SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis (original) (raw)
. Author manuscript; available in PMC: 2012 Mar 31.
Published in final edited form as: Nature. 2011 Mar 31;471(7340):637–641. doi: 10.1038/nature09814
Abstract
SHARPIN is a ubiquitin-binding and ubiquitin-like domain-containing protein which, when mutated in mice, results in immune system disorders and multiorgan inflammation1,2. Here we report that SHARPIN functions as a novel component of the Linear Ubiquitin Chain Assembly Complex (LUBAC) and that the absence of SHARPIN causes disregulation of NF-κB and apoptotic signalling pathways, explaining the severe phenotypes displayed by chronic proliferative dermatitis in SHARPIN deficient mice. Upon binding to the LUBAC subunit HOIP, SHARPIN stimulates the formation of linear ubiquitin chains in vitro and in vivo. Co-expression of SHARPIN and HOIP promotes linear ubiquitylation of NEMO, an adaptor of the IκB kinases (IKKs) and subsequent activation of NF-κB signalling, while SHARPIN deficiency in mice causes an impaired activation of the IKK complex and NF-κB in B cells, macrophages, and mouse embryonic fibroblasts (MEFs). This effect is further enhanced upon concurrent downregulation of HOIL-1L, another HOIP-binding component of LUBAC. In addition, SHARPIN deficiency leads to rapid cell death upon TNFα stimulation via FADD- and Caspase-8-dependent pathways. SHARPIN thus activates NF-κB and inhibits apoptosis via distinct pathways in vivo.
In addition to the established roles of K63- and K48-linked ubiquitin chains, linear ubiquitin chains have recently emerged as important regulators of the NF-κB pathway, controlling immune responses, as well as cell survival, proliferation, and development3,4,5,6,7,8. Linear ubiquitin conjugation relies on two RING-between-RING (RBR) domain-containing proteins, HOIL-1L (longer isoform of hem-oxidized iron-regulatory protein 2 ubiquitin ligase-1) and HOIP (HOIL-1L interacting protein), which together form the E3 ligase complex LUBAC (linear ubiquitin chain assembly complex)9. We became interested in SHARPIN (SHANK-associated RH domain-interacting protein) because of the significant sequence homology of its C-terminal region, enclosing a ubiquitin-like (UBL) domain and a putative ubiquitin-binding NPL4 zinc finger domain (NZF), with the N-terminal region of HOIL-1L9,10 (Fig. 1a). Initially, we established SHARPIN as a bona fide ubiquitin binding protein by mapping the ubiquitin binding site to the NZF domain (Supplementary Fig. 1). A direct NZF-dependent interaction of SHARPIN with mono-ubiquitin, linear, and K63 ubiquitin chains was confirmed by pull-down assays (Supplementary Fig. 1b–d) and isothermal titration calorimetry (ITC) (Fig. 1b). Mutagenesis of two key residues surrounding the zinc coordination site of the NZF (T358L, F359V) abolished ubiquitin binding (Supplementary Fig. 1a and 1c). The isolated NZFs of SHARPIN, HOIL-1L, and HOIP interact with monoubiquitin with similar affinities (Fig. 1b), recognizing the classical hydrophobic patch surrounding I44, as indicated by the absence of any interaction with mutant I44A ubiquitin (Fig. 1b) and display no specificity for different chain types (Fig. 1b and Supplementary Fig. 1c). These data are consistent with binding assays employing a panel of HOIP and HOIL-1L protein variants expressed in transfected cells or in in vitro ubiquitin binding experiments (Supplementary Fig. 1e–h).
Figure 1. SHARPIN is a novel component of the LUBAC complex.
a, Schematic representation of the domain architecture of SHARPIN (A), HOIL-1L (B), and HOIP (C). b, Quantitative assessment of the binding of SHARPIN, HOIL-1L, and HOIP to a panel of ubiquitin species using isothermal titration calorimetry (ITC). The obtained equilibrium dissociation constants of individual measurements are listed. c, Formation of a trimeric complex between SHARPIN, HOIL-1L, and HOIP in HEK293T cells transfected with Myc- HOIP, HA-HOIL-1L, and GFP-SHARPIN or d, in MEFs. Lysates were immunoprecipitated and analyzed by Western blot.
We next investigated whether SHARPIN directly associates with the LUBAC complex in cells. Tagged versions of HOIP, HOIL-1L, and SHARPIN were readily immunoprecipitated from lysates of transiently transfected HEK293T cells (Fig. 1c). Likewise, the interaction of endogenous SHARPIN, HOIP, and HOIL-1L was confirmed in primary mouse embryonic fibroblasts (MEFs) (Fig. 1d). The interaction between SHARPIN and HOIP is mediated by the UBL domain of SHARPIN, which recognizes the NZF2 domain of HOIP (Supplementary Fig. 1i and j). In agreement, the interaction between HOIP and SHARPIN was abolished in response to either deletion or mutation (I272A) of the UBL domain of SHARPIN, but was not affected in the ubiquitin-binding deficient mutant of SHARPIN NZF (Supplementary Fig. 2a). On the other hand, purified HOIL-1L did not interact with full-length SHARPIN in in vitro binding assays (Supplementary Fig. 2b). Thus, HOIP has the capacity to interact with either SHARPIN or HOIL-1L or both of them. Indeed, endogenous HOIP can form a complex with only HOIL-1L or SHARPIN in the absence of the other, since in SHARPIN or HOIL-1 deficient MEFs, HOIP can co-precipitate with residual HOIL-1L or SHARPIN, respectively (Fig. 1d). Interestingly, the deficiency of either SHARPIN or HOIL-1L led to a partial destabilization of the other protein (Fig. 1d). Together these findings indicate the existence of three putative LUBAC complexes: dimeric complexes, HOIP-HOIL-1L (LUBAC-I) and HOIP-SHARPIN (LUBAC-II), and a trimeric SHARPIN-HOIP-HOIL-1L complex (Supplementary Fig. 2c).
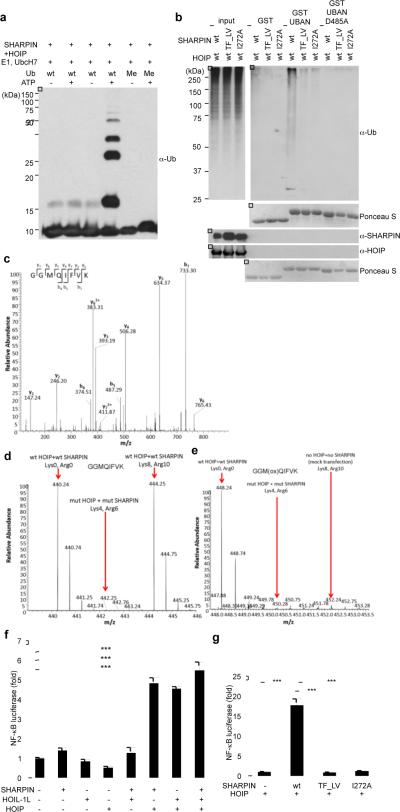
The purified HOIP-SHARPIN complex stimulated the in vitro assembly of linear ubiquitin chains (Fig. 2a), similar to the ligase activity of a HOIP-HOIL-1L complex (Supplementary Fig. 3a). We thus extended our studies to investigate whether SHARPIN and HOIP can induce linear ubiquitylation of proteins in cells. The UBAN domain of ABIN-1, known to preferentially bind linear ubiquitin chains11,12, was used as an affinity matrix to pull down cellular proteins modified by linear ubiquitylation upon overexpression of SHARPIN and HOIP (Fig. 2b and Supplementary Fig. 3f). Both the NZF and UBL domains of SHARPIN, were essential for this activity, since neither the SHARPIN-NZF (T358L, F359V), nor the SHARPIN-UBL (I272A) mutant showed any ability to induce linear ubiquitylation in vivo (Fig. 2b). Similarly, the ABIN1-UBAN D485A mutant, which is unable to bind linear polyubiquitin, did not pull down any ubiquitin conjugates (Fig. 2b, Supplementary Fig. 3d–e). A specific enrichment of linear ubiquitylation in cells overexpressing SHARPIN and HOIP was further confirmed by MS/MS analysis. In samples transfected with HOIP-SHARPIN as well as HOIP-HOIL-1L the linear ubiquitin peptide GGMQIFVK was detected (Fig. 2c). MS-based quantification (employing the AQUA approach described in Supplementary Fig. 4a) detected an increase in the absolute amounts of ubiquitin present in linear chains compared to the total pool of ubiquitin (measured by the TITLEVEPSDTIENVK peptide) in cells expressing wild type HOIP-SHARPIN, in contrast to cells expressing an inactive HOIP-SHARPIN complex (Supplementary Fig. 4b–e). We next performed a SILAC-based quantitative MS/MS analysis on immunoprecipitated NEMO upon co-transfection with either wild type or inactive HOIP-SHARPIN (experimental design is shown in Supplementary Fig. 5a and b). The measured intensity of linear ubiquitin peptides on immunoprecipitated NEMO was >25 times higher in cells transfected with wild type HOIP-SHARPIN as compared to cells expressing inactive HOIP-SHARPIN mutants (Fig. 2d and e). In the same sample we detected 13 ubiquitylation sites on NEMO, of which 11 were increased following transfection with wild type versus mutated HOIP-SHARPIN (Supplementary Fig. 5c), suggesting that NEMO is a substrate of LUBAC II.
Figure 2. SHARPIN and HOIP form a novel LUBAC complex with the ability to induce linear ubiquitylation and NF-κB activation.
a, In vitro linear ubiquitin chain synthesis by purified SHARPIN and HOIP. b, Stimulation of linear ubiquitylation by SHARPIN and HOIP in vivo. Immobilised GST-ABIN-1-UBAN domain was used to detect linear ubiquitylation of proteins. c, MS/MS spectrum of the prototypic linear ubiquitin peptide present in vivo. d, e, SILAC experiments comparing relative levels of linear ubiquitin on immunoprecipitated NEMO induced by HOIP and SHARPIN (d) or HOIP-mut (RING domain)-SHARPIN-mut( TF_LV)(Lys8,Arg10) (e). f, g, Stimulation of NF-κB transcriptional activity by SHARPIN and HOIP in vivo. NF-κB-luciferase assays using the indicated combinations of SHARPIN, HOIL-1L, and HOIP were performed. Results are shown as means and s.e.m. (n=4). *P < 0.0001, determined by the Student's _t_-test
SHARPIN deficient (Sharpincpdm/Sharpincpdm) mice exhibit diverse phenotypes in the immune system, which resemble the phenotypes of genetically engineered mouse models with impaired NF-κB activation7,8. As linear ubiquitylation is important for activation of the NF-κB pathway3,11,13,14 we speculated that the HOIP-SHARPIN LUBAC complex might play a critical role in regulation of the NF-κB pathway. Consistent with this hypothesis, cotransfection of SHARPIN and HOIP increased transcription of a luciferase-based NF-κB reporter, similar to the effect of HOIP and HOIL-1L co-expression (Fig. 2f), while no activation of NF-κB was detected when cells were transfected with SHARPIN, HOIL-1L, or HOIP alone (Fig. 2f), or a combination of mutant forms of NZF or UBL domains in SHARPIN together with HOIP (Fig. 2g).
Based on the finding that TNFα stimulated formation of a signalling complex between HOIP-SHARPIN and NEMO (Supplementary Fig. 6), we reasoned that the SHARPIN-HOIP complex might act as an upstream regulator of IKK activation. In vitro IKK kinase assay showed that TNFα rapidly enhanced IKK kinase activity in wild type MEFs, which was delayed and reduced in cells lacking SHARPIN (Fig. 3a). Upon additional abrogation of HOIL-1L expression in Sharpincpdm/Sharpincpdm MEFs by stably expressing shRNA against HOIL-1L (Sharpincpdm/Sharpincpdm shHOIL-1L) IKK activation was further inhibited (Fig. 3a). Moreover, downstream events of IKK activation, such as phosphorylation and subsequent degradation of IκBα, as well as nuclear translocation of p65, were also reduced in Sharpincpdm/Sharpincpdm MEFs and further impaired in response to shRNA-mediated downregulation of HOIL-1L (Fig. 3b, 3c and Supplementary Fig. 7a) or overexpression of an IκBα-super repressor mutant (Supplementary Fig. 7b).
Figure 3. SHARPIN and HOIL-1L are essential for full activation of IKK and NF-κB.
a, In vitro IKK-kinase activity in TNFα-treated Sharpincpdm/Sharpincpdm MEFs stably transduced with non-targeting (control) or HOIL-1L shRNA. b, c, Defect of TNFα-induced IκBα degradation and translocation of p65 in Sharpincpdm/Sharpincpdm MEFs stably transduced with HOIL-1L shRNA. (n = 19–23) d, e, Impaired NF-κB activation in primary B cells or macrophages of Sharpincpdm/Sharpincpdm stimulated with CD40L or LPS. e, f, g, Reduced capacity of Sharpincpdm/Sharpincpdm macrophages to secrete TNFα and MCP-1 in response to LPS. TNFα ELISA; n=5, MCP-1 ELISA; n=4. Results in c, f and g are shown as means and s.e.m. *P < 0.0001, determined by the Student's _t_-test.
Given the plethora of evidence presented above that SHARPIN modulates the NF-κB pathway, we further assessed the functional role of SHARPIN in NF-κB activation in vivo using cells isolated from the Sharpincpdm/Sharpincpdm mice. The absence of SHARPIN in Sharpincpdm/Sharpincpdm splenic B-cells stimulated with soluble CD40L resulted in impaired activation of the NF-κB pathway, evaluated by phosphorylation and degradation of IκBα, compared to wild type B-cells (Fig. 3d). Similar findings were obtained in thioglycollate-induced peritoneal macrophages from Sharpincpdm/Sharpincpdm mice, which in response to bacterial lipopolysacharide (LPS) stimulation failed to induce an efficient phosphorylation and degradation of IκBα (Fig. 3e), proper nuclear translocation of the NF-κB transcription factor p65 (Supplementary Fig. 8a) or secretion of TNFα and MCP-1 (Fig. 3f and g). Furthermore, IL-1β stimulation-induced NF-κB activation was delayed in SHARPIN deficient MEFs (Supplementary Fig. 8b). Similarly, we observed partial impairment of JNK activation in response to TNFα, CD40L, or LPS in Sharpincpdm/Sharpincpdm MEFs, B-cells, or macrophages, respectively, compared to wild type cells (Fig. 3e and Supplementary Fig. 8c–e).
An interesting feature of Sharpincpdm/Sharpincpdm mice is the combination of epidermal hyperplasia and inflammation with the presence of apoptotic bodies in keratinocytes and increased cell death in the skin15. Moreover, inhibition of TNFα-induced NF-κB signalling in keratinocytes results in inflammatory skin lesions as observed in genetically engineered mouse models, as well as in human diseases such as incontinentia pigmenti7. Using highly sensitive impedance based real-time cell analyzer, we found that Sharpincpdm/Sharpincpdm MEFs were extremely sensitive to rapid TNFα-induced cell death (Fig. 4a). This correlated with an appearance of typical apoptotic cell morphology (Supplementary Fig. 9a) and early signs (2–4 hours post TNFα stimulation) of apoptotic signalling, including proteolytic cleavage of the initiator caspase, caspase-8, the effector caspase, caspase-3, as well as PARP, a substrate of caspase-3 (Fig. 4b). In agreement with a requirement for caspase activity for the induction of apoptosis, pre-treatment of Sharpincpdm/Sharpincpdm cells with the general caspase inhibitor zVAD-FMK abolished the observed cell death in response to TNFα (Supplementary Fig. 9b). When performing the reciprocal experiments on MEFs derived from HOIL-1 null mice14, we observed a delayed and significantly weaker induction of the apoptotic response in cells stimulated with TNFα and cycloheximide (CHX) (Fig. 4a). In accordance, we were unable to detect rapid activation of caspase-3 and caspase-8 upon TNFα or TNFα + CHX treatment of HOIL-1L deficient MEFs (Supplementary Fig. 9c). Moreover, additional downregulation of HOIL-1L in Sharpincpdm/Sharpincpdm MEFs did not further increase TNFα-induced apoptosis or caspase activation (Supplementary Fig. 9d), whereas a combined lack of SHARPIN and HOIL-1L had a strong additive effect on the inhibition of the NF-κB pathway (Fig. 3a–c). Cell death in Sharpincpdm/Sharpincpdm MEFs was not upregulated by other stress stimuli, including Staurosporin, Brefeldin A, and Doxrubicin (Supplementary Fig. 9e–g).
Figure 4. Loss of SHARPIN sensitizes cells to FADD- and caspase-8-mediated apoptosis.
a, Increased susceptibility to TNFα-induced apoptosis in SHARPIN deficient MEFs. Cell detachment and death were measured continuously in real-time using an impedance based real-time cell analyzer (RTCA). b, Increased caspase activation by TNFα and CHX in SHARPIN deficient MEFs. c, d, Cell death caused by loss of SHARPIN via FADD and caspase-8 dependent apoptotic signalling. MEFs were retrovirally transduced with indicated proteins and treated with TNFα. e, Increased apoptotic cell death in the skin of Sharpincpdm/Sharpincpdm mice. Normal epidermis (A) and Sharpincpdm/Sharpincpdm (B) skin samples were immunostained for caspase-8 (C), caspase-9 (D) and caspase-3 (E). None of these markers were present in control mouse epidermis (F). H&E; hematoxylin and eosin. Bar = 2 μm. Results in a and c are shown as means and s.e.m. (n=3).
TNFα-induced cell death is known to proceed through the cytosolic TNFR1 signalling complex II that is composed of TRADD, TRAF2, RIP1, caspase-8/10, and the death domain containing protein FADD, which together serve as a cell death checkpoint control in TNF-sensitive cells16,17. The apoptotic cell death displayed by Sharpincpdm/Sharpincpdm MEFs occurs through the FADD and caspase-8-containing TNFR1 complex II, since retroviral transduction of either a dominant negative mutant of FADD or CrmA, a cowpox virus derived serine protease inhibitor (serpin) of caspase-8, markedly inhibited TNFα-induced cell death (Fig. 4c and 4d). Importantly, we have also detected activation of caspase-3, -8, and -9 in skin keratinocytes of Sharpincpdm/Sharpincpdm mice (Fig. 4e). Taken together, our results suggest that loss of SHARPIN expression sensitizes cells for TNFα-induced cell death, in part dependent on a caspase-8 pathway. This phenomenon might be an underlying mechanism for the development of inflammatory skin lesions in Sharpincpdm/Sharpincpdm mice.
In conclusion, this study provides physiological evidence for a novel LUBAC complex composed of HOIP, the major catalytic subunit, and the adaptor protein SHARPIN. Together, SHARPIN and HOIP mediate linear ubiquitylation of NEMO in vivo (Fig. 2a–e, Supplementary Fig. 4, 5) and activate the NF-κB pathway following overexpression in cells (Fig. 2f, g). The interaction between multiple ubiquitylated NEMO molecules, via their UBAN domains, may cause conformational changes promoting the activation of IKK kinases11. In such a model, the dynamics of NF-κB activation in response to different stimuli may be determined by the distinct ability of NEMO to recognize a variety of ubiquitin signals, including different linkages (linear, K63 and K11)4,6,18 and varying lengths of ubiquitin chains19. SHARPIN and HOIL-1L have overlapping roles in the function of LUBAC as a key activator of the IKK complex and NF-κB signalling downstream of several physiological stimuli (Fig. 3). On the other hand, SHARPIN, but not HOIL-1L, deficiency in mice results in an increased cell death that is dependent on caspase-mediated apoptosis, evident in MEFs and inflammatory skin lesions (Fig. 4). Given the identified anti-apoptotic function of SHARPIN, this study exposes a putative role for linear ubiquitylation in the regulation of cell death and strongly reinforces its impact on NF-κB signalling and immune responses.
Methods Summary
Protein purification, apoptosis and biochemical assays
Recombinant proteins were expressed in Rosetta cells (Novagen). Detailed descriptions of immunoprecipitation, immunoblotting, GST pull-downs and apoptotic assays are available as Supplementary Information. ITC measurements were performed using a VP-ITC or ITC200 (GE Healthcare), as described in Supplementary Methods.
Mice and isolation of primary cells
C57BNL/KaLawRij-Sharpincpdm/RijSunJ mice (JR7599, The Jackson Laboratory) were raised in a pathogen-free environment. Resting mature B cells were isolated using negative selection by CD43-coated magnetic beads (MiltenyiBiotec), thioglycollate-induced macrophages were harvested from the peritoneal cavity and MEFs were immortalized using SV40 large T-antigen transfection and dilution passaging.
Apoptosis assays
Quantitative apoptosis assays were performed in real time using a noninvasive impedance based real-time cell analyzer (Roche).
Full Methods and any associated references are available in the supplementary online material of the paper.
Methods
cDNA, Antibodies, and Cells
Full length SHARPIN, NZF (340–387) or UBL (233–314) from HeLa cDNA were subcloned into pGEX-4T1 (GE Healthcare), pEGFP-C1 (Novagen, Madison, WI) or pBabe-puro by standard PCR methods. Mutants of SHARPIN T358L, F359V, SHARPIN ΔNZF (Δ350–374), SHARPIN I272A, SHARPIN ΔC (1–232), SHARPIN ΔUBL, and insertion of 6XHis-tag in C-terminus of pGEX plasmid were introduced by site-directed mutagenesis. pcDNA3-Myc-human HOIP, pcDNA3-HA-human HOIL-1L ΔRBR-C, pcDNA3-HA-human HOIL-1L ΔUBL, pcDNA3-HA-human HOIL-1L ΔNZF, pGEX-6Pp1-HOIP NZF1 (aa350–379), pGEX-6P-1-HOIP NZF2 (aa 408–438), and pGEX-6P-1-HOIP NZF1+2 (aa 350–438) were described previously14. pGEX-4T1-Ubiquitin, pGEX-4T1-diUbiquitin, and pGEX-4T1-mouse NEMO-UBAN were described elsewhere11. pMALc2x-human NEMO was generated by a standard PCR method. pGEX-4T2-tetra-Ubiquitin was a kind gift from Caixia Guo and Errol Friedberg. pGEX-4T1-mouse ABIN1-UBAN was described previously12. pcDNA3-Myc-human HOIP RING mutant (C699/702S, C871/874S) and pGEX-4T1-mouse ABIN1-UBAN D485A were generated by standard site directed mutagenesis. pMSCV-GFP, pMSCV-Flag-DN-FADD-IRES-GFP, and pMSCV-Flag-CrmA-IRES-GFP were kind gifts from John Silke. Anti-His (Novagen, Madison, WI), anti-Ubiquitin (P4D1; Santa Cruz Biotechnology, Santa Cruz, CA), anti-GFP (B-2; Santa Cruz Biotechnology), anti-HA (HA.11; Covance, Berkeley, CA), anti-Myc (9E10; Santa Cruz Biotechnology), anti-Flag (M2, Covance), anti-Cleaved Caspase 3 (Cell Signaling), anti-Caspase 8 (Cell Signaling), anti-Tubulin (Sigma), anti-Vinculin (Sigma), anti-PARP (Cell Signaling), anti-p65 (Santa Cruz Biotechnology), anti-IκBα (Cell Signaling and Santa Cruz Biotechnology), anti-pIκBα (Cell Signaling), anti-IKKα (Imgenex), anti-NEMO (Santa Cruz Biotechnology), anti-pJNK (Cell Signaling), anti-pp38 (Cell signaling), anti-NUP62 (BD Transduction), anti-GAPDH (Abcam), and anti-Flag M2 agarose (Sigma) were purchased and used according to the manufacturers recommendations. Anti-HOIP and anti-HOIL-1L were described previously14. The anti-SHARPIN antibody was raised against a fragment representing the N-terminal region (1–162 amino acids) of human SHARPIN and affinity-purified using the antigen affinity matrix.
Human Embryonic Kidney (HEK) 293T cells (ATCC) were maintained at 37°C, in 5% CO2 condition in DMEM (Gibco) supplemented with 10% Fetal calf serum (Gibco) and 100 U/ml penicillin and streptomycin (Invitrogen, Carlsbad, CA). Murine TNFα, IL-1α and CD40L were purchased from PreproTech, while LPS was from Enzo Life Sciences. zVAD-FMK and cycloheximide were purchased from Bachem and R&D, respectively.
SILAC cells
HEK 293T cells were cultured in a custom-made SILAC-DMEM medium (lacking the amino acids lysine and arginine, PAA). The medium was supplemented with dialysed serum (PAA), L-Glutamine, penicillin, streptomycin and amino acids L-lysine and L-arginine. The “light” culture was supplemented with Lys0 and Arg0 (Sigma); the “medium-heavy” culture with Lys4 (2H4) and Arg6 (13C6); and the “heavy” culture with Lys8 (13C6 15N2) and Arg10 (13C6 15N4). All labelled amino acids were purchased from Cambridge Isotope Laboratories.
GST protein purification
Transformed E. coli were grown in LB/ampicillin at 37°C until OD = 0.3–0.5, induced with 1 mM IPTG and grown over night at 16°C. The bacteria were harvested by centrifugation and resuspended in GST buffer (20 mMTris, pH 8.0, 100 mMNaCl) supplemented with complete protease inhibitors (Roche Diagnostics), sonicated, and lysed in 1% Triton X-100. The cleared lysate was incubated with Glutathione Sepharose beads (GE Healthcare) for 2 hours at 4°C with continuous rolling. The beads were subsequently washed with buffer 1 (0.1 M Tris, pH 7.8, 0.5 M NaCl), GST-buffer containing 0.35% Triton X-100 and GST buffer alone. The purified GST-fusions were separated by SDS-PAGE and analyzed by Coomassie staining.
GST-Pull down and Immunoprecipitation assays
HEK293T cells were transfected with the indicated constructs using FuGene6 (Roche Diagnostics, Mannheim, Germany) or GeneJuice (Novagen) according to the manufacturer's protocol. Cells were treated before lysis (lysis buffer; 50 mM Hepes, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 25 mM NaF, 10 μM ZnCl2, 10% glycerol, 1% Triton X-100, supplemented with complete protease inhibitors (Roche)). Cleared lysates were subjected to immunoprecipitation by incubation with the indicated antibodies, followed by Protein A/G Agarose (Santa Cruz Biothechnology) or Protein A-agarose (Roche). Similarly, total cell lysates were incubated with GST-proteins conjugated to Glutathione sepharose beads at 4°C. After washing 3 times with lysis buffer, the immunoprecipitates were separated by SDS-PAGE and analyzed by Western blot.
Purification of proteins for ITC measurements
All constructs for ITC studies have been cloned into pGEX-4T1 or pGEX-6P1 and were expressed using E. coli BL21 as host strain. 5 to 10 L expression culture was grown to an OD of 0.8 in LB/ampicillin media supplemented with 100 μM ZnCl2 at 37°C prior to induction with 0.1 mM IPTG for 12 hours at 25°C. Cells were resuspended in buffer PX (100 mM HEPES pH 7.4, 500 mM NaCl, 1 mM DTT) and lysed by sonication. The lysate was cleared by centrifugation and loaded onto GSH Sepharose column (GE Healthcare). Bound GST fusion protein was eluted using buffer PX containing 20 mM glutathione, cleaved with thrombin or 3C protease, and further purified by anion exchange chromatography using a Q Sepharose column (GE Healthcare). Fractions containing the protein of interest were polished by size exclusion chromatography on a Superdex 75 or Superdex 200 column (GE Healthcare). Protein concentrations were determined by UV spectrometry at 280 nm using calculated extinction coefficients. K48 and K63-linked ubiquitin chains were prepared essentially as described20.
Isothermal titration calorimetry
ITC measurements were performed using a VP-ITC or ITC200 (GE Healthcare). Typically 50–100 μM of SHARPIN, HOIL-1L, the isolated NZF domains and the NZF1 of HOIP were loaded into the cell and 500–1000 μM of ubiquitin or ubiquitin chains into the syringe. Samples were dialyzed into buffer containing 50 mM HEPES pH 7.4, 150 mM NaCl, and 1 mM TCEP. SHARPIN titrations were performed at 10° C, HOIL-1L, and HOIP titrations at 20°C. ITC data were analyzed with Origin7 (Micro Cal) supplied by the manufacturer.
Luciferase assay
HEK293T cells were plated on 48-well plates at a density of 20,000 cells/well 24 hours prior to transfection. pNF-κB-Luc plasmid (Stratagene) and β-GAL plasmid were transfected together with the indicated plasmids using FuGene6. After 36 hours of transfection, lysates were prepared and subjected to luciferase assays following the manufacturer's protocol (Roche). Internal control was measured by β-Gal activity using its substrate (Roche). All experiments were done using quadruple samples.
In vitro ubiquitylation assay
Ubiquitin (10 μg), His-tag E1 (150 ng), UbcH7 (300 ng) (Boston Biochem), and the indicated E3 ligase complexes (1 μg) were incubated with or without 2 mM ATP (Sigma) for the indicated time at 37°C in ubiquitin assay buffer (20 mM Tris-HCl pH7.5, 5 mM MgCl2, 2 mM DTT). Methylated ubiquitin was used as a negative control. To stop the reaction, SDS-loading buffer was added and the samples were boiled at 95°C for 1 minute. The samples were subsequently analyzed by SDS-PAGE followed by Western blotting using a PVDF membrane.
In vitro kinase assay
MEFs of wt, Sharpincpdm/Sharpincpdm or Sharpincpdm/Sharpincpdm HOIL-1L shRNA were treated by TNFα (20 ng/ml) for indicated times. IKK complex was immunoprecipitated by anti-NEMO antibody from the total cell lysates prepared from treated MEFs. Kinase assays were performed using γ-(32P)ATP (5 μ-Ci), GST-IκBα 1–54 (1 μ-g), and the IKK complex, incubated in the kinase buffer (25 mM Tris-HCl pH 7.5, 10 mM MgCl2, 0.1 mM sodium orthovanadate, 5 mM β-glycerophosphate, 2 mM DTT, 20 μ-M ATP) at 30°C for 30 min.
Protein separation, digestion, and mass spectrometry
Complete protein eluates of the three pull-down experiments were combined and separated by 1D SDS PAGE (NuPAGE 12% precast Bis/Tris gels, Invitrogen). The proteins were visualized by staining using the Novex® Colloidal Blue Staining Kit (Invitrogen) according to the manufacturer's instructions and the corresponding gel lane was cut into eleven pieces. In gel trypsin digestion of the proteins was performed as described previously21.
Chloroacetamide was used for alkylation to prevent formation of lysine modifications isobaric to Gly-Gly22. Prior to LC-MS peptides were desalted using C18 StageTips23. LC-MS analysis was performed using a Proxeon Easy-LC system (ProxeonBiosystems) coupled to an LTQOrbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (ProxeonBiosystems), as described previously24. Peptides were eluted using a segmented gradient of 5–80% of solvent B (80% ACN in 0.5% acetic acid) with a constant flow of 200 nl/min over 118 minutes. Full scan MS spectra were acquired in a mass range from m/z 380 to 920 with a resolution of 60,000 in the Orbitrap mass analyzer using the lock mass option for internal calibration25. The ten most intense ions were sequentially isolated for CID fragmentation in the linear ion trap. Inclusion list containing 27 ions of special interest (ubiquitylated NEMO and ubiquitin peptides) was used and up to 500 precursor ion masses selected for MS/MS were dynamically excluded for 90s. Mass spectra were processed and quantified using the MaxQuant software suite26 (version 1.0.14.3) and the data were searched using Mascot search engine (Matrix Science) against a decoy human database (ipi.HUMAN.v3.64) containing 168,584 protein entries. Carbamidomethylation (Cys) was defined as fixed-, and protein N-terminal acetylation, oxidation (Met), Gly-Gly (Lys), and Gly-Gly (N-terminal) were defined as variable modifications. Initial mass tolerance for the precursor ions was set to 7 ppm, and for the fragment ions to 0.5 Da. The Gly-Gly (Lys) modification sites were considered localized if the localization probability (calculated by MaxQuant software) was higher than 0.75.
Absolute quantification of ubiquitin in whole cell lysate
For absolute quantification (AQUA11) proteins were precipitated from crude cell lysates of “light” (SHARPIN/HOIP) and “medium-heavy” (SHARPIN TF_LV/HOIP RING mutant) SILAC-labelled HEK293T cells by chloroform/methanol precipitation. Protein concentration was measured by Bradford before and after precipitation to determine protein recovery. Equal amounts of protein extracts were mixed and in-solution digested by trypsin as described previously24 and chloroacetamide was used for alkylation as described above. Four synthetic AQUA peptides (Thermo Fisher Scientific), labelled with either Lys8 or Arg10 on the C-terminus, were added to 100 μg of digested protein in the following amounts: GGM(ox)QIFVK (1 pmol), TLSDYNIQK (10 pmol), ESTLHLVLR (10 pmol), and TITLEVEPSDTIENVK (100 pmol). The peptide mixtures were further fractionated using isoelectric focusing on an OffGel 3100 Fractionator (Agilent). Focusing was done on 13 cm (12 well) ImmobilineDryStrips pH 3–10 (GE Healthcare) at a maximum current of 50 μA for 23 kVh. Peptide fractions were collected and desalted using C18 StageTips27. LC-MS analysis was performed as described above with the exception that full scan MS spectra were acquired in a mass range from m/z 420 to 920. Inclusion list containing 17 ions of special interest (ubiquitin peptides, “light”-, “medium-heavy”- and “heavy”-labeled) was applied and no additional masses were allowed for fragmentation. The five most intense ions from the inclusion list were sequentially isolated for CID fragmentation in the linear ion trap. Mass spectra were processed and searched as described above. The ESTLHLVLR peptide was discarded from final quantitation because of an interference caused by the overlapping isotopic distribution of another peptide. The final AQUA results are reported per 100 μg of the crude protein extract considering the protein loss during precipitation and substoichiometric oxidation of the GGMQIFVK peptide. The latter was estimated from the intensities of the modified (oxidized) and unmodified forms of the GGMQIFVK peptide.
Mouse, MEF and primary cells
Sharpincpdm/Sharpincpdm mice from the C57BL/KaLawRij colony were raised in the specific pathogen-free breeding facility at the Goethe University animal facility. Breeding was made by crossing heterozygous female and male mice. Immortalized mouse embryonic fibroblasts (MEFs) were established using SV40 large T-antigen transfection and serial passaging. Mature resting primary B-cells were isolated from the spleen of 12-week old mice of indicated genotypes by negative selection using CD43-coated magnetic beads (Miltenyi Biotec, Bergisch Gladbach, Germany). Peritoneal macrophages were isolated by washing the peritoneal cavity with growth media 5 days after intra-peritoneal injection of thioglycollate. MEFs and primary cells were treated with CD40L (250 ng/ml), LPS (100 ng/ml), TNFα (20 ng/ml) or IL-1β (1 ng/ml or 10 ng/ml) for the indicated times and subjected to SDS-PAGE and Western blot analysis. The supernatant of macrophages was assessed for secreted TNFα and MCP-1 by classical sandwich ELISA using ELISA kits from R&D systems.
Immunohistochemistry
Skin tissue samples from wild type and Sharpincpdm/Sharpincpdm (male, 10 weeks after birth) mice were fixed in Fekete's acid alcohol formalin solution and subjected to histological analysis by H&E staining, or immunohistochemical analysis using antibodies recognizing cleaved caspase 3, 8, and 9.
Retroviral production
pMSCV-control-GFP, pMSCV-Flag-DN-FADD IRES-GFP, and pMSCV-Flag-CrmA IRES-GFP vectors were transfected into ecotropic phoenix cells (ATCC) using Gene Juice (Novagen). After 36 hours of transfection, released retroviruses were filtered and used for infection of target cells using polybrene (4 μ-g/ml). Expression of infected protein was monitored by GFP expression.
Subcellular fractionation
To obtain nuclear and cytoplasmic fractions, cells were washed and sampled in 500 μl of cold PBS. Pelleted cells (500 × g) were resuspended in hypotonic buffer (10 mM Tris-HCl, pH 7.4, 10 mM sodium iodide, 5 mM MgCl2, 1 mM PMSF) and homogenized with 20 strokes in a tissue grinder. The supernatant (600 μ g) was designated the cytoplasmic fraction. The pellet, representing the nuclear fraction, was washed with hypotonic buffer containing 0.1% NP-40 (Calbiochem).
Apoptosis assays
Wild type, Sharpin+/Sharpincpdm or Sharpincpdm/Sharpincpdm MEFs were plated on 6-well or 24-well plates. After 24 hours of sub-culturing, cells were treated with CHX (1 μM) or TNFα (10 ng/ml) for the indicated times. Depending on the experimental set-up, the caspase inhibitor zVAD-FMK (50 nM) and retroviral infection were combined. After the treatment, cells were harvested for SDS-PAGE followed by Western blot analysis. In parallel, a highly sensitive, non-invasive and quantitative device, an impedance based real-time cell analyzer (Roche), was used to study the dynamics of cell death. This device continuously monitors the impedance to electric current flow by cells attaching on gold electrode-plated cell culture dishes. The area covered by cells is quantified and plotted as cell index as a function of time. 10,000 to 20,000 cells were plated per well of a 96-well E-plate. 24 to 36 hours later, cells were treated as indicated with TNFα (10 ng/ml) alone or in combination with cycloheximide (1 μ-g/ml), followed by continuous recordings of cell index in real-time and plotted by normalizing cell indices to one just prior to treatment of cells with the respective reagents.
Stable shRNA cell lines
Wild type or Sharpincpdm/Sharpincpdm MEFs were transduced with lentiviral particles (5 MOI) containing non-targeting (control) or HOIL-1L shRNA (sequence: CCGGCCTACCAGATACCTGCTTCATCTCGAGATGAAGCAGGTATCTGGTAGGTTT TTG) (Sigma). 48 hours post-transduction, cells were selected with medium containing 4 μg/ml puromycin. After two weeks, selected cells were maintained with medium containing 1 μg/ml puromycin.
Confocal fluorescence microscopy
Cells were grown on glass coverslips and treated as indicated in the figure legends. The cells were then fixed with 2% paraformaldehyde solution (Sigma), permeabilized with 0.2% Triton X-100 in PBS and immunostained with appropriate antibodies. Fluorophore-labeled secondary antibodies were from Jackson Immunoresearch (West Grove, PA). DAPI was used to stain the nuclei. The cells were mounted in Mowiol (Calbiochem, San Diego, CA) and examined by the laser scanning confocal microscope LSM 510 META (Carl Zeiss, Jena, Germany). Images were prepared and analyzed with the LSM Image Browser software (Carl Zeiss).
Supplementary Material
Supplementary Data
Acknowledgements
We thank Eunjoon Kim, Krishna Rajalingham and Hans-Jürgen Kreienkampfor reagents used in this study, Ivan Matic for initial MS analysis of HOIP/HOIL-1L samples, Silke Wahl for sample preparation, and Volker Dötsch and members of the Dikic lab for discussions and constructive comments. This work was supported by grants from Deutsche Forschungsgemeinschaft (DI 931/3-1), the Cluster of Excellence “Macromolecular Complexes” of the Goethe University Frankfurt (EXC115) to ID, Landesstiftung Baden-Württemberg to BM the Medical Research Council UK to KR and BS, JSPS Postdoctoral Fellowships for Research Abroad to FI, EMBO long-term fellowship to SS and The National Institutes of Health (AR049288 to JPS). VN was supported by the Unity Through Knowledge Fund, 3B Grant. CG acknowledges support from The International Human Frontier Science Program Organization.
Footnotes
Competing financial interests The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Grabbe C, Dikic I. Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins. Chem Rev. 2009;109(4):1481–1494. doi: 10.1021/cr800413p. [DOI] [PubMed] [Google Scholar]
- 2.Seymour RE, et al. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun. 2007;8(5):416–421. doi: 10.1038/sj.gene.6364403. [DOI] [PubMed] [Google Scholar]
- 3.Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132(3):344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- 4.Wertz IE, Dixit VM. Ubiquitin-mediated regulation of TNFR1 signaling. Cytokine Growth Factor Rev. 2008;19(3–4):313–324. doi: 10.1016/j.cytogfr.2008.04.014. [DOI] [PubMed] [Google Scholar]
- 5.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9(6):536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwai K, Tokunaga F. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO Rep. 2009;10(7):706–713. doi: 10.1038/embor.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasparakis M. Regulation of tissue homeostasis by NF-kappaBsignalling: implications for inflammatory diseases. Nat Rev Immunol. 2009;9(11):778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- 8.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 9.Kirisako T, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25(20):4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim S, et al. Sharpin, a novel postsynaptic density protein that directly interacts with the shank family of proteins. Mol Cell Neurosci. 2001;17(2):385–397. doi: 10.1006/mcne.2000.0940. [DOI] [PubMed] [Google Scholar]
- 11.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136(6):1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Wagner S, et al. Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene. 2008;27(26):3739–3745. doi: 10.1038/sj.onc.1211042. [DOI] [PubMed] [Google Scholar]
- 13.Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell. 2009;36(5):831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 14.Tokunaga F, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11(2):123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 15.Gijbels MJ, HogenEsch H, Blauw B, Roholl P, Zurcher C. Ultrastructure of epidermis of mice with chronic proliferative dermatitis. UltrastructPathol. 1995;19(2):107–111. doi: 10.3109/01913129509014610. [DOI] [PubMed] [Google Scholar]
- 16.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114(2):181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 17.Wilson NS, Dixit V, Ashkenazi A. Death receptor signal transducers: nodes of coordination in immune signaling networks. Nat Immunol. 2009;10(4):348–355. doi: 10.1038/ni.1714. [DOI] [PubMed] [Google Scholar]
- 18.Dynek JN, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29(24):4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda F, Crosetto N, Dikic I. What determines the specificity and outcomes of ubiquitin signaling? Cell. 2010;143(5):677–681. doi: 10.1016/j.cell.2010.10.026. [DOI] [PubMed] [Google Scholar]
- 20.Pickart CM, Raasi S. Controlled synthesis of polyubiquitin chains. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 21.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1(6):2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen ML, et al. Iodoacetamide-induced artifact mimics ubiquitination in mass spectrometry. Nat Methods. 2008;5(6):459–460. doi: 10.1038/nmeth0608-459. [DOI] [PubMed] [Google Scholar]
- 23.Ishihama Y, Rappsilber J, Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J Proteome Res. 2006;5(4):988–994. doi: 10.1021/pr050385q. [DOI] [PubMed] [Google Scholar]
- 24.Borchert N, et al. Proteogenomics of Pristionchuspacificus reveals distinct proteome structure of nematode models. Genome Res. 20(6):837–846. doi: 10.1101/gr.103119.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4(12):2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 27.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2(8):1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data