NEW EMBO MEMBER'S REVIEW: Acetylation: a regulatory modification to rival phosphorylation? (original) (raw)
Abstract
The fact that histones are modified by acetylation has been known for almost 30 years. The recent identification of enzymes that regulate histone acetylation has revealed a broader use of this modification than was suspected previously. Acetylases are now known to modify a variety of proteins, including transcription factors, nuclear import factors and α–tubulin. Acetylation regulates many diverse functions, including DNA recognition, protein–protein interaction and protein stability. There is even a conserved structure, the bromodomain, that recognizes acetylated residues and may serve as a signalling domain. If you think all this sounds familiar, it should be. These are features characteristic of kinases. So, is acetylation a modification analogous to phosphorylation? This review sets out what we know about the broader substrate specificity and regulation of acetyl– ases and goes on to compare acetylation with the process of phosphorylation.
Keywords: acetylase/acetylation/deacetylase/histones/phosphorylation
Introduction
The correlation between histone acetylation and increased transcription has been known for many years (Allfrey et al., 1964). The discovery of GCN5, the first nuclear acetylase (Brownwell et al., 1996), and HDAC1, the first deacetylase (Taunton et al., 1996), has verified that acetylation of histones is an important controlling step in transcription (reviewed in Grunstein, 1997). However, despite the many attempts to understand the role of acetylation in chromatin assembly, transcription factor accessibility and nucleosome remodelling, the fact remains that the precise mechanism by which acetylation of histones augments transcription remains elusive. The truth is likely to be complicated. Acetylation of histones most probably affects a combination of events, which allows for higher processivity of RNA polymerase II.
The discovery of nuclear acetylases has aided the discovery of novel nuclear targets for acetylation. Some of these targets are well known and extensively characterized transcription factors, so analysis of how acetylation affects their function has been more straightforward. Consequently, we now know a lot more about how non-histone proteins are affected by acetylation than we know about histones. Below, I summarize some of these data and ask the question: is acetylation a modification analogous to phosphorylation?
Acetylases and deacetylases
There are now several reported families of acetylases exemplified by PCAF/GCN5, p300/CBP, TAF250, SRC1 and MOZ (reviewed in Kouzarides, 1999). Of these enzymes, two families, PCAF/GCN5 and p300/CBP, are the most characterized and continue to receive the most attention. This bias partly reflects the many connections these enzymes have (both physical and functional) to transcription factors and signalling pathways. Perhaps a more practical contributory reason is the fact that these two families of enzymes are very potent acetylases. Other designated enzymes, such as SRC1 or TIP60 (Yamamoto and Horrikoshi, 1997), have very weak and often undetectable activity towards histones, whereas others such as BRCA2 (Siddique et al., 1998) have no detectable intrinsic activity under certain conditions (Fuks et al., 1998). One of the explanations for the low activity towards histones may be that the true target for these enzymes are non-histone proteins yet to be identified.
Until recently, there was a single family of human histone deacetylases (HDAC1, 2 and 3), which have a highly conserved catalytic domain and are related to the yeast deacetylase RPD3 (reviewed in Kouzarides, 1999). A new set of enzymes has now been identified (HDAC4, 5 and 6), which are more related to a distinct deacetylase in yeast called HDA1 (Fischle et al., 1999; Grozinger et al., 1999; Miska et al., 1999; Verdel and Khochbin, 1999).
Acetylation of non-histone proteins
Following the identification of nuclear histone acetylases, a number of non-histone proteins have been identified as substrates for PCAF and/or p300/CBP. Many of these substrates are involved in the regulation of transcription and include p53, E2F1, EKLF, TFIIEβ, TFIIF, TCF, GATA1, HMGI(Y) and ACTR (Gu and Roeder, 1997; Imhof et al., 1997; Boyes et al., 1998; Munshi et al., 1998; Waltzer and Bienz, 1998; Zhang and Bieker et al., 1998; Chen et al., 1999; Martínez-Balbás et al., 2000; Marzio et al., 2000). Even before the current boom in acetylation, it was clear that histones were not the only cellular proteins to enjoy the attention of acetylases. DNA-binding proteins such as HMG1 and even non-nuclear proteins such as α–tubulin were known to be modified by acetylation (Sterner et al., 1979; L'Hernault and Rosenbaum, 1985). Thus, substrates for acetylation now include DNA-binding proteins (histones and transcription factors), non-nuclear proteins (tubulin) and proteins that shuttle from the nucleus to the cytoplasm, such as the importin-α family of nuclear import factors (Bannister et al., 2000).
Specificity for acetylation
Acetylases modify very few lysines within a given protein, an indication of specificity. Alignment of sequences surrounding modified lysines and mutagenesis of Rch1 (human importin-α) suggest that GK may be part of a recognition motif (Bannister et al., 2000). Consistent with this, the recent crystal structure of GCN5 with a histone peptide identifies GKXXP as a possible recognition motif (Rojas et al., 1999). Less is known about the specificity and site selection of deacetylases. What is clear is that deacetylases such as HDAC1 can deacetylate not only histones but also a transcription factor, E2F1 (Martínez-Balbás et al., 2000). There is also evidence that a yeast enzyme, RPD3, is able preferentially to deacetylate Lys5 in histone H4, to repress transcription (Rundlett et al., 1998).
Acetylation and protein function
What is the consequence of acetylation? The answer to this depends on where within the protein acetylation takes place. In the case of four site-specific DNA-binding transcription factors, p53, E2F1, EKLF and GATA1, the acetylation site falls directly adjacent to the DNA-binding domain and acetylation results in stimulation of DNA binding (Gu and Roeder, 1997; Boyes et al., 1998; Zhang and Bieker, 1998; Martínez-Balbás et al., 2000). In contrast, the lysines acetylated within the HMGI(Y) transcription factor fall within the DNA-binding domain and result in disruption of DNA binding. Thus, the commonly held view that acetylation is stimulatory for transcription (originating with histones as the example) has not held true for transcription factors.
Besides affecting DNA binding, acetylation also regulates protein–protein interactions. In the case of Drosophila TCF, the binding of this transcription factor to its co-activator, armadillo, is disrupted by acetylation of TCF (Waltzer and Bienz, 1998). In a second, more recent example, the association of nuclear receptors with their co-activator ACTR is inhibited by acetylation (Chen et al., 1999). In a final example, acetylation of histones seems to generate a recognition site for the bromodomain, a structure conserved in many proteins, including acetylases (Dhalluin et al., 1999).
A third function regulated by acetylation is protein stability. Analysis of in vivo acetylated E2F1 shows that the acetylated version has a longer half-life (Martínez-Balbás et al., 2000). For α–tubulin also, the correlation has been made between acetylated α–tubulin and microtubule stability (Takemura et al., 1992).
The effect of acetylation on three other proteins involved in transcription, i.e. HMG1, TFIIEβ and TFIIF, remains to be established. Also uncharacterized is the observation that some acetylases, such as PCAF and p300, undergo autoacetylation (Herrera et al., 1997).
Targeting of enzyme to substrate
Targeting the enzyme to the substrate is likely to be very important since acetylated proteins are found associated with the enzymes in vivo. It may also be subject to regulation by other signalling pathways. This prediction arises from p53 whose phosphorylation stimulates acetylation, probably by increasing the association of p53 with CBP/p300 (Sakaguchi et al., 1998; Liu et al., 1999).
Another indication of the importance of enzyme targeting comes from the analysis of the deacetylase HDAC4 (Miska et al., 1999; Wang et al., 1999). This novel enzyme shuttles between the nucleus and the cytoplasm (a feature not exhibited by HDAC1, 2 or 3). Furthermore, HDAC4 can bind a myogenic factor MEF2A using unique sequences at its N-terminus that are not found in other deacetylases. Consequently, HDAC4 can repress MEF2A activity, but HDAC1 (and presumably the others too) cannot. In other words, accessing the substrate is crucial to enzyme function.
Regulation of acetyltransferase activity
A number of lines of evidence indicate that the enzymatic activity of acetylases is regulated by proliferation and differentiation signals. The regulatory signal can come via phosphorylation or via hormonal signalling.
Evidence that proliferation signals regulate acetylation comes from the finding that the HAT activity of CBP is stimulated at the G1–S boundary (Ait-Si-Ali et al., 1998). This effect is most likely to be mediated by a phosphorylation event carried out by a cyclin E–CDK complex. The HAT activity may also be responsive to DNA repair signals. This possibility is based on the observation that phosphorylation of GCN5 by the DNA-dependent protein kinase (DNA–PK) inhibits HAT activity (Barlev et al., 1998).
The hormone-induced stimulation of nuclear receptors is a well documented event. Recently, evidence was provided for a hormone-mediated repression of nuclear receptor function. The repression is transmitted by a hormone-induced acetylation of ACTR, the co-activator of nuclear receptors. This example provides a very clear indication that extracellular signals can modulate acetylase activity, which, given the role of nuclear receptors, will influence cell differentiation and development.
The binding of the viral oncoprotein E1A to p300/CBP and p/CAF may regulate the HAT activity of the enzyme, but here the literature is confusing as to the actual direction of regulation (up or down). What is in dispute is whether E1A can repress or stimulate this activity. One report shows that E1A elevates the HAT activity of CBP/p300 (Ait-Si-Ali et al., 1998). Another set of reports indicates that the HAT activity of CBP/p300 and p/CAF is decreased by E1A (Chakravarti et al., 1999; Hamamori et al., 1999), whereas another report suggests that E1A does not change the activity of p/CAF (Reid et al., 1998). The results may reflect differences in the in vivo versus in vitro analysis of the phenomenon. Alternatively, the discrepancy may well reflect the dual agenda of E1A to stimulate promoters required for S–phase and to repress those that prevent it. So, in principle, depending on the promoter, both modes of regulation may be operational.
Acetylation versus phosphorylation
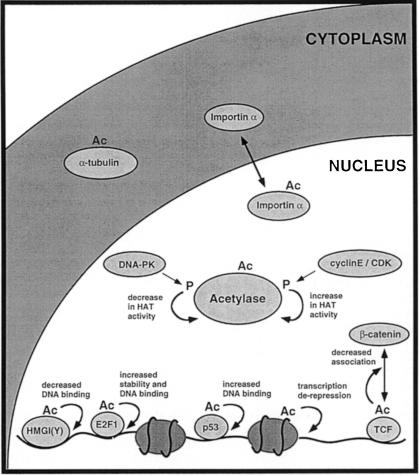
The first commonality between acetylation and phosphorylation is the diversity of substrates (Figure 1). Acetylation can occur on histones, DNA-binding transcription factors, acetylases, nuclear import factors and α–tubulin. As in the case of phosphorylation, substrates can be nuclear or cytoplasmic. Although clearly not as prevalent as phosphorylation at the moment, the list of acetylated proteins is increasing rapidly.
Fig. 1. Acetylation of a variety of proteins by acetylases affects their activity in different ways. The activity of acetylases is regulated, at least in vitro, by kinases involved in DNA repair (DNA–PK) and cell cycle progression (cyclin E–CDK). Acetylated targets include histones, nuclear acetylases (P/CAF and p300), transcription factors [e.g. HMGI(Y), E2F1, p53 and TCF], the nuclear import factor, importin-α and α–tubulin. Acetylation has many consequences, including effects on DNA binding, protein stability and protein–protein interaction. Ac, acetylation; p, phosphorylation.
Acetylation can regulate different functions, also a feature of kinases. It can modify the recognition of DNA, the stability of proteins and the interaction between proteins. Our limited knowledge of acetylated targets also suggests that they may regulate different cellular processes, such as microtubule function or nuclear import.
Thus, the analogy between acetylation and phosphorylation does hold true when we consider that both modifications affect multiple different proteins and regulate them in a variety of different ways. There are of course differences. For example, kinases do not necessarily associate with their substrates as avidly as acetylases. There are also far fewer acetylases than kinases, but this discrepancy may well reflect our current bias in defining these enzymes as histone acetylases. Perhaps if substrates other than histones were used in the screening for new acetylases, new families of enzymes might be identified that recognize only non-histone proteins. It is also true to say that homology within the catalytic domain of acetylases is not as high as, for example, between kinases, so we may be missing some obvious candidates.
Where the analogy with kinases breaks down is when we consider the signalling aspects of phosphorylation. There is no evidence as yet for an acetylation cascade, i.e. an acetylase modifying the enzymatic activity of a second acetylase in order to transmit a biological signal. However, the elements for the implementation of an acetylation cascade have been identified. The bromodomain, which recognizes acetyl-lysines, may be analogous to the SH2 domain, which recognizes phosphotyrosine and transmits the phosphorylation signal. A similar signalling function may be attributed to bromodomains, although this could be limited to nuclear events, given that bromodomains have not been identified in cytoplasmic proteins.
Many more questions remain regarding the similarity between the two modifications: is there a specific class of proteins that acts to inhibit acetylases (analogous to CDK inhibitors), is there a specifically cytoplasmic family of acetylases and deacetylases, is there synergy or antagonism between phosphorylation and acetylation?
Conclusion
Acetylation as a regulatory modification has come a long way since the identification of histone acetylases and deacetylases. Perhaps it is time to drop the ‘histone’ prefix for these enzymes given the multiplicity of other targets. It is this diversity of substrates that makes acetylation comparable to phosphorylation. However, acetylation lags behind phosphorylation in many ways, not least at the level of knowing which acetylase is the true in vivo enzyme for a given substrate. Specific inhibitors of acetylases would be very useful for dissecting different in vivo pathways. Such inhibitors have been invaluable to the kinase field.
Despite this discrepancy in knowledge and even given the eventuality that acetylation may not exactly parallel phosphorylation, the fact remains that both phosphorylation and acetylation can regulate key cellular processes in response to extracellular signals. So this evidence alone is sufficient to propose that we are witnessing the birth of a new biologically relevant regulatory modification to rival phosphorylation.
Acknowledgments
Acknowledgements
I am grateful to the Cancer Research Campaign for supporting the work on acetylation in my laboratory.
References
- Ait-Si-Ali A., et al. (1998)Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature, 396, 184–186. [DOI] [PubMed] [Google Scholar]
- Allfrey V.G., Faulkner, R. and Mirsky, A.E. (1964) Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA, 61, 786–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides, T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Miska,E., Gorlich,D. and Kouzarides,T. (2000) Nuclear import factors acetylated by CBP/p300. Curr. Biol., in press. [DOI] [PubMed] [Google Scholar]
- Barlev N.A., Poltoratsky,V., Owen-Hughes,T., Ying,C., Liu,L., Workman,J.L. and Berger,S.L. (1998) Repression of GCN5 histone acetyltransferase activity via bromodomain-mediated binding and phosphorylation by the Ku-DNA-dependent protein kinase complex. Mol. Cell. Biol., 18, 1349–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes J., Byfield, P., Nakatani, Y. and Ogryzko, V. (1998) Regulation of activity of the transcription factor GATA-1 by acetylation. Nature, 396, 594–598. [DOI] [PubMed] [Google Scholar]
- Brownell J.E., Zhou,J., Ranalli,T., Kobayashi,R., Edmondson,D.G., Roth,S.Y. and Allis,C.D. (1996) Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84, 843–851. [DOI] [PubMed] [Google Scholar]
- Chakravarti D., Ogryzko, V., Kao, H.Y., Nash, A., Chen, H., Nakatani, Y. and Evans, R.M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell, 96, 393–403. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin, J.R., Xie, W., Wilpitz, D. and Evans, M.R. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson, J.E., Zeng, L., He, C., Aggarwal, A. and Zhou, M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Fischle W., Emiliani, S., Hendzel, M.J., Nagase, T., Nomura, N., Voelter, W. and Verdin, E. (1999) A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J. Biol. Chem., 274, 11713–11720. [DOI] [PubMed] [Google Scholar]
- Fuks F., Milner, J. and Kouzarides, T. (1998) BRCA2 associates with acetyltransferase activity when bound to P/CAF. Oncogene, 17, 2531–2534. [DOI] [PubMed] [Google Scholar]
- Grozinger C.M., Hassig, C.A. and Schreiber, S.L. (1999) Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl Acad. Sci. USA, 96, 4868–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Gu W. and Roeder, R.G. (1997) Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell, 90, 595–606. [DOI] [PubMed] [Google Scholar]
- Hamamori Y., Sartorelli, V., Ogryzko, V., Puri, P.L., Wu, H.Y., Wang, J.Y.J., Nakatani, Y. and Kedes, L. (1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell, 96, 405–413. [DOI] [PubMed] [Google Scholar]
- Herrera J.E., Bergel, M., Yang, X.J., Nakatani, Y. and Bustin, M. (1997) The histone acetyltransferase activity of human GCN5 and PCAF is stabilized by coenzymes. J. Biol. Chem., 262, 27253–27258. [DOI] [PubMed] [Google Scholar]
- Imhof A., Yang, X.J., Ogryzko, V.V., Nakatani, Y., Wolffe, A.P. and Ge, H. (1997) Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol., 7, 689–692. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (1999) Histone acetylases and deacetylases in cell proliferation. Curr. Opin. Genet. Dev., 9, 40–48. [DOI] [PubMed] [Google Scholar]
- L'Hernault S.W. and Rosenbaum, J.L. (1985) Chlamydomonas α-tubulin is posttranslationally modified by acetylation on the ɛ-amino group of a lysine. Biochemistry, 24, 473–478. [DOI] [PubMed] [Google Scholar]
- Liu L., Scolnick, D.M., Trievel, R.C., Zhang, H.B., Marmorstein, R., Halazonetis, T.D. and Berger, S.L. (1999) p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol., 19, 1202–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Balbás M., Bauer, U.-M., Nielsen, S.J., Brehm, A. and Kouzarides, T. (2000) Regulation of E2F1 activity by acetylation. EMBO J., 19, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzio G., Wagener,C., Gutierrez,M.I., Cartwright,P., Helin,K. and Giacca,M. (2000) E2F family members are differentially regulated by reverse acetylation. J. Biol Chem., in press. [DOI] [PubMed] [Google Scholar]
- Miska E., Karlsson, C., Langley, E., Nielsen, S., Pines, J. and Kouzarides, T. (1999) HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J., 18, 5099–5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi N., Merika, M., Yie, J., Senger, K., Chen, G. and Thanos, D. (1998) Acetylation of HMG1 (Y) by CBP turns off IFNβ expression by disrupting the enhanceosome. Mol. Cell, 2, 457–467. [DOI] [PubMed] [Google Scholar]
- Reid J.L., Bannister, A.J., Zegerman, P., Martínez-Balbás, M. and Kouzarides, T. (1998) E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J., 17, 4469–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas J.R., Trievel, R.C., Zhou, J., Mo, Y., Li, X., Berger, S.L., Allis, C.D. and Marmorstein, R. (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature, 401, 93–97. [DOI] [PubMed] [Google Scholar]
- Rundlett S.E., Carmen, A.A., Suka, N., Turner, B.M. and Grunstein, M. (1998) Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392, 831–835. [DOI] [PubMed] [Google Scholar]
- Sakaguchi K., Herrera, J.E., Saito, S., Miki, T., Bustin, M., Vassilev, A., Anderson, C.W. and Appella, E. (1998) DNA damage activates p53 through a phosphorylation–acetylation cascade. Genes Dev., 12, 2831–2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddique H., Zou, J.P., Rao, V.N., Shyam, E. and Reddy, P. (1998) The BRCA2 is a histone acetyltransferase. Oncogene, 16, 2283–2285. [DOI] [PubMed] [Google Scholar]
- Sterner R., Vidali, G. and Allfrey, V.G. (1979) Studies of acetylation and deacetylation in high mobility group proteins. Identification of the sites of acetylation in HMG-1. J. Biol. Chem., 254, 11577–11583. [PubMed] [Google Scholar]
- Takemura R., Okabe, S., Umeyama, T., Kanai, Y., Cowan, N.J. and Hirokawa, N. (1992) Increased microtubule stability and α tubulin acetylation in cells transfected with microtubule-associated proteins MAP1B, MAP2 or τ. J. Cell Sci., 103, 953–964. [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig, C.A. and Schreiber, S.L. (1996) A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science, 272, 408–411. [DOI] [PubMed] [Google Scholar]
- Verdel A. and Khochbin, S. (1999) Identification of a new family of higher eukaryotic histone deacetylases. J. Biol. Chem., 274, 2440–2445. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz, M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- Wang A.H., Bertos,N.R., Vezmar,M., Pelletier,N., Crosato,M., Heng,H.H., Th'ng,J., Han,J. and Yang,X.-J. (1999) HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol. Cell. Biol., 19, 7816–7827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T. and Horikoshi, M. (1997) Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J. Biol. Chem., 272, 30595–30598. [DOI] [PubMed] [Google Scholar]
- Zhang W. and Bieker, J.J. (1998) Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl Acad. Sci. USA, 95, 9855–9860. [DOI] [PMC free article] [PubMed] [Google Scholar]