CpG methylation of half-CRE sequences creates C/EBPα binding sites that activate some tissue-specific genes (original) (raw)
Abstract
DNA methylation of the cytosine in the CpG dinucleotide is typically associated with gene silencing. Genomic analyses have identified low CpG promoters that are both methylated and transcriptionally active, but the mechanism underlying the activation of these methylated promoters remains unclear. Here we show that CpG methylation of the CRE sequence (TGACGTCA) enhances the DNA binding of the C/EBPα transcription factor, a protein critical for activation of differentiation in various cell types. Transfection assays also show that C/EBPα activates the CRE sequence only when it is methylated. The biological significance of this observation was seen in differentiating primary keratinocyte cultures from newborn mice where certain methylated promoters are both bound by C/EBPα and activated upon differentiation. Experimental demethylation by either 5-azacytidine treatment or DNMT1 depletion diminished both C/EBPα binding and activation of the same methylated promoters upon differentiation suggesting that CpG methylation can localize C/EBPα. Transfection studies in cell cultures using methylated tissue-specific proximal promoters identified half-CRE (CGTCA) and half-C/EBP (CGCAA) sequences that need to be methylated for C/EBPα mediated activation. In primary dermal fibroblasts, C/EBPα activates a different set of methylated tissue-specific promoters upon differentiation into adipocytes. These data identify a new function for methyl CpGs: producing DNA binding sites at half-CRE and half-C/EBP sequences for C/EBPα that are needed to activate tissue-specific genes.
Keywords: gene regulation, EMSA, transcription factor binding site
In mammalian genomes, CpG dinucleotides are rare but do occur in clusters called CpG islands that are often located in the proximal promoters of genes, particularly housekeeping genes (1–3). In the early embryo, there is little CpG methylation, but CpG dinucleotides outside of CpG islands typically become methylated during the blastula stage of development (4). Promoters with methylated CpGs are generally transcriptionally silent as occurs with X-chromosome inactivation and imprinting (4). CpG methylation both recruits repressive complexes (4) and prevents the DNA binding of many transcription factors (TFs) (5). In some cancers, methylation of tumor suppressor gene promoters is associated with gene repression (6) lending support to the suggestion that CpG methylation is a general repressive epigenetic mark (7). CpG methylation patterns are not as dynamic as previously thought (8), and it is mainly the regions outside of proximal promoters that become demethylated upon cellular differentiation (9–11). Genomic analyses have identified low CpG promoters that are both methylated and transcriptionally active (8, 12), but the mechanism underlying the activation of methylated promoters remains unclear.
Results and Discussion
C/EBPα Binds a Methyl CRE.
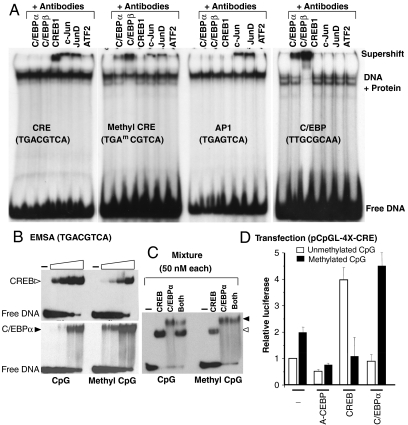
During a survey to evaluate if CpG methylation of classical proximal promoter elements affected DNA binding of nuclear extracts from keratinocytes (Fig. 1A) or mouse liver (SI Appendix, Fig. S1_A_), we observed that both unmethylated and methylated CRE sequences (TGACGTCA) were bound. This was surprising because methylation of the CRE has been reported to inhibit DNA binding of CREB, the protein known to bind the CRE (13). To identify the protein(s) that bind the unmethylated and methylated CRE, we added antibodies to B-ZIP proteins, including C/EBP family members and other proteins known to bind the CRE. These “supershift” experiments indicated that the unmethylated CRE was bound by CREB, c-Jun, JunD, and ATF2 as expected (14). In contrast, the methylated CRE was not bound by CREB but was bound by C/EBPα and C/EBPβ. c-Jun, JunD, and ATF2 showed a modest decrease in binding. Using pure CREB and C/EBPα B-ZIP domains, both EMSA (Fig. 1B) and circular dichroism thermal denaturations (SI Appendix, Fig. S1 B_–_C) recapitulated the nuclear extract results: C/EBPα preferentially binds the CRE when the CpG is methylated. Both methylated and unmethylated consensus C/EBP sequences (TTGCGCAA) were only bound in nuclear extracts by C/EBP family members (SI Appendix, Fig. S1_D_). More detailed studies using EMSA and circular thermal denaturation show pure C/EBPα protein preferentially binds when the central CpG is methylated (SI Appendix, Fig. S1 E and F). EMSA using an equimolar mixture of C/EBPα and CREB showed that C/EBPα preferentially binds to the methylated CRE even in the presence of CREB (Fig. 1C). We chose to study C/EBPα in more detail because it is involved in the activation of differentiation in several cell types (15), including keratinocytes (16–18) and adipocytes (19–21), potentially linking C/EBPα binding to methylated promoters with activation of methylated promoters upon differentiation (see below).
Fig. 1.
C/EBP family members bind to methylated consensus CRE site. (A) EMSA with primary keratinocyte nuclear extracts using antibodies to B-ZIP proteins and 28 bp oligonucleotides containing (i) an unmethylated CRE, (ii) a methylated CRE, (iii) an AP-1 consensus site, or (iv) a C/EBP consensus site identifies that CREB binds to an unmethylated CRE sequence, whereas C/EBP family members bind to a methylated CRE sequence. Only c-Jun and JunD bind the AP-1 sequence, and only C/EBP family members bind the C/EBP sequence. (B) EMSA using the CRE sequence used in A and increasing concentrations of pure CREB and C/EBPα B-ZIP protein domains (5-, 15-, 50-, and 150-nM dimer) show CREB preferentially binds the unmethylated CRE, whereas C/EBPα preferentially binds the methylated CRE. (C) EMSA showing a mixture of CREB and C/EBPα B-ZIP domains at equimolar concentrations (50-nM dimer). (D) pCpGL-4X-CRE, a luciferase reporter containing four consensus CRE sites, was constructed from pCpGL, a parental luciferase reporter plasmid without CpGs in the backbone. Cotransfection of pCpGL-4X-CRE into keratinocytes with CREB coding plasmid increases the activity of only the unmethylated reporter, whereas C/EBPα coding plasmid increases the activity of only the methylated reporter (T test, p < 0.001).
To evaluate whether the preferential binding of C/EBPα to a methylated CRE is biologically relevant, we did transient transfections using primary keratinocytes (Fig. 1D). Four copies of the CRE sequence were cloned into a reporter plasmid where all CpG dinucleotides have been deleted from the plasmid backbone (pCpGL) (22). The use of a reporter without any CpGs in the parental plasmid backbone allowed evaluation of the effect of CpG methylation within the insert on transcriptional activation. The CpGs in the pCpGL-4X-CRE plasmid insert were enzymatically methylated (SI Appendix, Fig. S1_G_) and transfected into keratinocytes where they maintained their CpG methylation status (SI Appendix, Fig. S1_H_). The methylated plasmid is more active and is preferentially inhibited when a dominant-negative C/EBP protein was expressed, suggesting that the methylation dependent activation of the methylated CRE plasmid could be due to the C/EBP family function. Cotransfection of methylated or unmethylated pCpGL-4X-CRE with an expression plasmid encoding C/EBPα or CREB showed that the methylated plasmid is activated by C/EBPα, whereas the unmethylated plasmid is activated by CREB (Fig. 1D and SI Appendix, Fig. S1_I_). Similar results were also obtained using an alternative approach to produce a luciferase reporter plasmid with methylated CRE sequences (SI Appendix, Fig. S1_J_). These data suggest that CpG methylation of the CRE sequence is needed to produce a transcription factor binding site (TFBS) for C/EBPα.
Keratinocyte Differentiation.
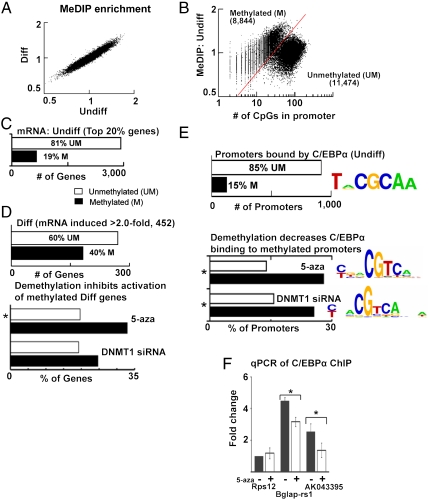
The observation that CpG methylation facilitates the DNA binding of a transcription factor involved in cellular differentiation is particularly interesting considering that demethylation using 5-azacytidine (5-aza), a cytosine analogue that cannot be methylated, or depletion of CpG methylation maintenance enzyme (DNA methyltransferase 1, DNMT1), can inhibit differentiation (23–25). One way this could happen is if CpG methylation is required for transcriptional activation of certain tissue-specific genes. Thus, we set out to investigate whether C/EBPα binds to any methylated promoters that are activated with differentiation. We used calcium to induce differentiation of primary keratinocyte cultures from newborn mice and identified 182 methylated promoters that become activated with differentiation (Fig. 2). We determined the methylation status of proximal promoters using methylated DNA immunoprecipitation (MeDIP) (26) and Nimblegen promoter microarrays and identified mRNAs that increased with differentiation using Affymetrix microarrays. No dramatic changes in CpG methylation at proximal promoters were observed with differentiation either for all genes or for differentiation specific genes (Fig. 2A and SI Appendix, Fig. S2_A_). Plotting CpG methylation in undifferentiated cells versus the number of CpGs in the promoter produced two distinct groups (Fig. 2B). One group is CpG sparse and these promoters are predominantly methylated with a steady increase in MeDIP enrichment as the number of CpGs in the promoter increases (8). The second group has more CpGs but lower MeDIP enrichment, suggesting that they are either partially methylated or completely unmethylated (12). These two groups of promoters are similar to those described in a recent study that adjusted the MeDIP enrichment for the CpG density of proximal promoters (27). We also examined the promoters classified into three groups based on their CpG density (12) (SI Appendix, Fig. S2_B_). In undifferentiated keratinocytes, the majority of the active genes have unmethylated promoters, and some (19%) active genes have methylated promoters, as expected (Fig. 2C and SI Appendix, Fig. S2_C_). With differentiation, the mRNAs for 452 genes increased more than 2-fold, and 40% of these genes have methylated promoters (Fig. 2D and SI Appendix, Fig. S2 D_–_H). Genes activated with differentiation also showed an increase in binding of two active marks [phosphorylated RNA polymerase II (p-RNAP) and H3K9acetyl] as assessed by chromatin immunoprecipitation followed by genome-wide promoter analysis (ChIP-chip) (SI Appendix, Fig. S2_I_). Immunocytochemistry showed that the increase in expression of differentiation specific proteins with differentiation was a general property of all cells in the culture (SI Appendix, Fig. S2 J_–_K).
Fig. 2.
C/EBPα binding activates methylated promoters in keratinocyte differentiation. (A) Scatter plot (log 2) of MeDIP enrichment before and after keratinocyte differentiation using Nimblegen mouse promoter arrays with each dot representing a promoter. (B) MeDIP enrichment in undifferentiated keratinocytes versus the number of CpG dinucleotides in the promoter (-1,000 bp to +500 bp). The red line demarcates two groups of promoters, unmethylated (UM) and methylated (M). (C) Methylation status of promoters for the most abundantly expressed genes (top 20%) in undifferentiated (Undiff) keratinocytes. (D) Number of UM and M promoters whose mRNA expression was doubled with differentiation (Diff, Upper). mRNA induction during differentiation is preferentially inhibited (induction decreases > 50%) for genes with methylated promoters by experimental demethylation using either 5-aza (*_χ_2 test, p < 0.01), or siRNA to DNMT1. Both demethylation methods decreased expression of the same methylated promoters (hypergeometric test, _p_ < 0.05). (_E_) Number of UM and M promoters bound by C/EBPα (binding enrichment > 1.2, T test, p < 0.03) in undifferentiated keratinocytes. Logo showing a C/EBP consensus site is the most enriched sequence in promoters bound by C/EBPα. C/EBPα binding is inhibited (< 0.8) preferentially at methylated promoters after treatment with 5-aza (*_χ_2 test, p < 0.001) or siRNA to DNMT1 (*_χ_2 test, p < 0.02). Both demethylation methods decreased C/EBPα binding to the same methylated promoters (hypergeometric test, p < 0.001). The half-CRE (CGTCA) is the most common DNA sequence occurring in promoters with decreased C/EBPα binding following either demethylation method. (F) qPCR of C/EBPα ChIP DNA from undifferentiated keratinocytes for the tissue-specific (Bglap-rs1 and AK043395) and housekeeping (Rps12) promoters with or without 5-aza treatment compared to input DNA. All values are expressed as fold change normalized to Rps12 (*T test, p < 0.05).
To determine if methylation is required for the activation of genes with methylated promoters during keratinocyte differentiation, we experimentally demethylated the CpG dinucleotides in the genome using two methods, namely, 5-aza treatment and siRNA mediated depletion of DNMT1 (SI Appendix, Fig. S3 A_–_E). Both demethylation methods caused ∼20% loss of global cytosine methylation as assayed by HPLC and ELISA (SI Appendix, Fig. S3 C_–_D). When we focused on activation of genes with differentiation, the 5-aza treated cultures preferentially inhibited the induction of the genes with methylated promoters. Similar but less statistically significant results were observed using DNMT1-depleted cultures. When both demethylation methods were compared, statistically significant similar genes with methylated promoters were inhibited suggesting that for these promoters, CpG methylation is critical for their activation (SI Appendix, Fig. S2 E and G).
We next determined the localization of C/EBPα in proximal promoters of undifferentiated keratinocytes (Fig. 2E and SI Appendix, Fig. S3 F_–_I). C/EBPα ChIP-chip showed that C/EBPα preferentially binds promoters containing the known C/EBP consensus site (28) and that 85% of bound promoters are unmethylated (Fig. 2E). However, 15% of bound promoters are methylated. C/EBPα ChIP-chip from either 5-aza-treated or DNMT1-depleted undifferentiated keratinocytes showed a preferential loss of binding to similar methylated promoters, suggesting that C/EBPα localization to these promoters is dependent on CpG methylation (Fig. 2E and SI Appendix, Fig. S3_G_), a result confirmed by qPCR (Fig. 2F). C/EBPα bound similar promoters in undifferentiated and differentiated keratinocytes, suggesting that the recruitment of C/EBPα to methylated promoters is not the signal for activation of these promoters with calcium (SI Appendix, Fig. S3_H_). Experimental demethylation of differentiating keratinocytes again preferentially inhibited C/EBPα binding to methylated promoters (SI Appendix, Fig. S3_I_), including the methylated promoters activated with differentiation (SI Appendix, Fig. S3 J and K). The sequence that is most enriched in promoters where C/EBPα binding is inhibited after demethylation by either 5-aza treatment or DNMT1 depletion is the half-CRE motif, suggesting that this methylated sequence may mediate the localization of C/EBPα to some methylated promoters (Fig. 2E and SI Appendix, Fig. S3_L_). The role of a methylated half-CRE in C/EBPα localization prompted us to determine the genomic localization of CREB, a transcription factor known to bind half-CRE sequences. CREB ChIP-chip in undifferentiated keratinocytes shows that the CRE consensus sequence is preferentially bound by CREB as expected, and 98% of bound promoters are unmethylated (SI Appendix, Fig. S4_A_). We observed a slight increase in CREB binding to some methylated promoters after 5-aza treatment, suggesting that CpG methylation may prevent CREB binding in vivo (SI Appendix, Fig. S4_B_) as it does in vitro.
The importance of C/EBPα function in keratinocyte differentiation was evaluated using two methods. First, we cultured primary keratinocytes from transgenic mice expressing a dominant-negative protein (A-C/EBP) that inhibits the DNA binding of C/EBP family members (18). Second, we used siRNA to deplete C/EBPα and C/EBPβ (SI Appendix, Fig. S4_C_). Global mRNA analysis indicated that both strategies suppressed keratinocyte differentiation and preferentially inhibited the activation of genes containing methylated promoters (SI Appendix, Fig. S4_D_). The same methylated genes where CpG demethylation inhibited induction with differentiation were also suppressed by C/EBPα inhibition, suggesting a connection between these two distinct events (SI Appendix, Fig. S4_E_).
Activity of Methylated Promoters.
We next turned to individual methylated promoters that are activated with keratinocyte differentiation to evaluate whether activation is mediated by C/EBPα binding to methylated DNA sequences. We examined four keratinocyte specific promoters that fit five criteria: (i) increased transcriptional activity upon differentiation, (ii) methylated as determined by MeDIP (SI Appendix, Fig. S4_F_), (iii) bound by C/EBPα, (iv) C/EBPα binding decreased by experimental demethylation, and (v) containing a half-CRE sequence. As a control, we examined two unmethylated housekeeping promoters.
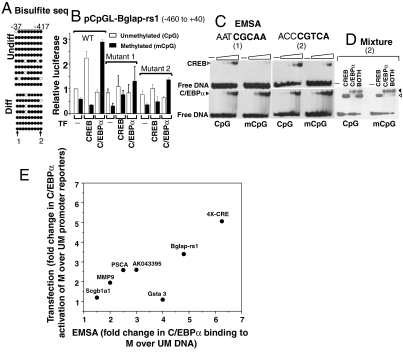
For promoters fitting the five criteria above, we determined the methylation status of CpGs in individual promoters before and after differentiation. We did bisulfite sequencing by clonal analyses for four promoters (Bglap-rs1, AK043395, Scgb1a1, and Gsta3). The promoters were predominantly methylated both before and after differentiation (Fig. 3A and SI Appendix, Figs. S4_G_ and S5). To evaluate the importance of C/EBPα and CpG methylation on promoter activity, these promoters were cloned into the pCpGL reporter, enzymatically methylated, and then transfected into keratinocytes where CpG methylation of the plasmid was maintained (SI Appendix, Fig. S4_H_). Fig. 3 shows data for the Bglap-rs1 promoter that has both a half-C/EBP site and a half-CRE site. C/EBPα activated the methylated promoter but not the unmethylated promoter. CREB, in contrast, activated the unmethylated promoter but not the methylated promoter (Fig. 3B). Mutation of either site (SI Appendix, Figs. S3_B_ and S4_I_) decreased C/EBPα activation of the methylated Bglap-rs1 reporter and CREB activation of the unmethylated promoter. Biochemical studies using circular dichroism thermal denaturation (SI Appendix, Fig. S6 A_–_D), DNase footprinting (SI Appendix, Fig. S6_E_), and EMSA with pure B-ZIP domains of C/EBPα and CREB (Fig. 3C) recapitulated what was observed in the transient transfections: C/EBPα bound either site better when they were methylated. Mixing C/EBPα and CREB together with DNA indicated that C/EBPα binds the methylated half-CRE site even in the presence of equimolar CREB, whereas CREB binds this site when it is unmethylated even in the presence of C/EBPα (Fig. 3D). These data suggest that methylation is essential for C/EBPα binding to methylated promoters, and a subset of these promoters is induced during keratinocyte differentiation. Methylation, however, is not the activation signal, as we do not see changes in methylation at these promoters upon calcium-mediated differentiation. The signal(s) that activate keratinocyte-specific gene expression can be potentially posttranslational modifications of C/EBPα (29) that recruit coactivators causing H3K9 acetylation and phosphorylation of RNAP (SI Appendix, Fig. S2_J_). As already shown, when C/EBPα was overexpressed in undifferentiated keratinocytes, we observed a methylation-dependent promoter activation.
Fig. 3.
C/EBPα activates methylated promoters in keratinocytes. (A) Bisulfite sequencing of the cloned keratinocyte specific Bglap-rs1 proximal promoter before and after differentiation shows that it is predominantly methylated. Filled circles denote methylated CpG and open circles denote unmethylated CpG. Half-C/EBP (-37 bp) (1) and half-CRE (-417 bp) (2) sequences are marked. (B) Luciferase activities of Bglap-rs1 promoter (pCpGL-Bglap-rs1) cotransfected with CREB or C/EBPα coding plasmid show that CREB activates the unmethylated plasmid, whereas C/EBPα activates the methylated plasmid (T test, p < 0.05). Promoters with the mutated half-C/EBP (1) or half-CRE (2) sequence have reduced activity. All reporter activities are expressed relative to the unmethylated plasmid. (C) EMSA showing CREB and C/EBPα (15-, 50-, 150-, and 500-nM dimer) binding to the unmethylated and methylated half-C/EBP (1) or half-CRE (2) sequences from Bglap-rs1. (D) Mixture of CREB and C/EBPα (each 500-nM dimer) shows C/EBPα binds preferentially to the methylated half-CRE sequence even in the presence of equimolar CREB. CREB and C/EBPα cannot bind the mutant half-CRE site of Bglap-rs1 (SI Appendix, Fig. S4_I_). (E) Fold difference in C/EBPα binding to methylated (M) compared to unmethylated (UM) DNA sequences determined by EMSA versus fold difference in C/EBPα transactivation of a methylated reporter compared to an unmethylated reporter. Seven sequences are plotted and show a positive correlation; i.e., preferential binding to the methylated sequence produces preferential transactivation of the methylated luciferase reporter.
Data for five additional methylated promoters activated with differentiation (AK043395, Scgb1a1, Gsta3, MMP9, and PSCA) and two housekeeping promoters (Kifc1 and Rps12) are shown in SI Appendix, Figs. S5, S7 A and B, and S8 A and B. Results for AK043395 are similar to those described for Bglap-rs1: C/EBPα preferentially activates the methylated plasmid via a half-CRE sequence and pure C/EBPα protein binds better to the methylated half-CRE sequence. In contrast, the Scgb1a1 and Gsta3 promoters are activated by C/EBPα via a half-CRE sequence independent of their methylation status. A plot of the effect of CpG methylation on C/EBPα binding to different half-CRE sequences versus transactivation in transient transfections shows a positive correlation: The more methylation enhances C/EBPα binding in EMSA, the more it enhances transactivation (Fig. 3E). An estimate of K D of CREB or C/EBPα and nine studied DNA sequences are shown in SI Appendix, Fig. S8_K_. The two unmethylated housekeeping promoters were almost totally inactivated by methylation supporting the general suggestion that methylation is transcriptionally suppressive (SI Appendix, Fig. S7 A and B).
Primary keratinocytes treated by 5-aza were used for C/EBPα ChIP to examine in vivo whether C/EBPα preferentially binds the methylated DNA in the presence of the same DNA that is unmethylated (SI Appendix, Fig. S7 C and D). Bisulfite sequencing of individual clones of genomic DNA from 5-aza-treated keratinocytes identified ∼20% decrease in CpG methylation of the Bglap-rs1 promoter (SI Appendix, Fig. S7_C_) as was observed globally by both ELISA and HPLC. C/EBPα ChIP from the 5-aza samples was enriched for the methylated sequences, demonstrating the importance of CpG methylation for C/EBPα localization to these proximal promoters. CREB in contrast preferentially binds to the unmethylated promoters (SI Appendix, Fig. S7 C and D).
Adipocyte Differentiation.
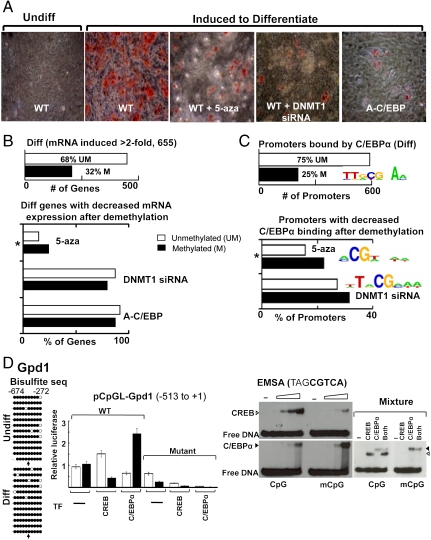
C/EBPα is also known to be important for adipocyte differentiation (21), and we investigated if C/EBPα binding to methylated CpG sequences is critical for differentiation (Fig. 4 and SI Appendix, Fig. S8_C_–S10). Dermal fibroblasts from newborn mice were differentiated into adipocytes and mRNA expression of ∼200 genes with methylated promoters increased by more than 2-fold (30). Global mRNA expression from dermal fibroblasts was similar to expression in adipocyte cultures differentiated from primary preadipocytes isolated from adult mice inguinal adipose tissue confirming that this unique differentiation system recapitulates adipocyte differentiation (SI Appendix, Fig. S8_C_). The CpG methylation profile did not markedly change with differentiation (SI Appendix, Fig. S8_D_) and was similar to what was observed in keratinocytes (SI Appendix, Fig. S8 E and F). Different methylated promoters were induced in adipocytes than in keratinocytes (SI Appendix, Fig. S8_G_). CpG demethylation by 5-aza treatment or DNMT1 depletion (SI Appendix, Fig. S9 A_–_C) inhibited the differentiation of dermal fibroblasts into adipocytes as measured by Oil-Red O staining and global mRNA analysis (Fig. 4 A and B and SI Appendix, Fig. S8_H_). Suppression of C/EBPα function by A-C/EBP also inhibited differentiation (Fig. 4 A and B). DNMT1 depletion or A-C/EBP almost completely inhibited differentiation, preventing us from observing a difference between the inhibition of methylated and unmethylated promoters. The 5-aza effect was more modest and the methylated promoters were preferentially inhibited, suggesting that methylation is critical for their activation. C/EBPα localization to methylated promoters was inhibited by CpG demethylation (Fig. 4C) as observed in keratinocytes. These data recapitulate what we observed in keratinocytes: Both methylation and C/EBPα function are needed for differentiation, and C/EBPα binding to methylated promoters is inhibited by demethylation (Fig. 4 and SI Appendix, Figs. S9_D_–S10).
Fig. 4.
C/EBPα binding activates methylated promoters in fibroblasts. (A) Oil-Red-O staining showing accumulation of neutral lipids in newborn dermal fibroblast cultures 8 d after induction for adipogenic differentiation. Differentiation is inhibited by 5-aza treatment, depletion of DNMT1, or expression of A-C/EBP, a dominant negative to C/EBP family members. (B) Number of UM and M promoters whose mRNA expression was doubled with differentiation (Upper). mRNA induction during differentiation is preferentially inhibited (induction decreases > 50%) for genes with methylated promoters by experimental demethylation using either 5-aza (*_χ_2 test, p < 0.05), or siRNA to DNMT1 or A-C/EBP. (_C_) Number of UM and M promoters bound by C/EBPα (binding enrichment > 1.2, T test, p < 0.03) in adipocytes. Logo showing a C/EBPα consensus site is the most enriched sequence in these promoters. (Lower) Decreased C/EBPα binding (< 0.8) after demethylation preferentially at methylated promoters by 5-aza (* _χ_2 test, p < 0.05) or siRNA to DNMT1. Both demethylation methods decreased C/EBPα binding to the same methylated promoters (hypergeometric test, p < 0.02). Logos containing a CpG are the most common DNA sequences occurring in promoters with decreased C/EBPα binding following either demethylation method. (D) Bisulfite sequencing of the adipocyte specific promoter, Gpd1, showing it is predominantly methylated before and after differentiation. Filled circles denote methylated CpG and open circles denote unmethylated CpG. The half-CRE sequence is marked by an arrow. Activity of a luciferase reporter containing the Gpd1 promoter (pCpGL-Gpd1), either unmethylated or enzymatically methylated, transiently transfected into differentiated fibroblasts. Cotransfection with expression plasmids encoding CREB or C/EBPα enhanced activity of unmethylated and methylated promoters, respectively (T test, p < 0.05). Mutation of half-CRE site reduced activity of the promoter. EMSA showing CREB and C/EBPα (15-, 50-, 150-, and 500-nM dimer) binding to the unmethylated and methylated half-CRE sequence from Gpd1 promoter. Mixture of CREB and C/EBPα (150-nM dimer each) shows C/EBPα binds preferentially to the methylated sequence even in the presence of equimolar CREB.
We next examined Gpd1, a gene with a methylated promoter that is induced during adipocyte differentiation (Fig. 4D and SI Appendix, Fig. S10 G and H). This promoter had the same five characteristics we used to select keratinocyte specific promoters for further studies. Bisulfite sequencing demonstrated that the Gpd1 promoter is primarily methylated before and after differentiation (Fig. 4D). A luciferase reporter assay showed that CREB can activate the unmethylated promoter, whereas C/EBPα can activate the methylated promoter. Again, the half-CRE in the promoter was critical for C/EBPα activation of the methylated promoter (Fig. 4D). EMSA also showed that C/EBPα binds the half-CRE sequence better than CREB only when the sequence is methylated (Fig. 4D). Thus, CpG methylation is critical for activation of a subset of adipocyte specific genes with methylated promoters.
Classically, the observation is that tissue-specific genes have unmethylated promoters in the tissue where they are expressed (31). We observed no change in methylation status of keratinocyte specific genes following 2 d of keratinocyte differentiation. When we made the same comparison for fibroblasts differentiated for 8 d, we observed a modest decrease in the methylation of the adipocyte specific genes with methylated promoters. We have extended this analysis and examined the methylation status of liver and heart in the adult mice. Here we observed that liver-specific promoters are demethylated in the liver compared to the heart (SI Appendix, Fig. S10 I_–_K). This suggests that tissue-specific genes with methylated promoters become activated with differentiation, and after activation, they may become demethylated as has been suggested previously (32, 33).
In summary, we show that CpG methylation produces C/EBPα binding sites at CRE-like sequences. These methylated sites localize C/EBPα to methylated proximal promoters allowing activation of a subset of differentiation specific genes in both keratinocytes and adipocytes. This may be a general property in other tissues where C/EBPα is involved in differentiation. Thus, in addition to the familiar role of CpG methylation in producing a binding site for proteins involved in gene repression, in some cases, CpG methylation can produce a binding site for proteins involved in gene activation.
Materials and Methods
EMSA used either nuclear extracts or pure CREB and C/EBPα B-ZIP domain. To produce plasmids where only the insert is methylated, a plasmid with no CpGs in the backbone (22) was used. Proximal promoters were cloned into pCpGL and enzymatically methylated using M. Sss I. Plasmids were transfected into keratinocytes or adipocytes and luciferase levels were measured. Primary keratinocyte cultures isolated from newborn mice were differentiated as described (34). Primary fibroblast cultures isolated from newborn mice were differentiated into adipocytes using a cocktail of hormones. MeDIP and ChIP-chip were performed using the indicated antibodies. DNA was isolated, PCR amplified, and hybridized to Nimblegen mouse promoter arrays. mRNA concentrations were determined using Affymetrix microarrays. Determination of the CpG methylation was determined by bisulfite conversion, cloning, and subsequent sequencing. Comprehensive details are provided in the SI Appendix.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Michael Rehli (University Hospital, Regensburg, Germany) for the pCpGL plasmid, Takeshi Tomita [National Cancer Institute (NCI), National Institutes of Health (NIH)] for tranactivator plasmids, Oksana Gavrilova (National Institute of Diabetes and Digestive and Kidney Diseases, NIH) for help with preadipocyte cultures, Xiaohui Yan (NCI, NIH) for bisulfite sequencing, Thomas A. Down (University of Cambridge, Cambridge, UK) for help with running BATMAN, and Thomas A. Johnson (NCI, NIH) for help with microarray experiments.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
References
- 1.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 2.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 3.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 5.Rozenberg JM, et al. All and only CpG containing sequences are enriched in promoters abundantly bound by RNA polymerase II in multiple tissues. BMC Genomics. 2008;9:67. doi: 10.1186/1471-2164-9-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird AP, Wolffe AP. Methylation-induced repression—Belts, braces, and chromatin. Cell. 1999;99:451–454. doi: 10.1016/s0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 8.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunk BP, Goldhamer DJ, Emerson CP., Jr Regulated demethylation of the myoD distal enhancer during skeletal myogenesis. Dev Biol. 1996;177:490–503. doi: 10.1006/dbio.1996.0180. [DOI] [PubMed] [Google Scholar]
- 10.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmidl C, et al. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009;19:1165–1174. doi: 10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 13.Iguchi-Ariga SM, Schaffner W. CpG methylation of the cAMP-responsive enhancer/promoter sequence TGACGTCA abolishes specific factor binding as well as transcriptional activation. Genes Dev. 1989;3:612–619. doi: 10.1101/gad.3.5.612. [DOI] [PubMed] [Google Scholar]
- 14.Ahn S, et al. A dominant-negative inhibitor of CREB reveals that it is a general mediator of stimulus-dependent transcription of c-fos. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birkenmeier EH, et al. Tissue-specific expression, developmental regulation, and genetic mapping of the gene encoding CCAAT/enhancer binding protein. Genes Dev. 1989;3:1146–1156. doi: 10.1101/gad.3.8.1146. [DOI] [PubMed] [Google Scholar]
- 16.Maytin EV, Habener JF. Transcription factors C/EBP alpha, C/EBP beta, and CHOP (Gadd153) expressed during the differentiation program of keratinocytes in vitro and in vivo. J Invest Dermatol. 1998;110:238–246. doi: 10.1046/j.1523-1747.1998.00123.x. [DOI] [PubMed] [Google Scholar]
- 17.Oh HS, Smart RC. Expression of CCAAT/enhancer binding proteins (C/EBP) is associated with squamous differentiation in epidermis and isolated primary keratinocytes and is altered in skin neoplasms. J Invest Dermatol. 1998;110:939–945. doi: 10.1046/j.1523-1747.1998.00199.x. [DOI] [PubMed] [Google Scholar]
- 18.Oh WJ, Rishi V, Orosz A, Gerdes MJ, Vinson C. Inhibition of CCAAT/enhancer binding protein family DNA binding in mouse epidermis prevents and regresses papillomas. Cancer Res. 2007;67:1867–1876. doi: 10.1158/0008-5472.CAN-06-2746. [DOI] [PubMed] [Google Scholar]
- 19.Umek RM, Friedman AD, McKnight SL. CCAAT-enhancer binding protein: A component of a differentiation switch. Science. 1991;251:288–292. doi: 10.1126/science.1987644. [DOI] [PubMed] [Google Scholar]
- 20.Zhang JW, Tang QQ, Vinson C, Lane MD. Dominant-negative C/EBP disrupts mitotic clonal expansion and differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA. 2004;101:43–47. doi: 10.1073/pnas.0307229101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefterova MI, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–2952. doi: 10.1101/gad.1709008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klug M, Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1:127–130. doi: 10.4161/epi.1.3.3327. [DOI] [PubMed] [Google Scholar]
- 23.Kopelovich L. 5-Azacytidine inhibits adipocytic conversion of human skin fibroblasts by Kirsten murine sarcoma virus and dexamethasone. Exp Cell Biol. 1987;55:276–280. doi: 10.1159/000163427. [DOI] [PubMed] [Google Scholar]
- 24.Jackson M, et al. Severe global DNA hypomethylation blocks differentiation and induces histone hyperacetylation in embryonic stem cells. Mol Cell Biol. 2004;24:8862–8871. doi: 10.1128/MCB.24.20.8862-8871.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikkelsen TS, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weber M, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 27.Down TA, et al. A Bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol. 2008;26:779–785. doi: 10.1038/nbt1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson PF. Identification of C/EBP basic region residues involved in DNA sequence recognition and half-site spacing preference. Mol Cell Biol. 1993;13:6919–6930. doi: 10.1128/mcb.13.11.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behre G, et al. Ras signaling enhances the activity of C/EBP alpha to induce granulocytic differentiation by phosphorylation of serine 248. J Biol Chem. 2002;277:26293–26299. doi: 10.1074/jbc.M202301200. [DOI] [PubMed] [Google Scholar]
- 30.Chen FG, et al. Clonal analysis of nestin(−) vimentin(+) multipotent fibroblasts isolated from human dermis. J Cell Sci. 2007;120:2875–2883. doi: 10.1242/jcs.03478. [DOI] [PubMed] [Google Scholar]
- 31.Razin A, Cedar H. DNA methylation and gene expression. Microbiol Rev. 1991;55:451–458. doi: 10.1128/mr.55.3.451-458.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.D’Alessio AC, Weaver IC, Szyf M. Acetylation-induced transcription is required for active DNA demethylation in methylation-silenced genes. Mol Cell Biol. 2007;27:7462–7474. doi: 10.1128/MCB.01120-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Dlugosz AA, Glick AB, Tennenbaum T, Weinberg WC, Yuspa SH. Isolation and utilization of epidermal keratinocytes for oncogene research. Methods Enzymol. 1995;254:3–20. doi: 10.1016/0076-6879(95)54003-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information