Ubiquitin makes its mark on immune regulation (original) (raw)
. Author manuscript; available in PMC: 2011 Dec 15.
Abstract
Ubiquitination, the covalent attachment of ubiquitin molecules to proteins, is emerging as a widely utilized mechanism for rapidly regulating cell signaling. Recent studies indicate that ubiquitination plays potent roles in regulating a variety of signals in both innate and adaptive immune cells. Here, we will review recent studies of ubiquitin ligases, ubiquitin chain linkages and ubiquitin binding proteins that highlight the diversity and specificity of ubiquitin dependent functions in immune cells. We will also review studies that shed light on how ubiquitination signals are integrated in cell-type specific fashion to regulate the immune system in vivo.
The ubiquitin system
The regulation of intracellular signals allows immune cells to integrate stimuli from their environment and to exhibit the dynamic plasticity characteristic of immune responses. The transduction of such signals requires rapid post-translational modifications of proteins via processes such as phosphorylation or ubiquitination. While phosphorylation events are generally binary, comprised of the presence or absence of a single phosphate group on selected amino acids of target proteins (e.g., serine, threonine, or tyrosine), ubiquitination events include the attachment of a variety of lengths and conformations of ubiquitin chains, mostly on lysine residues (Pickart and Fushman, 2004) Understanding how ubiquitination events are regulated, and how they regulate a diverse array of cellular responses (Table 1) requires an understanding of the components of the ubiquitin system.
Table 1.
E3 ubiquitin ligases are intergral mediators of immune regulation.
| Immune Cell Type | E3 Ubiquitin Ligase | Function | References |
|---|---|---|---|
| T cells | |||
| TRAF6 | TCR signaling, central T cell tolerance | King et al., 2006; Lin and Mak, 2007; King et al., 2008; Cejas et al., 2010 | |
| Cbl-B | Peripheral T cell tolerance | Lin and Mak, 2007; Venuprasad, 2010 | |
| ITCH (AIP4) | T cell tolerance; TH2 development | Venuprasad, 2010 | |
| GRAIL | T cell tolerance; Treg function | Nurieva et al., 2010 | |
| SOCS proteins | T cell maturation, differentiation, and function | Alexander and Hilton, 2004; Palmer and Restifo, 2009 | |
| B cells | |||
| TRAF2/3 | B cell development | Vallabhapurapu et al., 2008 | |
| TRAF6 | CD40 signaling; B cell development and TD and TI responses | Rowland et al., 2007; Kobayashi et al., 2009 | |
| A20 (TNFAIP3) | B cell tolerance; germinal center selection; CD40 signaling | Tavares et al., 2010; Chu et al., 2010 | |
| c-IAP-2 | GALT B cell homeostasis; noncanonical NF-kB signaling; | Conze et al., 2010 | |
| Act1 | B cell survival and tolerance | Qian et al., 2004; Qian et al., 2008 | |
| Innate Cells | |||
| TRAF2/5 | TNF signaling | Micheau and Tschopp, 2003 | |
| TRAF3 | TLR-mediated IRF3 signaling | Oganesyan et al., 2006 | |
| TRAF6 | TLR signaling | Deng et al., 2000 | |
| cIAPs | TNF signaling | Bertrand et al., 2008 | |
| ITCH | TNF signaling | Chang et al, 2006 | |
| A20 (TNFAIP3) | TNF, TLR, NLR signaling | Lee et al., 2000, Boone et al., 2004; Turer et al., 2008; Hitsomatsu et al., 2008; | |
| TRIM25 | RIG-I signaling; type I interferon production | Gack et al., 2007 | |
| HOIL-1/HOIP (LUBAC) | TNF, IL-1β signaling | Haas et al., 2009; Tokunaga et al., 2009 |
Ubiquitination involves a three step enzymatic reaction catalyzed by three different types of proteins, termed E1, E2, and E3 ubiquitin ligases (Pickart and Eddins, 2004). An E1 enzyme first “activates” ubiquitin by forming a thiol ester bond. Activated ubiquitin is transferred to an E2 ubiquitin conjugating enzyme. The E2- ubiquitin (Ub) interacts with an E3 ubiquitin ligase that facilitates transfer of the ubiquitin to the epsilon-amino group of a lysine (K) on substrate proteins. Together with ubiquitin binding proteins (or sensors) and proteases that function as de-ubiquitinating enzymes (DUBs), E1, E2, and E3 ubiquitin ligase complexes constitute the core biochemical machinery for building, editing and removing ubiquitin chains.
While two known E1 enzymes “charge” or activate ubiquitin molecules for virtually all ubiquitination events in the mammalian proteome, diverse combinations of E2 and E3 ubiquitin ligases attach distinct types of ubiquitin chains to specific substrate proteins. Approximately 38 E2 enzymes are predicted to exist (Ye and Rape, 2009). As there are many more E3 ubiquitin ligases (>600 predicted) than E2s, and most E2s functionally interact with many E3 ubiquitin ligases. In addition, at least some E3s can bind to multiple E2s. For example, the E3 ubiquitin ligase complex of BRCA1 and BARD1 can interact with 10 different E2s that display divergent functions; one E2 may effect ubiquitin initiation and other E2s may effect the elongation of various linkages depending on the E2 used (Christensen et al., 2007; Christensen and Klevit, 2009). Hence, a vast number of E2 and E3 combinations are available to specify the target proteins to be modified and the type of ubiquitin chains to be added.
E2 enzymes play a major role in determining the length and linkage type of ubiquitin chains that are formed (reviewed in Christensen and Kelvit, 2009; Ye and Rape, 2009). For example, the E2 enzyme Ubc13 preferentially builds K63-linked ubiquitin chains that support mitogen activated protein (MAP) kinase signal propagation (Yamamoto et al, 2006, Rodrigo-Brenni et al., 2010; David et al., 2010). Ubc13 is required for interleukin-1 (IL-1) and lipopolysaccharide (LPS) induced MAP kinase activation, but appears less important for Nuclear Factor-κB (NF-kB) signaling from these ligands in macrophages and fibroblasts (Yamamoto et al, 2006a). Ubc13 also appears to be dispensable for tumor necrosis factor (TNF) induced NF-kB signaling. In contrast, Ubc13 is important for T cell receptor (TCR) induced NF-kB signaling in thymocytes (Yamamoto et al., 2006b). It is possible that other E2 ligases, such as Ubc5, can support K63 ubiquitin dependent signals, depending on the cell type and stimulus. The selectivity of E2s for certain subsets of E3 enzymes (and hence substrates) and the predilection of E2s to form particular ubiquitin chain linkages combine to render these enzymes important regulators as well as mediators of ubiquitination.
E3 ubiquitin ligases confer substrate specificity to the ubiquitin reaction by binding to and mediating transfer of ubiquitin from E2 enzymes to target proteins such as signaling molecules. E3 ubiquitin ligases have been divided into two general types depending on the type of protein domain used to recognize substrates: Really interesting new gene (RING) and Homologous to E6-associated protein carboxyl terminus (HECT) E3 ubiquitin ligases. RING E3s make up the largest number by far, with over 600 predicted to be encoded in the human genome, while 28 HECT E3s are predicted to exist (Deshaies et al., 2009; Rotin and Kumar, 2009).
RING and HECT E3s mediate substrate ubiquitination by different mechanisms. RING E3s use their RING finger domain to direct the transfer of ubiquitin from the activated E2-Ub to the substrate, whereas HECT E3s accept the ubiquitin from the E2-Ub to form a covalent thioester bond intermediate before transferring ubiquitin to the target protein. Because HECT E3s are charged with a ubiquitin while bound to their target protein, they may also help determine the specificity of linkage chain formation (Ye and Rape, 2009). In addition, many RING E3s have ubiquitin binding domains (UBD) that may serve to orient the acceptor ubiquitin molecule for attack and thereby influence the type of linkage formed. Indeed, a subset of E3 ubiquitin ligases, occasionally termed E4 ligases, may preferentially extend ubiquitin chains on ubiquitinated substrates. Hence, E3 enzymes mediate target recognition and can also contribute to linkage specificity.
In addition to RING and HECT domain containing E3 ubiquitin ligases, other protein motifs, such as plant homology domains (PHD), U box domains, and a subset of zinc fingers, have been implicated in E3 ubiquitin ligase activity. The U box, so designated by the domain found in the yeast ubiquitination factor UFD2, is a modified RING finger that lacks the canonical cysteine residues for zinc binding but which can nevertheless mediate ubiquitin ligase activity (Aravind and Koonin, 2000; Hatakeyama et al., 2001). More recently, a novel zinc finger motif in the ubiquitin ligase A20 protein has been shown to mediate E3 ubiquitin ligase activity (Wertz et al, 2004). Although all proteins bearing such motifs have not been tested for E3 ubiquitin ligase activity, it is likely that the number of bona fide E3 ubiquitin ligases will grow significantly. Moreover, E3 ubiquitin ligases typically possess the ability to ubiquitinate multiple substrates, suggesting that a significant portion of the mammalian proteome undergoes ubiquitination.
Ubiquitin chains of diverse conformations regulate immune signals
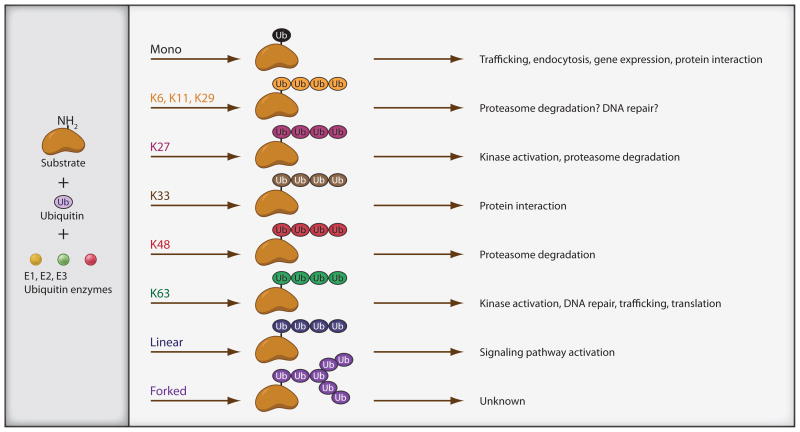
Ubiquitination events in immune cells mediate diverse cell signals and cellular responses. Part of this diversity is due to the fact that ubiquitin molecules can be attached to proteins as monomers or as polymers (Figure 1). Mono-ubiquitination events regulate DNA repair, receptor endocytosis, vesicle sorting, gene silencing (Sigismund et al., 2004). Ubiquitination of DNA repair proteins can impact immune processes such as class switch recombination (Li et al, 2010; Santos et al, 2010; Sun and Chen, 2004). Monoubiquitination has also been implicated in persistent NF-kB signaling that has implications for human T cell leukemia virus-1 (HTLV-1) infection and signaling thresholds mediating positive and negative thymic selection (Carter et al. 2005, Wada et al., 2009; Wang et al., 2010).
Figure 1. A diverse array of ubiquitin chain linkages can lead to different outcomes for the substrate protein and resulting in different cellular responses.
The colored circles denote monoubiquitin (black) or polyubiquitin chains denoted with various colors corresponding with different linkages, as defined by the arrows at left. Potential outcomes of substrate proteins conjugated with the given ubiquitin linkage are given in the box at right.
Polyubiquitin chains can be formed using any one of the seven internal lysine residues (K6, K11, K27, K29, K33, K48, and K63) or the N-terminal amino group of ubiquitin (Komander, 2009) to form distinct ubiquitin chain linkage types (Figure 1). Structural studies have revealed that different chain linkages adopt distinct conformations (Pickart and Fushman, 2004; Fushman and Walker, 2010). Hence, distinct chain types could be distinguished by ubiquitin dependent proteins.
K48-linked ubiquitin chains that are at least four ubiquitin molecules in length target misfolded or senescent proteins for recruitment to the proteasome for proteolytic degradation (Pickart and Fushman, 2004). In the context of cell signaling, K48-linked polyubiquitin chains facilitate degradation of signaling proteins, including both agonists and inhibitors of signal transduction. Signaling proteins probably do not exhibit the same biochemical features as misfolded proteins. E3 ubiquitin ligases that target signaling proteins often recognize modifications to these proteins that occur during their activation, such as phosphorylated residues. For example, phosphorylation of the NF-kB inhibitor IkBα leads to its recognition by a Skp1-Cul1-F box E3 complex called SCFβTrCP that adds K48 ubiquitin chains to IkBα and promotes its degradation (Skaug et al., 2009). Hence, regulated degradation of signaling inhibitors supports the propagation of canonical NF-kB signals. By contrast, K48 ubiquitination of agonist signaling proteins limits the duration of signals. An example of this type of ubiquitin regulation is the negative feedback inhibition of cytokine signaling by suppressors of cytokine signaling (SOCS) family proteins. SOCS proteins are E3 ubiquitin ligases that tag cytokine signaling proteins, such as Janus kinases (JAKs) and cytokine receptors, with ubiquitin, marking them for degradation (Alexander and Hilton, 2004). Regulated attachment of K48 ubiquitin chains to agonist signaling molecules has been increasingly recognized in restricting various immune signaling cascades.
Alternative (non-K48) ubiquitin chains can target modified proteins for nonproteolytic functions
A major revelation in cell signaling biology was the discovery that ubiquitin chains assembled in certain conformations can target proteins for outcomes other than proteosomal degradation (Deng et al, 2000; Pickart and Fushman, 2004). Structural studies showed that K63 ubiquitin chains are more flexible than K48 chains, providing a biochemical basis for selective recognition of K63-ubiquitinated proteins (Pickart and Fushman, 2004; Winget and Mayor, 2010; Fushman and Walker, 2010). A quantitative proteomics profile of polyubiquitin linkages in yeast showed that all the lysines in ubiquitin can form chains and, with the exception of lysine 63, can directly target proteins to the proteasome for degradation with varying efficiency (Xu et al., 2009b). While ubiquitin chains of distinct conformations had been defined in yeast, their importance in signaling pathways in metazoan organisms is now being more fully realized. We now highlight some of the recent discoveries that manifest how the diversity of ubiquitin signaling modalities impact immune cell signaling pathways.
The presence of four ubiquitin coding genes and the widespread use of ubiquitin modifications in multiple aspects of cell biology renders strategies for the genetic manipulation of ubiquitin linkages difficult. Chen and colleagues devised a novel ubiquitin replacement strategy for testing the requirements for specific ubiquitin chain linkage in cells. Using an inducible system for the coordinated knockdown of endogenous ubiquitin and expression of mutant ubiquitin in model cell lines, they showed that IL-1 induced NF-kB signaling requires K63 ubiquitin chains. By contrast, TNF induced NF-kB signaling can occur in the absence of K63 (Xu et al, 2009a). This strategy was also used to show that K63 ubiquitination is required for viral activation of interferon regulatory factor-3 (IRF3) (Zeng et al., 2010). These studies and others uncovered the physiological importance of K63 ubiquitin chains in immune signaling. In addition, functions of other non-K48 linkages have been described in immune cells.
K33 polyubiquitin chains and TCR signaling
Like K63 polyubiquitin chains, K33 linkages in yeast are relatively resistant to proteasomal degradation, and may thus support nondegradative functions (Xu et al., 2009b). K33 linkages have recently been described in T cells, where the E3 ubiquitin ligases Cbl-b and Itch appear to cooperatively promote K33-linked ubiquitination of TCRζ. This modification inhibits TCRζ’s phosphorylation and association with the tyrosine kinase Zap-70 and thereby restricts TCR signaling (Huang et al., 2010). Hence, K33 ubiquitination can disengage TCRζ from TCR signaling without inducing its proteosomal degradation, providing an additional mechanism by which ubiquitination can restrict signals. Utilization of this type of ubiquitin mediated restriction of signals, rather than proteosomal degradation, allows cells to reutilize TCRζ without spending energy on degrading and resynthesizing this protein.
K27 ubiquitination and IKKγ function
K27-linked ubiquitin chains have been described in several settings. The E3 ubiquitin ligase TRAF6, which helps build K63 ubiquitin chains, has been reported to promote K6, K27, and K29 ubiquitination of the Parkinson’s disease proteins DJ-1 and alpha-synucluin, stimulating their accumulation in cytoplasmic aggregates (Zucchelli et al., 2010). In the setting of host-pathogen interactions, two distinct K27 chain modifications of the NF-kB regulatory subunit IKKγ (also called NEMO) have been described. During viral infections, the attachment of K27 ubiquitin chains to IKKγ by host E3 ubiquitin ligase triparite motif protein 23 (TRIM23) supports IKKγ activation and production of anti-viral IFN-β (Arimoto et al, 2010). By contrast, during infection with the bacterium Shigella, the attachment of K27 linked ubiquitin chains to IKKγ by bacterial E3 ubiquitin ligase IpaH9.8 promotes its degradation and thus suppression of NF-kB signaling and host defense responses (Ashida et al., 2010). As bacteria do not possess a ubiquitin conjugation system, Shigella have apparently hijacked the eukaryotic E3 ubiquitin ligase system to suppress host immune defense mechanisms. This exemplifies how pathogens can usurp the ubiquitin system to affect host signaling cascades (Spallek et al., 2009). Moreover, these two studies identify K27 ubiquitination of two distinct lysines of IKKγ that lead to two different biochemical outcomes.
Linear ubiquitin chains are linked via the N-terminal amino group
In addition to forming ubiquitin chains using one of ubiquitin’s seven lysine residues, so-called “linear” ubiquitin chains can be built via the N-terminal amino group of ubiquitin. Linear chains are assembled by an E3 complex called linear ubiquitin chain assembly complex (LUBAC) (Kirisako et al., 2006), comprised of two E3 ubiquitin ligases, HOIL-1 and HOIP. A ubiquitin associated (UBA) domain in HOIP binds to the ubiquitin-like domain in HOIL-1 to form LUBAC. LUBAC activates the canonical NF-kB pathway by conjugating linear polyubiquitination chains to IKKγ, and HOIL-1 deficient mouse embryonic fibroblasts (MEFs) exhibit reduced TNF- or IL-1β-induced IKK kinase activity (Tokunaga et al., 2009). Hence, linear polyubiquitin chains appear to be a physiologically important form of ubiquitination. Of note, linear ubiquitin chains were not detected in a quantitative yeast proteomic study, so this aspect of the ubiquitin system may have evolved more recently than yeast (Xu et al., 2009b). Interestingly, the lysines of IKKγ that putatively undergo ubiquitination with linear ubiquitin chains overlap those identified to undergo K63 ubiquitination. Thus, ubiquitination of individual lysines on substrate proteins might cross-regulate ubiquitination of these same lysines by other chains.
Forked ubiquitin chains can be formed in vitro and in vivo
Most polyubuiqtuin chains are believed to be homogenous, i.e., contain only one type of isopeptide linkage. Recent studies have suggested that a proportion of polyubiquitin chains may be synthesized with mixed linkages, forming forked ubuiquitin chains (Kirkpatrick et al., 2005; Kirkpatrick et al., 2006). Forked polyubiquitin chains are resistant to proteasomal degradation and their formation in the cell can be regulated by association with a ubiquitin interacting motif (UIM) protein, suggesting that formation of forked chains may be important in the regulation of normal protein homeostasis, the cellular stress response, and/or in degenerative diseases (Kim et al., 2009).
Unanchored ubiquitin chains support NF-kB signals
“Anchored” ubiquitin chains refer to the typical polyubiquitin chains that are covalently attached to substrate proteins. “Unanchored” polyubiquitin chains, by contrast, are free chains that are not attached to any substrate. Unanchored chains can be built by various E3 ubiquitin ligases in vitro. As noted above, TRAF6 works with the E2 UBC13-UEV1A complex to build K63-linked ubiquitin chains. These chains are recognized by the TAK1-TAB1-TAB2 complex, a ubiquitin-dependent kinase complex that phoshorylates mitogen activated protein kinase kinase (MKK) and IκB kinase (IKK)γ. A recent study reported the startling discovery that unanchored K63 ubiquitin chains activate the transforming growth factor associated kinase 1(TAK1) by binding to TAK1 binding protein 2 (TAB2). The presence of unanchored chains in live cells was uncovered by treating immunoprecipitated cell lysates with isopeptidase T, a de-ubiquitinating enzyme that only cleaves unanchored chains (due to its requirement for access to the C-terminal carboxyl group of ubiquitin) (Xia et al, 2009).
Another example of the importance of unanchored chains in supporting signal transduction was discovered in the retinoic-acid-inducible gene-1 (RIG-1) pathway. RIG-1-like receptors (RLRs) are involved in viral recognition and trigger signal transduction cascades leading to type I interferon production, and TRIM-25 is an E3 ubiquitin ligase required for the activation of this pathway (Gack et al., 2007). Sequential binding of viral RNA to RIG-1’s C-terminal regulatory domain and unanchored K63 ubiquitin chain binding to RIG-1’s N-terminal Caspase recruiting domain (CARD) domain may lead to activation of RIG-1 (Zeng et al., 2010). The unanchored K63 chains that bind to and activate RIG-1 appear to be significantly shorter than the unanchored chains that bind TAK1. These observations raise several questions, including whether unanchored chains are generated as free chains, or whether they are cleaved after being initially built on substrates. If the latter occurs, then additional enzymatic steps must be required. More globally, how and why would unanchored chains, rather than anchored chains, be useful or sufficiently specific in propagating signals?
The answers to these questions should yield surprising insights into the mechanisms by which ubiquitin chains regulate signals.
In summary, a diverse array of physiological polyubiquitin chains provides nearly 10-fold greater biochemical variety than binary phosphorylation events. These varied polyubiquitin chains target modified proteins and signaling complexes for diverse protein-protein interactions such as proteasomal degradation, receptor recycling, signal complex localization, and/or recruitment of downstream signaling proteins. These interactions help support, restrict, or direct signaling cascades toward proper cellular responses. Precise construction and utilization of distinct polyubiquitin chains requires ubiquitin modifying enzymes that can build, edit and degrade these chains as well as ubiquitin sensors that recognize and bind specific chain types.
Ubiquitin modifying enzymes and sensors
De-ubiquitinating enzymes (DUBs) exhibit chain specificity
Like phosphorylation, ubiquitination is a reversible process. Over 100 deubiquitylating enzymes (DUBs) are predicted to exist in the human proteome. These enzymes provide an important layer of regulation for ubiquitination by enzymatically removing ubiquitin chains. Like E2 and E3 ubiquitin ligases, DUBs can exhibit ubiquitin chain specificity. For example, the DUB Otubain1 is highly specific for cleaving K48-linked chains (Wang et al., 2009). Meanwhile, the DUB Cezanne preferentially hydrolyzes K11 ubiquitin chains (Bremm et al., 2010). In addition to demonstrating chain specificity, DUBs may be able to remove ubiquitin molecules iteratively from chains or may cleave entire chains en masse from target proteins. Several excellent recent reviews have discussed the varied roles of DUBs in the immune system (Sun, 2008; Komander et al., 2009; Reyes-Turcu et al., 2009). While this review will focus on ubiquitin ligases, ligases and DUBs must be considered together as coordinated partners in regulating ubiquitination.
DUB and ubiquitin ligase activities can coordinately modify ubiquitination of individual proteins
The evidence of chain-specific ubiquitin ligases and DUB enzymes raise the possibility that individual target proteins may undergo conjugation with more than one type of ubiquitin chain, leading to different outcomes. One protein that appears to undergo ubiquitination with different ubiquitin chain types is TRAF3. TRAF3 is itself an E3 ubiquitin ligase that is required for Tir-domain-containing adaptor inducing interferon β (TRIF)-dependent type I interferon signaling, but may be a negative regulator of MAPK-dependent proinflammatory responses. TRAF3 undergoes ubiquitination with both K48 and K63 chains downstream of the TLR adaptor protein Myd88 and TRIF mediated TLR signals (Tseng et al., 2010). When macrophages respond to LPS, the E3 ubiquitin ligase cIAP1 (or cIAP2) builds K48 ubiquitin chains on TRAF3, leading to its degradation. This allows MAPK activation and proinflammatory cytokine production. In contrast, TRIF dependent signals lead to auto-ubiquitination of TRAF3 with K63 ubiquitin chains. This leads to activation of IRF3 and type I interferon responses. Selective degradation of cIAP prevents K48 but not K63 ubiquitination of TRAF3. This differential ubiquitination of TRAF3 inhibits the inflammatory response without affecting interferon production. These findings imply that different chain-specific E2 and E3 ubiquitin ligases regulate TRAF3. Moreover, the selective action of the SMAC mimetic may have clinical importance for the treatment of inflammatory diseases and cancer.
Secondly, IKKγ has also been described to undergo ubiquitination at a number of distinct lysines, depending upon the ligands and potentially E3 ubiquitin ligases involved (Table 2). While most of these events are thought to lead to IKKγ activation, at least one is thought to lead to proteosomal degradation of IKKγ (Ashida et al., 2010). Hence, conjugation of individual signaling proteins such as TRAF3, receptor interacting protein (RIP), and IKKγ with distinct ubiquitin chains regulates the character and duration of signaling cascades and the attendant biological responses.
Table 2.
NEMO Ubiquitination: Sites, Linkages, Outcomes
| Stimulus/receptor | E3 ligase | Site of Ub | Linkage | Outcome | Reference |
|---|---|---|---|---|---|
| TCR | TRAF6 | Lys 399 | K63 | Activation | Zhou et al., 2004; Sun et al., 2004 |
| MDP/Nod2 | Lys 285 | K63 | Activation | Abbott et al., 2004 | |
| LPS/TLR | Lys 285, 399 | K63 | Activation | Abbott et al., 2007 | |
| LUBAC | Lys 285, 309 | Linear | Activation | Tokunaga et al., 2009 | |
| Poly I:C, Sendai virus | TRIM23 | Lys 165, 309, 325, 326, 344 | K27 | Activation | Arimoto et al., 2010 |
| Shigella infection | Shigella IpaH9.8 | Lys 309, 321 | K27 | Degradation | Ashida et al., 2010 |
A third example of a signaling protein undergoing conjugation with two different types of polyubiquitin chains is RIP, a key mediator of TNF induced NF-kB signaling. RIP undergoes K63 ubiquitination, probably as a consequence of cIAPs. Ubiquitination of RIP at lysine 377 is required for the recruitment of the TAK1-TAB2 complex and IKKγ, which in turn leads to phosphorylation of IKKγ and IkBα and activation of NF-kB. Interestingly, RIP also undergoes ubiquitination with K48- linked chains, leading to RIP’s proteosomal degradation. These K48 chains are built by A20, which thereby restricts TNF induced NF-kB signaling (Wertz et al, 2004). A20 deficient cells exhibit prolonged TNF induced NF-kB signaling (Lee et al, 2000). How diverse ubiquitination events on individual proteins are coordinated is poorly understood. One remarkable example was unveiled by studies of A20’s modifications of RIP. In this case, A20’s N-terminal half harbors DUB activity which preferentially degrades anchored K63 chains, while the fourth of its seven C-terminal zinc fingers (ZF) exhibits E3 ubiquitin ligase activity, building K48 chains (Wertz et al., 2004). This complex ubiquitin editing activity is supported by structural data showing A20’s ZF4 ability to bind monoubiquitin and K63 polyubiquitin chains, and adjacent ZF5-7 binding to E2 (Bosanac et al., 2010). The net effect of A20’s dual biochemical activities is to function as a ubiquitin-editing enzyme, targeting K63-ubqiuitinated RIP molecules and exchanging the K63 chains for K48 chains. In this way, A20 both de-activates and degrades RIP. While A20 is currently the only protein known to exhibit this biochemical mechanism of regulating immune signals, it is intriguing to consider that other examples remain to be discovered, either embodied in single proteins or in higher order protein complexes.
Ubiquitin sensors can support or restrict immune signals
The ability of diverse polyubiquitin chains to trigger different biochemical outcomes for ubiquitinated substrates implies that ubiquitin binding proteins, or sensors, must exist that distinguish these chains. Four ubiquitin sensors have been described to date to play critical roles in immune signaling. TAB2 and TAB3 use a conserved zinc finger motif to bind K63 ubiquitin chains on RIP. IKKγ, the catalytic subunit of the IKK complex, harbors a novel ubiquitin binding domain, termed NUB for Nemo ubiquitin binding domain, that binds to K63 (Wu et al, 2006; Ea et al., 2006) and linear ubiquitin chains (Rahighi et al., 2009). Both of these ubiquitin binding functions are essential for TNF induced activation of IKKγ. A fourth ubiquitin sensor is called A20 Binding Inhibitor of NF-kB-1, or ABIN-1. ABIN-1 shares a NUB domain with IKKγ. Unlike TAB2, TAB3, or IKKγ, which use their ubiquitin binding motifs to support TNF induced NF-kB signaling, ABIN-1 uses its NUB domain to restrict TNF induced death signaling (Oshima et al, 2009). While the potential relationship between ABIN-1’s and IKKγ ‘s NUB domains is currently unclear, these domain are likely important for modulating the composition of signaling complexes.
How ubiquitin sensors distinguish between different types of ubiquitin chains is another important unanswered question. Initial clues have emerged from biochemical studies of chain-specific DUBs (Wang et al, 2009). These studies indicate how multi-point contacts between the K48 chain-specific DUB Otubain1 and K48 ubiquitin chains or between the K63 chain-specific DUB AMSH-LP and K63 chains determine the specificity of these enzymes (Wang et al, 2009; Sato et al, 2008). Linkage specific avidity is also accomplished by tandem ubiquitin binding sites in the Rap80 and ataxin 3 ubiquitin sensors (Sims et al, 2009a; Sims et al, 2009b). Whether and how tandem ubiquitin binding motifs may mediate immune signaling remains to be seen.
Together with ubiquitin modifying enzymes, ubiquitin sensors complete the core ubiquitin system that regulates immune signals. Growing appreciation of the biological diverse outcomes of biochemically distinct ubiquitin chains and of the proteins that build, bind, and/or degrade these chains provide the biochemical foundations for understanding how diverse ubiquitination events regulate immune signals.
Immune Functions of E3 ubiquitin ligases in mice
Cell-free biochemistry experiments and cell line based studies suggest that ubiquitin modifying enzymes and sensors regulate immune homeostasis and/or immune responses in intact mice. Mice lacking key components of the ubiquitin machinery, such as E2 and E3 ubiquitin ligases, provide a means to directly interrogate the in vivo function of individual ubiquitination proteins in different cellular contexts. The emerging roles of ubiquitination in innate and adaptive immunity (Table 1) have recently been reviewed (Lin and Mak, 2007; Bhoj and Chen, 2009; Skaug et al., 2009; Wertz and Dixit, 2010). Here, we will focus on selected gene targeted mice that have recently been reported and that illuminate how ubiquitin ligases regulate immune homeostasis, how ubiquitin dependent activation and survival signals can be integrated in specific immune cell types, and how phenotypes of globally deficient mice represent complex compilations of lineage-specific functions.
Lineage specific functions of E3 ubiquitin ligases
Many E3 ubiquitin ligases are expressed in multiple immune and nonimmune cell lineages and may regulate multiple pathways. Thus, complex and often lethal phenotypes result when E3 ubiquitin ligases or associated components of ubiquitination are globally deleted from mice. Deciphering intrinsic versus extrinsic phenotypes may be enigmatic, e.g., spontaneously activated innate immune cells can contribute to T and B lymphocyte activation, and vice versa. Hence, it is important to test their physiological roles in a cell specific context. Lineage specific deletions of ubiquitin modifying enzymes using LoxP-flanked alleles have recently begun to unveil cell-autonomous functions for these enzymes in mice. We discuss lineage specific deletions of the genes encoding TRAF6. A20, and ACT1 to illustrate the differing roles E3 ubiquitin ligases can play in different cell types.
Tumor necrosis factor receptor associated factors (TRAFs) are important signaling adaptors that can mediate signals from TNF super family receptors (TNFSFR), toll-like (TLR) and interleukin-1 (IL-1R) receptors, and receptor activator of NF-kB ligand (RANKL) to activate transcription factors NF-kB, NFAT, Akt, and MAP kinases. Mice deficient for TRAF2, TRAF3, or TRAF6 die in utero or perinatally from multiple organ abnormalities, demonstrating their non-overlapping roles in mouse development (Ha et al., 2009). TRAF6, the seminal E3 ligase shown to synthesize non-degradative K63 ubiquitin linkages and activate NF-kB signaling, is a good example of the divergent functions that one E3 ligase can play in cell specific contexts. Mice globally lacking TRAF6 develop osteopetrosis and Traf6−/− fibroblasts exhibit gross defects in IL-1, LPS and CD40 induced signaling (Lomaga et al, 1999). TRAF6 also supports NF-kB signals in B cells and dendritic cells (DC). Mice with B lineage specific deletion of Traf6 have impaired B cell maturation, particularly of the B-1a subset, and impaired B2 cell responses to T-dependent and T-independent antigens (Kobayashi et al., 2009). B cells deficient for both TRAF6 and TRAF2 exhibit defective CD40 signaling (Rowland et al., 2007). TRAF6 also appears to have an agonist function in DC maturation and development, as judged by chimeric mice bearing Traf6−/− hematopoietic cells (Kobayashi et al., 2003). A recent study suggested that TRAF6 auto-ubiquitination is not required for NF-kB and MAPK activation in response to IL-1 and RANKL (Walsh et al., 2008). Thus, TRAF6’s E3 ligase activity may be separable from its roles in supporting in these signaling pathways.
In contrast to TRAF6’s roles in supporting NF-kB and MAPK signals, mice lacking TRAF6 specifically in T cells develop multiorgan inflammatory disease, indicating that TRAF6 plays a T cell intrinsic role in preventing spontaneous inflammation. TRAF6 deficient T cells are both resistant to suppression by regulatory T cells and for induction of T cell anergy, and have an increased propensity for Th17 differentiation (King et al., 2006; King et al., 2008; Cejas et al., 2010). These surprising phenotypes are hard to reconcile with the known roles for TRAF6 in supporting activation signals and highlight the fact that we still understand this protein poorly.
Another E3 ubiquitin ligase with pleiotropic functions is A20. Mice lacking A20 (Tnfaip3−/− mice) exhibit multi-organ inflammation and perinatal death, which is largely, though not completely, abrogated in Tnfaip3−/−Myd88−/− compound mutant mice (Lee et al., 2000; Turer et al, 2008). Hence, A20 prevents lethal inflammation by restricting homeostatic MyD88-dependent signals (Boone et al, 2004; Turer et al., 2008).
A20 restricts both TNF induced NF-kB signals and TNF induced programmed cell death (PCD) signals in fibroblasts (Lee et al. 2000). Enterocyte specific deletion of Tnfaip3 (Tnfaip3Flox Vil1-Cre) causes hypersensitivity to dextran sodium sulfate (DSS)-induced colitis, which is associated with hypersensitivity of enterocytes to TNF induced PCD and is partially rescued by TNFR1 deficiency (Vereecke et al., 2010). By contrast, B cell specific loss of A20 surprisingly renders these cells resistant to PCD mediated by the TNFSFR member Fas. This resistance is potentially explained by the observation that A20-deficient B cells are hyper-sensitive to CD40 triggered NF-kB signaling, leading to exaggerated Bcl-x expression and resistance to Fas mediated PCD. The divergent fashions by which fibroblasts integrate TNF signals versus how activated B cell integrate Fas signals highlights the importance of understanding cell type-specific functions of such enzymes. This lesson is particularly important with proteins such as A20 that regulate both NF-kB activation signals and PCD signals.
Mice lacking A20 selectively in B cells (Tnfaip3flox/flox Cd19-Cre mice) also shed light on two important human diseases: systemic lupus erythematosus (SLE) and B cell lymphoma. Several single nucleotide polymorphisms (SNPs) near the human Tnfaip3 gene are associated with susceptibility to SLE in genome wide association studies (GWAS). Tnfaip3flox/flox Cd19-Cre mice exhibit increased numbers of germinal center B cells, accumulation of auto-reactive B cells, auto-antibodies, and spontaneous SLE-like disease (Tavares et al, 2010; Chu et al., 2010). These phenotypes are also observed in mice lacking only one copy of A20 in B cells (Tnfaip3flox/+ Cd19-Cre mice), demonstrating the biological importance of a full complement of this potent enzyme. Tnfaip3flox/+ Cd19-Cre mice more closely resemble the hypomorphic condition likely to occur in SLE patients bearing germline polymorphisms in the A20 gene. Meanwhile, somatic deletions and mutation of both A20 alleles are observed in up to 30% of human B cell lymphomas. Resistance to PCD, due at least in part to the increased expression of the antiapoptotic Bcl-2 family protein Bcl-x, in A20-deficient B cells provides a compelling molecular explanation for why A20 is a prevalent tumor suppressor in human B cells (reviewed in Malynn and Ma, 2009). As distinct human A20 single nucleotide polymorphisms (SNPs) are associated with susceptibilities to other autoimmune and inflammatory diseases (e.g., rheumatoid arthritis, psoriasis, celiac disease), lineage specific deletion or reduction of A20 expression may provide important insights into the biology underlying these genetic disease susceptibilities.
Act1, originally identified as an NF-kB activator, was subsequently shown to negatively regulate CD40 and BAFF-mediated B cell survival via its interaction with TRAF3 (Qian et al., 2004). Act1 deficient (Traf3ip2−/−) Balb/c mice develop Sjogren disease associated with lupus-like neprhitis (Qian et al., 2008). In contrast to its role in B cell signaling, Act1 is a positive signaling regulator of IL-17 mediated responses in T cells by recruiting TAK1 and TRAF6 (Qian et al., 2007). Act1 possesses a U box domain and functions as an E3 ubiquitin ligase that K63 polyubiquinates TRAF6. The ubiquinating activity mediated by the Act1 U box appears to be required for IL-17 dependent signaling (Liu et al., 2009). Traf3ip2−/− C57BL/6 mice are resistant to experimentally induced experimental autoimmune encephalitis (EAE) and inflammatory bowel disease (IBD), due to reduced expression of proinflammatory cytokines and chemokines in astrocytes and gut epithelial cells, respectively (Qian et al., 2007). The different functions of Act1 in B cells and T cells may be due to its different binding partners: TRAF3 in CD40 and BAFF signaling in B cells, and TRAF6 in T cells. It will be of interest to determine whether Act1’s E3 ubiquitin ligase activity is also involved in its regulatory activity in B cells.
III. Future directions
We are clearly in the very early stages of understanding how ubiquitination regulates immune signals. Future directions will likely include identifying the key ubiquitin modifying enzymes and sensors that regulate immune signaling cascades. In this regard, the recognition that most RING-containing proteins function as E3 ubiquitin ligases occurred as recently as 1999 (reviewed in Deshaies and Joazeiro, 2009). Bioinformatic analyses predict over 300 human genes encode RING domain proteins and functional evidence of ligase activity has been obtained for approximately half of them. Some RING proteins may not have intrinsic ligase activity but are subunits that enhance the activity of larger complexes. Other RING-like E3 ubiquitin ligases and HECT ligases bring the number of potential E3 complexes to over 600. Thus, it is likely that more E3 ubiquitin ligases will be found to impact immune signaling.
The finding that A20 possesses dual enzymatic activity raises the provocative notion that other proteins or protein complexes could exert similarly coordinated activities. The recent discoveries of novel domains that can mediate E3 ubiquitin ligase activity (e.g., A20’s fourth zinc finger) or ubiquitin binding (e.g., IKKγ/ABIN-1’s NUB domains) further suggest that additional biochemically related proteins could be important immune regulators.
Many of the basic principles and mechanisms of ubiquitin remain to be elucidated, e.g., how do E2 and E3 enzymes select each other, how do they build specific types of ubiquitin chains, and how do they select their substrates? It will be equally important to determine how unanchored chains function, how large ubiquitin-editing complexes are assembled and regulated, and how ubiquitin sensors are integrated into coupled signaling modalities such as kinase activity.
Identifying physiologically relevant ubiquitin chains in immune cells also poses a challenge. Unlike genetic targeting of ubiquitin modifying enzymes or sensors, mutating ubiquitin molecules in the germline will likely lead to early embryonic lethality as well as rapid and profound cellular adaptation. However, emerging technologies may assist with this challenging task. In addition to transient heterologous expression of mutant ubiquitins, semi-quantitative mass spectroscopic techniques that distinguish chain topology have also recently been developed (Kirkpatrick et al., 2005; Kirkpatrick et al., 2006). Moreover, the development of antibodies that recognize the epitope formed by specific ubiquitin linkages should facilitate dissection of ubiquitin bearing signaling complexes in cells (Newton et al., 2008; Wang et al., 2008; Matsumoto et al., 2010). Combined with structural and biochemical studies and molecular modeling, these approaches are slowly unveiling clues into how ubiquitin modifications are used to regulate signaling proteins and cascades (Winget and Mayor, 2010; Fushman and Walker, 2010).
The potent signaling activities of E3 ubiquitin ligases highlights the therapeutic potential of manipulating activation or cell death pathways by altering the activity of ubiquitin conjugating enzymes (e.g., Scheper et al., 2010). The allure of this area of drug discovery is enhanced by GWAS and other studies that implicate genes such as A20 in human autoimmunity and cancer. The successful development of SMAC mimetics that selectively induce auto-ubiquitination of cIAPs and kill cancer cells provides a tantalizing sample of this small molecule approach (reviewed in Chen and Huerta, 2009). The Nobel Prize in Chemistry in 2004 was awarded for the discovery of ubiquitin mediated degradation of proteins. Six years later, it is clear that we have only scratched the surface of the complex mechanisms by which ubiquitination regulates immune signaling and biology.
Acknowledgments
We apologize to the many authors whose publications are not cited due to space limitations. Work from our laboratory is supported by the National Institutes of Health, the Kenneth Rainin Foundation, and the Alliance for Lupus Research,
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Abbott DW, Wilkins A, Asara JM, Cantley LC. The Crohn’s disease protein, NOD2, requires RIP2 in order to induce ubiquitinylation of a novel site on NEMO. Curr Biol. 2004;14:2217–2227. doi: 10.1016/j.cub.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Abbott DW, Yang Y, Hutti JE, Madhavarapu S, Kelliher MA, Cantley LC. Coordinated regulation of Toll-like receptor and NOD2 signaling by K63-linked polyubiquitin chains. Mol Cell Biol. 2007;27:6012–6025. doi: 10.1128/MCB.00270-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander WS, Hilton DJ. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu Rev Immunol. 2004;22:503–529. doi: 10.1146/annurev.immunol.22.091003.090312. [DOI] [PubMed] [Google Scholar]
- Anandasabapathy N, Ford GS, Bloom D, Holness C, Paragas V, Seroogy C, Skrenta H, Hollenhorst M, Fathman CG, Soares L. GRAIL: an E3 ubiquitin ligase that inhibits cytokine gene transcription is expressed in anergic CD4+ T cells. Immunity. 2003;18:535–547. doi: 10.1016/s1074-7613(03)00084-0. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. The U box is a modified RING finger - a common domain in ubiquitination. Curr Biol. 2000;10:R132–134. doi: 10.1016/s0960-9822(00)00398-5. [DOI] [PubMed] [Google Scholar]
- Arimoto K, Funami K, Saeki Y, Tanaka K, Okawa K, Takeuchi O, Akira S, Murakami Y, Shimotohno K. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proc Natl Acad Sci USA. 2010;107:15856–61. doi: 10.1073/pnas.1004621107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashida H, Kim M, Schmidt-Supprian M, Ma A, Ogawa M, Sasakawa C. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKγ to dampen the host NF-kappaB-mediated inflammatory response. Nat Cell Biol. 2010;12:66–73. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand MJ, Milutinovic S, Dickson KM, Ho WC, Boudreault A, Durkin J, Gillard JW, Jaquith JB, Morris SJ, Barker PA. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30:689–700. doi: 10.1016/j.molcel.2008.05.014. [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Chen ZJ. Ubiquitylation in innate and adaptive immunity. Nature. 2009;458:430–437. doi: 10.1038/nature07959. [DOI] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- Bosanac I, Wertz IE, Pan B, Yu C, Kusam S, Lam C, Phu L, Phung Q, Maurer B, Arnott D, Kirkpatrick DS, Dixit VM, Hymowitz SG. Ubiquitin binding to A20 ZnF4 is required for modulation of NF-κB signaling. Mol Cell. 2010;40:548–557. doi: 10.1016/j.molcel.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–47. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter RS, Pennington KN, Arrate P, Oltz EM, Ballard DW. Site-specific monoubiquitination of IkappaB kinase IKKbeta regulates its phosphorylation and persistent activation. J Biol Chem. 2005;280:43272–43279. doi: 10.1074/jbc.M508656200. [DOI] [PubMed] [Google Scholar]
- Cejas PJ, Walsh MC, Pearce EL, Han D, Harms GM, Artis D, Turka LA, Choi Y. TRAF6 inhibits Th17 differentiation and TGF-beta-mediated suppression of IL-2. Blood. 2010;115:4750–4757. doi: 10.1182/blood-2009-09-242768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang L, Kamata H, Solinas G, Luo JL, Maeda S, Venuprasad K, Liu YC, Karin M. The E3 ubiquitin ligase itch couples JNK activation to TNFalpha-induced cell death by inducing c-FLIP(L) turnover. Cell. 2006;124:601–613. doi: 10.1016/j.cell.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Chen DJ, Huerta S. Smac mimetics as new cancer therapeutics. Anticancer Drugs. 2009;20:646–658. doi: 10.1097/CAD.0b013e32832ced78. [DOI] [PubMed] [Google Scholar]
- Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–8. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- Christensen DE, Klevit RE. Dynamic interactions of proteins in complex networks: identifying the complete set of interacting E2s for functional investigation of E3-dependent protein ubiquitination. FEBS J. 2009;276:5381–9. doi: 10.1111/j.1742-4658.2009.07249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Vahl JC, Kumar D, Heger K, Bertossi A, Wójtowicz E, Soberon V, Schenten D, Mack B, Reutelshöfer M, Beyaert R, Amann K, van Loo G, Schmidt-Supprian M. B cells lacking the tumor suppressor TNFAIP3/A20 display impaired differentiation, hyperactivation, cause inflammation and autoimmunity in aged mice. Blood. 2010 doi: 10.1182/blood-2010-09-306019. [DOI] [PubMed] [Google Scholar]
- Conze DB, Zhao Y, Ashwell JD. Non-canonical NF-κB activation and abnormal B cell accumulation in mice expressing ubiquitin protein ligase-inactive c-IAP2. PLoS Biol. 2010;8:e1000518. doi: 10.1371/journal.pbio.1000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David Y, Ziv T, Admon A, Navon A. The E2 ubiquitin-conjugating enzymes direct polyubiquitination to preferred lysines. J Biol Chem. 2010;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22:245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- Fushman D, Walker O. Exploring the linkage dependence of polyubiquitin conformations using molecular modeling. J Mol Biol. 2010;395:803–814. doi: 10.1016/j.jmb.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Ha H, Han D, Choi Y. TRAF-mediated TNFR-family signaling. Curr Protoc Immunol. 2009;Chapter 11(Unit11.9D) doi: 10.1002/0471142735.im1109ds87. [DOI] [PubMed] [Google Scholar]
- Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T, Koschny R, Komander D, Silke J, Walczak H. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S, Yada M, Matsumoto M, Ishida N, Nakayama KI. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- Hitotsumatsu O, Ahmad RC, Tavares R, Wang M, Philpott D, Turer EE, Lee BL, Shiffin N, Advincula R, Malynn BA, Werts C, Ma A. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–390. doi: 10.1016/j.immuni.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Jeon MS, Liao L, Yang C, Elly C, Yates JR, 3rd, Liu YC. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity. 2010;33:60–70. doi: 10.1016/j.immuni.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–8. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Kim HT, Kim KP, Uchiki T, Gygi SP, Goldberg AL. S5a promotes protein degradation by blocking synthesis of nondegradable forked ubiquitin chains. EMBO J. 2009;28:1867–1877. doi: 10.1038/emboj.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King CG, Kobayashi T, Cejas PJ, Kim T, Yoon K, Kim GK, Chiffoleau E, Hickman SP, Walsh PT, Turka LA, Choi Y. TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis. Nat Med. 2006;12:1088–1092. doi: 10.1038/nm1449. [DOI] [PubMed] [Google Scholar]
- King CG, Buckler JL, Kobayashi T, Hannah JR, Bassett G, Kim T, Pearce EL, Kim GG, Turka LA, Choi Y. Cutting edge: requirement for TRAF6 in the induction of T cell anergy. J Immunol. 2008;180:34–38. doi: 10.4049/jimmunol.180.1.34. [DOI] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Denison C, Gygi SP. Weighing in on ubiquitin: the expanding role of mass-spectrometry-based proteomics. Nat Cell Biol. 2005;7:750–757. doi: 10.1038/ncb0805-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Walsh PT, Walsh MC, Speirs KM, Chiffoleau E, King CG, Hancock WW, Caamano JH, Hunter CA, Scott P, Turka LA, Choi Y. TRAF6 is a critical factor for dendritic cell maturation and development. Immunity. 2003;19:353–363. doi: 10.1016/s1074-7613(03)00230-9. [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Kim TS, Jacob A, Walsh MC, Kadono Y, Fuentes-Pananá E, Yoshioka T, Yoshimura A, Yamamoto M, Kaisho T, Akira S, Monroe JG, Choi Y. TRAF6 is required for generation of the B-1a B cell compartment as well as T cell-dependent and -independent humoral immune responses. PLoS One. 2009;4:e4736. doi: 10.1371/journal.pone.0004736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10:550–563. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- Komander D. The emerging complexity of protein ubiquitination. Biochem Soc Trans. 2009;37:937–953. doi: 10.1042/BST0370937. [DOI] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science. 2000;289:2350–4. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Halayby MJ, Hakem A, Cardoso R, El Ghamrasni S, Harding S, Chan N, Bristow R, Sanchez O, Durocher D, Hakem R. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med. 2010;10:983–987. doi: 10.1084/jem.20092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AE, Mak TW. The role of E3 ligases in autoimmunity and the regulation of autoreactive T cells. Curr Opin Immunol. 2007;19:665–673. doi: 10.1016/j.coi.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Liu C, Qian W, Qian Y, Giltiay NV, Lu Y, Swaidani S, Misra S, Deng L, Chen ZJ, Li X. Act1, a U-box E3 ubiquitin ligase for IL-17 signaling. Sci Signal. 2009;2:ra63. doi: 10.1126/scisignal.2000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomaga MA, Yeh WC, Sarosi I, Duncan GS, Furlonger C, Ho A, Morony S, Capparelli C, Van G, Kaufman S, van der Heiden A, Itie A, Wakeham A, Khoo W, Sasaki T, Cao Z, Penninger JM, Paige CJ, Lacey DL, Dunstan CR, Boyle WJ, Goeddel DV, Mak TW. TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 1999;13:1015–1024. doi: 10.1101/gad.13.8.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn BA, Ma A. A20 takes on tumors: tumor suppression by an ubiquitin-editing enzyme. J Exp Med. 2009;206:977–980. doi: 10.1084/jem.20090765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, Kelley RF, Dixit VM. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Mol Cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- Newton K, Matsumoto ML, Wertz IE, Kirkpatrick DS, Lill JR, Tan J, Dugger D, Gordon N, Sidhu SS, Fellouse FA, Komuves L, French DM, Ferrando RE, Lam C, Compaan D, Yu C, Bosanac I, Hymowitz SG, Kelley RF, Dixit VM. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 2008;134:668–678. doi: 10.1016/j.cell.2008.07.039. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Zheng S, Jin W, Chung Y, Zhang Y, Martinez GJ, Reynolds JM, Wang SL, Lin X, Sun SC, Lozano G, Dong C. The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity. 2010;32:670–680. doi: 10.1016/j.immuni.2010.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, Perry A, Cheng G. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- Oshima S, Turer EE, Callahan JA, Chai S, Advincula R, Barrera J, Shifrin N, Lee B, Benedict Yen TS, Woo T, Malynn BA, Ma A. ABIN-1 is a ubiquitin sensor that restricts cell death and sustains embryonic development. Nature. 2009;457:906–909. doi: 10.1038/nature07575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer DC, Restifo NP. Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 2009;30:592–602. doi: 10.1016/j.it.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: stuctures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Qian Y, Qin J, Cui G, Naramura M, Snow EC, Ware CF, Fairchild RL, Omori SA, Rickert RC, Scott M, Kotzin BL, Li X. Act1, a negative regulator in CD40- and BAFF-mediated B cell survival. Immunity. 2004;21:575–587. doi: 10.1016/j.immuni.2004.09.001. [DOI] [PubMed] [Google Scholar]
- Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, Xiao J, Lu Y, Giltiay N, Liu J, Kordula T, Zhang QW, Vallance B, Swaidani S, Aronica M, Tuohy VK, Hamilton T, Li X. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol. 2007;8:247–256. doi: 10.1038/ni1439. [DOI] [PubMed] [Google Scholar]
- Qian Y, Giltiay N, Xiao J, Wang Y, Tian J, Han S, Scott M, Carter R, Jorgensen TN, Li X. Deficiency of Act1, a critical modulator of B cell function, leads to development of Sjogren’s syndrome. Eur J Immunol. 2008;38:2219–2228. doi: 10.1002/eji.200738113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D, Randow F, Wakatsuki S, Dikic I. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Foster SA, Morgan DO. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell. 2010;39:548–559. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Foster SA, Morgan DO. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell. 2010;39:548–559. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- Rowland SL, Tremblay MM, Ellison JM, Stunz LL, Bishop GA, Hostager BS. A novel mechanism for TNFR-associated factor 6-dependent CD40 signaling. J Immunol. 2007;179:4645–4653. doi: 10.4049/jimmunol.179.7.4645. [DOI] [PubMed] [Google Scholar]
- Santos MA, Huen MS, Jankovic M, Chen HT, Lopez-Contreras AJ, Klein IA, Wong N, Barbancho JL, Fernandez-Capetillo O, Nussenzweig MC, Chen J, Nussenzweig A. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J Exp Med. 2010;10:973–81. doi: 10.1084/jem.20092308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys63-linked polyubiquitin chains. Nature. 2008;455:358. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- Scheper J, Guerra-Rebollo M, Sanclimens G, Moure A, Masip I, González-Ruiz D, Rubio N, Crosas B, Meca-Cortés O, Loukili N, Plans V, Morreale A, Blanco J, Ortiz AR, Messeguer A, Thomson TM. Protein-protein interaction antagonists as novel inhibitors of non-canonical polyubiquitylation. PLoS One. 2010;5:e11403. doi: 10.1371/journal.pone.0011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigismund S, Polo S, Di Fiore PP. Signaling through monoubiquitination. Curr Top Microbiol Immunol. 2004;286:149–185. doi: 10.1007/978-3-540-69494-6_6. [DOI] [PubMed] [Google Scholar]
- Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009a;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JJ, Haririnia A, Dickinson BC, Fushman D, Cohen RE. Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat Struct Mol Biol. 2009b;16:883–889. doi: 10.1038/nsmb.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaug B, Jiang X, Chen ZJ. The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem. 2009;78:769–796. doi: 10.1146/annurev.biochem.78.070907.102750. [DOI] [PubMed] [Google Scholar]
- Spallek T, Robatzek S, Göhre V. How microbes utilize host ubiquitination. Cell Microbiol. 2009;11:1425–1434. doi: 10.1111/j.1462-5822.2009.01346.x. [DOI] [PubMed] [Google Scholar]
- Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Op Cell Biol. 2004;16:119–26. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Sun SC. Deubiquitylation and regulation of the immune response. Nat Rev Immunol. 2008;8:501–11. doi: 10.1038/nri2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares RM, Turer EE, Liu CL, Advincula R, Scapini P, Rhee L, Barrera J, Lowell CA, Utz PJ, Malynn BA, Ma A. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S, Yamamoto M, Akira S, Takao T, Tanaka K, Iwai K. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- Tseng PH, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11:70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, Lee B, Shifrin N, Malynn BA, Ma A. Homeostatic MyD88-dependent signals cause lethal inflammation in the absence of A20. J Exp Med. 2008;205:451–464. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, Vignali DA, Bergsagel PL, Karin M. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol. 2008;9:1364–1370. doi: 10.1038/ni.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad K. Cbl-b and itch: key regulators of peripheral T-cell tolerance. Cancer Res. 2010;70:3009–3012. doi: 10.1158/0008-5472.CAN-09-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereecke L, Sze M, Guire CM, Rogiers B, Chu Y, Schmidt-Supprian M, Pasparakis M, Beyaert R, van Loo G. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. J Exp Med. 2010;207:1513–1523. doi: 10.1084/jem.20092474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada K, Niida M, Tanaka M, Kamitani T. Ro52-mediated monoubiquitination of IKK{beta} down-regulates NF-{kappa}B signalling. J Biochem. 2009;146:821–832. doi: 10.1093/jb/mvp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh MC, Kim GK, Maurizio PL, Molnar EE, Choi Y. TRAF6 autoubiquitination-independent activation of the NFkappaB and MAPK pathways in response to IL-1 and RANKL. PLoS One. 2008;3:e4064. doi: 10.1371/journal.pone.0004064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Matsuzawa A, Brown SA, Zhou J, Guy CS, Tseng PH, Forbes K, Nicholson TP, Sheppard PW, Häcker H, Karin M, Vignali DA. Analysis of nondegradative protein ubiquitylation with a monoclonal antibody specific for lysine-63-linked polyubiquitin. Proc Natl Acad Sci USA. 2008;105:20197–20202. doi: 10.1073/pnas.0810461105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Yin L, Cooper EM, Lai MY, Dickey S, Pickart CM, Fushman D, Wilkinson KD, Cohen RE, Wolberger C. Evidence for bidentate substrate binding as the basis for the K48 linkage specificity of otubain 1. J Mol Biol. 2009;386:1011–1023. doi: 10.1016/j.jmb.2008.12.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Holst J, Woo SR, Guy C, Bettini M, Wang Y, Shafer A, Naramura M, Mingueneau M, Dragone LL, Hayes SM, Malissen B, Band H, Vignali DA. Tonic ubiquitylation controls T-cell receptor:CD3 complex expression during T-cell development. EMBO J. 2010;29:1285–1298. doi: 10.1038/emboj.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- Wertz IE, Dixit VM. Signaling to NF-kappaB: regulation by ubiquitination. Cold Spring Harb Perspect Biol. 2010;2:a003350. doi: 10.1101/cshperspect.a003350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winget JM, Mayor T. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol Cell. 2010;38:627–635. doi: 10.1016/j.molcel.2010.05.003. [DOI] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation. Nat Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- Xia ZP, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Skaug B, Zeng W, Chen ZJ. A ubiquitin replacement strategy in human cells reveals distinct mechanisms of IKK activation by TNFalpha and IL-1beta. Mol Cell. 2009a;36:302–14. doi: 10.1016/j.molcel.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009b;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Okamoto T, Takeda K, Sato S, Sanjo H, Uematsu S, Saitoh T, Yamamoto N, Sakurai H, Ishii KJ, Yamaoka S, Kawai T, Matsuura Y, Takeuchi O, Akira S. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat Immunol. 2006;7:962–970. doi: 10.1038/ni1367. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Sato S, Saitoh T, Sakurai H, Uematsu S, Kawai T, Ishii KJ, Takeuchi O, Akira S. Cutting Edge: Pivotal function of Ubc13 in thymocyte TCR signaling. J Immunol. 2006;177:7520–7524. doi: 10.4049/jimmunol.177.11.7520. [DOI] [PubMed] [Google Scholar]
- Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Wertz I, O’Rourke K, Ultsch M, Seshagiri S, Eby M, Xiao W, Dixit VM. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- Zucchelli S, Codrich M, Marcuzzi F, Pinto M, Vilotti S, Biagioli M, Ferrer I, Gustincich S. TRAF6 promotes atypical ubiquitination of mutant DJ-1 and alphasynuclein and is localized to Lewy bodies in sporadic Parkinson’s disease brains. Hum Mol Genet. 2010;19:3759–3770. doi: 10.1093/hmg/ddq290. [DOI] [PubMed] [Google Scholar]