Doublecortin and CaM Kinase-like-1 and Leucine-Rich-Repeat-Containing G-Protein-Coupled Receptor Mark Quiescent and Cycling Intestinal Stem Cells, Respectively (original) (raw)
. Author manuscript; available in PMC: 2011 Mar 7.
Published in final edited form as: Stem Cells. 2009 Oct;27(10):2571–2579. doi: 10.1002/stem.193
Abstract
It is thought that small intestinal epithelia (IE) undergo continuous self-renewal primarily due to their population of undifferentiated stem cells. These stem cells give rise to transit amplifying (daughter/progenitor) cells, which can differentiate into all mature cell types required for normal gut function. Identification of stem cells in IE is paramount to fully understanding this renewal process. One major obstacle in gastrointestinal stem cell biology has been the lack of definitive markers that identify small intestinal stem cells (ISCs). Here we demonstrate that the novel putative ISC marker doublecortin and CaM kinase-like-1 (DCAMKL-1) is predominantly expressed in quiescent cells in the lower two-thirds of intestinal crypt epithelium and in occasional crypt-based columnar cells (CBCs). In contrast, the novel putative stem cell marker leucine-rich-repeat-containing G-protein-coupled receptor (LGR5) is observed in rapidly cycling CBCs and in occasional crypt epithelial cells. Furthermore, functionally quiescent DCAMKL-1+ crypt epithelial cells retain bromo-deoxyuridine in a modified label retention assay. Moreover, we demonstrate that DCAMKL-1 is a cell surface expressing protein; DCAMKL-1+ cells, isolated from the adult mouse small intestine by fluorescence activated cell sorting, self-renew and ultimately form spheroids in suspension culture. These spheroids formed glandular epithelial structures in the flanks of athymic nude mice, which expressed multiple markers of gut epithelial lineage. Thus, DCAMKL-1 is a marker of quiescent ISCs and can be distinguished from the cycling stem/progenitors (LGR51). Moreover, DCAMKL-1 can be used to isolate normal small intestinal stem cells and represents a novel research tool for regenerative medicine and cancer therapy.
Keywords: Stem cell marker, Doublecortin and CaM kinase-like-1, Leucine-rich-repeat-containing G-protein-coupled receptor, GPR49, Label retention, Intestine, DCLK1
Introduction
The adult small intestinal epithelium is continuously and rapidly replaced by cell replication within the crypts of Lieberkühn and subsequent migration of their progeny onto the villus epithelium in the small intestine, or onto the surface epithelium in the colon [1]. Small intestinal epithelial cells are ultimately derived from multipotent stem cell(s) located near the base of each small intestinal crypt [2-5]. In the adult mouse small intestine, crypt stem cells divide to produce a daughter stem cell (self-renewal) as well as a more rapidly replicating transit amplifying (TA) cell. TA cells divide in the crypt proliferative zone, and their progeny ultimately differentiate into the mature small intestinal epithelial cell types [2, 6, 7]. Knowledge of the biological characteristics of intestinal stem cells (ISCs) has been largely acquired by inference from experiments using chimeric and transgenic mice [1, 4, 8]. Bjerknes and Cheng [9] originally proposed the existence of a stem cell-permissive microenvironment near the crypt base at positions 1-4 interspersed between Paneth cells. These cells, termed crypt base columnar cells (CBCs), were proposed as ISCs [10] and were found to give rise to mutant clones containing multiple cell types [11].
Traditionally, it was believed that adult stem cells in mammals existed either in a prolonged quiescent state or were extremely slow cycling [12]. On the basis of this feature, long-term label retention assays were developed to assist in the localization of putative stem cells [13, 14]. Using this technique, Potten et al. [15] localized label-retaining cells or putative ISCs to a position +4 from the crypt base, directly above the Paneth cell zone [16]. However, the +4 position is an average location and may vary depending on the crypt being analyzed. It is important to note that not all +4 cells are putative stem cells.
Recent work presented by Barker et al. [17] has identified a single marker, LGR5/GPR49 gene, a leucine-rich orphan G-protein-coupled receptor, that specifically labels stem cells in the mouse small intestine as well as other adult tissues. Furthermore, using mice generated from a LGR5-EGFP-IRES-Cre-ERT2 X RosaLacZ cross, they demonstrated that LGR5+ CBCs are multipotent for all mature intestinal epithelial cell types, undergo self-renewal, persist for at least 60 days based on LacZ expression, and are resistant to irradiation [17]. Furthermore, LGR5 marked ISCs that were rapidly cycling (dividing every 24 hours) under homeostatic conditions [17].
We have recently reported that doublecortin and Ca2+/calmodulin-dependent kinase-like-1 (DCAMKL-1), a microtubule-associated kinase expressed in post-mitotic neurons [18], is a novel putative ISC marker [19-22]. DCAMKL-1 was identified by the Jeff Gordon's group as a Gene Ontogeny-enriched transcript expressed in comparison with gastric epithelial progenitor and whole stomach libraries [23] and more recently in gastric stem cells [24]. Utilizing immunohistochemical analysis, we demonstrated cell-specific small intestinal DCAMKL-1 expression patterns in adult wild-type (WT) and in Apc Min/+ mice to visualize crypt epithelial stem cells at baseline and in response to radiation injury [19]. Immunoreactive DCAMKL-1 cells were found at or near position +4, at a frequency of one cell per five crypts. We also observed DCAMKL-1+ CBCs, albeit much less frequently.
In this report, we investigate the cell specific expression patterns of DCAMKL-1 and leucine-rich-repeat-containing G-protein-coupled receptor (LGR5) in the small intestinal epithelial cells in uninjured adult mice. DCAMKL-1 and LGR5 mark distinctly different cells. Moreover, DCAMKL-1 did not colocalize with enteroendocrine markers such as chromogranin A (ChrA), somatostatin, secretin, or other putative small intestinal stem cell markers such as phosphorylated PTEN (pPTEN) or phosphorylated AKT (pAKT). Furthermore, using a combination of a modified label retention assay (mLRA) and immunohistochemical analysis, we determined that DCAMKL-1 is expressed in quiescent label retaining cells within the intestinal crypt. LGR5 identifies proliferative CBCs and TA cells in the gut as evidenced by co-labeling with proliferating cell nuclear antigen (PCNA). Additionally, we demonstrate early glandular epithelial structures in nude mice isografts following fluorescence-activated cell sorting (FACS) of normal mouse small intestinal epithelial cells using DCAMKL-1. Thus, we propose that the original hypothesis of a +4 ISC should not yet be abandoned and contend that the DCAMKL-1 expressing cell represents a quiescent ISC.
Materials and Methods
Tissue Preparation and Immunohistochemistry
Heat-induced epitope retrieval was performed on formalin-fixed paraffin-embedded sections utilizing a pressurized decloaking chamber (Biocare Medical, Concord, CA, http://www.biocare.net) in citrate buffer (pH 6.0) at 99°C for 18 minutes. (a) Bright-field: Slides were incubated in 3% hydrogen peroxide, then normal serum and Bovine Serum Albumin (BSA) at room temperature for 20 minutes. After incubation with primary antibody [DCAMKL-1 C-terminal 1:100, LGR5 1:300, Msi-1 1:250, Chromogranin A 1:500, Secretin 1:100 (Abcam, Cambridge, MA, http://www.abcam.com), 5-bromo-2′-deoxyuridine (BrdUrd) 1:500, Math1 1:250 (Millipore, Billerica, MA, http://www.millipore.com), PCNA 1:150, L-FABP 1:300, Cytokeratin 14 1:100, phosphorylated-PTEN 1:100, phosphorylated-AKT 1:100, Somatostatin 1:100 (Santa Cruz Biotechnology, Santa Cruz, CA, http://www.scbt.com)], the slides were then incubated in peroxidase-conjugated EnVision+ polymer detection kit (DAKO, Glostrup, Denmark, http://www.dako.com). Slides were developed with diaminobenzidine (Sigma-Aldrich, St. Louis, MO, http://www.sigmaaldrich.com). (b) Fluorescence: Slides were first incubated in Image-iT FX signal enhancer (Invitrogen, Carlsbad, CA, http://www.invitrogen.com), followed by normal serum and Bovine Serum Albumin (BSA) at room temperature for 20 minutes. After incubation with primary antibody, slides were incubated in appropriate Alexa Fluor conjugated secondary [488 (green) and 568 (red)].
Microscopic Examination
Slides were examined utilizing the Nikon 80i microscope and DXM1200C camera for bright-field. Fluorescent images were taken with PlanFluoro objectives, utilizing CoolSnap ES2 camera (Nikon, Melville, NY, www.nikonusa.com). Images were captured utilizing NIS-Elements software (Nikon). Confocal imaging was performed using the Leica TCS NT microscope.
DCAMKL-1 Cellular Distribution
Cellular distribution of DCAMKL-1 on a positional basis was determined in adult C57BL/6 mice (n = 3). Longitudinal sections from the distal jejunum were prepared from each mouse, and the number of DCAMKL-1+ cells was determined by counting positive cells at the numbered positions (1-17), starting from the midpoint at the base of the crypt along the crypt–villus axis (total 500 crypts).
Modified Label Retention Assay
C57BL/6 mice (Jackson Labs, Bar Harbor, ME, http://www.jax.org) were subjected to 8 Gy whole-body gamma irradiation using a Nordion 137Cs γ-irradiator with a dose rate of 0.9 Gy per minute. Animals received 120 mg/kg intraperitoneal injections of BrdUrd in isotonic saline twice daily, beginning 24 and ending 84 hours after irradiation. This time period was chosen in order to maximize the potential of label incorporation during the crypt regeneration phase, following severe genotoxic injury. Animals were sacrificed at 7 and 10 days after the initial injury when restoration of crypt villus morphology was returning towards baseline. Co-immunostaining for BrdUrd and DCAMKL-1 was performed to identify label retaining stem cells. Additionally coimmunostaining for PCNA and DCAMKL-1 was performed to determine the proliferative status of the label retaining cells.
Stem Cell Isolation
On the basis of protocols developed in intestinal stem cell biology [25, 26], we isolated and propagated stem cells from fresh mouse small intestinal tissues. Intestines were opened longitudinally and cut into small strips, washed and incubated with 1 mM Dithiothreitol (DTT) (Sigma-Aldrich, St. Louis, MO) for 30 minutes at room temperature. Tissues were further incubated with 30 mmol/l EDTA (Sigma-Aldrich, St. Louis, MO) for 10 minutes at 37° C, lightly sonicated for 5 seconds, shaken vigorously in fresh Hank's Buffered Salt Solution (HBSS) (Cellgro, Manassas, VA, www.cellgro.com), and filtered through 400 _μ_m mesh (Spectrum Labs, Rancho Dominguez, CA, http://www.spectrapor.com) to separate the detached small intestinal crypt epithelial cells from the tissue. The filtrate was passed through 80 lm mesh (BD Biosciences, San Jose, CA, http://www.bdbiosciences.com) to retain the crypts and washed. The crypts were digested in trypsin at 37°C to create a single cell suspension.
FACS
The cells isolated from mouse intestine were incubated with 1:100 dilution of Alexa Fluor 568 (Invitrogen, Carlsbad, CA) conjugated DCAMKL-1 antibody for 30 minutes. The cells were washed twice with HBSS containing 10% serum and sorted using Influx-V cell sorter (Cytopeia, Seattle, WA, http://www.cytopeia.com). The cells collected were grown on Dulbecco's modified Eagle's medium containing epidermal growth factor (EGF) (25 ng/ml), fibroblast growth factor (20 ng/ml), and insulin (5 ng/ml) (Sigma-Aldrich, St. Louis, MO), on nonadherent/ultralow attachment plates (BD Biosciences, San Jose, CA).
Isotransplantation Assay
DCAMKL-1+ cells isolated from intestine were grown in suspension culture and formed spheroids by day 21. Mechanically dissociated spheroids (50-100 cells) were suspended in Matrigel and injected subcutaneously into the flanks of athymic nude mice (n = 3) (NCI, Fredrick, MD, http://web.ncifcrf.gov/) and monitored for the appearance of nodular growth.
Cell Surface Protein Isolation and Western Blot Analysis
SW480 colon cancer cells were grown and surface proteins were labeled with sulfo-NHS Biotin (Pierce Biotechnology Inc., Rockford, IL, http://www.piercenet.com). Cell lysates were prepared and the biotinylated proteins were separated from intracellular non-biotinylated proteins as per manufacturer's instructions (Pierce Biotechnology Inc., Rockford, IL). Protein concentration was determined by BCA protein assay kit (Pierce Biotechnology Inc., Rockford, IL). Forty lg of the protein was size separated in a 15% SDS polyacrylamide gel and transferred onto a nitro-cellulose membrane with a semidry transfer apparatus (Amersham-Pharmacia, Pittsburgh, PA, www.gelifesciences.com). The membrane was blocked in 5% nonfat dry milk for 1 hour and probed overnight with a rabbit anti-DCAMKL-1 antibody or with rabbit anti-EGFR antibody (Cell Signaling Technology, Danvers, MA, http://www.cellsignal.com). Subsequently, the membrane was incubated with anti-rabbit IgG horseradish peroxidase-conjugated antibodies (Amersham-Pharmacia, Pittsburgh, PA) for 1 hour at room temperature. The 82 kDa DCAMKL-1 and 175 kDa epithelial growth factor receptor (EGFR) proteins were detected using ECL Western Blotting detection reagents (Amersham-Pharmacia, Pittsburgh, PA).
Results
Small Intestinal DCAMKL-1 Expression Along the Crypt-Villus Axis
Out of 500 total crypts counted, we found 49% of DCAMKL-1 positive cells were located at position +4 (excluding the CBCs) (supporting information Fig. 1A, 1B). DCAMKL-1 was also expressed in rare CBCs (4% of total crypts counted). As previously reported, DCAMKL-1 cells were found in the villi [19]. However, we noted that DCAMKL-1 crypt with simultaneous villus expression was rare (<5% of total crypt villus units).
DCAMKL-1 Marks a Unique Small Intestinal Cell Type
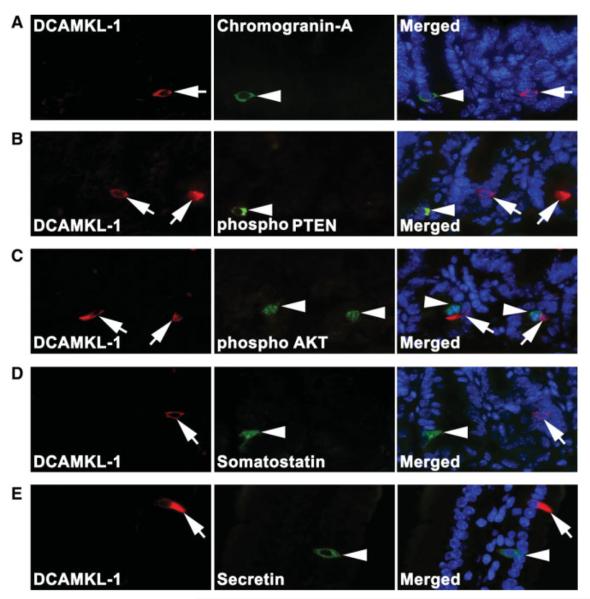
To determine whether DCAMKL-1 was co-expressed with other putative stem cell and enteroendocrine markers, we performed double-labeled immunofluorescence staining for DCAMKL-1 with ChrA, pPTEN, pAKT, somatostatin, and secretin. There was no colocalization observed for any of the markers tested (Fig. 1A–1E). These data suggest that DCAMKL-1 marks a unique cell within the mouse small intestine.
Figure 1.
Expression of DCAMKL-1 in the mouse small intestine. (A): Co-immunofluorescence staining for DCAMKL-1 (red, arrow, left panel) and Chromogranin A (ChrA) (green, arrow head, middle panel) in crypts. No colocalization was observed in the merged image (right panel). (B): DCAMKL-1 (red, arrow, left panel) and phosphorylated PTEN (green, arrow head, middle panel) in crypts. No colocalization was observed in the merged image (right panel). (C): DCAMKL-1 (red, arrow, left panel) and phosphorylated AKT (green, arrow head, middle panel) in crypts. No colocalization was observed in the merged image (right panel). (D): DCAMKL-1 (red, arrow, left panel) and somatostatin (green, arrow head, middle panel) in crypts. No colocalization was observed in the merged image (right panel). (E): DCAMKL-1 (red, arrow, left panel) and secretin (green, arrow head, middle panel) on villus. No colocalization was observed in the merged image (right panel). Abbreviations: DCAMKL-1, doublecortin and CaM kinase-like-1. *Nuclei in all merged images are stained blue with Hoechst 33342 DNA dye.
Small Intestinal LGR5 and DCAMKL-1 Mark Distinctly Different Cells
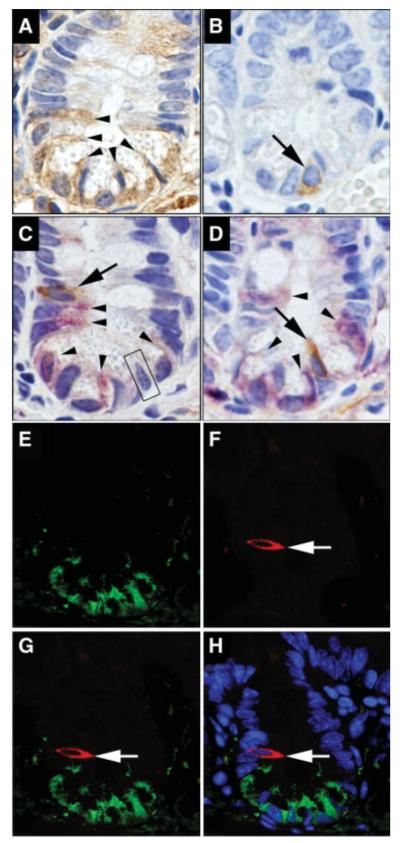
In the small intestine, LGR5 expression was observed in crypt epithelial and in CBCs as predicted (Fig. 2A). LGR5+ cells were also scattered throughout the mesenchyme and villus epithelial cells (supporting information Fig. 2). This was consistent with the LacZ expression patterns described in the original Clevers group LGR5 stem cell report [17], expression of LGR5 at the base of the crypt in normal human colon and small intestine [27], and the previously reported immunostaining for LGR5/GPR49 in colon and cancer tissues [28]. We have previously demonstrated DCAMKL-1 expression at position +4 and in rare CBCs [19] (Fig. 2B). On occasion, LGR5 expressing cells were immediately adjacent to DCAMKL-1+ cells (Fig. 2C, 2D). However, no DCAMKL-1 colocalization with LGR5 was observed in intestinal crypts (Fig. 2E–2H).
Figure 2.
Leucine-rich-repeat-containing G-protein-coupled receptor (LGR5) and doublecortin and CaM kinase-like-1 (DCAMKL-1) in the mouse small intestine. (A): Brown indicates LGR5+ cells (arrow-heads). (B): Brown indicates DCAMKL-1+ cell (arrow). (C and D): Co-immunostaining for LGR5 (purple, arrowhead) and DCAMKL-1 (brown, arrow). No colocalization of LGR5 and DCAMKL-1 was observed in the putative stem cell zone (C) or crypt-based columnar cells (CBCs) (D). Black box in (C) demonstrates a cell negative for both LGR5 and DCAMKL-1. (E–H): Co-immunofluorescence staining for LGR5 (green) (E) and DCAMKL-1 (red, arrow) (F). No colocalization of LGR5 and DCAMKL-1 was observed in merged images (G) and (H). *Nuclei in merged image (H) are stained blue with Hoechst 33342 DNA dye.
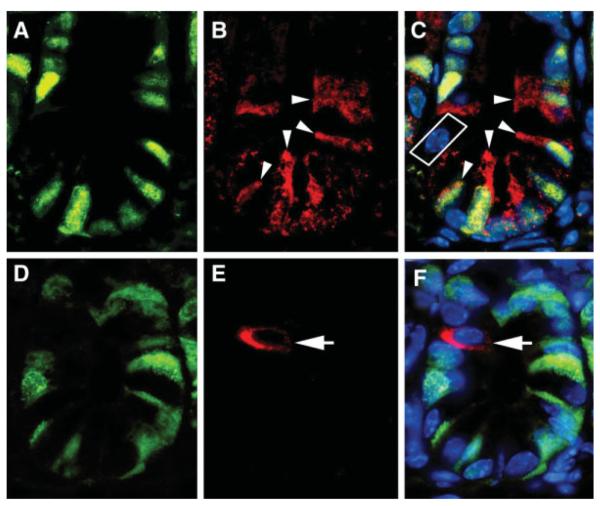
Proliferation Status of LGR5 and DCAMKL-1 Expressing Cells
PCNA staining was performed to assess the proliferative status of LGR5 and DCAMKL-1 expressing cells in the intestine. LGR5 expressing cells were invariably PCNA+ (actively cycling) (Fig. 3A–3C). We also occasionally noted cells at position +4 that did not express either PCNA or LGR5 (Fig. 3C, white box). PCNA− cells, particularly at position +4, were distinctly DCAMKL-1+ (Fig. 3D–3F), suggesting functional quiescence at baseline. Thus, DCAMKL-1 and LGR5 identify cell populations with differing proliferation status at baseline. These findings lend support to the longstanding +4 hypothesis, which suggests that a functionally quiescent or very slowly cycling cell is primarily anchored in the stem cell niche [15, 16, 29]. We contend that this quiescent cell is marked by DCAMKL-1.
Figure 3.
Leucine-rich-repeat-containing G-protein-coupled receptor (LGR5) and doublecortin and CaM kinase-like-1 (DCAMKL-1) mark proliferative and non-proliferative cells, respectively, in the mouse small intestine. Co-immunofluorescence staining for proliferating cell nuclear antigen (PCNA) (green) (A) and LGR5 (red, arrowheads) (B). PCNA+ LGR5+ cells are indicated with arrow-heads in the merged image (C). PCNA (green) (D) and DCAMKL-1 (red, arrow) (E). A PCNA− DCAMKL-1+ cell is indicated by the arrow in the merged image (F). *Nuclei in all merged images (C and F) are stained blue with Hoechst 33342 DNA dye.
DCAMKL-1 Label-Retaining Cells Are Functionally Quiescent
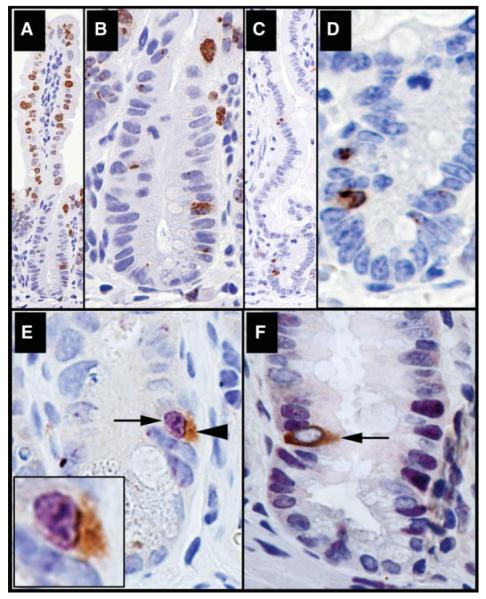
Although the “anchored stem cell” is often found at position +4, we suspect that under certain conditions this cell can exit the niche [30]. Indeed, we occasionally observe DCAMKL-1 staining outside of the crypt, particularly in APC min/+ mice [19]. We employed a modification of the traditional label retention assay (mLRA) [13-16] by utilizing 8 Gy as the inciting dose in adult WT mice. DCAMKL-1 expression is lost in regenerative crypts by 84 hours after lethal dose ionizing radiation (IR) (>8 Gy) but reappears 7 and 10 days following IR in regenerated intestine tissues [19]. This suggests that, by 7-10 days after IR, the normal crypt villus units and the niche related microenvironmental signals required for DCAMKL-1 expression are restored. We previously demonstrated that 24 hours after IR is a critical time point when DCAMKL-1 expressing cells undergo both mitosis and apoptosis [19]. Thus, we decided to pulse label (BrdUrd) throughout the entire 24-84-hour crypt regeneration cycle. Animals were allowed to recover and were sacrificed at 7 and 10 days [31]. This period of regeneration allows for BrdUrd incorporation into dividing stem cells that would otherwise be problematic under quiescent basal conditions. At 7 days post IR, residual BrdUrd labeled cells were detected in the upper crypt and throughout the villi (Fig. 4A, 4B). However at 10 days, BrdUrd labeling had essentially disappeared and only rare cells near the crypt base retained significant label (Fig. 4C, 4D).
Figure 4.
Doublecortin and CaM kinase-like-1 (DCAMKL-1) identifies the quiescent anchored stem cell. Following mLRA, mouse intestines (distal jejunum) were immunostained for BrdUrd (brown) at day 7 (A) magnified in (B) and at day 10 (C) magnified in (D). (E–F): Mouse intestines 10 days post 8 Gy IR were co-immunostained for DCAMKL-1 (brown) and BrdUrd (purple) or PCNA (purple). (E): Colocalization of BrdUrd (purple, arrow) and DCAMKL-1 (brown, arrowhead) in a single cell (inset is a magnified image). (F): Arrow indicates a PCNA− (quiescent) and DCAMKL-1+ cell.
We next sought to determine whether the cells retaining BrdUrd label following the mLRA also expressed DCAMKL-1. At 10 days post IR, we performed double-label immunohistochemistry, and distinct co-expression of BrdUrd and DCAMKL-1 at position +4 was observed (Fig. 4E). Whereas-this cell retains label, it does not necessarily mean that it was actively proliferating. We sought to answer this question by examining DCAMKL-1 expressing cells following the mLRA for the presence of PCNA activity. Interestingly, there was no PCNA expression in the nucleus of the DCAMKL-1+ cell, yet clear PCNA staining could be identified in many adjacent cells (Fig. 4F). Thus, the label-retaining DCAMKL-1 expressing “stem cells” are again quiescent at 7 and 10 days after IR.
DCAMKL-1 Is Expressed on the Cell Surface and Can Be Used to Isolate Stem Cells
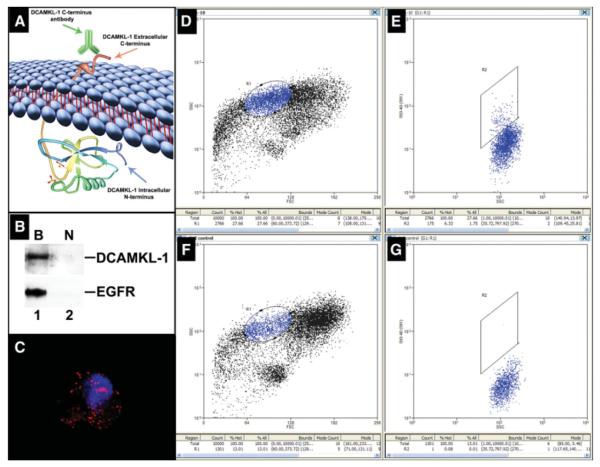
To further investigate the potential “stemness” of DCAMKL-1 expressing cells, we employed FACS using the modified protocol of Dekaney et al. [25]. Although originally considered to be a cytoplasmic protein [23], analysis of the DCAMKL-1 protein using TMPred program (http://www.ch.embnet.org/software/TMPRED_form.html) suggested that amino acids 534-560 constitute a transmembrane domain and that amino acids 561-729 are extracellular. Furthermore, it has been reported that DCAMKL-1 is expressed in the brains of adult humans and mice, with two transmembrane domains (amino acids 534-559 and 568-585), suggesting that it is a cell surface expressing protein with intra and extracellular domains [32, 33] (Fig. 5A). To confirm the cell surface expression of DCAMKL-1, we used the Pierce Cell Surface Protein Isolation Kit (Pierce, Rockford, IL) to isolate total cell surface expressing proteins from SW480 cells (supporting information Fig. 3). Western blot analyses demonstrated the presence of DCAMKL-1 in the avidin-bound fraction but not in the unbound fraction (Fig. 5B). This data demonstrates that DCAMKL-1 protein is indeed present on the cell surface. Epithelial growth factor receptor (EGFR), a cell surface expressing protein in the bound fraction, was used as a positive control.
Figure 5.
Isolation of small intestinal stem cells using DCAMKL-1 based fluorescence activated cell sorting (FACS). (A): Schematic diagram depicting the predicted cell surface expression and extracellular C-terminal domain of DCAMKL-1. (B): Western blot analyses demonstrating cell surface expression of DCAMKL-1 following biotinylation (Pierce Cell Surface Protein Isolation Kit). Biotinylated cell surface protein extract from intact cells* demonstrated the presence of DCAMKL-1 (Lane B), but not in the unbound non-biotinylated intracellular protein extract fraction (Lane N). As a positive control, EGFR, a known cell surface expressing protein, was detected only in the bound fraction. (C): A representative Alexa Fluor 568 conjugated DCAMKL-1+ cell following FACS (red); nucleus is stained blue with Hoechst 33342 DNA dye post-sorting. (D): FACS plot of side scatter (chosen based on previous sorting experiments) of cells stained with Alexa Fluor 568 conjugated DCAMKL-1 antibody. Gate R1 indicates localization of the DCAMKL-1+ fluorescing cell population. (E): These cells were further gated through R2 based on fluorescence intensity. (F): FACS plot of side scatter of unstained control cells. (G): No cells were detected within gate R2. Abbreviations: DCAMKL-1, doublecortin and CaM kinase-like-1; EGFR, epithelial growth factor receptor. *Please refer to supporting information Figure 3.
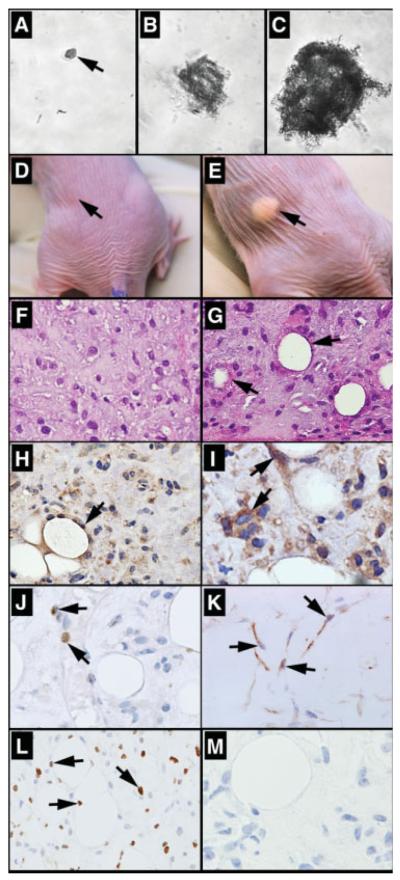
We conjugated anti-DCAMKL-1 antibody, which targets the extracellular C-terminal epitope [18, 32, 33] with Alexa Fluor 568 to label intact functional epithelial stem cells from the normal mouse small intestine following the mechanical disassociation (supporting information Fig. 4). These cells were then subjected to FACS analysis. For sorting, gate R1 was assigned based on our previous experiments [34], where the DCAMKL-1+ fluorescing cell population was found to be aggregated. These cells were further gated through R2 based on fluorescence intensity. We isolated approximately 1.75% of the total cells sorted using this method (Fig. 5D–5G). Sorted cells were examined by fluorescence microscopy to confirm the presence of DCAMKL-1 (Fig. 5C). Furthermore, we have stained the DCAMKL-1 +ve and −ve cell populations for LGR5 to demonstrate the purity of the isolation technique (supporting information Fig. 5). The cells were then grown in suspension culture with growth factor supplemented media using the method of Dontu et al. [35]. After 3 weeks, approximately 30% of single DCAMKL-1 +ve sorted cells formed spheroids in suspension culture (Fig. 6A–C), whereas single DCAMKL-1 −ve sorted cells did not demonstrate growth (data not shown). The spheroids containing 50-100 cells were mechanically dissociated and subsequently injected into contralateral flanks of nude mice. After 3 weeks, nodular structures were observed (Fig. 6D, 6E) in 11 of 12 spheroid injection sites (data not shown). Two weeks later, animals were sacrificed and the nodules were excised and subjected to immunohistochemical and histological analysis. In the control (Matrigel injected) nodules, we observed an inflammatory response including the presence of macrophages, but with no evidence of epithelial cells (Fig. 6F). In the spheroid injected nodules, however, there were single cells with oval nuclei and large nucleoli that lined up around central spaces and appeared to represent poorly formed glands (Fig. 6G). Cytokeratin 14 immunoreactivity demonstrates that these cells were of glandular epithelial origin [36, 37] (Fig. 6H). To determine whether they expressed stem and/or TA (progenitor) cell markers, we stained for the epithelial stem/progenitor cell marker Msi-1 [38, 39]. Significant Msi-1 immunoreactivity was observed in these epithelial structures, providing additional support for the epithelial and perhaps stem/progenitor cell origin of these cells (Fig. 6I). Moreover, several cells expressed Math1, indicating an early intestinal epithelial secretory lineage commitment (goblet, enteroendocrine, and Paneth cells) [40, 41] and _L_-type fatty acid binding protein (L-FABP) (marker of enterocyte lineage) [42] (Fig. 6J, 6K; supporting information Fig. 6). These studies demonstrate that DCAMKL-1 can be used as a cell surface marker to isolate stem cells from the normal mouse small intestine and investigate their lineage determination and viability in vivo.
Figure 6.
Characterization of isolated doublecortin and CaM kinase-like-1 (DCAMKL-1) cells. (A): A single DCAMKL-1 sorted cell in suspension culture at day 0. (B--C): Spheroid growth at day 14 and day 21 (containing 50-100 cells). Isotransplantation assays: (D): Matrigel alone injected control mouse, (E): spheroid injected mouse demonstrating nodular growth on the flank (arrow), H&E staining of excised nodules from (F) control mouse and (G) spheroid injected mouse (arrow indicates glandular formation). Spheroid injected nodule stained for (H): cytokeratin-14, (I): Msi-1, (J): Math1, (K): _L_-type fatty acid binding protein, (L): proliferating cell nuclear antigen, and (M): DCAMKL-1, with representative cells indicated by arrows.
Discussion
In this report, we have demonstrated that the novel stem/progenitor markers DCAMKL-1 and LGR5 identify small intestinal stem and progenitor cells, respectively. This distinction is primarily based on the proliferative status of the cells, because no in vivo genetic lineage tracing studies have yet been performed for DCAMKL-1. The major distinguishing feature presented here is that DCAMKL-1 identifies a slowly cycling or basally quiescent cell; whereas LGR5 identifies a more proliferative cell. It is important to note that these classifications do not necessarily address the question of multipotency, as it is clear that an early small intestinal progenitor cell is capable of repopulating the crypt with each of the four cell types expressed in the intestine [43]. Our data presented here suggest that there may be two different populations of stem cells in the gut. One population is at or near the traditional +4 position, is restricted primarily to the niche, and may have a functional role in gut homeostasis and injury response. The second population (CBCs) is interspersed between the Paneth cells and may be responsible for Paneth cell repopulation in response to bacterial mediated injury.
Although this hypothesis is speculative, we contend that it is reasonable given the recent report by Sangiorgi and Capecchi [44] identifying Bmi1 as yet another novel ISC marker. In that report using a knock-in transgenic mouse model, they presented data demonstrating that Bmi1 labels ISCs predominantly at the +4 position of the crypt. The authors suggest that Bmi1 and LGR5 label different states of ISCs. Bmi1 labels the more quiescent ISCs, whereas LGR5 labels ISCs more prone to enter proliferation [44]. Our hypothesis is feasible and is supported by reports that the putative stem/progenitor cell markers DCAMKL-1, LGR5, and Msi-1 [20-22] are all expressed in CBCs [17, 19, 39]. Occasionally, we found the presence of DCAMKL-1 and pAKT in different, but adjacent cells. We predict pAKT may well mark active stem or transit amplifying cells, but this is not clearly evidenced by the data presented in this manuscript.
One exciting outcome of this study is the use of FACS for isolation of cells expressing DCAMKL-1. Although originally considered to be a cytoplasmic protein [23], it has been reported that DCAMKL-1 is expressed in adult brain with two transmembrane domains (amino acids 534-559 and 568-585), making it a cell surface expressing protein with intraand extracellular domains [32, 33]. In this report, cell surface isolation experiments confirm that DCAMKL-1 is indeed expressed on the cell surface. Accordingly, we conjugated anti-DCAMKL-1 antibody with Alexa Fluor 568 for use in cell sorting experiments. Here we have demonstrated that putative stem cells isolated from the normal mouse intestine by FACS form spheroids in suspension culture and, upon injection into the flanks of nude mice, form early glandular epithelial structures. Moreover, these cells expressed Msi-1 [38, 39], Cytokeratin 14 [36, 37], Math1 [40, 41], and L-FABP [42], markers of intestinal epithelial lineage.
The data presented in this report taken together provide strong evidence that LGR5+ and DCAMKL-1+ cells are distinctly different and may even have different functions. However, we predict that both of these cell types are likely to have full multipotency and have the potential to regenerate a fully functional small intestinal tract following injury. Our data supports the contention that, for the first time, these critical cell types can be identified in situ based on the discovery of these two novel markers. In supporting information Figure 7, we propose a model for the specific expression patterns of the putative markers DCAMKL-1, Msi-1, and LGR5 in the intestinal crypts.
Conclusion
The importance of reliable markers for identifying both stem and progenitor cells goes well beyond their use as a tool for sorting. The unique expression of DCAMKL-1 in quiescent ISCs raises the question of whether functional quiescence is a requirement for gut homeostasis and what factors regulate these processes. Identification of DCAMKL-1 and LGR5 expressing cells will enable us, for the first time, to directly examine the gene expression profiles and molecular signatures of stem and progenitor cells, respectively. Furthermore, the effects of geno/cytotoxic injury, genetic manipulation, and pharmacologic agents on stem cell fate can be studied in vivo. These findings should accelerate progress towards the development of regenerative strategies to induce or replace normal stem cells following severe injury and perhaps identify and eliminate tumor stem cells in cancer.
Supplementary Material
Suplementary Fig 4
Suplementary Fig 5
Suplementary Fig 6
Suplementary Fig Legends
Suplementary Fig 1
Suplementary Fig 2
Suplementary Fig 3
Acknowledgments
This work was supported by National Institute of Health Grants DK-065887, DK-002822, and Veterans Administration Merit award to C.W.H. The authors wish to thank Dr. Michael Bronze, University of Oklahoma Health Sciences Center for his support; Oklahoma University Advanced Immunohistochemistry & Morphology Core and Jim Henthorn of The Flow and Image Cytometry Core Laboratory; and Dr. Susan Henning, University of North Carolina at Chapel Hill, for her helpful comments on this manuscript.
Footnotes
Author contributions: R.M., S.M.S.: conception and design, collection and assembly of data, data analysis and integration, manuscript writing, and final approval of the manuscript; T.E.R.: manuscript writing and final approval of the manuscript; S.A.L., R.R., J.W.H.: data analysis and integration and final approval of the manuscript; S.A.: conception and design, data analysis, and integration and final approval of the manuscript; C.W.H.: conception and design, financial support, collection and assembly of data, data analysis and integration, manuscript writing, and final approval of the manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
References
- 1.Gordon JI, Hermiston ML. Differentiation and self-renewal in the mouse gastrointestinal epithelium. Curr Opin Cell Biol. 1994;6:795–803. doi: 10.1016/0955-0674(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 2.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian Theory of the origin of the four epithelial cell types. Am J Anatomy. 1974;141:537–561. doi: 10.1002/aja.1001410407. [DOI] [PubMed] [Google Scholar]
- 3.Cohn SM, Simon TC, Roth KA, et al. Use of transgenic mice to map cis-acting elements in the intestinal fatty acid binding protein gene (Fabpi) that control its cell lineage-specific and regional patterns of expression along the duodenal-colonic and crypt-villus axes of the gut epithelium. J Cell Biol. 1992;119:27–44. doi: 10.1083/jcb.119.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmidt GH, Wilkinson MM, Ponder BA. Cell migration pathway in the intestinal epithelium: An in situ marker system using mouse aggregation chimeras. Cell. 1985;40:425–429. doi: 10.1016/0092-8674(85)90156-4. [DOI] [PubMed] [Google Scholar]
- 5.Winton DJ, Ponder BA. Stem-cell organization in mouse small intestine. Proc Biol Sci. 1990;241:13–18. doi: 10.1098/rspb.1990.0059. [DOI] [PubMed] [Google Scholar]
- 6.Potten CS, Loeffler M. A comprehensive model of the crypts of the small intestine of the mouse provides insight into the mechanisms of cell migration and the proliferation hierarchy. J Theor Biol. 1987;127:381–391. doi: 10.1016/s0022-5193(87)80136-4. [DOI] [PubMed] [Google Scholar]
- 7.Potten CS, Loeffler M. Stem cells: Attributes, cycles, spirals, pitfalls and uncertainties. Lessons for and from the Crypt. Development. 1990;110:1001–1020. doi: 10.1242/dev.110.4.1001. [DOI] [PubMed] [Google Scholar]
- 8.Hauft SM, Kim SH, Schmidt GH, et al. Expression of SV-40 T antigen in the small intestinal epithelium of transgenic mice results in proliferative changes in the crypt and reentry of villus-associated enterocytes into the cell cycle but has no apparent effect on cellular differentiation programs and does not cause neoplastic transformation. J Cell Biol. 1992;117:825–839. doi: 10.1083/jcb.117.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjerknes M, Cheng H. The stem-cell zone of the small intestinal epithelium. IV. Effects of resecting 30% of the small intestine. Am J Anatomy. 1981;160:93–103. doi: 10.1002/aja.1001600108. [DOI] [PubMed] [Google Scholar]
- 10.Cheng H, Leblond CP. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. I. Columnar cell. Am J Anatomy. 1974;141:461–479. doi: 10.1002/aja.1001410403. [DOI] [PubMed] [Google Scholar]
- 11.Bjerknes M, Cheng H. Clonal analysis of mouse intestinal epithelial progenitors. Gastroenterology. 1999;116:7–14. doi: 10.1016/s0016-5085(99)70222-2. [DOI] [PubMed] [Google Scholar]
- 12.Cheshier SH, Morrison SJ, Liao X, et al. In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc Natl Acad Sci USA. 1999;96:3120–3125. doi: 10.1073/pnas.96.6.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: Implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 14.Zhang J, Niu C, Ye L, et al. Identification of the haematopoietic stem cell niche and control of the niche size. Nature. 2003;425:836–841. doi: 10.1038/nature02041. [DOI] [PubMed] [Google Scholar]
- 15.Potten CS, Owen G, Booth D. Intestinal stem cells protect their genome by selective segregation of template DNA strands. J Cell Sci. 2002;115:2381–2388. doi: 10.1242/jcs.115.11.2381. [DOI] [PubMed] [Google Scholar]
- 16.Marshman E, Booth C, Potten CS. The intestinal epithelial stem cell. Bioessays. 2002;24:91–98. doi: 10.1002/bies.10028. [DOI] [PubMed] [Google Scholar]
- 17.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 18.Lin PT, Gleeson JG, Corbo JC, et al. DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J Neurosci. 2000;20:9152–9161. doi: 10.1523/JNEUROSCI.20-24-09152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.May R, Riehl TE, Hunt C, et al. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 2008;26:630–637. doi: 10.1634/stemcells.2007-0621. [DOI] [PubMed] [Google Scholar]
- 20.Quante M, Wang TC. Inflammation and stem cells in gastrointestinal carcinogenesis. Physiology. 2008;23:350–359. doi: 10.1152/physiol.00031.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samuel S, Walsh R, Webb J, et al. Characterization of putative stem cells in isolated human colonic crypt epithelial cells and their interactions with myofibroblasts. Am J Physiol Cell Physiol. 2009;296:C296–305. doi: 10.1152/ajpcell.00383.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Humphries A, Wright NA. Colonic crypt organization and tumorigenesis. Nature Rev. 2008;8:415–424. doi: 10.1038/nrc2392. [DOI] [PubMed] [Google Scholar]
- 23.Giannakis M, Stappenbeck TS, Mills JC, et al. Molecular properties of adult mouse gastric and intestinal epithelial progenitors in their niches. J Biol Chem. 2006;281:11292–11300. doi: 10.1074/jbc.M512118200. [DOI] [PubMed] [Google Scholar]
- 24.Giannakis M, Chen SL, Karam SM, et al. Helicobacter pylori evolution during progression from chronic atrophic gastritis to gastric cancer and its impact on gastric stem cells. Proc Natl Acad Sci USA. 2008;105:4358–4363. doi: 10.1073/pnas.0800668105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dekaney CM, Rodriguez JM, Graul MC, et al. Isolation and characterization of a putative intestinal stem cell fraction from mouse jejunum. Gastroenterology. 2005;129:1567–1580. doi: 10.1053/j.gastro.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 26.Grossmann J, Walther K, Artinger M, et al. Progress on isolation and short-term ex-vivo culture of highly purified non-apoptotic human intestinal epithelial cells (IEC) Eur J Cell Biol. 2003;82:262–270. doi: 10.1078/0171-9335-00312. [DOI] [PubMed] [Google Scholar]
- 27.Becker L, Huang Q, Mashimo H. Immunostaining of Lgr5, an intestinal stem cell marker, in normal and premalignant human gastrointestinal tissue. ScientificWorldJournal. 2008;8:1168–1176. doi: 10.1100/tsw.2008.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClanahan T, Koseoglu S, Smith K, et al. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther. 2006;5:419–426. doi: 10.4161/cbt.5.4.2521. [DOI] [PubMed] [Google Scholar]
- 29.Potten CS, Booth C, Pritchard DM. The intestinal epithelial stem cell: The mucosal governor. Int J Exp Pathol. 1997;78:219–243. doi: 10.1046/j.1365-2613.1997.280362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frye M, Gardner C, Li ER, et al. Evidence that Myc activation depletes the epidermal stem cell compartment by modulating adhesive interactions with the local microenvironment. Development. 2003;130:2793–2808. doi: 10.1242/dev.00462. [DOI] [PubMed] [Google Scholar]
- 31.Potten CS, Taylor Y, Hendry JH. The doubling time of regenerating clonogenic cells in the crypts of the irradiated mouse small intestine. Int J Radiat Biol. 1988;54:1041–1051. doi: 10.1080/09553008814552421. [DOI] [PubMed] [Google Scholar]
- 32.Sossey-Alaoui K, Srivastava AK. DCAMKL1, a brain-specific transmembrane protein on 13q12.3 that is similar to doublecortin (DCX) Genomics. 1999;56:121–126. doi: 10.1006/geno.1998.5718. [DOI] [PubMed] [Google Scholar]
- 33.Kim MH, Cierpicki T, Derewenda U, et al. The DCX-domain tandems of doublecortin and doublecortin-like kinase. Nature Struct Biol. 2003;10:324–333. doi: 10.1038/nsb918. [DOI] [PubMed] [Google Scholar]
- 34.Sureban SM, May R, Ramalingam S, et al. Selective blockade of DCAMKL-1 results in tumor growth arrest by a Let-7a microRNA-dependent mechanism. Gastroenterology. 2009;137:649–659. doi: 10.1053/j.gastro.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dontu G. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moll R, Divo M, Langbein L. The human keratins: Biology and pathology. Histochem Cell Biol. 2008;129:705–733. doi: 10.1007/s00418-008-0435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Purkis PE, Steel JB, Mackenzie IC, et al. Antibody markers of basal cells in complex epithelia. J Cell Sci. 1990;97(Pt 1):39–50. doi: 10.1242/jcs.97.1.39. [DOI] [PubMed] [Google Scholar]
- 38.Sureban SM, May R, George RJ, et al. Knockdown of RNA Binding Protein Musashi-1 Leads to Tumor Regression In Vivo. Gastroenterology. 2008;134:1448–1458. doi: 10.1053/j.gastro.2008.02.057. e1442. [DOI] [PubMed] [Google Scholar]
- 39.Potten CS. Identification of a putative intestinal stem cell and early lineage marker; musashi-1. Differentiation. 2003;71:28–41. doi: 10.1046/j.1432-0436.2003.700603.x. [DOI] [PubMed] [Google Scholar]
- 40.Yang Q, Bermingham NA, Finegold MJ, et al. Requirement of Math1 for secretory cell lineage commitment in the mouse intestine. Science. 2001;294:2155–2158. doi: 10.1126/science.1065718. [DOI] [PubMed] [Google Scholar]
- 41.Shroyer NF, Wallis D, Venken KJ, et al. Gfi1 functions downstream of Math1 to control intestinal secretory cell subtype allocation and differentiation. Genes Dev. 2005;19:2412–2417. doi: 10.1101/gad.1353905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubin DC, Swietlicki E, Roth KA, et al. Use of fetal intestinal isografts from normal and transgenic mice to study the programming of positional information along the duodenal-to-colonic axis. J Biol Chem. 1992;267:15122–15133. [PubMed] [Google Scholar]
- 43.Crossman MW, Hauft SM, Gordon JI. The mouse ileal lipid-binding protein gene: a model for studying axial patterning during gut morphogenesis. J Cell Biol. 1994;126:1547–1564. doi: 10.1083/jcb.126.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Suplementary Fig 4
Suplementary Fig 5
Suplementary Fig 6
Suplementary Fig Legends
Suplementary Fig 1
Suplementary Fig 2
Suplementary Fig 3