Identification of Genes Encoding tRNA Modification Enzymes by Comparative Genomics (original) (raw)
. Author manuscript; available in PMC: 2011 Feb 7.
Abstract
As the molecular adapters between codons and amino acids, transfer-RNAs are pivotal molecules of the genetic code. The coding properties of a tRNA molecule do not reside only in its primary sequence. Posttranscriptional nucleoside modifications, particularly in the anticodon loop, can modify cognate codon recognition, affect aminoacylation properties, or stabilize the codon-anticodon wobble base pairing to prevent ribosomal frameshifting. Despite a wealth of biophysical and structural knowledge of the tRNA modifications themselves, their pathways of biosynthesis had been until recently only partially characterized. This discrepancy was mainly due to the lack of obvious phenotypes for tRNA modification–deficient strains and to the difficulty of the biochemical assays used to detect tRNA modifications. However, the availability of hundreds of whole-genome sequences has allowed the identification of many of these missing tRNA-modification genes. This chapter reviews the methods that were used to identify these genes with a special emphasis on the comparative genomic approaches. Methods that link gene and function but do not rely on sequence homology will be detailed, with examples taken from the tRNA modification field.
1. Introduction
The availability of nearly 500 complete genomes (http://www.genomesonline.org/) has changed the manner by which experimental scientists can identify novel enzymes and pathways. Traditionally, linking genes and their functions started with protein purification or mutant isolation steps. Today, the bench scientist can make and validate functional predictions by combining genomic datamining with wet-laboratory experiments (see El Yacoubi et al. [2006]; Gerdes et al. [2006]; Loh et al. [2006]; and Xu et al. [2006] for recent examples). No programming skills are needed, because the genomic data and analysis tools are now freely accessible through web-based interfaces.
The sequencing effort of the past decade has revealed that 20–60% of the predicted proteins in any given genome are of unknown function (Osterman and Overbeek, 2003). Experimentalists have in-depth knowledge of specific metabolic and biological areas that most computer scientists lack. If they can harness the genomic data-mining tools, biologists and chemists are uniquely poised to predict the function of the “unknowns” and validate them in the laboratory.
The field of tRNA modification provides a good illustration of the combined power of comparative genomics and experimental validation. Even though most modifications present in tRNA molecules were discovered 20–30 years ago, many tRNA-modification genes were left unidentified (Björk, 1995; Hopper and Phizicky, 2003). The lack of knowledge about the pathways involved in nucleoside modification was largely due to their resistance to traditional biochemical and genetic characterization. Identification and purification of relevant enzyme activities from crude cell-free extracts was complicated by several factors: the difficulty of obtaining appropriate tRNA substrates, the presence of endogenous RNases that degrade the RNA substrates and products, a lack of appropriate assays, and the typically low abundance of the enzymes involved. Likewise, traditional genetic approaches were hindered by the lack of specific phenotypes in most cases. Finally, the unambiguous identification of a gene involved in tRNA modification ultimately depended on determining the presence or absence of the specific modified nucleoside in tRNA—a laborious and technically challenging process when working with large libraries of mutants (Grosjean et al., 2004). As a consequence, the identities of 50% of the tRNA modification genes were still unknown 5 years ago (de Crécy-lagard, 2004; Eastwood Leung et al., 1998). Some, such as the dihydrouridine synthesis genes, were “globally missing,” meaning they had not been identified in any organisms. Others, such as the gram-positive m5U54 methylase, were “locally missing” and identified only in a subset of organisms. This represented quite a large number of genes given that, in most organisms, ~1% of the genome is dedicated to encoding tRNA modification enzymes (Björk and Kohli, 1990; Hopper and Phizicky, 2003). Clearly, new approaches to identify tRNA modification genes were necessary.
2. Methods to Identify Missing tRNA Modification Genes
With the discovery of nearly 50 genes since 2002 (Tables 7.1-7.3), this gap in genetic understanding of tRNA modification has been nearly filled (at least for the model organisms Escherichia coli and Saccharomyces cerevisiae). Only a handful of these genes were identified by traditional genetic or biochemical methods (Table 7.1), whereas most were found by use of postgenomic experimental platforms (Table 7.1) or bioinformatic tools (Tables 7.2 and 7.3).
Table 7.1.
tRNA modification genes recently identified by use of experimentally driven approaches
| Functional role | Verified in | Protein name | Initial experimental method and reference |
|---|---|---|---|
| Postgenomic systematic approaches | |||
| Dihydrouridine synthases | S. cerevisiae | Dus1 Dus2 Dus3 Dus4 Dus5 | Biochemical profiling (Xing et al., 2002) |
| tRNA(His) guanylyltransferase | S. cerevisiae | Thg1 | Biochemical profiling (Gu et al., 2003) |
| tRNA m7G-methyltransferase | S. cerevisiae | Trm8 Trm82 | Biochemical profiling (Alexandrov et al., 2002) |
| 2-thiouridine synthesis | E. coli | TusA TusB TusC TusD | Systematic mutant analysis (Ikeuchi et al., 2006) |
| Wybutosine biosynthesis | S. cerevisiae | WyeA WyeB WyeC WyeD WyeE | Systematic mutant analysis (Noma et al., 2006) |
| Classical genetic or biochemical approaches | |||
| tRNA (uridine-5-oxyacetic acid methyl ester) 34 synthase | E. coli | CmoA | Genetic screen (Nasvall et al., 2004) |
| tRNA (5-methoxyuridine) 34 synthase | E. coli | CmoB | Genetic screen (Nasvall et al., 2004) |
| tRNA pseudouridine 13 synthase | E. coli | TruD | Protein purification (Kaya and Ofengand, 2003) |
| tRNA(cytosine32)-2-thiocytidine synthetase | E. coli | TtcA | Mapping of a previously identified mutation (Jager et al., 2004) |
| Queuosine biosynthesis | E. coli | QueC | Mutant complementation (Gaur and Varshney, 2005) |
Table 7.3.
tRNA modification genes identified by use of non-homology–based comparative genomics techniques
| Functional role | Verified in | Protein name | Key bionformatic evidence and reference |
|---|---|---|---|
| 5-Methylaminomethyl-2-thiouridine synthase | E. coli | MnmC | Clustering/fold recognition (Bujnicki et al., 2004b; de Crécy-lagard, 2004) |
| Flavin-dependent tRNA:m5U methyltransferase | B. subtilis | Gid | Occurrence profile (Urbonavicius et al., 2005) |
| tRNA lysidine synthase | E. coli | MesJ | Occurrence profile and essentiality data (Soma et al., 2003) |
| tRNA Carbamoyl-threonyl-adenosine synthase | S. cerevisiae | Sua5 | Occurrence profile/structurea |
| Wybutosine biosynthesis | S. cerevisiae | WyeA | Occurrence profile (Waas et al., 2005) |
| Bacterial tRNA dihydrouridine synthase | E. coli | DusA DusB DusC | Occurrence profile/operon (Bishop et al., 2002) |
| Queuosine/archeosine biosynthesis | Acinetobacter baylyi | QueE QueC QueD | Occurrence profile/operon (Reader et al., 2004) |
| PreQ0 reductase | E. coli B. subtilis | QueF | Occurrence profile/operon (Reader et al., 2004; Van Lanen et al., 2005) |
Table 7.2.
tRNA modification genes recently identified by use of homology-based bioinformatic approaches
| Functional role | Verified in | Protein name | Identification method and reference |
|---|---|---|---|
| tRNA-specific adenosine-34 deaminase (EC 3.5.4.-) | E. coli | TadA | BLAST with S. cerevisiae deaminase gene (Wolf et al., 2002) |
| Selenophosphate-dependent tRNA 2-selenouridine synthase | E. coli | YbbB | Search for proteins containing rhodanese domains (Wolfe et al., 2004) |
| tRNA pseudouridine synthase (position 55) | P. abyssi | PsuX | Gapped-BLAST with Euglena gracilis Cbf5p (Roovers et al., 2006; Watanabe and Gray, 2000) |
| tRNA m22G10 methyltransferase | S. cerevisiae | Trm112p Trm11p | Protein fold prediction (Purushothaman et al., 2005) |
| tRNA m1A58 methyltransferase | Themus thermophilus | TrmI | tBlastN with Rv2118c from M. tuberculosis (Droogmans et al., 2003) |
| tRNA (cytosine32/34–2′-O-)-methyltransferase | S. cerevisiae | Trm7p | BLAST with FtsJ of E. coli (Pintard et al., 2002) |
| tRNA:Cm32/Um32 methyltransferase | E. coli | YhfQ = TrmJ | SPOUT domain search (Purta et al., 2006) |
| tRNA (m7G46) methyltransferase | E. coli | YggH | Protein fold prediction (De Bie et al., 2003) |
| Wybutosine biosynthesis | S. cerevisiae | WyeC | Methylase Motif searches (Kalhor et al., 2005) |
| Mitochondrial tRNA-specific 2-thiouridylase 1 | Homo sapiens S. cerevisiae | MTU1 | BLAST with E. coli MnmA (Umeda et al., 2005) |
| tRNA (ribose 2′-O-methylase), position cytosine 56 | Pyrococcus abyssii | PAB1040 | SPOUT domain search (Renalier et al., 2005) |
The availability of whole-genome sequences has driven large-scale systematic experimental efforts such as structural genomics initiatives, systematic interaction mapping, or systematic gene disruption combined with phenotypic screenings (Huynen et al., 2004; Mittl and Grutter, 2001). For the purpose of identifying missing tRNA modification genes, these approaches have been quite effective. For example, nearly 10 genes (Table 7.1) have been identified by use of “biochemical profiling approaches” (discussed in Chapter 6). In these studies, all the proteins of S. cerevisiae (Martzen et al.,1999) and E. coli (Kitagawa et al., 2005) have been cloned and expressed and were tested in pools or individually for specific enzyme activities. In other studies, large-scale deletion mutant libraries have been completed for S. cerevisiae (Winzeler et al., 1999), B. subtilis (Kobayashi et al.,2003), and E. coli, and screening these libraries by LC-MS analysis of enzymatic digests of tRNA isolated from individual clones led to the identification of 10 other tRNA-modification gene families (Table 7.1).
Other systematic efforts that could lead to the discovery of missing tRNA-modification genes are the application of microarray technology to detect modifications (Hiley et al., 2005; Peng et al., 2003) and the availability of structural genomics data. To date, 2000 structures have been deposited by structural genomics programs in the Protein Data Bank (http://www.rcsb.org/pdb/), and more than 50,000 of these proteins have been cloned and expressed in the process (http://targetdb.pdb.org/statistics/sites/PSI.html). Structural proteomics has been quite efficient at predicting RNA/protein interactions, a first hint that a protein could be involved in tRNA or rRNA processing (see Yakunin et al. [2004] for review).
These postgenomic methods are still labor intensive and expensive. Starting with protein pools (Martzen et al., 1999) or with mutants carrying large deletions (Ikeuchi et al., 2006), reduces the quantity of assays to manageable numbers, but laboratories that use these systematic approaches to find tRNA-modification genes are still scarce, mainly because of the remaining complexity of the tRNA-modification enzyme assays. However, the availability of these postgenomic resources (clones and mutants) tremendously increases the speed at which bioinformatic-driven predictions can be tested. Hence, most tRNA-modification genes recently identified were found by combining an initial bioinformatic search with an experimental validation step (Tables 7.2 and 7.3). The bioinformatic tools used can be separated into homology based and non-homology based. The homology-based mining tools are known to most experimental scientists and will only be briefly discussed here in the context of tRNA-modification enzymes. The use of the less familiar non-homology–based genomic mining tools is the main focus of this review.
3. Homology-Based Genomic Data Mining Methods
Functional inferences based on comparative sequence analysis are well-established foundations of genomic annotation. The most significant advances in this field over the past decade are directly related to the dramatic increase in the number of sequenced genomes, as well as to the development of robust and sensitive search algorithms, such as FASTA, BLAST and their modifications (for an overview, see Koonin and Galperin [2003]). Domain analysis and grouping of putative orthologs (such as Cluster of Orthologous Groups or COGs [Tatusov et al., 2001]) play an important role in projection of functional assignments between diverse species. For well-studied gene families, in which the initial annotation has been experimentally verified, these homology-based methods are quite accurate in predicting function (Tian and Skolnick, 2003b). However, factors such as low sequence similarity (Tian and Skolnick, 2003b), multidomain proteins (Hegyi and Gerstein, 2001), gene duplications (Gerlt and Babbitt, 2000; Tian and Skolnick, 2003a), and nonorthologous displacements (Galperin and Koonin, 1998) have all contributed to incorrect or absent annotations. This has been a major problem in the field of tRNA-modification enzymes, because many are members of large paralogous families, and transferring functional annotations with BLAST scores alone can be very dangerous, particularly between kingdoms. Cases where the closest homologs in two genomes do not catalyze the same reaction are numerous in the tRNA-modification field with the added complication of having both tRNA and rRNA as potential substrates (see Jeltsch et al. [2006]; Motorin and Grosjean [1999]; Urbonavicius et al. [2005]; and Xing et al. [2004] for specific examples). That said, the use of sensitive search algorithms such as PSI-BLAST or Gapped-BLAST (Altschul et al., 1997), the development of protein fold–based methods and motifs to differentiate methylase subfamilies (Bujnicki et al., 2004a; Katz et al., 2003), and the identification of RNA binding domains such as THUMP, PUA, or SPOUT (Anantharaman et al., 2002a,b; Aravind and Koonin, 2001; Gustafsson et al., 1996; Kurowski et al., 2003) have led to many of the predictions and validations listed in Table 7.2. These methods are, however, limited to tRNA-modification enzymes that are members of superfamilies such as deaminases, methylases, or pseudouridine synthases. For the other “missing” tRNA-modification genes, the inherent limitations of homology-based approaches (only similar objects can be identified) require the use of non-homology–based comparative genomic methods.
4. Non-Homology–Based Genomic Data Mining Methods
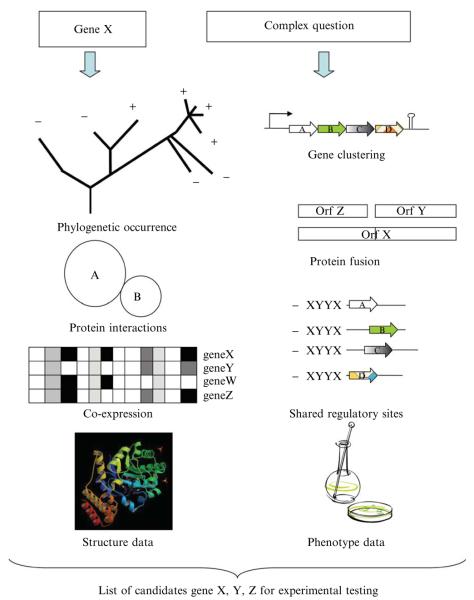
Integrating different types of genomic evidence to identify missing genes or predict the function of unknown genes started in the late 1990s just a few years after the first set of genomes was sequenced (see Bishop et al. [2002]; Bobik and Rasche [2001]; Daugherty et al. [2001]; Graham et al. [2001]; and Heath and Rock [2000] for early examples). Ten years later, the success stories are now plentiful, and several reviews have covered both the techniques and specific examples (Galperin and Koonin, 2000; Huynen et al., 2003; Kharchenko et al., 2006; Makarova and Koonin, 2003). The author recommends starting with the review by Osterman and Overbeek (Osterman and Overbeek, 2003) to grasp the core concepts of this field. These are summarized in Fig. 7.1. In short, analysis of gene clustering on the chromosome, gene fusions events, phylogenetic distribution profiles, interaction data, coexpression data, structural genomics data, phenomics data, and regulatory motifs can lead to non-homology–based predictions that can be then tested experimentally. Comparative genomics platforms in which the experimental scientist would input a gene name or sequence and all the possible functional association would be given as outputs or where one could ask complex questions integrating different types of data and genes answering these criteria would be found automatically are still not available. However, many tools have already been developed and partially integrated. Describing how to make predictions on gene function by use of these tools in a time-efficient manner with just a personal computer and Internet access is the focus of the rest of the review.
Figure 7.1.
Comparative genomic strategies used to make predictions on gene function.
With so many databases now available (Chen et al., 2007; Field et al., 2005), the “experimental” section cannot be exhaustive and reflects the personal preferences of the author (see Table 7.4 for the list of databases discussed in this review). However, a deliberate choice was made to include only resources that are available through a web interface and that are the most useful to make predictions on gene function. In the limits of the allocated space, it was impossible to walk the reader through all the query steps; however, most databases used here are straightforward to navigate. (Readers should consult the original description and/or help sections if they do not find the query processes intuitive.) One exception is the SEED database (Overbeek et al., 2005). To fully take advantage of all the possibilities of this comparative genomic platform requires an initial effort and a few hours of tutorial from a more experienced user. The derived National Microbial Pathogen Database Resource or NMPDR database (McNeil et al., 2007) is of easier access, and it is recommended to start with that interface before switching to SEED for more elaborate tasks.
Table 7.4.
Freely available databases and analysis platforms discussed in this reviewa
4.1. Predictions based on gene clustering on chromosomes
4.1.1. Overview
Genes of a given pathway have a high probability of being physically linked on the chromosome (Overbeek et al., 1999), particularly in prokaryotes. If a gene of unknown function is physically clustered with a gene of known function, a functional relationship can be inferred. The analysis of such clustering relationship is sometimes referred to as functional context analysis (Overbeek et al., 2005). The exponential growth in the number of sequenced genomes increases the chances of making inferences from clustering events at the cost of having to eliminate noninformative clustering information deriving from closely related genomes. Precomputed clustering relationships can be easily accessed through the “Search Tool for the Retrieval of Interacting Proteins” or STRING database (von Mering et al., 2003), the PhydBac database (Enault et al., 2004), or the Regulon tool of MicrobesOnline (Alm et al., 2005). A number of clustering tools are included in SEED and well described in the “functional context section” of the NMPDR tutorial (http://www.nmpdr.org/content/navigate.php). SEED is the only database that differentiates between direct (genes that cluster with a given input gene) and indirect functional coupling (genes that cluster with homologs of an input gene). These different databases will be compared in the case study that follows. The author recommends that readers try to follow the described queries in the different databases when reading the case studies presented in the review.
4.1.2. Case study
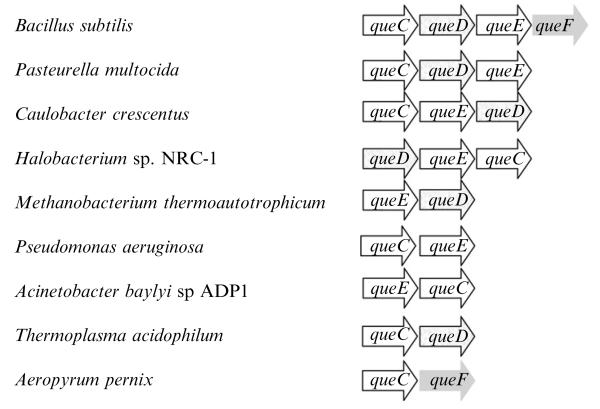
The newly discovered 7-aminomethyldeazaguanosine (preQ1) biosynthesis pathway will be used to compare the available clustering analysis platforms. This GTP-derived metabolite is the precursor of the modified base queuosine (Q) found at position 34 of tRNAHis,Tyr,Asp,Asn in most bacteria and many eukaryotes (Kersten and Kersten, 1990; Kuchino et al., 1976; Okada et al., 1978). The synthesis of preQ1 most certainly requires several genes, but none had been identified, a typical example of a globally missing pathway. By combining several comparative genomic methods with experimental validation, four new queCDEF genes involved in this pathway were identified (Reader et al., 2004). The B. subtilis queCDEF genes (ykvJKLM) are in an operon, whereas the E. coli homologs (ybaX, ygcM, ygcF, and yqcD, respectively) are scattered around the chromosome (Reader et al., 2004). Homologs of the four genes are often clustered in phylogenetically diverse genomes as shown in Fig. 7.2.
Figure 7.2.
Clustering of queCDEF genes in several genomes.
To test the different platforms, the following questions were asked. Had only one of the four queECDF gene families been identified, would the clustering tools allow the identification of the other three? Also, does the choice of the starting gene in a given gene family influence the results? Finally, are the different tools equivalent?
Each of the queCDEF genes from E. coli and B. subtilis (using the organism specific respective names) were used as initial inputs in the STRING, PhydBac, MicrobesOnline, and NMPDR databases to extract the corresponding clustered genes. The results are summarized in Table 7.5. Both the PhydBac and STRING clustering tools found that the four genes were highly clustered independently of the starting input gene. False-positive results were rare in both databases. However, the results were not strictly identical. For example, yhhQ, which is predicted to encode a preQ1/preQ0 transporter (see below), was identified only in PhydBac. SEED and GenomesOnline were both less efficient than STRING and PhydBac at detecting clustering relationships when the input genes were unclustered (as in E. coli). On the gene page in NMPDR, a “show functional coupling” link reveals direct clustering events. Clicking on the CL sign near the gene ID will lead to clustering detected with a homolog (indirect clustering events). When starting with the (unclustered) E. coli genes, no clustering was detected directly as expected. Only queE (ygcF) could be detected through the CL tool and only when starting with queC (ybaX)or queD (ygcM) not with queF (yqcD). When starting with the (clustered) B. subtilis genes, the clustering of queCDE (ykvJKL) was systematically detected (as expected), but the fourth gene of the operon queF (ykvM) failed to be identified. Results can be much improved and the clustering of the four genes identified if genes from different organisms are used as inputs (data not shown), confirming what most SEED users know from experience; it is absolutely necessary to check clustering starting from wide range of phylogenetically diverse orthologs.
Table 7.5.
Comparison of the STRING, PhydBac, NMPDR (SEED), and MicrobesOnline platforms to detect clustering events
| Predicted clustered genes | ||||
|---|---|---|---|---|
| Inputgene | String | PhydBac | NMPDR | Microbes-Online |
| QueCEc = ybaX | YgcF (0.938)a | YgcF | None detected by direct functional coupling. YbaX was detected through the CL tool. | YgcF |
| YgcM (0.844) | YgcM | YgcM | ||
| YqcD (0.675) | YbgF | |||
| YqcD | ||||
| Pal | ||||
| QueDEc = ygcM | YbaX (0.844) | YgcF | As above | YbaX |
| YgcF (0.804) | YbaX | |||
| YqcD (0.535) | YqcD | |||
| QueEEc = ygcF | YbaX (0.938) | YgcM | As above | YbaX |
| YgcM (0.804) | YqcD | |||
| YqcD (0.720) | YbaX | |||
| PyrG (0.519) | YbgF | |||
| Pal TolB | ||||
| QueFEc = yqcD | YgcF (0.720) | YgcF | None detected | None detected |
| YbaX (0.675) | YhhQ | |||
| YgcM (0.583) | YgcM | |||
| YgdH (0.440) | YbaX | |||
| QueCBs = ykvJ | YkvL (0.940) | NAb | YkvK (score 12)a | YkvJKLM |
| YkvM (0.840) | YkvL (score 12) | |||
| YkvK (0.804) | ||||
| QueDBs = ykvK | YkvL(0.926) | NA | YkvJ (score 12) | YkvJKLM |
| YkvJ (0.804) | YkvL (score 6) | |||
| YkvM (0.481) | ||||
| QueEBs = ykvL | YkvJ (0.940) | NA | YkvJ (score 12) | YkvJKLM |
| YkvK (0.926) | YkvK (score 6) | |||
| YkvM (0.792) | ||||
| QueFBs = ykvM | YkvJ (0.840) | NA | None detected by direct functional coupling. YkvJ and YkvL were detected through the CL tool. | YkvJKLM |
| YkvL (0.792) | ||||
| YkvK (0.481) |
In terms of visualization tools, all four databases have graphical summaries of the clustering, but they are all precomputed. One exception is the Genome Browser tool of MicrobesOnline that displays the regions surrounding a given gene in different genomes and allows the user to choose the genomes and the size of the regions in a format that can be exported in graphics. This feature can be very useful when preparing figures.
4.1.3. Conclusion
The user should not be faithful to any one database but should try them all when searching for clustering events. As a rule, several members of a given gene family should be used as inputs, particularly in the SEED database, because the strength of SEED lies more in detecting and representing clustering in the context of a subsystem analysis as discussed in the following.
4.2. Detecting protein fusion events
4.2.1. Overview
In a gene fusion event, two separate parent genes are encoded in a single multifunctional polypeptide. These fusions, which have been called Rosetta stone proteins, suggest a high probability of functional interaction between the two proteins (Enright et al., 1999; Pellegrini et al., 1999). As with the inferences driven by the physical clustering analysis described previously, if the function of one of the two genes is known and the other is not, detecting the fusion event can allow strong functional predictions (see Daugherty et al. [2002] and Levin et al. [2004] for examples). Several web-based platforms have been specifically designed to detect these fusion events, but the author has not found them very effective. One reason is that the best fusion hints often come from comparing eukaryotic and prokaryotic genomes, because fusions events are more frequent in eukaryotic genomes (Veitia, 2002). Unfortunately, the databases that are the most user-friendly and directly integrate the fusion data (FusionDB [Suhre and Claverie, 2004] and STRING) focus mainly on prokaryotic genomes and, therefore, miss many fusion events. Until better specialized databases are available, the author has found that databases that analyze protein domains such as CDART at NCBI (Geer et al., 2002)orPfam at the Sanger Center (Finn et al., 2006) are very effective at detecting protein fusion events. Both cover all known proteins from both prokaryotic and eukaryotic genomes. The output might not be very selective, particularly with protein domains that are ubiquitous; however, both CDART and Pfam present the results in graphic summaries that can be analyzed very effectively. Fusion events can also be easily detected in the SEED database with the color coding of the protein similarities table (see [http://theseed.uchicago.edu/FIG/Html/similarity_region_colors.html] for explanations).
4.2.2. Case study
To illustrate both the use of these fusion detection tools and the efficiency of the domain databases with a tRNA-related example, the PAB1506 protein from Pyrococcus abyssii that encodes a stand-alone PUA domain was analyzed. The PUA domain is found in many RNA binding proteins (Anantharaman et al., 2002a) and in several tRNA modifying enzymes such as archaeosine tRNA guanine transglycosidase (Tgt) (Ishitani et al., 2002) and pseudouridine synthase (TruB) (Hoang and Ferre-D'Amare, 2001). The function of PAB1506 is unknown. When the PhydBac database is queried with PAB1506, two fusion events can be detected linking PAB1506 to PAB2176 (annotated as an esterase) and to PAB0064 (annotated as a hydrolase). These two fusions were not detected in the STRING database by use of the exact same input protein nor in the CDART, Pfam or SEED databases. When analyzed in detail, the PhydBac result is most likely due to sequencing errors. As a rule, fusion events detected in only one genome should be carefully checked.
Neither STRING nor FusionDB detected the PUA fusions to the TGT and TruB domains. This result was expected, because their method is designed to eliminate hits from domains found in many different proteins in the same genome (explained in http://www.igs.cnrs-mrs.fr/FusionDB/methods.html). These were identified with both the CDART and Pfam domain analysis tool and the SEED color-coding homology tool. Additional fusions with the metabolic enzymes glutamate-5-kinase and 3′-phosphoadenosine 5′-phosphosulfate sulfotransferase (CysH domain) were also detected in CDART and Pfam. All these fusion events had previously been identified in a comprehensive graph-based analysis (Ye and Godzik, 2005).
The fusion with the glutamate 5-kinase is present in nearly all bacterial genomes, and the PUA domain has a role in activation of the enzyme and not in tRNA binding (Perez-Arellano et al., 2005). The fusion of the PUA and CysH domains is limited to the archaeal kingdom. CDART also detected proteins from methanogenic archaea that contained not only the PUA and CysH domains but also additional domains such as cysteine desulfurase domains or ferredoxin domains (data not shown). This observation is interesting, because the sulfur metabolism in methanogenic archaea is not fully understood, and it has been recently proposed that cysteine biosynthesis could occur mainly on the tRNACys molecule (Helgadottir et al., 2007; Sauerwald et al., 2005). The archaeal PUA-CysH family could, therefore, be involved in channeling the thiol from the charged tRNA to the target metabolites, and we are currently testing this hypothesis.
4.2.3. Conclusion
As the ultimate in genome clustering events, protein fusions are very powerful prediction tools. However, no database is available that is really efficient in detecting these events yet. The protein domain analysis tools CDART, Pfam or the SEED color-coding tools are the best default ones to date, with the caveat that one will retrieve not only true fusion events but also all proteins containing a given domain.
4.3. Searches based on phylogenetic distribution profiles
4.3.1. Overview
Another powerful tool that does not rely on any homology information is to query phylogenetic distribution profiles (Pellegrini et al., 1999). In this application, one needs to compute the proteins that are present in a given set of organisms and absent in another set. The initial COG database was the pioneer for such queries (Tatusov et al., 1997) but became outdated quickly because of its limited set of genomes (43 only). The new version of COG contains 66 genomes (Tatusov et al., 2003), but to the author's dismay, the phylogenetic query tool has disappeared (or is very difficult to find). Protein Links Explorer or PLEX (Date and Marcotte, 2005) has slightly more genomes than the COG platform (88). The strength of COG and PLEX is their speed, because all the phylogenetic patterns are precomputed, but this is also a limitation, because they do not get updated often. STRING and PhydBac have many more genomes and precomputed phylogenetic profiles. These databases are very powerful for identifying genes that follow the same profile as an initial query gene; however, the user cannot extract a list of genes that follow a phylogenetic distribution pattern.
The author is aware of three databases that combine constantly updated genomes with robust phylogenetic distribution query tools. CoGenT++ is part of an extensive computational genomics environment led by the European Bioinformatics Institute and has a phylogenetic profile tool (Goldovsky et al., 2005). The NMPDR database (McNeil et al., 2007) includes a “signature gene tool” with the added possibility of filtering the output list with keywords. Another valuable resource is the orthologous distribution tables of the MicroBial Genome Database for comparative analysis or MBGD (Uchiyama, 2007). A table containing all of the orthologous families in a given set of genomes can be generated and then queried for particular phylogenetic distribution patterns. There again filters can be used to sort the output data. Only 100 genomes can be analyzed, but this is sufficient for most queries.
4.3.2. Case study
To illustrate the power of the use of phylogenetic distribution profiles, we tested the strategy used to identify the gene encoding the missing wybutosine tricyclic guanosine-ring forming enzyme WyeA (Waas et al., 2005) in the different databases. Literature analysis extracted from the tRNA database (Sprinzl et al., 1999) suggested that this gene should be present in archaea, yeast, and Homo sapiens and absent in bacteria and fly. An input profile was generated to query the different databases. The gene family should be present in Methanococcus janaschii, Homo sapiens, S. cerevisiae, but absent in E. coli, B. subtilis, and Drosophila melanogaster. To query MBGD that lacks many eukaryotic genomes, Homo sapiens was eliminated from the query list and the fly genome was replaced by another insect, Encephalotazoon cuniculi. Remarkably the output from the PLEX search showed only the two protein families exemplified by the yeast proteins Ypl207w and Ygl050w. Experimental validation by several groups has shown that both these proteins are, indeed, involved in wybutosine biosynthesis (Kalhor et al., 2005; Noma et al., 2006; Waas et al., 2005). The output from the NMPDR and MBGD, CoGenT++ databases were less selective. Both protein families identified in the PLEX search were identified, but more false-positive hits obscured the result. Thirty-five protein families were extracted by use of MBGD, because fewer genomes were used in the query. Several hundred were found by use of CoGenT++, because the user is not given the choice of a genome for the output list, making the results quite difficult to analyze. The NMPDR analysis gave 435 output proteins, but the NMPDR signature tool also extracts gene families that do not follow exactly the query with perfect matches given the highest score of two.
4.3.3. Conclusion
A well-designed phylogenetic query is very efficient at identifying gene candidates for a given function, and several databases make these queries possible. Success will depend on: (1) the robustness of the initial biological information used to design the profile; (2) imposing the fewest possible query constraints but still being stringent enough so that the output is not too large, which usually means trying different combinations of genome choices as inputs; (3) trying several search databases; and (4) even if the output list of families is large, it can be reduced by combining with other criteria such as keywords or physical clustering as discussed later with the identification of the lysidine synthase gene (tilS).
4.4. Mining other types of “Omics” data
Inferences on gene functions can be derived from many types of associations. For example, genes in the same pathways are often regulated by a common protein recognizing a specific DNA sequence or by common riboswitches (Gelfand et al., 2000b). Finding genes that share regulatory sites is, therefore, a powerful method to link genes functionally (see Barrick et al. [2004]; Rodionov et al. [2006]; and Yang et al. [2006] for examples). In an example related to tRNA modification, a riboswitch was identified upstream of the B. subtilis ykvJKLM operon (Barrick et al., 2004). Genes under the control of the same riboswitch in other genomes include yhhQ (Barrick et al., 2004), a predicted transporter protein that also clusters with queuosine pathway genes (see Table 7.5). We are, therefore, currently testing the hypothesis that YhhQ is a preQ1/preQ0 transporter. Unfortunately, the algorithms to detect conserved DNA motifs over all sequenced genomes such as SignalX are not available yet as web-based applications (Gelfand et al., 2000a). The reader will have to wait before such queries can be performed without the use of specialized programs that require some computer programming skills.
Associations can also be derived from interaction data sets (results of systematic two-hybrid or Tap-Tag experiments), coexpression data sets (results of expression profiling on microarrays), or phenotype arrays. The rapid increase in the volume and quality of functional genomics data is expected to strongly impact functional gene characterization in the near future. Among the growing number of web resources are the Stanford Microarray Database (SMD) for expression data (http://genome-www5.stanford.edu/) and the Database of Interacting Proteins (DIP) for protein–protein interactions (http://dip.doe-mbi.ucla.edu/). For the purpose of using these resources to predict gene function, two main problems remain: (1) the great number of false-positive or noninformative associations; and (2) the difficulty of mining these data at the click of a mouse (particularly for microarray results). However, adding filters and/or combining with other types of information can solve the first problem, as does the STRING database that integrates results from interactions and array experiments with clustering and phylogenetic data (von Mering et al., 2007).
The information that the researcher hunting for a gene's function would like to extract from array data is the list of genes having the same expression profile as the input gene over all expression data available. The Program Array prospector (Jensen et al., 2004) was designed for this purpose, but, unfortunately, the web site did not seem to be working when tested. Organism-specific databases such as Saccharomyces Genome Database (SGD) for yeast (Nash et al., 2007) or TAIR for Arabidopsis thaliana (Rhee et al., 2003) that are constantly integrating all the available “omics” data will allow such queries and are obvious starting points when possible.
Finally, the availability of large-scale mutant libraries allows the implementation of phenomics approaches (consisting of phenotype arrays or multiplexed phenotype tests screening of all mutants). These data are also starting to be mined (Kahraman et al., 2005). One example is the Prophecy database that enables phenotypes of all available S. cerevisiae mutants to be accessed through different query formats (Fernandez-Ricaud et al., 2007). However, phenomic information is not yet integrated in comparative genomic databases except for essentiality data. Systematic mutant construction libraries or transposon library mapping (see Osterman and Begley [2006] for descriptions of techniques) allows the prediction of which genes are essential for growth in specific organisms. This information has been integrated in the SEED database.
4.4.1. Case study
One example of the use of essentiality information in the tRNA modification field is the discovery of tilS encoding lysidine synthase (Soma et al., 2003). This modification was predicted to be found only in bacteria and to be essential for survival, because in its absence, the minor tRNAIleCAU would be charged by methionine (Muramatsu et al., 1988). By use of the signature tool of the NMPDR database, the following query can be performed. Which genes are present in Bacillus subtilis 168, Buchnera aphidicola str. APS, Escherichia coli K12, Mycoplasma mycoides subsp. mycoides, Wolbachia sp. endosymbiont of Drosophila melanogaster absent in Arabidopsis thaliana, Methanocaldococcus jannaschii, Saccharomyces cerevisiae and essential for growth in E. coli. The output list of 91 genes contains only approximately 10 genes of unknown function; one of them encodes the lysidine synthase.
4.4.2. Conclusion
Both the postgenomic experimental data sets and the platforms to analyze them are constantly improving. We anticipate that if we update this review in a few years, examples of prediction driven by mining postgenomic data will be much more numerous than today.
4.5. Subsystem analysis
4.5.1. Overview
Very early in the genomic era it was apparent that a dramatic enhancement of the quality and utility of genomic annotations can be achieved with metabolic reconstruction technology in which genes encoding metabolic pathways are inventoried in given genome (Galperin and Brenner, 1998; Selkov et al., 1997). By placing genes in the context of metabolic pathways, metabolic reconstruction was a key component of the success of genome sequencing, because the physiology and metabolism of an organism can now be predicted from genomes (Galperin and Brenner, 1998; Overbeek et al., 1999).
Stemming from metabolic reconstruction technology is the possibility of analyzing metabolic pathways across all genomes by computing the presence or absence of pathway genes. The consequence of this type of analysis was the realization that the number of missing genes (both “locally” or “globally” missing) was much larger than expected, reflecting the diversity of metabolic solutions used by life. Many public resources support this approach such as KEGG/GenomeNet (Kanehisa et al., 2006), MetaCyc (Krieger et al., 2004), the CMR-genome properties (Haft et al., 2005), and MicrobesOnline (Alm et al., 2005). In all of these platforms, spreadsheets computing the distribution of the genes of specific pathways in all (or in a subset of ) genomes can be generated. MicrobesOnline has also developed very helpful graphical interfaces by use of the KEGG pathway maps as templates. The great limitation of these databases is that all of the pathways that can be profiled are precomputed, and all use the KEGG pathway database as template. This led Ross Overbeek and colleagues to develop the concept of the subsystem, first with the commercial ERGO database (Overbeek, 2003), then with the freely available SEED database (Overbeek et al., 2005). A subsystem is a collection of genes that is built by the user and in which the genes are analyzed as a group. It can consist of genes of a pathway or a complex but is not limited to these. Subsystems can be updated or modified at will. The tools to build and analyze subsystems are at the core of the SEED platform.
4.5.2. Case study
In the case of preQ1 biosynthesis, no pathway was present in KEGG, because the enzymes of the pathway had not been characterized. A queuosine subsystem was created, including the known Q biosynthesis enzymes such as tRNA-guanine transglycolase (TGT) (Noguchi et al., 1982), QueA (Reuter et al., 1991; Slany et al., 1994), and the newly discovered QueCDEF enzymes. After a step in which all orthologs of the subsystem families are annotated in all genomes by the user (this step should not be performed without adequate SEED training), the subsystem spreadsheet was generated. As shown in Table 7.6, for every genome in the database the presence or absence of the genes of the subsystem is visualized with a link to the corresponding protein. If two genes are physically clustered in a given genome, they will be highlighted in the same color. The clustering of the queCDEF genes becomes very apparent with the color coding (Table 7.6). Also rare clustering events such as the queD-tgt proximity in Synechocystis that had not been detected by any of the clustering tools discussed previously become easy to visualize. This is important, because in some cases, clustering occurring in just a few genomes can give the initial association clue.
Table 7.6.
Clustering of the queCDEF and tgt genes derived from subsystem analysisa
| Organism | Genomeid | QueD | QueC | QueE | QueF | Tgt |
|---|---|---|---|---|---|---|
| Bacteroides fragilis ATCC 25285 | 272559.3 | 3666 | 1310 | 3665 | 1311 | 2272 |
| Porphyromonas gingivalis W83 | 242619.1 | 913 | 1124 | 914 | 1154 | 436 |
| Synechocystis sp. PCC 6803 | 1148.1 | 2581 | 2844 | 1133 | 2787 | 2794 |
| Bacillus subtilis subsp. subtilis str. 168 | 224308.1 | 1375 | 1374 | 1376 | 1377 | 2774 |
| Staphylococcus aureus subsp. aureus COL | 93062.4 | 260 | 261 | 259 | 275 | 1875 |
| Bradyrhizobium japonicum USDA 110 | 224911.1 | 2482 | 4495 | 2483 | 4796 | 4683 |
| Oceanicaulis alexandrii HTCC2633 | 314254.3 | 1577 | 2212 | 2211 | 1787 | 1713 |
| Rickettsia felis URRWXCal2 | 315456.3 | 1093 | 51 | 185 | 539 | 25 |
| Zymomonas mobilis subsp. mobilis ZM4 | 264203.3 | 1550 | 1861 | 107 | 822 | 859 |
| Bordetella bronchiseptica RB50 | 257310.1 | 3059 | 417 | 3058 | 3322 | 1355 |
| Neisseria meningitidis FAM18 | 487.2 | 550 | 548 | 552 | 1595 | 1794 |
| Nitrosomonas europaea ATCC 19718 | 228410.1 | 1457 | 215 | 214 | 2184 | 1097 |
| Wolinella succinogenes DSM 1740 | 273121.1 | 1516 | 1515 | 1514 | 4 | 1394 |
| Buchnera aphidicola str. Sg (Schizaphis graminum) | 198804.1 | 382 | 435 | 381 | 273 | 122 |
| Escherichia coli K12 | 83333.1 | 2721 | 441 | 2733 | 2750 | 403 |
| Salmonella typhimurium LT2 | 99287.1 | 2845 | 440 | 2847 | 2864 | 391 |
| Yersinia pestis KIM | 187410.1 | 805 | 1016 | 804 | 3097 | 973 |
| Acinetobacter sp. ADP1 | 62977.3 | 2241 | 2377 | 2376 | 2161 | 128 |
| Psychrobacter sp. 273–4 | 259536.4 | 1015 | 1369 | 1335 | 219 | 1592 |
| Xanthomonas campestris pv. campestris str. 8004 | 314565.3 | 4169 | 1290 | 1289 | 3587 | 2464 |
| Magnetococcus sp. MC-1 | 156889.1 | 2790 | 1234 | 2789 | 928 | 3471 |
4.5.3. Conclusion
By focusing on a specific subsystem, the user can identify the globally and locally missing genes, visualize clustering, or see phylogenetic distribution patterns. In this way, SEED soon becomes for the user a virtual laboratory where specific hypotheses can be tested in silico.
5. General Conclusion: The Power of Integration
The possibility of asking complex queries that integrate many types of data is the next bioinformatic challenge. For example, to find the tRNA dihydrouridine synthase genes, one could ask the following question: what protein families are absent in Pyrococcus sp. but present in E. coli, B. subtilis, and S. cerevisiae, and are part of a dehydrogenase family, bind tRNA, and cluster with genes related to translation (Bishop et al.,2002)? The data allowing the correct prediction are available but have not yet been integrated in a database in a queriable form. Several platforms are working toward this goal, and a summary of the integrative capabilities of a few databases is presented in Table 7.7. These tools are constantly improving, and in a few years such complex queries might, indeed, become possible. One example of integration is the query toolbox of CMR-Genome properties that can filter searches by use of different types of characteristic such as MW, pI, or keywords in the annotations. Finally, although the examples in this review were taken from the tRNA modifications field, the techniques discussed here could be applied to any field of metabolism.
Table 7.7.
Comparison of integrative databases
| Databases | ||||||
|---|---|---|---|---|---|---|
| Properties | Entrez | STRING | SEED/MNPDR | CoGenT++ | Microbes-Online | CMR-Genome Properties |
| Chromosome clustering | No | Yes | Yes | No | Yes | No |
| Protein fusion analysis | CDARTa | Yes | Yesa | Yesb | Yesa | Yesa |
| Precomputed phylogenetic profiles | COG | Yes | No | Yes | No | No |
| Phylogenetic query tool | COG | No | Yes | Yes | No | No |
| Precomputed pathways analysis tools | No | No | Yes | Not yet | Yes | Yes |
| User-defined pathways analysis tool | No | No | Yes | No | No | No |
| Array data | GEO | Yes | No | No | Yes | No |
| Interaction data | No | Yes | No | No | No | No |
| Essentiality | No | No | Yes | No | No | No |
| Domain/family | CDART | No | SEED-FAM | Yes | No | TIGRFAM |
| Literature integration | PubMed | Yes | No | No | No | No |
| Elaborate queries | MyNCBI | No | No | No | No | Yes |
ACKNOWLEDGMENTS
This work was supported in part by The National Science foundation (MCB-05169448) and by the National Institutes of Health (R01 GM70641–01). The author thanks Madeline Rasche, Andrew Hanson, Basma El Yacoubi, Andrei Osterman, and Henri Grosjean for helpful discussions and critical reading of the manuscript.
REFERENCES
- Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA. 2002;8:1253–1266. doi: 10.1017/s1355838202024019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm EJ, Huang KH, Price MN, Koche RP, Keller K, Dubchak IL, Arkin AP. The MicrobesOnline Web site for comparative genomics. Genome Res. 2005;15:1015–1022. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucl. Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucl. Acids Res. 2002a;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. SPOUT: A class of methyl-transferases that includes spoU and trmD RNA methylase superfamilies, and novel super-families of predicted prokaryotic RNA methylases. J. Mol. Microbiol. Biotechnol. 2002b;4:71–75. [PubMed] [Google Scholar]
- Aravind L, Koonin EV. THUMP—a predicted RNA-binding domain shared by 4-thiouridine, pseudouridine synthases and RNA methylases. Trends Biochem. Sci. 2001;26:215–217. doi: 10.1016/s0968-0004(01)01826-6. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, Wickiser JK, Breaker RR. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AC, Xu J, Johnson RC, Schimmel P, de Crécy-Lagard V. Identification of the tRNA-dihydrouridine synthase family. J. Biol. Chem. 2002;277:25090–25095. doi: 10.1074/jbc.M203208200. [DOI] [PubMed] [Google Scholar]
- Björk GR. Biosynthesis and Function of Modified Nucleosides. In: RajBhandary UL, editor. tRNA: Structure, Biosynthesis, and Function. ASM Press; Washington D. C.: 1995. pp. 165–206. [Google Scholar]
- Björk GR, Kohli J. Synthesis and Function of Modified Nucleosides in tRNA. In: Gehrke C, Kuo K, editors. Chromatography and Modification of Nucleosides. Part B. Biological Roles and Function of Modification. Elsevier; Amsterdam: 1990. pp. B13–B67. [Google Scholar]
- Bobik TA, Rasche ME. Identification of the human methylmalonyl-CoA racemase gene based on the analysis of prokaryotic gene arrangements. Implications for decoding the human genome. J. Biol. Chem. 2001;276:37194–37198. doi: 10.1074/jbc.M107232200. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM, Droogmans L, Grosjean H, Purushothaman SK, Lapeyre B. Bioinformatics-guided identification of novel RNA methyltransferases. In: Bujnicki JM, editor. Practical Bioinformatics. Vol. 15. Springer-Verlag; Berlin Heidelberg: 2004a. pp. 139–168. [Google Scholar]
- Bujnicki JM, Oudjama Y, Roovers M, Owczarek S, Caillet J, Droogmans L. Identification of a bifunctional enzyme MnmC involved in the biosynthesis of a hypermodified uridine in the wobble position of tRNA. RNA. 2004b;10:1236–1242. doi: 10.1261/rna.7470904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-B, Chattopadhyay A, Bergen P, Gadd C, Tannery N. The Online Bioinformatics Resources Collection at the University of Pittsburgh Health Sciences Library System—a one-stop gateway to online bioinformatics databases and software tools. Nucl. Acids Res. 2007;35:D780–D785. doi: 10.1093/nar/gkl781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Date SV, Marcotte EM. Protein function prediction using the Protein Link EXplorer (PLEX) Bioinformatics. 2005;21:2558–2559. doi: 10.1093/bioinformatics/bti313. [DOI] [PubMed] [Google Scholar]
- Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, de Crécy-Lagard V, Osterman A. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J. Biol. Chem. 2002;277:21431–21439. doi: 10.1074/jbc.M201708200. [DOI] [PubMed] [Google Scholar]
- Daugherty M, Vonstein V, Overbeek R, Osterman A. Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol. 2001;183:292–300. doi: 10.1128/JB.183.1.292-300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bie LG, Roovers M, Oudjama Y, Wattiez R, Tricot C, Stalon V, Droogmans L, Bujnicki JM. The yggH gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase. J. Bacteriol. 2003;185:3238–3243. doi: 10.1128/JB.185.10.3238-3243.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crécy-lagard V. Bioinformatics leads the path to the identification of missing tRNA modification genes. In: Bujnicki JM, editor. Practical Bioinformatics. Vol. 15. Springer-Verlag; Berlin Heidelberg: 2004. pp. 169–190. [Google Scholar]
- Droogmans L, Roovers M, Bujnicki JM, Tricot C, Hartsch T, Stalon V, Grosjean H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucl. Acids Res. 2003;31:2148–2156. doi: 10.1093/nar/gkg314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood Leung H-C, G. HT, Björk GR, Winkler ME. Genetic locations and database accession numbers of RNA-modifying and -editing enzymes. In: Benne R, editor. Modification and Editing of RNA. ASM Press; Washington, D.C.: 1998. pp. 561–568. [Google Scholar]
- El Yacoubi B, Bonnett S, Anderson JN, Swairjo MA, Iwata-Reuyl D, de Crécy-Lagard V. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J. Biol. Chem. 2006;281:37586–37593. doi: 10.1074/jbc.M607114200. [DOI] [PubMed] [Google Scholar]
- Enault F, Suhre K, Poirot O, Abergel C, Claverie JM. Phydbac2: Improved inference of gene function using interactive phylogenomic profiling and chromosomal location analysis. Nucl. Acids Res. 2004;32:W336–W339. doi: 10.1093/nar/gkh365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86–90. doi: 10.1038/47056. [DOI] [PubMed] [Google Scholar]
- Fernandez-Ricaud L, Warringer J, Ericson E, Glaab K, Davidsson P, Nilsson F, Kemp GJL, Nerman O, Blomberg A. PROPHECY—a yeast phenome database, update 2006. Nucl. Acids Res. 2007;35:D463–D467. doi: 10.1093/nar/gkl1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field D, Feil EJ, Wilson GA. Databases and software for the comparison of prokaryotic genomes. Microbiology. 2005;151:2125–2132. doi: 10.1099/mic.0.28006-0. [DOI] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer ELL, et al. Pfam: Clans, web tools and services. Nucl. Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Brenner SE. Using metabolic pathway databases for functional annotation. Trends Genet. 1998;14:332–333. doi: 10.1016/s0168-9525(98)01523-6. [DOI] [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Sources of systematic error in functional annotation of genomes: Domain rearrangement, non-orthologous gene displacement and operon disruption. In Silico Biol. 1998;1:55–67. [PubMed] [Google Scholar]
- Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat. Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- Gaur R, Varshney U. Genetic analysis identifies a function for the queC (ybaX) gene product at an initial step in the queuosine biosynthetic pathway in Escherichia coli. J. Bacteriol. 2005;187:6893–6901. doi: 10.1128/JB.187.20.6893-6901.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer LY, Domrachev M, Lipman DJ, Bryant SH. CDART: Protein Homology by Domain Architecture. Genome Res. 2002;12:1619–1623. doi: 10.1101/gr.278202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Koonin EV, Mironov AA. Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucl. Acids Res. 2000a;28:695–705. doi: 10.1093/nar/28.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Novichkov PS, Novichkova ES, Mironov AA. Comparative analysis of regulatory patterns in bacterial genomes. Brief Bioinform. 2000b;1:357–371. doi: 10.1093/bib/1.4.357. [DOI] [PubMed] [Google Scholar]
- Gerdes SY, Kurnasov OV, Shatalin K, Polanuyer B, Sloutsky R, Vonstein V, Overbeek R, Osterman AL. Comparative genomics of NAD biosynthesis in Cyanobacteria. J. Bacteriol. 2006;188:3012–3023. doi: 10.1128/JB.188.8.3012-3023.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlt JA, Babbitt PC. Can sequence determine function? Genome Biol. 2000;1:1–10. doi: 10.1186/gb-2000-1-5-reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldovsky L, Janssen P, Ahren D, Audit B, Cases I, Darzentas N, Enright AJ, Lopez-Bigas N, Peregrin-Alvarez JM, Smith M, Tsoka S, Kunin V, Ouzounis CA. CoGenT++: An extensive and extensible data environment for computational genomics. Bioinformatics. 2005;21:3806–3810. doi: 10.1093/bioinformatics/bti579. [DOI] [PubMed] [Google Scholar]
- Graham DE, Graupner M, Xu H, White RH. Identification of coenzyme M biosynthetic 2-phosphosulfolactate phosphatase. A member of a new class of Mg2+-dependent acid phosphatases. Eur. J. Biochem. 2001;268:5176–5188. doi: 10.1046/j.0014-2956.2001.02451.x. [DOI] [PubMed] [Google Scholar]
- Grosjean H, Keith G, Droogmans L. Detection and quantification of modified nucleotides in RMA using thin-layer chromatography. In: Gott J, editor. Methods in Molecular Biology. Vol. 265. Humana Press; Totowa, NJ: 2004. pp. 357–391. [DOI] [PubMed] [Google Scholar]
- Gu W, Jackman JE, Lohan AJ, Gray MW, Phizicky EM. tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes Dev. 2003;17:2889–2901. doi: 10.1101/gad.1148603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson C, Reid R, Greene PJ, Santi DV. Identification of new RNA modifying enzymes by iterative genome search using known modifying enzymes as probes. Nucl. Acids Res. 1996;24:3756–3762. doi: 10.1093/nar/24.19.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut JD, Brinkac LM, Zafar N, White O. Genome Properties: A system for the investigation of prokaryotic genetic content for microbiology, genome annotation and comparative genomics. Bioinformatics. 2005;21:293–306. doi: 10.1093/bioinformatics/bti015. [DOI] [PubMed] [Google Scholar]
- Heath RJ, Rock CO. A triclosan-resistant bacterial enzyme. Nature. 2000;406:145–146. doi: 10.1038/35018162. [DOI] [PubMed] [Google Scholar]
- Hegyi H, Gerstein M. Annotation transfer for genomics: Measuring functional divergence in multi-domain proteins. Genome Res. 2001;11:1632–1640. doi: 10.1101/gr.183801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgadottir S, Rosas-Sandoval G, Soll D, Graham DE. Biosynthesis of phosphoserine in the Methanococcales. J. Bacteriol. 2007;189:575–582. doi: 10.1128/JB.01269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiley SL, Jackman J, Babak T, Trochesset M, Morris QD, Phizicky E, Hughes TR. Detection and discovery of RNA modifications using microarrays. Nucl. Acids Res. 2005;33:e2. doi: 10.1093/nar/gni002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang C, Ferre-D'Amare AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: Nucleotide flipping by an RNA modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Phizicky EM. tRNA transfers to the limelight. Genes Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Snel B, Mering C, Bork P. Function prediction and protein networks. Curr. Opin Cell. Biol. 2003;15:191–198. doi: 10.1016/s0955-0674(03)00009-7. [DOI] [PubMed] [Google Scholar]
- Huynen MA, Snel B, van Noort V. Comparative genomics for reliable protein-function prediction from genomic data. Trends Genet. 2004;20:340–344. doi: 10.1016/j.tig.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Ikeuchi Y, Shigi N, Kato J, Nishimura A, Suzuki T. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell. 2006;21:97–108. doi: 10.1016/j.molcel.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Ishitani R, Nureki O, Fukai S, Kijimoto T, Nameki N, Watanabe M, Kondo H, Sekine M, Okada N, Nishimura S, Yokoyama S. Crystal structure of archaeosine tRNA-guanine transglycosylase. J. Mol. Biol. 2002;318:665–677. doi: 10.1016/S0022-2836(02)00090-6. [DOI] [PubMed] [Google Scholar]
- Jager G, Leipuviene R, Pollard MG, Qian Q, Björk GR. The conserved Cys-X1-X2-Cys motif present in the TtcA protein is required for the thiolation of cytidine in position 32 of tRNA from Salmonella enterica serovar Typhimurium. J. Bacteriol. 2004;186:750–757. doi: 10.1128/JB.186.3.750-757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeltsch A, Nellen W, Lyko F. Two substrates are better than one: Dual specificities for Dnmt2 methyltransferases. Trends Biochem. Sci. 2006;31:306. doi: 10.1016/j.tibs.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Jensen LJ, Lagarde J, von Mering C, Bork P. ArrayProspector: A web resource of functional associations inferred from microarray expression data. Nucl. Acids Res. 2004;32:W445–W448. doi: 10.1093/nar/gkh407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahraman A, Avramov A, Nashev LG, Popov D, Ternes R, Pohlenz H-D, Weiss B. PhenomicDB: A multi-species genotype/phenotype database for comparative phenomics. Bioinformatics. 2005;21:418–420. doi: 10.1093/bioinformatics/bti010. [DOI] [PubMed] [Google Scholar]
- Kalhor HR, Penjwini M, Clarke S. A novel methyltransferase required for the formation of the hypermodified nucleoside wybutosine in eucaryotic tRNA. Bioch. Bioph. Res. Com. 2005;334:433. doi: 10.1016/j.bbrc.2005.06.111. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: New developments in KEGG. Nucl. Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JE, Dlakic M, Clarke S. Automated Identification of Putative Methyl-transferases from Genomic Open Reading Frames. Mol. Cell. Proteomics. 2003;2:525–540. doi: 10.1074/mcp.M300037-MCP200. [DOI] [PubMed] [Google Scholar]
- Kaya Y, Ofengand J. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA. 2003;9:711–721. doi: 10.1261/rna.5230603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersten H, Kersten W. Biosynthesis and Function of Queuine and Queuosine tRNAs. In: Kuo KCT, editor. Chromatography and Modification of Nucleosides Part B. Elsevier; Amsterdam: 1990. pp. B69–B108. [Google Scholar]
- Kharchenko P, Chen L, Freund Y, Vitkup D, Church G. Identifying metabolic enzymes with multiple types of association evidence. BMC Bioinformatics. 2006;7:177. doi: 10.1186/1471-2105-7-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): Unique Resources for Biological Research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, Boland F, Brignell SC, et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Galperin MY. SEQUENCE-EVOLUTION-FUNCTION. Computational approaches in comparative genomics. Kluwer Academic Publishers; Dordrecht, Netherlands: 2003. [PubMed] [Google Scholar]
- Krieger CJ, Zhang P, Mueller LA, Wang A, Paley S, Arnaud M, Pick J, Rhee SY, Karp PD. MetaCyc: A multiorganism database of metabolic pathways and enzymes. Nucl. Acids Res. 2004;32:D438–D442. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchino Y, Kasai H, Nihei K, Nishimura S. Biosynthesis of the Modified Nucleoside Q in Transfer RNA. Nucl. Acids Res. 1976;3:393–398. doi: 10.1093/nar/3.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurowski M, Sasin J, Feder M, Debski J, Bujnicki J. Characterization of the cofactor-binding site in the SPOUT-fold methyltransferases by computational docking of S-adenosylmethionine to three crystal structures. BMC Bioinformatics. 2003;4:9. doi: 10.1186/1471-2105-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin I, Giladi M, Altman-Price N, Ortenberg R, Mevarech M. An alternative pathway for reduced folate biosynthesis in bacteria and halophilic archaea. Mol. Microbiol. 2004;54:1307–1318. doi: 10.1111/j.1365-2958.2004.04339.x. [DOI] [PubMed] [Google Scholar]
- Loh KD, Gyaneshwar P, Markenscoff Papadimitriou E, Fong R, Kim KS, Parales R, Zhou Z, Inwood W, Kustu S. A previously undescribed pathway for pyrimidine catabolism. Proc. Natl. Acad. Sci. USA. 2006;103:5114–5119. doi: 10.1073/pnas.0600521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova K, Koonin E. Comparative genomics of archaea: How much have we learned in six years, and what's next? Genome Biol. 2003;4:115. doi: 10.1186/gb-2003-4-8-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martzen MR, McCraith SM, Spinelli SL, Torres FM, Fields S, Grayhack EJ, Phizicky EM. A Biochemical Genomics Approach for Identifying Genes by the Activity of Their Products. Science. 1999;286:1153–1155. doi: 10.1126/science.286.5442.1153. [DOI] [PubMed] [Google Scholar]
- McNeil LK, Reich C, Aziz RK, Bartels D, Cohoon M, Disz T, Edwards RA, Gerdes S, Hwang K, Kubal M, Margaryan GR, Meyer F, et al. The National Microbial Pathogen Database Resource (NMPDR): A genomics platform based on subsystem annotation. Nucl. Acids Res. 2007;35:D347–D353. doi: 10.1093/nar/gkl947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittl PR, Grutter MG. Structural genomics: Opportunities and challenges. Curr. Opin. Chem. Biol. 2001;5:402–408. doi: 10.1016/s1367-5931(00)00221-0. [DOI] [PubMed] [Google Scholar]
- Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: Identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T, Yokoyama S, Horie N, Matsuda A, Ueda T, Yamaizumi Z, Kuchino Y, Nishimura S, Miyazawa T. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J. Biol. Chem. 1988;263:9261–9267. doi: 10.1351/pac198961030573. [DOI] [PubMed] [Google Scholar]
- Nash R, Weng S, Hitz B, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Hong EL, Livstone MS, et al. Expanded protein information at SGD: New pages and proteome browser. Nucl. Acids Res. 2007;35:D468–D471. doi: 10.1093/nar/gkl931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasvall SJ, Chen P, Björk GR. The modified wobble nucleoside uridine-5-oxyacetic acid in tRNAPro(cmo5UGG) promotes reading of all four proline codons in vivo. RNA. 2004;10:1662–1673. doi: 10.1261/rna.7106404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi S, Nishimura Y, Hirota Y, Nishimura S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J. Biol. Chem. 1982;257:6544–6550. [PubMed] [Google Scholar]
- Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hypermodified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada N, Noguchi S, Nishimura S, Ohgi T, Goto T, Crain PF, McCloskey JA. Structure Determination of a Nucleoside Q Precursor Isolated from E. coli tRNA: 7-(aminomethyl)-7-deazaguanosine. Nucl. Acids Res. 1978;5:2289–2296. doi: 10.1093/nar/5.7.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman A, Begley T. A subsystems based approach to the identification of drug targets in bacterial pathogens. In: Boshoff HI, Barrry CEI, editors. Progress in Drug Research. Vol. 64. Birkhauser Verlag; Basel (Switzerland): 2006. pp. 133–170. [DOI] [PubMed] [Google Scholar]
- Osterman A, Overbeek R. Missing genes in metabolic pathways: A comparative genomics approach. Curr. Opin. Chem. Biol. 2003;7:238–251. doi: 10.1016/s1367-5931(03)00027-9. [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucl. Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crecy-Lagard V, Diaz N, Disz T, Edwards R, Fonstein M, Frank ED, et al. The ERGO Genome Analysis and Discovery System. Nucleic Acids Res. 2003;31:1–8. doi: 10.1093/nar/gkg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W-T, Robinson MD, Mnaimneh S, Krogan NJ, Cagney G, Morris Q, Davierwala AP, Grigull J, Yang X, Zhang W. A panoramic view of yeast noncoding RNA processing. Cell. 2003;113:919. doi: 10.1016/s0092-8674(03)00466-5. [DOI] [PubMed] [Google Scholar]
- Perez-Arellano I, Rubio V, Cervera J. Dissection of Escherichia coli glutamate 5-kinase: Functional impact of the deletion of the PUA domain. FEBS Lett. 2005;579:6903. doi: 10.1016/j.febslet.2005.11.037. [DOI] [PubMed] [Google Scholar]
- Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J. 2002;21:1811–1820. doi: 10.1093/emboj/21.7.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purta E, van Vliet F, Tkaczuk KL, Dunin-Horkawicz S, Mori H, Droogmans L, Bujnicki JM. The yfhQ gene of Escherichia coli encodes a tRNA:Cm32/Um32 methyltransferase. BMC Mol. Biol. 2006;7:23. doi: 10.1186/1471-2199-7-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. Trm11p and Trm112p Are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol. Cell. Biol. 2005;25:4359–4370. doi: 10.1128/MCB.25.11.4359-4370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reader JS, Metzgar D, Schimmel P, de Crécy-Lagard V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J. Biol. Chem. 2004;279:6280–6285. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- Renalier M-H, Joseph N, Gaspin C, Thebault P, Mougin A. The Cm56 tRNA modification in archaea is catalyzed either by a specific 2′-O-methylase, or a C/D sRNP. RNA. 2005;11:1051–1063. doi: 10.1261/rna.2110805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter K, Slany R, Ullrich F, Kersten H. Structure and Organization of E. coli Genes Involved in Biosynthesis of the Deazaguanine Derivative Queuine, a Nutrient Factor for Eukaryotes. J. Bacteriol. 1991;173:2256–2264. doi: 10.1128/jb.173.7.2256-2264.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, Miller N, Mueller LA, et al. The Arabidopsis Information Resource (TAIR): A model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucl. Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Hebbeln P, Gelfand MS, Eitinger T. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: Evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 2006;188:317–327. doi: 10.1128/JB.188.1.317-327.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers M, Hale C, Tricot C, Terns MP, Terns RM, Grosjean H, Droogmans L. Formation of the conserved pseudouridine at position 55 in archaeal tRNA. Nucl. Acids Res. 2006;34:4293–4301. doi: 10.1093/nar/gkl530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR, 3rd, Ibba M, Söll D. RNA-dependent cysteine biosynthesis in Archaea. Science. 2005;307:1969–1972. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- Selkov E, Maltsev N, Olsen GJ, Overbeek R, Whitman WB. A reconstruction of the metabolism of Methanococcus jannaschii from sequence data. Gene. 1997;197:GC11–GC26. doi: 10.1016/s0378-1119(97)00307-7. [DOI] [PubMed] [Google Scholar]
- Slany RK, Bosl M, Kersten H. Transfer and isomerization of the ribose moiety of AdoMet during the biosynthesis of queuosine tRNAs, a new unique reaction catalyzed by the QueA protein from Escherichia coli. Biochimie. 1994;76:389–393. doi: 10.1016/0300-9084(94)90113-9. [DOI] [PubMed] [Google Scholar]
- Soma A, Ikeuchi Y, Kanemasa S, Kobayashi K, Ogasawara N, Ote T, Kato J, Watanabe K, Sekine Y, Suzuki T, Muramatsu T, Nishikawa K, et al. An RNA-modifying enzyme that governs both the codon and amino acid specificities of isoleucine tRNA. Mol. Cell. 2003;12:689–698. doi: 10.1016/s1097-2765(03)00346-0. [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Vassilenko KS, Emmerich J, Bauer F. tRNA Compilation 2000. 1999 http://www.uni-bayreuth.de/departments/biochemie/trna/
- Suhre K, Claverie J-M. FusionDB: A database for in-depth analysis of prokaryotic gene fusion events. Nucl. Acids Res. 2004;32:D273–D276. doi: 10.1093/nar/gkh053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, Krylov D, Mazumder R, Mekhedov S, Nikolskaya A, Rao BS, Smirnov S, et al. The COG database: An updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Natale DA, Garkavtsev IV, Tatusova TA, Shankavaram UT, Rao BS, Kiryutin B, Galperin MY, Fedorova ND, Koonin EV. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucl. Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J. Mol. Biol. 2003a;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- Tian W, Skolnick J. How well is enzyme function conserved as a function of pairwise sequence identity? J. Mol. Biol. 2003b;333:863–882. doi: 10.1016/j.jmb.2003.08.057. [DOI] [PubMed] [Google Scholar]
- Uchiyama I. MBGD: A platform for microbial comparative genomics based on the automated construction of orthologous groups. Nucl. Acids Res. 2007;35:D343–D346. doi: 10.1093/nar/gkl978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda N, Suzuki T, Yukawa M, Ohya Y, Shindo H, Watanabe K, Suzuki T. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs: Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2005;280:1613–1624. doi: 10.1074/jbc.M409306200. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria—evolutionary implications. Nucl. Acids Res. 2005;33:3955–3964. doi: 10.1093/nar/gki703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lanen SG, Reader JS, Swairjo MA, de Crécy-Lagard V, Lee B, Iwata-Reuyl D. From cyclohydrolase to oxidoreductase: Discovery of nitrile reductase activity in a common fold. Proc. Natl. Acad. Sci. USA. 2005;102:4264–4269. doi: 10.1073/pnas.0408056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veitia R. Rosetta Stone proteins: “Chance and necessity”? Genome Biology. 2002;1001:1–3. doi: 10.1186/gb-2002-3-2-interactions1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: A database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–261. doi: 10.1093/nar/gkg034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C, Jensen LJ, Kuhn M, Chaffron S, Doerks T, Kruger B, Snel B, Bork P. STRING 7—recent developments in the integration and prediction of protein interactions. Nucl. Acids Res. 2007;35:D358–D362. doi: 10.1093/nar/gkl825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waas WF, Crécy-Lagard d., Schimmel P. Discovery of a gene family critical to wyosine base formation in a subset of phenylalanine-specific transfer RNAs. J. Biol. Chem. 2005;280:37616–37622. doi: 10.1074/jbc.M506939200. [DOI] [PubMed] [Google Scholar]
- Watanabe Y.-i., Gray MW. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: Cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucl. Acids Res. 2000;28:2342–2352. doi: 10.1093/nar/28.12.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Wolf J, Gerber AP, Keller W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J. 2002;21:3841–3851. doi: 10.1093/emboj/cdf362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MD, Ahmed F, Lacourciere GM, Lauhon CT, Stadtman TC, Larson TJ. Functional diversity of the rhodanese homology domain: The Escherichia coli ybbB gene encodes a selenophosphate-dependent tRNA 2-selenouridine synthase. J. Biol. Chem. 2004;279:1801–1809. doi: 10.1074/jbc.M310442200. [DOI] [PubMed] [Google Scholar]
- Xing F, Hiley SL, Hughes TR, Phizicky EM. The specificities of four yeast dihydrouridine synthases for cytoplasmic tRNAs. J. Biol. Chem. 2004;279:17850–17860. doi: 10.1074/jbc.M401221200. Epub 2004 Feb 16. [DOI] [PubMed] [Google Scholar]
- Xing F, Martzen MR, Phizicky EM. A conserved family of Saccharomyces cerevisiae synthases effects dihydrouridine modification of tRNA. RNA. 2002;8:370–381. doi: 10.1017/s1355838202029825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL. Biosynthesis of Selenocysteine on Its tRNA in Eukaryotes. PLoS Biol. 2006;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakunin AF, Yee AA, Savchenko A, Edwards AM, Arrowsmith CH. Structural proteomics: A tool for genome annotation. Curr. Opin. Chem. Biol. 2004;8:42–48. doi: 10.1016/j.cbpa.2003.12.003. [DOI] [PubMed] [Google Scholar]
- Yang C, Rodionov DA, Li X, Laikova ON, Gelfand MS, Zagnitko OP, Romine MF, Obraztsova AY, Nealson KH, Osterman AL. Comparative genomics and experimental characterization of N-acetylglucosamine utilization pathway of Shewanella oneidensis. J. Biol. Chem. 2006;281:29872–29885. doi: 10.1074/jbc.M605052200. [DOI] [PubMed] [Google Scholar]
- Ye Y, Godzik A. Multiple flexible structure alignment using partial order graphs. Bioinformatics. 2005;21:2362–2369. doi: 10.1093/bioinformatics/bti353. [DOI] [PubMed] [Google Scholar]