1995;268:738–40. doi: 10.1126/science.7632227. [DOI] [PubMed] [Google Scholar]
- [195].Jin YH, Garg P, Stith CM, Al-Refai H, Sterling JF, Murray LJ, et al. The multiple biological roles of the 3'-->5' exonuclease of Saccharomyces cerevisiae DNA polymerase delta require switching between the polymerase and exonuclease domains. Mol Cell Biol. 2005;25:461–71. doi: 10.1128/MCB.25.1.461-471.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Parsons JL, Preston BD, O'Connor TR, Dianov GL. DNA polymerase delta-dependent repair of DNA single strand breaks containing 3'-end proximal lesions. Nucleic Acids Res. 2007;35:1054–63. doi: 10.1093/nar/gkl1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Tseng SF, Gabriel A, Teng SC. Proofreading activity of DNA polymerase Pol2 mediates 3'-end processing during nonhomologous end joining in yeast. PLoS Genet. 2008;4:e1000060. doi: 10.1371/journal.pgen.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Iida T, Araki H. Noncompetitive counteractions of DNA polymerase ε and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:217–27. doi: 10.1128/MCB.24.1.217-227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Chao EC, Lipkin SM. Molecular models for the tissue specificity of DNA mismatch repair-deficient carcinogenesis. Nucleic Acids Res. 2006;34:840–52. doi: 10.1093/nar/gkj489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Sweasy JB, Lauper JM, Eckert KA. DNA polymerases and human diseases. Radiat Res. 2006;166:693–714. doi: 10.1667/RR0706.1. [DOI] [PubMed] [Google Scholar]
- [201].Loeb LA, Monnat RJ., Jr. DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- [202].Venkatesan RN, Hsu JJ, Lawrence NA, Preston BD, Loeb LA. Mutator phenotypes caused by substitution at a conserved motif A residue in eukaryotic DNA polymerase δ. J Biol Chem. 2006;281:4486–94. doi: 10.1074/jbc.M510245200. [DOI] [PubMed] [Google Scholar]
- [203].Venkatesan RN, Treuting PM, Fuller ED, Goldsby RE, Norwood TH, Gooley TA, et al. Mutation at the polymerase active site of mouse DNA polymerase δ increases genomic instability and accelerates tumorigenesis. Mol Cell Biol. 2007;27:7669–82. doi: 10.1128/MCB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Wang TS, Wong SW, Korn D. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989;3:14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- [205].Tahirov TH, Makarova KS, Rogozin IB, Pavlov YI, Koonin EV. Evolution of DNA polymerases: an inactivated polymerase-exonuclease module in Pol epsilon and a chimeric origin of eukaryotic polymerases from two classes of archaeal ancestors. Biol Direct. 2009;4:11. doi: 10.1186/1745-6150-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Daee DL, Mertz TM, Shcherbakova PV. A cancer-associated DNA polymerase δ variant modeled in yeast causes a catastrophic increase in genomic instability. Proc Natl Acad Sci USA. 2010;107:157–62. doi: 10.1073/pnas.0907526106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Monsees GM, Kraft P, Chanock SJ, Hunter DJ, Han J. Comprehensive screen of genetic variation in DNA repair pathway genes and postmenopausal breast cancer risk. Breast Cancer Res Treat. 2010 doi: 10.1007/s10549-010-0947-3. doi:10.1007/s10549-010-0947-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Wang SS, Gonzalez P, Yu K, Porras C, Li Q, Safaeian M, et al. Common genetic variants and risk for HPV persistence and progression to cervical cancer. PLoS One. 2010;5:e8667. doi: 10.1371/journal.pone.0008667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Matakidou A, el Galta R, Webb EL, Rudd MF, Bridle H, Eisen T, et al. Genetic variation in the DNA repair genes is predictive of outcome in lung cancer. Hum Mol Genet. 2007;16:2333–40. doi: 10.1093/hmg/ddm190. [DOI] [PubMed] [Google Scholar]
- [210].Bethke L, Murray A, Webb E, Schoemaker M, Muir K, McKinney P, et al. Comprehensive analysis of DNA repair gene variants and risk of meningioma. J Natl Cancer Inst. 2008;100:270–6. doi: 10.1093/jnci/djn004. [DOI] [PubMed] [Google Scholar]
- [211].Sigurdson AJ, Hauptmann M, Chatterjee N, Alexander BH, Doody MM, Rutter JL, et al. Kin-cohort estimates for familial breast cancer risk in relation to variants in DNA base excision repair, BRCA1 interacting and growth factor genes. BMC Cancer. 2004;4:9. doi: 10.1186/1471-2407-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Smith TR, Levine EA, Freimanis RI, Akman SA, Allen GO, Hoang KN, et al. Polygenic model of DNA repair genetic polymorphisms in human breast cancer risk. Carcinogenesis. 2008;29:2132–8. doi: 10.1093/carcin/bgn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Wu X, Gu J, Grossman HB, Amos CI, Etzel C, Huang M, et al. Bladder cancer predisposition: a multigenic approach to DNA-repair and cell-cycle-control genes. Am J Hum Genet. 2006;78:464–79. doi: 10.1086/500848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].da Costa LT, Liu B, el-Deiry W, Hamilton SR, Kinzler KW, Vogelstein B, et al. Polymerase δ variants in RER colorectal tumours. Nat Genet. 1995;9:10–1. doi: 10.1038/ng0195-10. [DOI] [PubMed] [Google Scholar]
- [215].Flohr T, Dai JC, Buttner J, Popanda O, Hagmuller E, Thielmann HW. Detection of mutations in the DNA polymerase δ gene of human sporadic colorectal cancers and colon cancer cell lines. Int J Cancer. 1999;80:919–29. doi: 10.1002/(sici)1097-0215(19990315)80:6<919::aid-ijc19>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- [216].Popanda O, Flohr T, Fox G, Thielmann HW. A mutation detected in DNA polymerase δ cDNA from Novikoff hepatoma cells correlates with abnormal catalytic properties of the enzyme. J Cancer Res Clin Oncol. 1999;125:598–608. doi: 10.1007/s004320050322. [DOI] [PubMed] [Google Scholar]
- [217].Popanda O, Fox G, Thielmann HW. DNA polymerases α, δ, and ε of Novikoff hepatoma cells differ from those of normal rat liver in physicochemical and catalytic properties. J Mol Med. 1995;73:259–68. doi: 10.1007/BF00189927. [DOI] [PubMed] [Google Scholar]
- [218].Fox G, Popanda O, Edler L, Thielmann HW. Preferential inhibition of DNA polymerases α, δ, and ε from Novikoff hepatoma cells by inhibitors of cell proliferation. J Cancer Res Clin Oncol. 1996;122:78–94. doi: 10.1007/BF01226265. [DOI] [PubMed] [Google Scholar]
- [219].Fox G, Popanda O, Thielmann HW. Evidence for reduced copying fidelity of DNA polymerases α, δ, and ε from Novikoff hepatoma cells. J Cancer Res Clin Oncol. 1997;123:659–68. doi: 10.1007/s004320050121. [DOI] [PubMed] [Google Scholar]
- [220].Kokoska RJ, Stefanovic L, DeMai J, Petes TD. Increased rates of genomic deletions generated by mutations in the yeast gene encoding DNA polymerase δ or by decreases in the cellular levels of DNA polymerase δ. Mol Cell Biol. 2000;20:7490–504. doi: 10.1128/mcb.20.20.7490-7504.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Nakamura M, Nabetani A, Mizuno T, Hanaoka F, Ishikawa F. Alterations of DNA and chromatin structures at telomeres and genetic instability in mouse cells defective in DNA polymerase alpha. Mol Cell Biol. 2005;25:11073–88. doi: 10.1128/MCB.25.24.11073-11088.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Lemoine FJ, Degtyareva NP, Lobachev K, Petes TD. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell. 2005;120:587–98. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- [223].Casper AM, Mieczkowski PA, Gawel M, Petes TD. Low levels of DNA polymerase alpha induce mitotic and meiotic instability in the ribosomal DNA gene cluster of Saccharomyces cerevisiae. PLoS Genet. 2008;4:e1000105. doi: 10.1371/journal.pgen.1000105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Lemoine FJ, Degtyareva NP, Kokoska RJ, Petes TD. Reduced levels of DNA polymerase δ induce chromosome fragile site instability in yeast. Mol Cell Biol. 2008;28:5359–68. doi: 10.1128/MCB.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Pavlov YI, Shcherbakova PV, Kunkel TA. In vivo consequences of putative active site mutations in yeast DNA polymerases α, ε, δ, and ζ. Genetics. 2001;159:47–64. doi: 10.1093/genetics/159.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Zhou Q, Elzagheid A, Jalava P, Sundfors C, Huang D, Syväoja JE, et al. Highly conserved region of the catalytic subunit of DNA polymerase epsilon in human breast cancer. Exp Oncol. 2003;25:206–10. [Google Scholar]
- [227].Zhou Q, Effati R, Talvinen K, Pospiech H, Syväoja JE, Collan Y. Genomic changes of the 55 kDa subunit of DNA polymerase epsilon in human breast cancer. Cancer Genomics Proteomics. 2008;5:287–92. [PubMed] [Google Scholar]
- [228].Zhou Q, Talvinen K, Sundstrom J, Elzagheid A, Pospiech H, Syvaoja JE, et al. Mutations/polymorphisms in the 55 kDa subunit of DNA polymerase epsilon in human colorectal cancer. Cancer Genomics Proteomics. 2009;6:297–304. [PubMed] [Google Scholar]
- [229].Zhou Q, Lehtonen T, Lehtonen J, Laakso S, Szymas J, Pospiech H, et al. Genomic analysis of the 55 kDa subunit of DNA polymerase epsilon in human intracranial neoplasms. Cancer Genomics Proteomics. 2010;7:143–6. [PubMed] [Google Scholar]
- [230].Boveri T. Über mehrpolige Mitosen als Mittel zur Analyze des Zellkerns. Verh Dtsch Zool Ges Würzburg. 1902;35:67–90. [Google Scholar]
- [231].Balmain A. Cancer genetics: from Boveri and Mendel to microarrays. Nat Rev Cancer. 2001;1:77–82. doi: 10.1038/35094086. [DOI] [PubMed] [Google Scholar]
- [232].Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–8. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- [233].Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347:1593–603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- [234].Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–7. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- [236].Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–90. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Fox EJ, Salk JJ, Loeb LA. Cancer genome sequencing--an interim analysis. Cancer Res. 2009;69:4948–50. doi: 10.1158/0008-5472.CAN-09-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: origin and consequences. Annu Rev Pathol. 2010;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Frank SA, Nowak MA. Problems of somatic mutation and cancer. Bioessays. 2004;26:291–9. doi: 10.1002/bies.20000. [DOI] [PubMed] [Google Scholar]