The Nucleolus (original) (raw)
Abstract
When cells are observed by phase contrast microscopy, nucleoli are among the most conspicuous structures. The nucleolus was formally described between 1835 and 1839, but it was another century before it was discovered to be associated with a specific chromosomal locus, thus defining it as a cytogenetic entity. Nucleoli were first isolated in the 1950s, from starfish oocytes. Then, in the early 1960s, a boomlet of studies led to one of the epochal discoveries in the modern era of genetics and cell biology: that the nucleolus is the site of ribosomal RNA synthesis and nascent ribosome assembly. This epistemologically repositioned the nucleolus as not merely an aspect of nuclear anatomy but rather as a cytological manifestation of gene action—a major heuristic advance. Indeed, the finding that the nucleolus is the seat of ribosome production constitutes one of the most vivid confluences of form and function in the history of cell biology. This account presents the nucleolus in both historical and contemporary perspectives. The modern era has brought the unanticipated discovery that the nucleolus is plurifunctional, constituting a paradigm shift.
Nucleoli are associated with specific DNA loci and act as sites for rRNA synthesis and nascent ribosome assembly. They also harbor numerous cell-cycle regulators and have been tantalizingly linked with stem cell biology.
FIRST SIGHTING
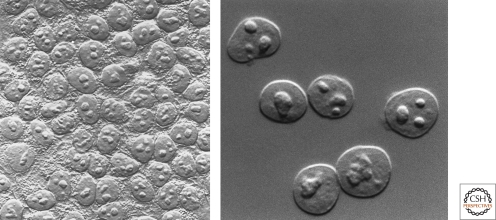
It is likely that some of the few lucky enough to have a microscope in the 18th century saw the nucleolus if they examined thin specimens of tissue in the mode of illumination that later became known as bright field, a century before phase contrast was discovered, for which a Nobel Prize, rare in microscopy, was conferred on Frits Zernike in 1953 (Fig. 1).
Figure 1.
Looking at the nucleolus. Nucleoli observed in HeLa cells (left) and isolated HeLa cell nuclei (right) by differential interference microscopy. (Images courtesy of David L. Spector [left] and Thoru Pederson [right].)
The first properly documented accounts of the nucleolus were made independently by Wagner (1835) and Valentin (1836, 1839). Beyond occasional studies in which nucleoli were mentioned in passing in the context of broader cytological work, no significant literature on the nucleolus ensued for another half century. Then, a monumental monograph on the nucleolus was published by Montgomery (1898), with an astonishing 346 hand-drawn color figures of nuclei and nucleoli from a vast array of biological material. But as comprehensive and elegant a piece of scholarship as it was, this treatise is primarily known to us today not because of what it revealed the nucleolus to be at the time, but because of what the nucleolus later became.
THE NUCLEOLUS BECOMES PART OF THE KARYOTYPE
For three decades after Montgomery’s treatise, there was little momentum until the discovery that the nucleolus arises at a specific chromosomal locus (Heitz 1931; McClintock 1934). While the intimacy of associations between nucleoli and heterochromatin had previously been noted, the independent findings of Heitz and McClintock established that the nucleolus actually forms at a discrete locus, termed the nucleolus organizer by McClintock. The importance of this advance cannot be overstated because it meant that rather than being mere nuclear anatomy, nucleoli are cytogenetic entities. From this perspective, the astonishing diversity of nucleolar shapes and sizes recorded by Montgomery began to be sensed as reflecting differences in the chromosomal sites at which they arise. The discovery of the nucleolus organizer meant that, whatever its function(s), the nucleolus is stationed at a genetic locus, although it would still be another decade before the discovery that the genetic material is DNA.
MONTEVIDEO: A HIGH ALTITUDE VIEW OF THE NUCLEOLUS
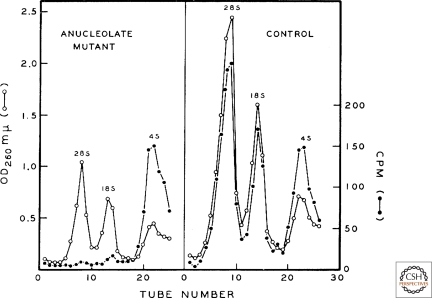
We have all attended meetings, and the luckiest of us know that sometimes, when the muses smile, we are witnesses to a renaissance. It just happened that leading up to a meeting on the nucleolus in Montevideo, Uruguay, in December 1965, several new findings had been made or were just about to be submitted for publication. Donald Brown presented a brilliant experiment, done with John Gurdon (Brown and Gurdon 1964), showing that anucleoate Xenopus embryos arrest in development, plausibly due to the inability of these embryos to make new ribosomes when the maternal stockpile becomes limiting (Fig. 2). Brown also reported the existence of amplified nucleoli in the germinal vesicle (nucleus) of Xenopus oocytes (a finding that had been made contemporaneously [Brown and Dawid 1968; Gall 1968]). Papers on the isolation of nucleoli were given by Walter Vincent (from starfish oocytes), and by Rachele Maggio and Harris Busch (from guinea pig and rat liver, respectively). The biosynthesis of rRNA via large precursor molecules was reported in talks by Joseph Gall, Sheldon Penman, Georgii Georgiev, and Robert Perry. But the most important discovery announced at the meeting was that reported in the talks by Max Birnstiel and Ferrucio Ritossa; namely, that nucleic acid hybridization revealed that DNA complementary to rRNA resides in the nucleolus, which, together with the results of Brown and Gurdon (1964), ushered in a new age in the history of the nucleolus. The proceedings of this conference, including the lively discussion exchanges after each talk (Vincent and Miller 1966), together with an exceptionally insightful synopsis of the meeting (Perry 1966), constitute a definitive archive of this exciting moment in the field.
Figure 2.
A portal to the modern era: the dependence of ribosomal RNA synthesis on the nucleolus. Anucleolate (left) or wild-type (right) Xenopus laevis embryos at the neurula stage were incubated with C14-labeled carbon dioxide and RNA was extracted 20 hours later, when both groups of embryos were still morphologically and physiologically indistinguishable. Shown are sucrose gradient sedimentation profiles of 28S and 18S rRNA and transfer RNA. This finding was a keystone in establishing the role of the nucleolus in the biosynthesis of ribosomes. (Reproduced from Brown DC and Gurdon JB 1964. Proc Natl Acad Sci USA 51: 139–146, by kind permission of Donald D. Brown, Carnegie Institution for Science, Washington, D.C.)
COMING DOWN FROM THE MOUNTAIN
Oscar Miller had presented EM pictures of spread nucleolar “cores” and stretched nucleolar DNA at the meeting, but they were his first attempts and not particularly revealing. But later, he and his talented research assistant Barbara Beatty showed the world what these genes really look like in full transcriptional action (Miller and Beatty 1969). These pictures earned their rightful place as among the most iconic of any in the history of cytology and cell biology. Incisive studies on the synthesis and processing of ribosomal RNA introduced by Sheldon Penman at the Montevideo meeting were subsequently refined by him and independently by the laboratory of James Darnell (reviewed by Lewin 1980). In contrast, the isolation of nucleoli presented at the meeting was to await several decades for further advances.
In his summary of the Montevideo conference, the Edinburgh embryologist C.H. Waddington said: “The nucleolus probably should not be considered a relatively simple organelle with a single function, comparable to a machine tool turning out a particular part of an automobile. It is not just ‘the organelle where the cell manufactures ribosomes.’ It is rather a structure through which materials of several different kinds are flowing, comparable more to a whole production line than to a single machine tool.” One cannot imagine a more prescient view. As we shall see, every atom of his statement has been borne out in subsequent research on the nucleolus.
INTO THE MODERN ERA
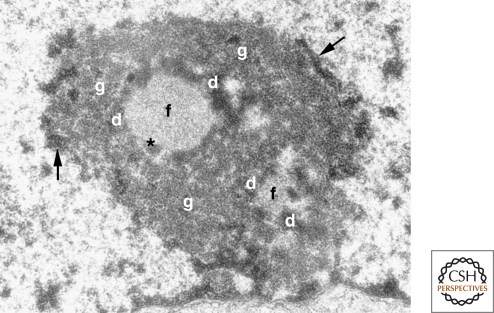
In the 1970s and 1980s, the nucleolus field addressed the details of ribosome biosynthesis. One axis was a troubled one: the goal of reconciling the stages of ribosome synthesis with the classically defined subcompartments of the nucleolus. Just as one commentator famously described (to every Latin student centuries later) the Roman conquest territory that would later become France, “Gallia est omnis divisa in partes tres” (Caesar, 40–50s BC), the nucleolus is also tripartite. Its three classical regions are defined by the different appearance of intra-nucleolar regions when viewed by electron microscopy (Fig. 3). These are the fibrillar centers, the surrounding dense fibrillar component, and the granular component. Few in the field anticipated the controversies that would ensue when several labs tried, logically enough, to link the sites of rRNA transcription and processing and assembly into nascent ribosomes with these three EM-defined nucleolar zones. These debates were waged with intense controversy and, in a few cases, frank boorishness on the part of some in this raging controversy on the floor at international meetings. In hindsight, this had to do with how short a pulse label one could introduce to tag nascent rRNA. Thankfully, recent advances have settled the issue. A key step in this resolution was a reconciliation of the “Christmas tree” EM images of spread nascent ribosomal RNA (Miller and Beatty 1969) with the high resolution detection of these genes and their transcripts in nucleoli in situ (Koberna et al. 2002; for further discussion of the earlier controversy and its resolution, see Raška et al. 2006). Recent work has indicated that one of the three nucleolar regions, the granular component, is itself composed of at least two distinct molecular domains.
Figure 3.
The tripartite organization of the nucleolus. Electron micrograph of a mouse fibroblast nucleolus. (f) fibrillar center, (d) dense fibrillar component, (g) granular component, (arrows) perinucleolar heterochromatin, (*) denotes the presence of dense fibrillar component material within the fibrillar center, which is occasionally observed. (Reprinted from Trends in Cell Biol 13: 517–525, Raška, I. © 2003, with permission from Elsevier.)
Meanwhile, the covalent steps in rRNA processing continued to be explored, revealing different nuances in yeast versus higher eukaryotes superimposed on a conserved foundation (reviewed by Fatica and Tollervey 2002). But another important axis was also emerging in the 1970s. The laboratory of Harris Busch, notably Ramachandra Reddy, came upon and sequenced several small nucleolar RNAs. One of these, U3 small nucleolar RNA, was subsequently shown to be base-paired with nucleolar rRNA in vivo (Calvet and Pederson 1981) and later was demonstrated to function in the initial cleavage of the 45S rRNA precursor (Kass et al. 1990). All of these studies were done, of course, with deproteinized nucleolar RNA. Soon thereafter, isolated nucleoli were subjected to various extraction conditions designed to preserve nascent rRNA–protein complexes, resulting in the characterization of the ribonucleoprotein forms of the rRNA precursor and processing intermediates that had previously been defined with deproteinized RNA (Warner and Soeiro 1967; Pederson and Kumar 1971; Kumar and Warner 1972).
Jumping ahead, we now enjoy a far more detailed picture of the covalent steps of rRNA maturation (Fatica and Tollervey 2002) as well as numerous accessory factors that support this pathway but are not part of the final ribosomes (reviewed by Granneman and Baserga 2004). Among the latter are more than 100 small nucleolar RNAs that guide the extensive site-specific nucleotide modifications (ribose 2′-_O_-methylation and pseudouridine formation) of rRNA during its processing (reviewed by Bachellerie et al. 2002). Like the initial rRNA precursor, processing intermediates, and products themselves, all these small nucleloar RNAs are complexed with proteins, thus instilling in the nucleolar landscape a high concentration of both RNA and protein, a point to which we shall return.
A PARADIGM SHIFT
By the 1960s and into the 1970s, the notion that the nucleolus is the seat of ribosome synthesis was as cogently established as anything in the modern canon of the molecular and cell biology of eukaryotes. Then, something unexpected happened.
In my laboratory, we had been studying the traffic of fluorescently labeled small nucleolar RNAs after microinjection into the nuclei of cultured mammalian cells and finding them to move into nucleoli, as expected, but extending this to define nucleolar targeting elements in these RNAs (Jacobson et al. 1995; 1997; Jacobson and Pederson 1998a). In the case of U3 and U8 (Jacobson and Pederson 1998a), we planned to expand the findings but first decided to submit an initial report to convey our novel experimental approach (reviewed in Pederson 2001a), something quite new at the time for the nucleolus, though we had previously applied it to study pre-mRNA traffic to nucleoplasmic domains (Wang et al. 1991). While writing this paper (Jacobson and Pederson 1998a), it occurred to us that we had only used a single RNA as a negative control. Although it did not display nucleolar localization after microinjection, we decided to try a second small RNA that presumably would also not localize in nucleoli, thus amplifying our case that the observed nucleolar localizations of U3 and U8 were specific. My post-doc Marty Jacobson suggested that we try the signal recognition particle RNA, a 300-nt pol III transcript, not only to fulfill the need for another negative control but also because, to his credit, he realized nothing was known about the nuclear phase of SRP biosynthesis. To our surprise, SRP RNA displayed as rapid and as quantitative a nucleolar localization as U3 and U8 snoRNAs. But looking into this further, we saw that SRP RNA trafficked into nucleoli only transiently. U3 and U8 on the other hand remained stably localized in nucleoli, as did other small nucleolar RNAs we had begun to study. SRP RNA then moved out into the cytoplasm, where it became associated with the endoplasmic reticulum, its expected final localization (Jacobson and Pederson 1998b).
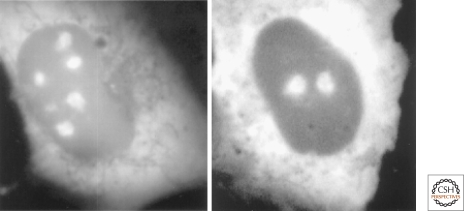
These findings on SRP RNA, together with a number of other contemporaneous results that were stirring, led to the formulation of the “plurifunctional nucleolus” hypothesis (Pederson 1998a). Subsequently, the link between the nucleolus and signal recognition particle biosynthesis was made in yeast (Ciufo and Brown 2000; Grosshans et al. 2001), and confirmed and extended in mammalian cells (Politz et al. 2000) (Fig. 4) and in Xenopus oocytes (Sommerville et al. 2005). A speculative essay suggested how a common nucleolar assembly site might have co-evolved for the four translational ribonucleoproteins (the two ribosomal subunits, the 5S rRNA–protein complex, and the SRP) (Pederson and Politz 2000). Notwithstanding how it came about in the evolution of eukaryotes, the fact that the nucleolus is a site of more than ribosome synthesis had clearly arrived once its role in the biosynthesis of the SRP had been discovered. Also embedded in the plurifunctional nucleolus hypothesis were provocative results linking the nucleolus to cell-cycle progression (Pederson 1998a,b). Both the SRP and cell-cycle aspects of the hypothesis were welcomed into reviews on the nucleolus (Scheer and Hock 1999; Olson et al. 2000; Olson et al. 2002; Visintin and Amon 2000), and the concept has continued to be well received (Alberts et al. 2002; Raška et al. 2006; Boisvert et al. 2007).
Figure 4.
The nucleolus and signal recognition particle biosynthesis. Nucleolar localization of GFP-tagged SRP19 (left) and SRP68 (right) proteins in rat NRK cells. Cytoplasmic localization is also observed, as expected from the known steady-state localization of the signal recognition particle. Similar results were obtained with GFP-tagged SRP72 protein, whereas the SRP54 protein assembles with the nascent signal recognition particle in the cytoplasm (Politz et al. 2000; Sommerville et al. 2005). Reproduced from Politz et al. 2000, with permission from the Proceedings of the National Academy of Sciences.
THE PARTS LIST, AND SEGUEING INTO THE CELL CYCLE
When nucleoli were first isolated (Vincent 1952), only assays for total protein and RNA, as well as tests for various enzymatic activities, were available (the latter scoring only one—acid phosphatase). Jumping ahead four and a half decades, we now speak of specific nucleolar molecules. As mentioned earlier, progress on chronicling all of the small RNAs present in nucleoli had proceeded through the 1980s and 1990s and on into the 2000 decade. As for nucleolar proteins (beyond the ribosomal structural proteins), early accounts (e.g., Soeiro and Basile 1973) were followed by numerous reports of individual, non-ribosomal proteins observed either in isolated nucleoli or, more typically, by immunofluorescence or GFP-tagged protein localization. These were useful findings on each protein’s individual merit but obviously did not reveal the molecular breadth of the nucleolar protein landscape. In 2002, two groups refined the classical nucleolar isolation methods (notably that developed by Maggio et al. 1963) and prepared high purity nucleoli from HeLa cells followed by proteomics (Andersen et al. 2002; Scherl et al. 2002; reviewed by Pederson 2002 and Leung et al. 2003).
These two contemporaneous proteomics studies revealed, in combination, a total of approximately 350 proteins. Excluding ribosomal structural proteins, there were many that had been or would be later linked to ribosome biosynthesis as chaperone-like mediators not ending up in mature ribosomes. This finding expanded and put more molecular definition on earlier research, showing that nucleolar pre-ribosomal particles have a higher protein:RNA mass ratio than mature ribosomes (Liau and Perry 1969; Pederson and Kumar 1971; Kumar and Warner 1972). Many others of the observed nucleolar proteins were ones involved in DNA replication or repair, cell-cycle progression, or its control, validating this aspect of the initial plurifunctional nucleolus hypothesis (Pederson 1998a) and findings being made at the time that had suggested such a link (reviewed by Pederson 1998b; Garcia and Pillus 1999; Vinistin and Amon 2000). Similar findings were later made in a proteomics analysis of purified nucleoli from Arabidopsis (Pendle et al. 2005).
Beyond proteomic analysis, and its emphasis on nonribosomal proteins, there has been recent work on ribosomal proteins themselves that has brought surprises. The first of these implicated ubiquitinylation, including polyubiquitinylation, of ribosomal proteins in the pathway of ribosome synthesis (Stavreva et al. 2006), a most unanticipated, counterintuitive result and one not immediately reconcilable with the “standard model” of ribosome biosynthesis. The second study revealed a surprisingly large pool of unassembled ribosomal proteins, exceeding the level needed to support ribosome assembly on the number of rRNA transcripts being produced (Lam et al. 2007). Obviously, one senses the likely possibility of a link between these two sets of observations. These findings also lead us to bear in mind the possibility that there may be further surprises ahead regarding the established lore of the nucleolar ribosome biosynthetic pathway, just as has been the case with respect to the nonribosomal aspects of nucleolar organization and function in the 12 years since the plurifunctional nucleolus hypothesis was advanced (Pederson 1998a).
THE NUCLEOLUS IS NEITHER AS STATIC NOR AS COMPACT AS FIRST THOUGHT
Between 2000 and 2001, a number of studies based on fluorescence recovery after photobleaching (FRAP), in one case employing as well the complementary method of fluorescence loss in photobleaching (FLIP), brought about a complete revision in thinking about the stasis of several nuclear proteins (for reviews, see Pederson 2000; 2001b; Leung and Lamond 2003). In the case of the nucleolar proteins studied, namely fibrillarin (Phair and Misteli 2000) and selected ribosomal structural proteins (Chen and Huang 2001), the dynamics were faster than would have been anticipated by many (perhaps most) nucleolus investigators. Fibrillarin functions as an rRNA modification enzyme in the dense fibrillar component and therefore might have been assumed to either be stably associated with the nuclelous or at least display a long residence time. A nucleolar stasis of ribosomal structural proteins was an even more entrenched notion, as they would have been assumed to enter nucleoli in stoichiometric amounts to assemble with nascent rRNA and yet they were found to be rapidly shuttling in and out of nucleoli. As mentioned above, later work revealed that ribosomal structural proteins undergo ubiquitinylation and degradation on the assembly pathway (Stavreva et al. 2006) and also enter the nucleus in supra-stoichiometric levels (Lam et al. 2007). Clearly, the FRAP results (Chen and Huang 2001) reflected, or at least were significantly influenced by, these later-recognized aspects of ribosomal protein instability, whereas the fast exchange of fibrillarin (Phair and Misteli 2000) was, and remains, even more surprising.
It has been more difficult to study the dynamics of nucleolar RNAs. The fact that fluorescent snoRNAs accumulate in nucleoli after microinjection into the nucleus does not address this question since the initial localization may either be filling unoccupied sites or replacing those formerly occupied by egressing snoRNAs. Stated differently, snoRNAs are always at high concentration in the nucleolus so the concentration of the putative molecules that have moved into the nucleoplasm in any experiment that examines the steady-state would be very low. Only methods like FRAP or FLIP can capture this and these are not applicable to RNA. However, endogenous RNAs can be tagged with fluorescent probes (Politz et al. 1998), including photoactivatable ones (Politz 1999; Politz et al. 2005). In a study using this approach to track 28S rRNA out of the nucleolus (Politz et al. 2003), we noted that some signal occasionally returned to nucleoli. We pondered that there is probably no barrier to a completed ribosomal subunit occasionally gaining access back into the nucleolus (which is surprisingly porous, vide infra) before it engages the nuclear export machinery. Notwithstanding this observation, how dynamic or static nucleolar RNAs are remains an open and important question.
How compact a bundle of mass is the nucleolus? Its appearance in differential interference microscopy (Fig. 1) indicates that it has higher mass per unit volume than the surrounding nucleolplasm, but how much higher? Fortunately, differential interference microscopy can be used in a quantitative mode. Its application to the nucleus of Xenopus oocytes revealed that the nucleoli are only about twice as dense as the surrounding nucleoplasm (Handwerger et al. 2005; discussed in Pederson 2010a.) In retrospect, it may be that a preferentially high affinity of some nucleolar proteins for the heavy metal atoms (uranium, osmium, tungsten) used in electron microscopy has led to a false perception of a higher (protein) density in the nucleolus, or in some of its regions, than is actually the case. The use of basic dyes in classical cytological work on the nucleolus that are taken preferentially by polyanions such as RNA, or silver stains that bind avidly to certain nucleolar proteins, may have contributed to this notion from cytochemistry. In any case, similar studies to those done in the nucleus of Xenopus oocyte nuclei are now needed in mammalian cells, where there are more data on nucleolar protein dynamics that can be used as parameters in computational models. A first step has been taken in this direction, in which the percolation of ovalbumin (molecular weight ∼43,000) in the nucleolus and nucleoplasm was measured in a mammalian cell (Spell and Kubitscheck 2009), revealing that this protein had the same mobility in both compartments. One is also reminded here that the use of photo-activatable GFP-tagged nucleolar proteins in such studies could be very revealing, as has been their use in studying another protein’s dynamics in the Xenopus oocyte nucleus (Deryusheva and Gall 2004).
A GROWING FAMILY OF CELL-CYCLE REGULATORS IN THE NUCLEOLUS
The nucleolus proteomics studies revealed numerous cell-cycle-related proteins among the residents (or visitors). A great many studies over the past decade have brought further insights into the nucleolar transit of cell-cycle regulatory proteins. These good endeavors have now engendered a large literature, much of which is more pertinent to cell-cycle progression control in general, whereas only a subset of this emerging work relates to the nucleolus; hence the brief, selective coverage that follows.
Before embarking on the relationship between nucleolar activity and cell-cycle progression, a caveat needs to be registered. There is no doubt that when ribosome production is impaired or limiting, cell growth slows or ceases. The obligatory relationship between ribosome biosynthesis and cell growth rate was established for bacteria in studies by Őle Maaloe and Karl Lark in the 1960s, and later was confirmed for eukaryotes. We can plausibly surmise that new ribosomes do count for a cell’s (or organism's) future because selection would disfavor cells, embryos, or adult organisms that are slow to build new proteins for either cell division (a necessary inheritance for G1 progeny) or for sustained vigor of tissues and organs. That said, we here take up the question of whether the nucleolus might play roles in cell-cycle progression beyond ribosome biosynthesis (see also Pederson 2007). This is not an idea that was pondered in the classical era, when ribosome production, protein synthesis rates, and cell growth were seen as the operation of a continuous, forward- and back-regulated production logic circuit.
The point of departure was the finding that the action of two cell-cycle regulators, p53 and Mdm2, are modulated by nucleolar sequestration (Weber et al. 1999; Kulikov et al. 2010). Another key finding was that p53 is stabilized upon DNA damage-triggered nucleolar disruption or elicited by other cellular stressors (Rubbi and Milner 2003). Subsequent work has defined an extensive interactome of nucleolus-nucleoplasmic shuttling cell-cycle progression proteins and it now appears that the action of several is based on their dynamic interplay between the nucleolus and the nucleoplasm. An instructive example is nucleostemin (reviewed by Ma and Pederson 2008a; Pederson and Tsai 2009). This protein was discovered in a study of neural stem cells in the subventricular zone of the adult rat brain (Tsai and McKay 2002). Nucleostemin is a p53-interactive protein (Tsai and McKay 2002; Ma and Pederson 2007) and the initial idea (Tsai and McKay 2005; reviewed by Misteli 2005) was that when it shuttles from the nucleolus into the nucleoplasm, it binds to and disables p53, and thus drives the cell cycle. But the continuing challenge in this field is to reconcile a protein’s intranuclear location with function. For example, although p53 is mostly nucleoplasmic, the small nucleolar fraction of p53 has different binding partners, measured by bimolecular fluorescence complemenatation (see Ma and Pederson 2008b), than those with which it consorts in the nucleoplasm (Pederson, unpubl.). It is usually assumed that the highest intranuclear concentration of a protein is where it executes its function. This assumption may be suspect in some cases, and in the case of nucleostemin, the jury is out as to whether it enacts its cell-cycle-driving function in the nucleolus (at high concentration) or in the nucleoplasm (at low concentration). A protein’s concentration in a region of the cell may not be a quantitative signal for its action, which should be kept in mind as systems biology approaches now arrive in cell (and nuclear) biology.
THE PERINUCLEOLAR COMPARTMENT: ANOTHER LINK BETWEEN THE NUCLEOLUS AND GROWTH CONTROL?
In some cells, there is a discoid structure situated on the nucleolus, covering a portion of its surface as a “cap.” Termed the perinucleolar compartment (PNC), it was discovered on the basis of its high concentration of a particular hnRNP protein (Ghetti et al. 1992; Matera et al. 1995; for reviews, see Huang 2000 and Pollock and Huang 2010). The PNC accretes, but is not the transcription site of, a number of small RNAs (Matera et al. 1995; Wang et al. 2003). The fact that PNCs are not seen in all cells but are noted particularly in mammalian transformed cell lines, tumor-derived cell lines, and tumor biopsy specimens suggests a key link between their appearance and tumor initiation and/or progression (see Pollock and Huang 2010). Although this is an encouraging new tool for the pathologist, the PNC’s harboring of certain proteins and RNAs presently offers no clues whatsoever as to why it appears more frequently in tumor cells. Clearly, further work on the PNC is warranted. In fact, it is the only presently known link between the nucleolus (in the form of a nucleolus-vicinal cytological entity) and a very high morbidity/mortality human disease, breast carcinoma.
THE NUCLEOLUS AND VIRAL REPLICATION, STEM CELL BIOLOGY, AND CELLULAR SENESCENCE: ONLY ZEPHYRS SO FAR
Beyond cell-cycle progression and growth control, the past decade has seen a surge of attention to the nucleolus in three other fields: viral replication, stem cell biology, and cellular senescence. These studies include a number of provocative findings, briefly discussed below, but the objective summary statement is that a mechanistic link to the nucleolus has not been made.
Regarding viral replication, some viruses display a nucleolar tropism, but the functional significance of this is generally not well understood. A surprising recent finding was that the nucleolus and Cajal bodies are required for the establishment of a systemic infection by a plant virus (Kim et al. 2007). In an intriguing retroviral therapeutic development, the fact that unspliced HIV RNA exits the nucleus via the nucleolus was exploited to design a nucleolus-targeting inhibitor of viral replication (Michienzi et al. 2002). The nucleolus has also been implicated in the replication of herpes viruses (Boyne and Whitehouse 2006.) Yet another recent case is the finding that the replication of the Dependovirus AAV2 (adenovirus-associated virus 2) takes place in the nucleolus (Sonntag et al. 2010). AAV2 is among the smallest animal cell viruses known (∼25 nm in diameter) and its assembly might thus be feasible within the relatively porous nucleolus, yet the biological significance of this nucleolar tropism is presently unclear (reviewed by Pederson 2010a).
The connection of the nucleolus to stem cell biology has received some attention, with a possible link hinted at both by classical and contemporary studies (e.g., Gonda et al. 2003; reviewed by Misteli 2003; Fléchon 2006). Thus, when a somatic nucleus from an adult vertebrate animal is placed into an enucleated egg, development can proceed up to a tadpole in the case of amphibians or even birth in the case of mammals. Much attention has been placed on the effects of the egg cytoplasm on the implanted nucleus in terms of epigenetic marks and the redirected program of gene expression. But, something else also happens. In the nucleolus of the adult, somatic nucleus transiently disappears and then comes back into view a few hours later. This intriguing behavior of the nucleolus in somatic nucleus transfer experiments has not been explored to the extent it should be. If properly investigated, this is likely to reveal profound clues as to what happens when newly combined nuclei and cytoplasm become molecularly familiar with themselves.
Another potential link between the nucleolus and stem cell biology has arisen from studies of the transcription factor Hand1, whose localization within and outside the nucleolus acts as a trigger for cell fate determination (Martindill et al. 2007). Another engaging recent study has implicated a U3 small nucleolar RNA-bound protein, Wcd, in a neural stem cell niche of Drosophila embryos (Fichelson et al. 2009). Wcd displays asymmetric distribution to daughter cells and thus has “stem cellness” written all over it. One hopes that these groups and/or other nucleolus investigators are pursuing these two findings, for they are among the most intriguing links between the nucleolus and stem cell biology to have been reported.
Studies linking the nucleolus to cellular senescence round out this body of “conceptually nucleolus-related” research in that, once again, some provocative clues have been uncovered, but there is no sense at present of a unifying concept or mechanism. For example, an influential study in yeast reported an acceleration of aging (whatever this means in fungi) in mutants displaying nucleolar fragmentation (Sinclair et al. 1997; reviewed in Guarente 1997). And, a number of studies have made a connection between the nucleolus and Werner syndrome, an inherited human-aging-like condition (Marciniak et al. 1998; Szekely et al. 2000; von Kobbe and Bohr 2002; Kyng et al. 2003), and this seems the best case so far for a role of the nucleolus in cellular senescence, though still far from being linked to aging of a higher eukaryotic organism.
A QUADRIPARTITE NUCLEOLUS, OR MORE?
After discovering that the SRP assembly occurs in the nucleolus (Jacobson and Pederson 1998b; Politz et al. 2000; Sommerville et al. 2005), the question arose as to the intranucleolar site. Probing for both SRP RNA and 28S rRNA in dual color in situ hybridization experiments revealed incomplete overlap, although both signals resided in the granular component (Politz et al. 2002). Given this evidence for molecular segregation even within a given nucleolar sub-compartment, we reasoned that probing for a molecule not involved in either ribosome or SRP biosynthesis would be the logical next step and chose nucleostemin. The results conformed and amplified the notion that the nucleolar granular component is molecularly territorialized (Politz et al. 2005). But, the most important finding in this study came from the application of electron spectroscopic imaging, which revealed the granular component to be an interspersed array of RNA-rich and RNA-deficient particles, rather than a uniform lawn of nascent ribosomes, as would have been anticipated (Fig. 5). It remains to be seen how many subdivisions exist within the granular component, or within the dense fibrillar component for that matter. The probes and optical resolution are now at hand to address this further.
Figure 5.
The nucleolar granular component contains both RNA-rich and RNA-deficient particles. Electron spectroscopic imaging (ESI) is a mode of energy loss measurement in electron microscopy that can be tuned to record beam collisions with orbital electrons of nitrogen or phosphorus in the specimen. Using the empirical chemical formulas of protein and nucleic acid with respect to these two elements, ESI allows one to construct a map of protein-rich versus nucleic-acid-rich objects. Shown is an ESI image from a portion of a human neuroblastoma cell nucleus spanning a region of the nucleoplasm and part of a nucleolus. The protein-rich and nucleic-acid-rich regions have been colored blue and yellow, respectively. (DCh) decondensed chromatin, (CCh) condensed chromatin, (PML) promyelocytic leukemia body, (GC) granular component of the nucleolus. The PML body (entirely blue, i.e., protein) serves as a valuable internal control, as it is known not to contain appreciable nucleic acid. The granular component of the nucleolus, once thought to be a uniform zone of nascent ribosomes, contains an interspersed landscape of nucleic-acid-rich versus nucleic-acid-deficient particles. ESI cannot distinguish between DNA and RNA, but since several independent methods have detected no DNA in the nucleolar granular component, the nucleic-acid-rich particles are plausibly nascent ribosomes, perhaps with a contribution from assembling signal recognition particles, whereas the nucleic-acid-deficient, protein-rich particles may be heterotypic complexes of cell-cycle regulatory proteins, as discussed. (Reprinted from Politz JC et al. 2005, Mol Biol Cell 16: 3401–3410.)
TO RESIST OR REVISIT HERESY? DOES THE NUCLEOLUS PARTICIPATE IN MESSENGER RNA BIOSYNTHESIS?
Beyond the possible roles of the nucleolus in cell-cycle control, viral replication, stem cell biology, and cellular senescence, there is another potential nucleolus function that warrants consideration, both for its historical precedence and its now reasonable plausibility based on a number of studies. This is the possibility that the nucleolus is involved, somehow, in the production of messenger RNA.
In the 1960s and 1970s, Henry Harris at the University of Oxford posited that the nucleolus was a way station for mRNA export from the nucleus (see Harris 1974). He had been a pioneer of somatic cell genetics, unsullied as such, but was also a constitutional contrarian. He had become something of an enfant terrible outside his immediate field by challenging the then-emerging (and prevailing) views on the meaning of high molecular weight nuclear RNA as regards the pathway of eukaryotic messenger RNA biosynthesis (at a time when the connection was admittedly still wobbly). But, in contrast to his mRNA biosynthesis contrarianism, Harris’ idea that the nucleolus might be involved in mRNA export was based on results, not mere speculation. He had fused chicken erythrocytes, containing a condensed, inactive nucleus, with HeLa cells and found that chicken-specific proteins appeared in these heterokaryons only after the erythrocyte nucleus had undergone chromatin decondensation and the appearance of nucleoli. From this, he elaborated, and for years promulgated, the idea that messenger RNA must obligatorily pass through the nucleolus. The contemporaneous work of Penman, Darnell, and other groups dispelled this notion for mRNA as a population, en masse, but in a few corners of the field the possibility lingered that a subset of mRNAs might visit the nucleolus prior to export.
Subsequently, there were reports of specific mRNAs in the nucleolus (reviewed in Pederson 1998a) and a recent study (Kim et al. 2009) has rekindled these earlier observations and carried them into the modern era of methodology. In addition, several microRNAs have now been localized in the nucleolus (Politz et al. 2006; Politz et al. 2009) and there is increasing evidence that some microRNAs may be derived from snoRNAs (discussed in Politz et al. 2009; see also Taft et al. 2009). There is also recent evidence for transfer RNA-derived microRNAs (reviewed by Pederson 2010b), and given that the nucleolus has been implicated in tRNA biosynthesis, this is yet another fascinating possibility for a link between the nucleolus and mRNA regulation via microRNAs. Although it is counterintuitive that negative regulators of mRNA function would be associating with nascent mRNAs before the latter even leave the nucleus, intuition has often been an unreliable docent in the field of gene expression. The possible role of the nucleolus in mRNA export and/or microRNA function and dynamics certainly warrants further investigation.
NEW FRONTIERS: WHAT LIES BENEATH
Among the remaining issues, the classical phenomenon of nucleolar dominance stands ready for renewed analysis, as has been happening productively (Earley et al. 2010; reviewed by Costa-Nunes et al. 2010). The role of intergenic transcripts in the control of ribosomal RNA transcription (Mayer et al. 2006; Santoro et al. 2010) is another fertile area of current investigation (and likely intersects with nucleolar dominance). Moreover, there is growing evidence that a gene’s positioning or repositioning near or at the nucleolus can regulate its expression (e.g., Royo et al. 2009) and that the rDNA can itself impact the genome globally (e.g., Parades and Maggert 2009). Understanding the process of nucleolar assembly is also a high priority and although there has been substantial progress on this from a cell biological perspective (reviewed by Hernandez-Verdun et al. 2002), a new system (Prieto and McStay 2007, 2008) promises molecular detail. A role for the nucleolus has recently emerged in studies of how a postsynaptic density protein of dendritic spines shuttles back to the cell body after neurotransmitter activation (reviewed by Richer and Fallon 2007), raising the possibility of an entire new field linking the nucleolus to neurobiology. Also foremost on the agenda is continued investigation of the extent to which the nucleolus directs the cell cycle (Pederson 2007), as well as the clinical utility of the PNC. A link between the nucleolus, p53, and innate immunity in C. elegans (Fuhrman et al. 2009) is also among the most provocative recent findings and warrants immediate exploration both in nematodes and in a wider phyletic range. Also standing before us is the entire question of mRNA and microRNA traffic through the nucleolus and its functional meaning. Yet, other new vistas on the nucleolus horizon will almost certainly appear in due course, but meanwhile, the agenda is both full and promising.
CONCLUSION
The nucleolus is unique in that the discovery of one of its functions took far longer, namely more than a century, than was the case for most other cell components. Another half-century has brought the realization that the nucleolus is also the site of signal recognition particle biosynthesis, serves as a regulatory zone of cell-cycle progression mediators, and is a locus of mRNA and microRNA traffic, the functional meaning of which remains to be discovered. The nucleolus is now known to be a more dynamic domain of nuclear organization than once thought, and with functions even beyond those first grandly recognized atop a Uruguay mountain in 1965.
ACKNOWLEDGMENTS
Cited work from the author’s laboratory was supported by grants R01 GM-21595 and R01 GM-60551 from the National Institutes of Health, grant MCB-0445841 from the National Science Foundation, and grant RGP0031 from the Human Frontier Scientific Program Organization (with co-investigators Jan Ellenberg, Angus Lamond, and Matthias Mann). Marty Jacobson in the author’s lab played a key role in the signal recognition particle work that was a catalyst in formulating the plurifunctional nucleolus hypothesis. Joan Politz creatively advanced and confirmed this hypothesis in her SRP work, amidst other pioneer research she led on mRNA and rRNA intranuclear dynamics in the author’s lab. I am greatly indebted to them both. I am also grateful to Joseph Gall, Susan Gerbi, and Masayasu Nomura for their longstanding support of my research on the nucleolus, encouraging my efforts into unsettled and unexplored terrain.
Footnotes
Editors: Tom Misteli and David L. Spector
REFERENCES
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P 2002. Molecular Biology of the Cell, 4th edition Garland Science, New York: p. 332 [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI 2002. Directed proteomic analysis of the human nucleolus. Curr Biol 12: 1–11 [DOI] [PubMed] [Google Scholar]
- Bachellerie JP, Cavaillé J, Hüttenhofer A 2002. The expanding RNA world. Biochimie 84: 775–790 [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI 2007. The multifunctional nucleolus. Nat Rev Mol Cell Biol 8: 574–585 [DOI] [PubMed] [Google Scholar]
- Boyne JR, Whitehouse A 2006. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc Natl Acad Sci USA 103: 15190–15195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Gurdon JB 1964. Absence of rRNA synthesis in the anucleolate mutant of X. laevis. Proc Natl Acad Sci USA 51: 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DD, Dawid IG 1968. Specific gene amplication in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science 160: 272–280 [DOI] [PubMed] [Google Scholar]
- Caesar J (40-50s BC). De Bello Gallico I: 1 [Google Scholar]
- Calvet JP, Pederson T 1981. Base-pairing interactions between snRNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell 26: 363–370 [DOI] [PubMed] [Google Scholar]
- Chen D, Huang S 2001. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol 153: 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciufo LF, Brown JD 2000. Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr Biol 10: 1256–1264 [DOI] [PubMed] [Google Scholar]
- Costa-Nunes P, Pontes O, Preuss SB, Pikaard CS 2010. Extra views on RNA-dependent DNA methylation and MBD6-dependent heterochromatin formation in nucleolar dominance. Nucleus 1: 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deryusheva S, Gall JG 2004. Dynamics of coilin in Cajal bodies of the Xenopus germinal vesicle. Proc Natl Acad Sci USA 101: 4810–4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Pontvianne F, Wierzbicki AT, Blevins T, Tucker S, Costa-Nunes P, Pontes O, Pikaard CS 2010. Mechanisms of HDA6-mediated rRNA gene silencing; suppression of intergenic pol II transcription and differential effects on maintenance versus siRNA-directed cytosine methylation. Genes Dev 24: 1119–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica A, Tollervey D 2002. Making ribosomes. Curr Opin Cell Biol 14: 313–318 [DOI] [PubMed] [Google Scholar]
- Fichelson P, Moch C, Ivanovitch K, Martin C, Sidor CM, Lepesant J-A, Bellaiche Y, Huynh J-R 2009. Live-imaging of single stem cells within their niche reveals that a U3 snoRNP component segregates asymmetrically and is required for self-renewal in Drosophila. Nat. Cell Biol 11: 685–693 [DOI] [PubMed] [Google Scholar]
- Fléchon JE 2006. Analysis of the nucleolar compartment of the nucleus as an indicator of nuclear reprogramming after nuclear transfer. Meth Mol Biol 325: 225–238 [DOI] [PubMed] [Google Scholar]
- Fuhrman LE, Goel AK, Smith J, Shianna KV, Aballay A 2009. Nucleolar proteins suppress Caenorhabditis elegans innate immunity by inhibiting p53/CEP-1. PLoS Genetics 5: e1000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG 1968. Differential synthesis of the genes for ribosomal RNA during amphibian oogenesis. Proc Natl Acad Sci USA 60: 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia SN, Pillus L 1999. Net results of nucleolar dynamics. Cell 97: 825–828 [DOI] [PubMed] [Google Scholar]
- Ghetti A, Pinol-Roma S, Michael WM, Morandi C, Dreyfuss G 1992. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res 20: 3671–3678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda K, Fowler J, Katoku-Kikyo N, Haroldson J, Wudel J, Kikyo N 2003. Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat Cell Biol 5: 205–210 [DOI] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ 2004. Ribosome biogensis: of knobs and RNA processing. Exp Cell Res 296: 43–50 [DOI] [PubMed] [Google Scholar]
- Grosshans H, Deinhert K, Hurt E, Somos G 2001. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP RNA, and Xpo1p-mediated export. J Cell Biol 153: 745–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L 1997. Link between aging and the nucleolus. Genes Dev 11: 2449–2455 [DOI] [PubMed] [Google Scholar]
- Handwerger KE, Cordero JA, Gall JG 2005. Cajal bodies, nucleoli and speckles have a low-density, sponge-like structure. Mol Biol Cell 16: 202–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris H 1974. Nucleus and Cytoplasm 3rd edition Clarendon Press, Oxford [Google Scholar]
- Heitz E 1931. Nukleolar und chromosomen in der gattung. Vicia Planta 15: 495–505 [Google Scholar]
- Hernandez-Verdun D, Roussel P, Gébrane-Younès J 2002. Emerging concepts of nucleolar assembly. J Cell Sci 115: 2265–2270 [DOI] [PubMed] [Google Scholar]
- Huang S 2000. Perinucleolar structures. J Struct Biol 129: 233–240 [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Wang YL, Pederson T 1995. Dynamic localization of RNase MRP RNA in the nucleolus observed by fluorescent RNA cytochemistry in living cells. J Cell Biol 131: 1649–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T 1997. Nuclear domains of the RNA subunit of RNase P. J Cell Sci 110: 829–837 [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T 1998a. A 7-methylguanosine cap commits U3 and U8 small nuclear RNAs to the nucleolar localization pathway. Nucleic Acids Res 26: 756–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson MR, Pederson T 1998b. Localization of signal particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci USA 95: 7981–7986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B 1990. The U3 small nucleolar ribonucleoprotein functions in the first step of pre-ribosomal RNA processing. Cell 60: 897–908 [DOI] [PubMed] [Google Scholar]
- Kim SH, Koroleva OA, Lewandowska D, Pendle AF, Clark GP, Simpson CG, Shaw PJ, Brown JW 2009. Aberrant mRNA transcripts and the nonsense-mediated decay proteins UPF2 and UPF3 are enriched in the Arabidopsis nucleolus. Plant Cell 21: 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Ryabov EV, Kalinina NO, Rakitina DV, Gillespie T, MacFarlane S, Haupt S, Brown JWS, Taliansky M 2007. Cajal bodies and the nucleolus are required for a plant virus systemic infection. EMBO J 26: 2169–2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koberna K, Malinsky J, Pliss A, Masata M, Vecerova J, Fiolova M, Bednar J, Raška I 2002. Ribosomal genes in focus: new transcripts label the dense fibrillar components and form clusters indicative of “Christmas trees”. J Cell Biol 157: 743–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulikov R, Letienne J, Kaur M, Grossman SR, Arts J, Blattner C 2010. Mdm2 facilitates the association of p53 with the proteasome. Proc Natl Acad Sci USA 107: 10038–10043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Warner JR 1972. Characterization of ribosomal precursor particles from HeLa cell nucleoli. J Mol Biol 63: 233–246 [DOI] [PubMed] [Google Scholar]
- Kyng KJ, May A, Kǿvraa S, Bohr VA 2003. Gene expression profiling in Werner syndrome closely resembles that of normal aging. Proc Natl Acad Sci USA 100: 12259–12264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YW, Lamond AI, Mann M, Andersen JS 2007. Analysis of nucleolar protein dynamics reveals the nuclear degradation of ribosomal proteins. Curr Biol 17: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL, Andersen JS, Mann M, Lamond AI 2003a. Bioinformatic analysis of the nucleolus. Biochem J 376: 553–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AKL, Lamond AI 2003. The dynamics of the nucleolus. Crit Rev Euk Gene Expr 13: 39–54 [DOI] [PubMed] [Google Scholar]
- Lewin B 1980. Gene Expression. Volume 2. Eukaryotic Chromosomes. 2nd edition John Wiley & Sons, New York [Google Scholar]
- Liau MC, Perry RP 1969. Ribosome precursor particles in nucleoli. J Cell Biol 42: 272–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Pederson T 2007. Depletion of the nucleolar protein nucleostemin causes G1 cell cycle arrest via the p53 pathway. Mol Biol Cell 18: 2630–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Pederson T 2008a. Nucleostemin: a multiplex regulator of cell-cycle progression. Trends Cell Biol 18: 575–579 [DOI] [PubMed] [Google Scholar]
- Ma H, Pederson T 2008b. Nucleophosphmin is a binding partner of nucleostemin in human osteosarcoma cells. Mol Biol Cell 19: 2870–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio R, Siekevitz P, Palade GE 1963. Studies on isolated nuclei. I. Isolation and chemical composition of nucleolar and nucleoplasmic subfractions. J Cell Biol 18: 293–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak RA, Lombard DB, Johnson FB, Guarente L 1998. Nucleolar localization of the Werner syndrome protein in human cells. Proc Natl Acad Sci USA 95: 6887–6892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martindill DMJ, Risebro CA, Smart N, Franco-Viseras MDM, Rosario CO, Swallow CJ, Dennis JW, Roliry PR 2007. Nucleolar release of Hand1 acts as a molecular switch to determine cell fate. Nat Cell Biol 9: 1131–1141 [DOI] [PubMed] [Google Scholar]
- Matera AG, Frey MR, Margelot K, Wolin SL 1995. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein. J Cell Biol 129: 1181–1193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R 2006. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 22: 351–361 [DOI] [PubMed] [Google Scholar]
- McClintock B 1934. The relationship of a particular chromosomal element to the development of the nucleoli in Zea mays. Z. Zellforsch Mikrosk 21: 294–398 [Google Scholar]
- Michienzi A, Li S, Zaia JS, Rossi JJ 2002. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc Natl Acad Sci USA 99: 14047–14052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller OL Jr, Beatty BR 1969. Visualization of nucleolar genes. Science 164: 955–957 [DOI] [PubMed] [Google Scholar]
- Misteli T 2003. A nucleolar disappearing act in somatic cloning. Nat Cell Biol 5; 183–184 [DOI] [PubMed] [Google Scholar]
- Misteli T 2005. Going in GTP cycles in the nucleolus. J Cell Biol 168: 177–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TH 1898. Comparative cytological studies, with especial regard to the morphology of the nucleolus. J Morphol 15: 265–582 [Google Scholar]
- Olson MO, Dundr M, Szebeni A 2000. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol 10: 189–196 [DOI] [PubMed] [Google Scholar]
- Olson MO, Hingorani K, Szebeni A 2002. Conventional and nonconventional roles of the nucleolus. Int Rev Cytol 219: 199–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parades S, Maggert KA 2009. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci USA 106: 17829–17834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T 1998a. The plurifunctional nucleolus. Nucleic Acids Res 26: 3871–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T 1998b. Growth factors in the nucleolus? J Cell Biol 143: 279–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T 2000. Diffusional protein transport within the nucleus: a message in the medium. Nat Cell Biol 2: E73–E74 [DOI] [PubMed] [Google Scholar]
- Pederson T 2001a. Fluorescent RNA cytochemistry: tracking gene transcripts in living cells. Nucleic Acids Res 29: 1013–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T 2001b. Protein mobility within the nucleus- what are the right moves? Cell 104: 635–638 [DOI] [PubMed] [Google Scholar]
- Pederson T 2002. Proteomics of the nucleolus: more proteins, more functions? Trends Biochem Sci 27: 111–112 [DOI] [PubMed] [Google Scholar]
- Pederson T 2007. Ribosomal protein mutations in Diamond-Blackfan anemia: might they operate upstream of protein synthesis? The FASEB J 21: 3442–3445 [DOI] [PubMed] [Google Scholar]
- Pederson T 2010a. “Compact” nuclear domains: reconsidering the nucleolus. Nucleus 1: 444–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T 2010b. Regulatory derived from transfer RNA? RNA 16: 1865–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, Kumar A 1971. Relationship between protein synthesis and ribosome assembly in HeLa cells. J Mol Biol 61: 655–668 [DOI] [PubMed] [Google Scholar]
- Pederson T, Politz JC 2000. The nucleolus and the four ribonucleoproteins of translation. J Cell Biol 148: 1091–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pederson T, Tsai RYL 2009. In search of nonribosomal protein function and regulation. J Cell Biol 184: 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ 2005. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry RP 1966. Nucleolus: Structure and function. Science 153: 214–219 [DOI] [PubMed] [Google Scholar]
- Phair RD, Misteli T 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404: 604–609 [DOI] [PubMed] [Google Scholar]
- Politz JC 1999. Use of caged fluorochromes to track macromolecular movement in living cells. Trends Cell Biol 9: 284–287 [DOI] [PubMed] [Google Scholar]
- Politz JC, Browne ES, Wolf DE, Pederson T 1998. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy. Proc Natl Acad Sci USA 95: 6043–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JCR, Hogan EM, Pederson T 2009. MicroRNAs with a nucleolar location. RNA 15: 1705–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Lewandowski LB, Pederson T 2002. Signal recognition particle RNA localization within the nucleolus differs from the classical sites of ribosome synthesis. J Cell Biol 159: 411–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JCR, Polena I, Trask I, Bazett-Jones DP, Pederson T 2005. A nonribosomal landscape in the nucleolus revealed by the stem cell protein nucleostemin. Mol Biol Cell 16: 3401–3410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JC, Tuft RA, Pederson T 2003. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell 14: 4805–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JCR, Tuft RA, Pederson T 2005. Photoactivation-based labeling and in vivo tracking of RNA molecules in the nucleus. Live Cell Imaging: A Laboratory Manual Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Politz JC, Yarovoi S, Kilroy S, Gowda K, Zwieb C, Pederson T 2000. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci USA 97: 55–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politz JCR, Zhang F, Pederson T 2006. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc Natl Acad Sci USA 103: 18957–18962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock C, Huang S 2010. The perinucleolar compartment. Cold Spring Harb Perspect Biol doi: 10.1101/cshperspect.a000679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J-L, McStay B 2007. Recruitment of factors linking transcription and processing of pre-rRNA to NOR chromatin is UBF-dependent and occurs independent of transcription in human cells. Genes Dev 21: 2041–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieto J-L, McStay B 2008. Pseudo-NORs; a novel method for studying nucleoli. Biochim et Biophys Acta 1783: 2116–2123 [DOI] [PubMed] [Google Scholar]
- Raška I 2003. Oldies but goldies: searching for Christmas trees within the nucleolar architecture. Trends Cell Biol 13: 517–525 [DOI] [PubMed] [Google Scholar]
- Raška I, Shaw PJ, Cmarko D 2006. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol 18: 325–334 [DOI] [PubMed] [Google Scholar]
- Richter JD. Fallon JR 2007. Synapses go nucle(ol)ar. Nat Neurosci 10: 399–400 [DOI] [PubMed] [Google Scholar]
- Royo F, Paz N, Espinosa L, McQueen PG, Vellón L, Parada LA 2009. Spatial link between nucleoli and expression of the ZacI gene. Chromosoma 118: 711–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stressors. EMBO J 22: 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Schmitz K-M, Sandoval J, Grummt I 2010. Intergenic transcripts originating from a subclass of ribosomal DNA repeats silence ribosomal RNA genes in trans. EMBO rep 11: 52–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer U, Hock R 1999. Structure and function of the nucleolus. Curr Opin Cell Biol 11: 385–390 [DOI] [PubMed] [Google Scholar]
- Scherl A, Couté Y, Déon C, Callé A, Kindbeiter K, Sanchez JC, Greco A, Hochstrasser D, Diaz JJ 2002. Functional proteomic analysis of human nucleolus. Mol Biol Cell 13: 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Milles K, Guarente L 1997. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science 277: 1313–1316 [DOI] [PubMed] [Google Scholar]
- Soeiro R, Basile C 1973. Non-ribosomal nucleolar proteins in HeLa cells. J Mol Biol 79: 507–519 [DOI] [PubMed] [Google Scholar]
- Sommerville J, Brumwell CL, Politz JCR, Pederson T 2005. Signal recognition particle assembly in relation to the function of amplified nucleoli of Xenopus oocytes. J Cell Sci 118: 1299–1307 [DOI] [PubMed] [Google Scholar]
- Sonntag F, Schmidt K, Kleinschmidt JA 2010. A viral assembly factor promotes AAV2 capsid assembly in the nucleolus. Proc Natl Acad Sci USA 107: 10220–10225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spell J, Kubitscheck U 2009. Single ovalbumin molecules exploring nucleoplasm and nucleoli of living cell nuclei. Biochim Biophys Acta 1803: 396–404 [DOI] [PubMed] [Google Scholar]
- Stavreva DA, Kawasaki M, Dundr M, Koberna K, Muller WG, Tsujimura-Takahashi T, Komatsu W, Hayano T, Isobe T, Raška I, et al. 2006. Potential roles for ubiquitin and the proteasome during ribosome biogenesis. Mol Cell Biol 26: 5131–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely AM, Chen Y-H, Zhang C, Oshima J, Weissman SM 2000. Werner protein recruits DNA polymerase δ to the nucleolus. Proc Natl Acad Sci USA 97: 11365–11370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, McKay RD 2002. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev 16: 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RY, McKay RD 2005. A multistep, GTP-driven mechanism driving the dynamic cycling of nucleostemin. J Cell Biol 168: 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin G 1836. Repertorium für Anatomie und Physiologie 1: 1–293 Verlag von Veit und Comp; Berlin [Google Scholar]
- Valentin G 1839. Repertorium für Anatomie und Physiologie 4: 1–275 Verlag von Veit und Comp; Berlin [Google Scholar]
- Vincent WS 1952. The isolation and chemical properties of the nucleoli of starfish oocytes. Proc Natl Acad Sci USA 38: 139–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent WS, Miller OL 1966. International symposium on the nucleolus. Its structure and function. Montevideo, Uruguay. J Natl Cancer Inst Monogr 23: 1–610 [Google Scholar]
- Vinistin R, Amon A 2000. The nucleolus: the magician’s hat for cell cycle tricks. Curr Opin Cell Biol 12: 372–377 [DOI] [PubMed] [Google Scholar]
- von Kobbe C, Bohr VA 2002. A nucleolar targetting sequence in the Werner syndrome protein resides within residues 949-1092. J Cell Sci 115: 3901–3907 [DOI] [PubMed] [Google Scholar]
- Wagner R 1835. Einige Bemerkungen und Fragen über das Keimbläschen (vesicular germinativa). Müller’s Archiv Anat Physiol Wissenschaft Med 373–377 [Google Scholar]
- Wang J, Cao LG, Wang YL, Pederson T 1991. Localization of pre-messenger RNA at discrete nuclear sites. Proc Natl Acad Sci 88: 7391–7395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Politz JC, Pederson T, Huang S 2003. RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol Biol Cell 14: 2425–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR, Soeiro R 1967. Nascent ribosomes from HeLa cells. Proc Natl Acad Sci USA 58: 1984–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol 1: 20–26 [DOI] [PubMed] [Google Scholar]