Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells (original) (raw)
. Author manuscript; available in PMC: 2012 Apr 22.
Abstract
The transcription factor FOXP3 is essential for the suppressive function of regulatory T cells that are required for maintaining self-tolerance. We have solved the crystal structure of the FOXP3 forkhead domain, as a ternary complex with the DNA-binding domain of nuclear factor of activated T cells-1 (NFAT1) and a DNA oligonucleotide from the interleukin-2 promoter. A striking feature of this structure is that FOXP3 forms a domain-swapped dimer that bridges two molecules of DNA. Structure-guided or autoimmune disease (IPEX)-associated mutations in the domain-swap interface diminished dimer formation by the FOXP3 forkhead domain without compromising FOXP3 DNA binding. These mutations also eliminated T cell suppressive activity conferred by FOXP3, both in vitro and in a murine model of autoimmune diabetes in vivo. We conclude that FOXP3-mediated suppressor function requires dimerization through the forkhead domain, and that mutations in the dimer interface can lead to the systemic autoimmunity observed in IPEX patients.
INTRODUCTION
FOXP3 is one of four members of the FOXP subfamily of Forkhead (FOX) transcription factors (Hannenhalli and Kaestner, 2009). FOXP3 is expressed in regulatory T (Treg) cells, a specialized subset of CD4+ T cells that suppress immune responses, particularly those of autoreactive T cells (Chatila, 2009; Fontenot et al., 2005; Josefowicz and Rudensky, 2009; Shevach, 2009; Workman et al., 2009; Ziegler, 2006). In humans, inactivating mutations in FOXP3 result in a severe autoimmune syndrome termed IPEX (immune dysregulation, polyendocrinopathy, enteropathy, X-linked), which manifests in affected male infants as lymphocyte infiltration and multi-organ inflammation (Bennett et al., 2001; Brunkow et al., 2001; Wildin et al., 2001). A similar pathology is observed in the naturally-occurring mouse mutant scurfy, which bears a nonsense mutation in the Foxp3 gene and lacks FOXP3 protein expression (Bennett et al., 2001; Brunkow et al., 2001; Wildin et al., 2001). Targeted deletion of Foxp3 in CD4+ T cells of neonatal or adult mice also results in severe autoimmunity, emphasizing the ongoing importance of Foxp3 for regulatory T cell function (Williams and Rudensky, 2007). Finally, ectopic expression of FOXP3 in conventional CD4+ T cells confers suppressor function, enabling the transduced T cells to suppress the proliferation of bystander T cells in vitro and limit the effector functions of autoreactive T cells in vivo (Fontenot et al., 2003; Hori et al., 2003). While numerous studies have elucidated the conditions and signaling pathways required for FOXP3 expression, the biochemical mechanisms by which FOXP3 induces T regulatory functions in cells in which it is expressed are still poorly understood.
Chromatin immunoprecipitation combined with microarray analyses revealed numerous transcriptional targets of human and murine FOXP3, including genes whose expression is upregulated (Cd25, Ctla4, Tnfrsf18 (Gitr)) or repressed (Il2, Ptpn22) (Marson et al., 2007; Zheng et al., 2007). FOXP3 cooperates with additional transcription factors, such as NFAT1 and RUNX1, to regulate gene expression (Wu et al., 2006) (Kitoh et al., 2009; Ono et al., 2007; Rudra et al., 2009). Transcriptional repression by FOXP3 may reflect its ability to recruit histone deacetylase (HDAC) family members through an N-terminal repressor domain (Li and Greene, 2007; Li et al., 2007a). However, the same N-terminal domain is also responsible for transcriptional activation by FOXP3 (Wu et al., 2006), and the stimulatory effects of FOXP3 on gene expression cannot be explained by recruitment of HDAC co-repressor complexes alone.
IPEX is associated with both nonsense and missense mutations in FOXP3 (van der Vliet and Nieuwenhuis, 2007). FOXP3 forms dimers through a leucine zipper (LZ) region implicated in homo- and hetero-oligomerisation (Chae et al., 2006; Li et al., 2007b; Lopes et al., 2006), and an IPEX deletion mutation (ΔE251) in this region is predicted to disrupt oligomerisation (Chae et al., 2006; Li et al., 2007b; Lopes et al., 2006). Because the forkhead (FKH) DNA-binding domain of FOXP3 (~80 amino acids) is located at the C-terminal end of the protein, all nonsense mutations compromise FOXP3 DNA binding. The majority of missense mutations in IPEX also localize to the FKH domain of FOXP3 (Ochs et al., 2007; Torgerson and Ochs, 2007); however, only a single IPEX mutation, A384T (Bennett et al., 2001; Wildin et al., 2001) lies within Helix H3 in the structural interface that mediates DNA contact (Wu et al., 2006). Thus the mechanisms by which other IPEX mutations in the FKH domain affect FOXP3 function remain to be explored.
To gain structural insights into the mechanisms by which FOXP3 modulates gene expression and confers suppressor functions, we solved the crystal structure of the FKH domain of FOXP3 bound to DNA in conjunction with the DNA-binding domain of its partner, nuclear factor of activated T cells-1 (NFAT1). A prominent feature we observed was that the FKH domain of FOXP3 formed a domain-swapped dimer, simultaneously engaging and bringing into close proximity two distant FOXP3 binding sites in DNA. Based on comparison with other FOX proteins, we introduced structure-guided mutations into the dimerization interface of the FKH domain; these mutations did not affect FOXP3 expression or DNA-binding, but caused dysregulated expression of a subset of FOXP3 target genes and substantially diminished the ability of FOXP3 to confer suppressive functions on retrovirally-transduced T cells in culture and in mice. Two known IPEX mutations in the domain-swap interface had similar biological effects. Our data highlight the importance of dimer formation for transcriptional regulation by FOXP3, and indicate that two distinct dimer interfaces of FOXP3, the LZ motif and the domain swap interface in the FKH domain, are required for FOXP3-mediated induction of T regulatory cell function.
RESULTS
Overall structure of the NFAT1:FOXP3:DNA complex
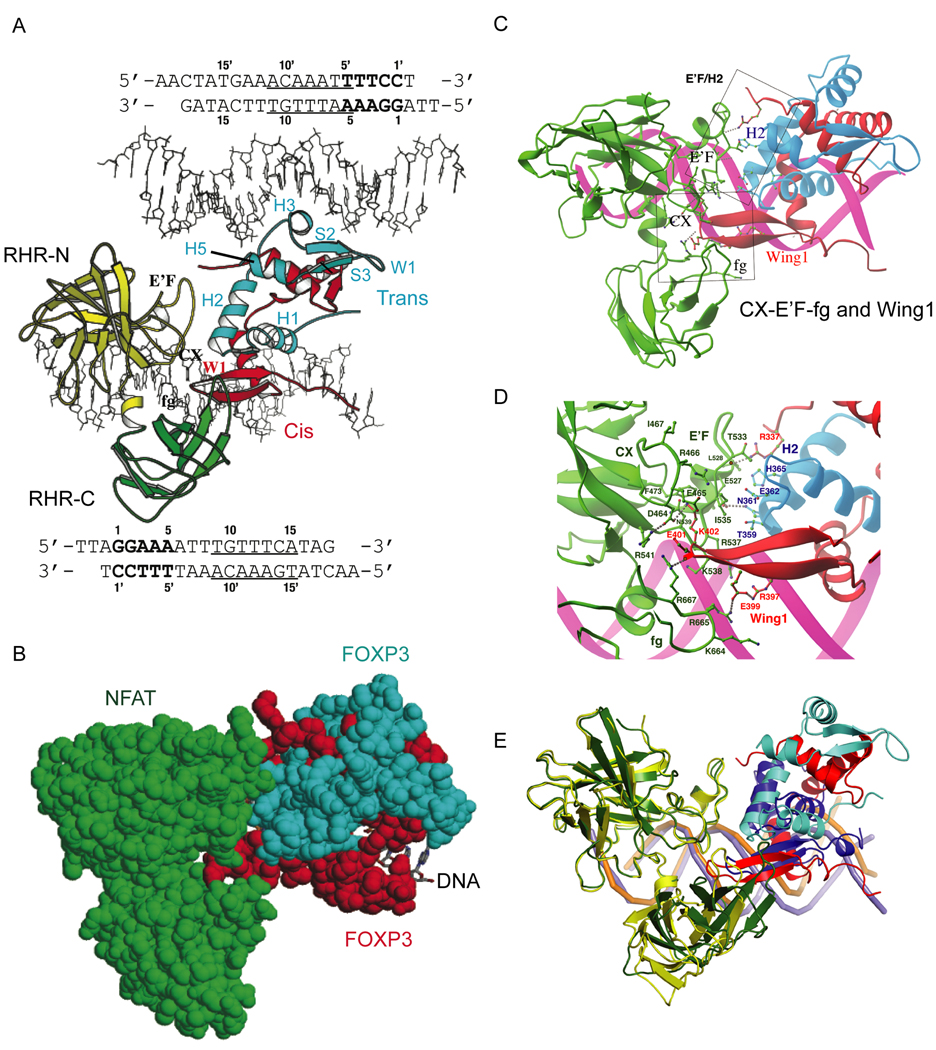
We solved a high-resolution crystal structure of a ternary complex containing the NFAT1 DNA-binding domain (Rel homology region, RHR) and the FOXP3 FKH domain bound to DNA (ARRE2 site of the interleukin-2 (IL-2) promoter) (Figure 1A, Figure S1). The asymmetric unit (Figure S1) contained two copies of the NFAT1 RHR, four copies of the FOXP3 FKH domain (i.e. two domain-swapped dimers) and two DNA molecules. These formed two sets of nearly identical protein-DNA complexes, each containing one ARRE2 DNA bound by one NFAT and one domain-swapped FOXP3 dimer. Figure 1A shows part of the asymmetric unit (NFAT1 and FOXP3 dimer bound to the upper DNA omitted for clarity). The FOXP3 dimer bound a different cognate FOXP site on each DNA, thus bridging two molecules of DNA (see below).
Figure 1. Overall structure of the NFAT1:FOXP3:DNA complex.
(A) Arrangement of protein and DNA components in the asymmetric unit. The N-terminal (RHR-N: yellow) and C-terminal domains (RHR-C: green) of NFAT1, and the cis- (red) and trans-monomer (cyan) of FOXP3 are shown. The sequence of each DNA molecule (stick model) is shown along its orientation in the crystal. The FOXP3 binding site of the cis-monomer (bottom DNA) and trans-monomer (top DNA) are underlined. The NFAT site is in bold. (B) Protein-protein interactions between NFAT1 and FOXP3. The NFAT1 (green) and FOXP3 dimer (red and cyan) are shown in space-filling model and DNA in stick model (C) Structural elements of each FOXP3 monomer contribute to protein-protein interactions with NFAT1. Helix H2 of the trans-monomer forms an extensive interaction interface with the E’F loop of NFAT1. The Wing1 of the cis-monomer inserts into a groove formed by the CX loop, the C-terminal stem of the E’F loop, and the fg loop of NFAT). (D) Detailed interactions between NFAT and the FOXP3 dimer. Interacting residues are coloured and labelled according to their respective proteins. (E) Superposition of the complex of NFAT1 (yellow), FOXP3 dimer (red and cyan) and DNA (purple) with the complex of NFAT1 (green), FOXP2 monomer (blue) and DNA (orange). The DNA (orange) in the NFAT1:FOXP2:DNA complex is clearly bent whereas DNA (purple) in the NFAT1:FOXP3:DNA complex is nearly straight. The NFAT-binding elements (helix H2 and wing W1) of FOXP2 and the FOXP3 dimer are aligned similarly toward NFAT but with a distinct shift.
The FOXP3 FKH domain has an optimized NFAT-interaction interface
Each domain-swapped dimer of FOXP3 contains two intertwined globular units with the typical structure of the FKH domain, engaged in intimate contacts with NFAT (Figure S1, Figure 1B, Figure 2). The details of the protein-protein interaction between NFAT and FOXP3 (Figures 1C, 1D) are similar to those in the monomeric NFAT:FOXP2 interface (Stroud et al., 2006; Wu et al., 2006), supporting our previous analysis (Wu et al., 2006) and confirming that Thr359, Asn361, His365, Glu399, Glu401 of FOXP3 are located in the centre of the NFAT1:FOXP3 interface and engage in extensive hydrogen bonding, van der Waals contacts, and electrostatic interactions with NFAT (Figure 1C, 1D).
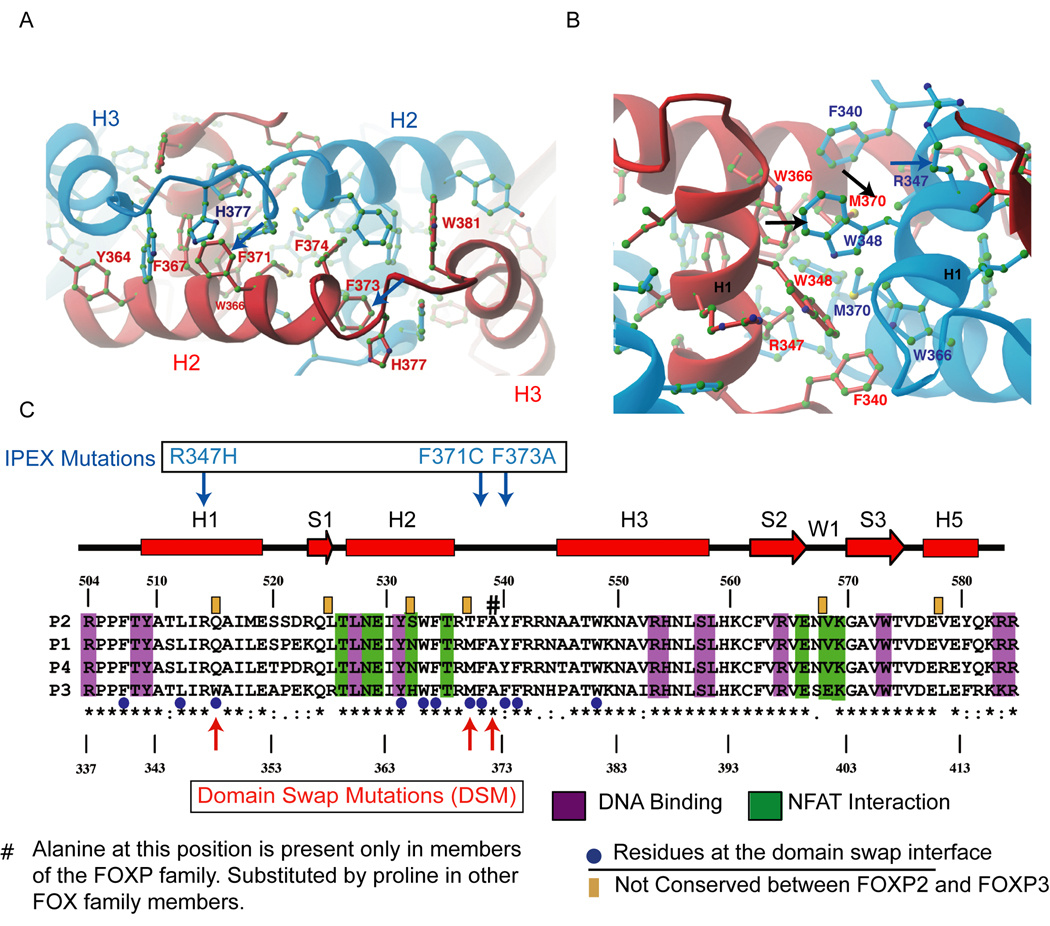
Figure 2. The dimerisation interface in the forkhead domain of FOXP3.
(A) An interaction network of aromatic residues at the domain-swapped dimer interface present in both FOXP2 and FOXP3 (see Figure S3, top left). The red and blue monomers of FOXP3 are shown with residues coloured according to the monomer to which they belong. The different conformations of His377 in the two monomers are induced by different DNA binding interactions at Site A and Site B (see Figure S2 for details) (B) An additional hydrophobic core (exclusive to FOXP3, see Figure S3, top right) that enhances the stability of the domain-swapped FOXP3 dimer. (C) Sequence alignment of FOXP family. The secondary structural elements are shown above the sequence. Numbering of FOXP2 is shown above whereas that of FOXP3 is shown below. The homology is indicated by standard convention below of the sequence alignment. Residues involved in DNA binding (magenta), NFAT interaction (green), and domain swapping (blue dots) are highlighted. The six residues that differ between FOXP2 and FOXP3 forkhead domains are indicated by orange boxes above the sequence alignment. The blue downward-pointing arrows indicate the positions of IPEX mutations R347H, F371C and F373A that occur at the domain-swap interface, and the red upward arrows and the pound sign depict the position of the structure-guided mutations that we engineered into the interface (see text )
Besides the similarities, there are some notable differences in the binding of FOXP2 and FOXP3 to NFAT. Compared to the NFAT1:FOXP2 interface, the NFAT:FOXP3 interface is a better fit and has better chemical complementarity. In particular, the NFAT1:FOXP3 interface is larger than that of NFAT1:FOXP2, burying about 1030 Å2 solvent-accessible surface area as compared with the 623 Å2 buried in the NFAT1:FOXP2 interface. Moreover, superposition of the backbone of the NFAT RHR N-terminal domain (RHR-N) of the two complexes shows that Wing1 of the cis-monomer of FOXP3 inserts more deeply into the CX-E’F-fg groove than Wing1 of FOXP2; as a result, the C-terminal domains of the NFAT RHR in the two complexes (RHR-C) show a distinct shift (Figure 1E). At E’F loop and H2 helix interface, His365 of FOXP3 made extensive van der Waals contacts with the E’F loop of NFAT (Figure 1D); the corresponding residue in FOXP2 is Ser532, which is too short to interact with the E’F loop directly (Wu et al., 2006). At the Wing1 and CX-E’F-fg loop interface, Glu399 and Glu401 form electrostatic interactions with the basic fg loop of NFAT that contains a string of positively charged residues, Lys664, Arg665, Lys666, and Arg667 (Figure 1D). In contrast in FOXP1, FOXP2 and FOXP4, Glu401 corresponds to a valine, which may not interact with the basic fg loop of NFAT as favorably (Wu et al., 2006) (see Figure 1C). Overall, therefore, our structural analyses indicate that FOXP3 has a better binding surface for NFAT than FOXP2.
The FKH domain of FOXP3 forms a domain-swapped dimer
A striking feature of the crystal structure is that the FOXP3 FKH domain exists in the complex as a stable domain-swapped dimer that bridges two molecules of DNA (Figure 1A, Figure S1). The domain-swapped dimer of FOXP3 is formed by an exchange of structural elements: Helix1 (H1), Strand1 (S1) and Helix2 (H2) of one monomer folded upon Helix3 (H3), Strand2 (S2) and Strand3 (S3) of the other (Figure 1A; secondary structure labels follow published conventions (Stroud et al., 2006)). The H2 helices are extended, displacing the DNA-binding helix H3 from its own monomer and causing it to be stabilized through interactions with helices H1 and H2 of the alternate monomer (Figure 1A).
One monomer (the cis-monomer, red in Figure 1A) bound to a FOXP site located toward the 3’ end of ARRE2: 5’-TGGAAAATTTGTTTCA-3’ (Figure 1A, site B, underlined; sequence in bold is the NFAT binding site). The other monomer (the trans-monomer, blue in Figure 1A) bound another FOXP site on a separate ARRE2 fragment: 5’-TGGAA AATTTGTTTCA-3’ (Figure 1A, site A, underlined). The detailed mechanisms of FOXP3-DNA recognition on site B (Figure S2A) and site A (Figure S2B) are similar to one another and to those observed in DNA complexes bound by monomeric FOXP2 (Stroud et al., 2006). Similarly the structure of the DNA-binding domain (RHR) of NFAT and its DNA-binding interactions are similar to those observed in previously characterized NFAT complexes (Bates et al., 2008; Chen et al., 1998; Giffin et al., 2003; Jin et al., 2003; Stroud and Chen, 2003)(data not shown).
Domain swapping in FOXP3 is mediated by an extended network of hydrophobic residues, including a series of aromatic residues (Tyr364, Trp366, Phe367, Phe371, Phe373, Phe374, and Trp381) that are highly conserved within the FOX superfamily (Figure 2A, highlighted by blue dots in the alignment of Figure 2C); two mutations in this interface, F371C and F373A, have been described in IPEX patients (Figure 2C). The stability of the domain-swapped dimer is enhanced by an additional hydrophobic interface, containing residues Phe340, Leu345, Trp348, Trp366 and Met370 (Figure 2B). At the centre of this hydrophobic interface are Trp348 and Met370, which correspond to Gln515 and Thr537 in FOXP2, respectively; the IPEX mutation R347H is also in this interface (Figure 2C). The sequence differences in the hydrophobic cores of FOXP2 and FOXP3 may explain the differences in structure and stability of their respective domain-swapped dimers, as further validated below.
The FOXP3 FKH domain is likely to form the most stable domain-swapped dimer in the FOXP family. The sequence of the 81-residue FKH domains is highly conserved in the FOXP family, with only six non-homologous residues (highlighted by brown bars above the sequence in Figure 2C). Two of these six residues (His365 and Glu401 of FOXP3) occur at the centre of the NFAT1:FOXP3 interface while another two (Trp348 and Met370) are located at the core of the domain-swapping dimer interface, suggesting that the limited variation of surface residues in the FOXP3 FKH domain confers unique functions on FOXP3 within the FOXP family. In particular, Trp348 in the domain-swap interface is conserved across species in FOXP3 but is absent in FOXP1, FOXP2 and FOXP4 (Figure 2C). Comparison of the FOXP2 (Stroud et al., 2006) and FOXP3 FKH domain structures shows that the domain-swapped FOXP2 dimer is much less arched than the corresponding FOXP3 dimer, and thus lacks one of the dimerisation interfaces present in FOXP3 (Figure S3A). Collectively these crystal-structure data reveal that among the different FOXP family members the FKH domain of FOXP3 is most optimized to form a domain-swapped dimer, and also to interact effectively with NFAT.
The isolated FKH domain of FOXP3 is a dimer in solution in the presence or absence of DNA
To rule out the possibility that dimerization of the FOXP3 FKH domain occurred only at the high concentrations required for crystallization, we monitored dimer formation by the FKH domain in solution (Figure 3A). In the presence of the protein cross-linker DSS, the FOXP3 FKH domain was predominantly a dimer (Figure 3A, lane 2). A ternary complex of FOXP3, NFAT and DNA was observed only in the presence of DNA containing the composite ARRE2 element (Figure 3A, lane 4): this complex was not formed in the absence of DNA (lane 6) or in the presence of non-specific DNA (lane 5). These data confirm that the FOXP3 dimer observed in the NFAT:FOXP3:DNA crystal structure is maintained in solution, and that the cooperative NFAT:FOXP3 complex binds to specific DNA sequences (Wu et al., 2006).
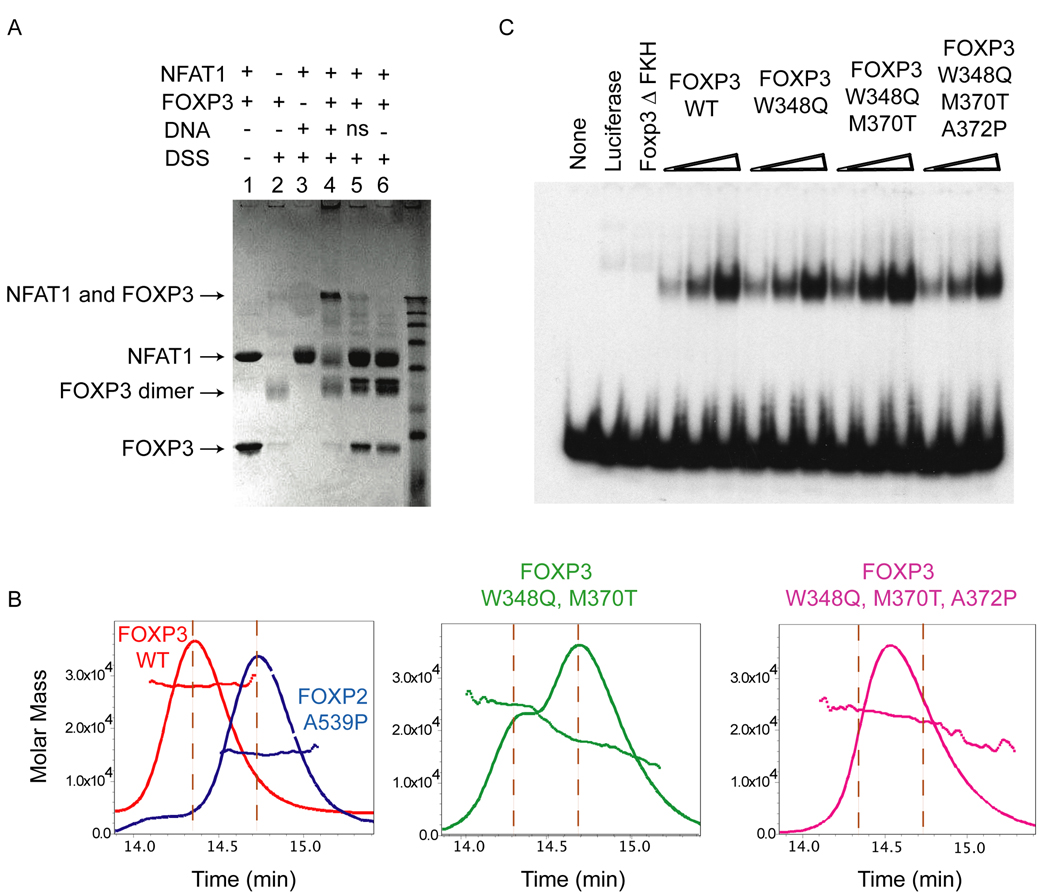
Figure 3. DSM mutations affect dimerization, but do not impair DNA binding by the forkhead domain of FOXP3.
(A) Denaturing SDS gel showing formation of a crosslinked FOXP3 dimer after DSS crosslinking (lanes 1 and 2), and sequence-dependent cross-linking of NFAT1 and FOXP3 on the ARRE2 DNA element (lanes 3–6). The composition of each reaction is indicated above the lane, and the concentrations of each component are described in Methods. ns, nonspecific DNA. (B) SEC-MALS analysis of the forkhead domains of WT-FOXP3 and the indicated mutants. FOXP3 exists as a stable dimer in solution and elutes as a single peak (~26 kDa, left panel); the double mutant; W348Q and M370T FOXP3 elutes as two peaks of 26 kDa and 13 kDa representing the dimeric and monomeric forms (middle panel); and the triple DSM mutant; W348Q, M370T and A372P elutes as a single polydisperse peak (right panel). (C) EMSA analysis depicting binding of WT-FOXP3 and indicated mutants to a DNA probe containing optimized FOXP3 binding sequences. The mutations do not impair DNA binding by FOXP3.
Corroborating the cross-linking studies, the FOXP3 FKH domain eluted as a single peak in size exclusion chromatography followed by multi-angle light scattering (SEC-MALS) analysis; the molecular weight measured across the entire peak corresponded to a dimer of 26 kDa, suggesting that the FOXP3 dimer does not exchange into monomer at the lower concentrations prevailing at the edge of the peak (Figure 3B, left panel). We also did not observe any FOXP3 monomer in size exclusion chromatography in dilute solutions (< 0.1 mg/ml, data not shown). In contrast, FOXP2 eluted in SEC-MALS experiments as a mixture of monomers and dimers ((Stroud et al., 2006); see Figure S3B, left panel), and mutation of the critical alanine (Ala539 in FOXP2) to proline, conserved in most other FOX proteins, rendered the FOXP2 FKH domain completely monomeric (Figure 3B, left panel; also shown in Figure S3B, right panel). Thus, the propensity to form dimers is much stronger in FOXP3 than in FOXP2, and the FOXP3 FKH domain exists primarily as a stable dimer in solution.
To test whether the unique hydrophobic residues Trp348 and Met370, which are centrally located at the domain-swap interface, imparted enhanced stability to the FOXP3 dimer, we replaced Trp348 and Met370 with Gln and Thr, the residues present at the equivalent positions in FOXP2 (see Figure 2B and 2C). As predicted by the crystal structure, these substitutions destabilized dimerisation of the FOXP3 forkhead domain. SEC-MALS analysis showed that the double mutant FOXP3 (W348Q and M370T) eluted as two peaks of 26 kDa and 13 kDa representing the dimeric and monomeric forms (Figure 3B, middle). An additional substitution of Ala372 to proline in FOXP3, analogous to the FOXP2 A539P substitution which renders FOXP2 completely monomeric (Stroud et al., 2006) (Figure 3B, left), caused the triple mutant (W348Q, M370T and A372P; hereafter referred to as FOXP3 Domain Swap Mutant, DSM) to elute as a single peak on a size exclusion column (Figure 3B, right). MALS analysis revealed that this peak was polydisperse, suggesting that the triple mutant still undergoes some degree of dynamic association in solution. Nevertheless, these biochemical analyses corroborate the crystal structure by confirming that residues Trp348 and Met370 participate in promoting dimerization of the FOXP3 FKH domain.
Disruption of the domain-swap interface of FOXP3 does not impair DNA binding
As predicted by the crystal structure, the DSM mutations did not affect FOXP3 DNA-binding. In electrophoretic mobility shift assays (Koh et al., 2009), wild-type ΔN-FOXP3 (amino acids 182–431) and ΔN-FOXP3 bearing the domain-swap mutations bound equivalently to the optimized probe (Figure 3C). Together these data confirm that the domain-swap interface is not involved in FOXP3 DNA contact.
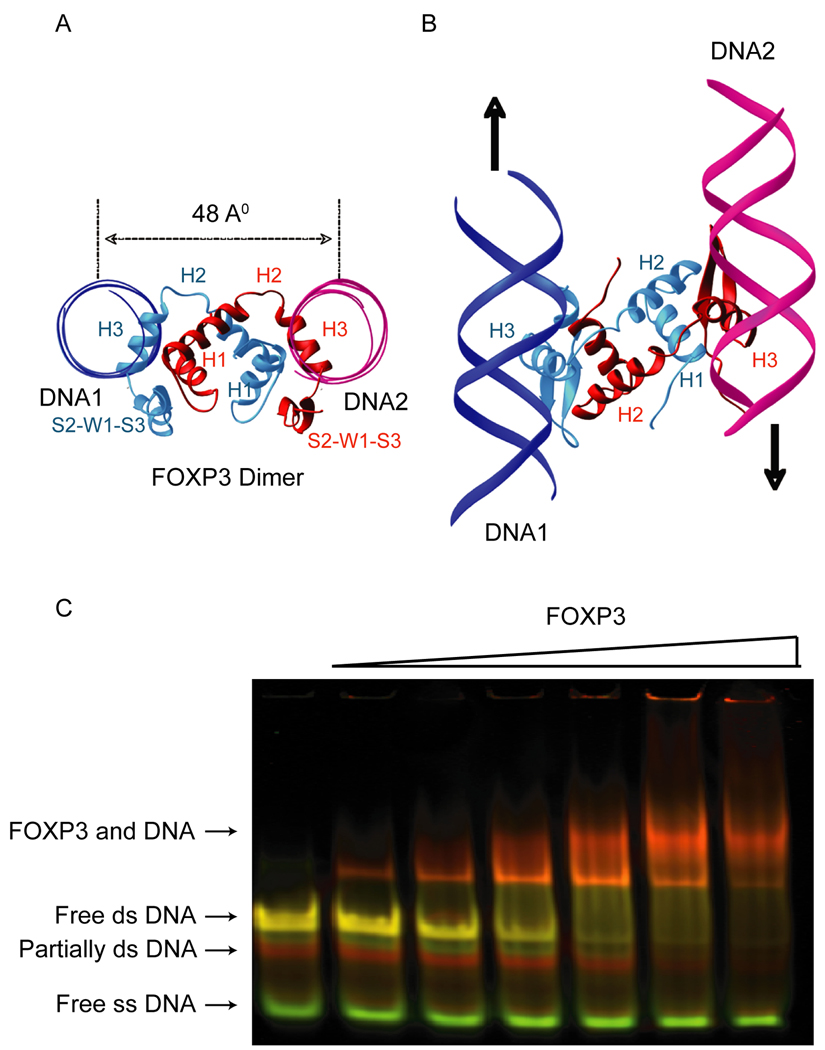
The FKH domain of FOXP3 can bring together two DNA binding elements in solution
Domain swapping caused each FOXP3 monomer in the crystal structure to assume an extended conformation that oriented the two DNA binding helices (H3) away from each other with a distance of 48 Å from the centre of one DNA helix to the other (Figure 4A). Furthermore, the two DNA-binding H3 helices bound DNA in anti-parallel orientation (Figure 4B) making it unlikely that the FOXP3 FKH dimer would be able to bind two closely-spaced FOXP3 binding sites. Keeping these structural constraints in mind, we tested the ability of the FOXP3 FKH domain to engage two distinct FOXP3-binding sites simultaneously in solution. We designed a specialized synthetic probe for use in electrophoretic mobility shift assays (EMSA), containing FOXP3 binding sites at the 5’ and 3’ ends that were labeled with the fluorescence energy transfer (FRET) donor-acceptor pair Cy3 and Cy5 respectively and were separated by a flexible 19-base single-stranded DNA linker. The probe was incubated with increasing concentrations of recombinant FOXP3 FKH domain in an in-gel FRET assay. Even at the lowest protein concentrations tested, a clear FRET signal was visible from the protein-DNA complex (Figure 4C). While these data do not distinguish whether the FOXP3 FKH domain bridges two DNA sites inter- or intra-molecularly, they provide clear evidence that the FOXP3 FKH dimer can simultaneously bind two distinct FOXP3-binding sites in solution and bring them into close approximation.
Figure 4. The FOXP3 forkhead domain can bring together two FOXP3 DNA binding elements in solution.
The domain-swapped FOXP3 dimer (red and cyan) binds two separate DNA molecules (magenta and blue, only backbones are shown in ribbons). (A) View along the DNA axis. (B) View from the top. (C) In-Gel FRET analysis: Pseudo-colored image showing an overlay of the fluorescence of the Cy3-donor (green) and the fluorescence of the Cy5-acceptor (red) fluorophores. Labels to the left indicate the respective DNA/ protein species. Note, unannealed probe (free ss DNA, green) is not bound by FOXP3 forkhead dimer even at high protein concentrations and remains unretarded during electrophoresis.
Mutations that affect domain swapping differentially affect the expression of FOXP3 target genes
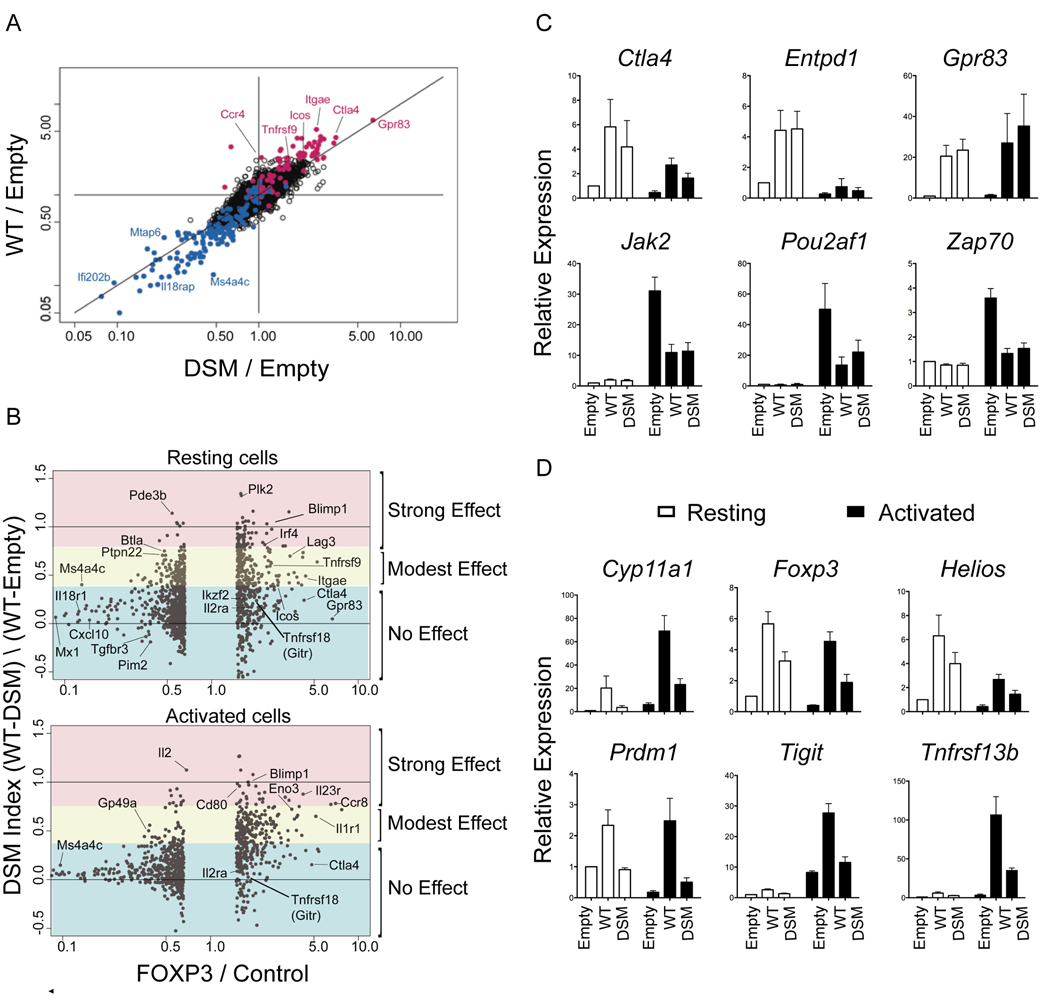
To determine whether the triple DSM mutation affected FOXP3 transcriptional activity, we performed microarray-based transcriptional profiling of T cells retrovirally transduced with wild type (WT) and DSM FOXP3. Naïve T cells, obtained from OT-II TCR-transgenic mice on a _Tcra_−/− background, were transduced with control (empty) MSCV IRES-Thy1.1 retrovirus or retroviruses encoding wild type WT or DSM FOXP3; flow cytometric analyses performed 72 hr after retroviral infection confirmed that WT and mutant FOXP3 were expressed at comparable levels in primary murine T cells (Figure S4). Successfully transduced T cells were sorted 72 hr after retroviral infection; RNA was isolated from a portion of the cells immediately, and from another portion after re-stimulation for 4 hours with phorbol myristate acetate (PMA) and ionomycin. The data (average of two experiments) showed that the transcriptional signature of FOXP3 was largely preserved: genes induced (red) or repressed (blue) by WT FOXP3 were also induced or repressed by the DSM mutant in both resting and activated cells (Figure 5A data not shown). However, the mutation did induce a clear off-diagonal shift for a subset of genes, indicating that for these genes, the DSM mutations diminished both the activating and repressive effects of FOXP3.
Figure 5. Mutations that disrupt domain swapping differentially affect regulation of FOXP3 target genes.
(A) Gene expression in transfected cells represented as FoldChange versus FoldChange (FcFc) plots comparing the effect of transduction of WT and DSM Foxp3 relative to mock-transfected cells (resting conditions; all microarray profiles performed in duplicate). (B) Changes in gene expression as a function of the DSM mutation represented as DSM index (calculated from expression values, (WT-DSM) / (WT-Empty)) versus FoldChange in expression obtained upon FOXP3 transduction in resting (top) and activated cells (bottom). (C and D) Expression of indicated FOXP3 target genes in WT and DSM transduced T cells as estimated by real-time PCR analysis. Error bars represent the SEM observed in three independent experiments.
To better visualise the impact of the domain swap mutations, we calculated a “DSM index” for each gene from the averaged expression values (WT-DSM / WT-Empty) (see Suppl. Methods). The denominator of this index is positive for FOXP3-activated genes, negative for FOXP3-repressed genes, and 0 for genes whose expression was unaffected by FOXP3. As the denominator tends towards zero (i.e. for genes whose expression was only slightly altered by FOXP3), the DSM index tends to infinity in either direction; to minimize this problem, we have omitted genes whose expression was unaltered or only mildly affected by FOXP3 (Figure 5B, middle section of the graphs).
For both resting (Figure 5B, top) and activated (Figure 5B, bottom) cells, the “DSM index” highlighted the spectrum of effects caused by the DSM mutations. A considerable number of FOXP3-repressed genes were unaffected by the mutations (indices between 0 and 0.2); this was particularly true for strongly repressed genes. There were clear exceptions, however: for instance FOXP3 modestly repressed Btla, Pde3b, Ptpn22 and Il2 expression, but repression was eliminated by the domain-swap mutations (Figure 5B, left half of each panel, DSM indices > 0.5; Figure S5). Genes activated by FOXP3 showed a more extensive but usually partial effect (indices around 0.5 for Itgae (CD103), Lag3 and Tnfrsf9 (4-1BB), indicating that the DSM mutations eliminated ~50% of the activation by WT FOXP3). Activation of some transcripts was unaffected or only slightly affected by the DSM mutations (Gpr83, Il2ra (CD25, Ctla4), whereas activation of others was more strongly affected (Plk2, Blimp1, Ifr4).
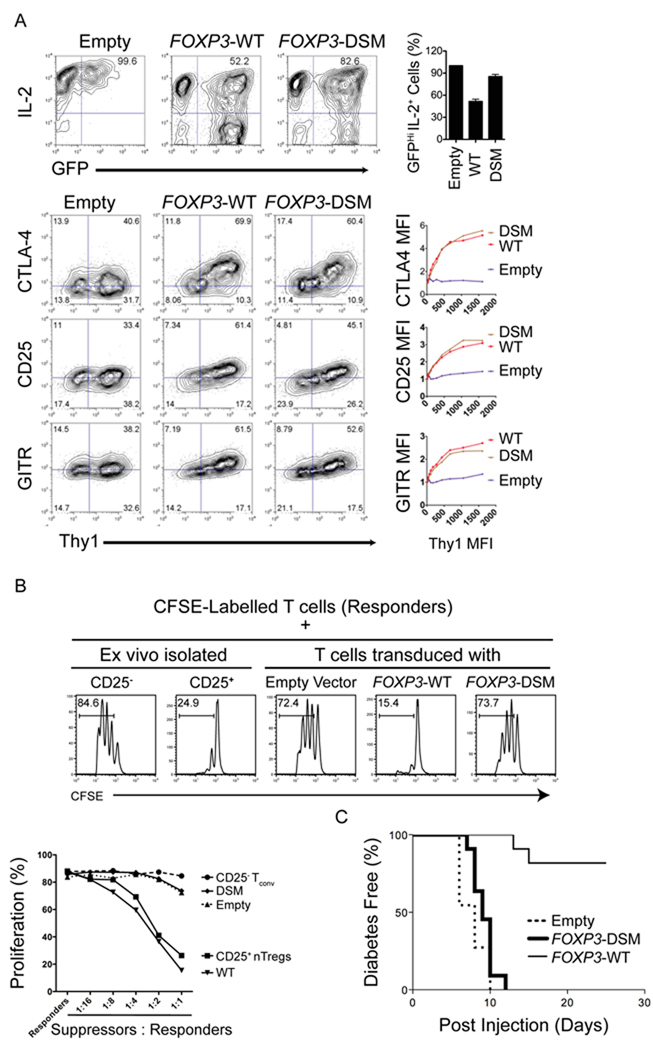
To confirm results obtained from the microarray analyses, we used real-time polymerase chain reaction (PCR) (Figures 5C, D) to quantify the expression of genes observed by transcriptional profiling to be either unaffected or dysregulated by the DSM mutations. For selected genes whose products could be assessed by flow cytometric analysis, we examined the concordance between mRNA and protein expression (Figure 6A). The DSM mutations did not substantially alter mRNA or protein expression of canonical FOXP3 target genes such as Cd25, Ctla4, Gitr, Entpd1 or Gpr83 (Figure 5C, 6A and data not shown), but considerably affected the expression of others (Il2, Cyp11a1, Foxp3 itself, Helios, Prdm1, Tigit, Tnfrsf13b; Figure 5D, 6A). For many genes, the transcriptional changes induced by WT and DSM FOXP3 were only apparent, or only striking, after activation of the transduced cells (Marson et al., 2007; Figures 5B–D).
Figure 6. The domain-swap interface is required for the function of FOXP3.
(A) Representative flow cytometry plots depicting expression of IL-2 (top panel), CTLA-4, GITR and CD25 (bottom panels) in CD4+ T cells transduced with an empty control vector encoding WT or DSM-FOXP3. The X-axis denotes GFP fluorescence, used as a proxy for FOXP3 expression; GFP expression is controlled by an IRES element within the bicistronic RNA encoding the FOXP3 coding sequence. The data are representative of three independent experiments. Bar graphs on right represent percent of IL-2hi cells within the GFP+ population (top panel) and fold increase in MFI of CTLA4 and CD25 in transduced cells (middle and bottom panels respectively). The data are representative of 3 independent experiments. (B) Cells expressing the FOXP3 domain swap mutant show decreased suppressor function in vitro. Top, Representative histograms depicting the CFSE dilution profile of responder CD4+ T cells cultured with either ex-vivo isolated CD4+ CD25+ T cells or transduced FOXP3-expressing cells (suppressors) at a ratio of 1:2 (suppressors:responders). Bottom, graph demonstrating relative proliferation of responder cells cultured with transduced FOXP3-expressing suppressor cells at different suppressor:respondor ratios. The data are representative of two independent experiments. (C) Cells transduced with DSM-FOXP3 fail to prevent induction of diabetes in NOD recipient mice. Neonatal BDC2.5 TCR transgenic NOD mice were injected with T cells retrovirally transduced with WT or DSM-FOXP3 together with Th1 cell effectors generated from BDC2.5 TCR transgenic mice. The graph represents the kinetics of diabetes induction in recipient mice: cells transduced with WT-FOXP3 prevent diabetes induction by the effector Th1 cells, whereas cells transduced with DSM-FOXP3 are ineffective. The data were obtained from three independent experiments.
Disruption of the domain-swap interface diminishes FOXP3-mediated suppressor functions
We next determined the impact of the DSM mutations on the suppressor functions of FOXP3. As expected, expression of WT-FOXP3 conferred potent regulatory activity on transduced T cells, read out as almost complete suppression of proliferation of the responder T cells (Figure 6B, top, compare third and fourth panels). In contrast, cells expressing DSM-FOXP3 exhibited no discernible regulatory ability, as judged by robust proliferation of the CFSE-labeled responder T cells (Figure 6B, top, last panel; quantified for different suppressor:responder ratios in Figure 6B, bottom). In control experiments, we confirmed that CD4+ CD25+ “natural” T regulatory cells, freshly-isolated ex vivo, displayed strong suppressive activity (Figure 6B, top, compare first and second panels). Together the data suggest that domain-swapping contributes critically to the ability of FOXP3 to induce suppressor function in retrovirally-transduced T cells in this co-culture system.
To ask if the effects of the DSM mutation on FOXP3 regulatory function were also apparent in vivo, we used a well-established mouse model of autoimmune diabetes (Katz et al., 1995) that we previously employed to demonstrate NFAT:FOXP3 cooperation (Wu et al., 2006). Naïve T cells from BDC2.5 T cell receptor (TCR) transgenic mice, which bear a transgenic TCR that recognize chromogranin A expressed by pancreatic β-cells (Stadinski et al., 2010), were differentiated to yield T helper-1 (Th1) cell-biased T cells. These cells are strongly diabetogenic when adoptively transferred into neonatal NOD mice, as read out by rapid onset of diabetes within a week of adoptive transfer. Co-transfer of CD4+ T cells expressing WT-FOXP3 suppressed the development of diabetes in the NOD mice as expected (Peng et al., 2007; Wu et al., 2006). In contrast, co-transfer of CD4+ T cells expressing mutant DSM- FOXP3 failed to protect the recipients: hyperglycemia and diabetes were observed within 10 days, as in mice that received only effector cells (Figure 6C). Nine days after transfer, comparable numbers of transferred cells expressing DSM-FOXP3 or WT-FOXP3 were detected in the spleens of recipient mice (Figure S6), indicating that cells expressing DSM-FOXP3 mutant were not compromised for survival.
An IPEX mutation in the FKH domain impairs domain swapping and abrogates FOXP3 suppressor function
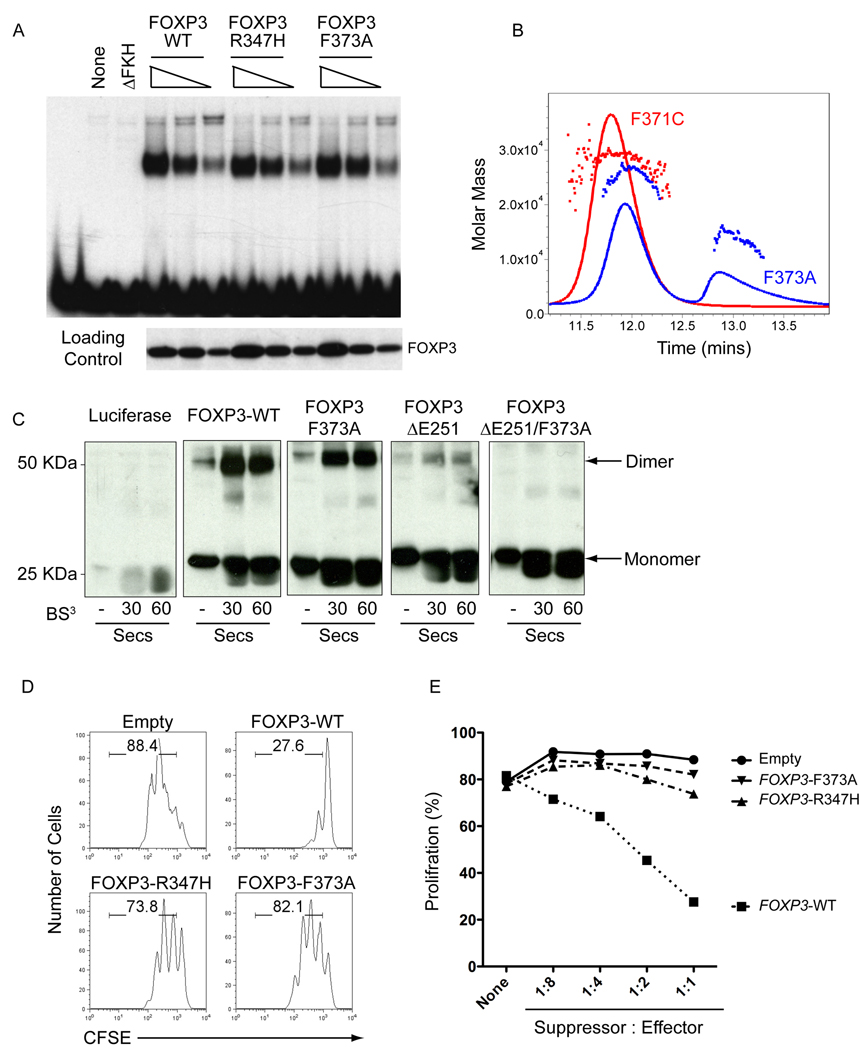
As described above, three IPEX mutations -- F371C, F373A and R347H – are found within or near the domain-swap interface of FOXP3 (Figure 2). Residues F371 and F373 are located at the interface between the two H2 helices: F373 is buried deeply within the hydrophobic core, whereas the aromatic ring of F371 is angled away from the domain-swap interface (Figure 2A). Consistent with a structural role for F373 in the center of the domain-swapped interface, the F373A mutant eluted as a mixture of monomer and dimer peaks on a size exclusion column (Figure 7B). Thus the F373A substitution in the FOXP3 FKH domain destabilizes dimer formation; since the SEC-MALS experiments are performed at high micromolar concentrations of recombinant proteins, it is likely that the equilibrium would be further shifted towards the monomer at physiological concentrations of the F373A protein. In contrast, as predicted by the structure, the F371C mutation did not disrupt dimerization of the FOXP3 FKH domain, since the mutant protein eluted as a single peak corresponding to a FOXP3 FKH dimer (Figure 7B). The dimerization status of the R347H mutant FKH domain could not be evaluated, as this recombinant protein was insoluble when expressed in bacteria; however, FOXP3 bearing both the F347H and F373A mutations bound as strongly as WT-FOXP3 to DNA (Figure 7B). Based on these findings, we focused on the biological effect of the F373A IPEX mutation.
Figure 7. IPEX mutations that disrupt domain swapping diminish FOXP3 regulatory activity.
(A) Forkhead mutations that lie at the domain swap interface do not disrupt DNA binding. EMSA analysis comparing the ability of WT and indicated FOXP3 forkhead mutants to bind DNA. (B) The F373A mutation at the domain swap interface destabilizes dimer formation by the FOXP3 forkhead domain. SEC-MALS analysis of the forkhead domains of FOXP3 F371C and F373A shows that the F371C mutant remains a dimer whereas the F373A mutants elutes as a mixture of monomer and dimer. (C) Denaturing SDS Gel of FOXP3 (aa 182–431) before and after brief crosslining with BS3, showing that FOXP3 contains two dimerisation interfaces in the leucine zipper and the forkhead domain. The combination of the ΔE251 mutation in the leucine zipper and the F373A mutation in the forkhead domain eliminates dimer formation. (D) IPEX mutations that destabilize domain swapping abrogate the ability of FOXP3 to induce regulatory functions. Shown is a representative histogram depicting the CFSE dilution profile of responder cells cultured with transduced WT-FOXP3 or IPEX mutant-expressing cells (suppressors) at a ratio of 1:2 (suppressors:responders). (E), Graph quantitating relative proliferation of CFSE-labelled responder cells cultured with transduced FOXP3-expressing suppressor cells at different suppressor: responder ratios. These data are representative of at least two independent experiments.
As an independent confirmation of dimerization through the FOXP3 FKH domain, we assessed dimer formation after limited cross-linking with BS3, a cross-linking agent with a short spacer arm (11.4 A0). For these experiments, we used the same larger fragment of FOXP3 (aa 182–431) used for the EMSA assays that contains both the FKH domain and the LZ motif. Into this fragment, we engineered two known IPEX mutations, ΔE51 in the LZ motif (Lopes et al., 2006) (Chae et al., 2006) and F373A in the domain-swap interface respectively. When expressed in a cell-free system, WT-FOXP3 formed a small amount of an sodium dodecyl sulfate (SDS)-resistant dimer even in the absence of cross-linker; addition of BS3 led rapidly, within 30 s, to the appearance of a ~ 50 kDa band corresponding to a FOXP3 dimer (Figure 7C, panel 2). The ΔE251 mutation substantially reduced but did not abrogate dimer formation (Figure 7C, panel 4). The residual dimer formed by the ΔE251 mutant involves the domain-swap interface of the FKH domain, since the combination of ΔE251 and F373A mutations eliminated formation of both SDS-resistant and cross-linked dimers (Figure 7C panel 5). Note that the FKH and LZ interfaces are distinct: the purified FKH domain of FOXP3 clearly formed dimers, either as the isolated domain (Figure 3A; Figure 3B, left) or in the context of a larger fragment bearing the ΔE251 mutation in the LZ interface.
Together these data show clearly that there are two distinct and independent dimerization interfaces in FOXP3, the LZ motif and the domain-swap interface in the FKH domain. Since the F373A mutation alone did not appreciably diminish dimer formation, whereas the ΔE251 mutation caused a substantial decrease (Figure 7C, compare panels 3 and 4), it is likely that the LZ makes a larger contribution to dimer formation. However, as shown below, the lower “affinity” FKH dimer interface is essential for the suppressive function of FOXP3.
We then asked whether the R347H and F373A IPEX mutations impaired the ability of FOXP3 to induce suppressor activity in retrovirally-transduced T cells. T cells expressing WT-FOXP3 effectively suppressed the proliferation of bystander T cells, whereas FOXP3 proteins bearing the F373A or R347H mutations were unable to suppress proliferation (Figure 7D; quantified for different suppressor:responder ratios in Figure 7E). Together these data suggest strongly that the R347H and F373A IPEX mutations induce their autoimmune phenotypes in humans by interfering not with DNA binding (Figure 7A), but rather with the ability of FOXP3 to form a domain-swapped dimer.
We asked whether we could rescue the defect in suppression caused by the F373A IPEX mutation in FOXP3 by overexpressing selected FOXP3 target genes whose expression was affected by the DSM mutation. T cells were simultaneously infected with retroviruses encoding F373A-FOXP3 (with IRES Thy1.1) and either CYP11A1 or TIGIT (with IRES-Gfp). Doubly-transduced cells were sorted based on expression of both markers (Thy1.1 and GFP), and their suppressive function was compared in co-culture assays with that of cells expressing F373A-FOXP3 and empty Thy1.1 vector. When expressed alone, however, neither CYP11A1 nor TIGIT rescued the defect in suppressive activity of F373A-FOXP3 (Suppl. Fig. 7B), consistent with the hypothesis that suppression depends on the simultaneous combined actions of several target gene products whose expression is disrupted by mutations in the domain-swap interface.
DISCUSSION
In this study, we have solved the crystal structure of a complex of the FOXP3 FKH domain and the DNA-binding domain of NFAT1 on a composite NFAT:AP-1 (FOXP) DNA element. Our data revealed that the FKH domain of FOXP3 is optimized relative to that of FOXP2, both for NFAT interaction and for the ability to form a domain-swapped dimer with the capacity to simultaneously engage two distinct molecules of DNA. Notably, despite their surprisingly limited effects on FOXP3-mediated gene expression, structure-guided DSM mutations in the domain-swapped dimer interface of FOXP3 effectively eliminated FOXP3-mediated suppressor functions, both in culture and in mouse models of autoimmune disease.
Our studies have important implications for how certain IPEX mutations in the FKH domain affect the suppressive functions of regulatory T cells. Based on its location in the FKH domain, the F373A IPEX mutation was originally proposed to impair DNA binding by FOXP3 (Bacchetta et al., 2006; Wildin et al., 2001). Our crystal structure reveals that the F373 residue is located in the domain-swap interface of FOXP3; as predicted from this structure, the F373A mutation disrupted dimer formation by the FKH domain but did not affect DNA-binding, and this disruption of dimer formation was associated with a dramatic loss of FOXP3-mediated suppressive function. These findings are consistent with those observed in the clinic: an infant harboring the F373A mutation presented with a severe form of IPEX and developed autoimmune insulin-dependent diabetes mellitus (IDDM) within 2 weeks of life, despite the fact that his CD4+ CD25+ T cells showed normal expression of FOXP3 protein as well as the cell markers CD25, CTLA4 and GITR (Bacchetta et al., 2006). For technical reasons associated with inability to purify the recombinant mutant protein, we were unable to evaluate the effect on dimer formation of a second mutation, R347H, that maps to the domain swap interface; however, this mutation also eliminated FOXP3 suppressor function while leaving DNA binding unimpaired. Notably, the severity of autoimmunity in patients with the F373A and R347H mutations is similar to that observed in patients with nonsense or frameshift mutations that result in complete loss of FOXP3 expression; and both these missense mutations are associated with neonatal development of insulin-dependent autoimmune diabetes IDDM (Kobayashi et al., 2001) (Wildin et al., 2001; Wildin et al., 2002) (Bacchetta et al., 2006). A third IPEX mutation in the domain swap interface, F371C, did not impair dimerisation of the FOXP3 FKH domain, most likely because this residue does not point inward into the interface; the autoimmunity observed in patients with the F371C could involve formation of inappropriate inter- or intramolecular disulphide bonds through the substituted cysteine. Together, our results highlight the domain-swap dimer interface as a novel structural feature of the FOXP3 forkhead domain, whose disruption underlies human autoimmune disease.
Our data emphasise that FOXP3 can form multimers in two distinct modes: through the domain-swap interface in the FKH domain or through the LZ which mediates FOXP3 homo-oligomerization and hetero-oligomerization with FOXP1 (Wang et al., 2003) (Li et al., 2007b). How is dimer formation by the FKH domain related to oligomerisation through the zinc finger (Znf) and LZ motifs? Modeling studies based on our structural data suggest that full-length FOXP3 most likely exists as a dimer of dimers, wherein a pair of FOXP3 molecules form a domain-swapped dimer through their FKH domains and the two dimers associate with each other through their Znf-LZ regions. This oligomerization mechanism is very similar to that of p53, where a tetramerization domain brings together two pairs of p53 DNA-binding domains, each binding to palindromic site as a dimer (Chen et al., 2010; Kitayner et al., 2006; Wells et al., 2008). However, unlike p53, which binds tandem palindromic sites as a tetramer (Joerger and Fersht, 2008; Wei et al., 2006), the two DNA binding surfaces on the domain-swapped FOXP3 dimer are located on the opposite faces of a well-folded protein body, thus preventing the FOXP3 dimer binding to adjacent sites. In the crystal structure, the FOXP3 dimer bound two separate DNA segments in an anti-parallel orientation. In cells, this interaction may facilitate the assembly of higher-order transcription complexes that depend on stereospecific arrangement of distal DNA elements bridged by the domain-swapped FOXP3 dimer.
It is unlikely that disruption of domain-swapping impairs the ability of FOXP3 to interact with its known binding partners NFAT1 (Wu et al., 2006), RUNX1 (Ono et al., 2007) and the HDAC and 60 kDa Tat-interactive protein (Tip60) complex (Li et al., 2007a). First, the domain-swap interface and the NFAT1-binding interface of FOXP3 are physically and functionally distinct: CD25, CTLA4 and GITR expression are not affected by the DSM mutations, but are abrogated by disruption of the NFAT1:FOXP3 interaction (Wu et al., 2006). Second, RUNX1:FOXP3 interactions diminish FOXP3 expression, not FOXP3-mediated suppressor function (Bruno et al., 2009; Kitoh et al., 2009; Klunker et al., 2009; Rudra et al., 2009); because we induce ectopic expression of FOXP3 using retroviruses, it is unlikely that the defects in suppressor functions induced by the DSM mutants are due to disruption of RUNX1:FOXP3 interactions. Finally, interaction of FOXP3 with the HDAC and Tip60 complex has been demonstrated to be essential for FOXP3-mediated repression of its target genes (Li et al., 2007a). Because repression of diverse FOXP3 target genes is unaffected by the DSM mutations, it seems unlikely that these mutations interfere with formation of the FOXP3:HDAC:Tip60 corepressor complex. An interesting possibility, however, is that the domain-swapped dimer provides a binding surface for a yet-unidentified partner of FOXP3.
It is not yet obvious how the limited transcriptional effects of domain-swap mutations translate into the strong observed impairment of suppressive activity. The functional impairment may reflect the combined effect of small changes in the expression of many target genes that play a critical role in Treg activity, as observed in Treg cells with ~80–90% decrease in expression of FOXP3 (Wan and Flavell, 2007); or it may reflect a requirement for a small number of specific target genes that have not yet been identified. An equally interesting possibility is that the domain-swapped FKH dimer recruits novel binding partners and/or transcriptional regulators to FOXP3-binding sites in DNA. Future structure-function analyses will expand our understanding of the mechanism through which FOXP3 induces a suppressor program in T cells, and may provide novel targets for the clinical manipulation of autoimmune and/or inflammatory diseases and cancer.
Experimental Procedures
Detailed experimental procedures can be found in supplementary methods.
Sample preparation and crystallization
Human FOXP3 (336–419) and NFAT1 RHR (392–678) were prepared as described (Stroud et al., 2006; Wu et al., 2006). Data collection, structure determination and analyses are described in supplementary methods. The coordinates and structural factors have been deposited in the RCSB protein database under accession code 3QRF.
Biochemistry
For chemical cross-linking experiments, recombinant proteins were incubated with DSS for one hour. Cross-linked dimers and complexes were visualized by Commassie staining after separation via denaturing SDS-PAGE. EMSA assays for FOXP3 DNA binding were performed as described (Koh et al., 2009). For in-gel FRET analyses a synthetic probe, containing two double-stranded FOXP3 binding sites separated by a flexible 19-base single-stranded DNA linker and labeled at the 5’ and 3’ ends with the FRET-pair Cy3 and Cy5 respectively, was incubated with increasing concentrations of recombinant FOXP3 forkhead fragment. DNA-protein complexes were resolved by non-denaturing PAGE and visualized by Typhoon 8610 variable mode imager. The Cy3 image (assigned in green) and the Cy5 (red) were overlaid.
T cell manipulations
T cells from _Tcra_−/− OT-II TCR-transgenic mice were transduced with WT or DSM FOXP3-IRES-GFP or Thy1.1 retroviruses. IL-2, CTLA-4 and FOXP3 levels were determined by intracellular staining, CD25 and GITR levels by cell surface staining (Wu et al., 2007). For suppression assays, sorted Thy1.1+ CD4+ T cells expressing WT or DSM FOXP3 (suppressors) were co-cultured for 72 hr in 96-well round-bottom plates with 104 mitomycin C treated splenocytes, anti-CD3 mAb, and 104 CFSE-labelled CD4+CD25− responder T cells isolated from spleens and lymph nodes of mice by positive selection using anti-CD4 magnetic beads (Dynal) followed by depletion of CD25+ cells with anti-CD25 microbeads (Miltenyi). Proliferation of the responders was monitored by CSFE dye dilution. RNA from sorted Thy1.1+ T cells expressing WT or DSM FOXP3 was analysed by real-time RT-PCR (StepOne plus thermal cycler, Applied Biosystems). For microarray analysis, RNA from the sorted cells was extracted, labeled, amplified and hybridized to Mouse Gene 1.0 ST Arrays (Affymetrix) (Feuerer et al., 2009)
Induction of diabetes
CD4+ T cells from BDC2.5/NOD TCR-transgenic mice were activated under Th1 cell polarizing conditions (Ansel et al., 2004) and infected with MSCV IRES-Thy1.1 retrovirus, empty or encoding WT or DSM FOXP3. Three days after infection, Thy1.1+ cells were sorted and transferred into neonatal NOD mice at a 1:2 ratio (0.5 × 105: 1 × 105) with untransduced BDC2.5-Th1 cells (“effector” cells). Recipient mice were monitored for diabetes (urinary glucose more than 300 mg/dl in two consecutive measurements) for 6 weeks. Detailed experimental procedures can be found in supplementary methods.
Supplementary Material
01
Acknowledgements
We thank M.K. Levings and A.N. McMurchy for discussions and for sharing their findings regarding the IPEX mutations investigated in this study, the USC NanoBiophysics Core facility for the MALS study, ALS BCSB staff members Corie Ralston, Peter Zwart, and Kevin Royal for help with data collection, and Nouri Neamati for help with FRET studies. This work was supported by NIH grants AI48213, AI44432, CA42471, AI40127 and Juvenile Diabetes Research Foundation grants 17-2010-421 and 16-2007-427 (to A.R.) and NIH grants RO1 GM077320 and GM064642 (to L.C). H.S.B. was supported by a postdoctoral fellowship from the Lady Tata Memorial Trust and currently by a postdoctoral fellowship from the GlaxoSmithKline-Immune Disease Institute Alliance. S.G. is supported by an NIH postdoctoral training grant (T32).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bacchetta R, Passerini L, Gambineri E, Dai M, Allan SE, Perroni L, Dagna-Bricarelli F, Sartirana C, Matthes-Martin S, Lawitschka A, et al. Defective regulatory and effector T cell functions in patients with FOXP3 mutations. J. Clin. Invest. 2006;116:1713–1722. doi: 10.1172/JCI25112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates DL, Barthel KK, Wu Y, Kalhor R, Stroud JC, Giffin MJ, Chen L. Crystal structure of NFAT bound to the HIV-1 LTR tandem kappaB enhancer element. Structure. 2008;16:684–694. doi: 10.1016/j.str.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat. Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat. Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Bruno L, Mazzarella L, Hoogenkamp M, Hertweck A, Cobb BS, Sauer S, Hadjur S, Leleu M, Naoe Y, Telfer JC, et al. Runx proteins regulate Foxp3 expression. J. Exp. Med. 2009;206:2329–2337. doi: 10.1084/jem.20090226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae WJ, Henegariu O, Lee SK, Bothwell AL. The mutant leucine-zipper domain impairs both dimerization and suppressive function of Foxp3 in T cells. Proc. Natl. Acad. Sci. U S A. 2006;103:9631–9636. doi: 10.1073/pnas.0600225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatila TA. Regulatory T cells: key players in tolerance and autoimmunity. Endocrinol. Metab. Clin. North. Am. 2009;38:265–272. doi: 10.1016/j.ecl.2009.01.002. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- Chen Y, Dey R, Chen L. Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer. Structure. 2010;18:246–256. doi: 10.1016/j.str.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuerer M, Shen Y, Littman DR, Benoist C, Mathis D. How punctual ablation of regulatory T cells unleashes an autoimmune lesion within the pancreatic islets. immunity. 2009;31:654–664. doi: 10.1016/j.immuni.2009.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Giffin MJ, Stroud JC, Bates DL, von Koenig KD, Hardin J, Chen L. Structure of NFAT1 bound as a dimer to the HIV-1 LTR kappa B element. Nat. Struct. Biol. 2003;10:800–806. doi: 10.1038/nsb981. [DOI] [PubMed] [Google Scholar]
- Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat. Rev. Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- Jin L, Sliz P, Chen L, Macian F, Rao A, Hogan PG, Harrison SC. An asymmetric NFAT1 dimer on a pseudo-palindromic kappa B-like DNA site. Nat. Struct. Biol. 2003;10:807–811. doi: 10.1038/nsb975. [DOI] [PubMed] [Google Scholar]
- Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- Josefowicz SZ, Rudensky A. Control of regulatory T cell lineage commitment and maintenance. Immunity. 2009;30:616–625. doi: 10.1016/j.immuni.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz JD, Benoist C, Mathis D. T helper cell subsets in insulin-dependent diabetes. Science. 1995;268:1185–1188. doi: 10.1126/science.7761837. [DOI] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. Structural basis of DNA recognition by p53 tetramers. Mol. Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, Kitabayashi I, Tsukada T, Nomura T, Miyachi Y, et al. Indispensable role of the Runx1-Cbfbeta transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity. 2009;31:609–620. doi: 10.1016/j.immuni.2009.09.003. [DOI] [PubMed] [Google Scholar]
- Klunker S, Chong MM, Mantel PY, Palomares O, Bassin C, Ziegler M, Ruckert B, Meiler F, Akdis M, Littman DR, Akdis CA. Transcription factors RUNX1 and RUNX3 in the induction and suppressive function of Foxp3+ inducible regulatory T cells. J. Exp. Med. 2009;206:2701–2715. doi: 10.1084/jem.20090596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi I, Shiari R, Yamada M, Kawamura N, Okano M, Yara A, Iguchi A, Ishikawa N, Ariga T, Sakiyama Y, et al. Novel mutations of FOXP3 in two Japanese patients with immune dysregulation, polyendocrinopathy, enteropathy, X linked syndrome (IPEX) J. Med. Genet. 2001;38:874–876. doi: 10.1136/jmg.38.12.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh KP, Sundrud MS, Rao A. Domain Requirements and Sequence Specificity of DNA Binding for the Forkhead Transcription Factor FOXP3. PLoS ONE. 2009 doi: 10.1371/journal.pone.0008109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Greene MI. FOXP3 actively represses transcription by recruiting the HAT/HDAC complex. Cell Cycle. 2007;6:1432–1436. [PubMed] [Google Scholar]
- Li B, Samanta A, Song X, Iacono KT, Bembas K, Tao R, Basu S, Riley JL, Hancock WW, Shen Y, et al. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc. Natl. Acad. Sci. U S A. 2007a;104:4571–4576. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Samanta A, Song X, Iacono KT, Brennan P, Chatila TA, Roncador G, Banham AH, Riley JL, Wang Q et al. FOXP3 is a homo-oligomer and a component of a supramolecular regulatory complex disabled in the human XLAAD/IPEX autoimmune disease. Int. Immunol. 2007b;19:825–835. doi: 10.1093/intimm/dxm043. [DOI] [PubMed] [Google Scholar]
- Lopes JE, Torgerson TR, Schubert LA, Anover SD, Ocheltree EL, Ochs HD, Ziegler SF. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J. Immunol. 2006;177:3133–3142. doi: 10.4049/jimmunol.177.5.3133. [DOI] [PubMed] [Google Scholar]
- Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, Fraenkel E, von Boehmer H, Young RA. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature. 2007;445:931–935. doi: 10.1038/nature05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs HD, Gambineri E, Torgerson TR. IPEX, FOXP3 and regulatory T-cells: a model for autoimmunity. Immunol. Res. 2007;38:112–121. doi: 10.1007/s12026-007-0022-2. [DOI] [PubMed] [Google Scholar]
- Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, Tsukada T, Sakaguchi S. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nature. 2007;446:685–689. doi: 10.1038/nature05673. [DOI] [PubMed] [Google Scholar]
- Peng J, Dicker B, Du W, Tang F, Nguyen P, Geiger T, Wong FS, Wen L. Converting antigen-specific diabetogenic CD4 and CD8 T cells to TGF-beta producing non-pathogenic regulatory cells following FoxP3 transduction. J. Autoimmun. 2007;28:188–200. doi: 10.1016/j.jaut.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Rudra D, Egawa T, Chong MM, Treuting P, Littman DR, Rudensky AY. Runx-CBFbeta complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol. 2009;10:1170–1177. doi: 10.1038/ni.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- Stadinski BD, Delong T, Reisdorph N, Reisdorph R, Powell RL, LArmstrong M, Piganelli JD, Barbour G, Bradley B, Crawford F, et al. Chromogranin A is an autoantigen in type 1 diabetes. Nat Immunol. 2010;11:225–231. doi: 10.1038/ni.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroud JC, Chen L. Structure of NFAT bound to DNA as a monomer. J. Mol. Biol. 2003;334:1009–1022. doi: 10.1016/j.jmb.2003.09.065. [DOI] [PubMed] [Google Scholar]
- Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14:159–166. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Torgerson TR, Ochs HD. Immune dysregulation, polyendocrinopathy, enteropathy, X-linked: forkhead box protein 3 mutations and lack of regulatory T cells. J. Allergy Clin. Immunol. 2007;120:744–750. doi: 10.1016/j.jaci.2007.08.044. quiz 751-742. [DOI] [PubMed] [Google Scholar]
- van der Vliet HJ, Nieuwenhuis EE. IPEX as a result of mutations in FOXP3. Clin. Dev. Immunol. 2007;2007:89017. doi: 10.1155/2007/89017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- Wang B, Lin D, Li C, Tucker P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J. Biol. Chem. 2003;278:24259–24268. doi: 10.1074/jbc.M207174200. [DOI] [PubMed] [Google Scholar]
- Wei CL, Wu Q, Vega VB, Chiu KP, Ng P, Zhang T, Shahab A, Yong HC, Fu Y, Weng Z, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wells M, Tidow H, Rutherford TJ, Markwick P, Jensen MR, Mylonas E, Svergun DI, Blackledge M, Fersht AR. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc. Natl. Acad. Sci. U S A. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildin RS, Ramsdell F, Peake J, Faravelli F, Casanova JL, Buist N, Levy-Lahad E, Mazzella M, Goulet O, Perroni L, et al. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nat. Genet. 2001;27:18–20. doi: 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- Wildin RS, Smyk-Pearson S, Filipovich AH. Clinical and molecular features of the immunodysregulation, polyendocrinopathy, enteropathy, X linked (IPEX) syndrome. J. Med. Genet. 2002;39:537–545. doi: 10.1136/jmg.39.8.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Rudensky AY. Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 2007;8:277–284. doi: 10.1038/ni1437. [DOI] [PubMed] [Google Scholar]
- Workman CJ, Szymczak-Workman AL, Collison LW, Pillai MR, Vignali DA. The development and function of regulatory T cells. Cell. Mol. Life Sci. 2009;66:2603–2622. doi: 10.1007/s00018-009-0026-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Josefowicz SZ, Kas A, Chu TT, Gavin MA, Rudensky AY. Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature. 2007;445:936–940. doi: 10.1038/nature05563. [DOI] [PubMed] [Google Scholar]
- Ziegler SF. FOXP3: of mice and men. Annu. Rev. Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01