Differential microRNA regulation of HLA-C expression and its association with HIV control (original) (raw)
. Author manuscript; available in PMC: 2011 Oct 28.
Published in final edited form as: Nature. 2011 Apr 17;472(7344):495–498. doi: 10.1038/nature09914
Abstract
The HLA-C locus is distinct relative to the other classical HLA class I loci in that it has relatively limited polymorphism1, lower expression on the cell surface2,3, and more extensive ligand-receptor interactions with killer cell immunoglobulin-like receptors (KIR)4. A single nucleotide polymorphism (SNP) 35Kb upstream of HLA-C (rs9264942; termed −35) associates with control of HIV5–7, and with levels of HLA-C mRNA transcripts8 and cell surface expression7, but the mechanism underlying its varied expression is unknown. We proposed that the −35 SNP is not the causal variant for differential HLA-C expression, but rather is marking another polymorphism that directly affects levels of HLA-C7. Here we show that variation within the 3′ untranslated region of HLA-C regulates binding of the microRNA Hsa-miR-148a to its target site, resulting in relatively low surface expression of alleles that bind this microRNA and high expression of HLA-C alleles that escape post-transcriptional regulation. The 3′UTR variant associates strongly with control of HIV, potentially adding to the effects of genetic variation encoding the peptide-binding region of the HLA class I loci. Variation in HLA-C expression adds another layer of diversity to this highly polymorphic locus that must be considered when deciphering the function of these molecules in health and disease.
MicroRNAs (miRNA) are a class of non-protein coding RNAs that are estimated to regulate 30% of all genes in animals9 by binding to specific sites in the 3′ untranslated regions (3′UTR), resulting in posttranscriptional repression, cleavage or destabilization10–12. The 3′UTR of the HLA-C gene is predicted to be a target for 26 distinct human miRNAs using three miRNA-target prediction programs (Supplementary Fig. 1), of which three (miR-148a and miR-148b, which bind the same target site, and miR-657) were shown to have the greatest likelihood of binding. We sequenced the 3′UTRs of the common HLA-C alleles (Supplementary Fig. 2) and show that the two binding sites of these three miRNAs are polymorphic (Supplementary Fig. 3a). The binding site for miR-148a/miR-148b contains a single base pair insertion/deletion at position 263 downstream of the HLA-C stop codon (rs67384697G representing the insertion [263ins] and rs67384697- representing the deletion [263del]) along with other precisely linked variants (259 C/T, 261T/C, 266C/T). These variants are likely to impose a restriction in miR-148a/miR-148b binding, as prediction algorithms indicate that the binding of these miRNAs to the alleles marked by 263ins (e.g. Cw*0702, a low expression allotype) is more stable than to alleles with 263del (e.g. Cw*0602, a high expression allotype) (Supplementary Fig. 3b). Similarly, alleles with 307C within the miR-657 target site are predicted to be better targets of miR-657 than those with 307T (Supplementary Fig. 4). Thus, variation in the 3′UTR of HLA-C may influence the interaction between these miRNAs and their putative binding sites in an allele specific manner, potentially leading to differential levels of HLA-C allotype expression.
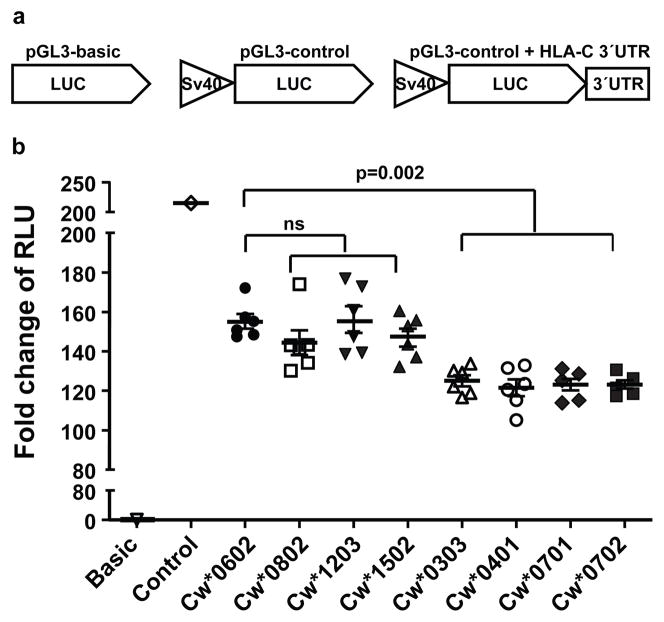
To test directly whether the variation in the HLA-C 3′UTR affects levels of protein expression, the full length 3′UTRs containing intact miR-148a/miR-148b and miR-657 binding sites (i.e. 263ins and 307C, respectively; Cw*0702, Cw*0303, Cw*0401, Cw*0701) and disrupted binding sites (i.e. 263del and 307T, respectively; Cw*0602, Cw*0802, Cw*1203, Cw*1502) were each cloned downstream of the luciferase gene in a pGL3 reporter construct (Fig. 1a). The constructs were then transfected into HLA class I negative B721.221 cells, and the level of luciferase activity was measured (fold increase of relative light units [RLU]). Although the Cw*0602 3′UTR repressed luciferase activity as compared to the control containing no 3′UTR, the constructs containing intact miRNA binding sites (i.e. 263ins and 307C; Cw*0702, Cw*0303, Cw*0401, Cw*0701) produced significantly lower luciferase activity relative to the construct containing the 3′UTR of Cw*0602, which contains 263del and 307T (Fig. 1b). However, 3′UTRs from other alleles with the 263del and 307T variants (Cw*0802, Cw*1203, Cw*1502) did not show significant variation in luciferase activity as compared to Cw*0602 (Fig. 1b). Psicheck2 reporter constructs containing 3′UTRs of Cw*0602 also produced significantly higher luciferase activity as compared to those with Cw*0702 3′UTR (Supplementary Fig. 5a), indicating that this effect was reproducible in a distinct reporter construct. Further, pGL3 constructs containing 3′UTRs of Cw*0602 and Cw*0702 in three additional cell lines showed the same pattern as that seen in B721.221, indicating a consistent difference of these 3′UTRs in the regulation of HLA-C expression that is independent of cell type (Supplementary Fig. 5b–e). Thus, HLA-C 3′UTR alleles characterized by variation at positions 263 and 307 within miRNA binding regions differentially regulate gene expression.
Figure 1. Variation in the HLA-C 3′UTR differentially affects the expression of a reporter gene.
Full length 3′UTRs of various HLA-C alleles cloned into luciferase reporter constructs were transfected into B721.221 cell lines and the stability of the mRNA was estimated by dual luciferase reporter assays. The normalized luciferase activity is presented as fold change of relative light units (RLU). The data represent six replicates in each experimental group, the Mean ±SE are depicted as horizontal and vertical bars for each group, respectively, and one of three comparable experiments performed is shown. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p values are indicated. ns = not significant. a. Schematic representations of the luciferase reporter constructs used in this study. b. Fold change in luciferase activity of 3′UTRs of HLA-C alleles as compared to that of Cw*0602.
The expression of endogenous mature miR-148b and miR-152, another miR-148 miRNA family member, was very low as compared to miR-148a, and miR-657 was undetectable in HLA-C homozygous BLCL and B721.221 (Supplementary Fig. 6). These data point to the involvement of miR-148a rather than miR-148b, miR-152, or miR-657 in regulation of HLA-C expression. Additionally, disruption of the miR-657 binding site by site directed mutagenesis had no effect on luciferase activity (Supplementary Fig. 7a & b), indicating that miR-657 does not affect HLA-C expression.
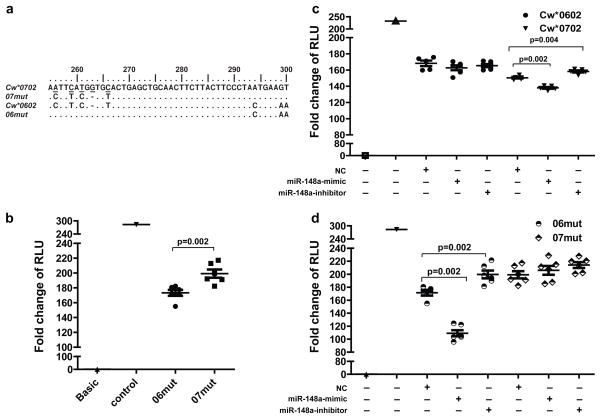
To test whether variants in the miR-148a binding site account for the differential gene expression patterns, we swapped positions 256-266 of the 3′UTR of Cw*0602 to match those of Cw*0702 and vice versa, thereby providing an intact miR-148a binding site to the 3′UTR of Cw*0602 (06mut) and disrupting the binding site for miR-148a in the Cw*0702 3′UTR (07mut), but leaving the remainder of the 3′UTR sequences intact (Fig. 2a). The luciferase activity of 06mut was significantly lower than that of 07mut (Fig. 2b), indicating that the polymorphisms between positions 256–266 in the miR-148a binding region account for the difference in luciferase expression between constructs containing the 3′UTRs of Cw*0602 vs. Cw*0702. Two other polymorphic sites, A256C and A267G, in the miR-148a binding site (Supplementary Fig. 3a) distinguish different sets of alleles as compared to 263del/ins, but these two variants had no effect on miRNA-mediated suppression (Supplementary Fig. 7c & d).
Figure 2. Disruption of miR-148a target site rescues suppression.
a. Partial sequence of mutated 3′UTRs of Cw*0602 and Cw*0702 (06mut and 07mut, respectively) are aligned to 3′UTR sequences of native Cw*0602 and Cw*0702. Identical nucleotides are shown as dots, altered nucleotides are underlined, and deletions are indicated by hyphens (−) for optimal alignment. b. Fold change in luciferase activity of the modified 3′UTR (06mut and 07mut). c. Fold change in luciferase activity of reporters containing wild type Cw*0602 or Cw*0702 3′UTR sequences upon introduction of miR-148a mimic and inhibitor. d. Fold change in luciferase activity of reporters containing 06mut and 07mut 3′UTR sequences upon introduction of miR-148a mimic and inhibitor. Presence (+) or absence (−) of each variable, including a negative control (NC) miRNA, a mimic of miR-148a, or an inhibitor of miR-148a is shown. The data represent six replicates in each experimental group, the Mean ±SE are depicted as horizontal and vertical bars for each group, respectively, and one of three comparable experiments performed is shown. Non-parametric Wilcoxon-Mann-Whitney tests were used for statistical comparisons and two tailed p values are indicated.
Further validation of the differential regulation of HLA-C alleles by miR-148a was achieved by co-transfection of B221.227 cell lines with either a mimic or an inhibitor of miR-148a along with a luciferase reporter construct that contained the 3′UTR with 263ins (Cw*0702 or 06mut) or with 263del (Cw*0602 or 07mut). The normalized luciferase activity in cells transfected with the constructs containing the 263del allele (Cw*0602 and 07mut 3′UTR) was not significantly altered by co-transfection with either the mimic or the inhibitor (Fig. 2c and 2d). However, the mimic of miR-148a further repressed luciferase activity in cells transfected with the 263ins allele (Cw*0702 and 06mut 3′UTR), whereas co-transfection with inhibitor rescued the suppression significantly (Fig. 2c and 2d). These data provide further support for allele-specific miR-148a targeting of the HLA-C 3′UTR.
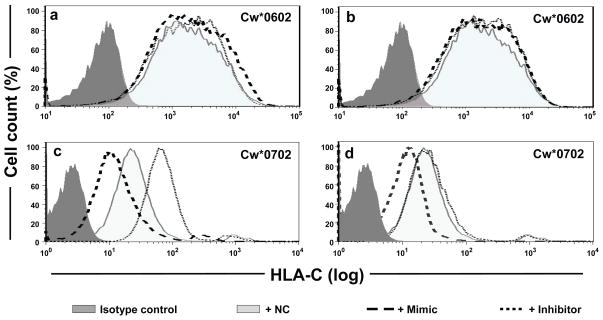
B Lymphoblastoid Cell Lines (BLCL) from individuals homozygous for either Cw*0602 (BLCL-Cw*0602Hom) or Cw*0702 (BLCL-Cw*0702Hom) were used to determine whether the variation in the miR-148a binding site affected endogenous HLA-C expression on the cell surface. As described previously, overall HLA-C expression on a Cw*0602 homozygous cell line was higher than that on a Cw*0702 homozygous cell line. As expected, transfection with mimics or inhibitors of miR-148a (Fig. 3a) and miR-148b (Fig. 3b) had no significant effect on cell surface expression of Cw*0602, an allele containing 263del in the 3′UTR that disrupts miR-148a/miR-148b binding. However, transfection of either miR-148a or miR-148b mimic resulted in decreased expression of Cw*0702 relative to cells transfected with a negative control (NC; Fig. 3c & d, respectively), indicating that increased levels of either of these miRNA can further downregulate HLA-C expression of alleles that contain an intact binding site for miR-148a/miR-148b (263ins), such as Cw*0702. The inhibitor of miR-148a significantly increased the level of endogenous Cw*0702 expression, but the inhibitor of miR-148b had no effect on expression of Cw*0702 (Fig. 3c & d, respectively), confirming the very low levels of miR-148b endogenous expression (Supplementary Fig. 6).
Figure 3. miR-148a affects cell surface expression of HLA-C.
Histograms of HLA-C cell surface expression on HLA-C homozygous BLCL using flow cytometry are illustrated. In each plot, a NC miRNA that does not bind to the 3′UTR of HLA-C was included. a–b. HLA-Cw*0602 homozygous cells (BLCL-Cw*0602Hom) transfected with either a mimic or an inhibitor of miR-148a (a) or miR-148b (b). c–d. HLA-Cw*0702 homozygous cells (BLCL-Cw*0702Hom) transfected with either a mimic or an inhibitor of miR-148a (c) or miR-148b (d).
The differential miR-148a regulation of expression across HLA-C alleles was precisely reflected in experiments involving: i) additional HLA-C homozygous BLCLs (Supplementary Fig. 8a & b), ii) use of another form of miR-148a inhibitor (peptide nucleic acid inhibitor; Supplementary Fig. 8c), and iii) analysis of total cellular HLA-C protein as determined by Western blot (Supplementary Fig. 9–10 and Supplementary note 1). We conclude that miR-148a regulates the expression of HLA-C in an allele-specific manner that is dependent on variation in the miR-148a binding site of the HLA-C 3′UTR.
The miR-148a binding site of the HLA-C 3′UTR is in strong LD with the -35 SNP that was shown to associate with control of HIV and HLA-C expression levels5–7 (D′=0.75, r2 =0.74; p<0.0001, N=1760). While there is no explanation for a direct causal effect of −35 variation on HLA-C expression7, the interaction between miR-148a and its polymorphic binding site in the 3′UTR of _HLA-C_ presents a clear rationale for variable levels of HLA-C expression. We determined the frequencies of the 263del/ins genotypes in a cohort of 2527 HIV infected European American (EA) individuals. Subjects with mean plasma viral loads of <2000 copies of viral RNA/ml of plasma (controllers) were enriched for 263del, whereas those with viral loads of >10,000 copies of viral RNA/ml of plasma (noncontrollers) had a significantly higher frequency of 263ins (Supplementary Table S1). Because of the strong LD across the HLA-C and HLA-B genes (Supplementary Table S2 and S3), we determined whether the 3′UTR variant has an effect on HIV control that is independent of individual HLA-A, -B, or -C alleles. A logistic regression approach with stepwise selection of the HLA-C 3′UTR 263 variant along with all HLA-A, -B, and -C alleles that have ≥1% frequency in our cohort (63 alleles) was employed. In this analysis, the 263del/del vs. 263ins/ins comparison remains significant along with five of the 63 HLA alleles (B*5701, B*5703, B*2705, B*5801 and Cw*1402; Table 1; for frequencies, see Supplementary Table S1). While these data suggest that the 3′UTR del/ins variant has an independent effect on HIV control (see Supplementary note 2 for potential mechanisms that could explain the association), we still cannot completely rule out the possibility that the strong LD in the region is confounding the results (Supplementary Table S4; Supplementary note 3).
Table 1.
Effect of HLA-C 3′UTR 263 on mean viral load using a logistic regression model by stepwise selection of HLA-A, -B and -C alleles
| Significant independent variables | p value | OR | 95% CI |
|---|---|---|---|
| del/del vs ins/ins | 2×10−14 | 0.33 | 0.25–0.43 |
| B*2705 vs others | 3×10−6 | 0.34 | 0.22–0.54 |
| B*5701 vs others | 1×10−12 | 0.21 | 0.14–0.32 |
| B*5703 vs others | 3×10−5 | 0.01 | 0.002–0.10 |
| B*5801 vs others | 9×10−4 | 0.27 | 0.12–0.59 |
| Cw*1402 vs others | 1×10−4 | 0.26 | 0.13–0.52 |
The extensive number of disease associations with HLA class I and II genes has largely been ascribed to the polymorphic peptide binding amino acid positions of these molecules. Some reports have speculated that gene expression13 and/or splicing patterns of the HLA genes14,15 may play a role, but convincing data is missing. Of interest, the HLA-G 3′UTR was shown to encode a polymorphic target site for miR-148a/b16,17. Levels of HLA-G have been suggested to alter risk of asthma in children of mothers with asthma17, though specificity of assays reporting HLA-G expression beyond implanting placental cells has been questioned18. Recently, a variant 35Kb upstream of the HLA-C gene was shown to associate with differential HLA-C mRNA expression, cell surface expression, and outcome after HIV infection5–8. We have now established a very convincing case that this −35 SNP is marking a functional insertion/deletion variant in the 3′UTR of HLA-C that directly determines expression of the various HLA-C allotypes differentially through miR-148a recognition. These data indicate another tier of diversity to the polymorphic HLA-C locus beyond that encoding variation in the peptide-binding region of the gene. We suggest that disease associated haplotypes may exert their effects through multiple mechanisms, including the type of peptides they bind and their level of expression, and that it is the combination of these that then determines the overall susceptibility status of the haplotype.
Expression levels of different HLA-C allotypes occur as a continuous gradient rather than the bimodal expression pattern that would be expected if miR-148a regulation were the sole mechanism involved. Thus, additional cis acting factors may fine tune HLA-C expression in an allotype-specific manner. Trans acting factors unlinked to the HLA-C locus may also affect expression levels in a manner that is independent of HLA-C allotype, leading to some degree of variation in expression levels of a given HLA-C allotype. Although the system regulating HLA-C expression is multifactorial, the significant involvement of miRNA in this process provides new approaches for manipulation of the immune system in the treatment of human disease.
Methods Summary
DNA from 2527 HIV+ patients of European descent was used to determine the effect of the HLA-C 3′UTR variation on control of HIV viral load. Viral load measurements were obtained from participants of the Multicenter AIDS cohort study (MACS)19, Swiss HIV Cohort (http://www.shcs.ch), the SCOPE cohort20, and the International HIV Controllers Study Cohort (www.hivcontrollers.org). Individuals were grouped into those who maintain mean VL<2000 (controllers) and those who have mean VL>10,000 (noncontrollers).
Complete HLA-C 3′UTR fragments were amplified, inserted into _Xba_I site downstream of the luciferase gene in a pGL3-control vector (Promega), and transfected into B721.221, BLCL and Jurkat cells using AMAXA nucleofector (Lonza) and into 293T cells using Fugene6 (Roche). Luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega) and presented as fold change of relative light units21. For studies of miR-148a/miR-148b mimics and inhibitors (Dharmacon), 20 pmole/well of oligonucleotide mimics or inhibitors of miR-148a and miR-148b were transfected into the cells. Surface expression of HLA-C on BLCLs was analyzed using staining with DT9 antibody (kindly provided by V. Braud)22.
Total RNA was extracted using the Total RNA Purification Kit (Norgen). Relative quantification of miR-148a and miR-148b was performed using a _Taq_man real time PCR assay (Applied Biosystems) and RNU48 served as an endogenous RNA control.
SAS9.1 (SAS Institute) was used for data management and statistical analyses.
Supplementary Material
1
2
Acknowledgments
This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contracts HHSN261200800001E, N02-CP-55504, R01-DA04334 and R01-DA12568. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and the Cancer Inflammation Program Project Award for the year 2009, a grant from the Bill & Melinda Gates Foundation as part of the Collaboration for AIDS Vaccine Discovery, and the Mark and Lisa Schwartz Foundation. We would also like to thank the patients and investigators involved in the Multicenter AIDS Cohort Study (the MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute and the National Heart, Lung and Blood Institute grants UO1-AI-35042, 5-MO1-RR-00722 [GCRC], UO1-AI-35043, UO1-AI-37984, UO1-AI-35039, UO1-AI-35040, UO1-AI-37613, and UO1-AI-35041), the Swiss HIV Cohort Study (see Supplementary note 4 for the list of members), supported by the Swiss National Science Foundation grant number 33CSC0-108787, and the SCOPE study, which was funded by the UL1 RR024131 (Clinical and Translational Sciences Award) and P30 AI27763 (Center for AIDS Research) grants. We thank Stephen Anderson, Geraldine O’Connor, and Rasmi Thomas for helpful advice, Anthony Kronfli and Karthika Ramakrishnan for assistance in plasmid and genomic DNA preparations, Adelle McFarland for western blots, Veronique Braud for the DT9 antibody, Randall Johnson and George Nelson for statistical advice and Teresa Covell for administrative assistance.
Footnotes
Author Contributions
S.K, R.S. performed and evaluated the microRNA experiments. S.K., R.S., H.A.Y, and M.C. designed the study. M.C. directed the study. S.K., R.S., and M.C. wrote the manuscript. X.G., Y.Y., S.B. and M.M. genotyped HLA. Statistical analysis was performed by Y.Q. The clinical samples and data were contributed by P.H., D.D., A.T., D. G, S.W., F.P. and B.W. Intellectual input was provided by all authors.
Author Information Reprints and permissions information is available at www.nature.com/reprets.
The authors declare no competing financial interests.
References
- 1.Zemmour J, Parham P. Distinctive polymorphism at the HLA-C locus: implications for the expression of HLA-C. J Exp Med. 1992;176:937–950. doi: 10.1084/jem.176.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCutcheon JA, Gumperz J, Smith KD, Lutz CT, Parham P. Low HLA-C expression at cell surfaces correlates with increased turnover of heavy chain mRNA. J Exp Med. 1995;181:2085–2095. doi: 10.1084/jem.181.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Snary D, Barnstable CJ, Bodmer WF, Crumpton MJ. Molecular structure of human histocompatibility antigens: the HLA-C series. Eur J Immunol. 1977;7:580–585. doi: 10.1002/eji.1830070816. [DOI] [PubMed] [Google Scholar]
- 4.Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 5.Fellay J, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–947. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International HIV Controllers Study. The Major Genetic Determinants of HIV-1 Control Affect HLA Class I Peptide Presentation. Science. 2010;310:1551–1557. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas R, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41:1290–1294. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stranger BE, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1:e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 10.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 11.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 12.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaefer MR, et al. A novel trafficking signal within the HLA-C cytoplasmic tail allows regulated expression upon differentiation of macrophages. J Immunol. 2008;180:7804–7817. doi: 10.4049/jimmunol.180.12.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kralovicova J, Vorechovsky I. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J Immunol. 2006;176:2381–2388. doi: 10.4049/jimmunol.176.4.2381. [DOI] [PubMed] [Google Scholar]
- 15.Krangel MS. Secretion of HLA-A and -B antigens via an alternative RNA splicing pathway. J Exp Med. 1986;163:1173–1190. doi: 10.1084/jem.163.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castelli EC, et al. In silico analysis of microRNAS targeting the HLA-G 3′ untranslated region alleles and haplotypes. Hum Immunol. 2009;70:1020–1025. doi: 10.1016/j.humimm.2009.07.028. [DOI] [PubMed] [Google Scholar]
- 17.Tan Z, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet. 2007;81:829–834. doi: 10.1086/521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Apps R, Gardner L, Moffett A. A critical look at HLA-G. Trends Immunol. 2008;29:313–321. doi: 10.1016/j.it.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Phair J, et al. Acquired immune deficiency syndrome occurring within 5 years of infection with human immunodeficiency virus type-1: the Multicenter AIDS Cohort Study. J Acquir Immune Defic Syndr. 1992;5:490–496. [PubMed] [Google Scholar]
- 20.Emu B, et al. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J Virol. 2005;79:14169–14178. doi: 10.1128/JVI.79.22.14169-14178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Wright PW, Anderson SK. Identification and analysis of novel transcripts and promoters in the human killer cell immunoglobulin-like receptor (KIR) genes. Methods Mol Biol. 2010;612:377–391. doi: 10.1007/978-1-60761-362-6_26. [DOI] [PubMed] [Google Scholar]
- 22.Braud VM, Allan DS, Wilson D, McMichael AJ. TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr Biol. 1998;8:1–10. doi: 10.1016/s0960-9822(98)70014-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2