ERCC1 and ERCC2 Polymorphisms Predict Clinical Outcomes of Oxaliplatin-based Chemotherapies in Gastric and Colorectal Cancer: A Systemic Review and Meta-analysis (original) (raw)
. Author manuscript; available in PMC: 2012 Mar 15.
Abstract
Purpose
Nucleotide excision repair (NER) modulates platinum-based chemotherapeutic efficacy by removing drug-produced DNA damage. To summarize published data on the association between polymorphisms of NER genes (ERCC1 and ERCC2) and responses to oxaliplatin-based chemotherapies, we performed a meta-analysis of gastric and colorectal cancer for commonly studied polymorphisms ERCC1 rs11615C>T and ERCC2 rs13181T>G.
Patients and methods
In 17 previous published studies, 1787 cancer patients were treated with the oxaliplatin-based regimen. Primary outcomes included therapeutic response (TR) (i.e., complete response + partial response vs. stable disease + progressive disease), progression-free survival (PFS) and overall survival (OS). We calculated odds ratio (OR) or hazard ratio (HR) with 95% confidence intervals (CI) to estimate the risk or hazard.
Results
We found consistent and clinically substantial risk or hazard for TR, PFS and OS in the oxaliplatin-treated gastric and colorectal cancer patients with an ethnic discrepancy. For ERCC1 rs11615C>T, the T allele was associated with reduced response, PFS and OS in Asians (TR: OR, 0.53 and 95% CI, 0.35–0.81; PFS: HR, 1.69 and 95% CI, 1.05–2.70; and OS: HR, 2.03 and 95% CI, 1.60–2.59). For ERCC2 rs13181T>G, the G allele was associated with reduced response, PFS and OS in Caucasians (TR: OR, 0.56 and 95% CI, 0.35–0.88; PFS: HR, 1.41 and 95% CI, 1.02–1.95; and OS: HR, 1.42 and 95% CI, 1.11–1.81).
Conclusions
NER ERCC1 rs11615C>T and ERCC2 rs13181T>G polymorphisms are useful prognostic factors in oxaliplatin treatment of gastric and colorectal cancer. Larger studies and further clinical trials are warranted to confirm these findings.
Keywords: Chemotherapy, DNA repair, Meta-analysis, Pharmacogenetics, Platinum
Introduction
Fluoropyrimidines are essential in the treatment of gastric and colorectal cancer in advanced stages and have shown survival benefit, compared with the best supportive care (1, 2). Oxaliplatin is the new generation of platinum drugs that improve response rate and survival after adding to the 5-Fu/leucovorin (LV) regimen. Combination treatment of 5-Fu/LV plus oxaliplatin (FOLFOX) is now considered the standard treatment for gastric and colorectal cancer, with a response rate of over 40% for the first-line treatment (3, 4). Despite the efficacy of combined chemotherapies, a large proportion of patients display varying levels of resistance, indicating that the therapeutic efficacy has a remarkable inter-individual variability. Since DNA kinking is the major feature of platinum-DNA adducts that block DNA replication and lead to cancer cell death (5, 6), which is recognized and repaired by the nucleotide excision repair (NER) pathway, it is conceivable that the inter-individual difference in the NER capacity may influence the efficacy of oxaliplatin-based chemotherapy and clinical outcomes of the treated cancer patients.
ERCC1 and ERCC2 proteins are major components of the NER complex, acting as the rate-limiting enzymes in the NER pathway. Several common and putatively functional single nucleotide polymorphisms (SNPs) of ERCC1 and ERCC2 have been identified, of which ERCC1 rs11615 and rs3212986 SNPs (C118T and C8092A) have some effects on ERCC1 mRNA expression (7), whereas ERCC2 rs1799793 and rs13181 SNPs (Asp312Asn [G>A] and Lys751Gln, [T>G], respectively) SNPs are associated with suboptimal DNA repair capacity (8, 9). Previous studies have suggested that ERCC1 is a promising predictive marker for response to the platinum-based chemotherapy, because of its low expression associated with increased chemotherapeutic sensitivity (10). Therefore, these ERCC1 and ERCC2 SNPs may be useful prognostic markers for treatment with platinum agents.
Because published reports of an association between NER SNPs and clinical outcome of platinum-based chemotherapy from individual studies are not consistent, we performed a systemic review and meta-analysis to assess the evidence of effects of ERCC1 rs11615C>T and ERCC2 rs13181T>G SNPs on the efficacy of oxaliplatin-based chemotherapy in gastric and colorectal cancer patients.
Patients and Methods
Study selection
We searched for relevant publications before June 1st, 2010 in English literature by using electronic MEDLINE and EMBASE databases with the following terms “ERCC1”, “ERCC2 or XPD” or “ERCC”, “gastric or stomach cancer”, “colon or colorectal cancer”, “polymorphism or variant”, and “treatment or chemotherapy”. References of the retrieved articles were further screened for earlier original studies. The inclusion criteria were as follows: advanced, recurrent, or metastatic gastric or colorectal cancer; treated purely by regimens of FOLFOX (oxaliplatin plus 5-Fu/leucovorin) or XELOX (oxaliplatin plus capecitabine, a drug which converts to 5-Fu in vivo) (excluding neoadjuvant chemotherapy); cancer histologically or pathologically confirmed; East Asian (China, Korea and Japan) or Caucasian (European descendents) ethnicities; and ERCC1 rs11615C>T and or ERCC2 rs13181T>G genotyped. The corresponding authors were contacted to obtain missing information, and some studies were excluded if critical missing information was not obtained by our repeated requests. Abstracts, unpublished reports and articles with sample size less than 45 or written in non-English language were also excluded.
Statistical methods
We estimated the odds ratio (OR) for objective response versus no response after platinum-based chemotherapy [CR+PR vs. PD+SD, using the WHO criteria (11) or the Response Evaluation Criteria in Solid Tumors criteria (RECIST) (12)]. Progression-free survival (PFS) and overall survival (OS) were evaluated by pooled Cox proportional hazard ratios (HRs) and 95% confidence intervals (CIs) using published methods (13) because a meta-analysis of summary results is statistically as efficient as a joint analysis of individual participant data (14). We assessed the between-study heterogeneity by using the Cochran’s Q test with a significance level of P < 0.05. We performed initial analyses with a fixed-effect model and confirmatory analyses with a random-effect model, if there was significant heterogeneity. We used inverted funnel plots and the Egger’s test to examine the effect of publication bias. We compared the difference in the effect estimates between subgroups as described previously (15). All P values were two-sided, and all analyses were performed using the Stata software (Stata Corporation, Texas) and Review Manager (v5.0; Oxford, England).
Results
We identified 65 related publications by initial screening (as of June 1st, 2010), of which 21 publications seemed to meet the inclusion criteria. We excluded one study, in which data were inestimable and authors were unreachable (16), two studies that used other chemotherapeutic agents (i.e. irinotecan and cetuximab) in addition to FOLFOX or XELOX (17, 18), and one study with study sample size less than 45 (19) (Fig. 1). As a result, the final data pool consisted of 17 studies, including 1787 cancer patients (Table 1).
Fig. 1.
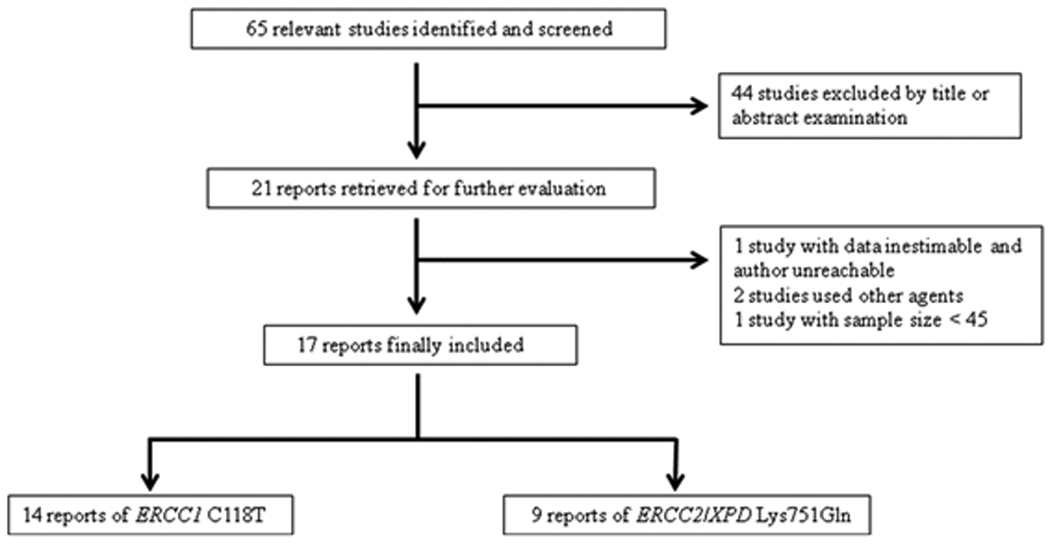
Study flow chart for the process of selecting the eligible publications.
Table 1.
Studies on oxaliplatin-based chemotherapy and ERCC1 (rs11615C>T) and ERCC2 (rs13181T>G) polymorphisms included in the meta-analysis.
| Study | Country | Tumor | Drug | No. | Biomarkers | SNPs | AlleleFreqa |
|---|---|---|---|---|---|---|---|
| Asians | |||||||
| Chang 2009 (21) | Taiwan | Colorectal | FOLFOX | 168 | TR, OS, PFS | rs11615 | T: 0.254 |
| Lai 2009 (34) | Taiwan | Colorectal | FOLFOX | 188 | TR, OS, PFS | rs13181 | G: 0.080 |
| Keam 2008 (22) | Korea | Gastric | FOLFOX | 73 | TR, OS, PFS | rs11615 rs13181 | T: 0.260G: 0.082 |
| Liang 2008 (35) | China | Colorectal | FOLFOX or XELOX | 99 | TR, PFS | rs11615 | T: 0.288 |
| Seo 2009 (36) | Korea | Gastric | FOLFOX | 75 | TR, OS, PFS | rs11615 | T: 0.240 |
| Huang 2008 (37) | China | Gastric | FOLFOX | 89 | OS, PFS | rs11615 | T: 0.281 |
| Liang 2009 (38) | China | Colorectal | FOLFOX or XELOX | 113 | OS | rs11615 | T: 0.323 |
| Caucasians | |||||||
| Le Morvan 2007 (39) | France | Colorectal | FOLFOX or XELOX | 59 | TR, OS, PFS | rs13181 | G: 0.381 |
| Paré 2008 (20) | Spain | Colorectal | FOLFOX | 126 | TR, OS, PFS | rs11615 rs13181 | T: 0.586G: 0.384 |
| Park 2001 (40) | USA | Colorectal | FOLFOX | 70 | TR | rs13181 | G: 0.421 |
| Chua 2009 (41) | Australia | Colorectal | FOLFOX | 115 | TR, OS, PFS | rs11615 | T: 0.635 |
| Spindler 2010 (42) | Denmark | Colorectal | XELOX | 66 | TR, PFSb | rs11615 | T: 0.652 |
| Viguier 2005 (43) | France | Colorectal | FOLFOX | 61 | TR | rs11615 | T: 0.557 |
| Ruzzo 2007 (44) | Italy | Colorectal | FOLFOX | 166 | PFS | rs11615 rs13181 | T: 0.557G: 0.443 |
| Stoehl-macher 2004 (45) | USA | Colorectal | FOLFOX | 106 | OS, PFS | rs11615 rs13181 | T: 0.505G: 0.373 |
| Martinez-Balibrea 2008 (46) | Spain | Colorectal | FOLFOX or XELOX | 96 | PFS | rs11615 rs13181 | T: 0.615G: 0.354 |
| Etienne-Grimaldi 2010 (47) | France | Colorectal | FOLFOX | 117 | TR, OS, PFS | rs11615 rs13181 | T: 0.538G: 0.385 |
| HapMapc | China | (normal) | 137136 | rs11615 rs13181 | T: 0.243G: 0.095 | ||
| Europe | (normal) | 113113 | rs11615 rs13181 | T: 0.642G: 0.332 |
ERCC1 rs11615C>T
Objective response
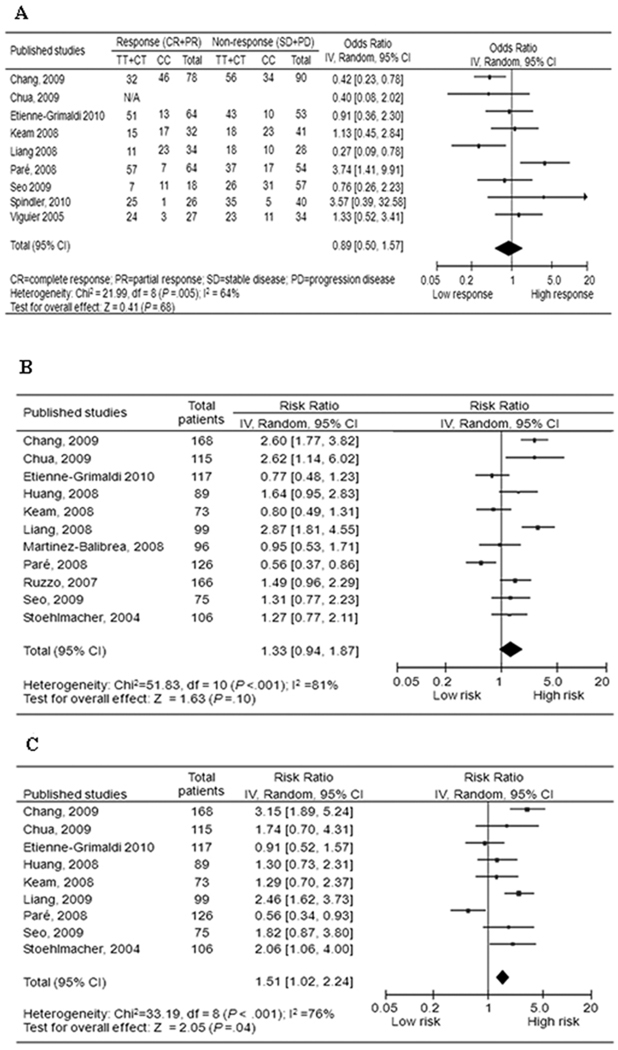
Nine studies including 855 patients were eligible for the final analysis. In the dominant model, the minor variant T allele was not associated with objective response in all patients (T/T+C/T versus C/C: OR, 0.89; 95% CI, 0.50–1.57) (Fig. 2A), and no single study altered the result substantially by the sensitivity test. However, stratified analysis by ethnicity showed a significant difference in the estimates of effect between Asians and Caucasians (P = 0.002), and the T allele was associated with a significantly lower objective response rate in Asians (OR, 0.53; 95% CI, 0.35–0.81). When only colorectal cancer was included, the OR was similar to that of the overall patients (OR, 0.88; 95% CI, 0.42–1.87) (Table 2). No publication bias was detected by either the funnel plot or Egger’s test (data not shown).
Fig. 2.
Forest plot of (A) objective response; (B) progression-free survival; and (C) overall survival in gastric and colorectal cancer patients treated with oxaliplatin-based therapies by ERCC1 rs11615C>T polymorphism (T/T+C/T vs. C/C, reference group = C/C).
Table 2.
Analysis of the association between ERCC1 rs11615C>T and ERCC2 rs13181T>G polymorphisms and objective response, PFS and OS
| Objective response | PFS | OS | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Studya | Fixed effect | Random effect | Study | Fixed effect | Random effect | Study | Fixed effect | Random effect | ||||
| (Cases) | T/T+T/C vs. C/C | T/T+T/C vs. C/C | Phetb | (Cases) | T/T+T/C vs. C/C | T/T+T/C vs. C/C | Phetb | (Cases) | T/T+T/C vs. C/C | T/T+T/C vs. C/C | Phetb | |
| ERCC1 rs11615C>T | ||||||||||||
| All | 9 (855) | 0.81 (0.58–1.12) | 0.89 (0.50–1.57) | 0.005 | 11(1230) | 1.33 (1.15–1.54) | 1.33 (0.94–1.87) | <0.001 | 9 (968) | 1.53 (1.27–1.85) | 1.51 (1.02–2.24) | <0.001 |
| Asian | 4 (378) | 0.53 (0.35–0.81) | 0.55 (0.31–0.98) | 0.158 | 5 (504) | 1.79 (1.45–2.21) | 1.69 (1.05–2.70) | <0.001 | 5 (504) | 2.03 (1.60–2.59) | 1.95 (1.37–2.78) | 0.078 |
| Caucasian | 5 (477) | 1.47 (0.89–2.43) | 1.44 (0.68–3.02) | 0.103 | 6 (726) | 1.00 (0.81–1.23) | 1.07 (0.72–1.59) | 0.004 | 4 (464) | 0.97 (0.72–1.32) | 1.10 (0.60–2.03) | 0.012 |
| Colorectal only | 7 (707) | 0.77 (0.54–1.12) | 0.88 (0.42–1.87) | 0.002 | 8 (993) | 1.38 (1.17–1.64) | 1.39 (0.89–2.17) | <0.001 | 6 (731) | 1.58 (1.27–1.98) | 1.55 (0.87–2.77) | <0.001 |
| ERCC2 rs13181T>G | G/G+G/T vs. T/T | G/G+G/T vs. T/T | G/G+G/T vs. T/T | G/G+G/T vs. T/T | G/G+G/T vs. T/T | G/G+G/T vs. T/T | ||||||
| All | 6 (625) | 0.53 (0.37–0.78) | 0.53 (0.36–0.78) | 0.588 | 8 (931) | 1.37 (1.15–1.63) | 1.41 (1.06–1.89) | 0.010 | 6 (669) | 1.61 (1.29–2.00) | 1.54 (0.96–2.50) | <0.001 |
| Asian | 2 (261) | 2 (261) | 2 (261) | |||||||||
| Caucasian | 4 (364) | 0.56 (0.35–0.88) | 0.56 (0.35–0.89) | 0.368 | 6 (670) | 1.33 (1.09–1.62) | 1.41 (1.02–1.95) | 0.022 | 4 (408) | 1.42 (1.11–1.81) | 1.42 (1.06–1.90) | 0.236 |
| Colorectal only | 5 (552) | 0.52 (0.35–0.77) | 0.52 (0.35–0.77) | 0.472 | 7 (858) | 1.42 (1.19–1.71) | 1.50 (1.11–2.02) | 0.008 | 5 (596) | 1.70 (1.36–2.13) | 1.77 (1.11–2.84) | 0.002 |
Progression-free survival
Eleven studies including 1230 patients were eligible for the final analysis. The T allele was associated with a non-significant increase of hazard for PFS in all patients (T/T+C/T versus C/C: HR, 1.33; 95% CI, 0.94 to 1.87) (Fig. 2B), and the single study by Paré et al. (20) showed substantial influence over the pooled result, the exclusion of which elevated the HR significantly (HR, 1.46; 95% CI, 1.07 to 1.99). Although stratified analysis by ethnicity showed a clinically substantial and statistically significant increase in the hazard of progression in Asian patients (HR, 1.69; 95% CI, 1.05 to 2.70), further comparison did not show significant difference in the estimates of effect between Asians and Caucasians (P = 0.147). When only colorectal cancer was included, the HR was similar to that of overall patients (HR, 1.39; 95% CI, 0.89 to 2.17) (Table 2). No publication bias was detected by either the funnel plot or the Egger’s test (data not shown).
Overall survival
Nine studies including 968 patients were eligible for the final analysis. There appeared a significant effect of ERCC1 rs11615C>T polymorphism on OS in all patients (T/T+C/T versus C/C: HR, 1.51; 95% CI, 1.02 to 2.24) (Fig. 2C). Further analysis demonstrated substantial influence from the single study of Chang et al. (21), the exclusion of which led to loss of significance of the pooled result (HR, 1.36; 95% CI, 0.92 to 2.02). Stratified analysis indicated a more pronounced effect in Asian patients (HR, 2.03; 95% CI, 1.60 to 2.59), compared with the Caucasian patients (HR, 1.10; 95% CI, 0.60 to 2.03), and a marginally significant difference existed in the estimates of effect between these two ethnicities (P = 0.064). When only colorectal cancer was included, the T allele was associated with a non-significant increased hazard of death (HR, 1.55; 95% CI, 0.87 to 2.77) (Table 2). No publication bias was detected by either the funnel plot or the Egger’s test (data not shown).
ERCC2 rs13181T>G
Objective response
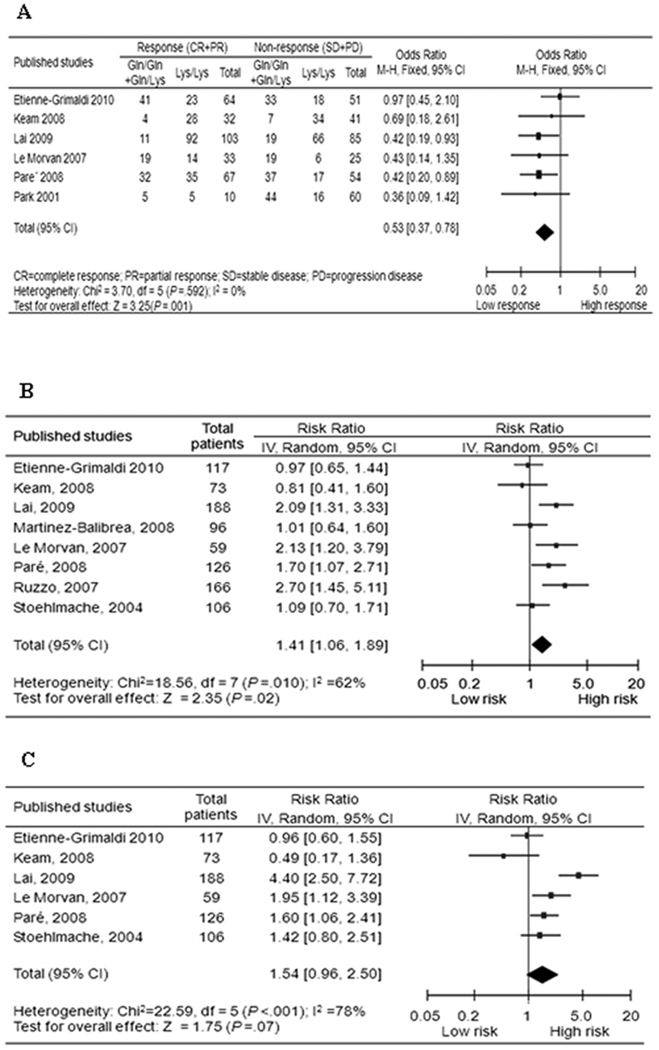
Six studies including 625 patients were eligible for the final analysis. The G allele was associated with a reduced objective response in all patients (G/G+G/T versus T/T: OR, 0.53; 95% CI, 0.37–0.78) (Fig. 3A), and no single study influenced the pooled result substantially. In stratified analyses, the association remained significant in subgroups of Caucasians (OR, 0.56; 95% CI, 0.35–0.88) and colorectal cancer (OR, 0.52; 95% CI, 0.35–0.77) (Table 2). No publication bias was detected by either the funnel plot or the Egger’s test (data not shown).
Fig. 3.
Forest plot of (A) objective response; (B) progression-free survival; and (C) overall survival in gastric and colorectal cancer patients treated with oxaliplatin-based therapies by ERCC2 rs13181T>G polymorphism (G/G+G/T vs. T/T, reference group = T/T).
Progression-free survival
Eight eligible studies of 931 patients were included in the final analysis, only two of which included Asians. Overall, there was a substantial effect of the G allele on progression hazard in all patients (G/G+G/T versus T/T: HR, 1.41; 95% CI, 1.06–1.89) (Fig. 3B and Table 2), and no single study influenced the pooled result substantially. In stratified analyses, the significance remained in subgroups of Caucasians (HR, 1.41; 95% CI, 1.02 to 1.95) and colorectal cancer (HR, 1.50; 95% CI, 1.11–2.02) (Table 2). No publication bias was detected by either the funnel plot or the Egger’s test (data not shown).
Overall survival
Six studies including 669 patients were eligible for the final analysis, again only two of which included Asians. The G allele was associated with a non-significant increase in the hazard of death in all patients (G/G+G/T versus T/T: HR, 1.54; 95% CI, 0.96 to 2.50) (Fig. 3C and Table 2), and the single study by Keam et al. (22) had a substantial influence over the pooled result, the exclusion of which elevated the HR significantly (HR, 1.77; 95% CI, 1.11 to 2.84). In stratified analyses, the significance remained in subgroups of Caucasians (HR, 1.42; 95% CI, 1.11–1.81), and colorectal cancer (HR, 1.77; 95% CI, 1.11–2.84). No publication bias was detected by either the funnel plot or the Egger’s test (data not shown).
Discussion
In this meta-analysis, we provided evidence of an association between ERCC1 rs11615C>T and ERCC2 rs13181T>G SNPs and clinical outcomes of Asians and Caucasians patients, respectively, with gastric and colorectal cancers that were treated by oxaliplatin-based chemotherapy.
Previous studies showed that clinical outcomes, measured as either tumor progression or survival, were better in patients susceptible to higher levels of platinum-induced DNA adduct (23, 24). Resistance to platinum may result from numerous mechanisms (25), among which NER is the predominant mechanism for moderate levels of platinum resistance seen clinically (26). There is evidence that cancer patients with congenital NER mutations are sensitive to platinum treatment and that hypersensitivity of testicular cancer to cisplatin is due to DNA repair deficiency (27, 28). ERCC1 and ERCC2 are two key rate-limiting enzymes in the multistep NER process. ERCC1, in collaboration with XPF protein, is involved in DNA lesion recognition, whereas ERCC2 is a subunit of human transcriptional initiation factor TFIIH with ATP-dependent helicase activity. Therefore, functional ERCC1 and ERCC2 SNPs may contribute directly to phenotypes of drug sensitivity by modifying related genes’ functions and reflect platinum sensitivity as an inborn trait.
Our meta-analysis used objective response, PFS and OS as primary parameters to assess the influence of NER SNPs on clinical outcomes of oxaliplatin-based chemotherapy, because these parameters are intrinsically correlated but not necessarily consistent with one another. Most often, a low objective response rate suggests tumor resistance to the chemotherapeutic regimen and a short PFS and OS is very likely the consequence. However, a high objective response rate may lead to an increased PFS and OS, or no survival benefit at all (29), demonstrating the necessity of including all three parameters to make a comprehensive assessment. In our meta-analysis, ERCC1 rs11615 T allele was a biomarker of low objective response, short PFS and OS in Asian patients, whereas ERCC2 rs13181 G allele showed significant or marginally significant association with low objective response, short PFS and OS in overall patients, Caucasians and colorectal cancer subgroups. Although some single studies may have influenced the significance of the pooled results, the association tendency was obvious with or without these studies. The consistent changes of three parameters strongly suggested that ERCC1 rs11615C>T and ERCC2 rs13181T>G both had an effect on oxaliplatin-based chemotherapy and that objective response could be a useful surrogate of survival in oxaliplatin-treated gastric and colorectal cancer patients.
Our results could be reasonably explained by the biological significance of these two SNPs. The rs11615 T allele of ERCC1 polymorphism was found to be associated with high mRNA expression of the corresponding gene (30), whereas the rs13181 G allele of ERCC2 polymorphism was found to be associated with a low number of X-ray induced chromatid aberrations (8). Functional studies confirmed a substantial influence of the ERCC1 rs11615C>T and ERCC2 rs13181T>G SNPs on the phenotype of NER capacity (7, 31, 32), and possessing the TT genotype of ERCC2 rs13181T>G SNP was associated with the risk of sub-optimal DNA repair up to seven fold, compared with the GG/GT genotype (8). Hence, patients carrying the ERCC1 rs11615 T or ERCC2 rs13181 G allele may have higher DNA repair capacity that could effectively reduce the anticancer effect of oxaliplatin, leading to poor prognosis of these patients.
Notably, there was an apparent ethnic discrepancy in the prognostic values between Asians and Caucasians, and statistical test also confirmed the existence of ethnical difference in the estimates of effect for the ERCC1 allele. As shown in Table 1, there was a remarkably lower prevalence of ERCC2 rs13181 G allele in Asians than in Caucasians, which might explain the lack of effect of ERCC2 rs13181T>G SNP in Asian patients. However, it is interesting to find that there was no predictive value of ERCC1 rs11615C>T SNP in Caucasians, even though the rs11615 T allele was much more common in Caucasians than in Asians. Although the underlying mechanisms are not clear, numerous factors may have played a role, such as gene-gene interaction from different genetic background, and gene-environmental interaction from different lifestyles. Additional large studies are warranted to investigate these possibilities.
Despite our efforts to make an accurate and comprehensive analysis, limitations of our meta-analysis need to be addressed. First, some data were excluded from our analysis because of loss of contact (16) or missing in the original study (33), which could cause some bias in our estimates but was unlikely to change our major conclusions, because Spindler et al. showed no association between ERCC1 rs11615C>T polymorphism and PFS in Caucasians (33), and Liu et al. showed no association between ERCC2 rs13181T>G polymorphism and OS in Asians (16), which were consistent with our findings. Second, most of the included studies were retrospective and differed significantly in study designs. In addition, the frequencies of ERCC1 rs11615 T and ERCC2 rs13181 G alleles were also substantially different among patient populations with different ethnicity. All these may have caused wide and significant heterogeneity between studies. Third, our analysis largely used unadjusted estimates, because not all published studies presented adjusted estimates or when they did, the estimates were not adjusted by the same potential confounders. However, when only those studies with the available adjusted estimates were used, the conclusions were not significantly changed (data now shown). Fourth, we were unable to analyze the association between ERCC1 and ERCC2 SNPs and platinum toxicities, because few studies provided this information or used different toxicity profiles. Finally, oxaliplatin is not used as a single compound but in combination with 5-Fu in the regimen, and unfortunately, we were unable to investigate potential gene-gene interactions between NER variants and folate metabolizing gene variants because of the limited publications available on this topic.
Overall, our meta-analysis demonstrated that ERCC1 rs11615C>T and ERCC2 rs13181T>G SNPs might be useful prognostic factors for assessing clinical outcomes of oxaliplatin-based chemotherapies (FOLFOX or XELOX) in gastric and colorectal cancers. However, future prospective studies with large sample sizes and better study designs are required to confirm our findings.
Translational Relevance
Combination treatment of oxaliplatin and fluoropyrimidines is the standard treatment for gastric and colorectal cancer, which improves patients’ response and overall survival. The nucleotide excision repair (NER) pathway is responsible for removal of DNA adducts caused by oxaliplatin and thus may influence chemotherapeutic efficacy. Our meta-analysis provided evidence of an association between NER ERCC1 rs11615C>T and ERCC2 rs13181T>G SNPs and clinical outcomes in gastric and colorectal cancer patients receiving oxaliplatin-based chemotherapy in both Asians and Caucasians. Our results suggest that it is feasible to use a pharmacogenomic approach to predict clinical outcomes of oxaliplatin-treated gastric and colorectal cancer patients.
Acknowledgement
We thank Bhumsuk Keam, Jim Paul, Albert Abad, Denis Smith, Valerie Le Morvan, Dionyssios Katsaros, Nick Thatcher and Anders Jakobsen for data coordination.
Funding sources:
This study was in part supported by the National Institutes of Health grants R01 CA131274 and R01 ES-011740 (Q. Wei) and P30 CA016672 (to M. D. Anderson Cancer Center). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
Role of the authors:
Ming Yin, Jingrong Yan, Qingyi Wei conceived the ideas, conducted literatures search and data collections; Eva Martinez-Balibrea, Francesco Graziano, Heinz-Josef Lenz, Hyo-Jin Kim, Jacques Robert, Seock-Ah Im, Wei-Shu Wang, Marie-Christine Etienne-Grimaldi provided the raw data of their original studies, and all authors contributed to the writing, revising, and approval for final submission.
Footnotes
Disclosure of Potential Conflicts of Interest:
The authors have declared no conflicts of interest.
References
- 1.Thirion P, Michiels S, Pignon JP, et al. Modulation of fluorouracil by leucovorin in patients with advanced colorectal cancer: an updated meta-analysis. J Clin Oncol. 2004;22:3766–3775. doi: 10.1200/JCO.2004.03.104. [DOI] [PubMed] [Google Scholar]
- 2.Wagner AD, Grothe W, Haerting J, Kleber G, Grothey A, Fleig WE. Chemotherapy in advanced gastric cancer: a systematic review and meta-analysis based on aggregate data. J Clin Oncol. 2006;24:2903–2909. doi: 10.1200/JCO.2005.05.0245. [DOI] [PubMed] [Google Scholar]
- 3.Ajani JA. Evolving chemotherapy for advanced gastric cancer. Oncologist. 2005;10 Suppl 3:49–58. doi: 10.1634/theoncologist.10-90003-49. [DOI] [PubMed] [Google Scholar]
- 4.Kelly H, Goldberg RM. Systemic therapy for metastatic colorectal cancer: current options, current evidence. J Clin Oncol. 2005;23:4553–4560. doi: 10.1200/JCO.2005.17.749. [DOI] [PubMed] [Google Scholar]
- 5.Faivre S, Chan D, Salinas R, Woynarowska B, Woynarowski JM. DNA strand breaks and apoptosis induced by oxaliplatin in cancer cells. Biochem Pharmacol. 2003;66:225–237. doi: 10.1016/s0006-2952(03)00260-0. [DOI] [PubMed] [Google Scholar]
- 6.Reed E. ERCC1 and clinical resistance to platinum-based therapy. Clin Cancer Res. 2005;11:6100–6102. doi: 10.1158/1078-0432.CCR-05-1083. [DOI] [PubMed] [Google Scholar]
- 7.Yu JJ, Lee KB, Mu C, et al. Comparison of two human ovarian carcinoma cell lines (A2780/CP70 and MCAS) that are equally resistant to platinum, but differ at codon 118 of the ERCC1 gene. Int J Oncol. 2000;16:555–560. doi: 10.3892/ijo.16.3.555. [DOI] [PubMed] [Google Scholar]
- 8.Lunn RM, Helzlsouer KJ, Parshad R, et al. XPD polymorphisms: effects on DNA repair proficiency. Carcinogenesis. 2000;21:551–555. doi: 10.1093/carcin/21.4.551. [DOI] [PubMed] [Google Scholar]
- 9.Duell EJ, Wiencke JK, Cheng TJ, et al. Polymorphisms in the DNA repair genes XRCC1 and ERCC2 and biomarkers of DNA damage in human blood mononuclear cells. Carcinogenesis. 2000;21:965–971. doi: 10.1093/carcin/21.5.965. [DOI] [PubMed] [Google Scholar]
- 10.Vilmar A, Sorensen JB. Excision repair cross-complementation group 1 (ERCC1) in platinum-based treatment of non-small cell lung cancer with special emphasis on carboplatin: a review of current literature. Lung Cancer. 2009;64:131–139. doi: 10.1016/j.lungcan.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 11.Miller AB, Hoogstraten B, Staquet M, Winkler A. Reporting results of cancer treatment. Cancer. 1981;47:207–214. doi: 10.1002/1097-0142(19810101)47:1<207::aid-cncr2820470134>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 12.Therasse P, Arbuck SG, Eisenhauer EA, et al. New guidelines to evaluate the response to treatment in solid tumors. European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst. 2000;92:205–216. doi: 10.1093/jnci/92.3.205. [DOI] [PubMed] [Google Scholar]
- 13.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin DY, Sullivan PF. Meta-analysis of genome-wide association studies with overlapping subjects. Am J Hum Genet. 2009;85:862–872. doi: 10.1016/j.ajhg.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Altman DG, Bland JM. Interaction revisited: the difference between two estimates. BMJ. 2003;326:219. doi: 10.1136/bmj.326.7382.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B, Wei J, Zou Z, et al. Polymorphism of XRCC1 predicts overall survival of gastric cancer patients receiving oxaliplatin-based chemotherapy in Chinese population. Eur J Hum Genet. 2007;15:1049–1053. doi: 10.1038/sj.ejhg.5201884. [DOI] [PubMed] [Google Scholar]
- 17.Zarate R, Rodriguez J, Bandres E, et al. Oxaliplatin, irinotecan and capecitabine as first-line therapy in metastatic colorectal cancer (mCRC): a dose-finding study and pharmacogenomic analysis. Br J Cancer. 2010;102:987–994. doi: 10.1038/sj.bjc.6605595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han SW, Oh DY, Im SA, et al. Epidermal growth factor receptor intron 1 CA dinucleotide repeat polymorphism and survival of advanced gastric cancer patients treated with cetuximab plus modified FOLFOX6. Cancer Sci. 2010;101:793–799. doi: 10.1111/j.1349-7006.2009.01447.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monzo M, Moreno I, Navarro A, et al. Single nucleotide polymorphisms in nucleotide excision repair genes XPA, XPD, XPG and ERCC1 in advanced colorectal cancer patients treated with first-line oxaliplatin/fluoropyrimidine. Oncology. 2007;72:364–370. doi: 10.1159/000113534. [DOI] [PubMed] [Google Scholar]
- 20.Pare L, Marcuello E, Altes A, et al. Pharmacogenetic prediction of clinical outcome in advanced colorectal cancer patients receiving oxaliplatin/5-fluorouracil as first-line chemotherapy. Br J Cancer. 2008;99:1050–1055. doi: 10.1038/sj.bjc.6604671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang PM, Tzeng CH, Chen PM, et al. ERCC1 codon 118 C-->T polymorphism associated with ERCC1 expression and outcome of FOLFOX-4 treatment in Asian patients with metastatic colorectal carcinoma. Cancer Sci. 2009;100:278–283. doi: 10.1111/j.1349-7006.2008.01031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keam B, Im SA, Han SW, et al. Modified FOLFOX-6 chemotherapy in advanced gastric cancer: Results of phase II study and comprehensive analysis of polymorphisms as a predictive and prognostic marker. BMC Cancer. 2008;8:148. doi: 10.1186/1471-2407-8-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van de Vaart PJ, Belderbos J, de Jong D, et al. DNA-adduct levels as a predictor of outcome for NSCLC patients receiving daily cisplatin and radiotherapy. Int J Cancer. 2000;89:160–166. [PubMed] [Google Scholar]
- 24.Reed E, Ozols RF, Tarone R, Yuspa SH, Poirier MC. Platinum-DNA adducts in leukocyte DNA correlate with disease response in ovarian cancer patients receiving platinum-based chemotherapy. Proc Natl Acad Sci U S A. 1987;84:5024–5028. doi: 10.1073/pnas.84.14.5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Shellard SA, Fichtinger-Schepman AM, Lazo JS, Hill BT. Evidence of differential cisplatin-DNA adduct formation, removal and tolerance of DNA damage in three human lung carcinoma cell lines. Anticancer Drugs. 1993;4:491–500. doi: 10.1097/00001813-199308000-00011. [DOI] [PubMed] [Google Scholar]
- 27.Furuta T, Ueda T, Aune G, Sarasin A, Kraemer KH, Pommier Y. Transcription-coupled nucleotide excision repair as a determinant of cisplatin sensitivity of human cells. Cancer research. 2002;62:4899–4902. [PubMed] [Google Scholar]
- 28.Kelland LR, Mistry P, Abel G, et al. Establishment and characterization of an in vitro model of acquired resistance to cisplatin in a human testicular nonseminomatous germ cell line. Cancer research. 1992;52:1710–1716. [PubMed] [Google Scholar]
- 29.Oye RK, Shapiro MF. Reporting results from chemotherapy trials. Does response make a difference in patient survival? JAMA. 1984;252:2722–2725. [PubMed] [Google Scholar]
- 30.Park D, Stoehlmacher J, Zhang W, Tsao-Wei D, Groshen S, H-J L. ERCC1 polymorphism is associated with differential ERCC1 gene expression. American Association for Cancer Research; San Francisco, CA. ASCO Proceedings; 2002. [Google Scholar]
- 31.Shi Q, Wang LE, Bondy ML, Brewster A, Singletary SE, Wei Q. Reduced DNA repair of benzo[a]pyrene diol epoxide-induced adducts and common XPD polymorphisms in breast cancer patients. Carcinogenesis. 2004;25:1695–1700. doi: 10.1093/carcin/bgh167. [DOI] [PubMed] [Google Scholar]
- 32.Qiao Y, Spitz MR, Shen H, et al. Modulation of repair of ultraviolet damage in the host-cell reactivation assay by polymorphic XPC and XPD/ERCC2 genotypes. Carcinogenesis. 2002;23:295–299. doi: 10.1093/carcin/23.2.295. [DOI] [PubMed] [Google Scholar]
- 33.Spindler KL, Andersen RF, Jensen LH, Ploen J, Jakobsen A. EGF61A>G polymorphism as predictive marker of clinical outcome to first-line capecitabine and oxaliplatin in metastatic colorectal cancer. Ann Oncol. 2010;21:535–539. doi: 10.1093/annonc/mdp336. [DOI] [PubMed] [Google Scholar]
- 34.Lai JI, Tzeng CH, Chen PM, et al. Very low prevalence of XPD K751Q polymorphism and its association with XPD expression and outcomes of FOLFOX-4 treatment in Asian patients with colorectal carcinoma. Cancer Sci. 2009;100:1261–1266. doi: 10.1111/j.1349-7006.2009.01186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang JLH, Yao R, Liang H, Wu G. ERCC1 Asn118Asn polymorphism as predictor for cancer response to oxaliplatin-based chemotherapy in patients with advanced colorectal cancer. Chinese-German Journal of Clinical Oncology. 2008;7:455–459. [Google Scholar]
- 36.Seo BG, Kwon HC, Oh SY, et al. Comprehensive analysis of excision repair complementation group 1, glutathione S-transferase, thymidylate synthase and uridine diphosphate glucuronosyl transferase 1A1 polymorphisms predictive for treatment outcome in patients with advanced gastric cancer treated with FOLFOX or FOLFIRI. Oncol Rep. 2009;22:127–136. [PubMed] [Google Scholar]
- 37.Huang ZH, Hua D, Du X, et al. ERCC1 polymorphism, expression and clinical outcome of oxaliplatin-based adjuvant chemotherapy in gastric cancer. World J Gastroenterol. 2008;14:6401–6407. doi: 10.3748/wjg.14.6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang J, Jiang T, Yao RY, Liu ZM, Lv HY, Qi WW. The combination of ERCC1 and XRCC1 gene polymorphisms better predicts clinical outcome to oxaliplatin-based chemotherapy in metastatic colorectal cancer. Cancer Chemother Pharmacol. 2009;66:493–500. doi: 10.1007/s00280-009-1186-3. [DOI] [PubMed] [Google Scholar]
- 39.Le Morvan V, Smith D, Laurand A, et al. Determination of ERCC2 Lys751Gln and GSTP1 Ile105Val gene polymorphisms in colorectal cancer patients: relationships with treatment outcome. Pharmacogenomics. 2007;8:1693–1703. doi: 10.2217/14622416.8.12.1693. [DOI] [PubMed] [Google Scholar]
- 40.Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei DD, Groshen S, Lenz HJ. A Xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res. 2001;61:8654–8658. [PubMed] [Google Scholar]
- 41.Chua W, Goldstein D, Lee CK, et al. Molecular markers of response and toxicity to FOLFOX chemotherapy in metastatic colorectal cancer. Br J Cancer. 2009;101:998–1004. doi: 10.1038/sj.bjc.6605239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spindler KL, Andersen RF, Jensen LH, Ploen J, Jakobsen A. EGF61A>G polymorphism as predictive marker of clinical outcome to first-line capecitabine and oxaliplatin in metastatic colorectal cancer. Ann Oncol. 21:535–539. doi: 10.1093/annonc/mdp336. [DOI] [PubMed] [Google Scholar]
- 43.Viguier J, Boige V, Miquel C, et al. ERCC1 codon 118 polymorphism is a predictive factor for the tumor response to oxaliplatin/5-fluorouracil combination chemotherapy in patients with advanced colorectal cancer. Clin Cancer Res. 2005;11:6212–6217. doi: 10.1158/1078-0432.CCR-04-2216. [DOI] [PubMed] [Google Scholar]
- 44.Ruzzo A, Graziano F, Loupakis F, et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first-line FOLFOX-4 chemotherapy. J Clin Oncol. 2007;25:1247–1254. doi: 10.1200/JCO.2006.08.1844. [DOI] [PubMed] [Google Scholar]
- 45.Stoehlmacher J, Park DJ, Zhang W, et al. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5-FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer. 2004;91:344–354. doi: 10.1038/sj.bjc.6601975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez-Balibrea E, Abad A, Aranda E, et al. Pharmacogenetic approach for capecitabine or 5-fluorouracil selection to be combined with oxaliplatin as first-line chemotherapy in advanced colorectal cancer. Eur J Cancer. 2008;44:1229–1237. doi: 10.1016/j.ejca.2008.03.025. [DOI] [PubMed] [Google Scholar]
- 47.Etienne-Grimaldi MC, Milano G, Maindrault-Goebel F, et al. Methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms and FOLFOX response in colorectal cancer patients. Br J Clin Pharmacol. 2010;69:58–66. doi: 10.1111/j.1365-2125.2009.03556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]