Polarity protein alterations in carcinoma: a focus on emerging roles for polarity regulators (original) (raw)
. Author manuscript; available in PMC: 2011 Feb 1.
Published in final edited form as: Curr Opin Genet Dev. 2010 Jan 21;20(1):41–50. doi: 10.1016/j.gde.2009.12.001
Abstract
In this review we discuss both gene expression and protein localization changes of polarity proteins in carcinoma. We highlight the importance of protein mislocalization and its possible role in cancer. We also discuss the emerging role of polarity proteins as regulators of proliferation, apoptosis, tissue polarity, epithelial mesenchymal transition, in addition to their known role in cell junction biogenesis.
Carcinomas are malignant cancers of epithelial origins and they occur in many organs of humans, such as liver, lung, breast, colon, prostate, ovary, pancreas, and esophagus. According to American Cancer Society, at least 80% of all cancers are carcinomas. Thus understanding the biology of epithelial cells will not only provide insights into mechanisms that regulate early events of cancer and steps that regulate cancer progression but also help to develop novel strategies for diagnosing and treating malignancy.
Cell and tissue polarity in epithelial organs
Epithelial organs are polarized both at the tissue level and at the cellular level. At the tissue level, epithelial cells organize to form multicellular structures with polarized organization to maintain tissue structure and perform normal physiological functions such as secretion of milk into the lumen, a property referred to as tissue polarity. At the cellular level, epithelial cells have asymmetric distribution of cytoplasmic and membrane proteins to regulate cell structure and transduce signals a property referred to as cell polarity. Both tissue polarity and cell polarity are lost early during the neoplastic process, which is exploited by pathologists for detecting cancer [1]. There are several types of cell polarity. Cells lining ductal structures exhibit apical-basal polarity where proteins asymmetrically distribute along the apical-basal axis [2]; fields of cells in a tissue exhibit planar cell polarity (PCP) which refers to the ability of cells to orient in a predetermined direction [3]; migrating cells exhibit front-back polarity where proteins asymmetrically distribute along the axis of migration and; lastly progenitor cells exhibit asymmetric cell division, a property that facilitates generation of two daughter cells with different cell fates [4]. Thus, the ability of cells to asymmetrically distribute proteins plays important roles during various physiological events. In this article, we will review cancer related changes in gene expression and localization patterns of polarity proteins with a focus on the emerging roles for polarity proteins as regulators of biological processes related to tumorigenesis in mammals.
Polarity proteins
Studies in model organisms such as yeast, worms and flies have led to the identification of a set of proteins that regulate various aspects of cell polarity, which includes scaffolding molecules, kinases and GTPases (for review see [5]). Polarity proteins function as multi-protein complexes and their primary function is to generate asymmetry within a cell. In epithelial cells with apical-basal polarity, polarity proteins are asymmetrically distributed along the apical–basal axis (see Figure 1 for localizations of several polarity proteins). For example, members of the Crumbs polarity complex (Crumbs/Protein associated with Lin seven 1(Pals1)/Pals1-associated tight junction protein (PATJ)) and the Partitioning defective (Par) polarity complex (Par3/Par6/atypical protein kinase C (aPKC)) are localized to the apical cortex (for review of those proteins, see [6]), whereas members of the Scribble polarity complex (Scribble/Discs large (Dlg)/Lethal giant Larvae (Lgl)) are localized at the basolateral regions of the cell [7-10].
Figure 1.
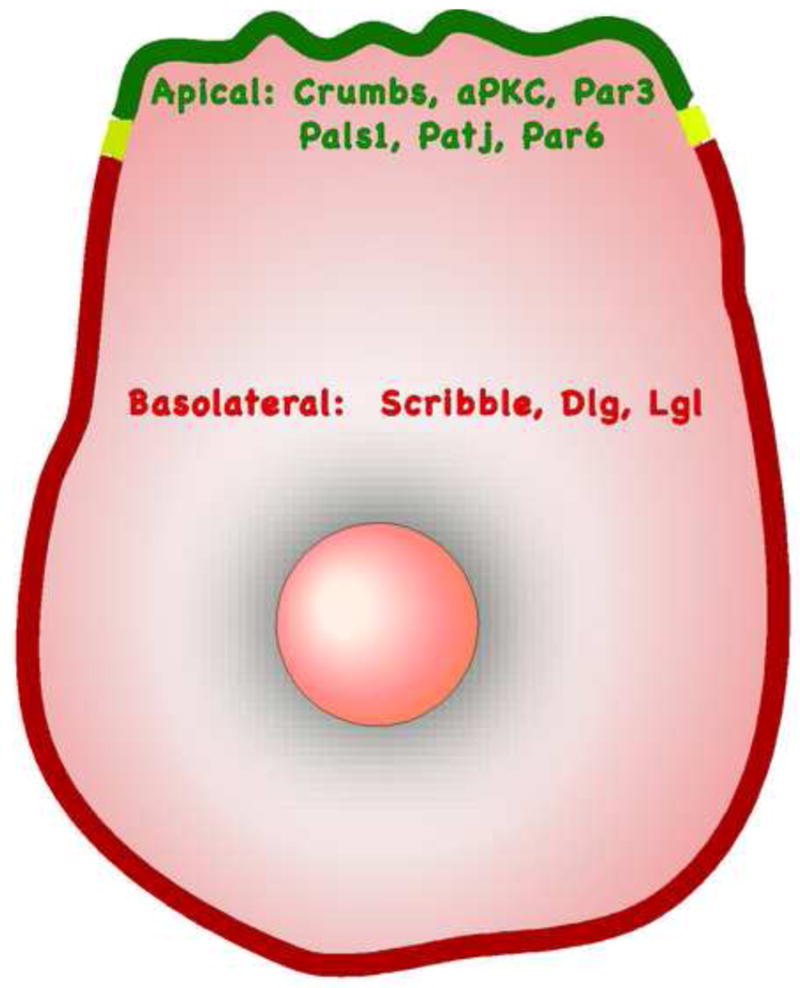
Spatial distribution of polarity regulators in epithelial cells polarized along the apical-basal axis.
Crumbs are transmembrane proteins that bind to PDZ domains of Pals1 and PATJ through C-terminal ERLI motif. Pals1 is a member of the membrane-associated guanylate kinase (MAGUK) family, which has a PDZ domain, two L27 domains, a SH3 domain and a guanylate kinase domain. PATJ has ten PDZ domains and one L27N domain. Par3 and Par6 interact with each other via their PDZ domains. Par3, through aPKC and Par6, can also interact with the Crumbs complex (see[11] for a detailed review). There are two aPKC isoforms in mammals: PKCζ and PKCλ/ι. The two isoforms have 72% sequence similarity and currently functional differences between the two are not well understood. Kinase activities of aPKC are calcium independent and negatively regulated by multiple proteins including Partitioning defective-4 (Par-4) protein (LKB1) kinase, the Par6 scaffolding protein as well as the inhibitory PKCζII protein [12].
Scribble, Dlg and Lgl were identified in Drosophila as tumor suppressors and their roles in mammalian carcinomas are starting to be revealed [9,13,14]. Scribble is a member of LAP family with leucine rich repeats and PDZ domains, while Dlg is a member of the MAGUK family containing PDZ domains. Lgl does not contain PDZ domains but has WD40 domains, which is thought to mediate interactions with phosphorylated serine and tyrosine [15]. Planar cell polarity (PCP) is orthogonal to apical-basal polarity. The core PCP proteins in vertebrates include transmembrane proteins Frizzled, Vang and Celsr1 as well as cytoplasmic proteins Dishevelled and Prickle [3]. In vertebrates, there are only a few events clearly involving PCP, such as the convergent extension and neurulation during embryonic development and orientation of the stereocilia in cochlea of the inner ear (for review, see [3]).
The apical polarity proteins can cross-regulate basolateral polarity proteins during establishment and maintenance of cell polarity. Lgl is a substrate of aPKC and its cellular localization is regulated by aPKCs in normal epithelia as well as in carcinoma tissues [16,17]. On the other hand, Scribble is required to restrict apical proteins such as Crumbs to the apical region of Drosophila epithelia [18]. The localization of Lgl is also regulated by PCP pathway, knockdown of Dishevelled mislocalizes Lgl from cell membrane and downregulates Lgl protein levels in Xenopus embryonic ectoderm [19].
A given polarity protein can regulate different types of cell polarity. For example, Scribble, is involved in all the types of cell polarity discussed above [18,20-24]. These common components in different polarity pathways as well as the mutual dependence of different polarity complexes highlight the fact that one needs to be careful in choosing model systems to study the role played by polarity proteins, because many polarity proteins are likely to function in a context-specific manner.
Alteration of polarity proteins in carcinoma
Tissue transformation in cancer is frequently associated with loss of cell and tissue polarity but the molecular mechanisms are not known. It is likely that the transformation process involves alteration in polarity genes expression and/or their protein subcellular localization resulting in functional inactivation of polarity pathways [25]. There is a significant body of evidence demonstrating that alteration of polarity protein genes are frequently observed in carcinomas (see Table 1 for summary).
Table 1.
Summary on expression and localization alternations of polarity proteins in carcinomas cell lines and primary tissues.
| Protein | Defects | Tissue/Cell line | Functions/Phenotype | Reference |
|---|---|---|---|---|
| Crumbs | Downregulated | Tumor derived cell lines from in vivo selection in mice | Cell junctions disrupted, increase of metastasis | [29] |
| Par3 | Gene deleted, or downregulated | Esophageal squamous carcinoma cell lines and primary tumor tissues | Cell junctions disrupted | [26] |
| Par6 | Overexpressed | ER positive breast cancer cell lines and primary tumor tissues | Cell hyperproliferation | [27] |
| Overexpressed, phosphorylated | Mouse mammary cell lines, BRCA1 associated tumor tissues | Lumen filling, cell junctions disrupted, increase of metastasis | [73] | |
| Overexpressed | Stromal cells in non small cell lung cancer tissues | Good prognosis | [28] | |
| PKC zeta | Downregulated | Rat prostatic carcinoma cell lines | Prostatic tumor | [31] |
| Overexpressed | Hepatocellular carcinoma | Liver cancer | [35] | |
| Overexpressed | Urinary bladder tumor cell lines and primary tumor tissues | Invasive tumor | [36] | |
| Overexpressed, Phosphorylated | Dysplastic oral epithelia tissues, squamous cell carcinoma of head and neck tissues and cell lines | Increase in cell proliferation, squamous carcinoma | [37] | |
| Overexpressed | Normal pancreas, pancreatic cancer tissues | Pancreatic cancers | [39] | |
| Overexpressed | Hyperplastic enlarged lobular units of precancerous breast lesion | Increase in cell proliferation | [27] | |
| PKC lambda/iota | Gene amplified, protein overexpressed and mislocalized | Ovarian cancer | Ovarian cancer, low survival rate | [16,32] |
| Overexpressed, Phosphorylated | Hepatocellular carcinoma tissues | E cadherin reduction, metastasis and invasion | [33] | |
| Overexpressed | Non small cell lung cancer cell lines, primary tumor tissues | Cell transformation, poor prognosis | [34] | |
| Overexpressed | Primary breast cancer tissues | Larger tumor, invasion, metastasis | [38] | |
| Organ structure specific regulation | Normal pancreas, pancreatic cancer tissues | Pancreatic cancers | [39] | |
| Scribble | Mislocalized or downregulated | HPV positive cervical high grade squamous intraepithelial lesions, invasive cervical cancer | Cervical cancers | [40] |
| Mislocalized or downregulated | Neoplastic colon mucosa | Colon adenocarcinoma | [41] | |
| Mislocalized or downregulated | Human breast cancer tissues | Breast cancer | [14] | |
| Dlg | Mislocalized or downregulated | High grade uterine cervical neoplasm | Cervical cancers | [42] |
| Mislocalized or downregulated | Neoplastic colon mucosa | Colon adenocarcinoma | [41] | |
| Upregulated | Progestin treated MCF7 cells | Cell growth inhibition | [78] | |
| Lgl | Downregulated | Tumor tissues of breast, prostate, lung, ovary | Carcinoma | [43] |
| Downregulated | Colon cancer lines, colon cancer primary tissues | Colon cancers, adenoma | [44] | |
| Aberrantly spliced tmRNA, truncated protein | Hepatocellular carcinoma cell line and primary tissues | Poor differentiation, larger tumor size | [46] | |
| Mislocalization, downregulation | Gastric epithelial dysplasia, adenocarcinoma | Tissue disruption, cancers | [45] | |
| Prickle-1 | Downregulated | hHepatocellular carcinoma cell line and primary tissues | High β-catenin activity, cell overproliferation | [47] |
| hNkd | Overexpressed | Colon cancer lines, colon cancer primary tissues | Not determined | [48] |
| Overexpressed | Hepatocellular carcinoma primary tissues | Antagonizes Wnt3a activation | [49] | |
| Wnt5a | Overexpressed | Thyroid cancer cell lines and primary tumor tissues | Overexpression inhibits cell proliferation, motility and invasions | [51] |
| Downregulated | Invasive ductal carcinoma and invasive lobular carcinoma of the breast | High tumor grade and tumor recurrence of invasive ductal carcinomas | [52] | |
| Overexpressed | Macrophages in breast cancers, | Increases MMP-7 expression in breast cancer cells, increase invasion | [79] | |
| LKB1 (Par-4) | Mutations (Rare) | Colorectal cancer, testicular tumor, cervical adenocarcinoma, lung adenocarcinoma, | Cancer | [80,81] |
| Promoter hypermethylation (Rare) | Colorectal cancer, testicular tumor, | Cancer | [82] | |
| Downregulation | Breast cancer | Breast cancer, poor prognosis | [83] |
Members of apical polarity complex are altered in multiple cancers. The Par3 gene is deleted in 15% of primary esophageal squamous cell carcinoma (ESCC) and the mRNA levels are downregulated in 23 of 33 ESCC tumor tissues compared to matched normal [26]. Par6β mRNA levels are upregulated in ER-positive primary breast tumors and in ER-positive breast tumor cell lines [27]. In non-small cell lung cancers (NSCLCs), Par6 is highly expressed in stromal cells and correlates with good prognosis [28]. Downregulation of Crumbs3 is required for transformation and tumorigenesis of immortalized baby rat kidney (BRK) cells and re-expression of Crumbs restores polarity and inhibits invasion of BRK cells [29] demonstrating that loss of Crumbs3 is causally linked to the transformation process. Cancer genome sequencing efforts are likely to identify additional cancer-associated genetic alterations in polarity proteins. For example, peering into data generated from a transcriptome sequencing study shows the presence of a novel fusion between the transcription factor ZNF667 and the polarity protein Par6β [30]. Careful mining into the data generated by the cancer genome atlas efforts is likely to identify novel genetic alterations associated with polarity proteins.
It is likely that aPKC isoforms play cell type specific roles in cancer. In androgen insensitive rat prostate cancer lines, PKCζ mRNA is downregulated [31] whereas in human liver cancer, ovarian cancer and non-small cell lung cancers cancer mRNA levels of PKCλ/ι are upregulated [32-34]. High levels of PKCζ proteins are observed in hepatocellular carcinomas, bladder transitional carcinomas, dysplastic oral epithelia and squamous cell carcinomas of the head and neck [35-37]. PKCλ/ι proteins are increased in breast cancers, ovarian cancer, liver cancer and non-small cell lung cancer [32-34,38]. In breast cancer, overexpression and mislocalization of PKCι from the apical membrane is correlated with invasive disease [38]. PKCι gene is amplified and the protein is mislocalized in ovarian cancers [16,32]. Atypical PKC also shows intra-tumor heterogeneity. For example, PKCζ is overexpressed in both ductal and ampullary pancreatic cancers, whereas, PKCι is overexpressed only in major ducts of ductal cancers and ampullary cancers but downregulated in minor ductules. Furthermore, in the stroma, PKCζ, but not PKCι, is overexpressed [39]. Together these studies suggest that the role played by polarity proteins in tumor progression is likely to be context-dependent.
Members of the basolateral polarity complex are also altered at the level of protein localization in cancers. The Scribble polarity protein is altered at multiple levels in human cancers. The mRNA levels are increased in 37.5% and reduced in 53.1% of breast cancer compared to normal tissue samples and the protein is mislocalized in almost 50% of DCIS lesions [14]. In HPV positive cervical high-grade squamous intraepithelial lesions and invasive cancers, Scribble protein levels are reduced and cellular distribution is disrupted [40]. In colon carcinomas, Scribble is mislocalized and expression downregulated in poorly differentiated adenocarcinomas [41]. Dlg expression levels and basolateral localizations are altered in colon cancers, high-grade premalignant cervical neoplasias and invasive squamous carcinomas [41,42]. Lgl-1 transcripts are lost in tumor tissues of the breast (76%), prostate (53%), lung (63%), ovarian (50%) and colon carcinomas (75%) [43]. Re-expression of Lgl-1in 293 cells increases cell adhesions [44] suggesting that loss of Lgl-1 is causally linked to loss of cell adhesion. Lgl2 is either negative or mainly cytoplasmic in gastric carcinomas [45]. In addition to changes in mRNA levels, polarity protein can also be regulated by alternative splicing. Lu et al report that aberrant splicing of Lgl-1 transcripts is observed in hepatocellular carcinomas [46]. The truncated Lgl-1 forms are correlated with large tumor sizes.
Members of the PCP complex are also altered in human cancers. In several hepatocellular carcinoma cell lines and primary tumors, Prickle-1 mRNA level is downregulated and correlates with increased expression of Dishevelled and β-catenin [47]. Human homolog of Drosophila Naked Cuticle (hNkd), an antagonist of the canonical Wnt signaling, is overexpressed in 72% of primary colon tumors [48] and all of 23 hepatocellular carcinoma samples analyzed [49]. Wnt5a, a regulator of PCP [50], is overexpressed in thyroid carcinoma cell lines and tumor tissues [51] but lost in 44% of invasive ductal carcinomas (IDC) and 24% of invasive lobular carcinomas (ILC) of the breast [52]. Moreover, in IDC but not in ILC, loss of Wnt5a is correlated with higher grade of cancer as well as tumor recurrence.
Together the data highlighted above demonstrate that functions of polarity proteins are regulated at several levels. They are amplified or overexpressed, deleted or underexpressed, mRNA alternatively spliced and protein mislocalized from their respective intracellular location. Thus, in addition to assessing genomic alterations and expression changes, monitoring changes in protein subcellular localization should be an important component of the studies aimed at analyzing changes in polarity proteins in cancer.
Cell polarity proteins as regulators of cancer relevant cell biological processes
In this section, we will discuss how polarity pathways can also regulate multiple biological processes related to tumor initiation and progression (see Figure 2), in addition to their known role in cell junction biogenesis.
Figure 2.
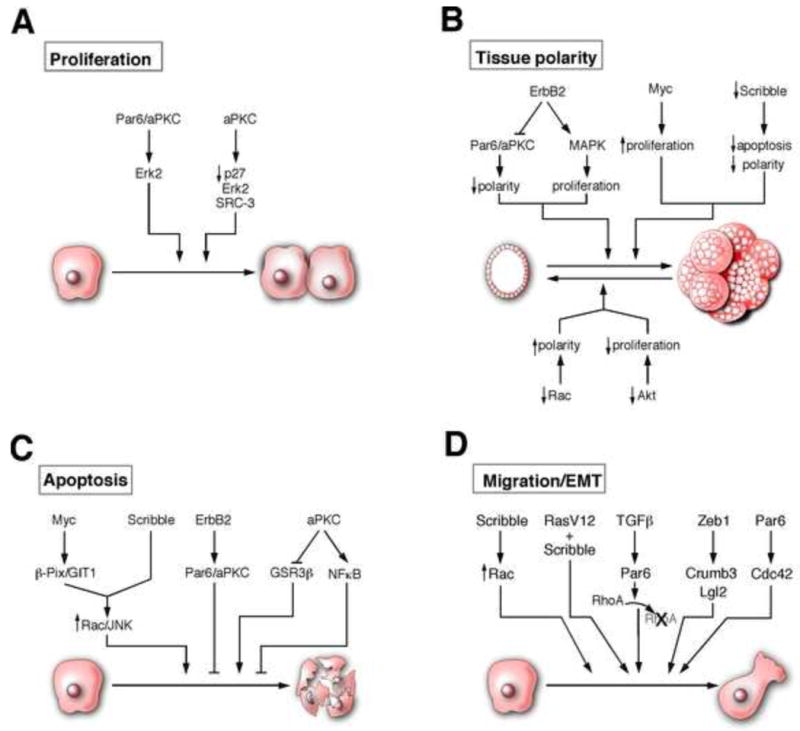
Emerging roles for polarity proteins: A, polarity pathways regulate cell proliferation [27,54,55]. B, polarity pathways regulate apoptosis [14,59,62]. C, polarity proteins regulate tissue polarity [58,63,64,66,68,77]. D, polarity pathways regulate cell migration and EMT [13,71-73,75].
Proliferation
There is increasing evidence to suggest that polarity proteins can also function as regulators of cell proliferation. Qiu et al discovered that Par6 mediates the interaction between Rac1/Cdc42 and PKCζ [53]. Par6 and PKCζ are required for Rac1 and Cdc42, but not Raf, induced transformation of NIH3T3 cells. We have shown that overexpression of Par6 induces growth factor-independent proliferation of human mammary epithelial cells by promoting sustained activation of Mitogen activated protein kinase (MAPK) in an aPKC and Cdc42/Rac-binding domain dependent manner [27]. Furthermore, downregulation of aPKC in MCF-7 breast cancer cell line inhibits cell proliferation demonstrating that the Par6/aPKC module is likely to be an important regulator of cell proliferation in both non-tumorigenic and tumor-derived mammary epithelial cells. Atypical PKCs can regulate cell cycle progression by multiple mechanisms: PKCζ is required for estrogen-induced nuclear transport of Erk2 and degradation of p27 [54]. The aPKC can also promote tumor growth by stabilizing the Steroid receptor coactivator 3 (SRC-3), known regulator of cell proliferation [55]. Phosphorylation of SRC-3 protein by PKCζ stabilizes the protein by inhibiting interaction between SRC-3 and the C8 subunit of 20s proteasome [55].
Scribble, Dlg and Lgl were shown to regulate G1-S transition in a Drosophila genetic suppressor screen for novel regulators of G1-S transition [56]. Expression of the PCP gene Prickle1 inhibits growth of hepatocellular carcinoma cells by promoting degradation of Dishevelled 3, a positive regulator of Wnt signaling [47]. However, further investigation will be necessary to understand if these polarity proteins directly regulate cell cycle.
Apoptosis
Evasion of apoptosis is a critical step during cancer progression [57]. We recently reported a novel role for Scribble as a regulator of apoptosis in mammary epithelial cells. Both downregulation of Scribble protein and mislocalization of Scribble from cell-cell junctions inhibited lumen formation during three-dimensional (3D) morphogenesis of mammary epithelial cells and during Myc-induced transformation and tumorigenesis. Thus, proper localization of Scribble at cell-cell junctions is critical for normal morphogenesis and to resist Myc-induced transformation [14]. However, clonal analysis of cells lacking Scribble in Drosophila eye imaginal disc demonstrates that loss of Scribble triggers a JNK-dependent cell death [56]. Although these results seem contradictory, they could be explained by differences in cell autonomous (in mammalian studies) versus non-autonomous (Drosophila clonal studies) effects of Scribble loss. These observations highlight the need for a better understanding of the relationship between Scribble and regulation of cell death.
Atypical PKC also regulates apoptosis in epithelial cells. We reported that the Par6/aPKC complex was required for ErbB2-induced inhibition of cell death [58] in MCF-10A cells. Inhibition of aPKC significantly increases apoptosis during 3D cysts morphogenesis of MDCK cells by activating GSK3β [59]. Atypical PKC also inhibits apoptosis by activating the NFκB pathway either by phosphorylating and activating IKKβ kinase or by phosphorylating RelA subunit of NFκB to activate transcription [60,61]. Atypical PKC can also function downstream of another protein Prostate Apoptosis Response-4 (Par-4, different from the Partitioning Defective proteins that regulate epithelial cell polarity) to regulate apoptosis [62]. Par-4 binds and inhibits aPKC activity resulting in low NFκB activation. Loss of Par-4 results in activation of aPKC and increased expression of an inhibitor of apoptosis, XIAP, an NFκB target. Thus polarity proteins are a novel class of regulators of apoptosis in cancer.
Tissue polarity
Changes in cell size, cell shape, and cell organization within the 3D space of tissue lead to a loss in tissue polarity and are known to occur very early and worsen during the progression of carcinoma. Homotypic interactions between epithelial cells and heterotypic interactions between epithelia and stromal cell types and interactions with surrounding extracellular matrix will influence the ability of epithelial cells to organize in a 3D space and develop tissue polarity. The molecular mechanisms that regulate establishment of normal tissue polarity and how they are disrupted in cancer are only beginning to be understood. Culture conditions where epithelial cells grown on 3D matrices to undergo morphogenesis and form cysts or acini are useful platforms to begin to understand how epithelial cells establish the normal multicellular organization and develop tissue polarity. Several studies have identified critical roles for cell junctions and cell-matrix interaction during the 3D morphogenesis of epithelial structures (For detailed reviews on this topic please refer to [6,63,64]). In addition, cell polarity proteins Crumbs, PATJ, Par3, Par6, aPKC, Cdc42, and Scribble have been demonstrated to regulate normal epithelial morphogenesis and establishment of tissue polarity (for details please see [59,65,66]). The precise mechanisms by which cell polarity pathways help regulate development of tissue polarity and how alterations in polarity pathways deregulate 3D tissue organization and polarity remain to be understood.
The mechanisms by which oncogenic events induce disruption of polarized, 3D organized epithelial structures are only beginning to be understood. Here again, use of epithelial cells grown in 3D matrices is beginning to reveal novel insights that were not apparent from using traditional cell culture platforms. For example, under three dimensional culture conditions, normal mammary epithelia form structures with normal tissue polarity and growth control whereas tumor derived cells grow as unorganized mass with no apparent tissue polarity or growth control [67]. We demonstrated that the oncogene ErbB2 requires an interaction with cell polarity proteins Par6/aPKC to disrupt tissue polarity of acini derived from normal mammary epithelia [58]. The interaction with Par6/aPKC is required for the ability of ErbB2 to disrupt apical-basal polarity but not required for ErbB2-induced cell proliferation [58]. The distinct role for polarity and proliferation is also true when one tries to restore tissue polarity to cancer-derived cell lines. Inhibition of both Rac mediated regulation of cell polarity and Akt mediated cell proliferation is required to make breast cancer-derived cells form normal acini-like structures in 3D matrix [68]. Taken together, the above observations demonstrate that polarity pathways are critical regulators of tissue polarity loss observed in carcinoma. It is likely that a deeper analysis of the mechanisms by which cell and tissue polarity pathways are altered in cancer and how they cooperate with oncogenes to transform polarized epithelia will identify novel diagnostic and therapeutic strategies.
Cell migration and epithelial-mesenchymal transition
Polarity proteins are regulators of front-back polarity in migrating cells. For example, Par6 is required for orienting microtubule organizing centers towards the leading edge in migrating astrocytes [69]. Several polarity proteins including Par3, Par6, aPKC, Scribble, Dlg and Lgl are localized to the leading edge of migrating cells and play critical roles during directional migration (for a detailed review see [70]).
Loss of epithelial characteristics and gain of mesenchymal properties are frequently associated with metastatic progression. Migration/invasion of cells is always associated with dramatic changes in cell architecture suggesting that cell polarity proteins are likely to play important roles during this process. Consistent with this notion, genetic screen in Drosophila identified mutation in several polarity proteins including Scribble, Dlg, Lgl, Baz, and Cdc42 that cooperate with RasV12 to promote invasion and metastasis [71]. The role played by polarity proteins during epithelial to mesenchymal transition (EMT) and invasion of mammalian epithelia is beginning to be unraveled. The Par6 polarity protein plays a critical role during TGFβ-induced EMT. TGFβ type II receptor phosphorylates Par6 to promote assembly of a protein complex containing Par6, Smurf1 (an ubiquitin ligase) and RhoA to promote localized degradation of RhoA [72]. Phosphorylation of Par6 and interaction with Smurf1 are required for TGFβ-induced disruption of tight junctions and migration/invasion of NMuMG cells in culture and in vivo [72,73] demonstrating a critical role for the polarity protein Par6 in breast cancer metastasis.
Transcriptional regulators of EMT target polarity proteins. In breast and colorectal cancer cells the transcriptional repressor ZEB1 inhibits expression of polarity proteins Crumbs, Lgl2 and PATJ to induce mesenchymal phenotypes [74], and re-expression of Lgl2 restores epithelial cell polarity and inhibits invasion and metastasis [75] in these cells. In addition to breast and colon, loss of Lgl2 is also associated with EMT in melanoma-derived cells and restoration of Lgl2 restores epithelial cell polarity and blocks cell migration [76]. These studies identify polarity proteins as important regulators of the EMT process in cells derived from multiple organs.
Conclusion and Perspective
We have attempted to summarize the changes of polarity protein observed in human carcinomas. As outlined above, polarity proteins are not only targets of genetic and epigenetic changes but are also regulated at the level of subcellular localization. These findings highlight the need for analyzing subcellular localization, in addition to gene expression changes, when studying polarity proteins in carcinoma. There is an emerging body of evidence implicating polarity proteins as regulators of multiple cell biological properties in addition to cell junction biogenesis. Thus, it is critical to use appropriate biological context for understanding the consequences of polarity protein alterations in cancer. A better and deeper analysis of changes in polarity proteins and the pathways they regulate is likely to identify a new class of biomarkers and drug targets for diagnosing and controlling cancer.
Acknowledgments
We would like to thank Jim Duffy (Cold Spring Harbor Laboratory) for assistance with graphics. SKM is funded by grants from NCI (CA098830), US Army DOD (BC075024) and Lee K. and Margaret Lau Chair in Breast Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kumar V, Abbsa A, Fausto N, editors. Pathologic Basis of Disease. 7th. Elsevier Saunders; 1999. Neoplasia. [Google Scholar]
- 2.Ivanov AI, Nusrat A, Parkos CA. Endocytosis of the Apical Junctional Complex: Mechanisms and Possible Roles in Regulation of Epithelial Barriers. Bioessays. 2005;27:356–365. doi: 10.1002/bies.20203. [DOI] [PubMed] [Google Scholar]
- 3.Simons M, Mlodzik M. Planar Cell Polarity Signaling: From Fly Development to Human Disease. Annu Rev Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Knoblich JA. Mechanisms of Asymmetric Stem Cell Division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Iden S, Collard JG. Crosstalk between Small Gtpases and Polarity Proteins in Cell Polarization. Nat Rev Mol Cell Bio. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- 6.Shin K, Fogg VC, Margolis B. Tight Junctions and Cell Polarity. Annu Rev Cell Dev Bi. 2006;22:207–235. doi: 10.1146/annurev.cellbio.22.010305.104219. [DOI] [PubMed] [Google Scholar]
- 7.Opper M, Schuler G, Mechler BM. Hereditary Suppression of Lethal (2) Giant Larvae Malignant Tumor Development in Drosophila by Gene Transfer. Oncogene. 1987;1:91–96. [PubMed] [Google Scholar]
- 8.Woods DF, Bryant PJ. The Discs-Large Tumor Suppressor Gene of Drosophila Encodes a Guanylate Kinase Homolog Localized at Septate Junctions. Cell. 1991;66:451–464. doi: 10.1016/0092-8674(81)90009-x. [DOI] [PubMed] [Google Scholar]
- 9.Bilder D, Li M, Perrimon N. Cooperative Regulation of Cell Polarity and Growth by Drosophila Tumor Suppressors. Science. 2000;289:113–116. doi: 10.1126/science.289.5476.113. [DOI] [PubMed] [Google Scholar]
- 10.Humbert PO, Dow LE, Russell SM. The Scribble and Par Complexes in Polarity and Migration: Friends or Foes? Trends Cell Biol. 2006;16:622–630. doi: 10.1016/j.tcb.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 11.Etienne-Manneville S, Hall A. Cell Polarity: Par6, Apkc and Cytoskeletal Crosstalk. Curr Opin Cell Biol. 2003;15:67–72. doi: 10.1016/s0955-0674(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 12.Parkinson SJ, Le Good JA, Whelan RDH, Whitehead P, Parker PJ. Identification of Pkczetaii: An Endogenous Inhibitor of Cell Polarity. EMBO J. 2004;23:77–88. doi: 10.1038/sj.emboj.7600023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dow LE, Elsum IA, King CL, Kinross KM, Richardson HE, Humbert PO. Loss of Human Scribble Cooperates with H-Ras to Promote Cell Invasion through Deregulation of Mapk Signalling. Oncogene. 2008;27:5988–6001. doi: 10.1038/onc.2008.219. [DOI] [PubMed] [Google Scholar]
- **14.Zhan L, Rosenberg A, Bergami KC, Yu M, Xuan Z, Jaffe AB, Allred C, Muthuswamy SK. Deregulation of Scribble Promotes Mammary Tumorigenesis and Reveals a Role for Cell Polarity in Carcinoma. Cell. 2008;135:865–878. doi: 10.1016/j.cell.2008.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate that loss or mislocalization of the polarity protein Scribble in mammary epithelial cells blocks tissue polarity during 3D morphogenesis. Cooperates with Myc during transformation of mammary epithelial cells and during mammary tumorigenesis by inhibiting Myc-induced apoptosis.
- 15.Yaffe MB, Smerdon SJ. The Use of in Vitro Peptide-Library Screens in the Analysis of Phosphoserine/Threonine-Binding Domain Structure and Function. Annu Rev Biophys Biomol Struct. 2004;33:225–244. doi: 10.1146/annurev.biophys.33.110502.133346. [DOI] [PubMed] [Google Scholar]
- 16.Grifoni D, Garoia F, Bellosta P, Parisi F, De Biase D, Collina G, Strand D, Cavicchi S, Pession A. Apkczeta Cortical Loading Is Associated with Lgl Cytoplasmic Release and Tumor Growth in Drosophila and Human Epithelia. Oncogene. 2007;26:5960–5965. doi: 10.1038/sj.onc.1210389. [DOI] [PubMed] [Google Scholar]
- 17.Plant PJ, Fawcett JP, Lin DCC, Holdorf AD, Binns K, Kulkarni S, Pawson T. A Polarity Complex of Mpar-6 and Atypical Pkc Binds, Phosphorylates and Regulates Mammalian Lgl. Nat Cell Biol. 2003;5:301–308. doi: 10.1038/ncb948. [DOI] [PubMed] [Google Scholar]
- 18.Bilder D, Perrimon N. Localization of Apical Epithelial Determinants by the Basolateral Pdz Protein Scribble. Nature. 2000;403:676–680. doi: 10.1038/35001108. [DOI] [PubMed] [Google Scholar]
- 19.Dollar GL, Weber U, Mlodzik M, Sokol SY. Regulation of Lethal Giant Larvae by Dishevelled. Nature. 2005;437:1376–1380. doi: 10.1038/nature04116. [DOI] [PubMed] [Google Scholar]
- 20.Albertson R, Chabu C, Sheehan A, Doe CQ. Scribble Protein Domain Mapping Reveals a Multistep Localization Mechanism and Domains Necessary for Establishing Cortical Polarity. J Cell Sci. 2004;117:6061–6070. doi: 10.1242/jcs.01525. [DOI] [PubMed] [Google Scholar]
- 21.Montcouquiol M, Rachel RA, Lanford PJ, Copeland NG, Jenkins NA, Kelley MW. Identification of Vangl2 and Scrb1 as Planar Polarity Genes in Mammals. Nature. 2003;423:173–177. doi: 10.1038/nature01618. [DOI] [PubMed] [Google Scholar]
- 22.Qin Y, Capaldo C, Gumbiner BM, Macara IG. The Mammalian Scribble Polarity Protein Regulates Epithelial Cell Adhesion and Migration through E-Cadherin. J Cell Biol. 2005;171:1061–1071. doi: 10.1083/jcb.200506094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun Y, Aiga M, Yoshida E, Humbert PO, Bamji SX. Scribble Interacts with Beta-Catenin to Localize Synaptic Vesicles to Synapses. Mol Biol Cell. 2009;20:3390–3400. doi: 10.1091/mbc.E08-12-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albertson R, Doe CQ. Dlg, Scrib and Lgl Regulate Neuroblast Cell Size and Mitotic Spindle Asymmetry. Nat Cell Biol. 2003;5:166–170. doi: 10.1038/ncb922. [DOI] [PubMed] [Google Scholar]
- 25.Lee M, Vasioukhin V. Cell Polarity and Cancer - Cell and Tissue Polarity as a Non-Canonical Tumor Suppressor. J Cell Sci. 2008;121:1141–1150. doi: 10.1242/jcs.016634. [DOI] [PubMed] [Google Scholar]
- 26.Zen K, Yasui K, Gen Y, Dohi O, Wakabayashi N, Mitsufuji S, Itoh Y, Zen Y, Nakanuma Y, Taniwaki M, et al. Defective Expression of Polarity Protein Par-3 Gene (Pard3) in Esophageal Squamous Cell Carcinoma. Oncogene. 2009 doi: 10.1038/onc.2009.148. [DOI] [PubMed] [Google Scholar]
- *27.Nolan ME, Aranda V, Lee S, Lakshmi B, Basu S, Allred DC, Muthuswamy SK. The Polarity Protein Par6 Induces Cell Proliferation and Is Overexpressed in Breast Cancer. Cancer Res. 2008;68:8201–8209. doi: 10.1158/0008-5472.CAN-07-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors report that Par6 cooperates with aPKC and Cdc42 to activated ERK signaling and increase cell proliferation in breast cancer cells.
- 28.Al-Saad S, Al-Shibli K, Donnem T, Persson M, Bremnes RM, Busund LT. The Prognostic Impact of Nf-Kappab P105, Vimentin, E-Cadherin and Par6 Expression in Epithelial and Stromal Compartment in Non-Small-Cell Lung Cancer. Br J Cancer. 2008;99:1476–1483. doi: 10.1038/sj.bjc.6604713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *29.Karp CM, Tan TT, Mathew R, Nelson D, Mukherjee C, Degenhardt K, Karantza-Wadsworth V, White E. Role of the Polarity Determinant Crumbs in Suppressing Mammalian Epithelial Tumor Progression. Cancer Res. 2008;68:4105–4115. doi: 10.1158/0008-5472.CAN-07-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors demonstrate that loss of Crumbs is selected for during transformation and tumorigenesis in mice and show that re-expression of Crumbs can restores cell junctions, induces contact inhibition growth and suppression of metastasis.
- *30.Maher CA, Kumar-Sinha C, Cao XH, Kalyana-Sundaram S, Han B, Jing XJ, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome Sequencing to Detect Gene Fusions in Cancer. Nature. 2009;458:97–U99. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors report the presence of a fusion between of Par6-beta and the transcription factor ZNF667 in human tumors.
- 31.Powell CT, Fair WR, Heston WD. Differential Expression of Protein Kinase C Isozyme Messenger Rnas in Dunning R-3327 Rat Prostatic Tumors. Cell Growth Differ. 1994;5:143–149. [PubMed] [Google Scholar]
- 32.Eder AM, Sui X, Rosen DG, Nolden LK, Cheng KW, Lahad JP, Kango-Singh M, Lu KH, Warneke CL, Atkinson EN, et al. Atypical Pkci Contributes to Poor Prognosis through Loss of Apical-Basal Polarity and Cyclin E Overexpression in Ovarian Cancer. P Natl Acad Sci USA. 2005;102:12519–12524. doi: 10.1073/pnas.0505641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du GS, Wang JM, Lu JX, Li Q, Ma CQ, Du JT, Zou SQ. Expression of P-Apkc-Iota, E-Cadherin, and Beta-Catenin Related to Invasion and Metastasis in Hepatocellular Carcinoma. Ann Surg Oncol. 2009;16:1578–1586. doi: 10.1245/s10434-009-0423-7. [DOI] [PubMed] [Google Scholar]
- 34.Regala RP, Weems C, Jamieson L, Khoor A, Edell ES, Lohse CM, Fields AP. Atypical Protein Kinase C Iota Is an Oncogene in Human Non-Small Cell Lung Cancer. Cancer Res. 2005;65:8905–8911. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- 35.Tsai JH, Hsieh YS, Kuo SJ, Chen ST, Yu SY, Huang CY, Chang AC, Wang YW, Tsai MT, Liu JY. Alteration in the Expression of Protein Kinase C Isoforms in Human Hepatocellular Carcinoma. Cancer Lett. 2000;161:171–175. doi: 10.1016/s0304-3835(00)00597-8. [DOI] [PubMed] [Google Scholar]
- 36.Langzam L, Koren R, Gal R, Kugel V, Paz A, Farkas A, Sampson SR. Patterns of Protein Kinase C Isoenzyme Expression in Transitional Cell Carcinoma of Bladder. Relation to Degree of Malignancy. Am J Clin Pathol. 2001;116:377–385. doi: 10.1309/1VKK-HWH7-YVJN-7UF7. [DOI] [PubMed] [Google Scholar]
- 37.Cohen EEW, Lingen MW, Zhu B, Zhu H, Straza MW, Pierce C, Martin LE, Rosner MR. Protein Kinase C Zeta Mediates Epidermal Growth Factor-Induced Growth of Head and Neck Tumor Cells by Regulating Mitogen-Activated Protein Kinase. Cancer Res. 2006;66:6296–6303. doi: 10.1158/0008-5472.CAN-05-3139. [DOI] [PubMed] [Google Scholar]
- 38.Kojima Y, Akimoto K, Nagashima Y, Ishiguro H, Shirai S, Chishima T, Ichikawa Y, Ishikawa T, Sasaki T, Kubota Y, et al. The Overexpression and Altered Localization of the Atypical Protein Kinase C Lambda/Iota in Breast Cancer Correlates with the Pathologic Type of These Tumors. Hum Pathol. 2008;39:824–831. doi: 10.1016/j.humpath.2007.11.001. [DOI] [PubMed] [Google Scholar]
- *39.Evans JD, Cornford PA, Dodson A, Neoptolemos JP, Foster CS. Expression Patterns of Protein Kinase C Isoenzymes Are Characteristically Modulated in Chronic Pancreatitis and Pancreatic Cancer. Am J Clin Pathol. 2003;119:392–402. doi: 10.1309/bkpc9dx98r781b87. [DOI] [PubMed] [Google Scholar]; The authors use immunohistochemistry to detect the expression levels and cellular localizations of two aPKC isoforms (PKC iota and PKC zeta) in pancreas. Expression and localization patterns of the two isoforms show heterogeneity with the pancreas.
- 40.Nakagawa S, Yano T, Nakagawa K, Takizawa S, Suzuki Y, Yasugi T, Huibregtse JM, Taketani Y. Analysis of the Expression and Localisation of a Lap Protein, Human Scribble, in the Normal and Neoplastic Epithelium of Uterine Cervix. Brit J Cancer. 2004;90:194–199. doi: 10.1038/sj.bjc.6601465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gardiol D, Zacchi A, Petrera F, Stanta G, Banks L. Human Discs Large and Scrib Are Localized at the Same Regions in Colon Mucosa and Changes in Their Expression Patterns Are Correlated with Loss of Tissue Architecture During Malignant Progression. Int J Cancer. 2006;119:1285–1290. doi: 10.1002/ijc.21982. [DOI] [PubMed] [Google Scholar]
- 42.Lin HT, Steller MA, Aish L, Hanada T, Chishti AH. Differential Expression of Human Dlg in Cervical Intraepithelial Neoplasias. Gynecol Oncol. 2004;93:422–428. doi: 10.1016/j.ygyno.2004.01.025. [DOI] [PubMed] [Google Scholar]
- *43.Grifoni D, Garoia F, Schimanski CC, Schmitz G, Laurenti E, Galle PR, Pession A, Cavicchi S, Strand D. The Human Protein Hugl-1 Substitutes for Drosophila Lethal Giant Larvae Tumour Suppressor Function in Vivo. Oncogene. 2004;23:8688–8694. doi: 10.1038/sj.onc.1208023. [DOI] [PubMed] [Google Scholar]; The authors observe the expression of human Lgl1 is lost in solid tumors from human breast, prostate, lung and ovary.
- 44.Schimanski CC, Schmitz G, Kashyap A, Bosserhoff AK, Bataille F, Schäfer SC, Lehr HA, Berger MR, Galle PR, Strand S, et al. Reduced Expression of Hugl-1, the Human Homologue of Drosophila Tumour Suppressor Gene Lgl, Contributes to Progression of Colorectal Cancer. Oncogene. 2005;24:3100–3109. doi: 10.1038/sj.onc.1208520. [DOI] [PubMed] [Google Scholar]
- 45.Lisovsky M, Dresser K, Baker S, Fisher A, Woda B, Banner B, Lauwers GY. Cell Polarity Protein Lgl2 Is Lost or Aberrantly Localized in Gastric Dysplasia and Adenocarcinoma: An Immunohistochemical Study. Mod Pathol. 2009;22:977–984. doi: 10.1038/modpathol.2009.68. [DOI] [PubMed] [Google Scholar]
- *46.Lu X, Feng X, Man X, Yang G, Tang L, Du D, Zhang F, Yuan H, Huang Q, Zhang Z, et al. Aberrant Splicing of Hugl-1 Is Associated with Hepatocellular Carcinoma Progression. Clin Cancer Res. 2009;15:3287–3296. doi: 10.1158/1078-0432.CCR-08-2078. [DOI] [PubMed] [Google Scholar]; The authors detected abnormal splicing of human Lgl1 in liver cancers and show that expressions of these aberrant spliced forms enhance the growth of hepatocellular tumors while wild type Lgl1 inhibits tumor growth.
- 47.Chan DW, Chan CY, Yam JWP, Ching YP, Ng IOL. Prickle-1 Negatively Regulates Wnt/Beta-Catenin Pathway by Promoting Dishevelled Ubiquitination/Degradation in Liver Cancer. Gastroenterology. 2006;131:1218–1227. doi: 10.1053/j.gastro.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 48.Yan D, Wiesmann M, Rohan M, Chan V, Jefferson AB, Guo L, Sakamoto D, Caothien RH, Fuller JH, Reinhard C, et al. Elevated Expression of Axin2 and Hnkd Mrna Provides Evidence That Wnt/Beta -Catenin Signaling Is Activated in Human Colon Tumors. P Natl Acad Sci USA. 2001;98:14973–14978. doi: 10.1073/pnas.261574498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koch A, Waha A, Hartmann W, Hrychyk A, Schüller U, Waha A, Wharton KA, Fuchs SY, Von Schweinitz D, Pietsch T. Elevated Expression of Wnt Antagonists Is a Common Event in Hepatoblastomas. Clin Cancer Res. 2005;11:4295–4304. doi: 10.1158/1078-0432.CCR-04-1162. [DOI] [PubMed] [Google Scholar]
- 50.Qian D, Jones C, Rzadzinska A, Mark S, Zhang X, Steel KP, Dai X, Chen P. Wnt5a Functions in Planar Cell Polarity Regulation in Mice. Dev Biol. 2007;306:121–133. doi: 10.1016/j.ydbio.2007.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kremenevskaja N, Von Wasielewski R, Rao AS, Schöfl C, Andersson T, Brabant G. Wnt-5a Has Tumor Suppressor Activity in Thyroid Carcinoma. Oncogene. 2005;24:2144–2154. doi: 10.1038/sj.onc.1208370. [DOI] [PubMed] [Google Scholar]
- 52.Jönsson M, Dejmek J, Bendahl PO, Andersson T. Loss of Wnt-5a Protein Is Associated with Early Relapse in Invasive Ductal Breast Carcinomas. Cancer Res. 2002;62:409–416. [PubMed] [Google Scholar]
- **53.Qiu RG, Abo A, Martin GS. A Human Homolog of the C. Elegans Polarity Determinant Par-6 Links Rac and Cdc42 to PKC Zeta Signaling and Cell Transformation. Curr Biol. 2000;10:697–707. doi: 10.1016/s0960-9822(00)00535-2. [DOI] [PubMed] [Google Scholar]; The authors show that Par6 physically links Cdc42 and Rac to aPKC and this interaction is required for cell transformation by Rac.
- 54.Castoria G, Migliaccio A, Di Domenico M, Lombardi M, De Falco A, Varricchio L, Bilancio A, Barone MV, Auricchio F. Role of Atypical Protein Kinase C in Estradiol-Triggered G1/S Progression of Mcf-7 Cells. Mol Cell Biol. 2004;24:7643–7653. doi: 10.1128/MCB.24.17.7643-7653.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yi P, Feng Q, Amazit L, Lonard DM, Tsai SY, Tsai MJ, O'Malley BW. Atypical Protein Kinase C Regulates Dual Pathways for Degradation of the Oncogenic Coactivator Src-3/Aib1. Mol Cell. 2008;29:465–476. doi: 10.1016/j.molcel.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *56.Brumby AM, Richardson HE. Scribble Mutants Cooperate with Oncogenic Ras or Notch to Cause Neoplastic Overgrowth in Drosophila. EMBO J. 2003;22:5769–5779. doi: 10.1093/emboj/cdg548. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that mosaic loss of Scribble results in apoptosis of mutant cells whereas expression of of Ras or Notch inhibits apoptosis and result in neoplastic tumors.
- 57.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- *58.Aranda V, Haire T, Nolan ME, Calarco JP, Rosenberg AZ, Fawcett JP, Pawson T, Muthuswamy SK. Par6-Apkc Uncouples Erbb2 Induced Disruption of Polarized Epithelial Organization from Proliferation Control. Nat Cell Biol. 2006;8:1235–1245. doi: 10.1038/ncb1485. [DOI] [PubMed] [Google Scholar]; The authors show that activation of ErbB2 induces interaction between ErbB2 and Par6/aPKC. This interaction is required for ErbB2 induced disruption of apical-basal polarity and inhibition of apoptosis but not for cell proliferation.
- *59.Kim M, Datta A, Brakeman P, Yu W, Mostov KE. Polarity Proteins Par6 and Apkc Regulate Cell Death through GSK-3beta in 3d Epithelial Morphogenesis. J Cell Sci. 2007;120:2309–2317. doi: 10.1242/jcs.007443. [DOI] [PubMed] [Google Scholar]; The authors show that aPKC phosphorylates GSK-3beta and inhibits GSK-3beta dependent apoptosis of MDCK cells grown as 3D cysts. This function of aPKC is regulated by Par6.
- 60.Lallena MJ, Diaz-Meco MT, Bren G, Paya CV, Moscat J. Activation of Ikappa B Kinase Beta by Protein Kinase C Isoforms. Mol Cell Biol. 1999;19:2180–2188. doi: 10.1128/mcb.19.3.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duran A, Diaz-Meco MT, Moscat J. Essential Role of Rela Ser311 Phosphorylation by [Zeta]Pkc in Nf-[Kappa]B Transcriptional Activation. EMBO J. 2003;22:3910–3918. doi: 10.1093/emboj/cdg370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcia-Cao I, Lafuente MJ, Criado LM, Diaz-Meco MT, Serrano M, Moscat J. Genetic Inactivation of Par4 Results in Hyperactivation of Nf-Kappab and Impairment of Jnk and P38. EMBO Rep. 2003;4:307–312. doi: 10.1038/sj.embor.embor769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nelson WJ. Adaptation of Core Mechanisms to Generate Cell Polarity. Nature. 2003;422:766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Feigin ME, Muthuswamy SK. Polarity Proteins Regulate Mammalian Cell-Cell Junctions and Cancer Pathogenesis. Curr Opin Cell Biol. 2009;21:694–700. doi: 10.1016/j.ceb.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martín-Belmonte F, Yu W, Rodríguez-Fraticelli AE, Ewald A, Werb Z, Alonso MA, Mostov K. Cell-Polarity Dynamics Controls the Mechanism of Lumen Formation in Epithelial Morphogenesis. Curr Biol. 2008;18:507–513. doi: 10.1016/j.cub.2008.02.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *66.Hurd TW, Gao L, Roh MH, Macara IG, Margolis B. Direct Interaction of Two Polarity Complexes Implicated in Epithelial Tight Junction Assembly. Nat Cell Biol. 2003;5:137–142. doi: 10.1038/ncb923. [DOI] [PubMed] [Google Scholar]; The authors demonstrate physical interaction between Par6 and Pals1 which is regulated by Cdc42. The interaction between the two polarity proteins is required for the formation of tight junctions in MDCK cells.
- **67.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with Basement Membrane Serves to Rapidly Distinguish Growth and Differentiation Pattern of Normal and Malignant Human Breast Epithelial Cells. P Natl Acad Sci USA. 1992;89:9064–9068. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that while normal mammary epithelial cells for growth arrested organized multicellular structures when cultured on 3D basement membrane, Matrigel, cancer cells form disorganized multicellular aggregates with high proliferation rates.
- *68.Liu H, Radisky DC, Wang F, Bissell MJ. Polarity and Proliferation Are Controlled by Distinct Signaling Pathways Downstream of Pi3-Kinase in Breast Epithelial Tumor Cells. J Cell Biol. 2004;164:603–612. doi: 10.1083/jcb.200306090. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show restoration of normal tissue polarity in tumorigenic mammary epithelial cells require inhibition of Rac mediated changes in cell polarity and Akt medicated increase in cell proliferation.
- 69.Etienne-Manneville S, Hall A. Cdc42 Regulates Gsk-3[Beta] and Adenomatous Polyposis Coli to Control Cell Polarity. Nature. 2003;421:753–756. doi: 10.1038/nature01423. [DOI] [PubMed] [Google Scholar]
- *70.Etienne-Manneville S. Polarity Proteins in Migration and Invasion. Oncogene. 2008;27:6970–6980. doi: 10.1038/onc.2008.347. [DOI] [PubMed] [Google Scholar]; A Recent review summarizes findings on the roles of polarity proteins in regulating tumor cell migration and invasion.
- **71.Pagliarini RA, Xu T. A Genetic Screen in Drosophila for Metastatic Behavior. Science. 2003;302:1227–1231. doi: 10.1126/science.1088474. [DOI] [PubMed] [Google Scholar]; The authors use Drosophila eye disc as a model to understand tumor invasion. They report that loss of polarity proteins such as Scribble and Dlg cooperate with Ras during tumor invasion.
- **72.Wang HR, Zhang Y, Ozdamar B, Ogunjimi AA, Alexandrova E, Thomsen GH, Wrana JL. Regulation of Cell Polarity and Protrusion Formation by Targeting Rhoa for Degradation. Science. 2003;302:1775–1779. doi: 10.1126/science.1090772. [DOI] [PubMed] [Google Scholar]; The authors show that TGFb-induced invasion requires an interaction with Par/aPKC. Par6/aPKC recruits Smurf1 to cellular protrusions to promote localized degradation of RhoA to promote disruption of tight junctions and induce epithelial to mesenchymal transition.
- 73.Viloria-Petit AM, David L, Jia JY, Erdemir T, Bane AL, Pinnaduwage D, Roncari L, Narimatsu M, Bose R, Moffat J, et al. A Role for the Tgfbeta-Par6 Polarity Pathway in Breast Cancer Progression. P Natl Acad Sci USA. 2009 doi: 10.1073/pnas.0906796106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *74.Aigner K, Dampier B, Descovich L, Mikula M, Sultan A, Schreiber M, Mikulits W, Brabletz T, Strand D, Obrist P, et al. The Transcription Factor Zeb1 ([Delta]Ef1) Promotes Tumour Cell Dedifferentiation by Repressing Master Regulators of Epithelial Polarity. Oncogene. 2007;26:6979–6988. doi: 10.1038/sj.onc.1210508. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors show that ZEB1 induced epithelial to mesenchymal transition requires a ZEB1-mediated repression of Lgl2 expression. They also show that re-expression of Lgl2 restores epithelial characteristics.
- *75.Spaderna S, Schmalhofer O, Wahlbuhl M, Dimmler A, Bauer K, Sultan A, Hlubek F, Jung A, Strand D, Eger A, et al. The Transcriptional Repressor ZEB1 Promotes Metastasis and Loss of Cell Polarity in Cancer. Cancer Res. 2008;68:537–544. doi: 10.1158/0008-5472.CAN-07-5682. [DOI] [PubMed] [Google Scholar]; see notes for citation 74
- 76.Kuphal S, Wallner S, Schimanski CC, Bataille F, Hofer P, Strand S, Strand D, Bosserhoff AK. Expression of Hugl-1 Is Strongly Reduced in Malignant Melanoma. Oncogene. 2005;25:103–110. doi: 10.1038/sj.onc.1209008. [DOI] [PubMed] [Google Scholar]
- 77.Martin-Belmonte F, Mostov K. Regulation of Cell Polarity During Epithelial Morphogenesis. Curr Opin Cell Biol. 2008;20:227–234. doi: 10.1016/j.ceb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 78.Purmonen S, Ahola TM, Pennanen P, Aksenov N, Zhuang YH, Tuohimaa P, Ylikomi T. Hdlg5/Kiaa0583, Encoding a Maguk-Family Protein, Is a Primary Progesterone Target Gene in Breast Cancer Cells. Int J Cancer. 2002;102:1–6. doi: 10.1002/ijc.10665. [DOI] [PubMed] [Google Scholar]
- 79.Pukrop T, Klemm F, Hagemann T, Gradl D, Schulz M, Siemes S, Trümper L, Binder C. Wnt 5a Signaling Is Critical for Macrophage-Induced Invasion of Breast Cancer Cell Lines. P Natl Acad Sci USA. 2006;103:5454–5459. doi: 10.1073/pnas.0509703103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Avizienyte E, Roth S, Loukola A, Hemminki A, Lothe RA, Stenwig AE, Fossa SD, Salovaara R, Aaltonen LA. Somatic mutations in LKB1 are rare in sporadic colorectal and testicular tumors. Cancer Res. 1998;58:2087–2090. [PubMed] [Google Scholar]
- 81.Avizienyte E, Loukola A, Roth S, Hemminki A, Tarkkanen M, Salovaara R, Arola J, Butzow R, Husgafvel-Pursiainen K, Kokkola A, et al. LKB1 somatic mutations in sporadic tumors. Am J Pathol. 1999;154:677–681. doi: 10.1016/S0002-9440(10)65314-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Esteller M, Avizienyte E, Corn PG, Lothe RA, Baylin SB, Aaltonen LA, Herman JG. Epigenetic inactivation of LKB1 in primary tumors associated with the Peutz-Jeghers syndrome. Oncogene. 2000;19:164–168. doi: 10.1038/sj.onc.1203227. [DOI] [PubMed] [Google Scholar]
- 83.Shen Z, Wen XF, Lan F, Shen ZZ, Shao ZM. The Tumor Suppressor Gene LKB1 Is Associated with Prognosis in Human Breast Carcinoma. Clin Cancer Res. 2002;8:2085–2090. [PubMed] [Google Scholar]