Core Transcriptional Regulatory Circuitry in Human Embryonic Stem Cells (original) (raw)
. Author manuscript; available in PMC: 2010 Dec 21.
Summary
The transcription factors OCT4, SOX2, and NANOG have essential roles in early development and are required for the propagation of undifferentiated embryonic stem (ES) cells in culture. To gain insights into transcriptional regulation of human ES cells, we have identified OCT4, SOX2, and NANOG target genes using genome-scale location analysis. We found, surprisingly, that OCT4, SOX2, and NANOG co-occupy a substantial portion of their target genes. These target genes frequently encode transcription factors, many of which are developmentally important homeodomain proteins. Our data also indicate that OCT4, SOX2, and NANOG collaborate to form regulatory circuitry consisting of autoregulatory and feedforward loops. These results provide new insights into the transcriptional regulation of stem cells and reveal how OCT4, SOX2, and NANOG contribute to pluripotency and self-renewal.
Introduction
Mammalian development requires the specification of over 200 unique cell types from a single totipotent cell. Embryonic stem (ES) cells are derived from the inner cell mass (ICM) of the developing blastocyst and can be propagated in culture in an undifferentiated state while maintaining the capacity to generate any cell type in the body. The recent derivation of human ES cells provides a unique opportunity to study early development and is thought to hold great promise for regenerative medicine (Pera and Trounson, 2004; Reubinoff et al., 2000; Thomson et al., 1998). An understanding of the transcriptional regulatory circuitry that is responsible for pluripotency and self-renewal in human ES cells is fundamental to understanding human development and realizing the therapeutic potential of these cells.
Homeodomain transcription factors are evolutionarily conserved and play key roles in cell-fate specification in many organisms (Hombria and Lovegrove, 2003). Two such factors, OCT4/POU5F1 and NANOG, are essential regulators of early development and ES cell identity (Chambers et al., 2003; Hay et al., 2004; Matin et al., 2004; Mitsui et al., 2003; Nichols et al., 1998; Zaehres et al., 2005). Several genetic studies in mouse suggest that these regulators have distinct roles but may function in related pathways to maintain the developmental potential of these cells (Chambers, 2004). For example, disruption of OCT4 or NANOG results in the inappropriate differentiation of ICM and ES cells to trophectoderm and extra-embryonic endoderm, respectively (Chambers et al., 2003; Mitsui et al., 2003; Nichols et al., 1998). However, overexpression of OCT4 in ES cells leads to a phenotype that is similar to loss of NANOG function (Chambers et al., 2003; Mitsui et al., 2003; Nichols et al., 1998; Niwa et al., 2000). Knowledge of the set of genes regulated by these two transcription factors might reveal why manipulation of OCT4 and NANOG results in these phenotypic consequences.
OCT4 is known to interact with other transcription factors to activate and repress gene expression in mouse ES cells (Pesce and Schöler, 2001). For example, OCT4, a member of the POU (PIT/OCT/UNC) class of homeodomain proteins, can heterodimerize with the HMG-box transcription factor, SOX2, to affect the expression of several genes in mouse ES cells (Botquin et al., 1998; Nishimoto et al., 1999; Yuan et al., 1995). The cooperative interaction of POU homeodomain and HMG factors is thought to be a fundamental mechanism for the developmental control of gene expression (Dailey and Basilico, 2001). The extent to which ES cell gene regulation is accomplished by OCT4 through an OCT4/SOX2 complex and whether NANOG has a role in this process are unknown.
OCT4, SOX2, and NANOG are thought to be central to the transcriptional regulatory hierarchy that specifies ES cell identity because of their unique expression patterns and their essential roles during early development (Avilion et al., 2003; Chambers et al., 2003; Hart et al., 2004; Lee et al., 2004; Mitsui et al., 2003; Nichols et al., 1998; Schöler et al., 1990). Studies in a broad range of eukaryotes have shown that transcriptional regulators that have key roles in cellular processes frequently regulate other regulators associated with that process (Guenther et al., 2005; Lee et al., 2002; Odom et al., 2004). It is likely that the key stem cell regulators bind and regulate genes encoding other transcriptional regulators, which in turn determine the developmental potential of these cells, but we currently lack substantial knowledge of the regulatory circuitry of ES cells and other vertebrate cells.
To further our understanding of the means by which OCT4, SOX2, and NANOG control the pluripotency and self-renewal of human ES cells, we have used genomescale location analysis (chromatin immunoprecipitation coupled with DNA microarrays) to identify the target genes of all three regulators in vivo. The results reveal that OCT4, SOX2, and NANOG co-occupy the promoters of a large population of genes, that many of these target genes encode developmentally important homeodomain transcription factors, and that these regulators contribute to specialized regulatory circuits in ES cells.
Results and Discussion
OCT4 Promoter Occupancy in Human ES Cells
DNA sequences occupied by OCT4 in human H9 ES cells (NIH code WA09; Supplemental Data) were identified in a replicate set of experiments using chromatin immunoprecipitation (ChIP) combined with DNA microarrays (Figure 1A and Supplemental Data). For this purpose, DNA microarrays were designed that contain 60-mer oligonucleotide probes covering the region from −8 kb to +2 kb relative to the transcript start sites for 17,917 annotated human genes. Although some transcription factors are known to regulate genes from distances greater than 8 kb, 98% of known binding sites for human transcription factors occur within 8 kb of target genes (Figure S1). The sites occupied by OCT4 were identified as peaks of ChIP-enriched DNA that span closely neighboring probes (Figure 1B). OCT4 was associated with 623 (3%) of the promoter regions for known protein-coding genes and 5 (3%) of the promoters for known miRNA genes in human ES cells (Table S2).
Figure 1. Genome-Wide Location Analysis in Human Embryonic Stem Cells.
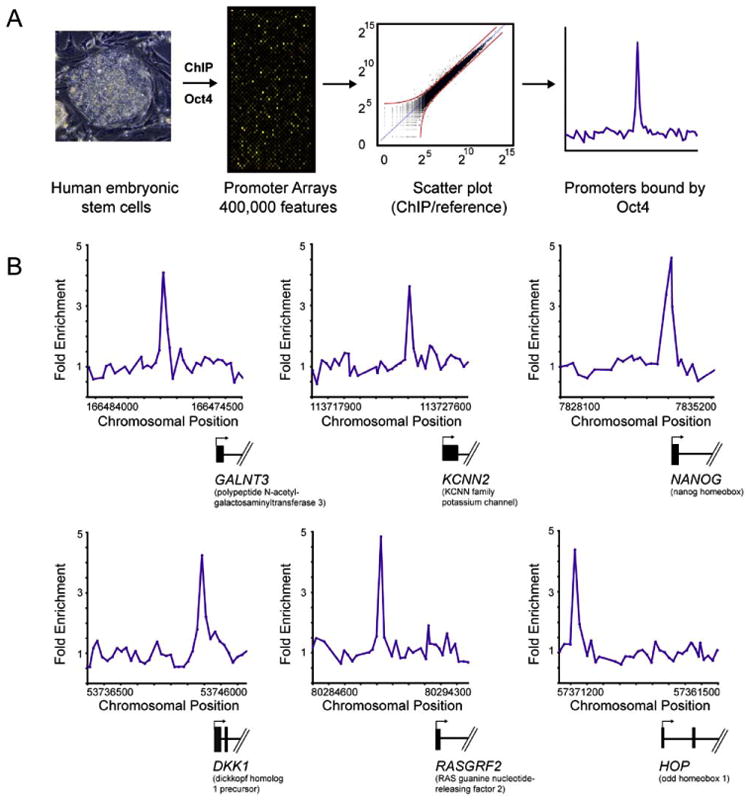
(A) DNA segments bound by transcriptional regulators were identified using chromatin immunoprecipitation (ChIP) and identified with DNA microarrays containing 60-mer oligonucleotide probes covering the region from −8 kb to +2 kb for 17,917 annotated transcription start sites for human genes. ES cell growth and quality control, ChIP protocol, DNA microarray probe design, and data analysis methods are described in detail in Experimental Procedures and Supplemental Data.
(B) Examples of OCT4 bound regions. Plots display unprocessed ChIP-enrichment ratios for all probes within a genomic region. Genes are shown to scale below plots (exons and introns are represented by thick vertical and horizontal lines, respectively), and the genomic region represented is indicated beneath the plot. The transcription start site and transcript direction are denoted by arrows.
Two lines of evidence suggested that this protein-DNA interaction dataset is of high quality. First, the genes occupied by OCT4 in our analysis included many previously identified or supposed target genes in mouse ES cells or genes whose transcripts are highly enriched in ES cells, including OCT4, SOX2, NANOG, LEFTY2/EBAF, CDX2, HAND1, DPPA4, GJA1/CONNEXIN43, FOXO1A, CRIPTO/TDGF1, and ZIC3 (Abeyta et al., 2004; Brandenberger et al., 2004; Catena et al., 2004; Kuroda et al., 2005; Niwa, 2001; Okumura-Nakanishi et al., 2005; Rodda et al., 2005; Sato et al., 2003; Wei et al., 2005) (Table S2). Second, we have used improved protocols and DNA microarray technology in these experiments (Supplemental Data) that should reduce false positive rates relative to those obtained in previous genome-scale experiments (Odom et al., 2004). By using this new technology with yeast transcription factors, where considerable prior knowledge of transcription factor binding sites has been established, we estimated that this platform has a false positive rate of <1% and a false negative rate of 20% (Supplemental Data).
OCT4, SOX2, and NANOG Co-Occupy Many Target Genes
We next identified, with location analysis, protein-coding and miRNA genes targeted by the stem cell regulators SOX2 and NANOG. SOX2 and NANOG were found associated with 1271 (7%) and 1687 (9%), respectively, of the promoter regions for known protein-coding genes in human ES cells (Tables S2–S4). It was immediately evident that many of the target genes were shared by OCT4, SOX2, and NANOG (Figure 2A). Examples of protein-coding genes that are co-occupied by the three regulators are shown in Figure 2B (Table S5). Control experiments showed that the set of promoters bound by the cell-cycle transcription factor E2F4 in these human ES cells did not overlap substantially with those bound by the three stem cell regulators (Tables S2 and S6). We found that OCT4, SOX2, and NANOG together occupy at least 353 genes in human ES cells.
Figure 2. OCT4, SOX2, and NANOG Target Genes in Human ES Cells.
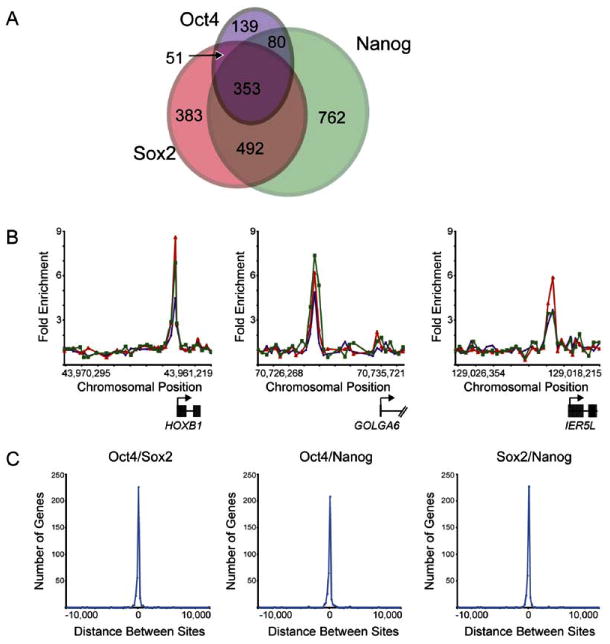
(A) Venn diagram representing the overlap of OCT4, SOX2, and NANOG promoter bound regions.
(B) Representative examples of protein-coding genes co-occupied by OCT4, SOX2, and NANOG. Plots display unprocessed ChIP-enrichment ratios for all probes within a genomic region. Genes are shown to scale relative to their chromosomal position. Exons and introns are represented by thick vertical and horizontal lines, respectively. The start and direction of transcription are denoted by arrows. Green, red, and purple lines represent NANOG, SOX2, and OCT4 bound regions, respectively.
(C) OCT4, SOX2, and NANOG bind in close proximity. The distances between the midpoint of bound regions for pairs of transcription factors was calculated for the 353 regions bound by all three transcription factors. Negative and positive values indicate whether the first factor is upstream or downstream of the second factor in relation to the gene. The frequency of different distances between the bound regions is plotted as a histogram.
Previous studies in murine ES cells have shown that SOX2 and OCT4 can interact to synergistically activate transcription of target genes and that this activity is dependent upon the juxtaposition of OCT4 and SOX2 binding sites (Ambrosetti et al., 1997; Remenyi et al., 2004). Our results revealed that approximately half of the promoter regions occupied by OCT4 were also bound by SOX2 in human ES cells (Figure 2A; Table S2). It was surprising, however, to find that >90% of promoter regions bound by both OCT4 and SOX2 were also occupied by NANOG. Furthermore, we found that OCT4, SOX2, and NANOG binding sites occurred in close proximity at nearly all of the genes that they co-occupied (Figure 2C). These data suggest that OCT4, SOX2, and NANOG function together to regulate a significant proportion of their target genes in human ES cells.
A class of small noncoding RNAs known as micro-RNAs (miRNA) play vital roles in gene regulation, and recent studies indicate that more than a third of mammalian protein-coding genes are conserved miRNA targets (Bartel, 2004; Lewis et al., 2005). ES cells lacking the machinery that processes miRNA transcripts are unable to differentiate (Kanellopoulou et al., 2005). Moreover, recent evidence indicates that microRNAs play an important role in organismal development through regulation of gene expression (Pasquinelli et al., 2005). OCT4, SOX2, and NANOG were found associated with 14 miRNA genes and co-occupied the promoters of at least two miRNA genes, mir-137 and mir-301 (Table 1). Our results suggest that miRNA genes are likely regulated by OCT4, SOX2, and NANOG in human ES cells and are important components of the transcriptional regulatory circuitry in these cells.
Table 1. miRNA Loci near OCT4, SOX2, and NANOG Bound Regions.
| Transcription Factor | |||
|---|---|---|---|
| miRNA | OCT4 | SOX2 | NANOG |
| mir-7-1 | + | ||
| mir-10a | + | ||
| mir-22 | + | + | |
| mir-32 | + | + | |
| mir-128a | + | ||
| mir-135b | + | + | |
| mir-137 | + | + | + |
| mir-196a-1 | + | ||
| mir-196b | + | ||
| mir-204 | + | + | |
| mir-205 | + | + | |
| mir-301 | + | + | + |
| mir-361 | + | ||
| mir-448 | + |
ES Cell Transcription Factors Occupy Active and Inactive Genes
OCT4 and SOX2 are known to be involved in both gene activation and repression in vivo (Botquin et al., 1998; Nishimoto et al., 1999; Yuan et al., 1995), so we sought to identify the transcriptional state of genes occupied by the stem cell regulators. To this end, the set of genes bound by OCT4, SOX2, and NANOG were compared to gene expression datasets generated from multiple ES cell lines (Abeyta et al., 2004; Brandenberger et al., 2004; Sato et al., 2003; Wei et al., 2005) to identify transcriptionally active and inactive genes (Table S2). The results showed that one or more of the stem cell transcription factors occupied 1303 actively transcribed genes and 957 inactive genes.
The importance of OCT4, SOX2, and NANOG for early development and ES cell identity led us to focus additional analyses on the set of 353 genes that are co-occupied by these regulators in human ES cells (Table S5). We first identified transcriptionally active genes. Transcripts were consistently detected in ES cells for approximately half of the genes co-bound by OCT4, SOX2, and NANOG. Among these active genes, several encoding transcription factors (e.g., OCT4, SOX2, NANOG, STAT3, ZIC3) and components of the Tgf-β (e.g., TDGF1, LEFTY2/EBAF) and Wnt (e.g., DKK1, FRAT2) signaling pathways were notable targets. Recent studies have shown that Tgf-β and Wnt signaling play a role in pluripotency and self-renewal in both mouse and human ES cells (James et al., 2005; Sato et al., 2004). These observations suggest that OCT4, SOX2, and NANOG promote pluripotency and self-renewal through positive regulation of their own genes and genes encoding components of these key signaling pathways.
Among transcriptionally inactive genes co-occupied by OCT4, SOX2, and NANOG, we noted a striking enrichment for transcription factor genes (p < 10−18; Table S7), many of which have been implicated in developmental processes. These included genes that specify transcription factors important for differentiation into extra-embryonic, endodermal, mesodermal, and ectodermal lineages (e.g., ESX1l, HOXB1, MEIS1, PAX6, LHX5, LBX1, MYF5, ONECUT1) (Table S5). Moreover, nearly half of the transcription factor genes that were bound by the three regulators and transcriptionally inactive encoded developmentally important homeodomain proteins (Table 2). These results demonstrate that OCT4, SOX2, and NANOG occupy a set of repressed genes that are key to developmental processes.
Table 2. Examples of Inactive Homeodomain Genes Co-occupied by OCT4, SOX2, and NANOG.
| Gene Symbol | Entrez Gene ID | Gene Name |
|---|---|---|
| ATBF1 | 463 | AT binding transcription factor 1 |
| DLX1 | 1745 | distal-less homeobox 1 |
| DLX4 | 1748 | distal-less homeobox 4 |
| DLX5 | 1749 | distal-less homeobox 5 |
| EN1 | 2019 | engrailed homolog 1 |
| ESX1L | 80712 | extraembryonic, spermatogenesis, homeobox 1-like |
| GBX2 | 2637 | gastrulation brain homeobox 2 |
| GSC | 145258 | goosecoid |
| HOP | 84525 | homeodomain-only protein |
| HOXB1 | 3211 | homeobox B1 |
| HOXB3 | 3213 | homeobox B3 |
| HOXC4 | 3221 | homeobox C4 |
| IPF2 | 3651 | insulin promoter factor 2 |
| ISL1 | 3670 | ISL1 transcription factor, LIM/homeodomain (islet-1) |
| LBX1 | 10660 | transcription factor similar to D. melanogaster homeodomain protein lady bird late |
| LHX2 | 9355 | LIM homeobox 2 |
| LHX5 | 64211 | LIM homeobox 5 |
| MEIS1 | 4211 | myeloid ecotropic viral integration site 1 homolog (mouse) |
| NKX2-2 | 4821 | NK2 transcription factor related, locus 2 (Drosophila) |
| NKX2-3 | 159296 | NK2 transcription factor related, locus 3 (Drosophila) |
| ONECUT1 | 3175 | one cut domain, family member 1 |
| OTP | 23440 | orthopedia homolog (Drosophila) |
| OTX1 | 5013 | orthodenticle homolog 1 (Drosophila) |
| PAX6 | 5080 | paired box gene 6 |
| TITF1 | 7080 | thyroid transcription factor 1 |
To determine which of the OCT4, SOX2, and NANOG bound genes were preferentially expressed in ES cells, we compared expression datasets (Abeyta et al., 2004; Sato et al., 2003) from ES cells and a compendium of differentiated tissues and cell types (Su et al., 2004) (Figure 3; Supplemental Data). It was notable that DPPA4, TDGF1, OCT4, NANOG, and LEFTY2 were at the top of the rank order list of genes that are bound and preferentially expressed in ES cells (Figure 3A). All five of these genes have been implicated in pluripotency (James et al., 2005; Mitsui et al., 2003; Chambers et al., 2003; Nichols et al., 1998; Bortvin et al., 2003). Moreover, several genes that encode developmentally important homeodomain proteins such as DLX5, HOXB1, LHX5, TITF1, LBX1, and HOP were at the bottom of this list, indicating that they are preferentially repressed in ES cells.
Figure 3. Expression of OCT4, SOX2, and NANOG Co-Occupied Genes.
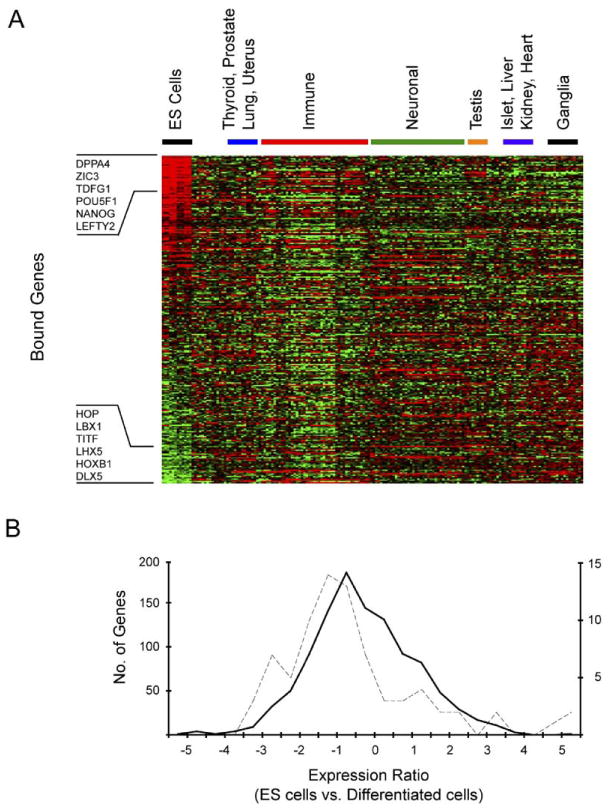
(A) Affymetrix expression data for ES cells were compared to a compendium of expression data from 158 experiments representing 79 other differentiated tissues and cell types (Supplemental data). Ratios were generated by comparing gene expression in ES cells to the median level of gene expression across all datasets for each individual gene. Genes were ordered by relative expression in ES cells, and the results were clustered by expression experiment using hierarchical clustering. Each gene is represented as a separate row and individual expression experiments are in separate columns. Red indicates higher expression in ES cells relative to differentiated cells. Green indicates lower expression in ES cells relative to differentiated cells. Examples of bound genes that are at the top and bottom of the rank order list are shown.
(B) Relative levels of gene expression in H9 ES cells compared to differentiated cells were generated and converted to log2 ratios. The distribution of these fold changes was calculated to derive a profile for different sets of genes. Data are shown for the distribution of expression changes between H9 ES cells and differentiated tissues for transcription factor genes that are not occupied by OCT4, SOX2, and NANOG (solid black line) and transcription factor genes occupied by all three (dotted line). The change in relative expression is indicated on the x axis, and the numbers of genes in each bin are indicated on the y axes (left axis for unoccupied genes, right axis for occupied genes). The shift in distribution of expression changes for genes occupied by OCT4, SOX2, and NANOG is significant (p value < 0.001 using a two-sampled Kolmogorov-Smirnov test), consistent with the model that OCT4, SOX2, and NANOG are contributing to the regulation of these genes.
The observation that OCT4, SOX2, and NANOG bound to transcriptionally active genes that have roles in pluripotency and transcriptionally inactive genes that promote development suggests that these binding events are regulatory. Two additional lines of evidence indicated that many of the binding events identified in this study contribute to regulation of their target genes. First, some of the genes identified here (e.g., OCT4, SOX2, and NANOG) were previously shown to be regulated by OCT4 and SOX2 in mouse ES cells (Catena et al., 2004; Kuroda et al., 2005, Okumura-Nakanishi et al., 2005; Rodda et al., 2005). Second, we further explored the hypothesis that bound genes are regulated by these transcription factors by taking advantage of the fact that OCT4 and NANOG are expressed in ES cells, but their expression is rapidly downregulated upon differentiation. We compared the expression of OCT4, SOX2, and NANOG occupied genes in human ES cells with expression patterns in 79 differentiated cell types (Su et al., 2004) (Supplemental Data) and focused the analysis on transcription factor genes because these were the dominant functional class targeted by the ES cell regulators (Figure 3B). We expected that for any set of genes, there would be a characteristic change in expression levels between ES cells and differentiated cells. If OCT4, SOX2, and NANOG do not regulate the genes they occupy, then these genes should have the same general expression profile as the control population. We found, however, a significant shift in the distribution of expression changes for genes occupied by OCT4, SOX2, and NANOG (p value < 0.001). Taken together, these data support the model that OCT4, SOX2, and NANOG functionally regulate the genes they occupy and suggest that loss of these regulators upon differentiation results in increased expression of genes necessary for development and reduced expression of a set of genes required for the maintenance of stem cell identity.
Our results suggest that OCT4, SOX2, and NANOG contribute to pluripotency and self-renewal by activating their own genes and genes encoding components of key signaling pathways and by repressing genes that are key to developmental processes. It is presently unclear how the three key regulators can activate some genes and repress others. It is likely that the activity of these key transcription factors is further controlled by additional cofactors, by the precise levels of OCT4, SOX2, and NANOG, and by posttranslational modifications.
Core Transcriptional Regulatory Circuitry in ES Cells
In order to identify regulatory network motifs associated with OCT4, SOX2, and NANOG, we assumed that regulator binding to a gene implies regulatory control and used algorithms that were previously devised to discover such regulatory circuits in yeast (Lee et al., 2002). The simplest units of commonly used transcriptional regulatory network architecture, or network motifs, provide specific regulatory capacities such as positive and negative feedback loops to control the levels of their components (Lee et al., 2002; Milo et al., 2002; Shen-Orr et al., 2002).
Our data indicated that OCT4, SOX2, and NANOG form feedforward loops that involve at least 353 protein coding and 2 miRNA genes (Figure 4A). Feedforward-loop motifs contain a regulator that controls a second regulator and have the additional feature that both regulators bind a set of common target genes. The feedforward loop has multiple regulatory capacities that may be especially useful for stem cells. When both regulators are positive, the feedforward loop can provide consistent activity that is relatively insensitive to transient changes in input (Mangan et al., 2003; Shen-Orr et al., 2002). If the regulators have positive and negative functions, the feedforward loop can act as a switch that enables a rapid response to inputs by providing a timesensitive delay where the downstream regulator acts to counter the effects of the upstream regulator in a delayed fashion (Mangan and Alon, 2003; Mangan et al., 2003). In ES cells, both regulatory capacities could be useful for maintaining the pluripotent state while retaining the ability to react appropriately to differentiation signals. Previous studies have shown that feedforward-loop architecture has been highly favored during the evolution of transcriptional regulatory networks in less complex eukaryotes (Lee et al., 2002; Ma et al., 2004; Milo et al., 2002; Resendis-Antonio et al., 2005; Shen-Orr et al., 2002). Our data suggest that feedforward regulation is an important feature of human ES cells as well.
Figure 4. Transcriptional Regulatory Motifs in Human ES Cells.
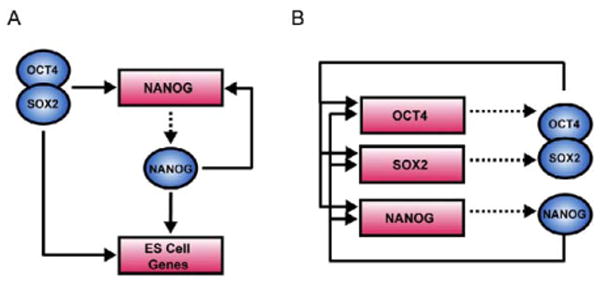
(A) An example of feedforward transcriptional regulatory circuitry in human ES cells. Regulators are represented by blue circles; gene promoters are represented by red rectangles. Binding of a regulator to a promoter is indicated by a solid arrow. Genes encoding regulators are linked to their respective regulators by dashed arrows.
(B) The interconnected autoregulatory loop formed by OCT4, SOX2, and NANOG.
Our results also showed that OCT4, SOX2, and NANOG together bound to the promoters of their own genes, forming interconnected autoregulatory loops (Figure 4B; see also Figure S2). Transcriptional regulation of OCT4, SOX2, and NANOG by the OCT4-SOX2 complex was recently described in murine ES cells (Catena et al., 2004; Kuroda et al., 2005; Okumura-Nakanishi et al., 2005; Rodda et al., 2005). Our data indicate that this autoregulatory loop is conserved in human ES cells and, more importantly, that NANOG is a component of the regulatory apparatus at these genes. Thus, it is likely that the expression and function of these three key stem cell factors are inextricably linked to one another. Autoregulation is thought to provide several advantages, including reduced response time to environmental stimuli and increased stability of gene expression (McAdams and Arkin, 1997; Rosenfeld et al., 2002; Shen-Orr et al., 2002; Thieffry et al., 1998).
The autoregulatory and feedforward circuitry described here may provide regulatory mechanisms by which stem cell identity can be robustly maintained yet permit cells to respond appropriately to developmental cues. Modifying OCT4 and NANOG levels and function can change the developmental potential of murine ES cells (Chambers et al., 2003; Mitsui et al., 2003; Nichols et al., 1998; Niwa et al., 2000), and this might be interpreted as being a consequence of perturbing independent regulatory pathways under the control of these two regulators. Our results argue that the levels and functions of these key stem cell regulators are tightly linked at both target genes and at their own promoters and thus provide an additional framework for interpreting the genetic studies. Changes in the relative stoichiometry of these factors would disturb the autoregulatory and feedforward circuitry, producing changes in global gene regulation and thus cell fate.
Expanded Transcriptional Regulatory Circuitry
An initial model for ES cell transcriptional regulatory circuitry was constructed by identifying OCT4, SOX2, and NANOG target genes that encode transcription factors and chromatin regulators and integrating knowledge of the functions of these downstream regulators in both human and mouse based on the available expression studies and literature (Figure 5). The model includes a subset of active and a subset of repressed target genes based on the extensive expression characterization of the 353 co-bound genes as described earlier. The active targets include genes encoding components of chromatin remodeling and histone-modifying complexes (e.g., SMARCAD1, MYST3, and SET), which may have general roles in transcriptional regulation, and genes encoding transcription factors (e.g., REST, SKIL, HESX1, and STAT3), which themselves are known to regulate specific genes. For instance, REST has recently been shown to be highly abundant in ES cells and functions in part to repress neuronal specific genes (Ballas et al., 2005). Previous studies have proposed that NANOG may function through the Tgf-β pathway in ES cells (Chambers, 2004). Our model suggests that this occurs through direct regulation of key components of this pathway (e.g., TDFG1, LEFTY2/EBAF) and through regulation of at least one transcription factor, SKIL, which controls the activity of downstream components of this pathway (SMAD2, SMAD4) (He et al., 2003). Our data also reveal that OCT4, SOX2, and NANOG co-occupy STAT3, a key regulator of self-renewal in mouse ES cells (Chambers, 2004), suggesting that STAT3 may also play a role in human ES cells.
Figure 5. Core Transcriptional Regulatory Network in Human ES Cells.
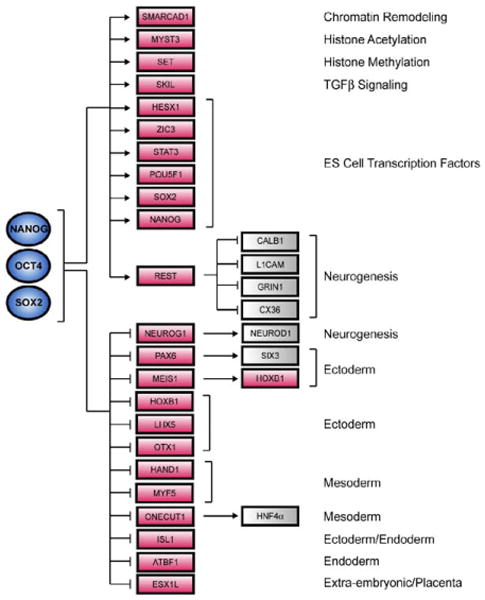
A model for the core transcriptional regulatory network was constructed by identifying OCT4, SOX2, and NANOG target genes that encode transcription factors and chromatin regulators and integrating knowledge of the functions of these downstream regulators based on comparison to multiple expression datasets (Supplemental Data) and to the literature. A subset of active and inactive genes co-occupied by the three factors in human ES cells is shown here. Regulators are represented by blue circles; gene promoters are represented by red rectangles; gray boxes represent putative downstream target genes. Positive regulation was assumed if the target gene was expressed whereas negative regulation was assumed if the target gene was not transcribed.
The model described in Figure 5 also depicts a subset of the genes bound by OCT4, SOX2, and NANOG that are inactive and that encode transcription factors that have key roles in differentiation and development. These include regulators with demonstrated roles in development of all embryonic lineages. This initial model for ES cell transcriptional regulatory circuitry is consistent with previous genetic studies in mice that suggest that OCT4 and NANOG maintain pluripotency through repression of differentiation programs (Chambers et al., 2003; Mitsui et al., 2003; Niwa et al., 2000). This model also provides a mechanistic framework for understanding how this is accomplished through regulation of specific sets of genes that control cell-fate specification.
Concluding Remarks
Discovering how gene expression programs are controlled in living cells promises to improve our understanding of cell biology, development, and human health. Identifying the target genes for key transcriptional regulators of human stem cells is a first critical step in the process of understanding these transcriptional regulatory networks and learning how they control cell identity. Mapping OCT4, SOX2, and NANOG to their binding sites within known promoters has revealed that these regulators collaborate to form in ES cells regulatory circuitry consisting of specialized autoregulatory and feedforward loops. Continued advances in our ability to culture and genetically manipulate human ES cells will allow us to test and manipulate this circuitry. Identification of the targets of additional transcription factors and chromatin regulators using the approaches described here should allow investigators to produce a more comprehensive map of transcriptional regulatory circuitry in these cells. Connecting signaling pathways to this circuit map may reveal how these pluripotent cells can be stimulated to differentiate into different cell types or how to reprogram differentiated cells back to a pluripotent state.
Experimental Procedures
Growth Conditions for Human Embryonic Stem Cells
Human embryonic stem (ES) cells were obtained from WiCell (Madison, Wisconsin; NIH Code WA09). Detailed protocol information on human ES cell growth conditions and culture reagents are available at http://www.mcb.harvard.edu/melton/hues. Briefly, passage 34 cells were grown in KO-DMEM medium supplemented with serum replacement, basic fibroblast growth factor (bFGF), recombinant human leukemia inhibitory factor (LIF), and a human plasma protein fraction. In order to minimize any MEF contribution in our analysis, H9 cells were cultured on a low density of irradiated murine embryonic fibroblasts (ICR MEFs) resulting in a ratio of approximately >8:1 H9 cell to MEF. The culture of H9 on low-density MEFs had no adverse effects on cell morphology, growth rate, or undifferentiated status as compared to cells grown under typical conditions. In addition, immunohistochemistry for pluripotency markers (e.g., OCT4, SSEA-3) indicated that H9 cells grown on a minimal feeder layer maintained the ability to generate derivates of ectoderm, mesoderm, and endoderm upon differentiation (Figures S3 and S4).
Antibodies
The NANOG (AF1997) and SOX2 (AF2018) antibodies used in this study were immunoaffinity purified against the human proteins and shown to recognize their target proteins in Western blots and by immunocytochemistry (R&D Systems Minneapolis, Minnesota). Multiple OCT4 antibodies directed against different portions of the protein (AF1759 R&D Systems, sc-8628 Santa Cruz, sc-9081 Santa Cruz), some of which were immunoaffinity purified, were used in this study and have been shown to recognize their target protein in Western blots and by immunocytochemistry. The E2F4 antibody used in this study was obtained from Santa Cruz (sc-1082) and has been shown to recognize E2F4-responsive genes identified in previous ChIP studies (Table S2) (Ren et al., 2002; Weinmann et al., 2002).
Chromatin Immunoprecipitation
Protocols describing all materials and methods can be downloaded from http://jura.wi.mit.edu/young/hESRegulation/.
Human embryonic stem cells were grown to a final count of 5 × 107−1 × 108 cells for each location analysis reaction. Cells were chemically crosslinked by the addition of one-tenth volume of fresh 11% formaldehyde solution for 15 min at room temperature. Cells were rinsed twice with 1 × PBS and harvested using a silicon scraper and flash frozen in liquid nitrogen and stored at −80°C prior to use. Cells were resuspended, lysed in lysis buffers, and sonicated to solubilize and shear crosslinked DNA. Sonication conditions vary depending on cells, culture conditions, crosslinking, and equipment. We used a Misonix Sonicator 3000 and sonicated at power 7 for 10 × 30 s pulses (90 s pause between pulses) at 4°C while samples were immersed in an ice bath. The resulting whole-cell extract was incubated overnight at 4°C with 100 μl of Dynal Protein G magnetic beads that had been preincubated with 10 μg of the appropriate antibody. Beads were washed five times with RIPA buffer and one time with TE containing 50 mM NaCl. Bound complexes were eluted from the beads by heating at 65°C with occasional vortexing, and crosslinking was reversed by overnight incubation at 65°C. Whole-cell extract DNA (reserved from the sonication step) was also treated for crosslink reversal. Immunoprecipitated DNA and whole-cell extract DNA were then purified by treatment with RNaseA, proteinase K, and multiple phenol:chloroform: isoamyl alcohol extractions. Purified DNA was blunted and ligated to linker and amplified using a two-stage PCR protocol. Amplified DNA was labeled and purified using Invitrogen Bioprime random primer labeling kits (immunoenriched DNA was labeled with Cy5 fluorophore, whole-cell extract DNA was labeled with Cy3 fluorophore). Labeled DNA was combined (5–6 μg each of immunoenriched and whole-cell extract DNA) and hybridized to arrays in Agilent hybridization chambers for 40 hr at 40°C. Arrays were then washed and scanned (Supplemental Data).
Array Design and Data Extraction
The design of the 10-slide oligo-based promoter arrays used in this study and data extraction methods are described in detail in Supplemental Data. Arrays were manufactured by Agilent Technologies (http://www.agilent.com).
Supplementary Material
Supplementary Data
Acknowledgments
We would like to thank Bioinformatics and Research Computing (BaRC) and the Center for Microarray Technology (CMT) at the Whitehead Institute for computational and technical support. We would also like to thank members of the Young lab as well as Chad Cowan and Kevin Eggan for helpful discussions. L.A.B. was supported by NRSA postdoctoral fellowship CA094664, and H.L.M. by NRSA postdoctoral fellowship GM068273. R.M.K. was supported by a fellowship from the American Cancer Society. This work was supported by NHGRI grant HG002668 to D.K.G. and R.A.Y. and NIH grant GM069400 to R.A.Y. T.I.L., D.K.G., and R.A.Y. consult for Agilent Technologies.
Footnotes
Accession Numbers
All microarray data from this study are available at ArrayExpress at the EBI (http://www.ebi.ac.uk/arrayexpress) under the accession designation E-WMIT-5.
References
- Abeyta MJ, Clark AT, Rodriguez RT, Bodnar MS, Pera RA, Firpo MT. Unique gene expression signatures of independently derived human embryonic stem cell lines. Hum Mol Genet. 2004;13:601–608. doi: 10.1093/hmg/ddh068. [DOI] [PubMed] [Google Scholar]
- Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R, Page DC, Jaenisch R. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- Botquin V, Hess H, Fuhrmann G, Anastassiadis C, Gross MK, Vriend G, Scholer HR. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 1998;12:2073–2090. doi: 10.1101/gad.12.13.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenberger R, Khrebtukova I, Thies RS, Miura T, Jingli C, Puri R, Vasicek T, Lebkowski J, Rao M. MPSS profiling of human embryonic stem cells. BMC Dev Biol. 2004;4:10–25. doi: 10.1186/1471-213X-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catena R, Tiveron C, Ronchi A, Porta S, Ferri A, Tatangelo L, Cavallaro M, Favaro R, Ottolenghi S, Reinbold R, et al. Conserved POU binding DNA sites in the Sox2 upstream enhancer regulate gene expression in embryonic and neural stem cells. J Biol Chem. 2004;279:41846–41857. doi: 10.1074/jbc.M405514200. [DOI] [PubMed] [Google Scholar]
- Chambers I. The molecular basis of pluripotency in mouse embryonic stem cells. Cloning Stem Cells. 2004;6:386–391. doi: 10.1089/clo.2004.6.386. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Dailey L, Basilico C. Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J Cell Physiol. 2001;186:315–328. doi: 10.1002/1097-4652(2001)9999:9999<000::AID-JCP1046>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci USA. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Ibrahim M, Robb L. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev Dyn. 2004;230:187–198. doi: 10.1002/dvdy.20034. [DOI] [PubMed] [Google Scholar]
- Hay DC, Sutherland L, Clark J, Burdon T. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 2004;22:225–235. doi: 10.1634/stemcells.22-2-225. [DOI] [PubMed] [Google Scholar]
- He J, Tegen SB, Krawitz AR, Martin GS, Luo K. The transforming activity of Ski and SnoN is dependent on their ability to repress the activity of Smad proteins. J Biol Chem. 2003;278:30540–30547. doi: 10.1074/jbc.M304016200. [DOI] [PubMed] [Google Scholar]
- Hombria JC, Lovegrove B. Beyond homeosis–HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. TGFbeta/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development. 2005;132:1273–1282. doi: 10.1242/dev.01706. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicerdeficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Hart SR, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004;38:32–38. doi: 10.1002/gene.10250. [DOI] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Ma HW, Kumar B, Ditges U, Gunzer F, Buer J, Zeng AP. An extended transcriptional regulatory network of Escherichia coli and analysis of its hierarchical structure and network motifs. Nucleic Acids Res. 2004;32:6643–6649. doi: 10.1093/nar/gkh1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feedforward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Zaslaver A, Alon U. The coherent feedforward loop serves as a sign-sensitive delay element in transcription networks. J Mol Biol. 2003;334:197–204. doi: 10.1016/j.jmb.2003.09.049. [DOI] [PubMed] [Google Scholar]
- Matin MM, Walsh JR, Gokhale PJ, Draper JS, Bahrami AR, Morton I, Moore HD, Andrews PW. Specific knockdown of Oct4 and beta2-microglobulin expression by RNA interference in human embryonic stem cells and embryonic carcinoma cells. Stem Cells. 2004;22:659–668. doi: 10.1634/stemcells.22-5-659. [DOI] [PubMed] [Google Scholar]
- McAdams HH, Arkin A. Stochastic mechanisms in gene expression. Proc Natl Acad Sci USA. 1997;94:814–819. doi: 10.1073/pnas.94.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milo R, Shen-Orr S, Itzkovitz S, Kashtan N, Chklovskii D, Alon U. Network motifs: simple building blocks of complex networks. Science. 2002;298:824–827. doi: 10.1126/science.298.5594.824. [DOI] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H. Molecular mechanism to maintain stem cell renewal of ES cells. Cell Struct Funct. 2001;26:137–148. doi: 10.1247/csf.26.137. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Odom DT, Zizlsperger N, Gordon DB, Bell GW, Rinaldi NJ, Murray HL, Volkert TL, Schreiber J, Rolfe PA, Gifford DK, et al. Control of pancreas and liver gene expression by HNF transcription factors. Science. 2004;303:1378–1381. doi: 10.1126/science.1089769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr Opin Genet Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Pera MF, Trounson AO. Human embryonic stem cells: prospects for development. Development. 2004;131:5515–5525. doi: 10.1242/dev.01451. [DOI] [PubMed] [Google Scholar]
- Pesce M, Schöler HR. Oct-4: gatekeeper in the beginnings of mammalian development. Stem Cells. 2001;19:271–278. doi: 10.1634/stemcells.19-4-271. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Schöler HR, Wilmanns M. Combinatorial control of gene expression. Nat Struct Mol Biol. 2004;11:812–815. doi: 10.1038/nsmb820. [DOI] [PubMed] [Google Scholar]
- Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, Dynlacht BD. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16:245–256. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendis-Antonio O, Freyre-Gonzalez JA, Menchaca-Mendez R, Gutierrez-Rios RM, Martinez-Antonio A, Avila-Sanchez C, Collado-Vides J. Modular analysis of the transcriptional regulatory network of E. coli. Trends Genet. 2005;21:16–20. doi: 10.1016/j.tig.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of Nanog by Oct4 and Sox2. J Biol Chem. 2005;18 doi: 10.1074/jbc.M502573200. in press. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Sato N, Sanjuan IM, Heke M, Uchida M, Naef F, Brivanlou AH. Molecular signature of human embryonic stem cells and its comparison with the mouse. Dev Biol. 2003;260:404–413. doi: 10.1016/s0012-1606(03)00256-2. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Schöler HR, Dressler GR, Balling R, Rohdewohld H, Gruss P. Oct-4: a germline-specific transcription factor mapping to the mouse t-complex. EMBO J. 1990;9:2185–2195. doi: 10.1002/j.1460-2075.1990.tb07388.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieffry D, Salgado H, Huerta AM, Collado-Vides J. Prediction of transcriptional regulatory sites in the complete genome sequence of Escherichia coli K-12. Bioinformatics. 1998;14:391–400. doi: 10.1093/bioinformatics/14.5.391. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Wei CL, Miura T, Robson P, Lim SK, Xu XQ, Lee MY, Gupta S, Stanton L, Luo Y, Schmitt J, et al. Transcriptome profiling of human and murine ESCs identifies divergent paths required to maintain the stem cell state. Stem Cells. 2005;23:166–185. doi: 10.1634/stemcells.2004-0162. [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipiation and CpG island microarray analysis. Genes Dev. 2002;16:235–244. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Zaehres H, Lensch MW, Daheron L, Stewart SA, Itskovitz-Eldor J, Daley GQ. High-efficiency RNA interference in human embryonic stem cells. Stem Cells. 2005;23:299–305. doi: 10.1634/stemcells.2004-0252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data