Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives (original) (raw)
Abstract
Human induced pluripotent stem (hiPS) cells provide therapeutic promises, as well as a potent in vitro model for studying biological processes that take place during human embryonic development and subsequent differentiation in normal and disease states. The epigenetic characteristics of iPS cells are reprogrammed to the embryonic state at which they acquire pluripotency. In addition, telomeres in hiPS cell must elongate sufficiently to provide the necessary replicative potential. Recent studies have demonstrated that the epigenetic characteristics of telomeric and subtelomeric regions are pivotal in regulating telomere length. Here we study telomere length, subtelomeric DNA methylation and telomeric-repeat-containing RNA (TERRA) expression in several hiPS cell clones derived from normal neonatal foreskin fibroblasts. We find that telomeres lengthen significantly in hiPS cells in comparison to the parental fibroblast source, and progressively shorten after differentiation back into fibroblast-like cells, concomitantly with telomerase activation and downregulation, respectively. Subtelomeres in hiPS cells were found to be generally hypermethylated in comparison to the parental source. However, bisulfite analysis revealed that at several subtelomeres examined, methylation levels differed between hiPS clones and that both de novo methylation and demethylation processes occurred during telomere reprogramming. Notably, although subtelomeres were in general very highly methylated, TERRA levels were elevated in hiPS cells, albeit to different degrees in the various clones. TERRA elevation may reflect enhanced stability or impaired degradation in hiPS cells, and/or alternatively, increased transcription from the hypomethylated subtelomeres. We suggest that TERRA may play a role in regulation of appropriate telomere function and length in hiPS cells.
Key words: human induced pluripotent stem cells, telomeres, subtelomeres, telomerase, DNA methylation, TERRA
Introduction
Induced pluripotent stem (iPS) cells are derived from somatic cells, mainly fibroblasts, and possess the capacity, similar to embryonic stem (ES) cells, to differentiate into cell derivatives of all three germ layers (reviewed in ref. 1 and 2). During the process of iPS cell generation, epigenetic reprogramming occurs, which involves mainly changes in DNA methylation and histone modifications. These modifications erase the fibroblast-specific epigenetic marks and establish a new repertoire of marks, similar to those of ES cells.3–8
iPS cells have been generated from somatic cells derived from aged organisms, both mouse9 and human.6,10 The studies in hiPS cells indicate that short telomere lengths in the parental cells do not limit the replicative capacity of the derived iPS cells. Telomeres are the specialized nucleoprotein complexes that maintain the integrity and stability of eukaryotic chromosome ends.11–13 In multicellular organisms, in the absence of telomerase or alternative mechanisms, telomeric DNA shortens with each round of cell division14 a process that eventually leads to replicative senescence.15 The replicative capacity of iPS cells suggests that telomeres are reprogrammed during the process of iPS cell generation and indeed telomerase activity has been shown to be present in iPS cells6,7,9,16 similar to the situation in ES cells.16,17 In accordance with telomerase upregulation, telomeres are elongated in mouse iPS cells;9 however, in hiPS cells conflicting findings were reported.10,16,18
In mammalian cells, the telomeres and adjacent subtelomeric regions are packaged as constitutive heterochromatin, facilitating the stabilization and capping of chromosome ends.19 The histone modifications in telomeric regions include enrichment of tri-methylation of histone H3 at Lysine 9 and histone H4 at Lysine 20 and low levels of acetylation of histones H3 and H4. In addition, Heterochromatin Protein 1 α binds these regions and subtelomeric DNA is heavily methylated during early development by DNA methyltransferase 3B (DNMT3B) in concert with additional factors (reviewed in ref. 20–22). Although devoid of genes, the subtelomeric and telomeric regions have the potential to transcribe telomeric repeat-containing RNA (TERRA), which is also designated Telomeric RNA (TelRNA).23,24
Studies from recent years have shown that the epigenetic characteristics of telomeric and subtelomeric regions are pivotal in regulating telomere length (reviewed in ref. 20–22). Subtelomeric regions extend 50–500 kb from telomere ends and contain an abundance of repetitive sequences.25 Subtelomeric DNA methylation has been shown to be associated with telomere length both in mice and in humans; however conflicting findings have been presented regarding the correlation between these two parameters.26–31 In mouse iPS cells, telomeres acquire the histone modifications typical of mouse ES cells indicating that the chromatin is more loosely packaged. However, the methylation of subtelomeric regions, as well as other repetitive regions, does not vary during the process of mouse iPS cell formation.8,9 TERRA levels increase in mouse iPS cells in comparison to the parental mouse embryonic fibroblasts (MEFs), but do not reach the same levels detected in mouse ES cells.9
Telomere length differs between humans and laboratory mice, with murine chromosomes harboring significantly longer telomeres. In addition, while telomerase expression in human somatic cells, with the exception of adult stem cells, is absent or present at levels insufficient to maintain telomere length,32 mouse telomerase is active, albeit at low levels, in a large repertoire of somatic tissues.33,34 These differences suggest that the process of telomere reprogramming during iPS cell generation may differ between human and mouse.16 Here we characterize the dynamics of telomere reprogramming during generation of human iPS cells and subsequent differentiation into fibroblast-like (FL) cells and demonstrate that this process involves changes both in subtelomeric methylation and in TERRA levels.
Results
Generation of hiPS cell clones from neonatal foreskin fibroblasts.
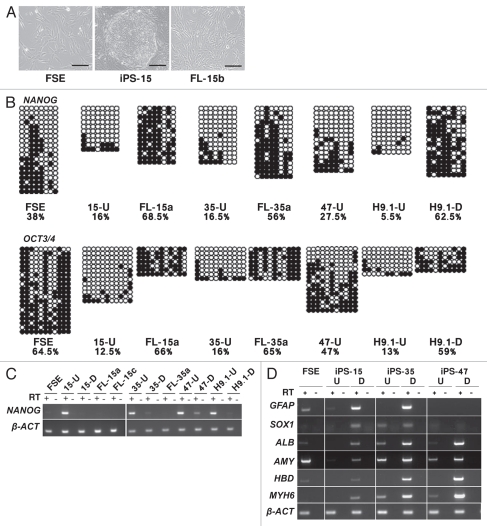
Neonatal foreskin fibroblasts, FSE (Fig. 1A), were utilized to generate hiPS cell lines iPS-15, iPS-35 and iPS-47, as described in Materials and Methods. Several clones, which morphologically resembled hES cells (Fig. 1A), were isolated and vital staining with TRA-1-81 or TRA-1-60 was performed. All screened colonies stained positively (Caspi et al. submitted) and were expanded and further characterized. Analysis of promoter regions of the NANOG and OCT3/4 genes was carried out by bisulfite genome sequencing (Fig. 1B) and these regions, as expected, demonstrated low CpG methylation levels. Concomitantly, NANOG was shown to be expressed by reverse transcription PCR (RT-PCR) only in the hiPS cell clones, but not in the parental FSE cells (Fig. 1C). Immunofluorescence (IF) staining with antibodies for NANOG and OCT3/4 confirmed protein expression of these genes in the hiPS cell clones, as well as expression of SSEA-4 (Caspi et al. submitted). The isolated iPS-15 and iPS-35 cell lines were capable of forming teratomas following injection into nude mice (Sup. Fig. S1).
Figure 1.
Generation of human iPS cells from primary fibroblasts. (A) Morphology of FSE fibroblasts, an undifferentiated colony of iPS-15 and FL-15b cells. Bar represents 1,000 µM. (B) Bisulfite sequencing analysis of NANOG and OCT3/4 promoters in FSE, undifferentiated iPS-15 (15-U), iPS-35 (35-U) and iPS-47 (47-U) cell lines, fibroblast-like cells: FL-15a and FL-35a and human H9.1 ES cells both in the undifferentiated (H9.1-U) and differentiated (H9.1-D) state. Open circles represent unmethylated CpG dinucleotides and closed circles represent methylated CpG dinucleotides. Each row is derived from an individual subclone. The value below the cell name indicates the total percentage of methylated CpG of all the tested clones. (C) RT-PCR analysis of NANOG in the indicated cell types. Expression of β-actin (β-ACT) was examined as a positive control. + and − indicate PCR reactions following RT either with (+) or without (−) reverse transcriptase addition. (D) RT-PCR analysis of in vitro differentiation of hiPS cells. RNA was extracted from undifferentiated (U) and differentiated (D) hiPS cells and analyzed for expression of genes representing the three germ layers. Ectoderm: glial fibrillary acidic protein (GFAP) and sex determining region Y-box 1 (SOX1), Endoderm: liver albumin (ALB) and pancreas amylase (AMY), Mesoderm: δ-globin (HBD) and cardiac myosin heavy chain 6 (MYH6). β-actin (β-ACT) was amplified as a control. + and − indicate PCR reactions following RT either with (+) or without (−) reverse transcriptase addition.
Several different hiPS clones were subjected to differentiation conditions in vitro, as described in Materials and Methods. Clones iPS-15 and iPS-35 demonstrated prominent cardiomyocyte differentiation (Caspi et al. submitted), while iPS-35 also demonstrated neuronal differentiation (A. Lange, personal communication). RT-PCR analysis of undifferentiated and differentiated iPS clones 15, 35 and 47 demonstrated upregulation of tissue-specific markers characteristic of all three embryonic germ layers (Fig. 1D).
Establishment of fibroblast-like cell lines from differentiated hiPS cells.
Following differentiation of iPS-15 and iPS-35, FL cells were isolated and cell lines were established. These cells resemble morphologically the parental FSE cell line (Fig. 1A) and were cultured under conditions appropriate for fibroblasts. FL cells demonstrated high levels of CpG methylation at NANOG and OCT3/4 promoters (Fig. 1B) and NANOG was no longer expressed (Fig. 1C), indicating that these cells are indeed differentiated derivatives of iPS-15 and -35.
Telomerase activity and telomere length dynamics during the process of hiPS cell generation and differentiation.
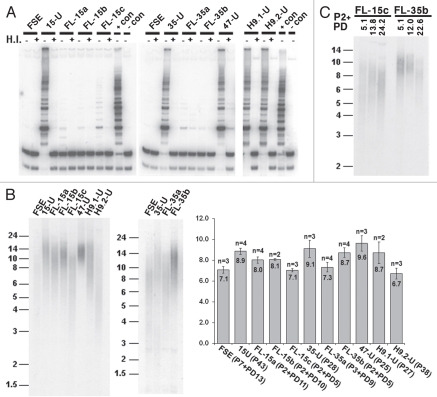
Previously it had been shown that telomerase is activated in hiPS cells,6,35 albeit to varying degrees16 and telomeres are elongated in most, but not all, hiPS clones.10,16,18 In order to confirm that telomerase is induced in the hiPS cells we generated and downregulated in the FL derivatives, we studied telomerase activity in the parental FSE fibroblasts, hiPS cells and the FL derivatives. In the hiPS cell clones a TRAP assay demonstrated high telomerase activity, similar to the activity detected in two hES cells, while the parental FSE cells exhibited no such activity. Telomerase activity was very low and, in some cases, undetectable, in the FL derivatives (Fig. 2A).
Figure 2.
Telomerase is activated and telomeres are elongated in the hiPS cells. (A) Detection of telomerase activity by the TRAP assay in FSE, undifferentiated iPS-15 (15-U), iPS-35 (35-U) and iPS-47 (47-U) cell lines, fibroblast-like cells: FL-15a, FL-15b, FL-15c, FL-35a and FL-35b, undifferentiated human ES cell lines H9.1 (H9.1-U) and H9.2 (H9.2-U) and positive and negative controls provided in the kit (+ con, − con). H.I., heat inactivated. (B) TRF analysis performed on DNA samples extracted from the various cell types. Size markers in kb appear to the left of the blots. Cell type, passage (P) and population doublings (PD) are indicated under the bar chart. Mean telomere length (MTL) in kb appears in the bar. Number of repeated hybridizations (n) appears above the bar. MTL of all three hiPS cell lines were significantly elongated in comparison to FSE MTL (p < 0.05, unpaired Student's t-test). MTL of FL-derivatives were significantly shortened in comparison to the parental hiPS in the case of FL-15a, b and c (p < 0.05) and FL-35a (p < 0.01). (C) TRF analysis of DNA extracted from FL-15c and FL-35b cell lines at gradually advancing PDs, as depicted above the lanes.
We next proceeded to determine whether telomere length was modified during the process of hiPS cell generation and subsequent differentiation. Telomere Restriction Fragment (TRF) analysis clearly demonstrated significant telomere elongation accompanying hiPS cell generation and subsequent shortening in FL-cell lines. At the earliest examinable PD (Fig. 2B), the shortening was significant in the case of the FL-derivatives of iPS-15 and in FL-35a, but not in the case of FL-35b (Fig. 2B). The number of PDs that took place prior to the establishment of the FL-cell line cannot be determined experimentally; therefore, no information can be derived from comparison of telomere lengths between different FL-lines. However, following establishment of each FL-line, PDs were recorded and TRF analysis demonstrated progressive shortening of telomeres with cell divisions (Fig. 2C). At P2 + PD22.6, the telomere length had shortened in FL-35b by 1.5 kb.
Previously it has been shown that telomeres are elongated in early mouse embryos by telomere recombination,36 and that high rates of telomere sister chromatid exchange (T-SCE) occur in mouse iPS cells.9 We therefore sought to determine whether telomere elongation in hiPS cells is mediated by telomere recombination, in addition to telomerase activity. Since telomere recombination takes place in promyelocytic leukemia (PML) bodies,37 we first determined whether telomeres and PML bodies co-localize in hiPS cells. Using IF with antibodies for TRF2 and PML protein38 there was no colocalization of these two proteins in iPS-35 and -47 (Sup. Fig. S2), suggesting that telomere elongation does not involve T-SCE in these cells. To strengthen this conclusion, we also subjected mitotic chromosomes from iPS-15 and -35 to chromatid orientation fluorescent in situ hybridization (CO-FISH) staining and T-SCE analysis31 (Sup. Fig. S3). Over 1,000 telomere ends from each hiPS cell line were scored for T-SCE, which should be manifested by “split” hybridization signals on both sister chromatids. In contrast to mouse iPS cells, no elevated rates of T-SCE were apparent in the hiPS cells. These findings, in concert with the IF results, indicate that at the stage at which the hiPS cell lines were examined, no recombination at telomeric regions was evident and are consistent with a predominant telomerase mediated mechanism for telomere elongation.
Dynamics of DNA methylation at subtelomeres and satellite 2 repeats during hiPS cell generation and differentiation.
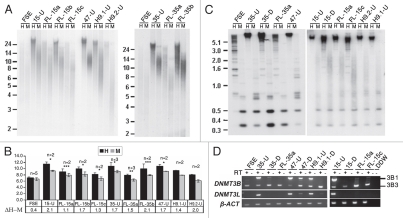
The epigenetic characteristics of subtelomeric and telomeric regions are pivotal in regulating telomere length. Among these features, DNA methylation has been shown to be an important regulator of telomere length, both in human and mouse,20–22 though subtelomeric DNA methylation is unaltered during the process of mouse iPS cell generation.9 To determine whether DNA methylation is reprogrammed at subtelomeres during human iPS cell formation, we initially determined subtelomeric DNA methylation using a modified methylation sensitive HpaII/MspI TRF analysis (Fig. 3A and B) and Southern analyses with subtelomeric repeat probes which hybridize to a large number of subtelomeres31 (Fig. 3C and Sup. Figs. S4 and S5). The mean telomere length (MTL) difference between HpaII and MspI digests (ΔH–M) increased significantly in all hiPS cells in comparison to the parental FSE cells, followed by a evident, though not statistically significant, decrease in most of the FL cell lines (Fig. 3A and B). These findings indicate that in comparison to the parental fibroblasts, subtelomeres are hypermethylated in hiPS cells, and following differentiation the methylation levels tend to decrease. Similar findings were found when analysis was carried out with the NBL-1 probe (Fig. 3C). The hypermethylation was especially evident when examining the ladder of bands generated by HpaII digestion of the FSE DNA, which is consolidated into a high-molecular signal in the hiPS cell-DNA, indicating the loss of HpaII sites in subtelomeric regions adjacent to telomeres due to de novo CpG methylation. Following differentiation, the FL-DNA showed reduction of this methylation, as demonstrated by the reappearance of the HpaII band ladder. Methylation analysis with the Hutel and TelSau2.0 probes (Sup. Figs. S4 and S5) displayed subtelomeric hypermethylation as well. However, the regions detected by these probes were initially highly methylated in FSE subtelomeres, therefore these analyses were less informative with regard to de novo methylation during hiPS cell generation. Analysis with the Hutel probe clearly illustrated the telomere length dynamics which accompanies the hiPS cell generation and subsequent differentiation, as shown previously (Fig. 2B and C).
Figure 3.
Subtelomeric regions are hypermethylated during the process of hiPS cell generation. (A) DNA extracted from FSE, undifferentiated iPS-15 (15-U), iPS-35 (35-U) and iPS-47 (47-U) cell lines, fibroblast-like cells: FL-15a, FL-15b, FL-15c, FL-35a and FL-35b and undifferentiated human ES cell lines H9.1 (H9.1-U) and H9.2 (H9.2-U) was subjected to TRF analysis following restriction digestion with both HpaII (H) and MspI (M). Size markers in kb appear on the left of the blots. (B) MTL in kb as determined by several repeats (n) of _HpaII/MspI_-TRF analysis, as shown in (A). The difference in MTL determined by HpaII and MspI digestions (ΔH–M), which indicates the degree of subtelomeric methylation, is displayed below the graph for each cell type in kb. Significant differences in ΔH–M were found between FSE and all three hiPS cell lines (p < 0.005 for each of three comparisons, unpaired Student's t-test). Comparison of ΔH–M values between each hiPS clone and its FL-derivatives did not show significant differences. (C) Methylation analysis with the subtelomeric NBL-1 probe following digestion of DNA samples with HpaII (H) and MspI (M). (D) RT-PCR analysis of DNMT3B, DNMT3L and β-actin (β-ACT) expression in FSE, hiPS cell lines, FL-derivatives and hES cell lines. + and − above the PCR results indicate PCR reactions following RT either with (+) or without (−) reverse transcriptase addition. 3B1 and 3B3 which appear to the right of the upper gel, designate the two most common alternative transcripts of DNMT3B.
DNMT3B is responsible for de novo methylation of subtelomeric regions, as well as additional repetitive sequences, during early development.31,39–41 Several alternative splice forms are generated from DNMT3B, with splice form 3B1 being the dominant transcript expressed during early embryonic development.42 In many non-embryonic cells, splice form 3B3, which lacks catalytic activity, is the most prevalent transcript.43 As expected, in FSE cells the dominant splice isoform present is 3B3, while in all three hiPS cell lines the 3B1 isoform is highly expressed (Fig. 3D). Following differentiation into FL cells, the 3B1 isoform was downregulated and expression of 3B3 persisted (Fig. 3D). This pattern of DNMT3B isoform expression is similar to that detected in undifferentiated and differentiated hES cells (Fig. 3D). hiPS-47, which failed to differentiate into cardiomyocytes, neuronal cells and FL cells, continued to express high levels of DNMT3B1 as long as three weeks following transfer to differentiation conditions, suggesting that indeed this clone is defective in its capability to fully differentiate.
DNMT3L is a member of the DNMT family, which lacks a catalytic domain, but is important for stimulating the catalytic activity of DNMT3A and DNMT3B.44 Similar to DNMT3B1, its expression is upregulated during early embryonic development at stages in which de novo methylation occurs.42 Accordingly, the expression pattern of DNMT3L at the various stages of hiPS cell generation and differentiation was similar to that of DNMT3B1 (Fig. 3D).
In addition to subtelomeric DNA, DNMT3B methylates various repetitive sequences such as satellite 2 repeats.45 We studied the methylation of satellite 2 repeats and found that they were hypermethylated during the process of hiPS cell generation and following differentiation the methylation levels were reduced (Sup. Fig. S6), indicating that hypermethylation during hiPS cell formation followed by hypomethylation after differentiation, occurs at various targets of DNMT3B.
Bisulfite analysis reveals a dynamic pattern of changes in subtelomeric methylation during hiPS cell generation.
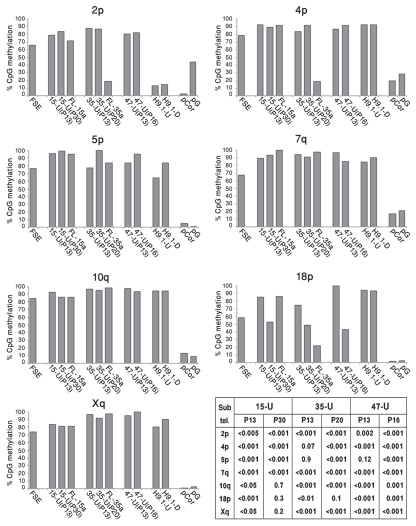
Southern analyses provide a general overview of the methylation status of all or a large fraction of subtelomeric regions, depending on the probe utilized for hybridization.31,46 In order to additionally gain a view of specific subtelomeric regions, whose exact distance from the telomere is defined and for which the CpG repeats can be individually examined, we performed bisulfite genome sequencing at seven subtelomeric regions (2p, 4p, 5p, 7q, 10q, 18p and Xq) all localized within several hundred nucleotides from the telomerichexamer track and containing between 12–32 CpG sites. These seven subtelomeric regions displayed varying degrees of methylation in the parental FSE fibroblasts ranging between 59–84%, and in control fibroblasts from two ICF (Immunodeficiency, Centromeric instability and Facial anomalies) syndrome patients (pCor and pG) the methylation levels are very low (Fig. 4), as shown previously.31 In all cases (with the exception of subtelomeres 4p and 5p in iPS-35) the percentage of methylated CpG sites was elevated significantly at early passage (P13) hiPS cells (Fig. 4), similar to the findings derived from Southern analyses (Fig. 3A–C). In addition, we analyzed the subtelomeric methylation pattern in two FL cell lines, FL-15a and FL-35a, and found that subtelomeric methylation changes which occur following differentiation varied for different subtelomeres and between the two FL cell lines. In FL-35a the methylation levels were reduced in comparison to iPS-35 in three out of the seven subtelomeres (Fig. 4); however, in the case of FL-15a almost no changes in subtelomeric methylation were observed following differentiation at any of the studied subtelomeres. Human ES H9.1 was methylated to a similar degree as the hiPS cells at five out of the seven subtelomeric regions, to a lesser degree at subtelomere 5p and in the case of subtelomere 2 was extremely hypomethylated in comparison to the hiPS cells, both in the undifferentiated and differentiated states.
Figure 4.
Bisulfite sequence analysis of subtelomeric regions indicates that subtelomeres undergo de novo methylation during the process of hiPS cell generation. Bisulfite sequence analysis was carried out for subtelomeric regions 2p, 4p, 5p, 7q, 10q, 18p and Xq in FSE, undifferentiated iPS-15 (15-U), iPS-35 (35-U) and iPS-47 (47-U) cell lines at two passages, fibroblast-like cells: FL-15a and FL-35a, human ES cell line H9.1 both in the undifferentiated (H9.1-U) and differentiated (H9.1-D) state and fibroblasts of two ICF patients (pCor and pG31). The percentage of methylated CpGs (% CpG methylation) for each of the cell types was determined by analysis of a minimum of eight clones (Sup. Figs. S7–S13). The table summarizes the p values obtained from comparisons of percentage of subtelomeric methylation in FSE and in the hiPS cells at the designated passages. p values were obtained by χ2 test with Yate's correction.
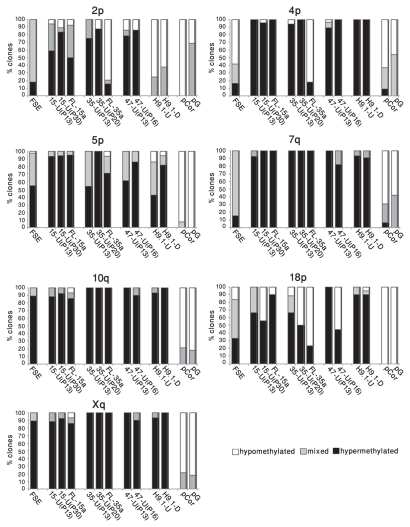
The individually sequenced clones could be classified into three categories (Fig. 5): hypermethylated—those that were methylated at a minimum of 80% of the CpG sites (designated by black bars), hypomethylated—those that were methylated at a maximum of 20% of the CpG sites (designated by white bars) and mixed clones with methylation levels ranging between 20–80% of the CpG sites (designated by gray bars). Examination of these methylation patterns revealed that while in FSE cells, a large percentage of the clones had a mixed methylation pattern, following hiPS cell generation, the percentage of the mixed clones was reduced at all seveb subtelomeres and in several subtelomeres only hypermethylated clones were detected. At a later passage, the percentage of mixed clones was, in most cases, even further reduced and interestingly, in three out of the seven subtelomeres (2p, 4p and 18p) hypomethylated clones appeared (Fig. 5 and Sup. Figs. S7–S13). Clones with the hypomethylated pattern persisted in several cases also in FL cells. The two extreme patterns of methylation suggest that during the process of hiPS formation, methylation dynamics vary in different cells, or perhaps even between the two alleles in the same cell. In addition, these findings, together with those emerging from Southern analyses, emphasize the advantage of utilizing both methods (methylation sensitive enzyme restriction analysis and bisulfite analysis) for methylation level assessment, which in concert enable analysis at different levels of resolution.
Figure 5.
Subtelomeric regions undergo both de novo methylation and demethylation during the process of hiPS cell generation. The individual bisulfite-analyzed clones were categorized according to the following criteria: hypermethylated (black blocks)-at least 80% of the CpG sites are methylated, hypomethylated (white blocks)-at least 80% of the CpG sites are not methylated, mixed (gray blocks)-between 20–80% of CpG sites are methylated. For each cell type, the percentage of individual clones that belong to each of the three categories appears in the bar graph, amounting together to 100%.
Telomeric-repeat-containing RNA (TERRA) levels are enhanced in human iPS cells.
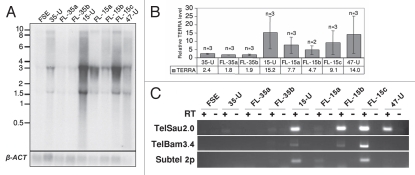
Telomeric regions produce TERRA or TelRNA transcripts, which initiate at subtelomeric regions and extend into telomeric sequence, with the telomere leading strand serving as the template for transcription.23,24 In mouse iPS cells TERRA levels were elevated in comparison to differentiated cells, but lower in comparison to the situation in mouse ES cells.9 Northern analysis of iPS-15, -35 and -47 revealed TERRA levels to be significantly elevated in comparison to FSE; however, the degree of TERRA levels varied among the different hiPS cell lines (Fig. 6A and B). Following differentiation, all FL cell lines demonstrated diminished TERRA levels in comparison to the hiPS cells from which they were derived. However the reduction varied among the various FL cell lines, and was not statistically significant in comparison to the hiPS cells (Fig. 6A and B).
Figure 6.
TERRA levels are elevated in hiPS cells. (A) Northern analysis of TERRA expression in FSE, undifferentiated iPS-15 (15-U), iPS-35 (35-U) and iPS-47 (47-U) cell lines and fibroblast-like cells: FL-15a, FL-15b, FL-15c, FL-35a and FL-35b. Size markers in Kb appear on the left. In the lower panel appear hybridization signals of the same blot to a β-actin (β-ACT) probe. (B) TERRA levels relative to the levels in FSE cells. TERRA signals were first calibrated according to the intensity of the β-actin hybridization signals and then levels were compared to those in FSE cells. TERRA levels were significantly elevated in hiPS cells in comparison to FSE (p < 0.0005 in 35-U, p < 0.05 in 15-U and 47-U, unpaired Student's t-test). No significant changes were found between TERRA levels in each hiPS clone and its FL-derivatives. (C) RT-PCR analysis of TERRA levels. RT-PCR analysis was carried out for subtelomeres 10q (TelSau2.0), Xq (TelBam3.4) and 2p (Subtel 2p). + and − indicate PCR reactions following RT either with (+) or without (−) reverse transcriptase addition.
In order to examine the relationship between the total TERRA levels, as demonstrated by the northern analysis, with TERRA expression from individual subtelomeres, we analyzed by RT-PCR TERRA expression from 3 subtelomeric regions: 2p, 10q and Xq as described previously.23,46 Subtelomere 10q was analyzed with primers located in the TelSau2.0 subtelomeric repeat and Xq with primers located in the TelBam 3.4 subtelomeric repeat46 (further description of the relevant primers appears in Sup. Figs. S4 and S5 and Sup. Table 1). Similar to the finding by northern analysis, TERRA levels detected by RT-PCR were the highest in iPS-15 and iPS-35 demonstrated no RT-PCR products from the three subtelomeres. TERRA was expressed from subtelomere 10q in iPS-47, but no expression was detected from subtelomeres Xq and 2p. These findings indicate that TERRA is expressed from different subtelomeric regions in various hiPS cell lines.
Discussion
hiPS cells are emerging as a potential new source for human pluripotent stem cells, circumventing obstacles related to hES cells such as alloimmune rejection.2 In addition to the applicative avenues, these cells may serve as a tool for studying various developmental processes at the very earliest embryonic stages both in normal and genetically perturbed states.
Pluripotency is achieved in iPS cells by erasure of the somatic donor cell epigenetic signature and establishment of a new repertoire of epigenetic marks which are characteristic of a pluripotent embryonic stem cell (reviewed in ref. 47). In addition to the foregoing, telomere length must be restored in the iPS cells in order to provide the appropriate replicative potential and following differentiation, the cells are expected to again downregulate hTERT expression and thus lose the potential for immortality and associated risk for tumorigenesis. Here we show, similarly to previous reports,10,18 that telomeres are elongated significantly during the generation of hiPS cells, albeit to lengths shorter than those reported by Shur et al.10 The MTL of the FSE fibroblasts, at the time point at which the hiPS cells were generated, was approximately 7 kb and telomeres were lengthened to approximately 9–9.5 kb in the hiPS cells at passages 25 and above, in contrast to lengths of 14–15 kb reported at passage five by Shur et al.10 On the contrary, Mathew et al.16 demonstrated that in two hiPS cell clones at passages 5 and 22, telomeres had shortened in comparison to the parental cells, a finding explained by low levels of telomerase activity in these hiPS clones. Vaziri et al.18 recently demonstrated that the initial telomere elongation in hiPS cells was variable and moderate, even in isogenic lines and following continuous passaging in culture, telomere shortening was evident in all hiPS, with the exception of one line, that lengthened. Collectively, these findings suggest that, similar to hES cells (reviewed in ref. 48–50 and our own findings regarding hES cell lines H9.1 and H9.2), telomere length may vary among hiPS cell lines.
In the current study, telomerase activity, as demonstrated by a TRAP assay, was undetectable in FSE cells, but was high in all three hiPS cell lines, at levels similar to those in two hES clones (Fig. 2A), suggesting that telomerase is the mediator of telomere elongation in hiPS cells. We found no evidence for elongation via telomere recombination in hiPS cells based on lack of colocalization between PML bodies and telomeres and very low levels of T-SCE. Telomere elongation via telomere recombination was previously ruled out as a contributor to telomere elongation in mouse iPS cells, even though elevated rates of T-SCE were detected in these cells.9 These findings also agree with studies on telomere dynamics following nuclear transfer, which demonstrate telomerase-dependent telomere elongation (reviewed in ref. 47). Due to the inability to experimentally analyze cells at the very initial stages of hiPS cell generation, one cannot rule out completely an early stage contribution of recombination to telomere elongation; however, the relative compacted telomere hybridization smear detected in the TRF blots (Fig. 2B) does not support such a scenario.
Following differentiation, little if any telomerase activity was detected in the various FL-lines and in concordance, telomere shortening was evident. The FL clones exhibited varying telomere lengths at the earliest examinable point, and this is most likely related to the number of PDs that occurred between the point at which hTERT expression was downregulated until the point at which the FL cells were isolated. Following establishment of each FL line, gradual telomere shortening was evident. This finding is encouraging for the prospect of therapeutic exploitation of iPS cells, since telomerase expression in terminally differentiated derivatives of hiPS cells would render them unsafe for in vivo utilization in patients.
Telomere elongation in hiPS cells was accompanied by a significant elevation in general subtelomeric methylation levels, as determined both by Southern analyses and bisulfite sequencing analysis. However, the latter analysis provided a more detailed picture of the subtelomeric methylation dynamics during hiPS cell generation. Individual clones from three subtelomeres obtained from the various hiPS cells demonstrated a striking phenomenon of two contrasting types of categorical methylation changes—extreme hyper- or hypomethylation. In addition, when following the patterns for two subtelomeres (2p and 18p) at two different passages, we observed that while a quarter to a third of the clones presented the “mixed” pattern at the earlier passage (P13), this pattern was absent from the later passages and all clones were either heavily methylated on all the CpG sites or completely unmethylated. In the case of subtelomere 2p the methylation levels were enhanced at the later passage, while in subtelomeres 18p the levels were reduced. These results indicate that at least up to passage 30, dynamic changes in subtelomeric methylation still take place. Given that during a culturing period of 20 passages, no significant change in MTL was evident in iPS-15 (results not shown), the dynamic changes in methylation do not seem to be associated with MTL changes. The epigenetic and telomere length dynamics during the very initial stages of iPS cell formation cannot be studied since the colonies cannot be detected until several rounds of cell division occur.
The dual patterns of hyper- and hypomethylation that were generated in hiPS cells in 3 of the 7 studied subtelomeric regions, and which persisted in the differentiated clones, are intriguing and may suggest either a process of selection or stochastic events. Interestingly, a similar phenomenon, also involving subtelomeric regions 2p and 18p, was observed in two Alternative Lengthening of Telomeres (ALT) cancer cell lines,28 suggesting that these regions may be prone to undergo this pattern of methylation changes. These contrasting patterns require both de novo methylation and demethylation events since both clone types differ significantly from the methylation pattern observed in FSE fibroblasts. The de novo methylation at subtelomeres is most probably associated with the upregulation of DNMT3B1 and DNMT3L in the hiPS cells, as DNMT3B has been demonstrated as the enzyme responsible for de novo methylation of subtelomeric repeats during development.31,39,41,51 Additional repetitive sequences that are targets of DNMT3B were hypermethylated as well during the process of hiPS cell generation, although this was not the case in mouse iPS cells.8 The mechanism for hypomethylation is unclear and may be either passive or active. If active, either DNMT3B may serve as a candidate for mediating active demethylation in addition to de novo methylation (reviewed in ref. 52) or activation-induced cytidine deaminase may be involved.53 Whether the dual patterns of hyper- and hypomethylation play a role in telomeric function in hiPS cells is unclear and is a subject for further studies.
Do subtelomeric methylation levels influence telomerase activity? Whether a correlation exists between subtelomeric methylation and telomerase levels is controversial, with findings either not supporting such a correlation30 and other findings suggesting a correlation between high levels of subtelomere methylation and telomerase activation in cancer cells.28 In the current study, at the point at which telomerase activity was examined, hypermethylated clones were already evident; however, it is unclear whether this reported correlation bespeaks a causative relation, and thus the question of whether telomerase activation and telomere lengthening are dependent on subtelomeric hypermethylation remains to be studied in hiPS cells in which genetic defects prevent de novo methylation at these sites.
Since TERRA has been proposed to regulate telomerase activity24,54,55 and has been correlated both with telomere length24,31 and subtelomeric methylation levels,31,46 we studied TERRA dynamics during the process of hiPS cells formation and differentiation. In the three hiPS cell clones analyzed in the current study we observed significantly elevated levels of TERRA, albeit to variable degrees. Clone iPS-15 demonstrated significantly higher levels in comparison to the other two clones. While it has been previously shown that subtelomeric methylation levels negatively regulate TERRA,26,31,46 in the current study it was interesting to find that TERRA was elevated despite the high degree of subtelomeric methylation. TERRA levels were also reported to be elevated in mouse iPS cells in comparison to the parental source, while no significant changes in subtelomeric methylation were detected in these cells.9 When individual subtelomeric regions of 2p, 10q and Xq were examined for both DNA methylation and TERRA levels, no clear correlation was detected between the two parameters. Similarly to our findings in hiPS cells, high levels of TERRA have been previously reported in ALT cells that display high subtelomeric methylation at regions 2p, 4p and 18p.28 The high levels of TERRA may be the consequence of increased stability or impaired degradation of these transcripts in hiPS cells, or alternatively, enhanced transcription from a subset of subtelomeres. In this regard, the presence of extremely hypomethylated clones, as observed by the bisulfite analysis, in three out of the seven examined subtelomeric regions, and possibly in additional unexamined subtelomeres, may explain the elevated TERRA levels, as suggested also in the case of the ALT cancer cell lines.28 Regarding the potential function of TERRA in hiPS cells, if TERRA indeed inhibits telomerase activity,24,54,55 we would expect TERRA levels to remain low during the initial stages of iPS cell formation. The hypermethylation of subtelomeric regions, may play a role in suppression of TERRA at early stages of hiPS generation, and thus enable maximal telomerase mediated elongation. However, following initial elongation, telomeres should be subjected to length regulation as in other telomerase positive cells and in particularly stem cell populations.48 Indeed, TERRA has been suggested as a means to control telomere length in mouse iPS cells once telomeres reach a certain telomere length.9,47 Similarly, in hiPS cells, elevated TERRA levels in established hiPS cell lines may contribute to telomere length homeostasis.
Materials and Methods
Cell culture of fibroblast cells.
Primary foreskin fibroblasts, FSE56 were grown in fibroblast media (DMEM media supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin) at standard conditions.
Generation of pluripotent stem cells from foreskin human fibroblasts.
Generation of human iPS cells was carried out according to reference 6 with several modifications as noted below. The mouse ecotrophic receptor, Slc7a1 (Addgene plasmid 17224)6 was introduced by lentiviral infection into FSE cells at approximately PD 10. Following selection with 1 µg/ml Blasticidin, a cell line stably expressing Slc7a1 was established, which served as the source for generating several iPS clones. In order to produce viral particles, the pCL-Eco plasmid (gag/pol/env; Addgene plasmid 12371,57) was co-transfected into the HEK293GP cell line separately with each of the following Moloney-based retroviral vectors (pMXs) containing human cDNAs of Oct4, Sox2 and Klf4 (Addgene plasmids 17964,58 17218 and 17219,7 respectively). Viral particle-containing medium was collected from the three co-transfections and used collectively to infect the Slc7a1-expressing-FSE cell line. The infection was carried out twice in an interval of 24 h, utilizing medium collected 48 and 72 h after transfection of the HEK293GP cells. Three days following infection, the cells were replated on a mitotically-inactivated MEF feeder layer in fibroblast media, at a density of 5 × 104 cells per well (six-well plate). The following day the media was converted to SR-high bFGF medium (Knockout DMEM supplemented with 20% Knockout serum replacement, 10 ng/ml bFGF, 1% nonessential amino acids, 0.1 mM β-mercaptoethanol and 1 mM glutamine) supplemented with 0.9 mM Valproic acid, as described59 and cells were propagated in this media for the following two weeks after which media was converted to regular SR medium (Knockout DMEM supplemented with 20% Knockout serum replacement, 4 ng/ml bFGF, 1% nonessential amino acids, 0.1 mM β-mercaptoethanol and 1 mM glutamine). iPS colonies, which started to emerge at this point, were isolated and expanded on mitotically-inactivated MEF feeder layers in SR media and colony propagation proceeded as previously described.60 A ROCK inhibitor, Y-27632,59 was added (5 µM) for the first 2–3 passages following colony isolation in order to increase seeding efficiency.
Identification of hiPS cell colonies.
iPS colonies were selected according to vital TRA-1-81 or TRA-1-60 staining. iPS colonies growing in six-well plates were incubated without fixation with TRA-1-81 (MAB4381, Millipore) or TRA-1-60 (sc-21705, Santa Cruz Biotechnology, Inc.) antibodies diluted 1:400 in SR medium at 37°C for 1 hour, and detected with a secondary antibody Cy3-anti mouse IgM (#715-165-140, Jackson ImmunoResearch Laboratories, Inc.) at a 1:200 dilution, for an additional hour, as described.58 Colonies, whose morphology resembled that of hES and were TRA-1-81 or TRA-1-60 positive, were isolated. These colonies were further expanded and assessed by molecular and functional parameters, as described below and in Caspi et al. (submitted). Teratoma generation and tissue processing were carried out as described previously.61
Differentiation of hiPS cells.
To induce differentiation, hiPS cells were dispersed into small clumps following incubation for 45 min with 300 U/ml of collagenase-IV and cultivated in suspension in ES media which is composed of knockout DMEM supplemented with 20% FBS (HyClone), 1% nonessential amino acids, 0.1 mM β-mercaptoethanol and 1 mM glutamine, as described previously.60 Under these culturing conditions, cells aggregated to form embryoid bodies (EBs). Seven to ten days later, EBs were plated on gelatin-coated six-well culture dishes in ES media [DMEM supplemented with 20% defined FBS (HyClone, Logan, UT), 1% nonessential amino acids, 0.1 mM β-mercaptoethanol, 100 U/ml penicillin, 100 µg/ml streptomycin and 1 mM glutamine]. Differentiating cultures were grown for at least an additional 2 weeks. During this period contracting EBs and FL cells, in addition to morphologically variable cell types, became apparent in the culture dishes. Further evaluation of differentiation potential was determined by RT-PCR analysis of genes representing the three germ layers.
Establishment of fibroblast-like cultures from differentiated hiPS cells.
Tissue culture wells containing differentiating hiPS cells that displayed a high concentration of morphologically FL cells were chosen to generate FL cell lines. Non-FL cells were removed mechanically by suction, while FL cells remained adherent to the culture dish. These remaining cells were detached from the culture dish by trypsinization. Following dispersion to single cell suspensions, cells were replated on standard tissue culture plates and fed with fibroblast media. From this point on the PDs of the FL cells were documented.
TRF analysis and DNA methylation studies by Southern analysis.
hiPS cells cultured for TRF analysis were transferred from growth on MEFs to growth on fibronectin coated plates and fed with conditioned media.62 The cells were passaged on fibronectin twice in order to eliminate the presence of MEFs in the harvested cells. Genomic DNA was extracted according to standard procedures. Purified DNA was subjected to restriction enzyme digestion and gel electrophoresis. DNA was transferred to a MagnaGraph nylon transfer membrane (Water and Process Technologies) and hybridized with the appropriate probes at conditions as described below.
Telomere length was determined by TRF analysis following analysis with MATELO software.31 Subtelomeremethylation analysis was carried out by a modified TRF analysis,31 by Southern analysis utilizing the NBL-1 and Hutel subtelomeric-repeat probes,31 and with a probe generated by the TelSau2.0 primers (reviewed in ref. 46 and described further in Sup. Fig. S5) on HpaII/MspI digested DNA separated on a 2% agarose gel.
The methylation status of satellite 2 repeats was determined by digestion of DNA with the restriction enzyme BstBI and hybridization with a satellite 2 oligonucleotide probe, as described.63
Bisulfite sequencing.
Genomic DNA (1 µg) was bisulfite-converted with the Methylamp DNA Modification kit (EPIGENTEK, NY) according to the manufacturer's instructions. After bisulfite conversion, DNA was amplified using Faststart Taq polymerase (Roche) as follows: 95°C-4 min; 5 cycles of: 95°C-30 sec, low annealing temperature-3 min, 72°C-3 min; 35 cycles of: 95°C-30 sec, high annealing temperature-30 sec, 72°C-30 sec; 72°C-10 min. Primer sequences and high and low annealing temperatures for each primer set appear in Supplemental Table 2. PCR products were TA-cloned into pCR2.1 plasmid (Invitrogen). Inserts were sequenced with M13 universal primers.
The chromosomal positions of the previously unpublished primers for bisulfite analysis are as following: subtelomere 5p-nucleotides: 12,129–12,350 (chromosome 5 contig, GRCh37), subtelomere 7q-nucleotides: 159,127,886–159,128,157 (chromosome 7 contig, GRCh37), subtelomere 10q-nucleotides 135,523,983–135,524,221 (chromosome 10 contig, GRCh37) and subtelomere Xq-nucleotides 155,259,277–155,259,530 (chromosome X contig, GRCh37). All these regions are located within 400 bp from the telomere tract.
Measurement of telomerase activity by TRAP assay.
Telomerase activity was measured in 600 ng protein extracts using the TRAPEZE® Telomerase Detection Kit (S7700, Chemicon International, Canada). The TS primer was radio-labeled and TRAP assay products were visualized by autoradiography following separation on a 12.5% polyacrylamide gel.
RNA isolation, RT -PCR and northern blotting.
HiPS cells cultured for the purpose of RNA extraction were transferred to growth on fibronectin-coated plates in conditioned media, as described above. RNA was extracted with TRI REAGENT (Molecular Research Center, Inc.,) and treated with DNase I (#79254, Qiagen). For northern analysis, RNA was concentrated on columns (#74204, Qiagen). For RT-PCR analysis of all transcripts, with exception of TERRA, 1 µg of DNaseI-treated RNA was converted to cDNA using iScript Synthesis Kit (170-8890, BIO-RAD). For RT-PCR analysis of TERRA expression, 1 µg of DNaseI-treated RNA was converted to cDNA using SuperScript III reverse transcriptase (Invitrogen life technologies) with the use of a (TTAGGG)5 oligo, as described previously.46 Primers and amplification conditions used for PCR following RT appear in Supplemental Table 1.
For northern analysis, 20 µg of RNA were subjected to electrophoresis on a 1.2% agarose gel and transferred to a MagnaGraph nylon transfer membrane. To detect transcripts from telomeric regions, membranes were hybridized to a 32P-end-labeled (TAA CCC)3 and relative levels of TERRA transcripts were determined as described previously.31
Immunofluorescence and microscopy analysis.
Cells were fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100. TRF-2 was detected by mouse anti-TRF2 (IMG-124, IMGENEX) diluted 1:300, and a secondary anti-mouse-Cy3 antibody. PML protein was detected by rabbit anti-PML (AB1370, Chemicon International) diluted 1:1,000 and a secondary anti-rabbit-FITC antibody. Nuclei were stained with DAPI. Fluorescence signals from stained cells were analyzed using a laser scanning confocal microscopy (LSM 510 Meta Confocal, Carl Zeiss Inc., Germany).
Chromosome orientation-fluorescence in situ hybridization and telomere sister-chromatid exchange analysis.
iPS cells growing in culture were subjected to 2.5 × 10−4 M BrdU for 17 hours. This incubation period covers the entire S + G2 phases in the hiPS cells. Colcemid was added at a concentration of 0.75 µg/ml during the last 45 min of the BrdU-incubation period. Metaphase spreads were prepared by standard techniques. CO-FISH was performed as described previously.31 T-SCE was determined by scoring the percentage of telomere ends displaying hybridization signals on both chromatids as described previously.31
Acknowledgements
We are grateful to Gil Arbel, Amira Gepstein and Yeela Shamai for help in tissue culturing, to Michal Levy for help with bisulfite sequence analysis, to Liron Berger for help with confocal microscopy, to Rita Fuhrer-Mor for her excellent service in DNA sequencing and to Noa Gil for her help in bisulfite analysis. We thank Daniel Kornitzer for comments on the manuscript. This research was supported in part by The Legacy Heritage Bio-Medical Program of the Israel Science Foundation (grant No. 725/09-S.S. and grant No. 1225/09-L.G.), Arthur and Rosalinde Gilbert Foundation of the American Technion Society and Richard D. Satell Fund of the American Technion Society (K.S.).
Abbreviations
ALT
alternative lengthening of telomeres
CO-FISH
chromatid orientation fluorescent in situ hybridization
DNMT
DNA methyltransferase
EBs
embryoid bodies
ES cells
embryonic stem cells
FL
fibroblast-like
ICF
immunodeficiency, centromeric instability and facial anomalies
IF
immunofluorescence
iPS cells
induced pluripotent stem cells
hiPS cells
human induced pluripotent stem cells
MTL
mean telomere length
MEFs
mouse embryonic fibroblasts
PDs
population doublings
PML
promyelocytic leukemia
RT-PCR
reverse transcription PCR
TERRA
telomeric-repeat-containing RNA
TRF
terminal restriction fragment
TelRNA
telomeric RNA
T-SCE
telomere sister chromatid exchange
Footnotes
Supplementary Material
Supplementary Material
References
- 1.Amabile G, Meissner A. Induced pluripotent stem cells: current progress and potential for regenerative medicine. Trends Mol Med. 2009;15:59–68. doi: 10.1016/j.molmed.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Zhao R, Daley GQ. From fibroblasts to iPS cells: induced pluripotency by defined factors. J Cell Biochem. 2008;105:949–955. doi: 10.1002/jcb.21871. [DOI] [PubMed] [Google Scholar]
- 3.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, et al. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park IH, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 9.Marion RM, Strati K, Li H, Tejera A, Schoeftner S, Ortega S, et al. Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell. 2009;4:141–154. doi: 10.1016/j.stem.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 10.Suhr ST, Chang EA, Rodriguez RM, Wang K, Ross PJ, Beyhan Z, et al. Telomere dynamics in human cells reprogrammed to pluripotency. PLoS One. 2009;4:8124. doi: 10.1371/journal.pone.0008124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hug N, Lingner J. Telomere length homeostasis. Chromosoma. 2006;115:413–425. doi: 10.1007/s00412-006-0067-3. [DOI] [PubMed] [Google Scholar]
- 12.LeBel C, Wellinger RJ. Telomeres: what's new at your end? J Cell Sci. 2005;118:2785–2788. doi: 10.1242/jcs.02394. [DOI] [PubMed] [Google Scholar]
- 13.Zakian VA. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 14.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 15.Allsopp RC, Harley CB. Evidence for a critical telomere length in senescent human fibroblasts. Exp Cell Res. 1995;219:130–136. doi: 10.1006/excr.1995.1213. [DOI] [PubMed] [Google Scholar]
- 16.Mathew R, Jia W, Sharma A, Zhao Y, Clarke LE, Cheng X, et al. Robust activation of the human but not mouse telomerase gene during the induction of pluripotency. FASEB J. 2010;24:2702–2715. doi: 10.1096/fj.09-148973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carpenter MK, Rosler E, Rao MS. Characterization and differentiation of human embryonic stem cells. Cloning Stem Cells. 2003;5:79–88. doi: 10.1089/153623003321512193. [DOI] [PubMed] [Google Scholar]
- 18.Vaziri H, Chapman KB, Guigova A, Teichroeb J, Lacher MD, Sternberg H, et al. Spontaneous reversal of the developmental aging of normal human cells following transcriptional reprogramming. Reg Med. 2010;5:345–363. doi: 10.2217/rme.10.21. [DOI] [PubMed] [Google Scholar]
- 19.Chan SW, Blackburn EH. New ways not to make ends meet: telomerase, DNA damage proteins and heterochromatin. Oncogene. 2002;21:553–563. doi: 10.1038/sj.onc.1205082. [DOI] [PubMed] [Google Scholar]
- 20.Blasco MA. Telomere epigenetics: a higher-order control of telomere length in mammalian cells. Carcinogenesis. 2004;25:1083–1087. doi: 10.1093/carcin/bgh185. [DOI] [PubMed] [Google Scholar]
- 21.Blasco MA. The epigenetic regulation of mammalian telomeres. Nat Rev Genet. 2007;8:299–309. doi: 10.1038/nrg2047. [DOI] [PubMed] [Google Scholar]
- 22.Schoeftner S, Blasco MA. A ‘higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 2009;28:2323–2336. doi: 10.1038/emboj.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Azzalin CM, Reichenback P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 24.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 25.Ambrosini A, Paul S, Hu S, Riethman H. Human subtelomeric duplicon structure and organization. Genome Biol. 2007;8:1511–1513. doi: 10.1186/gb-2007-8-7-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Z, Campbell AE, Lieberman PM. TERRA, CpG methylation and telomere heterochromatin: lessons from ICF syndrome cells. Cell Cycle. 2010;9:69–74. doi: 10.4161/cc.9.1.10358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee ME, Rha SY, Jeung HC, Chung HC, Oh BK. Subtelomeric DNA methylation and telomere length in human cancer cells. Cancer Lett. 2009;281:82–91. doi: 10.1016/j.canlet.2009.02.031. [DOI] [PubMed] [Google Scholar]
- 28.Ng LJ, Cropley JE, Pickett HA, Reddel RR, Suter CM. Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 2009;37:1152–1159. doi: 10.1093/nar/gkn1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tilman G, Loriot A, Van Beneden A, Arnoult N, Londono-Vallejo JA, De Smet C, et al. Subtelomeric DNA hypomethylation is not required for telomeric sister chromatid exchanges in ALT cells. Oncogene. 2009;28:1682–1693. doi: 10.1038/onc.2009.23. [DOI] [PubMed] [Google Scholar]
- 30.Vera E, Canela A, Fraga MF, Esteller M, Blasco MA. Epigenetic regulation of telomeres in human cancer. Oncogene. 2008;27:6817–6833. doi: 10.1038/onc.2008.289. [DOI] [PubMed] [Google Scholar]
- 31.Yehezkel S, Segev Y, Viegas-Pequignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17:2776–2789. doi: 10.1093/hmg/ddn177. [DOI] [PubMed] [Google Scholar]
- 32.Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, et al. Telomerase maintains telomere structure in normal human cells. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 33.Forsyth NR, Wright WE, Shay JW. Telomerase and differentiation in multicellular organisms: turn it off, turn it on, and turn it off again. Differentiation. 2002;69:188–197. doi: 10.1046/j.1432-0436.2002.690412.x. [DOI] [PubMed] [Google Scholar]
- 34.Greenberg RA, Allsopp RC, Chin L, Morin GB, DePinho RA. Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene. 1998;16:1723–1730. doi: 10.1038/sj.onc.1201933. [DOI] [PubMed] [Google Scholar]
- 35.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 36.Liu L, Bailey SM, Okuka M, Munoz P, Li C, Zhou L, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- 37.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 38.Cerone MA, Londono-Vallejo JA, Bacchetti S. Telomere maintenance by telomerase and by recombination can coexist in human cells. Hum Mol Genet. 2001;10:1945–1952. doi: 10.1093/hmg/10.18.1945. [DOI] [PubMed] [Google Scholar]
- 39.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 41.Ueda Y, Okano M, Williams C, Chen T, Georgopoulos K, Li E. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development. 2006;133:1183–1192. doi: 10.1242/dev.02293. [DOI] [PubMed] [Google Scholar]
- 42.Huntriss J, Hinkins M, Oliver B, Harris SE, Beazley JC, Rutherford AJ, et al. Expression of mRNAs for DNA methyltransferases and methyl-CpG-binding proteins in the human female germ line, preimplantation embryos, and embryonic stem cells. Mol Reprod Dev. 2004;67:323–336. doi: 10.1002/mrd.20030. [DOI] [PubMed] [Google Scholar]
- 43.Robertson KD, Uzvolgyi E, Liang G, Talmadge C, Sumegi J, Gonzales FA, et al. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suetake I, Shinozaki F, Miyagawa J, Takeshima H, Tajima S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J Biol Chem. 2004;279:27816–27823. doi: 10.1074/jbc.M400181200. [DOI] [PubMed] [Google Scholar]
- 45.Xu GL, Bestor TH, Bourc'his D, Hsieh CL, Tommerup N, Bugge M, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- 46.Nergadze SG, Farnung BO, Wischnewski H, Khoriauli L, Vitelli V, Chawla R, et al. CpG-island promoters drive transcription of human telomeres. RNA. 2009;15:2186–2194. doi: 10.1261/rna.1748309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marion RM, Blasco MA. Telomere rejuvenation during nuclear reprogramming. Curr Opin Genet Dev. 2010;20:1–7. doi: 10.1016/j.gde.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Allen ND, Baird DM. Telomere length maintenance in stem cell populations. Biochim Biophys Acta. 2009;1792:324–328. doi: 10.1016/j.bbadis.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Amit M, Carpenter MK, Inokuma MS, Chiu CP, Harris CP, Waknitz MA, et al. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev Biol. 2000;227:271–278. doi: 10.1006/dbio.2000.9912. [DOI] [PubMed] [Google Scholar]
- 50.Rosler ES, Fisk GJ, Ares X, Irving J, Miura T, Rao MS, et al. Long-term culture of human embryonic stem cells in feeder-free conditions. Dev Dyn. 2004;229:259–274. doi: 10.1002/dvdy.10430. [DOI] [PubMed] [Google Scholar]
- 51.Okano M, Takebayashi S, Okumura K, Li E. Assignment of cytosine-5 DNA methyltransferases Dnmt3a and Dnmt3b to mouse chromosome bands 12A2-A3 and 2H1 by in situ hybridization. Cytogenet Cell Genet. 1999;86:333–344. doi: 10.1159/000015331. [DOI] [PubMed] [Google Scholar]
- 52.Ooi SK, Bestor TH. The colorful history of active DNA demethylation. Cell. 2008;133:1145–1148. doi: 10.1016/j.cell.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luke B, Panza A, Redon S, Iglesias N, Li Z, Lingner J. The Rat1p 5′ to 3′ exonuclease degrades telomeric repeat-containing RNA and promotes telomere elongation in Saccharomyces cerevisiae. Mol Cell. 2008;32:465–477. doi: 10.1016/j.molcel.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 55.Luke B, Lingner J. TERRA: telomeric repeat-containing RNA. EMBO J. 2009;28:2503–2510. doi: 10.1038/emboj.2009.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yalon M, Gal S, Segev Y, Selig S, Skorecki KL. Sister chromatid separation at human telomeric regions. J Cell Sci. 2004;117:1961–1970. doi: 10.1242/jcs.01032. [DOI] [PubMed] [Google Scholar]
- 57.Naviaux RK, Costanzi E, Haas M, Verma IM. The pCL vector system: rapid production of helper-free, high-titer, recombinant retroviruses. J Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lowry WE, Richter L, Yachechko R, Pyle AD, Tchieu J, Sridharan R, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc Natl Acad Sci USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 60.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–414. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tzukerman M, Rosenberg T, Reiter I, Ben-Eliezer S, Denkberg G, Coleman R, et al. The influence of a human embryonic stem cell-derived microenvironment on targeting of human solid tumor xenografts. Cancer Res. 2006;66:3792–3801. doi: 10.1158/0008-5472.CAN-05-3467. [DOI] [PubMed] [Google Scholar]
- 62.Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, et al. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol. 2001;19:971–974. doi: 10.1038/nbt1001-971. [DOI] [PubMed] [Google Scholar]
- 63.Jiang YL, Rigolet M, Bourc'his D, Nigon F, Bokesoy I, Fryns JP, et al. DNMT3B mutations and DNA methylation defect define two types of ICF syndrome. Hum Mutat. 2005;25:56–63. doi: 10.1002/humu.20113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material