TLR signaling is required for virulence of an intracellular pathogen (original) (raw)
. Author manuscript; available in PMC: 2012 Mar 4.
Summary
Toll-like receptors (TLRs) contribute to host resistance to microbial pathogens and drive the evolution of virulence mechanisms. We have examined the relationship between host resistance and pathogen virulence using mice with a functional allele of the Nramp-1 gene and lacking combinations of TLRs. Mice deficient in both TLR2 and TLR4 were highly susceptible to the intracellular bacterial pathogen Salmonella typhimurium, consistent with reduced innate immune function. However, mice lacking additional TLRs involved in S. typhimurium recognition were less susceptible to infection. In these TLR-deficient cells, bacteria failed to upregulate Salmonella pathogenicity island 2 (SPI-2) genes and did not form a replicative compartment. We demonstrate that TLR signaling enhances the rate of acidification of the Salmonella containing phagosome, and inhibition of this acidification prevents SPI-2 induction. Our results indicate that S. typhimurium requires cues from the innate immune system to regulate virulence genes necessary for intracellular survival, growth, and systemic infection.
Introduction
During early stages of infection the innate immune system is essential for limiting microbial replication and spread before an adaptive response is mounted. Accordingly, pathogens have evolved virulence strategies to antagonize innate immune function (Hedrick, 2004; Rausher, 2001; Woolhouse et al., 2002). The interplay between host innate immune function and pathogen virulence mechanisms largely determines the outcome of most infections. Despite the logic of this conceptual framework, our understanding of the molecular interactions driving the emergence of virulence mechanisms remains relatively poor.
Innate immune receptors detect infection by recognizing conserved microbial features common to broad classes of microbes (Janeway, 1989; Medzhitov, 2007). The Toll-like receptors (TLRs) target a range of microbial ligands, including lipopolysaccharide (TLR4), lipoproteins (TLR2), flagellin (TLR5), unmethylated CpG motifs in DNA (TLR9), double-stranded RNA (TLR3), and single-stranded RNA (TLR7 and TLR8) (Akira et al., 2001; Kawai and Akira, 2005). Expression of TLRs on innate immune cells links microbial recognition to induction of antimicrobial mechanisms, such as production of reactive oxygen and nitrogen species and expression of antimicrobial peptides (AMPs). In addition, TLR activation can promote adaptive immunity through control of dendritic cell (DC) maturation (Iwasaki and Medzhitov, 2004).
To study the evolution of pathogen virulence and its relationship to innate immunity, we have focused on TLR-mediated recognition of Salmonella enterica serovar typhimurium. S. typhimurium is a gram-negative bacterium that can survive and replicate within host macrophages (Coburn et al., 2007). Survival within macrophages requires a set of genes, many of which are encoded within Salmonella pathogenicity island 2 (SPI-2) (Galan, 2001; Shea et al., 1996; Waterman and Holden, 2003). SPI-2 encodes a type 3 secretion system (T3SS) that is expressed after the bacterium is phagocytosed (Cirillo et al., 1998; Pfeifer et al., 1999; Valdivia and Falkow, 1997). Translocation of SPI-2 effectors into the host cell transforms the phagosome into a compartment that supports bacterial replication, the Salmonella containing vacuole (SCV) (Marcus et al., 2000). Multiple signals have been implicated in the transcriptional induction of SPI-2, including cation deprivation, phosphate starvation, and low pH (Chakravortty et al., 2005; Cirillo et al., 1998; Deiwick et al., 1999; Kim and Falkow, 2004; Rappl et al., 2003). Most of the studies implicating these signals have been performed on bacteria grown in vitro; whether the same signals are responsible for induction of SPI-2 genes within the phagosome remains unclear.
Recognition of S. typhimurium is largely mediated by TLR2, TLR4, and TLR5 (Feuillet et al., 2006; Hapfelmeier et al., 2005; O’Brien et al., 1980; Royle et al., 2003; Smith et al., 2003; Uematsu et al., 2006; Vazquez-Torres et al., 2004). Consistent with a central role for these receptors, S. typhimurium has evolved mechanisms to subvert this recognition or to avoid the consequences of TLR activation. For example, modification of lipid A by pagP reduces recognition by TLR4, although this modification is probably most relevant for resistance to AMPs (Bader et al., 2005; Detweiler et al., 2003; Guo et al., 1997; Guo et al., 1998). Mice lacking TLRs, especially TLR4, are more susceptible to S. typhimurium (Weiss et al., 2004). To circumvent the problem of redundancy, many studies have used mice lacking the common TLR adaptor MyD88 or lacking both MyD88 and another adaptor, TRIF (Hapfelmeier et al., 2005; Weiss et al., 2004). While these mice are very susceptible to S. typhimurium, these studies suffer from the caveat that MyD88 is also required for signaling by members of the IL-1 receptor (IL-1R) family. Because mice deficient in IL-1R are more susceptible to infection, the phenotype of MyD88-KO mice cannot be unequivocally attributed to TLRs (Mayer-Barber et al., 2010; Raupach et al., 2006).
In the studies described here, we sought to eliminate TLR-based recognition of S. typhimurium and examine the effect on pathogen virulence, while avoiding the caveats associated with MyD88-KO mice. In addition, we were concerned that the extreme susceptibility of C57Bl/6 mice (the genetic background on which most studies with TLR-KO mice have been performed) to S. typhimurium infection might mask any relationships between TLRs and bacterial virulence strategies. Many inbred mouse strains, including C57Bl/6, possess a nonfunctional allele of the nramp-1 gene. Nramp-1 encodes a multipass transmembrane protein that localizes to lysosomes and functions as a transporter of divalent cations, and mice with the non-functional allele are extremely susceptible to a number of intracellular pathogens (Forbes and Gros, 2001; Govoni et al., 1996; Vidal et al., 1995; Vidal et al., 1993; Vidal et al., 1996).
To avoid the caveats associated with nonfunctional Nramp-1 and TLR-independent functions of MyD88, we generated mice with a functional allele of nramp1 that lack individual or multiple TLRs. Studies in these mice led to a striking finding. While mice lacking a subset of the TLRs involved in S. typhimurium recognition showed increased susceptibility to infection, a lack of additional TLRs resulted in reduced susceptibility. The loss of virulence correlated with an inability of bacteria to survive and replicate within macrophages. We show that TLR signaling leads to rapid acidification of the SCV, and this signal is required for regulation of virulence gene expression. In the absence of this contextual cue, S. typhimurium is unable to survive and replicate intracellularly. Altogether, this work describes the molecular interactions underlying a bacterial pathogen’s dependence on the innate immune system for virulence.
Results
Multiple TLRs are involved in recognition of S. typhimurium
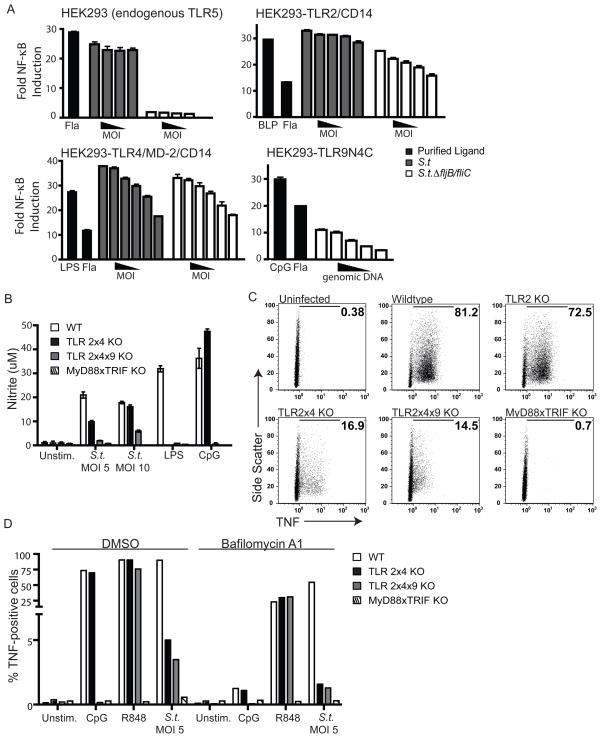
To identify which TLRs are relevant for innate recognition of S. typhimurium, we utilized HEK293 reporter cell lines expressing an NF-κB-luciferase reporter construct. Stimulation of these cells with heat-killed bacteria resulted in robust induction of NF-κB, which we attributed to endogenous TLR5 expressed by these cells (Figure 1A). This response was abrogated when cells were stimulated with bacteria lacking flagellin. To measure activation of other TLR family members, HEK293 reporter cells stably expressing individual TLRs were stimulated with bacteria lacking flagellin (to eliminate the contribution of endogenous TLR5). Using this approach, we observed activation of TLR2 and TLR4 by S. typhimurium (Figure 1A). Furthermore, S. typhimurium genomic DNA was capable of activating a surface localized version of TLR9 (Figure 1A).
Figure 1. Multiple TLRs recognize products of S. typhimurium.
(A) HEK293 cells expressing the indicated TLRs or TLR accessory proteins together with an NF-κB luciferase reporter were treated with heat-killed wildtype LT2 S. typhimurium (S.t.), flagellin-deficient LT2 (_S.t.Δ_fljB/fliC), or genomic DNA isolated from flagellin-deficient LT2. Luciferase activity was measured after 8h. Relative MOI range: 100 to 3.125, DNA concentration: 375 ng/mL to 12.5 ng/mL. Data are representative of 2 independent experiments and shown as mean +/− SEM. LPS, lipopolysaccharide; Fla, flagellin; BLP, bacterial lipopeptide. (B) BMMs differentiated from the indicated mice were treated overnight with 100 U/mL recombinant IFN-γ and infected the next morning with wildtype S. typhimurium (SL1344) at the indicated MOI. Nitrite production was measured 36h post-infection by Griess assay. Data are representative of 3 independent experiments and presented as mean +/− SD. (C) BMMs were infected as in (B) at an MOI of 5 for 8h, followed by intracellular cytokine staining for TNF. Percent TNF-positive cells are indicated in each panel. Data are representative of 3 independent experiments. (D) BMMs pre-treated for 2h with bafilomycinA1 or DMSO vehicle were infected with S. typhimurium (SL1344) at an MOI of 5 for 6h. Cells were processed and stained for intracellular cytokine staining as in (C).
While these results confirmed that TLR2, TLR4, TLR5, and TLR9 may play a role in recognition of S. typhimurium, they did not address the relative importance of each TLR during infection. To this end, we infected bone marrow-derived macrophages (BMMs) lacking combinations of TLRs and measured production of nitric oxide (NO). In agreement with previously published studies, BMMs lacking both TLR2 and TLR4 (TLR2x4-KO) produced much less NO than wildtype BMMs (Figure 1B). The remaining response was partially dependent on TLR9, as BMMs lacking TLR2, TLR4, and TLR9 (TLR2x4x9-KO) produced even less NO. Similar results were observed when tumor necrosis factor alpha (TNF) production was measured (Figure 1C). Importantly, all genotypes of BMMs responded equivalently to the TLR7 ligand R848 indicating that the cells were otherwise equivalent (Figure 1D and S1). The small amount of TNF and NO produced in TLR2x4x9-KO BMMs was dependent on other TLRs, as BMMs lacking both MyD88 and TRIF (and therefore all TLR-dependent signaling) did not respond to S. typhimurium (Figure 1B & C). As TLR5 is not expressed in murine BMMs, we reasoned that the residual TNF and NO produced by TLR2x4x9-KO BMMs was most likely due to TLR7 or TLR3 signaling. To address this possibility directly, we pretreated TLR2x4x9-KO BMMs with bafilomycinA1, an inhibitor of the vacuolar ATPase (V-ATPase) that prevents activation of endosomal TLRs. BafilomycinA1 treatment inhibited TNF production in TLR2x4-KO and TLR2x4x9-KO BMMs to almost background levels, suggesting that TLR7 and/or TLR3 are responsible for the remaining TNF production in response to S. typhimurium (Figure 1D). Collectively, these data indicate that TLR2, TLR4, TLR9, and TLR7 (and/or TLR3) each contribute to the recognition of S. typhimurium in infected BMMs.
TLR signaling is required for S. typhimurium virulence
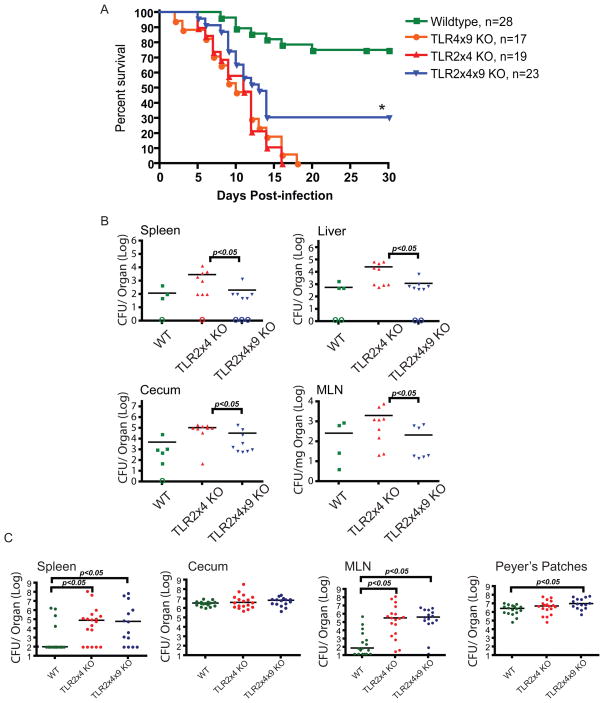
Having established which TLRs respond to ligands derived from S. typhiumurium in BMMs, we sought to test the effect of TLR deficiency on bacterial virulence in vivo. We crossed a functional allele of the nramp1 gene onto the C57BL/6 background and generated TLR-deficient or TLR adaptor-deficient mice with functional Nramp1 (see Extended Experimental Procedures). We expected that reduced TLR function would lead to greater susceptibility to infection. Indeed, all TLR2x4-KO mice died within 16 days when challenged orally with S. typhimurium, while 75% of the wildtype mice survived for the duration of the experiment (Figure 2A). By contrast, TLR2x4x9-KO mice were less susceptible to infection than TLR2x4-KO mice, despite a greater impairment in TLR function (Figure 2A). This increased survival was not a consequence of reduced immunopathology due to reduced TLR function. In fact, TLR2x4x9-KO mice had lower numbers of bacteria 4 days post-infection in spleens, livers, ceca and mesenteric lymph nodes (MLNs) relative to TLR2x4-KO mice (Figure 2B). Thus, despite less robust innate immune function, S. typhimurium was less virulent in TLR2x4x9-KO mice.
Figure 2. TLR2x4x9-KO mice are less susceptible to S. typhimurium than TLR2x4-KO mice.
(A) Survival plots of mice orally inoculated with 1.6 × 108 CFU S. typhimurium (SL1344) are shown. * p<0.005 by log-rank curve comparison test. Data are representative of at least 2 independent experiments (B) Groups of 8–10 week-old mice were orally inoculated with 1 × 109 CFU S. typhimurium (SL1344). Four days post-infection organs were harvested and homogenized for colony enumeration. Data are representative of at least 3 independent experiments. (C) Groups of mice of the indicated genotype were orally inoculated with 2 × 108 Y. entericolitica and CFU were measured in the indicated organs 3 days post-infection. Data presented are the combined results from 2 independent experiments. For (B) and (C), bars represent mean CFU of all mice, with data significance determined by Mann-Whitney U test. Open circles indicate mice for which no colonies were detected. MLN, mesenteric lymph node. See also Figure S2.
The difference in susceptibility between TLR2x4-KO and TLR2x4x9-KO mice could indicate that TLR9 plays a negative role in immunity to S. typhimurium. To test this possibility, we challenged mice lacking TLR4 and TLR9 (TLR4x9-KO). We reasoned that if TLR9 were playing a negative role in immunity, then any genotype lacking TLR9 would be resistant to infection. Instead, TLR4x9-KO mice were as susceptible to infection as TLR2x4-KO mice, indicating that lack of TLR9 by itself does not confer increased resistance to infection (Figure 2A). Thus, the data presented suggest that overall TLR signaling is in some way required for S. typhiumurium virulence. Despite this apparent requirement, MyD88-KO and MyD88xTRIF-KO mice (with wildtype Nramp1) were highly susceptible to S. typhimurium infection (Figure S2). As discussed earlier, the extreme sensitivity of these mice relative to TLR2x4x9-KO mice is likely due to the role of MyD88 downstream of the IL-1, IL-18, and IL-33 receptors (Mayer-Barber et al., 2010; Raupach et al., 2006). Thus, to examine the role for TLR signaling in S. typhimurium virulence we must use TLR-deficient mice, not mice lacking common signaling adaptors.
One potential caveat of these in vivo studies is that the commensal flora may be different between TLR2x4-KO and TLR2x4x9-KO mice. Recent studies have reported alterations in commensal communities in mice lacking certain TLRs or TLR signaling adaptors (Vijay-Kumar et al., 2008; Wen et al., 2008). To address this possibility, we challenged mice with a different gram-negative enteric pathogen, Yersinia entericolitica (Y. entericolitica), which shares a similar route of intestinal colonization but remains extracellular after crossing the intestinal epithelia. In contrast to our experiments with S. typhimurium, TLR2x4x9-KO mice were equally, if not more, susceptible relative to TLR2x4-KO mice (Figure 2C). The differential sensitivity of TLR2x4x9-KO mice to these two enteric bacteria argues that alterations in commensal flora are not contributing to the phenotypes of TLR2x4-KO and TLR2x4x9-KO mice. Instead, the reduction in TLR signaling in TLR2x4x9-KO mice appears to specifically impact the virulence of S. typhimurium.
TLR signaling is required for intracellular growth of bacteria
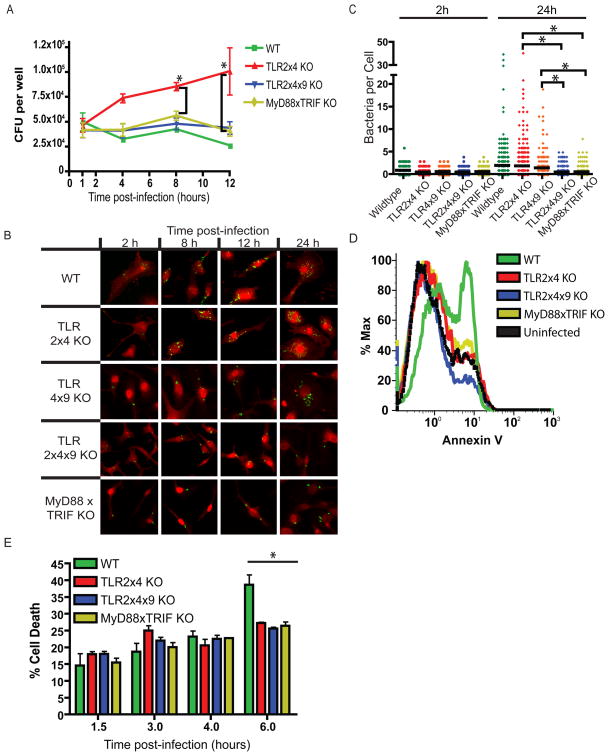
Because survival within macrophages is required for systemic infection (Fields et al., 1986; Leung and Finlay, 1991), we next examined survival and replication in BMMs lacking various TLRs by gentamicin protection assay. Consistent with our in vivo experiments, S. typhimurium was able to replicate in TLR2x4-KO but not TLR2x4x9-KO BMMs (Figure 3A). When we counted bacteria in individual BMMs by immunofluorescence microscopy (IF), the number of bacteria per cell in TLR2x4-KO BMMs accumulated over time, while the number of bacteria per cell in TLR2x4x9-KO BMMs remained constant, indicating that bacterial replication was responsible for the differences in CFU between genotypes (Figure 3B, C). We observed a similar lack of bacterial replication in MyD88xTRIF-KO BMMs (Figure 3A). Unlike our in vivo experiments, the phenotype of MyD88xTRIF-KO BMMs is most likely due to a deficiency in TLR signaling, as the IL-1 receptor family is not involved in the initial recognition of S. typhimurium within BMMs in vitro. Furthermore, TLR4x9-KO BMMs supported bacterial replication similarly to TLR2x4-KO BMMs, corroborating the conclusions from our in vivo experiments (Figure 3B, C). TLR2x4x9-KO and MyD88xTRIF-KO BMMs did support replication of Listeria monocytogenes and Legionella pneumophila (Figure S3). In addition, S. typhimurium replicated well in MyD88xTRIF-KO BMMs lacking functional Nramp1 (Figure S3). These results indicate that phagosomes of TLR-deficient cells are formally capable of supporting bacterial growth, but the combination of functional Nramp1 and lack of TLR signaling prevents S. typhimurium replication.
Figure 3. S. typhimurium is unable to replicate in TLR2x4x9-KO and MyD88xTRIF-KO BMMs.
(A) BMMs derived from mice of the indicated genotypes were infected with S. typhimurium (SL1344) at an MOI of 1, and intracellular CFU were measured by gentamicin protection assay at the indicated timepoints. Data are presented as the average of 3 independent experiments +/− SEM. * p<0.05 by student t test comparing TLR2x4-KO to MyD88xTRIF-KO and TLR2x4x9-KO. (B) BMMs infected as described in (A) were fixed and permeablized at the indicated times post-infection followed by staining with anti-Salmonella LPS antibody (green) and wheat germ agglutinin (red). (C) Intracellular bacteria per cell were counted in random fields at the 2 and 24h time-point from z-stacked images as shown in (B). Data are representative of 2 independent experiments, p value determined by student t test (* p<0.05). (D) BMMs of the indicated genotypes were infected with S. typhimurium (SL1344) at an MOI of 5. 8h post-infection, cells were harvested and stained with Annexin V. Data are representative of 2 independent experiments. (E) BMMs were infected at an MOI of 10 with S. typhimurium (SL1344), and release of lactate dehydrogenase (LDH) was measured in supernatants at the indicated time-points. Data are presented as mean +/− SD and are representative of at least 2 independent experiments; p value determined by student t test (* p<0.05) comparing wildtype to all other genotypes. See also Figure S3.
Collectively, these data suggest that S. typhimurium requires TLR signaling for replication in macrophages. However, the lack of replication in wildtype BMMs would appear to contradict this conclusion, as TLR function is normal in these cells. When we counted the number of bacteria per cell by IF, though, we observed a similar increase in bacteria per cell over time as in TLR2x4-KO BMMs (Figure 3C). This contradiction was resolved when we measured cell death of BMMs after infection. Wildtype BMMs exhibited greater cell death relative to each of the other genotypes (Figure 3D, E). Because only wildtype BMMs express functional TLR4, the increased death of these cells seems likely due to a previously described TLR4-dependent cell death that occurs in S. typhimurium infected cells (Cook et al., 2007; Hsu et al., 2004; Park et al., 2002). Thus, the apparent lack of replication as measured by CFU in wildtype BMMs is the result of macrophage death followed by gentamicin-mediated killing of the bacteria. In contrast, the inability of S. typhimurium to replicate in TLR2x4x9-KO or MyD88xTRIF-KO BMMs is due to a different mechanism.
TLR signaling is required for establishment of the SCV
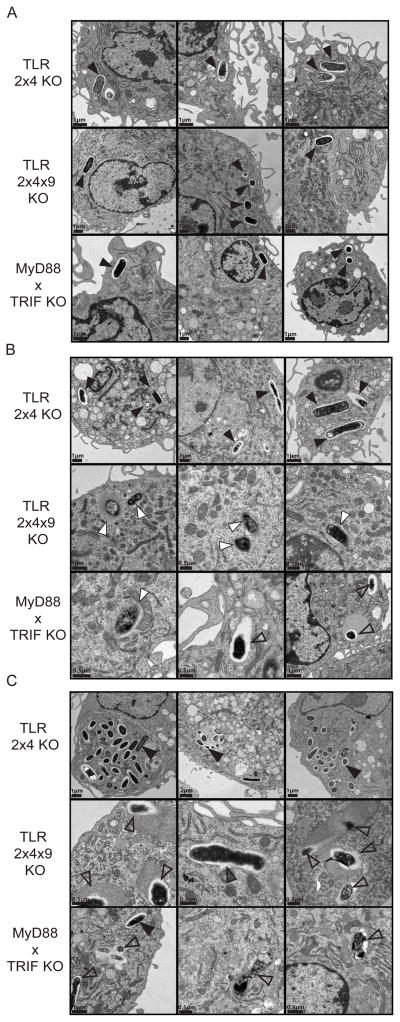
To better understand why S. typhimurium is unable to replicate in TLR2x4x9-KO and MyD88xTRIF-KO BMMs, we investigated the fate of bacteria in infected BMMs by transmission electron microscopy (EM). At 2h post-infection, bacteria were clearly visible in well-defined vacuoles in BMMs of all three genotypes (Figure 4A, black triangles). By 8h and 22h post-infection, bacteria in TLR2x4-KO BMMs remained largely unchanged, although evidence of replication was evident, especially at 22h (Figure 4B, C). In contrast, phagosomes containing bacteria in TLR2x4x9-KO and MyD88xTRIF-KO BMMs were quite distinct. The bacteria often appeared mottled or irregular in shape, and in many cases bacteria were surrounded by electron dense staining material consistent with lysosomal fusion (Figure 4, open triangles). In some instances, bacteria were no longer surrounded by membrane, suggesting they entered the cytosol (Figure 4, white triangles). Cytosolic bacteria have been described when bacteria fail to secrete certain SPI-2 effectors (Beuzon et al., 2000). In total, the images clearly demonstrate a defect in the ability of S. typhimurium to establish a replicative compartment in TLR2x4x9-KO and MyD88xTRIF-KO BMMs.
Figure 4. S. typhimurium fails to form an SCV in TLR2x4x9-KO and MyD88xTRIF-KO BMMs.
TLR2x4-KO, TLR2x4x9-KO, or MyD88xTRIF-KO BMMs were infected at an MOI of 10 with S. typhimurium (SL1344), and cells were fixed and processed for electron microscopy at 2h (A), 8h (B), and 22h (C) post-infection. Bacteria in intact vacuoles are shown with filled black arrowheads, cytosolic bacteria with filled white arrowheads, and bacteria that are degraded or have fused with lytic compartments are indicated with open black arrowheads. Micron bars are in the lower left corner of each panel. Three representative images are shown from different sections and independent infections.
Induction of SPI-2 genes by TLR signaling
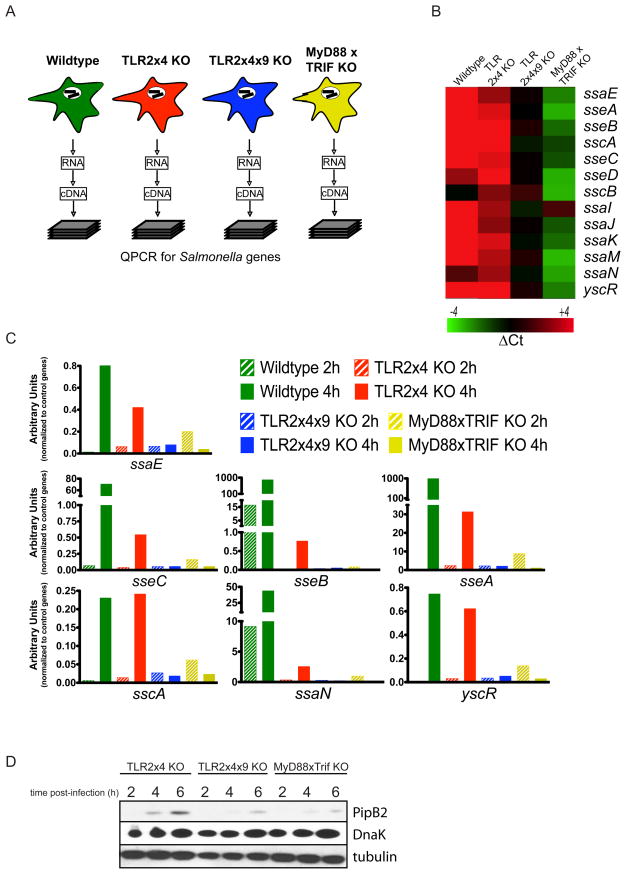
Our studies thus far indicate that intracellular growth of S. typhimurium is impaired in TLR2x4x9-KO and MyD88xTRIF-KO BMMs and suggest that this defect may be related to inefficient SCV formation. We next sought to define the underlying basis for impaired growth in BMMs lacking TLR function by profiling gene expression of bacteria isolated from BMMs of each genotype. To overcome the lack of sensitivity of microarray-based approaches, we performed quantitative RT-PCR to measure expression of all genes in the S. typhimurium genome (Figure 5A).
Figure 5. S. typhimurium fails to upregulate and secrete SPI-2 effectors in TLR2x4x9-KO and MyD88xTRIF-KO BMMs.
(A) Schematic of quantitative expression analyses for S. typhimurium genes. Total RNA was isolated from infected BMMs of the indicated genotypes followed by processing for quantitative RT-PCR (see Experimental Procedures). (B) Heat map of normalized expression data for SPI-2 genes in bacteria within BMMs of the indicated genotypes. For each BMM genotype, data are shown relative to the 2h timepoint. The gene designation is to the right of each row. (C) Relative induction of individual SPI-2 genes in bacteria isolated from BMMs of the indicated genotypes. Data are normalized to the average expression values of a set of control genes. The data presented in (B) and (C) represent the mean of 2 independent experiments (D) BMMs of the indicated genotypes were infected with an S. typhimurium strain (12032) expressing an HA-tagged allele of pipB2 expressed from the endogenous pipB2 locus. The presence of PipB2 in BMM lysates was detected by immunoprecipitation and immunoblot with anti-HA antibodies. Controls for number of BMM (tubulin) and bacteria (DnaK) are also shown. Data are representative of 3 independent experiments.
Using K-means clustering analysis, we identified subsets of genes with differential expression profiles between the BMM genotypes. Genes within the SPI-2 locus were upregulated in bacteria in wildtype and TLR2x4-KO BMMs but not in bacteria in TLR2x4x9-KO or MyD88xTRIF-KO BMMs. For validation, we reanalyzed expression of each gene within the SPI-2 locus and adjacent to the locus (as controls), using independent RNA samples from infected BMMs of all genotypes. As shown in Figure 5B, 13 genes within the SPI-2 locus were upregulated in wildtype and TLR2x4-KO BMMs but not in TLR2x4x9-KO and MyD88xTRIF-KO BMMs. These 13 genes most likely underestimate the extent to which the entire SPI-2 locus is differentially expressed between BMM genotypes, as many genes were statistically excluded due to extremely low levels of message in TLR2x4x9-KO or MyD88xTRIF-KO samples. For most SPI-2 genes, induction was higher in wildtype BMMs relative to TLR2x4-KO BMMs (Figure 5C), suggesting that induction correlates with the strength of TLR signaling. Thus, the lack of intracellular replication in TLR-deficient cells may be due to a failure to upregulate SPI-2 genes.
These expression-profiling studies indicated that transcription of SPI-2 genes within BMMs depends on signals downstream of TLR activation. To view SPI-2 induction at the protein level, we utilized a strain of S. typhimurium (12023) with an HA-tagged allele of pipB2, a SPI-2 effector. 12023 displays the same dependence on TLR signaling for intracellular growth as SL1344 (Figure S3D). PipB2 was strongly induced and secreted in infected TLR2x4-KO BMMs (Figure 5D). In contrast, the levels of PipB2 were significantly reduced in TLR2x4x9-KO BMMs and barely detectable in MyD88xTRIF-KO BMMs, despite equivalent numbers of bacteria in all samples (indicated by DnaK levels). These data are consistent with our transcriptional analyses and indicate that TLR signaling is required for the induction of SPI-2 genes.
SPI-2 genes are required for intracellular growth
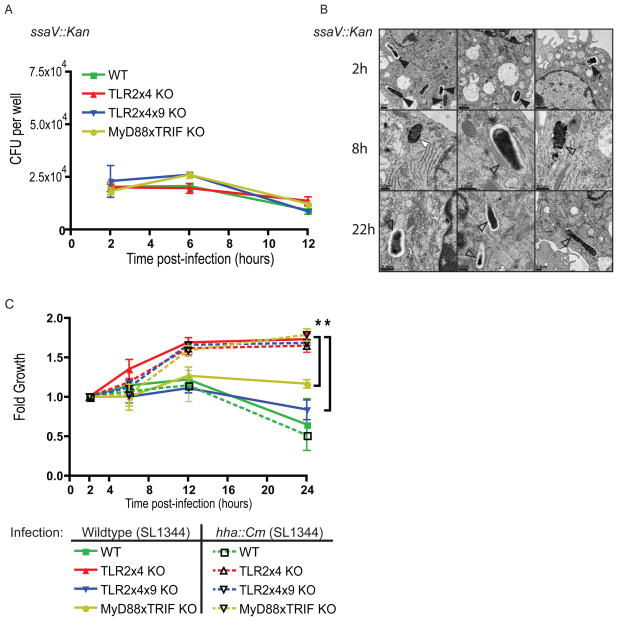
We hypothesized that the impaired induction of SPI-2 genes in bacteria isolated from TLR2x4x9-KO and MyD88xTRIF-KO BMMs was responsible for the defect in SCV formation and intracellular replication in these cells. To test this hypothesis, we compared the fate of bacteria lacking a functional SPI-2 secretion system (ssaV::Kan) in BMMs of each genotype. As expected, SPI-2 mutant bacteria were unable to replicate in BMMs of any genotype (Figure 6A). Moreover, EM analysis of SPI-2 mutant bacteria in TLR2x4-KO BMMs revealed the same lack of SCV formation observed for wildtype bacteria in TLR2x4x9-KO and MyD88xTRIF-KO BMMs (Figure 6B).
Figure 6. TLR-signaling is necessary for SPI-2 induction and intracellular growth.
(A) BMMs of the indicated genotypes were infected (MOI of 1) with SPI-2 mutant S. typhimurium (SL1344 ssaV::Kan). Intracellular CFU were measured by gentamicin protection assay. (B) TLR2x4-KO BMMs were infected as in (A) followed by fixation and processing for electron microscopy. Bacteria in intact vacuoles are indicated by filled black triangles, cytosolic bacteria by filled white triangles, and bacteria that are degraded or have fused with lytic compartments by open black triangles. Three representative images from different sections and from independent infections are shown. (C) BMMs of the indicated genotypes were infected (MOI of 1) with wildtype S. typhimurium or a strain with constitutive SPI-2 expression (SL1344 hha::Cm). Intracellular CFU were measured via gentamicin protection assay at the indicated timepoints post-infection. Data are presented as mean fold over the first time point (to control for minor inoculum differences between strains) +/− SEM and are representative of 3 independent experiments. * p<0.05 by student t test comparing TLR2x4-KO to MyD88xTRIF-KO and TLR2x4x9-KO at the indicated timepoint.
If the lack of intracellular growth in TLR2x4x9-KO and MyD88xTRIF-KO BMMs is due to failure to induce SPI-2 genes, then a S. typhimurium strain with constitutive expression of SPI-2 genes should regain the ability to grow in these cells. To test this possibility directly, we constructed a strain lacking hha (Δ_hha, hha::Cm_), a negative regulator of SPI-2 genes. Previous work has demonstrated that Δ_hha_ mutant strain expresses SPI-2 genes constitutively (Silphaduang et al., 2007). Remarkably, hha mutant bacteria replicated equivalently in BMMs of all genotypes, except wildtype cells where the lack of growth is due to TLR4-dependent cell death (Figure 6C). While Hha likely negatively regulates additional S. typhimurium virulence genes, restoration of growth in TLR-deficient BMMs is consistent with the conclusion that constitutive expression of SPI-2 genes can bypass the requirement for TLR signaling.
Induction of SPI-2 genes requires TLR-dependent acidification of the SCV
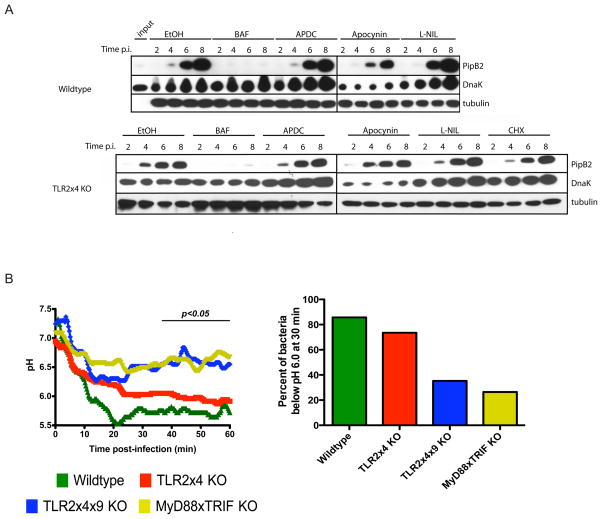
Our results thus far indicate that TLR signaling provides a cue used by S. typhimurium to regulate SPI-2 expression. TLR activation induces host transcription as well as more proximal effects, such as production of ROS and RNS and phagosome maturation and acidification, although this last aspect remains controversial. Using pharmacological inhibitors to block each of these potential signals we measured the effect on PipB2 induction and secretion. Treatment of TLR2x4-KO BMMs with cyclohexamide (CHX) had no effect on PipB2 induction, indicating that host translation was not required for generation of the signal sensed by S. typhimurium (Figure 7A, bottom panel). Similarly, blocking ROS or RNS production did not prevent PipB2 induction. However, inhibition of the V-ATPase with bafilomycinA1 blocked S. typhimurium induction of PipB2 in both TLR2x4-KO and wildtype BMMs. The block in TLR2x4-KO cells could be due to an inhibition of TLR signaling, as bafilomycinA1 almost completely inhibits the residual response to S. typhimurium (Figure 1D). In wildtype cells, though, TLR2 and TLR4 signaling is largely unaffected by bafilomycinA1, suggesting that TLR-dependent acidification of the SCV may be the signal required by S. typhimurium for SPI-2 gene induction (Figure 7A, top panel). Experiments analyzing the induction of SPI-2 genes at the transcriptional level also indicated a requirement for phagosome acidification (data not shown).
Figure 7. TLR-dependent phagosomal acidification is required for SPI-2 expression.
(A) BMMs pre-treated with each of the indicated inhibitors, were infected with an S. typhimurium strain (12032) expressing an HA-tagged allele of pipB2. PipB2 levels in BMM lysates were detected by immunoprecipitation and immunoblot with anti-HA antibodies at 2, 4, 6 and 8 hours post-infection. Immunoblots for tubulin and DnaK serve as loading controls for BMMs and bacteria, respectively. Data are representative of 2 independent experiments for wildtype cells and 3 independent experiments for TLR2x4-KO BMMs. (B) BMMs were infected with FITC-labeled S. typhimurium (SL1344) and the fluorescence intensities of individual bacteria excited at 490 nm or 440 nm were measured over time by live cell imaging. The fluorescence intensity ratio (490/440) reflects the pH within the phagosome (see Experimental Procedures). The plots presented represent the mean pH calculated from at least 35 independent bacteria in multiple imaging fields. The right panel shows the percent of bacteria at or below pH 6 at 30 minutes post-infection. See Experimental Procedures and Figure S4.
Based on these data, we hypothesized that the lack of SPI-2 induction in TLR2x4x9-KO and MyD88xTRIF-KO BMMs is due to failure of SCVs to acidify. The issue of whether TLR signaling influences the kinetics of phagosomal maturation remains controversial (Blander and Medzhitov, 2004, 2006a, b; Russell and Yates, 2007; Yates and Russell, 2005). To investigate this issue, we used ratiometric imaging to measure the pH of Salmonella containing phagosomes in BMMs of each genotype. While the mean pH of SCVs in wildtype and TLR2x4-KO BMMs dropped below 6 within 60 minutes post-infection, SCVs in TLR2x4x9-KO and MyD88xTRIF-KO BMMs failed to acidify to the same extent and exhibited slower acidification kinetics (Figure 7B). By 30 minutes post-infection, over 70% of SCVs in wildtype and TLR2x4-KO BMMs had reached pH 6, while less than 35% of SCVs in TLR2x4x9-KO and MyD88xTRIF-KO BMMs had similarly acidified (Figure 7B). Consistent with the lower transcriptional induction of SPI-2 genes in TLR2x4-KO BMMs (relative to wildtype), the rate of acidification in TLR2x4-KO cells was slower than in wildtype cells, despite ultimately reaching pH 6 by 60 minutes. Collectively, these data support a model in which TLR signaling accelerates phagosomal acidification, which is used by S. typhimurium as a cue for SPI-2 gene induction.
Discussion
Biological interactions are strong drivers of evolution, and the dynamics of host-pathogen interactions provide some of the clearest examples of this principle. Hosts have evolved resistance mechanisms, such as TLRs, that work by reducing pathogen fitness and drive the evolution of pathogen virulence (Hedrick, 2004; Rausher, 2001; Woolhouse et al., 2002). While virulence genes provide a fitness advantage, they can be energetically costly and often serve as targets of host sensors (Miao et al., 2010; Vance et al., 2009). Therefore, the ability to regulate expression of virulence genes based on changing environments is a key feature of microbial pathogenesis. In this study, we report the requirement of TLR signaling for S. typhimurium to establish a successful infection and cause disease. We demonstrate that this requirement stems, at least in part, from the need for TLR-dependent phagosome acidification to induce SPI-2 genes, resulting in replication and virulence of the microbe. These data demonstrate that a pathogen can evolve to require innate immune signaling for full virulence.
Previous studies have demonstrated that host genetic variation can result in prolonged survival upon infection (Raberg et al., 2007). However, these phenotypes are generally attributable to reduced inflammation and immunopathology, suggesting that the host is more tolerant to an increased pathogen burden (Gowen et al., 2006; Wang et al., 2004). By contrast, our work demonstrates that mice lacking sufficient TLR signaling are less susceptible to a S. typhimurium infection due to reduced bacterial growth. A similar relationship has been described in Drosophila, where mutations in the melanization arm of the innate immune response render flies less susceptible to Streptococcus pneumoniae with reduced levels of bacteria, but the mechanism behind this observation remains unclear (Ayres and Schneider, 2008). Our work clearly shows that S. typhimurium fails to induce virulence genes when deprived of innate immune signals.
Cross-talk between Nramp-1 and TLR signaling
Two aspects of our approach were crucial for our ability to observe the requirement for TLR-dependent signals in Salmonella virulence. First, by using mice that are deficient in multiple TLRs, as opposed to mice lacking MyD88 and TRIF, we were able to circumvent the susceptibility associated with lack of IL-1R family function. Indeed, the difference in susceptibility between MyD88xTRIF-KO and TLR2x4x9-KO mice that we observe underscores the importance of the IL-1R family in defense against infection. We were somewhat surprised by the role for nucleic acid sensing TLRs in innate recognition of Salmonella, although TLR9 and TLR7 have been implicated in recognition of bacterial nucleic acid (Bafica et al., 2005; Mancuso et al., 2009). While the simplest explanation for this observation is that some bacteria are degraded, it is also possible that a nucleic acid ligand is secreted by Salmonella or present on the bacterial surface (Whitchurch et al., 2002; Woodward et al., 2010).
A second critical aspect of our study is that we used mice with a functional allele of nramp1. Why the lack of this protein renders mice so susceptible to intracellular pathogens remains unclear, but this heightened sensitivity may simply obviate any requirement for TLR-dependent SPI-2 induction. The presence of functional Nramp1 has been shown to enhance SPI-2 expression as well as TLR-dependent responses (Fritsche et al., 2003; Valdez et al., 2008; Zaharik et al., 2002). Regardless of the precise mechanism responsible for the strong TLR-dependence when Nramp1 is functional, it is important to recognize that infection of cells with functional Nramp1 represents the “wildtype” scenario. Indeed, mutations in the human Nramp1 gene are associated with increased susceptibility to several intracellular pathogens (Bellamy et al., 1998; Malik et al., 2005). Therefore, examining virulence in the presence of functional Nramp1 most accurately reflects the host-pathogen interactions between S. typhimurium and the mammalian immune system.
TLR signaling alters the pH of the Salmonella containing vacuole
Our studies indicate that the difference in susceptibility of TLR-deficient mice is due to lack (or substantial delay) of SPI-2 induction. We show that SPI-2 induction requires phagosome acidification, and our measurements of phagosomal pH indicate that acidification is impaired and/or delayed in TLR-deficient cells. The extent to which TLR signaling influences phagosomal maturation (including increasing phagolysosomal fusion, acidification, and proteolytic activity) has remained a contentious issue (Blander and Medzhitov, 2004, 2006a, b; Russell and Yates, 2007; Yates and Russell, 2005). While our studies were not designed to address this controversy, we clearly show that TLR signaling is required for rapid acidification of the SCV and has profound implications for the fate of intracellular bacteria and disease outcome. The mechanism is likely similar to the acidification of lysosomes during DC maturation, when TLR signaling leads to recruitment of the V1 subunit of the vacuolar ATPase to the lysosomal membrane (Trombetta et al., 2003). The precise signaling pathways that lead to assembly of this machinery are unknown. Moreover, whether bacteria sense pH directly or utilize other phagosomal features that require acidic pH remains unclear.
Importantly, we are not suggesting that phagosome maturation cannot occur without TLR signaling. Indeed, our EM images of infected TLR2x4x9-KO and MyD88xTRIF-KO BMMs at late time points show bacteria within electron dense compartments, suggestive of phagolysosomal fusion. Due to technical limitations we have not extended our pH measurements beyond 60 minutes post-infection, but our images suggest that phagosomes in TLR-deficient cells eventually mature. In fact, we do observe a small percentage of SCV in TLR2x4x9-KO and MyD88xTRIF-KO cells with significant reductions in pH within 1h (Figure S4). The lack of bacterial replication in TLR-deficient cells suggests that the eventual maturation of phagosomes is not sufficient to induce SPI-2 genes or that the induction occurs too late to prevent bacterial killing by lysosomal contents. It is also possible that the phagosome breaks down in the absence of SPI-2 function, and bacteria enter the cytosol where they are unable to replicate (Beuzon et al., 2000). Our EM analyses suggest that both of these possibilities may contribute to the lack of bacterial replication.
Innate immune signaling as an environmental cue for virulence gene regulation
These findings have important implications for our understanding of the evolution of host-pathogen interactions and virulence mechanisms. Many pathogens trigger these mechanisms by utilizing signals downstream of innate receptors, most likely as a reliable mechanism to properly induce genes necessary for survival in the presence of antimicrobial mechanisms. For example, the PhoP/PhoQ two-component system, when activated by AMPs, induces expression of genes that modify lipidA and render the bacterial membrane more resistant to AMPs (Bader et al., 2005; Guo et al., 1998). By a potentially similar mechanism, prior activation of cells with TLR ligands can increase replication of S. typhimurium (Wong et al., 2009). Our finding that S. typhimurium has evolved to require host resistance signals for proper expression of virulence genes is conceptually distinct from these previously described antagonistic strategies. Notably, S. typhimurium is unable to replicate in TLR-deficient cells, despite the absence of the antimicrobial mechanisms normally induced by TLRs. One implication of this remaining dependence is that the virulence genes induced by TLR signaling are required for purposes other than simply evading TLR-induced antimicrobial mechanisms to promote S. typhimurium fitness.
Why would Salmonella use signals downstream of TLRs to broadly coordinate expression of virulence genes required for intracellular growth? These signals may be the most reliable contextual cues that Salmonella can use to sense its presence within a macrophage phagosome. In general, a fundamental problem faced by Salmonella is the need to interact with multiple cell types through the course of an infection. Unique sets of virulence genes are required to survive each of these stages and Salmonella must recognize its environment and induce the appropriate genes. For example, Salmonella must detect when it has encountered a macrophage and induce SPI-2 genes, which are necessary for formation of the SCV and maintenance of the integrity of the phagosome. Precise regulation of such virulence genes is clearly essential for optimal growth, as mutant bacteria with constitutive expression of SPI-2 genes (e.g., hha mutants) are attenuated in vivo (Coombes et al., 2005; Silphaduang et al., 2007). Inappropriate expression of certain virulence genes could result in decreased fitness due to recognition by innate sensors or may disrupt proper regulation of other virulence genes required at specific stages of infection. Therefore, Salmonella utilizes TLR-dependent signals within the phagosome to detect its presence within a macrophage. Linking the induction of virulence genes (including SPI-2) to phagosomal signals downstream of TLRs may be an efficient way of coordinating multiple virulence mechanisms in response to a unifying contextual cue.
Experimental Procedures
Cell Culture
BMMs were differentiated from bone marrow for 5 days using M-CSF as previously described (Ewald et al., 2008). See Extended Experimental Procedures for details.
Bacterial Strains and Infections
Overnight cultures of S. typhimurium were opsonized for infection. BMMs were spin-infected, incubated at 37 °C, then washed with PBS before the addition of 10 μg/mL gentamicin media. For intracellular CFU determination, cells were washed with PBS and lysed in 1% Triton-X100 in PBS. Lysates were plated on LB agar plates containing 200 ug/mL streptomycin (Life Technologies). See Extended Experimental Procedures for strain details and descriptions of assays with L. monocytogenes and L. pneumophila.
Measurement of Cell Death
Lactate dehydrogenase (LDH) release was quantified using the CytoTox 96 Non-radioactive cytotoxicity kit (Promega) according to manufacturer’s instructions. Annexin V staining was performed using Annexin V-FITC (BD Pharmingen) in Annexin V staining buffer according to manufacturer’s instructions.
Measurement of BMM activation
NO was quantified in supernatants from BMMs treated overnight with 100 μg/mL recombinant IFNγ (R&D Systems) using the Griess assay (all reagents from Sigma Aldrich). TNFα production was measured by intracellular cytokine staining using anti-TNFα antibody (eBioscience) according to manufacturer’s instructions (eBioscience). All steps prior to fixation were performed in the presence of 10 μg/mL gentamicin. Cells were analyzed on an FC500 flow cytometer (Beckman Coulter).
S. typhimurium Effector Secretion
BMMs were infected (MOI of 25) with _S. typhimurium pipB2_-2xHA (12032). At the indicated timepoints, cells were washed with PBS, lysed in 1% NP-40 in PBS with protease-inhibtor cocktail (Roche) and EDTA (Fisher), and lysates were subjected to immunoprecipitation with rat anti-HA agarose beads (Roche). Cells were pretreated with inhibitors for 1h before infection. See Extended Experimental Procedures for a more detailed explanation of sample processing and detection of effector secretion.
Mice and in vivo Infections
All animal experiments were performed in accordance with University of California Animal Care and Use Committee guidelines. See Extended Experimental Procedures for descriptions of strains and backcrossing analyses. For survival and CFU enumeration experiments, age-matched mice were fasted for 14h followed by oral gavage with 100 μL S. typhimurium (SL1344) or Yersinia enterocolitica (8081) in PBS (see Figure Legends for CFU). For CFU enumeration, organs were harvested, homogenized in PBS using a Polytron PT2100 homogenizer (Kinematica), diluted and plated on streptomycin (for Salmonella) or irgasan (1 μg/mL) (for Yersinia) LB-agar plates.
Gene expression analyses
RNA from infected BMMs (MOI of 5) was extracted with Trizol RNA reagent, purified using PureLink Micro-to-Midi total RNA purification system, DNase-treated, reverse transcribed using random hexamers (all reagents from Life Technologies), and processed for quantitative PCR. Sample processing, primer sequences/design, and description of data analysis can be found in Extended Experimental Procedures and Table S1.
Microscopy
For IF, BMMs on coverslips were stained with FITC-conjugated mouse anti-Salmonella antibody (clone 1E6, Santa Cruz Biotechnology) and Cy3-conjugated wheat germ agglutinin (Life Technologies) at the indicated timepoints. Cells were imaged on a Nikon E800 fluorescent microscope and bacteria per cell were counted in random, blinded, z-stacked images.
For pH determination and video microscopy, BMMs plated on 4-chamber slides (Nunc) were infected with FITC-labeled bacteria. Following infection, chambers were incubated at 37 °C for 5 min, washed extensively with PBS, incubated with phenol-free DMEM containing 10 μg/mL gentamicin, then visualized on a Nikon TE2000 inverted fluorescent microscope with environmental control. Images were collected with excitation at both 440 nm and 490 nm and analyzed using Imaris Scientific 3D/4D image processing and analysis software (Bitplane) to track individual intracellular bacteria. Background-subtracted fluorescence intensity values were used to determine the 490/440 ratios for each bacterium at each timepoint. Absolute pH values were determined by generating a standard curve using buffered pH solutions (see Extended Experimental Procedures).
For EM studies, cells were infected (MOI of 10) with wildtype or ssaV::Kan SL1344 (as described above). At each timepoint, cells were fixed, embedded, sectioned and stained for EM (see Extended Experimental Procedures).
Supplementary Material
01
02
03
04
Acknowledgments
We thank T. Machen, P. Herzmark, J. Ross, and E. Peled for assistance with ratiometric imaging; members of the Barton lab, J. Ayres, and R. Vance for critical reading of this manuscript; C. Rae and N. Meyer-Morse for assistance with L. monocytogenes; M. Fontana and K. Monroe for assistance with L. pneumophila; R. Zalpuri and K. McDonald for assistance with EM; H. Nolla for assistance with flow cytometry. This work was supported in part by grants from the NIH (P01-AI063302 to DMM and GMB, Y1-AI-8401 to SNP), the Burroughs Wellcome Fund (DMM), the University of California Cancer Research Coordinating Committee (GMB), and a NIH NRSA Trainee appointment on grant T32-GM007232 (NA).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol. 2001;2:675–680. doi: 10.1038/90609. [DOI] [PubMed] [Google Scholar]
- Ayres JS, Schneider DS. A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS Biol. 2008;6:2764–2773. doi: 10.1371/journal.pbio.0060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell. 2005;122:461–472. doi: 10.1016/j.cell.2005.05.030. [DOI] [PubMed] [Google Scholar]
- Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202:1715–1724. doi: 10.1084/jem.20051782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- Beuzon CR, Meresse S, Unsworth KE, Ruiz-Albert J, Garvis S, Waterman SR, Ryder TA, Boucrot E, Holden DW. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. Embo J. 2000;19:3235–3249. doi: 10.1093/emboj/19.13.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander JM, Medzhitov R. Regulation of phagosome maturation by signals from toll-like receptors. Science. 2004;304:1014–1018. doi: 10.1126/science.1096158. [DOI] [PubMed] [Google Scholar]
- Blander JM, Medzhitov R. On regulation of phagosome maturation and antigen presentation. Nat Immunol. 2006a;7:1029–1035. doi: 10.1038/ni1006-1029. [DOI] [PubMed] [Google Scholar]
- Blander JM, Medzhitov R. Toll-dependent selection of microbial antigens for presentation by dendritic cells. Nature. 2006b;440:808–812. doi: 10.1038/nature04596. [DOI] [PubMed] [Google Scholar]
- Chakravortty D, Rohde M, Jäger L, Deiwick J, Hensel M. Formation of a novel surface structure encoded by Salmonella Pathogenicity Island 2. EMBO J. 2005;24:2043–2052. doi: 10.1038/sj.emboj.7600676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo DM, Valdivia RH, Monack DM, Falkow S. Macrophage-dependent induction of the Salmonella pathogenicity island 2 type III secretion system and its role in intracellular survival. Mol Microbiol. 1998;30:175–188. doi: 10.1046/j.1365-2958.1998.01048.x. [DOI] [PubMed] [Google Scholar]
- Coburn B, Grassl GA, Finlay BB. Salmonella, the host and disease: a brief review. Immunol Cell Biol. 2007;85:112–118. doi: 10.1038/sj.icb.7100007. [DOI] [PubMed] [Google Scholar]
- Cook P, Tötemeyer S, Stevenson C, Fitzgerald KA, Yamamoto M, Akira S, Maskell DJ, Bryant CE. Salmonella-induced SipB-independent cell death requires Toll-like receptor-4 signalling via the adapter proteins Tram and Trif. Immunology. 2007;122:222–229. doi: 10.1111/j.1365-2567.2007.02631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes BK, Wickham ME, Lowden MJ, Brown NF, Finlay BB. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:17460–17465. doi: 10.1073/pnas.0505401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deiwick J, Nikolaus T, Erdogan S, Hensel M. Environmental regulation of Salmonella pathogenicity island 2 gene expression. Mol Microbiol. 1999;31:1759–1773. doi: 10.1046/j.1365-2958.1999.01312.x. [DOI] [PubMed] [Google Scholar]
- Detweiler CS, Monack DM, Brodsky IE, Mathew H, Falkow S. virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol Microbiol. 2003;48:385–400. doi: 10.1046/j.1365-2958.2003.03455.x. [DOI] [PubMed] [Google Scholar]
- Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi GP, Chapman HA, Barton GM. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456:658–662. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuillet V, Medjane S, Mondor I, Demaria O, Pagni PP, Galán JE, Flavell RA, Alexopoulou L. Involvement of Toll-like receptor 5 in the recognition of flagellated bacteria. Proc Natl Acad Sci USA. 2006;103:12487–12492. doi: 10.1073/pnas.0605200103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83:5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes JR, Gros P. Divalent-metal transport by NRAMP proteins at the interface of host-pathogen interactions. Trends Microbiol. 2001;9:397–403. doi: 10.1016/s0966-842x(01)02098-4. [DOI] [PubMed] [Google Scholar]
- Fritsche G, Dlaska M, Barton H, Theurl I, Garimorth K, Weiss G. Nramp1 functionality increases inducible nitric oxide synthase transcription via stimulation of IFN regulatory factor 1 expression. J Immunol. 2003;171:1994–1998. doi: 10.4049/jimmunol.171.4.1994. [DOI] [PubMed] [Google Scholar]
- Galan JE. Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol. 2001;17:53–86. doi: 10.1146/annurev.cellbio.17.1.53. [DOI] [PubMed] [Google Scholar]
- Govoni G, Vidal S, Gauthier S, Skamene E, Malo D, Gros P. The Bcg/Ity/Lsh locus: genetic transfer of resistance to infections in C57BL/6J mice transgenic for the Nramp1 Gly169 allele. Infect Immun. 1996;64:2923–2929. doi: 10.1128/iai.64.8.2923-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen BB, Hoopes JD, Wong MH, Jung KH, Isakson KC, Alexopoulou L, Flavell RA, Sidwell RW. TLR3 deletion limits mortality and disease severity due to Phlebovirus infection. J Immunol. 2006;177:6301–6307. doi: 10.4049/jimmunol.177.9.6301. [DOI] [PubMed] [Google Scholar]
- Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- Hapfelmeier S, Stecher B, Barthel M, Kremer M, Müller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol. 2005;174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- Hedrick S. The Acquired Immune System A Vantage from Beneath. Immunity. 2004 doi: 10.1016/j.immuni.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Hsu LC, Park JM, Zhang K, Luo JL, Maeda S, Kaufman RJ, Eckmann L, Guiney DG, Karin M. The protein kinase PKR is required for macrophage apoptosis after activation of Toll-like receptor 4. Nature. 2004;428:341–345. doi: 10.1038/nature02405. [DOI] [PubMed] [Google Scholar]
- Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Immunol. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Pathogen recognition with Toll-like receptors. Curr Opin Immunol. 2005;17:338–344. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Kim CC, Falkow S. Delineation of Upstream Signaling Events in the Salmonella Pathogenicity Island 2 Transcriptional Activation Pathway. The Journal of Bacteriology. 2004;186:4694–4704. doi: 10.1128/JB.186.14.4694-4704.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung KY, Finlay BB. Intracellular replication is essential for the virulence of Salmonella typhimurium. Proc Natl Acad Sci USA. 1991;88:11470–11474. doi: 10.1073/pnas.88.24.11470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Abel L, Tooker H, Poon A, Simkin L, Girard M, Adams GJ, Starke JR, Smith KC, Graviss EA, et al. Alleles of the NRAMP1 gene are risk factors for pediatric tuberculosis disease. Proc Natl Acad Sci U S A. 2005;102:12183–12188. doi: 10.1073/pnas.0503368102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G, Gambuzza M, Midiri A, Biondo C, Papasergi S, Akira S, Teti G, Beninati C. Bacterial recognition by TLR7 in the lysosomes of conventional dendritic cells. Nat Immunol. 2009;10:587–594. doi: 10.1038/ni.1733. [DOI] [PubMed] [Google Scholar]
- Marcus SL, Brumell JH, Pfeifer CG, Finlay BB. Salmonella pathogenicity islands: big virulence in small packages. Microbes Infect. 2000;2:145–156. doi: 10.1016/s1286-4579(00)00273-2. [DOI] [PubMed] [Google Scholar]
- Mayer-Barber KD, Barber DL, Shenderov K, White SD, Wilson MS, Cheever A, Kugler D, Hieny S, Caspar P, Nunez G, et al. Caspase-1 independent IL-1beta production is critical for host resistance to mycobacterium tuberculosis and does not require TLR signaling in vivo. J Immunol. 2010;184:3326–3330. doi: 10.4049/jimmunol.0904189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci U S A. 2010;107:3076–3080. doi: 10.1073/pnas.0913087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien AD, Rosenstreich DL, Scher I, Campbell GH, MacDermott RP, Formal SB. Genetic control of susceptibility to Salmonella typhimurium in mice: role of the LPS gene. J Immunol. 1980;124:20–24. [PubMed] [Google Scholar]
- Park JM, Greten FR, Li ZW, Karin M. Macrophage apoptosis by anthrax lethal factor through p38 MAP kinase inhibition. Science. 2002;297:2048–2051. doi: 10.1126/science.1073163. [DOI] [PubMed] [Google Scholar]
- Pfeifer CG, Marcus SL, Steele-Mortimer O, Knodler LA, Finlay BB. Salmonella typhimurium virulence genes are induced upon bacterial invasion into phagocytic and nonphagocytic cells. Infect Immun. 1999;67:5690–5698. doi: 10.1128/iai.67.11.5690-5698.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raberg L, Sim D, Read AF. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science. 2007;318:812–814. doi: 10.1126/science.1148526. [DOI] [PubMed] [Google Scholar]
- Rappl C, Deiwick J, Hensel M. Acidic pH is required for the functional assembly of the type III secretion system encoded by Salmonella pathogenicity island 2. FEMS Microbiol Lett. 2003;226:363–372. doi: 10.1016/S0378-1097(03)00638-4. [DOI] [PubMed] [Google Scholar]
- Raupach B, Peuschel SK, Monack DM, Zychlinsky A. Caspase-1-mediated activation of interleukin-1beta (IL-1beta) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect Immun. 2006;74:4922–4926. doi: 10.1128/IAI.00417-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausher MD. Co-evolution and plant resistance to natural enemies. Nature. 2001;411:857–864. doi: 10.1038/35081193. [DOI] [PubMed] [Google Scholar]
- Royle MC, Tötemeyer S, Alldridge LC, Maskell DJ, Bryant CE. Stimulation of Toll-like receptor 4 by lipopolysaccharide during cellular invasion by live Salmonella typhimurium is a critical but not exclusive event leading to macrophage responses. J Immunol. 2003;170:5445–5454. doi: 10.4049/jimmunol.170.11.5445. [DOI] [PubMed] [Google Scholar]
- Russell DG, Yates RM. Toll-like receptors and phagosome maturation. Nat Immunol. 2007;8:217. doi: 10.1038/ni0307-217a. author reply 217–218. [DOI] [PubMed] [Google Scholar]
- Shea JE, Hensel M, Gleeson C, Holden DW. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silphaduang U, Mascarenhas M, Karmali M, Coombes BK. Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J Bacteriol. 2007;189:3669–3673. doi: 10.1128/JB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, Cookson BT, Aderem A. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- Trombetta ES, Ebersold M, Garrett W, Pypaert M, Mellman I. Activation of lysosomal function during dendritic cell maturation. Science. 2003;299:1400–1403. doi: 10.1126/science.1080106. [DOI] [PubMed] [Google Scholar]
- Uematsu S, Jang MH, Chevrier N, Guo Z, Kumagai Y, Yamamoto M, Kato H, Sougawa N, Matsui H, Kuwata H, et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol. 2006;7:868–874. doi: 10.1038/ni1362. [DOI] [PubMed] [Google Scholar]
- Valdez Y, Diehl GE, Vallance BA, Grassl GA, Guttman JA, Brown NF, Rosenberger CM, Littman DR, Gros P, Finlay BB. Nramp1 expression by dendritic cells modulates inflammatory responses during Salmonella Typhimurium infection. Cell Microbiol. 2008;10:1646–1661. doi: 10.1111/j.1462-5822.2008.01155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia RH, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Torres A, Vallance BA, Bergman MA, Finlay BB, Cookson BT, Jones-Carson J, Fang FC. Toll-like receptor 4 dependence of innate and adaptive immunity to Salmonella: importance of the Kupffer cell network. J Immunol. 2004;172:6202–6208. doi: 10.4049/jimmunol.172.10.6202. [DOI] [PubMed] [Google Scholar]
- Vidal S, Tremblay ML, Govoni G, Gauthier S, Sebastiani G, Malo D, Skamene E, Olivier M, Jothy S, Gros P. The Ity/Lsh/Bcg locus: natural resistance to infection with intracellular parasites is abrogated by disruption of the Nramp1 gene. J Exp Med. 1995;182:655–666. doi: 10.1084/jem.182.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal SM, Malo D, Vogan K, Skamene E, Gros P. Natural resistance to infection with intracellular parasites: Isolation of a candidate for Bcg. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- Vidal SM, Pinner E, Lepage P, Gauthier S, Gros P. Natural resistance to intracellular infections: Nramp1 encodes a membrane phosphoglycoprotein absent in macrophages from susceptible (Nramp1 D169) mouse strains. J Immunol. 1996;157:3559–3568. [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Kumar A, Neish AS, Uematsu S, Akira S, Gewirtz AT. Toll-Like Receptor 5-Deficient Mice Have Dysregulated Intestinal Gene Expression and Nonspecific Resistance to Salmonella-Induced Typhoid-Like Disease. Infect Immun. 2008;76:1276–1281. doi: 10.1128/IAI.01491-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Town T, Alexopoulou L, Anderson JF, Fikrig E, Flavell RA. Toll-like receptor 3 mediates West Nile virus entry into the brain causing lethal encephalitis. Nat Med. 2004;10:1366–1373. doi: 10.1038/nm1140. [DOI] [PubMed] [Google Scholar]
- Waterman SR, Holden DW. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 2003;5:501–511. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Raupach B, Takeda K, Akira S, Zychlinsky A. Toll-Like Receptors Are Temporally Involved in Host Defense. The Journal of Immunology. 2004;172:4463–4469. doi: 10.4049/jimmunol.172.7.4463. [DOI] [PubMed] [Google Scholar]
- Wen L, Ley RE, Volchkov PY, Stranges PB, Avanesyan L, Stonebraker AC, Hu C, Wong FS, Szot GL, Bluestone JA, et al. Innate immunity and intestinal microbiota in the development of Type 1 diabetes. Nature. 2008;455:1109–1113. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Wong CE, Sad S, Coombes BK. Salmonella enterica Serovar Typhimurium Exploits Toll-Like Receptor Signaling during the Host-Pathogen Interaction. Infect Immun. 2009;77:4750–4760. doi: 10.1128/IAI.00545-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward JJ, Iavarone AT, Portnoy DA. c-di-AMP Secreted by Intracellular Listeria monocytogenes Activates a Host Type I Interferon Response. Science (New York, NY) 2010 doi: 10.1126/science.1189801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhouse ME, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genet. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- Yates RM, Russell DG. Phagosome maturation proceeds independently of stimulation of toll-like receptors 2 and 4. Immunity. 2005;23:409–417. doi: 10.1016/j.immuni.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Zaharik ML, Vallance BA, Puente JL, Gros P, Finlay BB. Host-pathogen interactions: Host resistance factor Nramp1 up-regulates the expression of Salmonella pathogenicity island-2 virulence genes. Proc Natl Acad Sci USA. 2002;99:15705–15710. doi: 10.1073/pnas.252415599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04