Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution (original) (raw)
. Author manuscript; available in PMC: 2011 May 1.
Published in final edited form as: Nat Genet. 2010 Oct 10;42(11):949–960. doi: 10.1038/ng.685
Abstract
Waist-hip ratio (WHR) is a measure of body fat distribution and a predictor of metabolic consequences independent of overall adiposity. WHR is heritable, but few genetic variants influencing this trait have been identified. We conducted a meta-analysis of 32 genome-wide association studies for WHR adjusted for body-mass-index (up to 77,167 participants), following up 16 loci in an additional 29 studies (up to 113,636 subjects). We identified 13 novel loci in or near RSPO3, VEGFA, TBX15-WARS2, NFE2L3, GRB14, DNM3-PIGC, ITPR2-SSPN, LY86, HOXC13, ADAMTS9, ZNRF3-KREMEN1, NISCH-STAB1, and CPEB4 (P 1.9 × 10−9 to 1.8 × 10−40), and the known signal at LYPLAL1. Seven of these loci exhibited marked sexual dimorphism, all with a stronger effect on WHR in women than men (P for sex-difference 1.9 × 10−3 to 1.2 × 10−13). These findings provide evidence for multiple loci that modulate body fat distribution, independent of overall adiposity, and reveal powerful gene-by-sex interactions.
Keywords: genome-wide association, waist-hip-ratio, body fat distribution, central obesity, meta-analysis, genetics, visceral adipose tissue, metabolism, body composition, Expression Quantitative Trait Loci, sex difference
Central obesity and body fat distribution, as measured by waist circumference (WC) and waist-hip-ratio (WHR), are associated with individual risk of type 2 diabetes (T2D)1,2 and coronary heart disease3, and with all-cause mortality4. These effects are independent of overall adiposity as measured by body mass index (BMI). WHR is of particular interest as a measure of body fat distribution, since it integrates the adverse metabolic risk associated with increasing WC with the more protective role of gluteal fat deposition with respect to diabetes, hypertension, and dyslipidemia5,6.
There is abundant evidence that body fat distribution is influenced by genetic loci distinct from those regulating BMI and overall adiposity. First, even after accounting for BMI, individual variation in WHR is heritable7,8, with estimates ranging from 22–61%7–10. Second, the striking abnormalities of regional fat deposition associated with lipodystrophic syndromes demonstrate that genetic variation can have dramatic effects on the development and maintenance of specific fat depots11,12. Third, in a previous genome-wide association analysis, we identified a locus near LYPLAL1 strongly associated with WHR independent of any effects on BMI13, providing proof-of-principle for the genetic control of body fat distribution, distinct from that of overall adiposity.
Within the GIANT (Genetic Investigation of Anthropometric Traits) consortium, we performed a large-scale meta-analysis of genome-wide association (GWA) studies informative for WHR, using adjustment for BMI to focus discovery towards genetic loci associated with body fat distribution rather than overall adiposity14–16.
RESULTS
Genome-wide significant association of WHR with 14 SNPs
We conducted a two-stage study among individuals of European descent (Supplementary Table 1 and Online Methods). In the discovery stage, up to 2,850,269 imputed and genotyped single nucleotide polymorphisms (SNPs) were examined in 32 GWA studies comprising up to 77,167 participants informative for anthropometric measures of body fat distribution. We performed a fixed-effects meta-analysis of WHR, employing study-specific linear regression adjusted for BMI and age, stratified by gender, and using an additive genetic model. After genomic control adjustment per study and in the meta-analysis, these analyses revealed a substantial excess of low p-values (Figure 1 a, b).
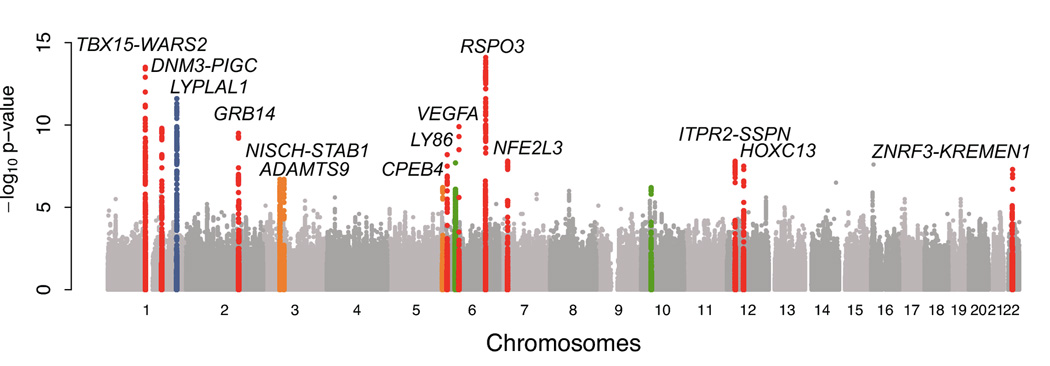
Figure 1. Genome-wide association analyses for WHR in discovery studies.
A. Manhattan plot shows results of the WHR association meta-analysis in discovery studies (P on the y-axis and SNP genomic position on the x-axis). Colored genomic loci indicate significant association (P < 5 × 10−8) detected previously (blue)13, in our GWA stage (red), and after the meta-analysis combining GWA and follow-up studies (orange). Two loci tested in the follow-up stage did not achieve genome-wide significance (green).
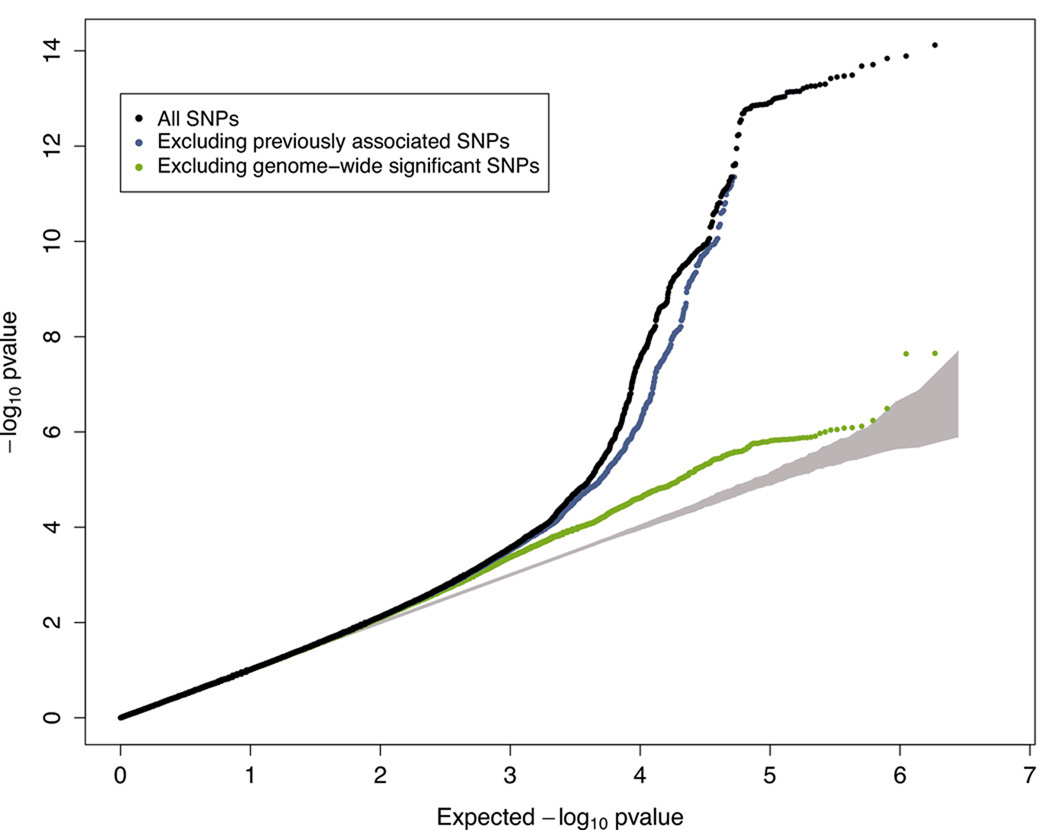
B. Quantile-quantile (QQ) plot of SNPs for the discovery meta-analysis of WHR (black) and after removing SNPs within 1 Mb of either the recently reported LYPLAL1 signal (blue) or the 14 significant associations (green). The grey area represents the 95% confidence interval around the test statistic under the null distribution.
We selected SNPs representing the top 16 independent (> 1 Mb distance) regions of association (discovery P < 1.4 × 10−6, Table 1) and evaluated them in 29 additional, independent studies (up to 113,636 individuals) using a mixture of in silico data and de novo genotyping. In these follow-up studies, 14 of the 16 showed strong directionally-consistent evidence for replication (P < 1.0 × 10−3) and ten reached genome-wide significance (P < 5.0 × 10−8). Joint analysis of the discovery and follow-up results revealed genome-wide significant associations for 14 signals (P between 1.9 × 10−9 and 1.8 × 10−40, Table 1).
Table 1.
Fourteen SNPs associated with WHR at genome-wide significance levels
| SNP | Chr | Position(b36) | Nearby genes | EA a | EAFb | Discovery | Follow-up | Combined | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P | Beta | N | P | Beta | N | P | Beta | ||||||
| SNPs evaluated in follow-up achieving genome-wide significance | |||||||||||||
| rs9491696 | 6 | 127,494,332 | RSPO3 | G | 0.520 | 2.10E-14 | 0.037 | 77,164 | 3.27E-28 | 0.045 | 113,582 | 1.84E-40 | 0.042 |
| rs6905288 | 6 | 43,866,851 | VEGFA | A | 0.562 | 4.72E-10 | 0.033 | 77,129 | 1.18E-16 | 0.039 | 95,430 | 5.88E-25 | 0.036 |
| rs984222 | 1 | 119,305,366 | TBX15-WARS2 | G | 0.365 | 3.81E-14 | 0.037 | 77,167 | 1.56E-12 | 0.031 | 109,623 | 8.69E-25 | 0.034 |
| rs1055144 | 7 | 25,837,634 | NFE2L3 | T | 0.210 | 1.49E-08 | 0.034 | 77,145 | 3.26E-18 | 0.043 | 113,636 | 9.97E-25 | 0.040 |
| rs10195252 | 2 | 165,221,337 | GRB14 | T | 0.599 | 3.23E-10 | 0.031 | 77,119 | 3.18E-16 | 0.036 | 102,449 | 2.09E-24 | 0.033 |
| rs4846567 | 1 | 217,817,340 | LYPLAL1 | G | 0.283 | 2.37E-12 | 0.037 | 77,167 | 3.15E-10 | 0.032 | 91,820 | 6.89E-21 | 0.034 |
| rs1011731 | 1 | 170,613,171 | DNM3-PIGC | G | 0.572 | 1.72E-10 | 0.031 | 77,094 | 7.47E-09 | 0.026 | 92,018 | 9.51E-18 | 0.028 |
| rs718314 | 12 | 26,344,550 | ITPR2-SSPN | G | 0.741 | 2.41E-08 | 0.031 | 77,167 | 1.49E-10 | 0.030 | 107,503 | 1.14E-17 | 0.030 |
| rs1294421 | 6 | 6,688,148 | LY86 | G | 0.387 | 6.31E-09 | 0.029 | 77,154 | 2.69E-10 | 0.028 | 102,189 | 1.75E-17 | 0.028 |
| rs1443512 | 12 | 52,628,951 | HOXC13 | A | 0.239 | 3.33E-08 | 0.031 | 77,165 | 2.92E-10 | 0.030 | 112,353 | 6.38E-17 | 0.031 |
| rs6795735 | 3 | 64,680,405 | ADAMTS9 | C | 0.406 | 2.47E-07 | 0.025 | 77,162 | 6.75E-08 | 0.026 | 84,480 | 9.79E-14 | 0.025 |
| rs4823006 | 22 | 27,781,671 | ZNRF3-KREMEN1 | A | 0.569 | 4.47E-08 | 0.027 | 77,086 | 2.41E-05 | 0.019 | 93,911 | 1.10E-11 | 0.023 |
| rs6784615 | 3 | 52,481,466 | NISCH-STAB1 | T | 0.941 | 3.18E-07 | 0.052 | 76,859 | 1.56E-04 | 0.036 | 109,028 | 3.84E-10 | 0.043 |
| rs6861681 | 5 | 173,295,064 | CPEB4 | A | 0.340 | 1.40E-06 | 0.026 | 77,164 | 2.13E-04 | 0.019 | 85,722 | 1.91E-09 | 0.022 |
| Further SNPs evaluated in follow-up but not achieving genome-wide significant in the combined analysis | |||||||||||||
| rs2076529 | 6 | 32,471,933 | BTNL2 | C | 0.570 | 2.22E-08 | 0.041 | 34,532 | 0.012 | 0.011 | 92,778 | 3.71E-07 | 0.020 |
| rs7081678 | 10 | 32,030,629 | ZEB1 | A | 0.085 | 5.76E-07 | 0.045 | 76,270 | 0.094 | 0.013 | 100,527 | 5.57E-06 | 0.027 |
Between-study heterogeneity was low (I2 < 30%) for all but two signals (GRB14 and LYPLAL1, see Supplementary Note) and all 14 associations remained genome-wide significant in a random-effects meta-analysis (Supplementary Table 2).
One of these SNPs, rs4846567, is in linkage disequilibrium (LD, r2 = 0.64, D’ = 0.84; HapMap CEU) with the previously reported WHR-associated variant (rs2605100) near the LYPLAL1 gene13. The remaining 13 loci were in or near genes not previously associated with WHR or other measures of adiposity: RSPO3, VEGFA, TBX15-WARS2, NFE2L3, GRB14, DNM3-PIGC, ITPR2-SSPN, LY86, HOXC13, ADAMTS9, ZNRF3-KREMEN1, NISCH-STAB1, and CPEB4 (Figure 2). These 14 loci explain 1.03% of the variance in WHR (after adjustment for BMI, age, and sex), with each locus contributing from 0.02% (ZNRF3-KREMEN1) to 0.14% (RSPO3) based on effect estimates in the follow-up stage.
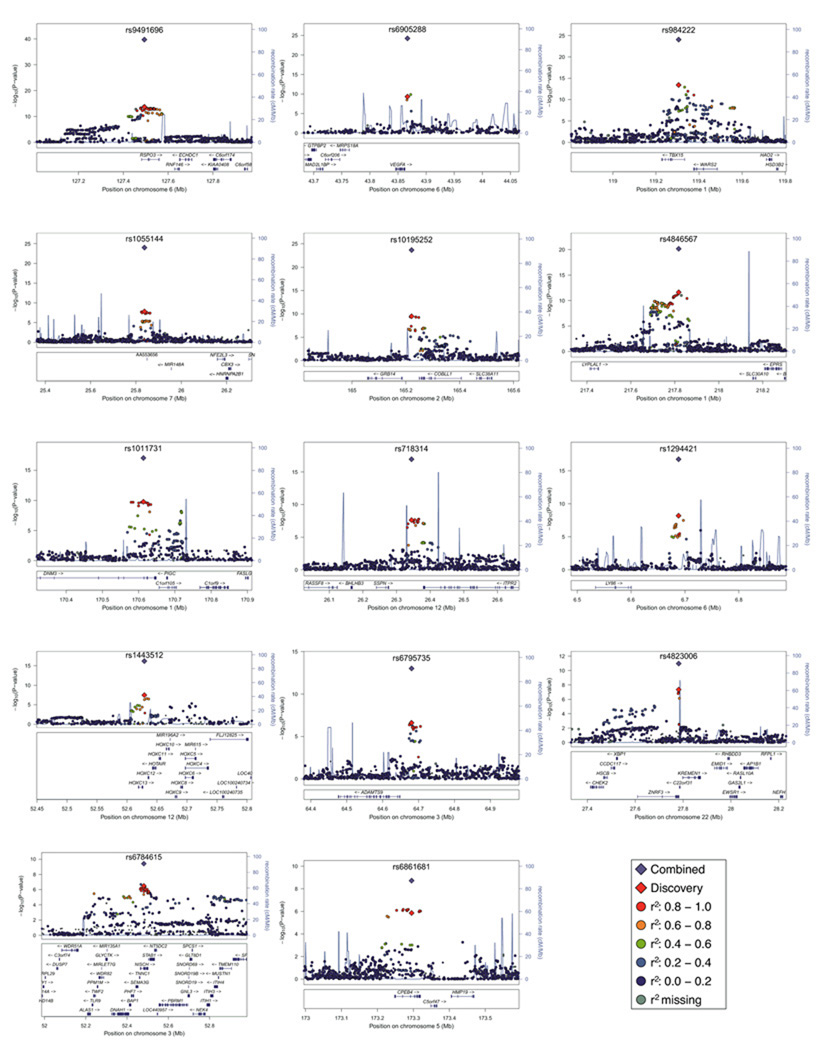
Figure 2. Regional plots of 14 loci with genome-wide significant association.
SNP association with WHR in meta-analysis of discovery studies for 14 loci (−log10 P on the y-axis and SNP genomic position on the x-axis). In each panel, an index SNP is denoted with a purple diamond and plotted using the P attained across discovery and follow-up data (Table 1). Estimated recombination rates are plotted in blue. SNPs are colored to reflect LD with the index SNP (pair-wise r2 values from HapMap CEU). Gene and microRNA annotations are from the UCSC genome browser.
Sexual dimorphism at several of the WHR loci
Given the known sexual dimorphism of WHR and evidence from variance decomposition studies that this reflects sex-specific genetic effects17, we performed sex-specific meta-analyses for the 14 WHR associated SNPs. These analyses included up to 108,979 women (42,735 discovery, 66,244 follow-up) or 82,483 men (34,601 discovery, 47,882 follow-up). In joint analysis of discovery and follow-up data, 12 of the 14 SNPs reached genome-wide significance in women, but only 3 in men (Table 2). At all but one locus (TBX15-WARS2), effect-size estimates were numerically greater in women. At seven of the loci (those near RSPO3, VEGFA, GRB14, LYPLAL1, HOXC13, ITPR2-SSPN and ADAMTS9), there were marked differences in sex-specific beta-coefficients (P ranging from 1.9 × 10−3 to 1.2 × 10−13). All loci displayed consistent patterns of sex-specific differences in both discovery and follow-up studies (Table 2). These 14 loci explain 1.34% of the variance in WHR (after adjustment for BMI and age) in women, but only 0.46% in men.
Table 2.
Evidence of sex-differences in the WHR association at seven of the 14 associated loci
| SNP | NearbyGenes | Men | Women | Sexdifference | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Discovery | Follow-up | Combined | Discovery | Follow-up | Combined | Com-bined | ||||||||
| P | Beta | P | Beta | P | Beta | P | Beta | P | Beta | P | Beta | P | ||
| rs9491696 | RSPO3 | 1.68E-04 | 0.026 | 6.97E-09 | 0.036 | 1.05E-11 | 0.031 | 1.62E-12 | 0.047 | 8.84E-22 | 0.053 | 1.93E-32 | 0.050 | 1.94E-03 |
| rs6905288 | VEGFA | 0.066 | 0.013 | 2.09E-04 | 0.025 | 7.38E-05 | 0.020 | 7.72E-13 | 0.052 | 3.14E-15 | 0.051 | 2.27E-26 | 0.052 | 5.20E-06 |
| rs984222 | TBX15-WARS2 | 3.32E-09 | 0.041 | 2.43E-05 | 0.029 | 9.41E-13 | 0.035 | 1.21E-07 | 0.036 | 1.33E-08 | 0.033 | 1.02E-14 | 0.034 | 0.951 |
| rs1055144 | NFE2L3 | 6.00E-04 | 0.029 | 5.67E-08 | 0.040 | 2.52E-10 | 0.035 | 2.34E-06 | 0.040 | 7.13E-12 | 0.046 | 1.41E-16 | 0.044 | 0.270 |
| rs10195252 | GRB14 | 0.201 | 0.009 | 0.114 | 0.011 | 0.043 | 0.010 | 6.33E-15 | 0.053 | 4.95E-21 | 0.054 | 3.84E-34 | 0.054 | 1.41E-11 |
| rs4846567 | LYPLAL1 | 0.191 | 0.010 | 0.982 | 0.000 | 0.358 | 0.005 | 4.84E-18 | 0.064 | 8.12E-17 | 0.055 | 4.95E-33 | 0.059 | 1.18E-13 |
| rs1011731 | DNM3-PIGC | 4.88E-07 | 0.034 | 1.95E-03 | 0.022 | 7.81E-09 | 0.028 | 2.13E-05 | 0.028 | 7.03E-07 | 0.030 | 6.90E-11 | 0.029 | 0.855 |
| rs718314 | ITPR2-SSPN | 0.177 | 0.010 | 2.02E-03 | 0.022 | 1.41E-03 | 0.017 | 8.29E-10 | 0.047 | 4.21E-09 | 0.038 | 2.41E-17 | 0.042 | 4.67E-04 |
| rs1294421 | LY86 | 4.18E-03 | 0.020 | 7.00E-06 | 0.030 | 1.63E-07 | 0.025 | 3.44E-08 | 0.038 | 7.32E-06 | 0.026 | 2.40E-12 | 0.031 | 0.357 |
| rs1443512 | HOXC13 | 0.184 | 0.011 | 9.74E-04 | 0.024 | 9.45E-04 | 0.018 | 1.43E-09 | 0.048 | 3.09E-08 | 0.035 | 6.38E-16 | 0.040 | 2.23E-03 |
| rs6795735 | ADAMTS9 | 0.011 | 0.017 | 0.614 | 0.004 | 0.027 | 0.011 | 7.85E-07 | 0.033 | 2.95E-11 | 0.042 | 1.92E-16 | 0.038 | 8.50E-05 |
| rs4823006 | ZNRF3-KREMEN1 | 6.87E-03 | 0.019 | 0.094 | 0.012 | 1.94E-03 | 0.015 | 6.86E-08 | 0.037 | 3.81E-05 | 0.024 | 3.24E-11 | 0.030 | 0.032 |
| rs6784615 | NISCH-STAB1 | 1.51E-03 | 0.045 | 0.033 | 0.032 | 1.68E-04 | 0.039 | 6.23E-05 | 0.057 | 1.72E-03 | 0.039 | 6.01E-07 | 0.047 | 0.574 |
| rs6861681 | CPEB4 | 1.88E-03 | 0.023 | 0.045 | 0.015 | 3.03E-04 | 0.019 | 2.14E-04 | 0.027 | 1.58E-03 | 0.021 | 1.55E-06 | 0.024 | 0.555 |
Association with other anthropometric measures
By focusing on WHR after adjustment for BMI, our goal was to detect effects on body fat distribution independent of those influencing overall adiposity. As expected, we found very little evidence that known BMI-associated variants were detected in our WHR analysis. Of the 10 loci identified shown to be associated with BMI in previous genome-wide association studies 14,15,18, only two showed nominally significant (P < 0.05) associations for BMI-adjusted WHR in the discovery analysis (FTO: rs805013614, P = 0.03, N = 77,074; TMEM18: rs654823815, P = 3.0 × 10−3, N = 77,016).
We also tested the 14 WHR-associated SNPs for their effect on BMI using data from up to 242,530 participants available from the GIANT consortium (including most of the studies available for WHR association). Of the 14 WHR loci, four (near TBX15-WARS2, CPEB4, LYPLAL1 and GRB14) also showed evidence of association with BMI (4.1 × 10−3 ≤ P ≤ 3.2×10−6) with the WHR-increasing allele associated with decreased BMI (Supplementary Table 3). When adding an interaction term of SNP and BMI into the model, we observed that BMI modified the WHR association at the LY86 locus (P for interaction = 9.5 × 10−5) with a larger WHR effect among the obese compared to the non-obese (see Supplementary Note).
To determine whether the WHR-associated signals exert their effects primarily through an effect on waist (WC) or hip circumference (HIP), we performed meta-analyses for these specific phenotypes in the discovery and follow-up studies (Supplementary Table 1 and 3). Overall, we observed stronger associations for HIP than for WC. Effect-size estimates were numerically greater for HIP than for WC at eleven of the 14 loci, and there were nominal associations (P < 0.05) with HIP for twelve of the WHR-associated loci but only four associations with WC. In both sexes, the WHR-associated loci displaying nominal association with HIP always featured the WHR-increasing allele associated with reduced HIP. In contrast, we observed sexual dimorphism in the pattern of WC associations. In women, the WHR-increasing allele at all 14 loci was associated with increased WC, whereas this was only true for 6 of these loci in men (Figure 3). At GRB14, for example, the WHR-increasing allele was associated with increased WC in women (P = 3.6 × 10−4) but decreased WC in men (P = 6.8 × 10−3). These differences in the relationships between WC, HIP and WHR underlie some of the sexual dimorphism in the patterns of WHR association.
Figure 3. Association of the 14 WHR loci with waist and hip circumference.
Beta-coefficients for waist circumference (WC, x-axis) and hip circumference (HIP, y-axis) in women and men derived from the joint discovery and follow-up analysis. P for WC and HIP are represented by color. In men, grey gene labels refer to those SNPs that were not significant in the male-specific WHR analysis. More details can be found in Supplementary Table 3.
Enrichment of association with metabolic traits
We evaluated the 14 WHR-associated loci for their relationships with related metabolic traits using GWA data provided by trait-specific consortia19–21 as well as our de novo genotyped follow-up studies. As expected, given the sample overlap between this GWA data with our WHR GWA data as well as known trait correlations (Supplementary Table 4), we observed directionally consistent enrichment of associations (P < 0.05) between the 14 WHR-associated alleles and increased triglycerides, LDL-cholesterol, fasting insulin, and HOMA-derived measures of insulin resistance (binomial P from 3.2 × 10−4 to 1.8 × 10−8; Table 3 and Supplementary Table 5a). For example, the WHR-increasing allele at GRB14 shows strong associations with increased triglycerides (P = 7.4 × 10−9), fasting insulin levels (P = 5.0 × 10−6) and insulin resistance (P = 1.9 × 10−6). Eleven of 14 WHR-associated loci showed directionally consistent associations with T2D, three of these (ADAMTS9, NISCH-STAB1, and ITPR2-SSPN) reaching nominal significance (P < 0.05) (Table 3 and Supplementary Table 5a,b). Because the association signals for correlated traits in this analysis are vulnerable to overestimation given the overlap in GWAS samples examined, we repeated these analyses restricted to our de novo genotyped follow-up studies. Although this also resulted in lower sample size, similar patterns of enrichment were still observed (Supplementary Table 5c).
Table 3.
WHR signals show enrichment of association with other traits related to metabolic disorders
| Trait | Samplesizea | SNPs in concordantdirectionb | SNPs in concordantdirection with P<.05c | ||
|---|---|---|---|---|---|
| # SNPs | P | # SNPs | P | ||
| Triglycerides | 43,826 | 14 | 6.10E-5 | 7 | 1.79E-8 |
| HDL-C | 45,561 | 13 | 9.16E-4 | 4 | 3.20E-4 |
| LDL-C | 43,889 | 10 | 0.090 | 1 | 0.298 |
| Fasting glucose | 63,849 | 10 | 0.090 | 1 | 0.298 |
| Fasting insulin | 54,883 | 13 | 9.16E-4 | 5 | 1.62E-5 |
| HOMA-IR | 53,625 | 13 | 9.16E-4 | 6 | 6.17E-7 |
| 2-hr glucose | 27,011 | 7 | 0.605 | 0 | 1.000 |
| Type 2 diabetes | 10,128f | 11 | 0.029 | 3 | 4.62E-3 |
Pathway analysis of the WHR-associated loci and potential biological role
To identify potential functional connections and pathway relationships between genes mapping at the WHR-associated loci, we focused on the 95 genes located in a 2 Mb interval centered around each of the 48 independent SNPs that attained a P < 1.0 × 10−5 in the WHR discovery studies.
First, we performed a survey of the published literature using GRAIL22, to search for connectivity between the genes and specific keywords that describe these functional connections (see Online Methods). Although there was no evidence, after correcting for multiple testing, that the connectivity between these genes was greater than chance, we identified 8 genes with nominal significance (P < 0.05) for potential functional connectivity (PLXND, HOXC10, TBX15, RSPO3, HOXC4, HOXC6, KREMEN1 and HOXC11). The keywords associated with these connections included “vegf”, “homeobox”, “patterning”, “mesenchyme”, “embryonic”, “development” and “angiogenesis”.
Additionally, we performed pathway analyses using the PANTHER database23 based on the same set of 95 genes (Online methods and Supplementary Note). This analysis generated some evidence for over-representation of “developmental processes” (P = 5.8 × 10−8) and “mRNA transcription regulation” (P = 2.7 × 10−6), but neither retained nominal significance after adjustment for bias (e.g. due to non-random SNP coverage in relation to genes) and the number of biological processes tested (Supplementary Note, Supplementary Table 6).
Finally, we examined the described functional roles of some of the most compelling candidates based on either proximity to the signal or other analyses described in this paper. These uncovered possible roles in adipocyte development (TBX15), pattern formation during embryonic development (HOXC13), angiogenesis (VEGFA, RSPO3, STAB1), Wnt/beta-catenin signaling (RSPO3, KREMEN1), insulin signaling (ADAMTS9, GRB14, NISCH), lipase activity (LYPLAL1), lipid biosynthesis (PIGC) and intracellular calcium signaling (ITPR2) (see Supplementary Note for details).
Evaluation of copy number variants (CNVs) and nonsynonymous changes
Both common and rare CNVs have been reported to be associated with overall adiposity14,15,24,25, but the impact of CNVs on fat distribution has not been evaluated previously. To examine the potential contribution of common CNVs to variation in WHR, we looked for evidence of association in our GWA discovery meta-analysis, using a set of 6,018 CNV-tagging SNPs, which collectively capture >40% of common CNVs >1 kb 26,27 (Online Methods, Supplementary Note).
One CNV-tagging SNP (rs1294421, LY86) was observed amongst our 14 WHR-associated loci. This SNP is in strong LD (r2 = 0.98) with a 2,832 bp duplication variant (CNVR2760.1)27, located 12 kb from an expressed sequence tag (BC039678) and 87 kb from LY86, such that the duplication allele is associated with reduced WHR. The duplicated region consists entirely of noncoding sequence but includes part of a predicted enhancer sequence (E.5552.1)28.
To identify other putatively causal variants in our associated regions, we searched for non-synonymous coding SNPs in strong LD (r2>0.7) with the most strongly associated SNPs at each locus using data from the HapMap (Build 21) and 1000 Genomes Project (April and August 2009 releases). In this search, one lead SNP (rs6784615, at the NISCH-STAB1 locus) was correlated with non-synonymous changes in two nearby genes, DNAH1 (Val441Leu, Arg1285Trp and Arg3809Cys) and GLYCTK (Leu170Val). Fine-mapping and functional studies will be required to determine whether the DNAH1 or GLYCTK SNPs or the LY86 CNV are causal for the WHR-associations at these loci.
Evaluation of effect of the WHR associations on expression in relevant tissues
Expression-QTL (eQTL) data can implicate regional transcripts that mediate trait-associations, and we therefore examined the 14 WHR-associated loci using eQTL data from human subcutaneous adipose tissue (SAT)29 (two separate sample sets, N=610 and N=603), omental fat30 (N=740), liver30 (N=518), blood29 (N=745), and lymphocytes31 (N=830) (Online methods, Supplementary Note).
At six of the loci, the WHR-associated SNP was either the strongest SNP associated with significant (P < 1.0 × 10−5) expression of a local (within 1 Mb) gene transcript or explained the majority of the association between the most significant eQTL SNP and the gene transcript in conditional analyses (adjusted _P_ > 0.05; Table 4). For example, the WHR-associated SNP rs1011731 (near DNM3-PIGC) was strongly-associated with expression of PIGC in lymphocytes (P = 5.9 × 10−10); furthermore, rs1011731 is in high LD (r2 = 1.00, D’ = 1.00, HapMap CEU) with the SNP with the strongest effect on PIGC expression (rs991790), and this _cis_-eQTL association is abolished by conditioning on rs1011731. These analyses therefore indicate that these two signals are coincident and that PIGC is a strong candidate for mediating the WHR-association at rs1011731. We found similar evidence for coincidence of the WHR signal with expression for rs984222 (TBX15 in omental fat), rs1055144 (expressed sequence tag AA553656 in SAT), rs10195252 (GRB14 in SAT), rs4823006 (ZNRF3 in SAT and omental fat), and rs6784615 (STAB1 in blood)(Table 4). Taken together, the overlap between trait association and gene expression at these loci suggests that the WHR associations are driven through altered expression of PIGC, TBX15, AA553656, GRB1, ZNRF3 and STAB1.
Table 4.
Expression quantitative trait locus analysis for 11 out of the 14 WHR signals.
| WHR SNP | Tissue | Gene | Effecta | WHR SNPassociationwith transcript (P) | Transcriptpeak SNP b | r2c | Peak SNP associationwith transcript (P) | ||
|---|---|---|---|---|---|---|---|---|---|
| Unadj. | Adj. forpeak SNP | Unadj. | Adj. forWHR SNP | ||||||
| rs9491696 | SAT-D | RSPO3 | + | 1.10E-07 | 0.03 | rs1936795 | 0.26 | 2.20E-13 | 7.40E-08 |
| rs984222 | Omental | TBX15 | + | 7.90E-10 | 1.00 | rs984222 | 1.00 | 7.90E-10 | 1.00 |
| Omental | WARS2 | + | 5.11E-36 | 0.03 | rs10802075 | 0.27 | 1.31E-163 | 1.33E-88 | |
| Subcutaneous fat | WARS2 | + | 1.67E-25 | 0.01 | rs10802075 | 0.22 | 3.88E-110 | 1.01E-63 | |
| Lymph. | WARS2 | − | 4.30E-18 | 5.47E-05 | rs2645305 | 0.27 | 5.57E-40 | 6.88E-26 | |
| Liver | WARS2 | + | 2.57E-17 | 0.07 | rs1057990 | 0.26 | 6.69E-59 | 1.97E-32 | |
| SAT-D | WARS2 | + | 1.10E-18 | 0.51 | rs1057990 | 0.26 | 5.80E-130 | 5.80E-100 | |
| Blood | WARS2 | + | 6.10E-17 | 0.11 | rs1057990 | 0.26 | 6.30E-75 | 1.10E-54 | |
| rs1055144 | SAT-D | AA553656d | − | 1.20E-11 | 0.96 | rs7798002 | 0.95 | 7.20E-12 | 0.32 |
| SAT-M | AA553656d | − | 2.46E-07 | 0.65 | rs1451385 | 0.77 | 5.93E-08 | 0.38 | |
| rs10195252 | SAT-D | GRB14 | + | 4.40E-11 | 1.00 | rs10195252 | 1.00 | 4.40E-11 | 1.00 |
| SAT-M | GRB14 | + | 5.51E-06 | 1.00 | rs10184004 | 1.00 | 5.51E-06 | 1.00 | |
| Omental | GRB14 | + | 1.02E-13 | 1.00 | rs10195252 | 1.00 | 1.02E-13 | 1.00 | |
| SAT-M | SLC38A11 | − | 3.93E-06 | 0.66 | rs10184126 | 0.18 | 7.76E-44 | 8.57E-34 | |
| SAT-D | SLC38A11 | − | 3.70E-09 | 0.35 | rs10184126 | 0.18 | 2.40E-94 | 7.40E-82 | |
| rs1011731 | Blood | C1orf105 | + | 3.80E-16 | 0.20 | rs2157451 | 0.28 | 1.30E-33 | 8.20E-18 |
| Lymph. | PIGC | − | 5.87E-10 | 1.00 | rs991790 | 1.00 | 5.65E-10 | 1.00 | |
| rs718314 | Lymph. | ITPR2 | + | 1.79E-09 | 0.98 | rs7976877 | 0.45 | 2.21E-18 | 1.91E-06 |
| Blood | ITPR2 | − | 2.40E-09 | 0.20 | rs2570 | 0.41 | 2.40E-37 | 1.80E-28 | |
| rs1294421 | SAT-M | BC039678 | − | 2.43E-07 | 0.38 | rs1294404 | 0.64 | 1.89E-16 | 3.42E-04 |
| Omental | BC039678 | − | 1.09E-06 | 0.33 | rs912056 | 0.71 | 8.28E-17 | 4.26E-05 | |
| rs6795735 | SAT-D | ADAMTS9 | − | 1.50E-06 | 0.04 | rs7372321 | 0.11 | 1.10E-09 | 2.30E-05 |
| Omental | AK022320 | − | 7.99E-15 | 0.64 | rs4521216 | 0.02 | 5.15E-42 | 1.49E-19 | |
| SAT-D | AK022320 | − | 2.24E-10 | 0.98 | rs4521216 | 0.02 | 9.62E-37 | 7.58E-19 | |
| rs4823006 | SAT-D | ZNRF3 | − | 2.40E-08 | 0.63 | rs3178915 | 0.81 | 6.70E-11 | 8.90E-04 |
| SAT-M | ZNRF3 | − | 1.08E-18 | 0.93 | rs6005975 | 0.79 | 1.59E-19 | 0.50 | |
| Omental | ZNRF3 | − | 9.13E-18 | 0.98 | rs6005975 | 0.79 | 6.07E-21 | 0.27 | |
| rs6784615 | Blood | STAB1 | + | 2.80E-09 | 0.32 | rs9846089 | 0.83 | 9.40E-10 | 0.08 |
| rs6861681 | Lymph. | CPEB4 | + | 3.79E-22 | 0.89 | rs7705502 | 0.87 | 4.95E-29 | 2.00E-03 |
| Blood | HMP19 | + | 1.60E-16 | 0.97 | rs10516107 | 0.83 | 1.10E-21 | 4.30E-06 |
Differential RNA expression of gluteal compared to abdominal fat tissue
To determine whether genes within the WHR-associated loci showed evidence of differential transcription in distinct fat-depots, we compared expression levels in gluteal or abdominal SAT in 49 individuals. We focused on the 15 genes with the strongest credentials for causal involvement (on the basis of proximity to the lead SNP and/or other biological or functional data: Table 1) for which expression data were available. Five of these (RSPO3, TBX15, ITPR2, WARS2 and STAB1) were differentially expressed between the two tissues (F-test, corrected for false discovery rate across the 15 expressed genes, P < 0.05; Supplementary Table 7). This supported the hypothesis that, at some loci at least, the association with WHR reflects depot-specific differences in expression patterns.
DISCUSSION
Overall, our findings demonstrate that the genetic regulation of body fat distribution involves loci and processes that are largely distinct from those that influence BMI and risk of obesity. This finding is consistent with the evidence that WHR displays substantial heritability even after adjustment for BMI. The loci that emerge from this study display no overlap with those shown to be associated with BMI, either in previous reports14,15,16 or in the expanded meta-analysis recently completed by the GIANT consortium32.
Another point of distinction between our findings and those for BMI relates to the evidence for sexual dimorphism that we observed at several of the WHR-associated loci. Sex differences in the regulation of body fat distribution have long been acknowledged without a clear understanding of the underlying molecular mechanisms. These differences become apparent during puberty and are generally attributed to the influence of sex hormones33. Consistent with our findings, variance decomposition studies have shown that the genetic contribution to the overall variance in WHR, waist or hip circumference is greater in women17. While there is some evidence for loci with differential sex effects influencing lipids34, uric acid levels35 and risk of schizophrenia36, we are unaware of prior reports indicating such strong enrichment of female-specific associations for any other phenotype, including BMI32.
The primary objective of genetic discovery efforts is to characterize the specific mechanisms involved in the regulation of the trait of interest. Despite the considerable challenges associated with moving from common variant association signals to definition of the causal alleles and pathways, we have identified strong candidates at several of the loci. For example, the _cis_-eQTL data implicate GRB14 as a compelling candidate for the WHR-association on chromosome 2, and we were able to show that the same GRB14 variants are also associated with triglyceride and insulin levels, consistent with previous association of this locus with HDL-cholesterol 37. These inferences about the role of GRB14 are supported by evidence that _Grb14_-deficient mice exhibit improved glucose homeostasis despite lower circulating insulin levels, and enhanced insulin signaling in liver and skeletal muscle38. The signal near ADAMTS9 overlaps a previously-reported T2D locus39, and the lead SNP for WHR in our study is identical to the SNP displaying the strongest T2D association in an expanded T2D meta-analysis40. Given evidence that ADAMTS9 T2D-risk alleles are associated with insulin resistance in peripheral tissues41, these findings are consistent with a primary effect of ADAMTS9 variants on body fat distribution. At the chromosome 6 locus, VEGFA is the most apparent biological candidate, given the presumed role of VEGFA as a mediator of adipogenesis42 and evidence that serum levels of VEGFA are correlated with obesity43,44. Finally, at the TBX15-WARS2 locus, TBX15 emerges as the strongest candidate based on the _cis_-eQTL data in omental fat, marked depot-specific differences in adipose tissue expression in mice and humans, and associations between TBX15 expression in visceral fat and WHR45,46.
Our efforts to use pathway- and literature-mining approaches to look for functional enrichment of the genes mapping to associated regions met with only limited success, but did provide some support for overrepresentation of developmental processes. Developmental genes have been implicated in fat accumulation and distribution45,46, and recent evidence supports a link between developmental genes and body fat distribution, including HOXC1347 and TBX1545,48. Developmental genes may in part determine the adipocyte-specific expression patterns that have been observed in different fat depots45. Taken together, our findings point to a set of genes influencing body fat distribution that have their principal effects in adipose tissue. This is in contrast to the predominantly central (hypothalamic) processes that are involved in the regulation of body mass index and overall adiposity49.
By providing novel insights into the regulation of body fat distribution, the present study raises a number of issues for future investigation. From the genetic perspective, re-sequencing, dense-array genotyping and fine-mapping approaches will be required to characterize causal variants at the loci we have identified, and to support further discoveries that may account for the substantial proportion of genetic variance unexplained by our findings. From the clinical perspective, it will be important to explore the relationship of these variants to more refined measures of body fat distribution derived from detailed imaging studies, to use the variants identified to characterise the causal relationships between body fat distribution and related metabolic and cardiovascular traits, and to explore ethnic differences in patterns of body fat distribution. Efforts to tackle overall obesity through therapeutic or lifestyle-based modulation of overall energy balance have proved extremely challenging to implement, and the manipulation of processes associated with more beneficial patterns of fat distribution offers an alternative perspective for future drug discovery.
Supplementary Material
1
2
3
Figure 4.
ACKNOWLEDGEMENTS
Academy of Finland (104781, 120315, 129269, 117797, 121584, 126925, 129418, 129568, 77299, 124243, 213506, 129680, 129494, 10404, 213506, 129680, 114382, 126775, 127437, 129255, 129306, 130326, 209072, 210595, 213225, 216374); ADA Mentor-Based Postdoctoral Fellowship grant; Affymetrix, Inc for genotyping services (N02-HL-6-4278); ALF/LUA Gothenburg; Althingi (the Icelandic Parliament); Amgen; AstraZeneca AB; Augustinus Foundation; Becket Foundation; Biocentrum Helsinki; Biomedicum Helsinki Foundation; Boston Obesity Nutrition Research Center (DK46200); British Diabetes Association (1192); British Diabetic Association Research; British Heart Foundation (97020, PG/02/128); Busselton Population Medical Research Foundation; Cambridge NIHR Comprehensive Biomedical Research Centre; CamStrad; Chief Scientist Office of the Scottish Government; Contrat Plan Etat Région de France; Danish Centre for Health Technology Assessment; Danish Diabetes Association; Danish Ministry of Internal Affairs and Health; Danish Heart Foundation; Danish Pharmaceutical Association; Danish Research Council; DIAB Core (German Network of Diabetes); Diabetes UK; Donald W. Reynolds Foundation; Dr Knut Krohn, Microarray Core Facility of the Interdisciplinary Centre for Clinical Research (IZKF), University of Leipzig, Germany; Dresden University of Technology Funding Grant, Med Drive; EMGO+ institute; Emil and Vera Cornell Foundation; Erasmus Medical Center and Erasmus University, Rotterdam; Estonian Government SF0180142s08; European Commission (2004310, 212111, 205419, 245536, DG XII, HEALTH-F4-2007-201413, FP7/2007-2013, QLG1-CT-2000-01643, QLG2-CT-2002-01254, LSHG-CT-2006-018947, LSHG-CT-2006-01947, LSHG-CT-2004-512066, LSHM-CT-2007-037273, EU/WLRT-2001-01254, LSHG-CT-2004-518153, SOC 95201408 05F02, Marie Curie Intra-European Fellowship); Federal Ministry of Education and Research, Germany (01ZZ9603, 01ZZ0103, 01ZZ0403, 03ZIK012, 01 EA 9401); Federal State of Mecklenburg- West Pomerania; Finnish Diabetes Research Foundation; Finnish Diabetes Research Society; Finnish Foundation for Pediatric Research; Finnish Foundation of Cardiovascular Research; Finnish Medical Society; Finska Läkaresällskapet; Finnish Ministry of Education; Folkhälsan Research Foundation; Fond Européen pour le Développement Régional; Fondation LeDucq; Foundation for Life and Health in Finland; GEN-AU "GOLD" from Austria; German Bundesministerium fuer Forschung und Technology (#01 AK 803 A-H, # 01 IG 07015 G); German National Genome Research Net NGFN2 and NGFNplus (01GS0823, FKZ 01GS0823); German Research Council (KFO-152); GlaxoSmithKline; Göteborg Medical Society; Gyllenberg Foundation; Health Care Centers in Vasa, Närpes and Korsholm; Healthway, Western Australia; Helmholtz Center Munich; Helsinki University Central Hospital; Hjartavernd (the Icelandic Heart Association); Ib Henriksen Foundation; IZKF (B27); Jalmari and Rauha Ahokas Foundation; Juho Vainio Foundation; Juvenile Diabetes Research Foundation International (JDRF); Karolinska Institute and the Stockholm County Council (560183); Knut and Alice Wallenberg Foundation; Lundbeck Foundation Centre of Applied Medical Genomics for Personalized Disease Prediction, Prevention and Care; Lundberg Foundation; MC Health; Ministry of Cultural Affairs of the Federal State of Mecklenburg-West Pomerania, Germany; South Tyrol Ministry of Health; Ministry of Science, Education and Sport of the Republic of Croatia (216-1080315-0302); Medical Research Council UK (G0000649, G0601261, G9521010D, G0000934, G0500539, G0600331, PrevMetSyn); Montreal Heart Institute Foundation; MRC Centre for Obesity related Metabolic disease; Municipal Health Care Center and Hospital in Jakobstad; Municipality of Rotterdam; Närpes Health Care Foundation; National Health and Medical Research Council of Australia and the Great Wine Estates Auctions; Netherlands Centre for Medical Systems Biology (SPI 56-464-1419); Netherlands Ministry for Health, Welfare and Sports; Netherlands Ministry of Education, Culture and Science; Netherlands Genomics Initiative; Netherlands Consortium for Healthy Aging (050-060-810); Netherlands Organisation of Scientific Research NWO Investments (175.010.2005.011, 911-03-012, 904-61-090,904-61-193,480-04-004,400-05-717); National Institute on Aging Intramural Research Program; National Institutes of Health (CA047988, CA65725, CA87969, CA49449, CA67262, CA50385, , DK075787, DK062370, DK58845, DK072193, K23-DK080145, K99HL094535, N01-HC85079 through N01-HC85086, N01-HG-65403; N01-AG-12100, N01-HC-25195, N01-HC35129, N01-HC15103, N01-HC55222, N01-HC75150, N01-HC45133, N01-HC55015, N01-HC55016, N01-HC-55018, N01-HC-55019, N01-HC-55020, N01-HC-55021, N01-HC-55022, NO1-AG-1-2109, HL71981, HG005581, HG002651, HL084729, HL043851, HHSN268200625226C, K23-DK080145; MH084698, P30-DK072488, R01-DK075787, R01 HL087652, R01-HL087641, R01-HL59367, R01-HL086694, R01-HL087647, R01-HL087679, R01-HL087700, R01-AG031890, R01-HL088119, R01-DK068336, R01-DK075681, R01-DK-073490, R01-DK075787, R01-MH63706; U01-HL72515, U01-GM074518, U01-HL084756, U01-HG004399, UO1-CA098233, UL1-RR025005, UL1-RR025005, U01-HG004402, U01-DK062418; U01 HL080295, T32-HG00040, 263-MA-410953; 1RL1-MH083268-01, intramural project 1Z01-HG000024); National Institute for Health Research (NIHR); Neuroscience Campus Amsterdam; Novo Nordisk Foundation; Novo Nordisk Inc., Research Foundation of Copenhagen County; Ollqvist Foundation; Paavo Nurmi Foundation; Päivikki and Sakari Sohlberg Foundation; Pew Scholarship for the Biomedical Sciences); Perklén Foundation; Petrus and Augusta Hedlunds Foundation; Research Institute for Diseases in the Elderly (014-93-015, RIDE, RIDE2); Sahlgrenska Center for Cardiovascular and Metabolic Research (CMR, A305:188); Siemens Healthcare, Erlangen, Germany; Signe and Ane Gyllenberg Foundation; Sigrid Juselius Foundation; Social Insurance Institution of Finland; Social Ministry of the Federal State of Mecklenburg-West Pomerania, Germany; South Tyrolean Sparkasse Foundation; State of Bavaria, Germany; Support for Science Funding programme; Swedish Cultural Foundation in Finland; Swedish Foundation for Strategic Research (SSF); Swedish Heart-Lung Foundation; Swedish Medical Research Council (8691, K2007-66X-20270-01-3, K2010-54X-09894-19-3); Swedish Society of Medicine; Swiss National Science Foundation (33CSCO-122661); the Royal Society; the Royal Swedish Academy of Science; Torsten and Ragnar Söderberg's Foundation; Turku University Hospitals; UK Department of Health Policy Research Programme; University and Research of the Autonomous Province of Bolzano; University Hospital Medical funds to Tampere; University Hospital Oulu, Biocenter, University of Oulu, Finland (75617); Västra Götaland Foundation; Wellcome Trust (077016/Z/05/Z, 068545/Z/02, 072960, 076113, 083270, 085301, 079557, 081682, 075491, 076113/B/04/Z, 091746/Z/10/Z, 079895, WT086596/Z/08/Z, WT Research Career Development Fellowship; WT Career Development Award); Western Australian Genetic Epidemiology Resource and the Western Australian DNA Bank (both National Health and Medical Research Council of Australia Enabling Facilities); Yrjö Jahnsson Foundation.
Abbreviations
BMI
Body-mass-index
WC
waist circumference
WHR
waist-hip ratio
HIP
hip circumference
GWA
genome-wide association
SNP
single-nucleotide polymorphism
LD
linkage disequilibrium
eQTL
Expression Quantitative Trait Loci
Footnotes
AUTHOR CONTRIBUTION
Writing group: IB, CSF, IMH (lead), CML (lead), MIM, KLMoh, LQ, VSte, GT, MCZ
Waist phenotype working group: TLA, NB, IB, LAC, CMD, CSF, TBH, IMH, AUJ, CML (lead), RJFL, RM, MIM, KLMoh, LQ, JCR, EKS, VSte, KSte, GT, UT, CCW, TW, TWW, HEW, MCZ
Data cleaning & analysis: SIB, IMH (lead), EI, AUJ, HL, CML (lead), RJFL (lead), JL, RM, LQ, JCR, EKS, GT, SV, MNW, EW, CJW, TWW, TW
Sex-specific analyses: SIB, TE, IMH, AUJ, TOK, ZK, SL, CML, RJFL, RM, KLMon, KEN, LQ, JCR (lead), VSte, GT, TWW (lead)
eQTL and expression analyses: SIB, ALD, CCH, JNH, FK, LMK, CML, LL, RJFL, JL, MFM, JLM, CM, GN, EES, EKS, VSte, GT, KTZ
Pathway and CNV analyses: CML, SAM, MIM, JN, VSte, GT, BFV
Secondary analyses: SIB, IBB, NC, KE, TMF, MFF, TF, MEG, JNH, EI, GL, CML, HL, RM, MMas, MIM, KLMon, DRN, JRO, SP, JRBP, JCR, AVS, EKS, PMV, MNW, CJW, RJW, EW, ARW, JY
Study-specific analyses: GRA, DA, NA, TA, TLA, NB, CC, PSC, LC, LAC, DIC, MNC, CMD, TE, KE, EF, MFF, TF, APG, NLG, MEG, CHay, NLH, IMH, JJH, AUJ, ÅJ, TJoh, JOJ, JRK, MKaa, KKap, SKet, JWK, PKra, ATK, ZK, JKet, CLam, RJFL, CLec, HL, MFL, CML, JL, RWL, RM, MMas, BM, KLMon, APM, NN, KEN, DRN, JRO, KKO, CO, MJP, OPol, IPro, NP, MP, LQ, JCR, NWR, SR, FR, NRR, CS, LJS, KSil, EKS, KSta, SS, AVS, NS, US, VSte, DPS, IS, MLT, TMT, NJT, AT, GT, AU, SV, VVit, LV, PMV, RMW, RW, RJW, SW, MNW, CCW, CJW, TWW, ARW, JY, JHZ, MCZ
Study-specific genotyping: DA, TLA, LDA, NB, IB, AJB, EB, LLB, IBB, HC, DIC, INMD, MDei, MRE, PE, KE, NBF, MF, APG, HG, CG, EJCG, CJG, THan, ALH, NH, CHay, AAH, JJH, FBH, DJH, JH, WI, MRJ, ÅJ, JOJ, JWK, PKov, ATK, HKK, JKet, PKra, RNL, CML, RJFL, JL, MLL, MAM, MMas, WLM, MIM, JBJM, MJN, MN, DRN, KKO, CO, OPed, LP, MJP, GP, ANP, NP, LQ, NWR, FR, NRR, CS, AJS, NS, ACS, MT, BT, AU, GU, VVat, PMV, HW, PZ
Study-specific phenotyping: HA, PA, DA, AMA, TLA, BB, SRB, RB, EB, IBB, JPB, MDör, CMD, PE, MFF, CSF, TMF, MF, SG, JG, LCG, THan, ASH, CHen, ALH, ATH, KHH, AHof, FBH, DJH, BI, TI, TJor, PJ, MRJ, ÅJ, AJ, ALJ, JOJ, FK, LK, JKuu, KKva, RK, SKet, JWK, IK, SKos, VK, MKäh, PKov, OL, RNL, BL, JL, GML, RJFL, TL, MMas, MIM, CO, BMP, OPed, CGPP, JFP, IPic, KP, OPol, AP, LQ, MR, IR, OR, VSal, JSar, PEHS, KSil, NJS, JHS, TDS, DPS, RS, HMS, JSin, TT, AT, MU, PV, CBV, LV, JV, DRW, GBW, SHW, GW, JCW, AFW, LZ, PZ
Study-specific management: GRA, AMA, BB, YBS, RNB, HB, JSB, SB, MB, EB, DIB, IBB, JPB, MJC, FSC, LAC, GD, CMD, SE, GE, PF, CSF, TMF, LCG, VG, UG, MEG, THan, CHen, KH, AHam, TBH, ATH, AHof, FBH, DJH, BI, TI, CI, TJor, MRJ, ALJ, FK, KTK, WHLK, RK, JKap, MKäh, ML, DAL, LJL, CML, RJFL, TL, MMar, TM, AMET, KM, MIM, KLMoh, PBM, KEN, MSN, DRN, BO, CO, OPed, LP, BWP, PPP, BMP, LJP, TQ, AR, IR, OR, PMR, VSal, PS, DS, ARS, NS, TDS, KSte, DPS, ACS, MS, TT, JT, UT, AT, MU, AU, TTV, PV, HV, JV, PMV, NJW, HEW, JFW, JCW, AFW
Steering committee: GRA, TLA, IB, SIB, MB, IBB, PD, CMD, CSF, TMF, LCG, THar, JNH (Chair), DJH, EI, RK, RJFL, MIM, KLMoh, KEN, JRO, LP, DS, DPS, UT, HEW
FINANCIAL DISCLOSURES
The authors declare competing financial interests. Details can be found in the Supplementary Note.
REFERENCES
- 1.Carey VJ, et al. Body fat distribution and risk of non-insulin-dependent diabetes mellitus in women. The Nurses' Health Study. Am J Epidemiol. 1997;145:614–619. doi: 10.1093/oxfordjournals.aje.a009158. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Rimm EB, Stampfer MJ, Willett WC, Hu FB. Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. Am J Clin Nutr. 2005;81:555–563. doi: 10.1093/ajcn/81.3.555. [DOI] [PubMed] [Google Scholar]
- 3.Canoy D. Distribution of body fat and risk of coronary heart disease in men and women. Curr Opin Cardiol. 2008;23:591–598. doi: 10.1097/HCO.0b013e328313133a. [DOI] [PubMed] [Google Scholar]
- 4.Pischon T, et al. General and abdominal adiposity and risk of death in Europe. N Engl J Med. 2008;359:2105–2120. doi: 10.1056/NEJMoa0801891. [DOI] [PubMed] [Google Scholar]
- 5.Snijder MB, et al. Associations of hip and thigh circumferences independent of waist circumference with the incidence of type 2 diabetes: the Hoorn Study. Am J Clin Nutr. 2003;77:1192–1197. doi: 10.1093/ajcn/77.5.1192. [DOI] [PubMed] [Google Scholar]
- 6.Snijder MB, et al. Trunk fat and leg fat have independent and opposite associations with fasting and postload glucose levels: the Hoorn study. Diabetes Care. 2004;27:372–377. doi: 10.2337/diacare.27.2.372. [DOI] [PubMed] [Google Scholar]
- 7.Mills GW, et al. Heritability estimates for beta cell function and features of the insulin resistance syndrome in UK families with an increased susceptibility to type 2 diabetes. Diabetologia. 2004;47:732–738. doi: 10.1007/s00125-004-1338-2. [DOI] [PubMed] [Google Scholar]
- 8.Souren NY, et al. Anthropometry, carbohydrate and lipid metabolism in the East Flanders Prospective Twin Survey: heritabilities. Diabetologia. 2007;50:2107–2116. doi: 10.1007/s00125-007-0784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose KM, Newman B, Mayer-Davis EJ, Selby JV. Genetic and behavioral determinants of waist-hip ratio and waist circumference in women twins. Obes Res. 1998;6:383–392. doi: 10.1002/j.1550-8528.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 10.Selby JV, et al. Genetic and behavioral influences on body fat distribution. Int J Obes. 1990;14:593–602. [PubMed] [Google Scholar]
- 11.Agarwal AK, Garg A. Genetic disorders of adipose tissue development, differentiation, and death. Annu Rev Genomics Hum Genet. 2006;7:175–199. doi: 10.1146/annurev.genom.7.080505.115715. [DOI] [PubMed] [Google Scholar]
- 12.Garg A. Acquired and inherited lipodystrophies. N Engl J Med. 2004;350:1220–1234. doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]
- 13.Lindgren CM, et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000508. e1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thorleifsson G, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 15.Willer CJ, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loos RJ, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zillikens MC, et al. Sex-specific genetic effects influence variation in body composition. Diabetologia. 2008;51:2233–2241. doi: 10.1007/s00125-008-1163-0. [DOI] [PubMed] [Google Scholar]
- 18.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kathiresan S, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saxena R, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 42:142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000534. e1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochukova EG, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2009 doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walters RG, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrad DF, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2009 doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature. 464:713–7420. doi: 10.1038/nature08979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I. Predicting tissue-specific enhancers in the human genome. Genome Res. 2007;17:201–211. doi: 10.1101/gr.5972507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emilsson V, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 30.Schadt EE, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060107. e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dixon AL, et al. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 32.Speliotes EK, et al. Association analyses of 249,796 individuals reveal eighteen new loci associated with body mass index. Nat Genet. doi: 10.1038/ng.686. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wells JC. Sexual dimorphism of body composition. Best Pract Res Clin Endocrinol Metab. 2007;21:415–430. doi: 10.1016/j.beem.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Aulchenko YS, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doring A, et al. SLC2A9 influences uric acid concentrations with pronounced sex-specific effects. Nat Genet. 2008;40:430–436. doi: 10.1038/ng.107. [DOI] [PubMed] [Google Scholar]
- 36.Shifman S, et al. Genome-wide association identifies a common variant in the reelin gene that increases the risk of schizophrenia only in women. PLoS Genet. 2008;4:e28. doi: 10.1371/journal.pgen.0040028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ridker PM, et al. Polymorphism in the CETP gene region, HDL cholesterol, and risk of future myocardial infarction: Genomewide analysis among 18 245 initially healthy women from the Women's Genome Health Study. Circ Cardiovasc Genet. 2009;2:26–33. doi: 10.1161/CIRCGENETICS.108.817304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooney GJ, et al. Improved glucose homeostasis and enhanced insulin signalling in Grb14-deficient mice. EMBO J. 2004;23:582–593. doi: 10.1038/sj.emboj.7600082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. doi: 10.1038/ng.609. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boesgaard TW, et al. Variant near ADAMTS9 known to associate with type 2 diabetes is related to insulin resistance in offspring of type 2 diabetes patients--EUGENE2 study. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007236. e7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nishimura S, et al. Adipogenesis in obesity requires close interplay between differentiating adipocytes, stromal cells, and blood vessels. Diabetes. 2007;56:1517–1526. doi: 10.2337/db06-1749. [DOI] [PubMed] [Google Scholar]
- 43.Silha JV, Krsek M, Sucharda P, Murphy LJ. Angiogenic factors are elevated in overweight and obese individuals. Int J Obes (Lond) 2005;29:1308–1314. doi: 10.1038/sj.ijo.0802987. [DOI] [PubMed] [Google Scholar]
- 44.Garcia de la Torre N, et al. Effects of weight loss after bariatric surgery for morbid obesity on vascular endothelial growth factor-A, adipocytokines, and insulin. J Clin Endocrinol Metab. 2008;93:4276–4281. doi: 10.1210/jc.2007-1370. [DOI] [PubMed] [Google Scholar]
- 45.Gesta S, et al. Evidence for a role of developmental genes in the origin of obesity and body fat distribution. Proc Natl Acad Sci U S A. 2006;103:6676–6681. doi: 10.1073/pnas.0601752103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Lanctot C, Kaspar C, Cremer T. Positioning of the mouse Hox gene clusters in the nuclei of developing embryos and differentiating embryoid bodies. Exp Cell Res. 2007;313:1449–1459. doi: 10.1016/j.yexcr.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 48.Candille SI, et al. Dorsoventral patterning of the mouse coat by Tbx15. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020003. E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462:307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 50.Krotkiewski M, Bjorntorp P, Sjostrom L, Smith U. Impact of obesity on metabolism in men and women. Importance of regional adipose tissue distribution. J Clin Invest. 1983;72:1150–1162. doi: 10.1172/JCI111040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fox CS, et al. Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation. 2007;116:39–48. doi: 10.1161/CIRCULATIONAHA.106.675355. [DOI] [PubMed] [Google Scholar]
- 52.Scherzer R, et al. Simple anthropometric measures correlate with metabolic risk indicators as strongly as magnetic resonance imaging-measured adipose tissue depots in both HIV-infected and control subjects. Am J Clin Nutr. 2008;87:1809–1817. doi: 10.1093/ajcn/87.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vega GL, et al. Influence of body fat content and distribution on variation in metabolic risk. J Clin Endocrinol Metab. 2006;91:4459–4466. doi: 10.1210/jc.2006-0814. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 56.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cox DR, Hinkley DV. Theoretical Statistics. London: Chapman and Hall; 1979. [Google Scholar]
- 58.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 59.Stouffer SA, Suchman EA, De Vinney LC, Star SA, Williams RM. Adjustment during army life. Princeton, NJ: Princeton University Press; 1949. [Google Scholar]
- 60.Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. Journal of Evolutionary Biology. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 61.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2
3