GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage (original) (raw)
Abstract
Chromatin structure is known to be a barrier to DNA repair and a large number of studies have now identified various factors that modify histones and remodel nucleosomes to facilitate repair. In response to ultraviolet (UV) radiation several histones are acetylated and this enhances the repair of DNA photoproducts by the nucleotide excision repair (NER) pathway. However, the molecular mechanism by which UV radiation induces histone acetylation to allow for efficient NER is not completely understood. We recently discovered that the E2F1 transcription factor accumulates at sites of UV-induced DNA damage and directly stimulates NER through a non-transcriptional mechanism. Here we demonstrate that E2F1 associates with the GCN5 acetyltransferase in response to UV radiation and recruits GCN5 to sites of damage. UV radiation induces the acetylation of histone H3 lysine 9 (H3K9) and this requires both GCN5 and E2F1. Moreover, as previously observed for E2F1, knock down of GCN5 results in impaired recruitment of NER factors to sites of damage and inefficient DNA repair. These findings demonstrate a direct role for GCN5 and E2F1 in NER involving H3K9 acetylation and increased accessibility to the NER machinery.
INTRODUCTION
Exposure to UV radiation from the sun is responsible for the DNA mutations that lead to the development of most human skin cancers (1,2). The major forms of DNA damage caused by UV radiation are the cyclobutane pyrimidine dimer (CPD) and the pyrimidine–pyrimidone (6-4) adduct, otherwise known as the (6-4) photoproduct [(6-4)PP]. The nucleotide excision repair (NER) pathway is responsible for repairing DNA damage caused by UV radiation. NER consists of two sub-pathways: global genome repair (GG-NER) that is responsible for the removal of lesions from the entire genome; and transcription-coupled repair (TCR) that preferentially repairs damage on an actively transcribed DNA strand. The importance of properly repairing UV-induced DNA damage is exemplified by patients with the rare autosomal disease Xeroderma Pigmentosum (XP). XP is caused by the inheritance of mutations in genes encoding NER proteins and is characterized by extreme sensitivity to the sun and strong predisposition to skin cancers. In addition to repairing UV-induced damage, NER is also important for repairing other types of lesions involving bulking DNA adducts and strand distortions.
XP can be broken down into seven complementation groups, XPA through XPG, with each representing a different gene encoding a protein involved in NER. Cloning of XP genes and the purification of repair proteins has lead to a detailed understanding of the biochemical events of NER [for review see (3)]. The first step in NER is the recognition of distortions in damaged DNA by the XPC complex (4). For some DNA lesions, such as CPD, recognition of DNA distortions may also require another factor termed DNA damage-binding protein (DDB) that is a heterodimer of DDB2 (XPE, p48) and DDB1 (p127) (5,6). Binding of the XPC complex results in further alterations to the DNA structure, which facilitates the recruitment of XPA, replication protein A (RPA) and the basal transcription factor complex TFIIH (7–9). The final steps of NER involve unwinding the DNA around the lesion, cleavage of the damaged strand by 3′ and 5′ incisions and gap filling by a DNA polymerase followed by ligation.
While in vitro studies with purified proteins and substrates have shed considerable light on the biochemical events of the NER reaction, a complete understanding of how NER is regulated in the context of chromatin is lacking. Previous studies demonstrated that packaging of DNA into nucleosomes inhibits NER (10,11). Moreover, older studies showed there is an increase in histone acetylation and a relaxation of chromatin structure in response to UV radiation that enhances NER (12–14). Several factors have been implicated in stimulating the repair of UV-induced DNA damage by increasing chromatin accessibility, including p53, p300 and p33ING (15–18). These factors appear to function in a common pathway that responds to UV damage and results in increased histone H4 acetylation and chromatin relaxation throughout the nucleus (16–18).
We recently found that the E2F1 transcription factor can also stimulate NER by enhancing the recruitment of DNA repair factors to sites of UV-induced DNA damage (19). It was previously shown that E2F1 is stabilized in response to various forms of DNA damage and that this involves the phosphorylation of E2F1 on serine 31 by the ataxia telangiectasia mutated (ATM) or ATM and Rad3-related (ATR) kinases (20). In the case of DNA double-strand breaks, phosphorylation of E2F1 by ATM results in the transcriptional activation of pro-apoptotic target genes, such as p73 and the induction of apoptosis (20–22). On the other hand, UV-induced DNA damage, while resulting in E2F1 stabilization, does not lead to the induction of E2F1 pro-apoptotic target genes (21). In fact, we have demonstrated that E2F1 has an anti-apoptotic function in response to UV that is likely related to its ability to stimulate NER (23,24). Mutational analysis of E2F1 demonstrates that this DNA-repair function is independent of E2F1-transcriptional activity but dependent on E2F1 serine 31 and the ATR kinase (19). In addition to stabilizing E2F1, phosphorylation of E2F1 at serine 31 also results in the recruitment of E2F1 to sites of both double-strand breaks and UV-induced DNA damage through a phospho-specific interaction with the TopBP1 protein (19,25). However, the function of E2F1 at sites of DNA damage is currently unknown.
Here we demonstrate that E2F1 associates with the GCN5 histone acetyltransferase in response to UV radiation and recruits GCN5 to sites of damage. In yeast, GCN5 is known to stimulate NER at the MFA2 locus by acetylating histone H3 (26–28) but a role for GCN5 in human NER has not been established. We now show that depletion of GCN5 or E2F1 in human cells results in impaired H3K9 acetylation in response to UV, decreased recruitment of NER factors to sites of damage and inefficient DNA repair.
MATERIALS AND METHODS
Cell culture
Primary dermal normal human fibroblasts (NHF, GM08399) were obtained from Coriell Institute. Cells were maintained in DMEM (Hyclone) supplemented with 10% fetal bovine serum (FBS, Atlanta).
UV treatment
For general UV treatment, UVB was delivered by Westinghouse FS20 sunlamps filtered through cellulose acetate (Kodacel from Kodak, St. Louis, MO, USA) with a wavelength cut-off of 290 nm. Dosimetry was determined with a IL1400 photometer coupled to a SCS 280 probe (International Light, Newburyport, MA, USA). Because filters absorb ∼90% of the UV radiation (29), it was necessary to use UVC for the co-localization assays since UVC is more efficient at inducing DNA damage compared to the more physiological relevant UVB radiation. UVC was delivered by Phillips Sterilamp G8T5 bulbs emitting predominantly 254 nm. The dose was measured using an IL-1400A Photometer coupled to SEL 240 detector.
Antibodies, western blot and co-IP
Cells were mock treated or treated with 500 J/m2 of UVB and whole-cell lysates were obtained using 1X lysis buffer (Cell signaling). For co-IP, E2F1 antibody conjugated Protein A/G agarose (Santa Cruz) was used for IP and precipitate was analyzed by western blot using E2F1 or GCN5 antibodies. Antibodies/antisera for western blot and IF staining were obtained from the following sources: E2F1 (C-20 and KH95), XPA polyclonal, CHK1, PCAF and p62, Santa Cruz; CPD and (6-4)PP, MBL; GCN5, Biolegend; XPA monoclonal (12F5), Lab Vision; total H3, Cell Signaling. Rabbit polyclonal antisera to the CPD and (6-4)PP photoproducts were developed by Dr David Mitchell. Antibodies specific to acetylated histones were obtained from Abcam (H4K16Ac, ab23352), Cell signaling (H3K9Ac, 9671) and Sigma (H3K9Ac, H0913). Western blot band densities were quantified using ImageJ software.
Filtered UV irradiation/IF staining
Co-localization of proteins with UV-induced DNA damage was performed as previously described (29–31). Briefly, cells grown on chamber slides (Nunc) were rinsed in PBS leaving a thin layer of buffer on top. Sterile isopore polycarbonate membrane filters (Milipore) containing pores of 8 µm in diameter were placed on top of the cells and the slides were irradiated from above with UVC. The filter was then removed and cells were incubated for designated time points before a cytoskeleton extraction procedure. After washing, cells were fixed in PBS containing 2% formaldehyde and 0.2% Triton X-100. For IF staining, fixed cells were incubated with 3% BSA, washed and treated with 2M HCL for 5 min at 37°C to denature the DNA. Washed cells were then incubated with appropriate primary antibodies (e.g. specific for CPD and E2F1) followed by incubation with appropriate fluorescent secondary antibodies (Alexa 488 or Alexa 594, Invitrogen). Cells were then stained with DAPI and sealed in mounting media (Vector Lab) with cover slips. The images were captured, digitally recorded and analyzed using a Nikon eclipse 80i microscope equipped with an X-cite 120 fluorescence illumination system and Metamorph image analysis software. Percent co-localization was determined by scoring 100 randomly selected cells for each experimental group.
Slot-blot DNA repair assay
UV-induced DNA damage was detected as described (32) with minor modifications. Briefly, cells were treated with 500 J/m2 of UVB and incubated for the designated time periods before isolating genomic DNA using the GenElute kit (Sigma). DNA was quantified and 0.5 mg of DNA was spotted on nitrocellulose membranes (Genemate) using a slot-blot transfer apparatus (Bio-Rad). The UV-induced DNA damage was detected by an immuno-blot procedure using antibodies against (6-4)PP (1:1000) and CPD (1:2000). The membrane was re-probed with specific antibody against single-stranded DNA (Chemicon, MAB3034) for loading control. The densities of the bands were quantified by ImageJ and the graphs were plotted accordingly after normalization with the loading control.
Small interference RNA
Small interference RNAs (siRNA) against human _E2F1_-coding region (E2F-1 siRNA sc29297: strand 5169 CACCUGAUGAAUAUCUGUA; strand 5170 CCUGAUGAAUAUCUGUACU; strand 5171 GAGUCUGUGUGGUGUGUAU) and human _GCN5_-coding region (GCN5 siRNA sc37946: duplex 1 strand CCAAGCAGGUCUAUUUCUA; duplex 2 strand GGAAAUGCAUCCUGCAGAU; duplex 3 strand GAGGCCUCAUUGACAAGUA) were purchased from Santa Cruz. Transfections were performed with Lipofectamine 2000 reagent (Invitrogen) following the manufacturer’s recommended protocol.
RESULTS
GCN5 co-localizes with sites of UV-induced DNA damage in an E2F1 dependent manner
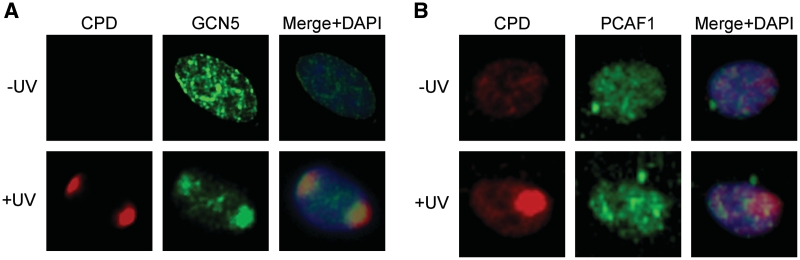
The local UV irradiation assay (29–31) was used to examine the recruitment of GCN5 to sites of DNA damage. For these experiments, normal human fibroblasts were covered with polycarbonate filters and exposed to 50 J/m2 of UVC. Cells were fixed 30 min following exposure and UV-induced DNA damage was detected by indirect immunofluorescence (IF) staining using a monoclonal antibody to CPD (Figure 1A). Cells were co-stained with polyclonal antisera to GCN5 followed by a fluorescently labeled secondary antibody. GCN5 staining in the absence of UV treatment, while punctuate, was relatively uniform throughout the nucleus. Following UV irradiation, GCN5 was found to redistribute to discrete areas of the nucleus that overlapped with CPD staining (Figure 1A). In contrast, the staining pattern of the related PCAF acetyltransferase did not change following UV irradiation and no co-localization with UV-damaged sites was observed (Figure 1B).
Figure 1.
GCN5 co-localizes with sites of UV-induced DNA damage. NHFs were untreated (upper panels) or irradiated with 50 J/m2 of UVC through a 8 µm pore filter (lower panels) and 30 min later stained for CPD (red) and (A) GCN5 or (B) PCAF (green). Cells were counter-stained with DAPI and images were digitally recorded.
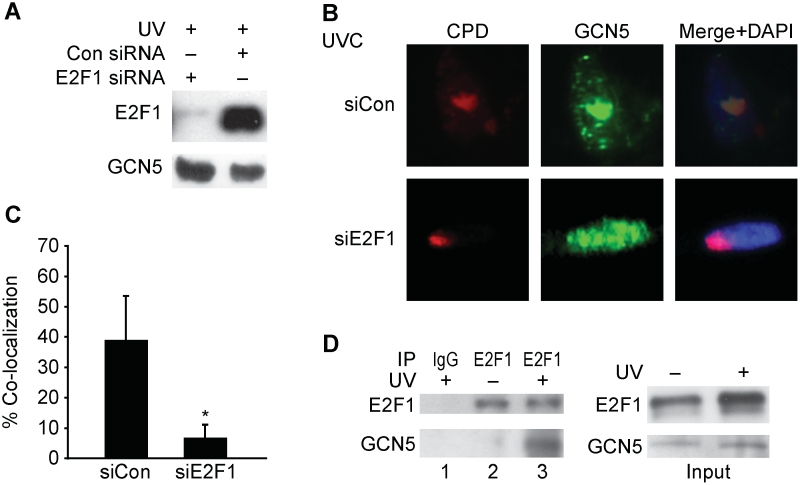
We recently discovered that the E2F1 transcription factor accumulates at sites of UV-induced DNA damage and functions to stimulate NER through a non-transcriptional mechanism (19). Given that GCN5 can partner with E2F factors in the context of transcription (33), we asked whether E2F1 might be involved in the localization of GCN5 to sites of UV damage. Depletion of E2F1 with siRNA did not affect the expression of GCN5 but did significantly decrease the co-localization of GCN5 with sites of damage (Figure 2A–C). Moreover, UV radiation induced a stable association between the endogenous GCN5 and E2F1 proteins (Figure 2D).
Figure 2.
E2F1 is required for GCN5 localization to sites of UV damage. (A) NHFs were transfected with siRNA to E2F1 or control siRNA and 48 h later exposed to 500 J/m2 of UVB 30 min prior to harvest. Western blot analysis was performed using whole-cell lysates and antibodies to E2F1 and GCN5. (B) NHFs transfected with control siRNA (upper panels) or siRNA to E2F1 (lower panels) were irradiated with 50 J/m2 of UVC through a filter. Thirty minute post-irradiation, cells were fluorescently stained for CPD (red) or GCN5 (green) and counter-stained with DAPI. (C) Co-localization of GCN5 with CPD were scored from three independent experiments as described. Asterisk indicates statistically significant difference (P < 0.05). (D) NHFs were exposed to 500 J/m2 of UVB 30 min prior to harvest (lanes 1 and 3) or untreated (lane 2) as indicated. Immunoprecipitation was performed using control IgG (lane 1) or antibody to E2F1 (lanes 2 and 3) and the precipitate was subjected to western blot analysis for E2F1 (top panel) and GCN5 (bottom panel). An input control western blot is shown at right.
Depletion of GCN5 with siRNA did not affect E2F1 protein expression levels or its induction in response to UV radiation (Supplementary Figure S1A). Knock down of GCN5 also did not affect the co-localization of E2F1 with sites of DNA damage (Supplementary Figure S1B and C). Taken together, these findings indicate that E2F1 is upstream of GCN5 and recruits GCN5 to sites of UV damage through a direct or indirect physical interaction induced by UV radiation.
E2F1 and GCN5 are required for the rapid acetylation of H3K9 at sites of UV damage
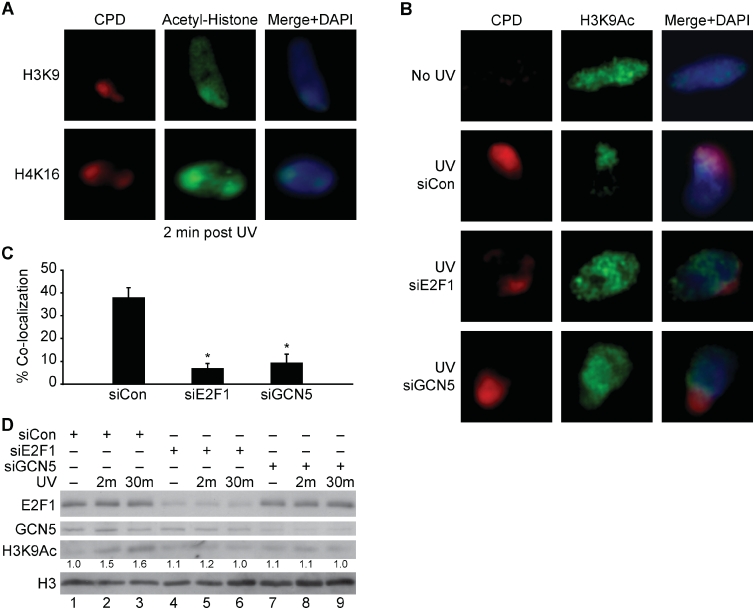
The local UV assay was next used to examine the acetylation of specific lysines on histones H3 and H4 at sites of UV damage. Antibodies to acetylated H3K9 and H4K16 were found to give staining patterns that co-localized with sites of UV damage (Figure 3A). Co-localization of acetylated H3K9 and H4K16 was observed within 2 min of UV exposure but this staining pattern dissipated by 30 min (Supplementary Figure S2). Depletion of E2F1 or GCN5 prevented the increased H3K9 acetylation at sites of damage but did not significantly affect the co-localized staining pattern of acetylated H4K16 with CPD (Figure 3B and C and Supplementary Figure S3). Global H3K9 acetylation levels were also increased in response UV radiation and this was inhibited by depletion of either E2F1 or GCN5 (Figure 3D).
Figure 3.
E2F1 and GCN5 promote the rapid acetylation of H3K9 at sites of UV-induced DNA damage. (A) NHFs were irradiated with 50 J/m2 of UVC through a filter and 2 min later cells were fixed and fluorescently stained for CPD (red) and acetylated H3K9 or H4K16 (green). Cells were counter stained with DAPI and images recorded as previously described. (B) NHFs were mock treated (upper panels) or transfected with control siRNA (middle panels) or siRNAs to E2F1 and GCN5 (lower panels). Transfected cells were irradiated with 50 J/m2 of UVC through a filter (middle and lower panels) and 2 min later fluorescently stained for CPD (red) and H3K9Ac (green) (C) Co-localization H3K9Ac with CPD shown in (B) were scored from two independent experiments as described. Asterisk indicates statistically significant difference from siCon (P < 0.05). (D) NHFs were transfected with control siRNA (lanes 1–3), or siRNA to E2F1 (lanes 4–6) or GCN5 (lanes 7–9), and 24 h later mock treated (lanes 1, 4, and 7) or irradiated with 500 J/m2 of UVB and harvested 2 min (lanes 2, 5 and 8) or 30 min (lanes 3, 6 and 9) post-irradiation. Western blot analysis was performed on whole-cell extracts using antibodies to E2F1, GCN5, acetylated H3K9 and total H3. Numbers below the H3K9Ac row represent band density values after setting the untreated, siCon sample (lane 1) as 1.0 and normalizing to total H3 values.
GCN5 promotes the recruitment of NER factors to sites of damage to enhance NER efficiency
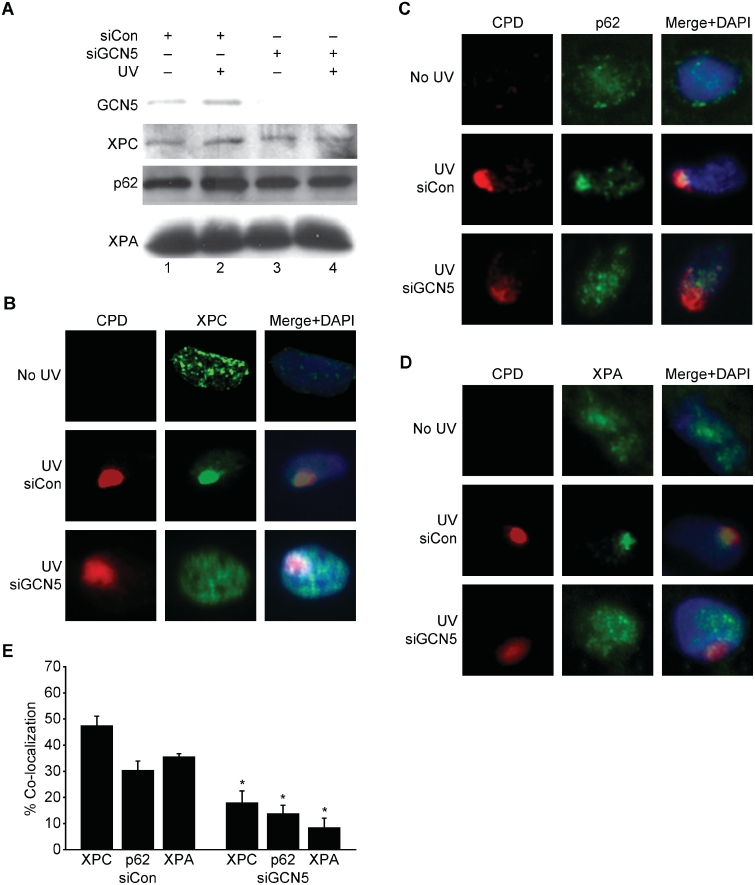
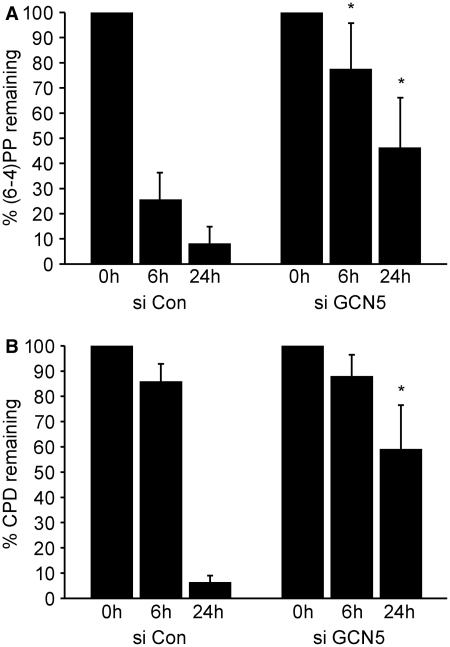
To determine if GCN5-dependent H3K9 acetylation regulates damage recognition by the NER machinery, GCN5 was depleted in NHFs and co-localization of NER factors with sites of UV damage was examined. Knock down of GCN5 did not affect the expression levels of XPC, p62 (part of the TFIIH complex), or XPA but did significantly impair their co-localization with sites of UV damage (Figure 4). To determine if GCN5 was important for NER efficiency, the removal of (6-4)PP and CPD from the genomic DNA of UV-irradiated cells was monitored by slot-blot analysis. After quantification of the band intensities and normalization with total single-stranded DNA, the percentage of photoproduct remaining was calculated. Depletion of GCN5 inhibited the repair of (6-4)PP at both early and late time points (Figure 5A). While the removal of CPD is less efficient and delayed compared with (6-4)PP, a significant difference in CPD repair was also observed at the later time point (Figure 5B). These findings are consistent with the impaired recruitment of NER factors to sites of damage and demonstrate that GCN5 is important for timely and efficient NER.
Figure 4.
GCN5 deficiency impairs the recruitment of NER factors to sites of UV damage. (A) NHFs were transfected with control siRNA or siRNA to GCN5 and 24 h later untreated (lanes 1 and 3) or treated with 500 J/m2 of UVB (lanes 2 and 4). Thirty-minute post-irradiation, western blot analysis was performed using whole-cell extracts and antibodies to GCN5, XPC, XPA or p62. (B–D) NHFs were mock treated (upper panels) or transfected with a control siRNA (middle panels) or siRNA to GCN5 (lower panels). Transfected cells were irradiated with 50 J/m2 of UVC through a filter (middle and lower panels). Thirty-minute post-irradiation, cells were fluorescently stained for CPD (red) and XPC, XPA or p62 (green). Cells were counter stained with DAPI and recorded as previously described. (E) Co-localization of XPC, XPA and p62 with CPD shown in (B–D) were scored from three independent experiments as described. Asterisk indicates statistically significant difference between siCon and siGCN5 transfected cells (P < 0.05).
Figure 5.
GCN5 deficiency impairs NER. NHFs were transfected with control siRNA or siRNA to GCN5. Cells were exposed 48 h after transfection to 500 J/m2 of UVB and harvested immediately (0 h) or 6- and 24-h post-irradiation. Genomic DNA was extracted and immuno-slot-blot analysis was performed for (6-4)PP (A) and CPD (B). Band intensities were quantified and the results are plotted after normalization with total single-strand DNA levels. The average from three independent experiments is presented. Asterisk indicates statistically significant difference between control and GCN5 siRNA transfected cells treated similarly.
DISCUSSION
It was suggested over 30 years ago that chromatin structure is altered to facilitate the repair of UV-induced DNA damage (34). Indeed, chromatin structure is known to be a barrier for NER (10,11) and previous studies demonstrated that increased histone acetylation in response to UV radiation stimulates repair (13,14). More recently, p53 was shown to function as a chromatin accessibility factor for GG-NER independent of its role in regulating transcription (17). The p33ING family of proteins also participates in p53-dependent chromatin relaxation following UV exposure and, like p53, the absence of p33ING impairs the recruitment of NER factors to sites of damage and reduces repair efficiency (15,16,18). Stimulation of NER by the p53-p33ING pathway is associated with increased acetylation of histone H4 (16,18), which may explain our finding that H4K16 acetylation in response to UV is independent of E2F1 and GCN5. We propose that E2F1 and GCN5 function in a pathway that is parallel to the p53-p33ING pathway to coordinate histone acetylation and increase accessibility of the NER machinery to sites of damage.
It was previously demonstrated that E2F1 accumulates at sites of DNA double-strand breaks and that this involves the phosphorylation of E2F1 at serine 31 by ATM and binding to the TopBP1 protein (25). We have found that E2F1 is also recruited to sites of UV damage through a similar mechanism, requiring phosphorylation on serine 31 by the ATR kinase and likely the same phospho-specific interaction with TopBP1 (19). Through an as yet unknown mechanism, UV radiation also induces an interaction between E2F1 and GCN5 and this is likely important for GCN5 localization and H3K9 acetylation at sites of UV damage. The co-localized staining pattern for acetylated H3K9 and UV damage is observed at 2 min but absent by 30 min after UV irradiation. This suggests that E2F1- and GCN5-mediated H3K9 acetylation is a rapid and perhaps transient event. However, global H3K9 acetylation continues to increase at 30 min post-UV and this is also dependent on E2F1 and GCN5. One possibility to explain these results is that H3K9 acetylation initiates at chromatin regions containing DNA damage but then spreads to the rest of the nucleus, thus leading to a loss of co-localization.
Jackson and co-workers (35) recently reported that global H3K9 acetylation levels decrease in response to a variety of DNA damaging agents, including UV. This is in contrast to our findings here and previous findings by others demonstrating an increase in H3 acetylation in response to UV (12,13,27,28). The reason for this discrepancy is unclear but may be due to differences in the time following exposure that H3K9 acetylation was examined or the cell type used.
Previous studies have implicated GCN5 in the repair of UV-induced DNA damage. In yeast, GCN5 was shown to stimulate the repair of UV damage at the MFA2 locus by acetylating histone H3 and increasing chromatin accessibility (27,28). Moreover, a purified human complex containing GCN5 was found to preferentially acetylate histone H3 in reconstituted nucleosomes-containing DNA photoproducts (36,37). Although this GCN5 complex has increased affinity for UV-damaged DNA in vitro, our findings suggest that E2F1 is required for efficient localization of GCN5 to sites of DNA damage, at least in human cells. This GCN5-containing complex, termed STAGA, was recently shown to harbour at least one other chromatin modifying enzyme, a histone H2B deubiquitinase (38,39). If E2F1 recruits GCN5 as part of a larger complex, then it is quite possible that E2F1 promotes additional chromatin modifications at sites of UV damage that might also contribute to efficient DNA repair.
This proposed role for E2F1 in GG-NER is not unlike the mechanism used by E2F1 to activate transcription. In the context of transcription, E2F1 recruits histone acetyltransferases and other co-activators to promoter regions of target genes to modify chromatin and facilitate the recruitment of the general transcription machinery. In the case of DNA repair, E2F1 recruits GCN5 to sites of UV damage to acetylate H3K9, which may increase access to the NER machinery. It is also possible that E2F1-dependent recruitment of GCN5 to sites of damage promotes the acetylation of other histone sites and additional proteins that may also influence DNA repair. While this process is not absolutely required for GG-NER, it is important for the timely and efficient repair of UV damage. Since NER not only repairs UV-induced DNA damage, but also other types of lesions, including damage caused by many chemotherapeutic drugs, this non-transcriptional function for E2F1 may also be important for the response to other carcinogens as well as cancer therapies.
E2F1 is also recruited to sites of DNA double-strand breaks (25) that also require chromatin modifications for efficient repair. Recently, it was demonstrated that GCN5 is involved in acetylating several sites on histone H3, including H3K9, in γH2AX-containing nucleosomes induced by ionizing radiation (40). GCN5-mediated acetylation of H3 helps to recruit the SWI/SNF chromatin remodeling complex via the bromo domains of BRG1 or hBrm. This in turn promotes additional γH2AX formation through a feedback activation loop (40). The mechanism by which GCN5 is recruited to sites of DNA double-strand breaks is unclear but our findings here suggest that it could involve E2F1. It will also be of interest to determine if E2F1- and GCN5-dependent H3 acetylation at sites of UV damage recruits the SWI/SNF complex to remodel nucleosomes to facilitate NER.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Data
FUNDING
The University of Texas Graduate School of Biomedical Sciences at Houston (in partial fulfillment of the requirements for the PhD degree); University of Texas M. D. Anderson Cancer Center, Houston, TX; National Institutes of Health (Grant numbers CA079468 to D.G.J., and CA105345, ES07784, ES07247 and CA016672). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Pam Blau, Jen Smith and Lakshmi Paniker for technical assistance, Chris Brown for assistance with figures and Becky Brooks for preparation of the manuscript. We also thank Karen Vazquez, Rick Wood, Snow Shen, Mark Bedford, Sharon Dent and Lei Li for helpful comments.
REFERENCES
- 1.Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Ponten J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl Acad. Sci. USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ziegler A, Jonason AS, Leffell DJ, Simon JA, Sharma HW, Kimmelman J, Remington L, Jacks T, Brash DE. Sunburn and p53 in the onset of skin cancer [see comments] Nature. 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 3.Friedberg EC. How nucleotide excision repair protects against cancer. Nat. Rev. Cancer. 2001;1:22–33. doi: 10.1038/35094000. [DOI] [PubMed] [Google Scholar]
- 4.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 5.Fitch ME, Nakajima S, Yasui A, Ford JM. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 2003;278:46906–46910. doi: 10.1074/jbc.M307254200. [DOI] [PubMed] [Google Scholar]
- 6.Wakasugi M, Kawashima A, Morioka H, Linn S, Sancar A, Mori T, Nikaido O, Matsunaga T. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 2002;277:1637–1640. doi: 10.1074/jbc.C100610200. [DOI] [PubMed] [Google Scholar]
- 7.Araujo SJ, Nigg EA, Wood RD. Strong functional interactions of TFIIH with XPC and XPG in human DNA nucleotide excision repair, without a preassembled repairosome. Mol. Cell. Biol. 2001;21:2281–2291. doi: 10.1128/MCB.21.7.2281-2291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reardon JT, Sancar A. Recognition and repair of the cyclobutane thymine dimer, a major cause of skin cancers, by the human excision nuclease. Genes Dev. 2003;17:2539–2551. doi: 10.1101/gad.1131003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokoi M, Masutani C, Maekawa T, Sugasawa K, Ohkuma Y, Hanaoka F. The xeroderma pigmentosum group C protein complex XPC-HR23B plays an important role in the recruitment of transcription factor IIH to damaged DNA. J. Biol. Chem. 2000;275:9870–9875. doi: 10.1074/jbc.275.13.9870. [DOI] [PubMed] [Google Scholar]
- 10.Hara R, Mo J, Sancar A. DNA damage in the nucleosome core is refractory to repair by human excision nuclease. Mol. Cell. Biol. 2000;20:9173–9181. doi: 10.1128/mcb.20.24.9173-9181.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang ZG, Wu XH, Friedberg EC. Nucleotide excision repair of DNA by human cell extracts is suppressed in reconstituted nucleosomes. J. Biol. Chem. 1991;266:22472–22478. [PubMed] [Google Scholar]
- 12.Ramanathan B, Smerdon MJ. Changes in nuclear protein acetylation in u.v.-damaged human cells. Carcinogenesis. 1986;7:1087–1094. doi: 10.1093/carcin/7.7.1087. [DOI] [PubMed] [Google Scholar]
- 13.Ramanathan B, Smerdon MJ. Enhanced DNA repair synthesis in hyperacetylated nucleosomes. J. Biol. Chem. 1989;264:11026–11034. [PubMed] [Google Scholar]
- 14.Smerdon MJ, Lan SY, Calza RE, Reeves R. Sodium butyrate stimulates DNA repair in UV-irradiated normal and xeroderma pigmentosum human fibroblasts. J. Biol. Chem. 1982;257:13441–13447. [PubMed] [Google Scholar]
- 15.Cheung KJ, Jr, Mitchell D, Lin P, Li G. The tumor suppressor candidate p33(ING1) mediates repair of UV-damaged DNA. Cancer Res. 2001;61:4974–4977. [PubMed] [Google Scholar]
- 16.Kuo WH, Wang Y, Wong RP, Campos EI, Li G. The ING1b tumor suppressor facilitates nucleotide excision repair by promoting chromatin accessibility to XPA. Exp. Cell Res. 2007;313:1628–1638. doi: 10.1016/j.yexcr.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 17.Rubbi CP, Milner J. p53 is a chromatin accessibility factor for nucleotide excision repair of DNA damage. EMBO J. 2003;22:975–986. doi: 10.1093/emboj/cdg082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Chin MY, Li G. The novel tumor suppressor p33ING2 enhances nucleotide excision repair via inducement of histone H4 acetylation and chromatin relaxation. Cancer Res. 2006;66:1906–1911. doi: 10.1158/0008-5472.CAN-05-3444. [DOI] [PubMed] [Google Scholar]
- 19.Guo R, Chen J, Zhu F, Biswas AK, Berton TR, Mitchell DL, Johnson DG. E2F1 localizes to sites of UV-induced DNA damage to enhance nucleotide excision repair. J. Biol. Chem. 2010;285:19308–19315. doi: 10.1074/jbc.M110.121939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM- dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 21.Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell Biol. 2003;5:552–558. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- 22.Wang B, Liu K, Lin FT, Lin WC. A role for 14-3-3 tau in E2F1 stabilization and DNA damage-induced apoptosis. J. Biol. Chem. 2004;279:54140–54152. doi: 10.1074/jbc.M410493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berton TR, Mitchell DL, Guo R, Johnson DG. Regulation of epidermal apoptosis and DNA repair by E2F1 in response to ultraviolet B radiation. Oncogene. 2005;24:2449–2460. doi: 10.1038/sj.onc.1208462. [DOI] [PubMed] [Google Scholar]
- 24.Wikonkal NM, Remenyik E, Knezevic D, Zhang W, Liu M, Zhao H, Berton TR, Johnson DG, Brash DE. Inactivating E2f1 reverts apoptosis resistance and cancer sensitivity in Trp53-deficient mice. Nat. Cell Biol. 2003;5:655–660. doi: 10.1038/ncb1001. [DOI] [PubMed] [Google Scholar]
- 25.Liu K, Lin FT, Ruppert JM, Lin WC. Regulation of E2F1 by BRCT domain-containing protein TopBP1. Mol. Cell. Biol. 2003;23:3287–3304. doi: 10.1128/MCB.23.9.3287-3304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teng Y, Liu H, Gill HW, Yu Y, Waters R, Reed SH. Saccharomyces cerevisiae Rad16 mediates ultraviolet-dependent histone H3 acetylation required for efficient global genome nucleotide-excision repair. EMBO Rep. 2008;9:97–102. doi: 10.1038/sj.embor.7401112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teng Y, Yu Y, Waters R. The Saccharomyces cerevisiae histone acetyltransferase Gcn5 has a role in the photoreactivation and nucleotide excision repair of UV-induced cyclobutane pyrimidine dimers in the MFA2 gene. J. Mol. Biol. 2002;316:489–499. doi: 10.1006/jmbi.2001.5383. [DOI] [PubMed] [Google Scholar]
- 28.Yu Y, Teng Y, Liu H, Reed SH, Waters R. UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc. Natl Acad. Sci. USA. 2005;102:8650–8655. doi: 10.1073/pnas.0501458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mone MJ, Volker M, Nikaido O, Mullenders LH, van Zeeland AA, Verschure PJ, Manders EM, van Driel R. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 2001;2:1013–1017. doi: 10.1093/embo-reports/kve224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katsumi S, Kobayashi N, Imoto K, Nakagawa A, Yamashina Y, Muramatsu T, Shirai T, Miyagawa S, Sugiura S, Hanaoka F, et al. In situ visualization of ultraviolet-light-induced DNA damage repair in locally irradiated human fibroblasts. J. Invest. Dermatol. 2001;117:1156–1161. doi: 10.1046/j.0022-202x.2001.01540.x. [DOI] [PubMed] [Google Scholar]
- 31.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 32.Wani AA, D’Ambrosio SM, Alvi NK. Quantitation of pyrimidine dimers by immunoslot blot following sublethal UV-irradiation of human cells. Photochem. Photobiol. 1987;46:477–482. doi: 10.1111/j.1751-1097.1987.tb04798.x. [DOI] [PubMed] [Google Scholar]
- 33.Lang SE, McMahon SB, Cole MD, Hearing P. E2F transcriptional activation requires TRRAP and GCN5 cofactors. J. Biol. Chem. 2001;276:32627–32634. doi: 10.1074/jbc.M102067200. [DOI] [PubMed] [Google Scholar]
- 34.Smerdon MJ, Lieberman MW. Nucleosome rearrangement in human chromatin during UV-induced DNA- repair synthesis. Proc. Natl Acad. Sci. USA. 1978;75:4238–4241. doi: 10.1073/pnas.75.9.4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. EMBO J. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brand M, Moggs JG, Oulad-Abdelghani M, Lejeune F, Dilworth FJ, Stevenin J, Almouzni G, Tora L. UV-damaged DNA-binding protein in the TFTC complex links DNA damage recognition to nucleosome acetylation. EMBO J. 2001;20:3187–3196. doi: 10.1093/emboj/20.12.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez E, Palhan VB, Tjernberg A, Lymar ES, Gamper AM, Kundu TK, Chait BT, Roeder RG. Human STAGA complex is a chromatin-acetylating transcription coactivator that interacts with pre-mRNA splicing and DNA damage-binding factors in vivo. Mol. Cell Biol. 2001;21:6782–6795. doi: 10.1128/MCB.21.20.6782-6795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, Suzuki E, Le Guezennec X, Stunnenberg HG, Krasnov A, et al. A TFTC/STAGA module mediates histone H2A and H2B deubiquitination, coactivates nuclear receptors, and counteracts heterochromatin silencing. Mol. Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 40.Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. EMBO J. 2010;29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data