Essential role for ubiquitin–ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate (original) (raw)
. Author manuscript; available in PMC: 2012 Apr 8.
Summary
During ubiquitin conjugation, the thioester bond that links ‘donor’ ubiquitin to ubiquitin-conjugating enzyme (E2) undergoes nucleophilic attack by the ε-amino group of an acceptor lysine, resulting in formation of an isopeptide bond. Models of ubiquitination have envisioned the donor ubiquitin to be a passive participant in this process. However, we show here that the I44A mutation in ubiquitin profoundly inhibits its ability to serve as a donor for ubiquitin chain initiation or elongation, but can be rescued by computationally-predicted compensatory mutations in the E2 Cdc34. The donor defect of ubiquitin-I44A can be partially suppressed either by using a low pKa amine (hydroxylamine) as the acceptor or by performing reactions at higher pH, suggesting that the discharge defect arises in part due to inefficient deprotonation of the acceptor lysine. We propose that interaction between Cdc34 and the donor ubiquitin organizes the active site to promote efficient ubiquitination of substrate.
Introduction
Attachment of ubiquitin (i.e. ubiquitination) to intracellular proteins regulates nearly all aspects of cellular function in eukaryotes. Ubiquitination involves covalent attachment of ubiquitin to the target protein via an isopeptide bond between the C-terminus of ubiquitin and, most often, a lysine residue of the acceptor substrate. Additional ubiquitins can be conjugated to any of the seven lysine residues of ubiquitin to form a polyubiquitin chain on substrate. A chain of four or more ubiquitins linked together via lysine 48 or lysine 11 targets modified proteins for degradation by the 26S proteasome (Chau et al., 1989; Matsumoto et al., 2010; Song and Rape, 2010; Thrower et al., 2000), whereas chains of ubiquitin linked via lysine 63 or the N-terminus have been implicated in signaling (Dikic et al., 2009; Hicke et al., 2005).
Protein ubiquitination is carried out by a cascade of ubiquitin-activating (E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3) enzymes (Deshaies and Joazeiro, 2009; Dye and Schulman, 2007). First, E1 forms a thioester bond between its active site cysteine and the C-terminus of ubiquitin in an ATP-dependent manner. This charged E1 then transfers the ubiquitin to an E2 protein, resulting in an E2~Ub thioester wherein the ubiquitin C-terminus is attached to the catalytic cysteine residue of the E2. Subsequent binding of both E2~Ub and substrate to an E3 enzyme enables attack of the thioester bond of E2~Ub by the ε-amino group of a lysine residue in the substrate, leading to isopeptide bond formation. The nucleophillic lysine can reside either within the primary sequence of the substrate, or in an ubiquitin or ubiquitin-like protein previously conjugated to the substrate. A key point is that for the ε-amino group of lysine to engage in a nucleophilic attack on the thioester bond, it must be in the deprotonated (i.e. NH2) state. Given that the pKa of this group is ~10.5, the vast majority of lysine is in the NH3+ form at physiological pH and thus must be deprotonated as a prelude to catalysis.
The pairing of E2s and substrates by E3s determines specificity in ubiquitination. E3s are conserved among eukaryotes, with nearly 600 E3s potentially expressed in human cells (K. Hoffman, personal communication). There are two major types of E3s in eukaryotes, defined by the presence of either a HECT domain or a RING or RING-like domain (Pickart, 2001). HECT domain E3s form a thioester intermediate with ubiquitin, whereas RING or RING-like domains (hereafter referred to as RING domain for simplicity) facilitate direct transfer of ubiquitin from the E2 to the substrate. Greater than 95 percent of E3s are of the RING domain type, of which nearly half are cullin–RING ligases (CRLs). Befitting their numbers, CRLs play a substantial role in protein turnover; greater than 20 percent of all protein degradation by the 26S proteasome is blocked upon inhibition of the pan-CRL activator, NAE (Soucy et al., 2009).
CRLs are modular multi-subunit complexes organized by a cullin scaffold. CRLs recruit E2 via a RING domain subunit, which is bound at one end of the cullin scaffold, and recruit substrate via a receptor tethered by an adapter protein to the other end of the cullin scaffold (Cardozo and Pagano, 2004; Deshaies and Joazeiro, 2009; Willems et al., 2004). SCF, the prototype of the CRLs, consists of the cullin Cul1, the RING subunit Rbx1/Roc1/Hrt1, the adaptor protein Skp1, and an F-box protein such as Skp2 or β-TrCP that binds substrates. Recruitment of substrate and E2 to SCF and the subsequent transfer of ubiquitin have been studied in considerable detail, making SCF the best understood ubiquitin ligase in terms of specificity, regulation, and mechanism of action. Crystal structures for several substrate–F-box pairs have elucidated binding of phosphorylated substrate to the F-box (Hao et al., 2005; Orlicky et al., 2003; Wu et al., 2003). Meanwhile, the interaction between the E2 Cdc34 and SCF has been characterized biochemically. Interaction between a basic ‘canyon’ in the Cul1 subunit and the acidic tail of Cdc34 enables rapid dynamics of Cdc34–SCF complex formation and dissolution, which underlies rapid and processive polymerization of a K48-linked ubiquitin chain on substrate (Kleiger et al., 2009a; Kleiger et al., 2009b). The K48 linkage specificity is dictated by the acidic loop in Cdc34, which presumably positions the lysine 48 of the attacking ubiquitin towards the Cdc34~Ub thioester (Petroski and Deshaies, 2005).
Regarding the function of the donor ubiquitin thioesterified to E2, relatively little is known about its role in the catalysis of protein ubiquitination, and it has been generally assumed that the donor ubiquitin serves as a passive ‘passenger’. Currently, there are five structural models of an E2~Ub thioester intermediate. However, in each of these models the ubiquitin interacts differently with the E2. In the case of Ubc1, a truncated catalytic domain was used to build an NMR-based docking model of the E2~Ub thioester (Hamilton et al., 2001). The model predicts complementary interaction surfaces involving conserved residues in the E2 and ubiquitin proteins that could be important for catalysis. In a subsequent study, the crystal structure of an Mms2–Ubc13~Ub complex was solved in which ubiquitin is joined to Ubc13 by a stable oxyester bond due to replacement of Ubc13’s catalytic cysteine with a serine (Eddins et al., 2006). In that structure, the donor ubiquitin attached to one Mms2–Ubc13 heterodimer is bound to the acceptor binding site of Mms 2 in an adjacent Mms2–Ubc13 heterodimer within the crystal, and so this is not likely to reflect the normal binding site for the donor ubiquitin. More recently, the structure of an UbcH8-Ub intermediate was solved by NMR by replacing the thioester bond with a more stable disulphide linkage between the C-terminal cysteine of a G76C mutant ubiquitin and the active site cysteine of UbcH8 (Serniwka and Shaw, 2009). In this complex, the donor ubiquitin makes multiple contacts with residues near the active site cysteine in the E2. Additionally, the crystal structure of ubiquitin ligase Nedd4L bound to UbcH5~Ub reveals that the esterified ubiquitin makes extensive contact with the E3 (Kamadurai et al., 2009). Finally, the crystal structure of UbcH5b–Ub oxyester reveals that Ub esterified to one UbcH5b binds the back-side of a second UbcH5b to form an infinite spiral (Brzovic et al., 2006; Sakata et al., 2010). Collectively, these studies reveal that donor ubiquitin can form interactions with E2, but the exact nature of the interaction is variable and the functional role remains poorly understood.
Here, we provide direct biochemical evidence that interaction between donor ubiquitin and Cdc34 is critical for discharge of ubiquitin from Cdc34~Ub to substrate. A combination of computational modeling and mutagenesis leads us to propose a model for the structure of a discharge-competent Cdc34~Ub complex.
Results
Ubiquitin-I44A mutant is defective in substrate ubiquitination by Cdc34–SCFβ-TrCP
In the course of performing studies on the mechanism of action of Cdc34–SCF, we made the unexpected observation that a mutant ubiquitin bearing an I44A substitution was poorly discharged from Cdc34. This suggested to us that the ‘hydrophobic patch’ centered about isoleucine 44, which is known to play a critical role in ubiquitin recognition by ubiquitin-binding domains (Beal et al., 1998), might also play an important role in the transfer of ubiquitin from E2 to substrate, possibly through interaction with the E2 to which it is thioesterified, with the nucleophilic acceptor, or with the E3.
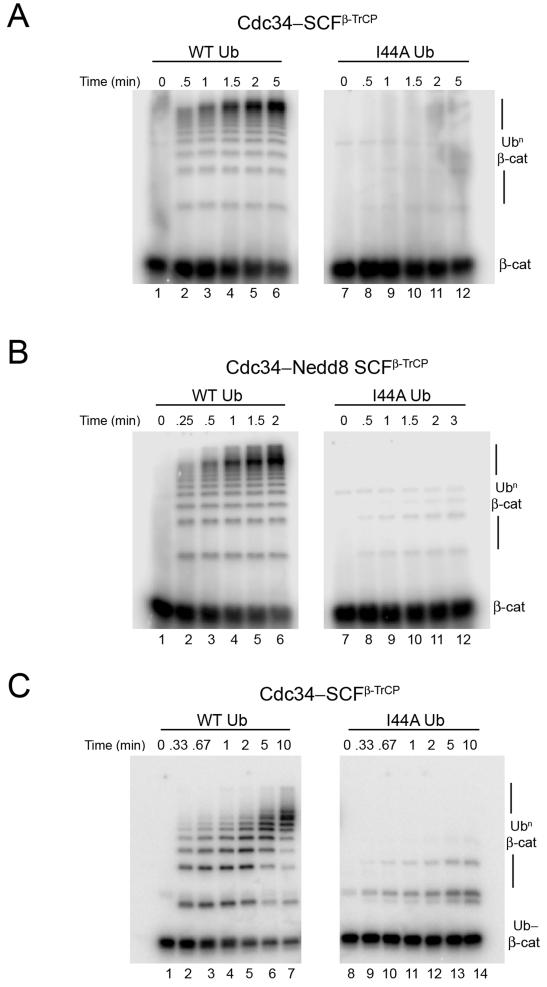
To better understand the molecular nature of the defect, we sought to quantify the effect of the I44A substitution using a previously established reconstituted system to measure transfer of ubiquitin to a 32P-labeled substrate peptide derived from β-catenin (Saha and Deshaies, 2008). In these single turnover experiments, SCF was present in stoichiometric excess of substrate and Cdc34~Ub was saturating SCF. With wild type (WT) ubiquitin, we observed robust ubiquitination of the peptide substrate with _k_cat ~ 0.3 min−1 (Figure 1A, lanes 1-6; _k_cat refers to the rate of consumption of substrate, irrespective of the number of ubiquitins transferred per molecule of substrate). By contrast, reactions carried out with ubiquitin-I44A yielded a severe decrease in β-catenin ubiquitination with ~ 20 fold lower _k_cat (Figure 1A, lanes 7-12). The decreased ubiquitination was reflected both in a lower fraction of modified substrate and fewer ubiquitin transfers per substrate. This was not due to defect in charging of E1 or Cdc34, since under the conditions of our assay, similar amounts of thioester were formed with both WT ubiquitin and ubiquitin-I44A (e.g. see lanes 1 and 7 in Figures 2A and 2B). A similar discharge defect of ubiquitin-I44A was seen at 26 μM Cdc34 (not shown), implying that it was a kcat effect and not due to a reduced KM for SCFβ-TrCP. This is further confirmed by studies shown in Figure 2.
Figure 1.
Ubiquitin-I44A cannot serve as a donor for ubiquitin chain initiation or elongation catalyzed by Cdc34–SCFβ-TrCP complex
(A) 32P-labeled β-catenin peptide (β-cat; 100 nM) was incubated with WT ubiquitin (lanes 1-6) or ubiquitin-I44A (lanes (7-12) in the presence of E1, ATP, 10 μM Cdc34 and 300 nM SCFβ-TrCP. Aliquots were removed at the indicated times and analyzed for substrate ubiquitination (Ubn β-cat) by SDS-PAGE followed by autoradiography.
(B) Same as in (A) but using 300 nM neddylated SCFβ-TrCP complex.
(C) Same as in (A) but using 32P-labeled monoubiquitinated β-catenin peptide substrate (Ub-β-cat; 1.5 μM) and 300 nM SCFβ-TrCP.
Figure 2.
The discharge defect of ubiquitin-I44A is intrinsic to the E2~Ub-I44A thioester complex.
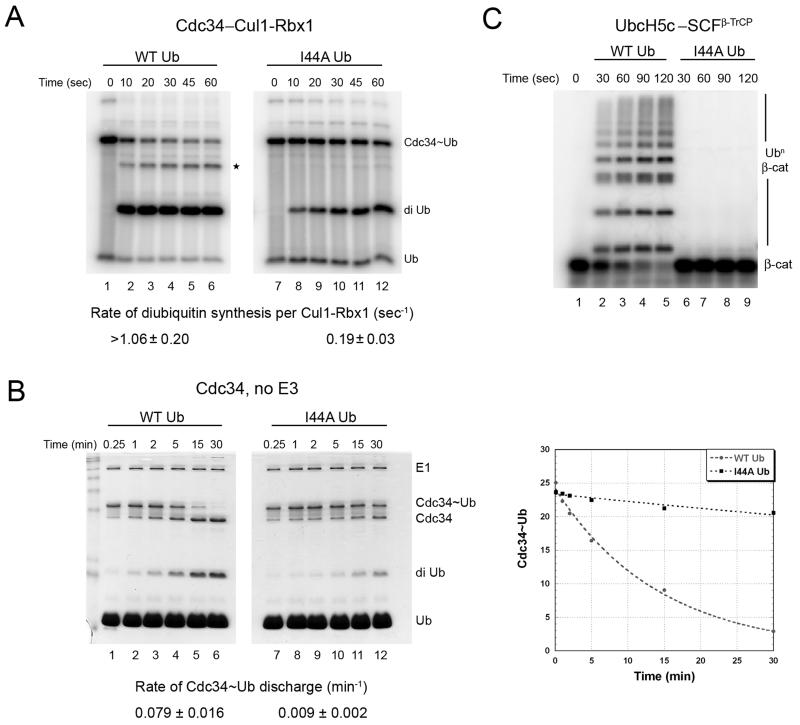
(A) Cdc34 (10 μM) was pre-incubated with equimolar 32P-labeled WT ubiquitin (lanes 1-6) or ubiquitin-I44A (lanes 7-12) in the presence of 1 μM E1 and ATP for 10 min at 23°C, followed by 5 min incubation with apyrase to terminate charging. Discharge of Cdc34~Ub was initiated by adding 75 μM ubiquitin-D77 plus 300 nM Cul1–Rbx1 and aliquots were removed at the indicated times and quenched with non-reducing sample buffer, resolved by SDS-PAGE, and analyzed by autoradiography. Rate of diubiquitin synthesis was estimated per unit Cul1–Rbx1. * denotes triubiquitin product.
(B) Same as in (A), except no Cul1–Rbx1 was added. Also, 120 μM ubiquitin-D77 was used, and the gel was stained with coomassie blue to reveal amounts of charged and discharged Cdc34. A Phosphor image of the stained gel was quantified and plotted, from which the rate of Cdc34~Ub discharge was estimated by fitting to a single exponential decay (right panel). A representative experiment is shown and the values reported (mean ± SD) are an average of 3 independent experiments.
(C) 32P-labeled β-catenin peptide (2 μM) was incubated with WT ubiquitin (lanes 2-5) or ubiquitin-I44A (lanes 6-9) in the presence of E1, ATP, 10 μM UbcH5c and 200 nM SCFβ-TrCP. Aliquots were removed at the indicated times and analyzed by SDS-PAGE followed by autoradiography.
The activity of CRLs is regulated by covalent modification of a conserved lysine residue on the cullin subunit with the ubiquitin-like protein, Nedd8. Nedd8 conjugation causes a major conformational change in the ligase that substantially increases the rate at which bound E2~Ub reacts with substrate (Duda et al., 2008; Saha and Deshaies, 2008; Yamoah et al., 2008). The ubiquitin-I44A mutant still showed a severe defect in substrate ubiquitination when assayed with neddylated SCFβ-TrCP (Figure 1B).
The pattern of ubiquitin conjugates observed with ubiquitin-I44A suggested that it is defective in both the first transfer to naïve substrate (i.e. chain initiation) as well as in subsequent transfers to the initiator ubiquitin (i.e. chain elongation). However, the chain elongation defect could be due to the inability of the initiator ubiquitin-I44A to serve as an acceptor for subsequent ubiquitin transfers, because ubiquitin-I44A is a poor acceptor in Cdc34~Ub discharge reactions (Petroski and Deshaies, 2005). To test directly whether ubiquitin-I44A is a poor donor during chain elongation, we performed a two-stage reaction. In the first stage, β-catenin peptide was modified with a single WT ubiquitin. This product was then purified and used as substrate in the second-stage reaction (Saha and Deshaies, 2008). The _k_cat for transfer of ubiquitin-I44A to monoubiquitinated β-catenin peptide was ~25-fold slower that for WT ubiquitin (Figure 1C). Thus, the I44A mutant is not competent to serve as a donor ubiquitin during either chain initiation or elongation. The defect observed with ubiquitin-I44A in transfer to two very different acceptors implies that the mutation does not perturb specific binding between the donor ubiquitin and the acceptor.
Ubiquitin-I44A is defective in discharge from Cdc34~Ub and UbcH5~Ub
To more incisively address the role of donor ubiquitin, we sought to strip down the ubiquitination reaction to a single-turnover discharge reaction wherein ubiquitin is transferred from Cdc34~Ub to free ubiquitin, resulting in formation of a diubiquitin conjugate (Petroski and Deshaies, 2005). This reaction involves charging of Cdc34 with equimolar ubiquitin in the presence of E1 and ATP, and then treating the reaction with apyrase to remove ATP and eliminate subsequent charging. Discharge of Cdc34~Ub is then initiated by addition of acceptor ubiquitin and formation of diubiquitin is monitored. In these reactions, the donor ubiquitin is 32P-labeled so that both the rate of thioester discharge and diubiquitin formation can be monitored by autoradiography.
We first examined the effect of the I44A mutation on discharge of Cdc34~Ub in the presence of Cul1–Rbx1. Both WT ubiquitin and ubiquitin-I44A formed similar levels of thioester intermediates under our reaction conditions (Figure 2A, compare lanes 1 and 7). Addition of unlabeled acceptor ubiquitin in the presence of Cul1–Rbx1 resulted in rapid discharge of the thioester to form diubiquitin. Both the rate of thioester discharge as well as the rate of product formation was ≥5 fold faster with WT ubiquitin compared to the I44A mutant (Figure 2A). This is a minimum estimate, because the initial rate of SCF-catalyzed discharge of WT ubiquitin was too rapid to be accurately measured under these conditions.
We next addressed whether the I44A mutation compromises interaction of donor ubiquitin with Cdc34 or with SCF by further simplifying the discharge reaction to measure SCF-independent discharge of the Cdc34~Ub thioester to free ubiquitin. In this experiment, reactants and products were monitored by staining with Coomassie Blue, followed by quantification by autoradiography. As for the previous experiments, ubiquitin-I44A thioester was severely compromised in its ability to discharge, resulting in diminished diubiquitin product formation (Figure 2B). To quantify the defect in the discharge rate we measured the difference in the rate of single exponential decay of the thioester. This provides a more accurate estimate of the discharge rate because it is independent of the starting amount of the thioester. This revealed that ubiquitin-I44A was discharged to acceptor ubiquitin ~ 9 fold more slowly than WT ubiquitin. Thus, the donor ubiquitin plays a critical role in discharge that is independent of SCF. We conclude that association of the hydrophobic patch of the donor ubiquitin with the surface of Cdc34 plays an important role in ubiquitin discharge.
Interestingly, in the modeled structure of Ubc1~Ub thioester, I44 is amongst the residues that are at the interface with Ubc1 (Hamilton et al., 2001). Therefore, we next sought to test whether the role of ubiquitin’s hydrophobic patch in discharge is unique to Cdc34~Ub, or if this is a more general phenomenon that applies to other E2s. To address this question, we performed β-catenin peptide ubiquitination assays with UbcH5c–SCFβ-TrCP. Similar to Cdc34, we observed that ubiquitin-I44A exhibited a ≥20 fold defect in substrate ubiquitination by UbcH5c (Figure 2C).
Charge swap in Cdc34 rescues thioester discharge defect of R42E ubiquitin
If the discharge defect of ubiquitin-I44A is due to loss of interaction with the E2 to which it is charged, it should be possible to identify compensatory mutations on the surface of the E2 that restore proper interaction and suppress the discharge defect. To gain further insight into the interface between the thioesterified donor ubiquitin and Cdc34, we used the modeled Ubc1~Ub thioester structure as a guide (Hamilton et al., 2001). In this model, I44 is adjacent to the α3 helix of Ubc1 (Figure S1). Another residue at the E2~Ub interface is arginine 42 of ubiquitin. Based on this model we introduced a charge-swap mutation in ubiquitin, R42E, that could potentially disrupt the interaction with Cdc34. To ensure that we were monitoring exclusively effects on E2-ubiquitin interaction, we performed discharge reactions in the absence of SCF. We found that ubiquitin-R42E was charged onto Cdc34 by E1 enzyme, but was compromised in both discharge from Cdc34 and diubiquitin formation (Figure 3A, lanes 1-6 compared to Figure 2B, lanes 1-6). Quantification based on single exponential decay of the thioester indicated that the ubiquitin-R42E thioester was ~ 4 fold defective in discharge. Defects of similar magnitude were observed when ubiquitin-R42E discharge to free ubiquitin (Figure 3B) or β-catenin peptide (Figure 3C) were measured in the presence of Cul1–Rbx1 or SCFβ-TrCP, respectively.
Figure 3.
E133R mutation in Cdc34 rescues thioester discharge defect of ubiquitin-R42E.
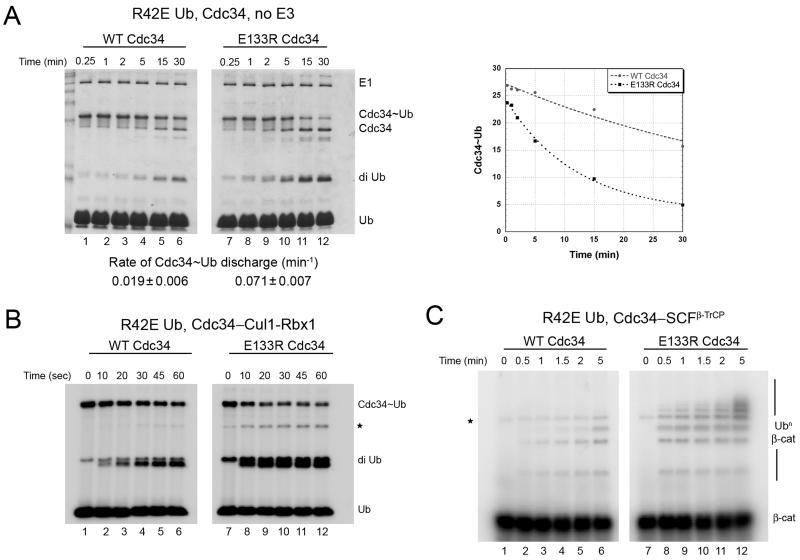
(A) 10 μM WT Cdc34 (lanes 1-6) or E133R mutant (lanes 7-12) was pre-incubated with equimolar 32P-labeled R42E Ub in the presence of 1 μM E1 and ATP for 10 min at 23°C, followed by 5 min incubation with apyrase. Discharge of Cdc34~Ub was initiated by adding 120 μM ubiquitin-D77 and aliquots were removed at the indicated times, quenched, fractionated by SDS-PAGE, and analyzed by coomassie blue staining followed by autoradiography. The image shown is that of the stained gel, and the phosphor image was quantified and plotted. The rate of Cdc34~Ub discharge was estimated by fitting to a single exponential decay (right panel). Shown is a representative experiment and the values reported (mean ± SD) are the average of 2 independent experiments.
(B) Same as in (A), but with the addition of 300 nM Cul1–Rbx1 complex and 75 μM ubiquitin-D77 (autorad image shown). * denotes triubiquitin product.
(C) 32P-labeled β-catenin peptide (1.5 μM) was ubiquitinated using WT Cdc34 (lanes 1-6) or Cdc34-E133R (lanes 7-12) in the presence of E1, ATP, 30 μM ubiquitin-R42E and 300 nM SCFβ-TrCP. Aliquots were removed at the indicated times and substrate ubiquitination analyzed by SDS-PAGE followed by autoradiography. * denotes contaminating band from the substrate.
To identify residues in Cdc34 that might counteract the effect of the R42E substitution in ubiquitin, we superimposed the sequence of Cdc34 on the Ubc1~Ub structure model. This implicated glutamate 133 of Cdc34 as being a potential interacting partner with residue 42 of ubiquitin. To evaluate this possibility, we generated and assayed a Cdc34-E133R mutant. The E133R mutation by itself had little effect on Cdc34 activity (Figure S2). Strikingly, however, Cdc34-E133R almost completely rescued the SCF-independent thioester discharge defect of ubiquitin-R42E. The rate of discharge of ubiquitin-R42E from Cdc34-E133R (0.071 min−1; Figure 3A, lanes 7-12) was nearly the same as the rate of discharge of WT ubiquitin from WT Cdc34 (0.079 min−1; Figure 2B). We also observed suppression of the discharge defect of ubiquitin-R42E by Cdc34-E133R when ubiquitin discharge/diubiquitin synthesis assays were carried out in the presence of Cul1–Rbx1 (Figure 3B) or when ubiquitination of β-catenin peptide was assayed in the presence of SCFβ-TrCP (Figure 3C). It is worth noting that if R42 of ubiquitin makes multiple contacts with Cdc34 besides E133 (but not vice versa), then the E133R mutation by itself would not necessarily inactivate Cdc34. The rescue of the ubiquitin-R42E mutant by a complementary charge swap in Cdc34 argues strongly that productive interaction between the donor ubiquitin and Cdc34 is critical for efficient discharge of the thioester.
Rescue of ubiquitin-I44A defect by compensatory mutation in Cdc34 predicted from a docked thioester model
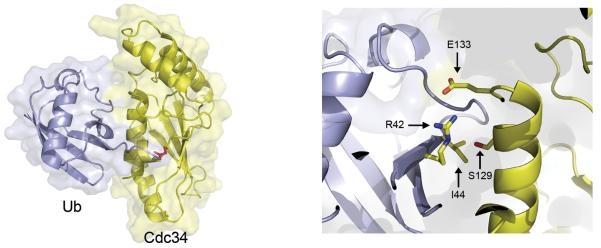
To further explore the interface between Cdc34 and thioesterified ubiquitin and seek deeper support for our hypothesis, we created a docking model from Cdc34 (2OB4) and ubiquitin (1UBQ) using a custom protocol written in Rosetta3 (Das and Baker, 2008; Leaver-Fay et al., 2011). Besides the thioester bond between the C-terminus of ubiquitin and Cdc34’s catalytic cysteine, the following two constraints were satisfied: (1) one of the charged atoms on the side chain of R42 of ubiquitin must be in close proximity with the charged atom on the side chain of E133 of Cdc34, and (2) I44 of ubiquitin must be close enough to at least one side chain on the surface of Cdc34 to engage in a van der Waals interaction. Whereas models lacking constraints predicted a wide variety of low energy structures (Figure S3), the imposition of these constraints enabled generation of a docked structure that allowed us to predict that serine 129 of Cdc34 is in close proximity to I44 of ubiquitin (Figure 4).
Figure 4.
Serine 129 of Cdc34 is in close proximity to isoleucine 44 of ubiquitin in a constrained Cdc34~Ub model.
Rosetta docking model for Cdc34~Ub thioester was generated by constraining the position of R42 of ubiquitin to be in close proximity to E133 of Cdc34. The thioester bond as well as the key residues for interaction are indicated. The figure was made in PYMOL.
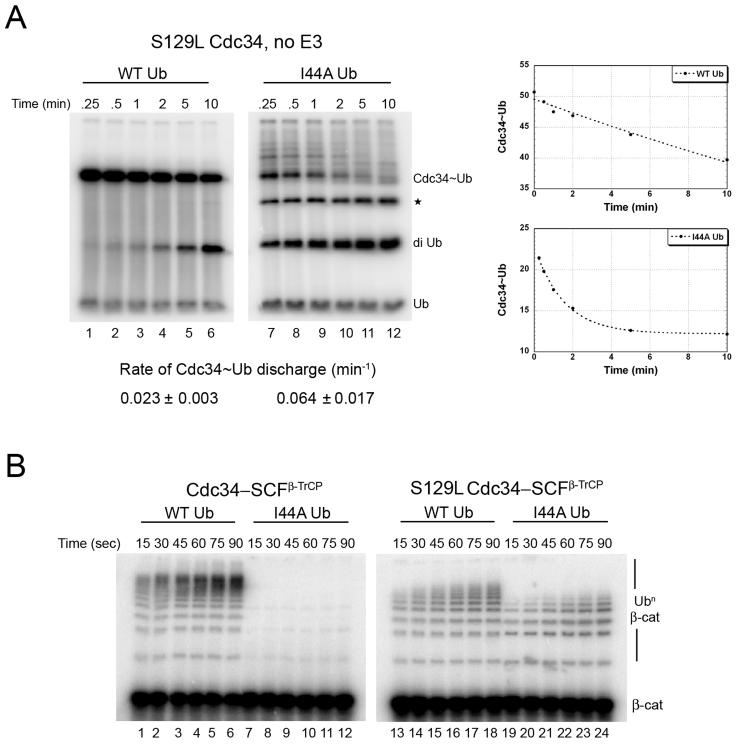
Based on the docking model, we predicted that an S129L mutation in Cdc34 should cause steric clash with I44 of ubiquitin due to the larger side chain of leucine relative to serine, rendering Cdc34-S129L defective in thioester discharge. To test this prediction we measured the rate of thioester discharge of Cdc34-S129L~Ub thioester in an SCF-independent diubiquitin synthesis assay and found it to be ~3.5-fold defective compared to WT Cdc34 (Figure 5A, lanes 1-6 compared to Figure 2B, lanes 1-6). Likewise, Cdc34-S129L was also defective in sustaining SCFβ-TrCP-dependent ubiquitination of β-catenin peptide (Figure 5B, lanes 1-6 compared to lanes 13-18). Based on our docking model, the defect in discharge of ubiquitin from Cdc34-S129L~Ub should be compensated by relieving the steric clash by placing a smaller side chain at position 44 of ubiquitin. We therefore reasoned that the I44A mutant, which by itself was profoundly defective in discharge, should relieve the steric clash and restore normal activity to Cdc34-S129L. Remarkably, ubiquitin-I44A and Cdc34-S129L rescued each other quite strongly: ubiquitin-I44A was discharged from Cdc34-S129L at a rate (0.064 min−1; measured as a single exponential decay) that was 80% the rate of discharge of WT ubiquitin from WT Cdc34 (0.079 min−1). Ubiquitin-I44A did not efficiently form a thioester with Cdc34-S129L (Figure 5A and Figure S4), but that did not interfere with our measurement because the rate of discharge was estimated based on a single exponential decay of the input thioester. Cdc34-S129L was also tested in an SCFβ-TrCP-dependent β-catenin peptide ubiquitination assay with ubiquitin-I44A (Figure 5B, lanes 19-24). Although ubiquitin-I44A failed to improve the activity of Cdc34-S129L, Cdc34-S129L rescued the defect of ubiquitin-I44A. The failure of ubiquitin-I44A to rescue Cdc34-S129L in a multi-turnover reaction was most likely due to the low efficiency of thioester formation, which is not a problem in a single-turnover chase experiment such as the one in Figure 5A. Taken together, these experiments indicate that serine 129 of Cdc34 packs against the I44 residue of Ub and this interaction is important for efficient thioester discharge.
Figure 5.
S129L mutation in Cdc34 suppresses the defect in thioester discharge of ubiquitin-I44A.
(A) Cdc34-S129L (10 μM) was pre-incubated with equimolar 32P-labeled WT ubiquitin (lanes 1-6) or ubiquitin-I44A (lanes 7-12) in the presence of 1 μM E1 and ATP for 10 min at 23°C, followed by 5 min incubation with apyrase. Discharge of Cdc34~Ub was initiated by adding 120 μM ubiquitin-D77 and aliquots were removed at the indicated times and quenched and analyzed by SDS-PAGE followed by autoradiography. The rate of Cdc34~Ub discharge was estimated by fitting to a single exponential decay. Shown is a representative experiment and the values reported (mean ± SD) are average of 2 independent experiments. * denotes triubiquitin product.
(B) 32P-labeled β-catenin peptide (100 nM) was incubated with WT Cdc34 (lanes 1-12) or the S129L mutant (lanes 13-24) in the presence of E1, ATP, 300 nM SCFβ-TrCP, and 30 μM WT ubiquitin (lanes 1-6 and 13-18) or ubiquitin-I44A (lanes 7-12 and 19-24). Aliquots were removed at the indicated times and substrate ubiquitination was analyzed by SDS-PAGE followed by autoradiography.
Cdc34 interaction with donor ubiquitin facilitates specific steps in catalysis of isopeptide bond formation
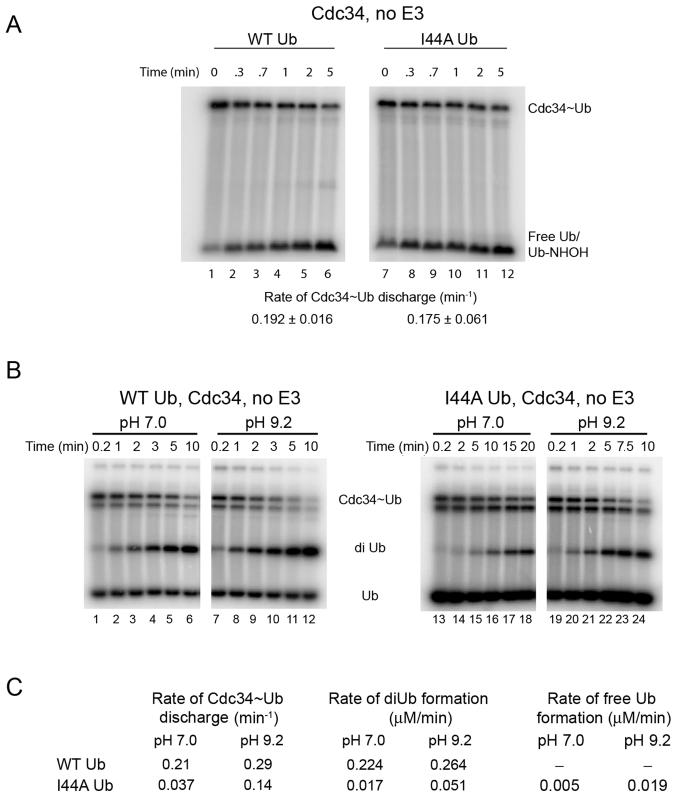
To gain insight into the biochemical basis of the ubiquitin-I44A defect, we employed an assay that monitors discharge of ubiquitin to the freely diffusing nucleophile hydroxylamine (Saha and Deshaies, 2008). Unexpectedly, in contrast to all of the other assays presented above, ubiquitin-I44A was discharged to hydroxylamine with normal kinetics (Figure 6A).
Figure 6.
Cdc34~I44A–ubiquitin complex is defective in nucleophilic activation
(A) Cdc34 (10 μM) was pre-incubated with equimolar 32P-labeled WT ubiquitin (lanes 1-6) or ubiquitin-I44A (lanes 7-12) in the presence of 1 μM E1 and ATP for 10 min at 23°C, followed by 5 min incubation with apyrase. Discharge of Cdc34~Ub was initiated by adding 4 mM hydroxylamine and aliquots were removed at the indicated times and quenched with non-reducing sample buffer. Rate of Cdc34~Ub discharge to form Ub-NHOH (which has been verified by mass spectrometry; Saha and Deshaies, 2008) was estimated and reported (mean ± SD).
(B) The rate of donor ubiquitin discharge to ubiquitin-D77 acceptor was measured for Cdc34 (6 μM) thioesterified with WT ubiquitin (lanes 1-12) or I44A ubiquitin (lanes 13-24). Discharge rates were measured at pH 7.0 (lanes 1-6 and 13-18) and pH 9.2 (lanes 7-12 and 19-24). Quantification of the gel is shown in Figure S5. *For unknown reasons, thioesterified Cdc34 resolved as two bands in this set of experiments.
(C) Estimates of reaction rates from Figure 6B.
Hydroxylamine differs from the other acceptors employed in this study in that it has a lower molecular weight and the pKa of the primary amine is far lower (6.0 vs. 10.5 for the ε-amino group of lysine). The low pKa of hydroxylamine should reduce dependence on catalytic elements that mediate deprotonation of the attacking lysine. To determine whether the low pKa of hydroxylamine might account, at least in part, for the suppression of the I44A mutation, we performed diubiquitin synthesis assays at pH 7.0 and 9.2. The idea behind this experiment is that the fraction of lysine that is protonated is ~100-fold less at pH 9.2 compared to pH 7, therefore a ubiquitination reaction carried out at the high pH should be less dependent on catalytic elements that mediate lysine deprotonation. To eliminate confounding effects on ubiquitin activation, this experiment was carried out under single-turnover discharge conditions similar to Figure 2B. Reactions carried out at higher pH narrowed the gap between reaction rates measured for WT and I44A ubiquitin (Figures 6B, C). The rate of diubiquitin synthesis with wild type donor was 12-fold faster than mutant at pH 7 but only 5-fold faster at pH 9.2 (Figure 6C, S5). Meanwhile, discharge of WT ubiquitin from Cdc34 was 6-fold faster than I44A at pH 7 and only 2-fold faster at pH 9.2. The better activity of I44A in the discharge reaction at higher pH was partly attributable to a higher fraction of the mutant thioester being hydrolyzed to regenerate free ubiquitin, at the expense of forming diubiquitin.
Discussion
Models of ubiquitination have typically assumed that the donor ubiquitin thioesterified to the E2 enzyme is a passive participant in the transfer reaction. We provide strong evidence that the isoleucine 44 residue of ubiquitin, which has been implicated in interactions with numerous ubiquitin-binding proteins, is also critical for transfer of donor ubiquitin from Cdc34~Ub thioester to substrate. Isoleucine 44 on the donor ubiquitin is important for transfer at both the initiation and elongation steps of ubiquitin chain assembly upon SCF substrates. Isoleucine 44 is also important for discharge of ubiquitin from Cdc34~Ub to free ubiquitin in the presence or absence of SCF. Finally, I44 is also important for transfer of ubiquitin from UbcH5~Ub to substrate.
The simplest model to explain our results is that the ubiquitin thioesterified to an E2 engages in interactions with the surface of the E2 that favor attack of the thioester bond by a nucleophilic acceptor. This hypothesis is consistent with structural models of Ubc1~Ub and UbcH8~Ub complexes that have been developed based on NMR data (Hamilton et al., 2001; Serniwka and Shaw, 2009). In each case, the thioesterified ubiquitin engages in interactions with surface residues on the E2, but the functional significance of these interactions was not explicitly demonstrated.
To probe the interaction between Cdc34 and its thioesterified ubiquitin cargo, we pursued two lines of experimentation. In the first test, we used homology modeling based on the Ubc1~Ub structure to generate charge-swap mutations in residues of Cdc34 (E133) and ubiquitin (R42) that were predicted to be in close proximity. In agreement with the homology model, an R42E mutation interferes with discharge of ubiquitin from wild type Cdc34, but this mutation is efficiently suppressed by compensatory introduction of an E133R mutation in Cdc34. Remarkably, when assayed in combination the two mutants exhibited nearly as much activity as was observed in reactions with wild type components. In the second test of our hypothesis, we used the information gathered in the first experiment as a constraint to model a Cdc34~Ub complex using Rosetta3 (Leaver-Fay et al., 2011). This model led to the prediction that an S129L mutation in Cdc34 should generate a steric clash with I44 of ubiquitin. Indeed, Cdc34-S129L showed reduced activity when assayed with wild type ubiquitin, but the Cdc34-S129L and ubiquitin-I44A mutations mutually suppressed each other. The simplest interpretation of our results is that the hydrophobic patch of ubiquitin comprising I44 binds in the vicinity of S129 on the surface of the Cdc34 to which it is thioesterified.
How does interaction between ubiquitin and the E2 to which it is thioesterified enhance its discharge? Formation of an isopeptide bond involves attack of a nucleophile at the thioester linkage to form an oxyanion transition state, which then resolves to yield product. Given that the pKa of the ε-amino group of lysine is ~10.5, at physiological pH the vast majority of lysine side chains are protonated (NH3+) and thus incompetent to engage in nucleophilic attack. With the exception of Ubc9 (Yunus and Lima, 2006), the mechanism underlying nucleophilic activation of lysine is poorly understood. We found that either using a low pKa acceptor (hydroxylamine; pKa ~6.0) or conducting reactions at pH 9.2 considerably improved the ability of ubiquitin–I44A to serve as a donor, suggesting that Cdc34~ubiquitin–I44A complexes are deficient in nucleophilic activation. Presumably, docking of donor ubiquitin to Cdc34 influences the positioning of S138, which has been proposed to play a critical role in nucleophilic activation (Yunus and Lima, 2006). Interestingly, the ubiquitin–I44A thioester exhibited appreciable hydrolysis in single-turnover diubiquitin synthesis assays. We suggest that interaction between the donor ubiquitin and Cdc34 helps to organize the active site to favor efficient formation of isopeptide bonds at the expense of other side reactions such as hydrolysis. A deeper mechanistic understanding of how ubiquitin-Cdc34 interaction organizes the active site to facilitate nucleophilic activation and isopeptide bond formation awaits the crystal structure of an intermediate in the transfer process.
Results in other systems support the concept that an active role for the donor ubiquitin in the transfer process is a conserved feature of ubiquitination pathways. As noted above, the NMR-based structural models for Ubc1~Ub and UbcH8~Ub point to a cis interaction between the proteins. In addition, the crystal structure of an UbcH5b~Ub complex with the HECT domain E3 Nedd4L revealed that the thioesterified ubiquitin engages in interactions with Nedd4L (Kamadurai et al., 2009). Finally, while the revised version of this manuscript was under review, (Wickliffe et al., 2011) reported that association of donor ubiquitin with Ube2S is important for its transfer to substrate. Using NMR chemical shift data, these authors derived a computational model for the structure of Ube2S~Ub. The authors used this model as a guide to generate mutations in Cdc34 (T122E, L125A, I128E) that disrupted the discharge of donor ubiquitin. These mutations map to the same helix as the S129L substitution described in this work (Figure S7), suggesting that the donor ubiquitin–E2 interaction described here is a conserved feature of E2 function. We propose that interaction of the hydrophobic patch of ubiquitin with the surface of E2 can serve at least two different purposes in addition to promoting ubiquitin discharge. First, it helps to maintain ubiquitin-like protein (UBL) specificity by requiring an interaction between UBL and ubiquitination enzyme at multiple steps in the transfer cascade. Second, it shields the hydrophobic patch from interacting with the many proteins in eukaryotic cells that recognize this surface. These results have important implications for attempts to reengineer or modulate the activity of ubiquitination pathways with mutants or small molecules.
Experimental procedures
Expression and purification of recombinant proteins
All proteins were recombinantly expressed in either E. coli or Hi5 insect cells and purified using standard procedures as outlined in Table S1.
Ubiquitination assay
All β-catenin substrate ubiquitination reactions (20 μl) were performed as described previously at 23°C in a buffer containing 30 mM Tris-Cl (pH 7.3), 100 mM NaCl, 5 mM MgCl2, 2 mM ATP, 2 mM DTT and typically contained 1 μM E1, 10 μM E2, E3 (as indicated), and 30 μM ubiquitin (Saha and Deshaies, 2008). Reactions were initiated by mixing E2~Ub to the preformed E3–Substrate complex. All reactions were quenched in reducing SDS sample buffer, resolved by SDS-PAGE, phosphor imaged, and quantified using Image Quant (G. E. Healthcare).
Diubiquitin synthesis reactions (20 μl) were performed as described previously at 23°C in a buffer containing 30 mM Tris-Cl (pH 7.3), 100 mM NaCl, 5 mM MgCl2, 2 mM ATP, 2 mM DTT (Saha and Deshaies, 2008). Typically 10 μM Cdc34 was pre-incubated with equimolar 32P-labeled ubiquitin in the presence of 1 μM E1 and ATP for 10 min at 23°C, followed by 5 min incubation with 2U apyrase (Sigma). Discharge of Cdc34~Ub was initiated by adding 120 μM ubiquitin-D77 (E3-independent) or 75 μM ubiquitin-D77 (Cul1–Rbx1 dependent, 300 nM). The addition of aspartate 77 following glycine 76 of ubiquitin ensures that it can only act as an acceptor ubiquitin in a steady-state diubiquitin synthesis assays (Petroski and Deshaies, 2005). For reactions in Figure 6 at pH 7.0 and 9.2, charging was done in a buffer containing 200 μM ATP, in the presence of 2.5 μM E1 and 15 mM 32P-labeled ubiquitin. Charging reactions were quenched by incubating with 2U apyrase for 5 min, followed by addition of 5 mM EDTA (final) (Figure S6). 5 μl of the thioester reaction was diluted in 20 μl of chase buffer with 50 mM bis-Tris-propane and 240 μM ubiquitin-D77 (final). Aliquots were removed at the indicated times and quenched with non-reducing sample buffer supplemented with 5 mM NEM. Samples were resolved by SDS-PAGE, analyzed by autoradiography and quantified using Image Quant (G. E. Healthcare). Rates of thioester discharge was fitted to a single exponential decay (y = (m1-m3)*exp(−m2*m0) + m3) using KaleidaGraph software. Rates were estimated for each replicates and averaged. All values reported are the average of at least two independent experiments.
Rosetta docking model
To model the Cdc34–ubiquitin interaction, a protocol was written as a part of the Rosetta3 suite (Leaver-Fay et al., 2011). It was designed to search the conformational space available to ubiquitin chemically conjugated via thioester to Cdc34 by sampling rotation about torsion angles near the thioester bond. For input structure, ubiquitin (1UBQ) and Cdc34 (2OB4) were used. To bias sampling towards conformations relevant to the experimental data, constraints were introduced and the top 20 hits based on the score was analyzed, which strongly suggested interaction between ubiquitin I44 and Cdc34 S129. Detailed procedure is outlined in the supplemental section.
Supplementary Material
01
Acknowledgement
We thank P. Douglas Renfrew for assisting with generation of thioester torsion parameters. We also thank Michael Rape for generously sharing data prior to publication. AS was supported in part by a senior postdoctoral fellowship from the Leukemia and Lymphoma Society. RJD is an HHMI Investigator and this work was supported in part by HHMI and NIH grant GM065997.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beal RE, Toscano-Cantaffa D, Young P, Rechsteiner M, Pickart CM. The Hydrophobic Effect Contributes to Polyubiquitin Chain Recognition. Biochemistry. 1998;37:2925–2934. doi: 10.1021/bi972514p. [DOI] [PubMed] [Google Scholar]
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Molecular cell. 2006;21:873–880. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nature reviews. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- Das R, Baker D. Macromolecular modeling with rosetta. Annual review of biochemistry. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annual review of biochemistry. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Dikic I, Wakatsuki S, Walters KJ. Ubiquitin-binding domains - from structures to functions. Nature reviews. 2009;10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye BT, Schulman BA. Structural Mechanisms Underlying Posttranslational Modification by Ubiquitin-Like Proteins. Annual Review of Biophysics and Biomolecular Structure. 2007;36:131–150. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- Hamilton KS, Ellison MJ, Barber KR, Williams RS, Huzil JT, McKenna S, Ptak C, Glover M, Shaw GS. Structure of a conjugating enzyme-ubiquitin thiolester intermediate reveals a novel role for the ubiquitin tail. Structure. 2001;9:897–904. doi: 10.1016/s0969-2126(01)00657-8. [DOI] [PubMed] [Google Scholar]
- Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Molecular cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nature reviews. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- Kamadurai HB, Souphron J, Scott DC, Duda DM, Miller DJ, Stringer D, Piper RC, Schulman BA. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Molecular cell. 2009;36:1095–1102. doi: 10.1016/j.molcel.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Hao B, Mohl DA, Deshaies RJ. The acidic tail of the Cdc34 ubiquitin-conjugating enzyme functions in both binding to and catalysis with ubiquitin ligase SCFCdc4. The Journal of biological chemistry. 2009a;284:36012–36023. doi: 10.1074/jbc.M109.058529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009b;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods in enzymology. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto ML, Wickliffe KE, Dong KC, Yu C, Bosanac I, Bustos D, Phu L, Kirkpatrick DS, Hymowitz SG, Rape M, et al. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Molecular cell. 2010;39:477–484. doi: 10.1016/j.molcel.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annual review of biochemistry. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Molecular cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata E, Satoh T, Yamamoto S, Yamaguchi Y, Yagi-Utsumi M, Kurimoto E, Tanaka K, Wakatsuki S, Kato K. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure. 2010;18:138–147. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Serniwka SA, Shaw GS. The structure of the UbcH8-ubiquitin complex shows a unique ubiquitin interaction site. Biochemistry. 2009;48:12169–12179. doi: 10.1021/bi901686j. [DOI] [PubMed] [Google Scholar]
- Song L, Rape M. Regulated degradation of spindle assembly factors by the anaphase-promoting complex. Molecular cell. 2010;38:369–382. doi: 10.1016/j.molcel.2010.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. The EMBO journal. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems AR, Schwab M, Tyers M. A hitchhiker’s guide to the cullin ubiquitin ligases: SCF and its kin. Biochimica et biophysica acta. 2004;1695:133–170. doi: 10.1016/j.bbamcr.2004.09.027. [DOI] [PubMed] [Google Scholar]
- Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Molecular cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01