RNA polymerase II kinetics in polo polyadenylation signal selection (original) (raw)
Abstract
Regulated alternative polyadenylation is an important feature of gene expression, but how gene transcription rate affects this process remains to be investigated. polo is a cell-cycle gene that uses two poly(A) signals in the 3′ untranslated region (UTR) to produce alternative messenger RNAs that differ in their 3′UTR length. Using a mutant Drosophila strain that has a lower transcriptional elongation rate, we show that transcription kinetics can determine alternative poly(A) site selection. The physiological consequences of incorrect polo poly(A) site choice are of vital importance; transgenic flies lacking the distal poly(A) signal cannot produce the longer transcript and die at the pupa stage due to a failure in the proliferation of the precursor cells of the abdomen, the histoblasts. This is due to the low translation efficiency of the shorter transcript produced by proximal poly(A) site usage. Our results show that correct polo poly(A) site selection functions to provide the correct levels of protein expression necessary for histoblast proliferation, and that the kinetics of RNA polymerase II have an important role in the mechanism of alternative polyadenylation.
Keywords: alternative polyadenylation, cell cycle, RNA polymerase II kinetics
Introduction
Transcription by RNA polymerase II (Pol II) is a complex and multistep process that consists of promoter binding, initiation, elongation and termination. For most eukaryotic genes, the nascent transcript is recognized by RNA processing enzymes, as it is being synthesized by Pol II. For instance, excision of non-coding introns to generate faithfully spliced messenger RNA (mRNA) occurs long before the polymerase has completed transcription of the whole gene (Moore and Proudfoot, 2009). Similarly, the nascent transcript is released from the transcribing polymerase by the action of a large processing protein complex (Shi et al, 2009) coupled to the addition of a poly(A) (pA) tail to the terminal 3′ end of the pre-mRNA and to transcriptional termination (for reviews, see Hirose and Manley (2000), Proudfoot (2004), Bentley (2005), Buratowski (2005) and Richard and Manley (2009)).
Although it was previously believed that the critical step for regulating transcription was at the stage of promoter binding and transcription initiation, recent advances have revealed that many genes are regulated at the phase that follows the promoter-paused Pol II entering into elongation (Core et al, 2008; Gilmour, 2009). At this stage, the speed at which Pol II transcribes a transcriptional unit may have an impact on the regulation of gene expression and may determine pre-mRNA processing (Caceres and Kornblihtt, 2002). In Drosophila, the C4 mutation in the RpII215 gene, which encodes the largest subunit of Pol II, changes amino acid 741 from Arg to His (Chen et al, 1993). In vivo this mutation induces Pol II α-amanitin resistance with a 50% slower transcription elongation rate than its wild-type counterpart (Coulter and Greenleaf, 1985; Chen et al, 1993) and reads less efficiently through DNA pause elements (Chen et al, 1996). In human cells, a mutation equivalent to C4 has been shown to affect the alternative splicing pattern of the human fibronectin (FN) exon 33 (E33) EDI. Here, the ‘slow’ Pol II acts in concert with splicing factors to induce the inclusion of the alternative E33 (de la Mata et al, 2003, 2010). Additionally, in Drosophila C4 embryos resplicing of the Hox gene Ultrabithorax is stimulated (de la Mata et al, 2003, 2010). Thus, a ‘kinetic coupling’ model in which the transcription elongation rate determines the outcome of the alternative splice site selection was put forward (Caceres and Kornblihtt, 2002).
The 3′ end processing complex is precisely regulated and specifically recognizes sequences in the nascent transcript that comprise the pA signal of the gene, which is typically AAUAAA, and a GU- or U-rich downstream RNA sequence element (for reviews, see Colgan and Manley (1997), Minvielle-Sebastia and Keller (1999), Wahle and Ruegsegger (1999), Zhao et al (1999), Edmonds (2002) and Shi et al (2009)). Additional sequences positioned either upstream (Carswell and Alwine, 1989; Gilmartin et al, 1992; Chen and Wilusz, 1998; Moreira et al, 1998; Hall-Pogar et al, 2005; Hu et al, 2005; Venkataraman et al, 2005; Danckwardt et al, 2007) or downstream of the pA signal (Bagga et al, 1995; Arhin et al, 2002; Dalziel et al, 2007) may act as auxiliary elements by binding to components of the 3′ end processing complex or to proteins that can influence the efficiency of this complex and consequently modulate the overall efficiency of mRNA 3′ end formation.
Poly(A) sites are thus integral parts of the gene, signalling where in the precursor RNA cleavage and polyadenylation should occur. However, a large percentage of genes possess multiple pA sites that can be alternatively utilized. It is estimated that over half of human genes contain more than one pA signal and that alternative polyadenylation may be a major player in gene regulation (Lutz, 2008; Neilson and Sandberg, 2010; Lutz and Moreira, 2011). In some cases the choice of alternative pA sites is coupled with alternative splicing, which results in the production of different transcripts that are translated into different protein products with diverse cellular functions, for example, the membrane or secreted forms of immunoglobulin in B cells (Peterson and Perry, 1986; Lou et al, 1996; Takagaki et al, 1996; Edwalds-Gilbert et al, 1997; Peterson, 2007). In contrast, the presence of alternative pA sites in the 3′-most exon (tandem pA signals) directs the synthesis of mRNAs that share the same coding sequence but have different 3′ untranslated regions (UTRs). This is usually coupled with the presence of regulatory elements in the 3′UTR that interact with RNA binding proteins or miRNAs, modulating mRNA stability, translation, mRNA transport and/or mRNA localization (Lewis et al, 1995; Huang and Carmichael, 1996; Jacobson and Peltz, 1996; Wickens et al, 1997; Preiss et al, 1998; Ghosh et al, 2008; Merritt et al, 2008; Newnham et al, 2010). Importantly, two recent genome-wide studies, in tumour cells and in activated versus resting T cells, have found that in general shorter 3′UTRs are indicative of enhanced cell proliferation (Sandberg et al, 2008; Mayr and Bartel, 2009). The shortening of the 3′UTRs resulting from alternative polyadenylation leads to the loss of regulatory elements in this region, such as miRNA target sites. Conversely, during mouse embryonic development, alternative polyadenylation was shown to be responsible for a progressive lengthening of mRNA 3′UTR (Ji and Tian, 2009). It was also recently shown that differences in the 3′UTR due in part to alternative polyadenylation, define molecular signatures that can distinguish tumour subtypes (Singh et al, 2009). In Drosophila the enhancer of rudimentary (e(r)) (Gawande et al, 2006) and the suppressor of forked (su(f)) are two examples of genes regulated by alternative polyadenylation (Audibert and Simonelig, 1998). e(r) produces two mRNAs due to two pA signals present in the 3′-most exon, but only the longer e(r) mRNA is specifically expressed in the female germline, which requires co-expression of the female-specific Sex-lethal (Wojcik et al, 1994; Gawande et al, 2006). su(f) contains three pA signals, the first one within intron 4, leading to the production of a truncated transcript, and the other two in the 3′UTR. It was suggested that the Su(f) protein regulates its own accumulation via a negative feedback loop that stimulates 3′ end formation of the truncated su(f) RNA (Audibert and Simonelig, 1998).
Polo was the first described member of an important family of cell-cycle kinases, the Polo-like kinases (Plks), and is involved in many critical steps in the cell cycle, including mitotic entry, centrosome organization, spindle formation, chromosome segregation and cytokinesis (Sunkel and Glover, 1988; Llamazares et al, 1991; Moutinho-Santos et al, 1999; Glover, 2005). polo contains a proximal and a distal pA signal in the 3′UTR (referred herein as pA1 and pA2, respectively). Despite evidence that pA site selection in the 3′ most exon of eukaryotic genes has a role in gene expression, the physiological impact of this mechanism on the development of an organism and the role of Pol II kinetics on alternative pA site choice is still poorly understood.
In this study, we show that in RpII215 mutant flies, which display reduced Pol II elongation kinetics, Pol II occupancy along polo is altered and polo pA1 is used 3.5-fold more efficiently than in wild-type flies. An increase in proximal pA site usage was also observed for five other alternatively polyadenylated transcripts in Drosophila. Moreover, we show that in flies carrying a deletion of the polo pA2 signal, Polo is translated at low levels and as a result precursor cells of the abdominal epidermis do not proliferate during metamorphosis. This causes a defect in abdomen development and lethality. Consequently, we demonstrate that in transgenic flies polo pA2 signal is necessary for proper protein levels production, cell proliferation and viability in the living organism. Importantly, Pol II kinetics has a critical role in the selection of alternative pA sites in Drosophila.
Results
Pol II kinetics affects polo pA site usage
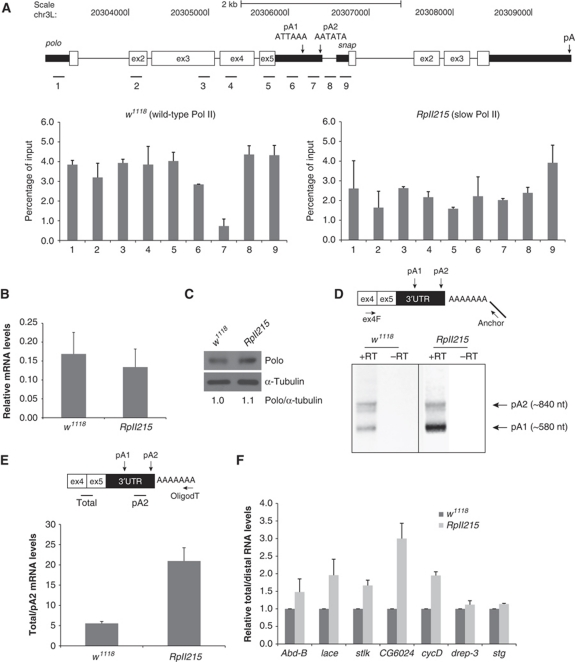
The polo gene, represented in Figure 1A, produces two mRNA species as previously described (Llamazares et al, 1991). The 3′ end of these two mRNAs was mapped by sequencing cDNAs isolated from a Poly(A)+ 0–24 h embryonic mRNA library, and they were shown to differ in their 3′UTR due to alternative usage of two pA signals localized in this region (Llamazares et al, 1991). The pA1 signal is the fly variant ATTAAA present in 10–32% of the Drosophila genes analysed, while the pA2 signal is the variant AATATA present in 5–10% of the Drosophila genes (Figure 1A; Graber et al, 1999; Retelska et al, 2006).
Figure 1.
RNA Pol II kinetics with a slower transcription elongation rate affects pA site selection. (A) Pol II ChIP across polo and polo-snap intergenic region. In the top panel, a schematic diagram is shown where polo and snap genes are depicted to scale (adapted from genome.ucsc.edu). ChIP was performed using α-Rpb3 antibody on wild-type (w 1118) and slow Pol II (RpII215 mutant, C4 mutation) adult flies chromatin. Numbers below each graph bar represent the position of real-time PCR primers. (B) RpII215 mutant generates similar levels of polo mRNA as compared with w 1118. Graph represents RT–qPCR quantification of polo mRNAs in adult flies, relative to rp49 mRNA. (C) RpII215 mutant presents similar levels of Polo protein as w 1118. Western blot from total protein extracts of w 1118 and RpII215 third instar larvae brains. MA294 Polo and DM1A α-tubulin antibodies were used. Quantification was made by densitometry (see Materials and methods) and Polo/α-tubulin ratio was set at 1 for w 1118. (D) Representative gel of fractionated 3′RACE products of polo mRNA from w 1118 and RpII215 adult flies is shown. Arrows indicate the bands corresponding to polo pA1 and pA2; −RT is a control reaction without reverse transcriptase. In both panels, phased-anchored oligodT was used for reverse transcription, as depicted in the diagram. (E) RpII215 shows an increase in the ratio of total/pA2 mRNAs. Diagram shows primer positions for qPCR analysis. Levels of total polo mRNAs and pA2 mRNAs were measured by real-time PCR and their ratio (total/pA2) in adult flies is shown. (F) RpII215 shows an increase in proximal site usage in several genes. RT–qPCR quantification was performed as in (E). cDNA synthesis was performed with random primers (see Materials and methods). The ratio for w 1118 was set at 1. For all the panels, error bars show s.e.m. from at least three independent experiments.
To investigate the role of Pol II kinetics on pA site usage of polo, we used the RpII215 Drosophila homozygous strain (referred herein as slow Pol II), which contains a C4 mutation in the largest subunit of Pol II reducing its transcription rate by 50% (Chen et al, 1996). We performed ChIP analysis on these flies, using an α-Rpb3 Pol II antibody to investigate Pol II distribution along the polo gene. Pol II occupancy was measured at nine positions within the polo transcription unit, the intergenic region and the promoter region of the next downstream gene, snap (Figure 1A), from chromatin sheared to an average size of 200–300 bp. Each position corresponds to a 125- to 150-bp amplicon separated by a minimum of 60 nucleotides (probes 7–8 and 8–9); the remaining probes are separated by >160 nucleotides. Quantification of the chromatin immunoprecipitation (ChIP) was done by real-time PCR using SybrGreen fluorescence (see Materials and methods). In wild-type flies, Pol II occupancy is reduced after pA1, suggesting that more pA1 than pA2 transcripts are being produced in adult flies (Figure 1A, left panel). This is in agreement with the quantification made in adult flies by real-time PCR (Figure 1E, w 1118) as well as with previously published results (Llamazares et al, 1991). The drop in Pol II occupancy that is seen after pA1 is indicative of polo termination, before it increases again over the closely spaced tandem gene snap (Figure 1A, left panel). However, in RpII215 flies this pattern is altered, with lower but more even Pol II levels across the polo-snap locus (Figure 1A, right panel), which may be due to the mutation in the RNA polymerase II. In particular, no drop in signal was observed following pA1 (probe 7). The lower levels of Pol II over polo-snap do not reflect a decrease in the levels of polo transcripts (Figure 1B) or Polo protein (Figure 1C) produced. We then determined whether this altered Pol II distribution affects polo pA site selection, by 3′RACE and RT–qPCR in RpII215 adult flies. 3′RACE analysis suggests that pA1 is used more efficiently than pA2 in RpII215 flies (Figure 1D). In order to quantify this effect, we measured the ratio between the total levels of polo transcripts (polo pA1+polo pA2) and polo pA2 mRNA levels by RT–qPCR. We observed that in RpII215 adult flies, this ratio is increased 3.5-fold in comparison with the wild-type flies (Figure 1E), indicating that pA1 is used more efficiently than pA2 in the RpII215 mutant. Our results clearly establish that in vivo the Drosophila slow Pol II affects polo pA site selection.
To extend this observation to other alternatively polyadenylated genes in Drosophila, we tested seven other previously described (GenBank, NCBI) or in silico predicted (Joel Graber, personal communication) genes, with a similar genomic structure to polo, that is containing two pA signals in the 3′UTR: abd-B (Abdominal-B, a member of the bithorax complex in Drosophila; Sanchez-Herrero et al, 1985), lace (a homologue of the LCB2 subunit of serine palmitoyltransferase; Adachi-Yamada et al, 1999), stlk (Ste20-like kinase), CG6024 (Artero et al, 2003), cycD (Cyclin D; Finley et al, 1996), drep-3 (DNA fragmentation factor-related protein 3; Inohara and Nunez, 1999) and stg (String, a Cdc5-like phosphatase; Edgar and O’Farrell, 1989). Using RT–qPCR with specific probes for these genes, we measured the ratio between total RNA levels and longer transcript levels (produced by distal pA signal selection) in RpII215 and w 1118 flies. Five out of the seven genes analysed showed an increase in total/distal RNA levels in RpII215 flies in comparison with the wild type similar to that found for polo (Figure 1F). CG6024 shows a three-fold increase and _Abd_-b, stlk and lace show a 1.5- to 2-fold increase. _Drep_-3 and stg show a slight increase in this ratio. These results clearly show that Pol II elongation kinetics affects alternative pA site selection in other genes and suggests that it may be a general effect in Drosophila.
polo pA2 is required for Drosophila viability and abdominal development
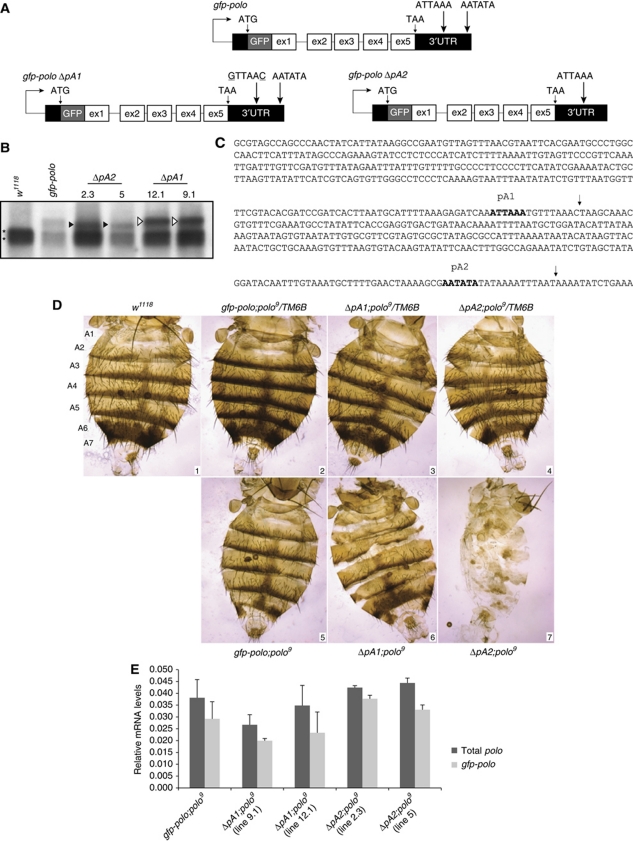
Having established that slow Pol II kinetics correlate with increased pA1 site usage, we then addressed the question of the function of the polo distal pA2 site in vivo. We therefore determined the consequence of mutation/deletion of each of the two tandem pA sites located in the polo 3′UTR (Figure 2C). Flies that express polo tagged with gfp at the N-terminus containing either one of the two pA signals were generated, using a previously described transgene gfp-polo (Moutinho-Santos et al, 1999). Point mutations were introduced in the pA1 signal (ATTAAA to GTTAAC), thus generating the transgene Δ_pA1_ that allows the expression of the longer polo transcript (Figure 2A). A second transgene, Δ_pA2_, was generated by deletion of pA2 signal and the entire downstream region (Figure 2A). The point mutations in pA1 and the deletion of pA2, inactivating pA1 and pA2, respectively, were therefore sufficient to allow the selective expression of only the longer or shorter polo mRNA in each transgenic line as shown by northern blot analysis in Figure 2B (open and black arrows, respectively). In this northern blot, polo pA1 and polo pA2 mRNAs are detected at similar levels in brains of third instar larvae in a wild-type background (Figure 2B) in contrast to what was observed in adult flies. This difference may reflect the nature of the developmental stage used and is in agreement with previous results (Llamazares et al, 1991). All transgenic lines express GFP–Polo protein throughout fly tissues (Supplementary Figure S1).
Figure 2.
Deletion of polo distal pA2 signal causes abnormal formation of the fly abdomen. (A) Schematic representation of the gfp-polo, Δ_pA1_ and Δ_pA2_ transgenes driven by the endogenous polo promoter. The two pA signals ATTAAA and ATATAA are indicated. In the Δ_pA1_ transgene, the proximal pA signal was mutated to GTTAAC. (B) Δ_pA1_ and Δ_pA2_ transgenes utilize pA2 and pA1 signals, respectively, as indicated by the arrows. Northern blot of w 1118, gfp-polo, Δ_pA2_ (lines 2.3 and 5) and Δ_pA1_ (lines 12.1 and 9.1) from third instar larvae brains total RNA. The asterisks indicate the two polo mRNAs present in w 1118, the black arrows the shorter transcripts present in Δ_pA2_ lines and the open arrows the longer transcripts present in Δ_pA1_ lines. All fly lines analysed contain the endogenous polo mRNAs that serve as internal controls (denoted by *). (C) polo 3′UTR sequence. Bold sequence indicates proximal (pA1) and distal (pA2) pA signals. The arrows indicate the cleavage site of the transcripts produced by usage of pA1 and pA2. (D) polo pA2 is required for abdominal development. Dorsal view of female abdomens from w 1118 control (panel 1); the seven tergites of the adult abdomen are indicated as A1–A7. Panels 2–4: gfp-polo, Δ_pA1_ (line 12.1) and Δ_pA2_ (line 2.3) individuals in a polo 9/TM6B background. Panels 5–7: gfp-polo, Δ_pA1_ (line 12.1) and Δ_pA2_ (line 2.3) individuals in a polo 9 background. (E) polo mRNA quantification by RT–qPCR in the transgenic fly lines in a polo 9 background. Total RNA was extracted from two adult flies of each genotype (for Δ_pA2_;polo 9, two escapers were used) in a polo 9 background, and from w 1118 flies. After cDNA synthesis with random hexamers, RNA levels were quantified using _gfp_-specific primers for amplification of the transgene-derived transcripts (light grey bars) or primers for exons 4 and 5 for amplification of the total polo RNAs present (dark grey bars). In all genotypes, rp49 was used for normalization. The difference between total polo (dark grey bars) and gfp-polo (light grey bars) corresponds to polo 9 contribution. Error bars show s.e.m. from at least three independent experiments.
To study the role of polo pA signals in vivo, the viability of homozygous lines for the transgenes was analysed in a polo 9 mutant background, which has been described as the strongest hypomorphic allele identified for polo (Donaldson et al, 2001). This mutant was generated by insertional mutagenesis of a P-element into the 5′UTR of polo, 176 nucleotides upstream of the ATG codon, where it interferes with polo transcription (Donaldson et al, 2001). polo 9 homozygous third instar larval brains have barely detectable levels of Polo protein that are not sufficient for the larvae to develop beyond this developmental stage, and consequently die (Donaldson et al, 2001). We first determined if the polo mRNA and protein levels contributed by the transgenes were sufficient to rescue the third instar larval lethal phenotype displayed by polo 9 individuals in the transgenic flies gfp-polo;polo 9, Δ_pA1;polo_ 9 and Δ_pA2;polo_ 9. As can be seen, the gfp-polo and Δ_pA1_ transgenes rescue polo 9 phenotype and the flies reach adulthood (Table I). In contrast, nearly all Δ_pA2;polo_ 9 individuals died during pupariation (Table I). These results clearly show that the longer polo mRNA produced using pA2 is required for normal Drosophila development beyond the late third instar larval stage, more precisely, during metamorphosis. Strikingly, analysis of the few Δ_pA2;polo_ 9 adult escapers indicated that these individuals have strong defects in abdominal morphology, with the complete absence or incorrect formation of tergites (Figure 2D, panel 7) while all other adult structures appear normal. Panels 2–4 show abdomens prepared from transgenic flies, in a polo 9 /TM6B background, containing the transgenes plus one copy of polo (TM6B). Panels 5–7 show transgenic flies in a polo 9 homozygous background. Transgenic gfp-polo;polo 9 adult flies have normal tergites as expected, while Δ_pA1;polo_ 9 present a mild abdominal phenotype (Figure 2D, panels 5 and 6). The possibility that the phenotype may arise as a result of the insertion of transgenes into the same genomic region in both transgenic lines was excluded (Supplementary Figure S2). In spite of the almost undetectable endogenous Polo in polo 9 (Donaldson et al, 2001), we quantified the remaining polo mRNA contribution by this allele, as well as the levels produced by each transgene, in all the transgenic polo 9 homozygous lines, by RT–qPCR (gfp-polo;polo 9, Δ_pA1;polo_ 9 and Δ_pA2;polo_ 9; Figure 2E). Using specific primers for the gfp transgene (light grey bars) or primers that amplify both endogenous polo and gfp_-polo mRNAs (dark grey bars), it is clear that the transgenes are principally responsible for polo mRNA levels (Figure 2E). The small difference between the bars corresponds to the endogenous polo contribution. Moreover, Δ_pA2;polo 9 flies show higher mRNA levels than Δ_pA1;polo_ 9 flies. This clearly indicates that the phenotype presented by Δ_pA2;polo_ 9 individuals (Figure 2D, panel 7) is due to the lack of polo pA2 transcript and that selection of the pA2 signal is essential for normal development of the abdomen and viability of the fly.
Table 1. Viability—deletion of polo distal pA2 causes high lethality levels.
| Strain | % Flies homozygous for polo9 carrying each of the transgenes |
|---|---|
| gfp-polo; polo 9 | 91 |
| Δ_pA1; polo_ 9 (line 9.1) | 100 |
| Δ_pA1; polo_ 9 (line 12.1) | 63 |
| Δ_pA2; polo_ 9 (line 2.3) | 0.1a |
| Δ_pA2; polo_ 9 (line 5) | 0.6a |
| Quantification of the viability of the transgenic lines Δ_pA1;polo_ 9, lines 9.1 and 12.1 and Δ_pA2;polo_ 9, lines 2.3 and 5 (presented as the percentage of flies that reached adulthood). For each strain, crosses were performed to rescue the polo 9 /polo 9 allelic combination. | |
| aMost individuals die as pharate pupae. |
pA2 is required for abdominal histoblast proliferation at metamorphosis onset and for correct polo protein expression
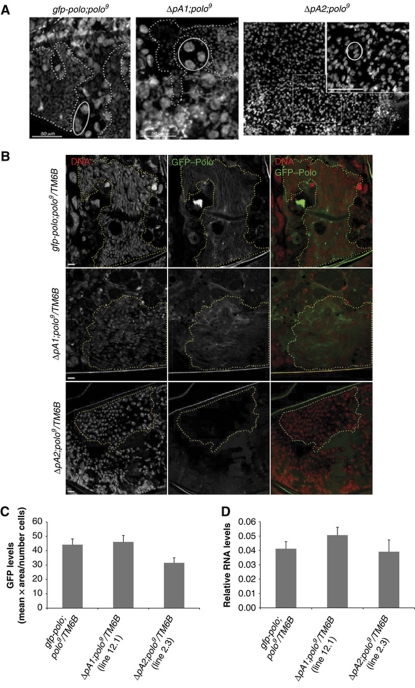
The high levels of lethality for Δ_pA2;polo_ 9 observed during the pupa stage suggested that polo pA2 was essential at this stage of development. Additionally, the phenotype presented by Δ_pA2;polo_ 9 is in agreement with that found in mutants for arrowhead (awh) and escargot (esg) genes, two other genes previously reported as necessary for the formation of the adult abdomen (Hayashi et al, 1993; Curtiss and Heilig, 1995). In esg mutants, the histoblasts—small diploid cells that arise during the last stages of embryogenesis (Roseland and Schneiderman, 1979; Madhavan and Madhavan, 1980)—undergo DNA endoreplication resulting in polyploid cells which fail to proliferate, leading to adult flies with an abdominal phenotype very similar to Δ_pA2_;polo 9 flies (Hayashi et al, 1993). Upon entering metamorphosis, the histoblasts undergo very rapid proliferation and from 15 h after pupa formation they expand, replacing all larval polytene epidermis cells (Ninov et al, 2007), starting to form a complete adult epidermis layer. Therefore, we analysed the histoblasts in pupae collected 26–27 h after pupa formation (Figure 3A). At this stage, histoblasts are already proliferating, replacing larval epidermis cells and forming over 50% of the adult abdomen epidermis (Roseland and Schneiderman, 1979). As expected, in gfp-polo;polo 9 and Δ_pA1;polo_ 9 pupae normal histoblast proliferation can be clearly seen: histoblasts are the small cells (delimited by the dotted line in Figure 3A). Larval epidermis cells (delimited by solid line in Figure 3A) are larger and easily distinguished from histoblasts. However, in Δ_pA2;polo_ 9 pupa epidermis only the large polytene larval cells are observed and histoblasts could not be detected in pupae at this stage (Figure 3A). Moreover, this phenotype is only observed in a polo 9 homozygous background (Supplementary Figure S3). In conclusion, our results show that in the absence of the polo pA2 transcripts, histoblasts are not detected during metamorphosis.
Figure 3.
Δ_pA2_ transgene causes a failure in abdominal histoblasts proliferation and produces lower Polo protein levels in histoblasts. (A) Pupa epidermis from gfp-polo, Δ_pA1_ or Δ_pA2_ pupae transgenes in a polo 9 homozygous background was dissected at 26–27 h after pupa formation and stained with DAPI. gfp-polo;polo 9 pupa epidermis exhibited large polyploid nuclei (circled), the larva epidermis cells (LECs). LECs were surrounded by smaller cells (highlighted by dotted lines), which are the proliferating abdominal histoblasts. Δ_pA1_;polo 9 pupa epidermis show a pattern very similar to that observed in gfp-polo;polo 9 tissues. In Δ_pA2;polo_ 9, only LECs are observed, indicating that at this stage, pupa epidermis is mainly composed of larval cells. Scale bar is 50 μm. (B) GFP–Polo expression (green) in abdominal histoblasts in pupae epidermis (26–27 h after pupa formation). Pupae from gfp-polo, Δ_pA1_ (line 12.1) and Δ_pA2_ (line 2.3) transgenic flies in a polo 9 /TM6B background were collected and the epidermis was dissected. The DNA (red) was stained with DAPI. The yellow dotted surrounding area encompasses some of the histoblast cells present in each image. Scale bar is 10 μm. (C) The levels of GFP–Polo were determined in the abdomen histoblasts, by quantification of the GFP fluorescence levels in gfp-polo, Δ_pA1_ (line 12.1) and Δ_pA2_ (line 2.3) pupae in a polo 9/TM6B background, 26–27 h after pupa formation. (D) The levels of gfp-polo transcripts were determined by RT–qPCR in gfp-polo, Δ_pA1_ (line 12.1) and Δ_pA2_ (line 2.3) whole pupae in a polo 9 /TM6B background, 26–27 h after pupa formation. The levels of gfp-polo RNAs were normalized to rp49 RNA. Error bars represent s.e.m. from three independent experiments.
The absence or incorrect formation of the tergites in Δ_pA2;polo_ 9 flies could result either from a failure in the specification/formation of abdominal histoblasts before the entrance to the pupal stage, or from the inability of these cells to proliferate during this developmental stage. To distinguish between these two possibilities, Δ_pA2;polo_ 9 third instar larvae were dissected and immunostained with anti-Headcase (Hdc) antibody, used as a marker for abdominal histoblasts, showing that histoblasts are clearly present in gfp-polo;polo 9, Δ_pA1;polo_ 9 and Δ_pA2;polo_ 9 larvae (Supplementary Figure S4A). Furthermore, all histoblast cells are strongly stained for Cyclin B, as expected from G2-arrested cells (Supplementary Figure S4A). Moreover, quantification of the number of histoblasts in each nest shows that these larvae have similar number of cells in each histoblast nest for the different abdominal segments (Supplementary Figure S4B). Taken together, these results indicate that Δ_pA2_ abdominal histoblasts are correctly formed during embryogenesis and are able to maintain the G2 arrest until the larvae stage. Consequently, the longer polo transcript derived from pA2 usage is essential for the proliferation of these cells at the onset of metamorphosis.
Having determined that deletion of polo pA2 was detrimental to the fly, we went on to investigate the expression of GFP–Polo obtained from the different transgenes in late pupal development, using pupae in a heterozygous background (polo 9/TM6B), to allow proliferation of the histoblasts. We observed a significant decrease of the GFP–Polo signal in histoblasts of Δ_pA2_ pupae (Figure 3B; histoblast cells are delimited by a dotted yellow line). Quantification of the fluorescent levels throughout the whole pupa epidermis, using the same individuals, showed that while gfp-polo and the Δ_pA1_ transgenes express similar levels of GFP–Polo a 30% decrease in fluorescence is observed for the Δ_pA2_ transgene (Figure 3C). To elucidate if this is due to a decrease in the mRNA produced, we quantified the mRNA by RT–qPCR. As can be seen in Figure 3D, there is no significant difference between the mRNA levels produced by each transgene; thus, the decrease in protein levels observed in Δ_pA2_ flies is unlikely to be due to a simple reduction in the mRNA levels. These results thus suggest that the appropriate levels of Polo protein in abdominal histoblasts may only be achieved when the distal pA2 site is used.
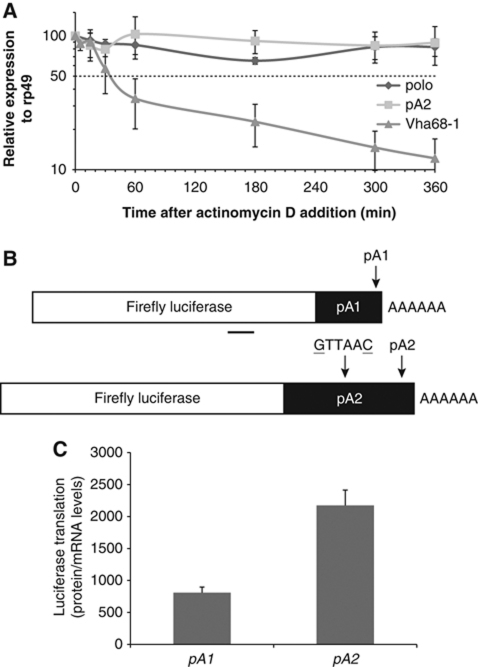
To identify the molecular mechanism responsible for the decrease in Polo protein when pA2 is deleted, we used a Drosophila cell line to assay for the stability of the endogenous polo mRNAs. Upon treatment with actinomycin D and measurement of RNA decay by RT–qPCR, we observe that both polo RNAs and polo pA2 RNA levels are maintained for at least 6 h (Figure 4A), suggesting that both transcripts are equally stable. As a control for the actinomycin D treatment, we used the _Vha68_-1 (vacuolar H+ATPase subunit 68-1) RNA which displays a half-life of ∼30 min, as previously shown (Behm-Ansmant et al, 2006). We then tested if the different 3′UTRs present in each transcript were responsible for the different levels of protein produced. The 3′UTR ending at pA1 was fused downstream of a Luciferase reporter gene to produce the pA1 plasmid (Figure 4B). In a construct containing the whole 3′UTR fused to luciferase, the pA1 signal was mutated (ATTAAA–GTTAAC) to produce plasmid pA2 (Figure 4B); the resultant plasmids, pA1 and pA2, were transfected into Kc cells. The mRNA 3′ends formed from each construct were confirmed by 3′RACE and to distinguish translational output from mRNA turnover, luciferase translational activity was normalized to _luciferase_-reporter mRNA levels to obtain the translation efficiency (defined in Supplementary Figure S5). Significantly, we found that pA1 causes a three-fold decrease in translation as compared with pA2 (Figure 4C). These results clearly indicate that the longer polo mRNA is more efficiently translated than the shorter transcript. This suggests that the correct levels of Polo are defined by pA2 signal selection, in agreement with the in vivo results obtained with the transgenic fly lines.
Figure 4.
polo pA2 RNA is stable and the corresponding 3′UTR confers higher translability to a reporter gene in Drosophila cells. (A) Endogenous polo pA2 RNA is stable. The decay of the endogenous polo RNAs and Vha68-1 RNA was monitored at the indicated time points after addition of actinomycin D (5 μg/ml). The levels of polo RNAs were normalized to rp49 RNA in three independent experiments and are plotted against time. RNA half-lives (_t_1/2), calculated from the decay curves, are longer than 6 h, except for Vha68-1 that is ∼30 min. (B) Representation of the firefly luciferase reporter constructs with the shorter polo 3′UTR (pA1) or the longer polo 3′UTR (pA2) (pA1 signal mutation: ATTAAA to GTTAAC). The region for specific qPCR primers is underlined. (C) pA1 produces three-fold less protein than pA2. Kc cells were transfected with the pA1 and pA2 reporters and 3 days later firefly luciferase activity and the corresponding mRNA levels were measured. Y axis represents luciferase values normalized to the mRNA levels. Error bars represent s.e.m. from at least three independent experiments.
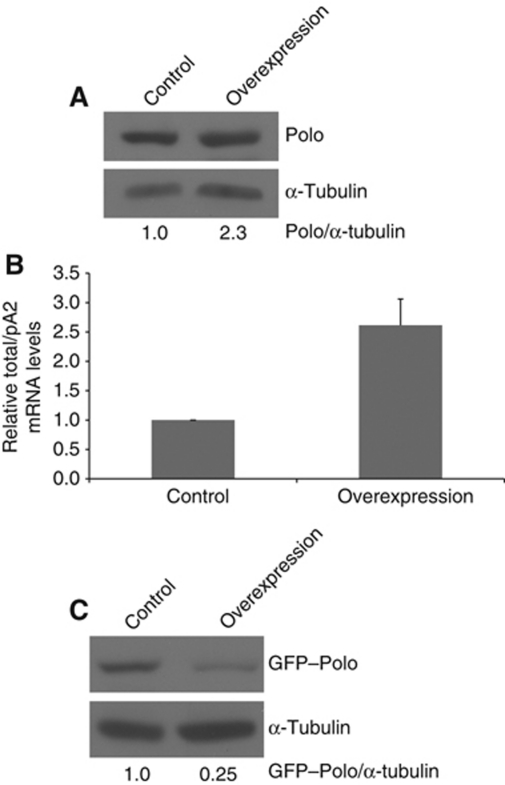
Finally, we asked if the levels of utilization of pA1 and pA2 could be regulated by in vivo overexpression of Polo using the previously established GAL4/UAS system (Brand and Perrimon, 1993). Using this system, it was previously shown that Polo is overexpressed by 2.5-fold which does not affect its physiological function in mitosis (Martins et al, 2009). Thus, using a neuroblast driver (MZ1061-Gal4) we tested the correlation between the expression of each transcript and protein levels in a tissue-specific manner. For this, MZ1061-Gal4; gfp-polo:polo 9 /TSTL females were crossed with MZ1061-Gal4;UAS-polo:polo 9 /TSTL males and the polo 9 homozygous brain progeny was analysed; in these flies, polo cDNA with the full 3′UTR is transcribed from an upstream inducible element (Mirouse et al, 2006). As shown in Figure 5A, Polo is overexpressed 2.3-fold relative to the wild-type control, levels that are in agreement with previous studies (Martins et al, 2009). Using RT–qPCR, we then measured the ratio of pA1 to pA2 mRNAs in the polo 9 homozygous F1 generation taking strain MZ1061;gfp-polo:polo 9 /TSTL as a control. Significantly, overexpression of Polo results in a 2.5-fold increase in the total/pA2 mRNA ratio (Figure 5B). We further determined the amount of GFP–Polo protein levels by western blot analysis and observed a four-fold decrease in GFP–Polo in the flies where polo was overexpressed (Figure 5C). Taken together, these results show that overexpression of polo induces a switch from pA2 to pA1 mRNA and a consequent decrease in the protein levels. These results suggest the existence of a potential auto-regulatory feedback loop mechanism.
Figure 5.
Overexpression of polo leads to an increase in pA1 utilization and a reduction in GFP–Polo protein levels. (A) MZ1061-Gal4;UAS-polo:polo 9 overexpresses Polo. L3 brains were analysed by western blot and a 2.3-fold increase in Polo was observed in comparison with the control—w 1118. Quantification was made by densitometry (see Materials and methods) and Polo/α-tubulin ratio was set at 1. Polo was detected with MA294 and α-tubulin with DM1A antibodies. (B) Overexpression of Polo causes a switch in the pAs ratio. Total RNA was extracted from L3 larvae brains of the MZ1061-Gal4;gfp-polo:polo 9/UAS-polo:polo 9 progeny (labelled as overexpression in the figure). The strain MZ1061-Gal4;gfp-polo:polo 9 was used as control. Total/pA2 mRNA levels were quantified by RT–qPCR with specific primers for both transcripts in the coding sequence and primers between the two pAs. A 2.5-fold change was detected in comparison with the control. (C) GFP–Polo protein expression was analysed by western blot, using the same conditions as in (A). When polo is overexpressed, a four-fold decrease in GFP–Polo is observed.
Recent studies have shown that in cancer cells alternative polyadenylation in general selects shorter 3′UTRs and consequently there is an increase of protein production (Mayr and Bartel, 2009). This was shown to be partly due to the presence of miRNA target sites in the extended 3′UTRs. We have searched in the available databases and prediction programs for miRNAs that could potentially target polo transcripts and found dme-mir-8 and dme-mir-1016 (Stark et al, 2007). The predicted miRNAs were quantified and tested in Kc cells. However, we could rule out an effect of dme-mir-8 and dme-mir-1016 overexpression on polo 3′UTR (Supplementary Figure S6).
Discussion
Expression levels of the essential cell-cycle regulatory gene polo are critical to Drosophila development. We show in these studies that Polo protein levels are regulated by the relative usage of two alternative pA sites (pA1 and pA2) present in the polo gene terminal exon. Unexpectedly, we show that pA2 is essential for fly viability whereas the pA1 site is non-essential. The molecular explanation for this specificity is that polo mRNA utilizing pA2 is translated more efficiently than polo pA1 mRNA thereby generating sufficient protein for cell proliferation. Thus, we show in these studies that deletion of pA2 displays a lethal phenotype (Table I; Figure 2D). Our studies further suggest that levels of Polo protein are auto-regulated by the selective usage of these two pA sites. Thus, polo overexpression results in a switch in pA site selection from pA2 to pA1 with consequent downregulation of Polo expression (Figure 5).
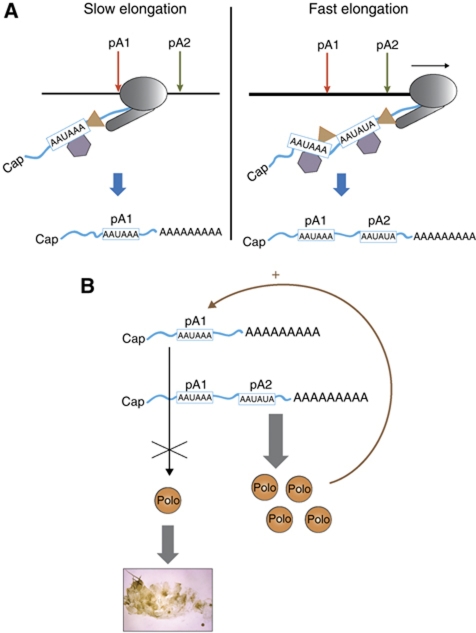
Since many genes in flies and other eukaryotes possess alternative pA sites in terminal exons, we investigated the possibility that general transcription kinetics might also have a role in alternative polyadenylation. To do this, we employed the well-characterized C4 fly mutant, which has a 50% decrease in Pol II elongation rate (Greenleaf et al, 1979). Interestingly, we show that this results in increased utilization of the polo pA1 site (Figure 1D and E). In particular, in C4 flies, polo pA1 is used 3.5-fold more efficiently than in the wild type. We further demonstrate that the effect of Pol II kinetics in alternative pA site selection is likely to be general as analysis of five additional Drosophila genes possessing alternative terminal exon pA sites showed a similar switch to upstream pA site usage in the C4 mutant. This clearly indicates that Pol II kinetics has an important role in alternative polyadenylation site selection. Significantly, human Pol II carrying the equivalent mutation to the Drosophila C4, when transfected into human cells, affects alternative splicing of a FN minigene. Inclusion of the alternative EDI exon in this system increases ∼4-fold (de la Mata et al, 2003), a similar level to that obtained for alternative polo pA signal selection (Figure 1E). C4 flies also show a difference in Pol II occupancy across the polo gene (Figure 1A). Thus, in wild-type flies Pol II levels are reduced downstream of pA1, while in C4 flies this pattern is disrupted. Presumably, the slow Pol II enables the pA1 signal on the nascent transcript to be exposed to the polyadenylation machinery for a longer time before Pol II transcribes pA2. Therefore, pA1 will be processed before pA2 is transcribed pointing to a mechanism that relies on the rule of ‘first come, first served’ (Figure 6A). When the Pol II elongation rate is higher, as in wild-type flies, it transcribes through pA1 and pA2 more efficiently, so that the polyadenylation machinery processes both pA signals on the nascent transcript (Figure 6A). This resembles the mechanism described above for EDI alternative splicing where slow Pol II preferentially includes the alternative EDI exon which is normally excluded, because it allows the machinery time to assemble on the spliceosome (de la Mata et al, 2003, 2010). Our results now indicate that both alternative polyadenylation and alternative splicing depend on Pol II kinetics. In view of recent findings that highlight the importance of alternative pA signal selection (Sandberg et al, 2008; Ji and Tian, 2009; Mayr and Bartel, 2009; Singh et al, 2009), our results now suggest a general molecular mechanism for this process.
Figure 6.
Proposed working model for the role of Pol II kinetics on pA site selection. (A) When Pol II elongation rate is low, Pol II spends more time transcribing polo allowing the pA factors (represented by a triangle and an hexagon) to assemble on the pA1 signal presented by the nascent pre-mRNA, leading to an increase in pA1 utilization, and therefore producing four-fold more polo pA1 mRNA. In conditions where Pol II processivity is high, Pol II transcribes more rapidly through pA1 and pA2. Under these conditions, both pA1 and pA2 signals are present on the nascent transcript and exposed to the polyadenylation machinery at the same time. Consequently, both polo pA1 and pA2 mRNAs are formed. (B) The longer polo mRNA, produced by pA2 signal selection and containing a longer 3′UTR, is more efficiently translated into protein than the shorter pA1 mRNA. Consequently, selection of pA2 is important for the viability of the fly. Levels of Polo protein are controlled by a negative feedback loop mechanism via pA site selection; in the presence of high levels of Polo, pA1 signal is predominantly selected, causing a decrease in the levels of Polo protein produced.
The physiological consequences of correct polo pA signal choice in vivo as shown in this study are profound: pA2 is essential for abdominal histoblast proliferation, development of the adult epidermis and viability of the transgenic flies. The lethality and strong abdominal phenotype observed in gfp-polo_Δ_pA2;polo 9 flies are due to the fact that these flies lack polo pA2 mRNA and consequently cannot express sufficient levels of Polo protein (from polo pA1 mRNA) for flies to survive the pupa stage. This phenotype is in agreement with earlier studies showing that polo 1 /polo 2 individuals express low levels of Polo protein (Herrmann et al, 1998) and present an abnormal development of the abdomen (Sunkel and Glover, 1988). We anticipated that the C4 mutant would display a similar phenotype to gfp-polo Δ_pA2_ flies since it downregulates pA2 usage. However, we observe a developmental defect similar to the so-called ‘Ubx effect’ (Greenleaf et al, 1980; Mortin et al, 1988). We note that slow Pol II elongation will impact on many genes so that any phenotype observed is likely to derive from complex genetic effects as previously reported (Greenleaf et al, 1980). We also note that polo pA2 mRNA is still produced at significant levels in the C4 mutant (Figure 1D and E), which may produce sufficient protein for development of the abdomen, as opposed to the Δ_pA2_ transgenic flies that lack polo pA2 mRNA (Figure 2B) and show a decrease in Polo protein production in the histoblasts (Figure 3B and C).
Two genes in humans (hap and Bzw1) where alternative distal pA site usage results in increased levels of protein production have been previously described (Qu et al, 2002; Yu et al, 2006). In T cells and cancer cell lines, proximal pA site selection in the 3′UTR results in a relief from microRNA repression with the same final effect of an increase in protein production (Sandberg et al, 2008). However, a considerable proportion of genes do not follow this pattern, pointing to the existence of other regulatory elements in the different 3′UTRs (Mayr and Bartel, 2009). Presumably, polo is one such gene as we show that its expression is not regulated by dme-mir-8 and dme-mir-1016 overexpression (Supplementary Figure S6). Instead regulated polo expression relies on the fact that the choice of pA1 by the transcriptional/processing machinery leads to a decrease in the translation of Polo (Figure 4). This indicates that Polo protein expression is modulated by pA signal selection and by translational control through the 3′UTR, suggesting the presence of regulatory elements in the different 3′UTRs (P Pinto and A Moreira, unpublished results). As Polo is a master regulator of the cell cycle, we predict that the consequences of this type of control will be critical to the cell.
Histoblasts have a very high proliferation rate and go through very rapid cell cycles at the onset of metamorphosis (Madhavan and Schneiderman, 1977; Roseland and Schneiderman, 1979; Madhavan and Madhavan, 1980; Ninov et al, 2007, 2009), therefore would be predicted to be especially sensitive to Polo levels. The lack of polo pA2 mRNA in Δ_pA2_ flies leads to a reduction in Polo protein levels with the consequent block of histoblast proliferation. This will in turn result in a subsequent failure to correctly develop the adult epidermis. Consistent with our studies, loss of Polo was previously shown to cause G2 arrest (Rehwinkel et al, 2005).
Interestingly, we also show that, in flies where Polo is overexpressed, the shorter mRNA produced by pA1 usage is more abundant than the longer mRNA produced by pA2 selection (Figure 5A and B). This suggests that Polo controls its own expression levels by an auto-regulatory loop, where higher levels of Polo lead to preferential recognition of pA1. As this mRNA is not efficiently translated (Figure 4C), this will lead to a decrease in the protein levels produced (Figure 6B). It was previously shown that upregulation of su(f) leads to an auto-regulatory feedback loop through the recognition of a weak intronic pA site (Audibert and Simonelig, 1998). This results in the production of a truncated polypeptide leading to a shut down in gene expression. In the case of polo, a more subtle process is evident, as pA1 selection generates a functional transcript, even though it is not efficiently translated into protein.
Taken together, our results suggest that Pol II kinetics have an important role in pA site selection. Moreover, our results reinforce the view that tight regulation at the level of pA signal selection is necessary for the cell. The importance of precisely choosing the correct pA signal to control cell viability during development is underlined by our studies.
Materials and methods
Plasmid constructs
Δ_pA1_ transgene: pXB7ΔpA1 was constructed by site directed mutagenesis (adapted from Makarova et al (2000)) using pXB7 (Moutinho-Santos et al, 1999) as a template. pXB7ΔpA1 was digested with _Bst_API and _Eco_RI and the 1576-bp fragment was subcloned into pCR-gfp-polo to generate the pCR-_gfp-polo_ΔpA1 plasmid. pCR-_gfp-polo_ΔpA1 was digested with Cci_NI and Xho_I and the 7717-bp DNA fragment was subcloned into pW8 to generate pW8-gfp-polo_ΔpA1 (named Δ_pA1). Δ_pA2 transgene: constructed by PCR amplification with the pshA1 genomic and not pA1 oligonucleotides. The PCR product was subcloned into pW8_-gfp-polo. For luciferase assays, polo 3′UTR (ΔpA1 and ΔpA2) was PCR amplified from the above constructs, subcloned into pCR®II-TOPO (Invitrogen) and subsequently subcloned into the _Eco_RI restriction site immediately downstream of the firefly coding region in the pAc5.1C-F-Luc plasmid. miRNA coding regions were PCR amplified from genomic DNA (∼400 bp surrounding the mature miRNA) and subcloned into pCR®II-TOPO (Invitrogen). miRNA reporters were subcloned into _Kpn_I and _Xho_I restriction sites of pAc5.1A vector. All constructs used in this study were fully sequenced.
Total RNA isolation and northern blot analysis
Total RNA was extracted with Trizol (Invitrogen) according to the manufacturer's protocol. Northern blot was performed as described (Sambrook and Russell, 2001). Quantifications were made by densitometry using a GS-800 densitometer (Bio-Rad).
Drosophila stocks
w 1118 and RpII215, carrying the RNA Pol II 215 C4 allele, were obtained from the Bloomington Stock Centre. UAS-polo and polo 9 /TM6C flies were kindly given by Jean-Louis Couderc (INSERM, France) and David Glover (University of Cambridge, UK), respectively. For the overexpression studies, MZ1061;gfp-polo:polo 9 /TSTL females were crossed with males MZ1061;UAS-polo:polo 9 /TSTL and the polo 9 homozygous progeny was analysed (Mirouse et al, 2006). The strain MZ1061;gfp-polo:polo 9 /TSTL was used as a control. All stocks were grown at 25°C using standard culture conditions and media.
Viability quantification
For the gfp-polo; polo 9/TM6B, Δ_pA1; polo_ 9/TM6B and Δ_pA2_; polo 9/TM6B crosses were performed to rescue the polo 9/polo 9 allelic combination. From these crosses, flies homozygous for polo 9 were quantified over the total number of flies.
Adult abdomen preparation
Flies were cut between the thorax and the abdomen and all appendages were removed. Abdomens were incubated in a lactic acid:ddH2O (3:1) solution overnight at 60°C, mounted in a fresh solution and incubated overnight at 60°C. The images were acquired with a Leica DM LB microscope (Leica Microsystems) with a 3 CCD color camera vision module (DONPISHA) and treated with the Adobe Photoshop 7.0 Software.
Pupa epidermis preparation
The dissection of 26–27 h pupae was performed according to Gompel and Carroll (2003). The epidermis was fixed in 1.85% (v/v) formaldehyde in fixation buffer (0.1 M PIPES pH 6.9, 1 mM EGTA pH 6.9, 1% (v/v) Triton X-100 and 1 mM MgCl2) for 10 min at RT. After fixation, the epidermis was washed twice for 5 min at RT in PBT (1 × PBS, 0.03% (v/v) Triton X-100) and mounted in 6 μl of VectaShield with DAPI (Vector Laboratories). The images were acquired with the AxioImager Z1 microscope (Carl Zeiss), with a b/w CCD camera Axiocam MR ver. 3.0 (Carl Zeiss) and treated using the Adobe Photoshop 7.0 Software. The number of histoblasts present in each nest represents the average number of cells in two pupae of each genotype.
Immunostaining of third instar larvae abdominal histoblasts
The protocol used was adapted from Weaver and White (1995) and Sullivan et al (2000). Third instar larvae were incubated in 1 × PBS at 55°C for 10 s, cut along the dorsal–ventral axis and all internal tissues were carefully removed. The cuticles were then fixed in 100 μl of 3.7% (v/v) formaldehyde in PBT (1 × PBS, 0.1% (v/v) Triton X-100) for 20 min at RT and washed three times in 100 μl of PBT. Blocking was performed for 1 h, at RT, in 200 μl of PBT with 10% (v/v) FBS. The cuticle was incubated ON at 4°C with 200 μl of α-Hdc antibody (1:5), kindly provided by Robert White (Weaver and White, 1995), and α-CycB antibody (Jacobs et al, 1998) (1:3000) in PBT with 10% (v/v) FBS. The following day, the cuticles were washed three times for 10 min in 200 μl of PBT and mounted in 20 μl of VectaShield with DAPI (Vector Laboratories). The images were acquired with an inverted Laser Scanning Confocal Microscope Leica SP2 AOBS SE (Leica Microsystems) and deconvolved and projected onto a single plan using the Huygens Deconvolution Software (SVI). The images were treated using the Adobe Photoshop 7.0 Software.
Quantification of GFP levels in 26–27 h after pupa formation pupae epidermis
The images were acquired using a Laser Scanning Confocal Microscope Leica SP2 AOBS SE (Leica Microsystems) and GFP fluorescence measured using the Leica Confocal Software, version 2.61 Build 1538 LCS Lite (Leica Microsystems). For each genotype, two pupae were analysed. For each sample, one plane was chosen and several GFP measurements were made throughout the epidermis. The GFP levels were quantified using the following formula: mean × area/number of cells, where the mean is the average level of GFP fluorescence measured for each sample.
cDNA synthesis and RT–qPCR
cDNA synthesis was performed using either oligodT or random hexamers with Superscript III following the manufacturer's instructions (Invitrogen). cDNA was then quantified in an iQ5 Bio-Rad real time PCR machine with iQ™ Sybr® Green Supermix. rp49 was used for normalization in all assays.
Western blot
Western blotting was performed as previously described (Moutinho-Santos et al, 1999). For Polo, MA294 antibody was diluted 1:80 and α-tubulin was detected using mAB DM1A (Sigma) diluted 1:8000. Quantifications were made by densitometry using a GS-800 densitometer (Bio-Rad).
mRNA stability assays
Kc cells were treated with actinomycin D (5 μg/ml), total RNA was isolated at designated time points and cDNA synthesis was performed using random hexamer primers with SuperScript III (Invitrogen) following the manufacturer's guidelines, followed by qPCR. mRNA half-life was calculated from the decay curves.
3′UTR and miRNA functional assays
Kc cells were transfected in 6-well plates using Effectene transfection reagent (Qiagen) with 100 ng of the Firefly luciferase reporter plasmid (F-Luc) containing the SV40 late polyadenylation signal (control) or the whole polo 3′UTR fused downstream to the Firefly luciferase reporter, 400 ng of the Renilla luciferase transfection control plasmid (R-Luc) and 500 ng of the plasmid (pAc5.1/V5-HisA, Invitrogen) expressing the miRNA primary transcripts. Three days after transfection, Firefly and Renilla activities were measured using a Dual-Luciferase reporter assay system (Promega). Total RNA was also isolated and northern blot (miRNA assay) or RT–qPCR (3′UTR assay) on oligodT synthesized cDNA was performed with Quantace Sensimix NoRef in a Rotor-Gene 6000 (Corbett LifeScience) using Firefly- and Renilla-specific primers as described above.
Chromatin immunoprecipitation
ChIP was performed as previously described (Schwartz et al, 2003). Each value for ChIP experiments was derived from IPs of at least three independent adult fly samples. Extracts were sonicated at 4°C on the medium-sonication setting of Bioruptor® (Diagenode) with 20 s bursts/30 s cool off, to achieve average fragment sizes of 200–300 bp. For the immunoprecipitations, 4 μl of rabbit anti-Rpb3 was used (Adelman et al, 2006). Primer sequences are available upon request.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information
Review Process File
Acknowledgments
We are grateful to John Lis (Cornell University, USA) for the α-Rpb3 Pol II antibody, David Glover (University of Cambridge, UK) for the polo 9 allele, Jean-Louis Couderc for the UAS-polo flies (INSERM, Clermont-Ferrand, France), Robert White (University of Cambridge, UK) for the Headcase antibody and Elisa Izaurralde (Tübingen, Germany) for the Luc plasmid. We are grateful to Joel Graber for sharing unpublished data, Fatima Gebauer, Martine Simonelig, Simon Lee and James Castelli-Gair for critical reading of the manuscript and to Fernando Casares, Enrique Martin-Blanco and Paulo Pereira for useful discussions. We thank J. Saraiva for technical support and all the members of the laboratories for their valuable suggestions on the work. The Wellcome Trust UK funds the NJP laboratory. AM and CES laboratories are funded by European Regional Development Fund (FEDER), Programa Operacional Ciência, Tecnologia e Inovação 2010 (POCI and POCTI 2010) and the Fundação para a Ciência e Tecnologia (FCT). PABP, TM, MOF and TH are recipients of studentships from FCT-POCTI. PAC is supported by a postdoctoral grant from FCT.
Author contributions: The overall study was conceived, designed and coordinated by AM, with important contributions from NJP, CES, PABP, TH, PAC and AMC. PABP, TH, MOF, PSW, TM and RD performed the experiments. AM, CES and NJP analysed the data. AM and PABP wrote the paper with substantial contribution from NJP.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adachi-Yamada T, Gotoh T, Sugimura I, Tateno M, Nishida Y, Onuki T, Date H (1999) De novo synthesis of sphingolipids is required for cell survival by down-regulating c-Jun N-terminal kinase in Drosophila imaginal discs. Mol Cell Biol 19: 7276–7286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Wei W, Ardehali MB, Werner J, Zhu B, Reinberg D, Lis JT (2006) Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol 26: 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arhin GK, Boots M, Bagga PS, Milcarek C, Wilusz J (2002) Downstream sequence elements with different affinities for the hnRNP H/H’ protein influence the processing efficiency of mammalian polyadenylation signals. Nucleic Acids Res 30: 1842–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artero R, Furlong EE, Beckett K, Scott MP, Baylies M (2003) Notch and Ras signaling pathway effector genes expressed in fusion competent and founder cells during Drosophila myogenesis. Development 130: 6257–6272 [DOI] [PubMed] [Google Scholar]
- Audibert A, Simonelig M (1998) Autoregulation at the level of mRNA 3′ end formation of the suppressor of forked gene of Drosophila melanogaster is conserved in Drosophila virilis. Proc Natl Acad Sci USA 95: 14302–14307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga PS, Ford LP, Chen F, Wilusz J (1995) The G-rich auxiliary downstream element has distinct sequence and position requirements and mediates efficient 3′ end pre-mRNA processing through a trans-acting factor. Nucleic Acids Res 23: 1625–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E (2006) mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev 20: 1885–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DL (2005) Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17: 251–256 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Buratowski S (2005) Connections between mRNA 3′ end processing and transcription termination. Curr Opin Cell Biol 17: 257–261 [DOI] [PubMed] [Google Scholar]
- Caceres JF, Kornblihtt AR (2002) Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet 18: 186–193 [DOI] [PubMed] [Google Scholar]
- Carswell S, Alwine JC (1989) Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol 9: 4248–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Wilusz J (1998) Auxiliary downstream elements are required for efficient polyadenylation of mammalian pre-mRNAs. Nucleic Acids Res 26: 2891–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chafin D, Price DH, Greenleaf AL (1996) Drosophila RNA polymerase II mutants that affect transcription elongation. J Biol Chem 271: 5993–5999 [PubMed] [Google Scholar]
- Chen Y, Weeks J, Mortin MA, Greenleaf AL (1993) Mapping mutations in genes encoding the two large subunits of Drosophila RNA polymerase II defines domains essential for basic transcription functions and for proper expression of developmental genes. Mol Cell Biol 13: 4214–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DF, Manley JL (1997) Mechanism and regulation of mRNA polyadenylation. Genes Dev 11: 2755–2766 [DOI] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322: 1845–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter DE, Greenleaf AL (1985) A mutation in the largest subunit of RNA polymerase II alters RNA chain elongation in vitro. J Biol Chem 260: 13190–13198 [PubMed] [Google Scholar]
- Curtiss J, Heilig JS (1995) Establishment of Drosophila imaginal precursor cells is controlled by the Arrowhead gene. Development 121: 3819–3828 [DOI] [PubMed] [Google Scholar]
- Dalziel M, Nunes NM, Furger A (2007) Two G-rich regulatory elements located adjacent to and 440 nucleotides downstream of the core poly(A) site of the intronless melanocortin receptor 1 gene are critical for efficient 3′ end processing. Mol Cell Biol 27: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danckwardt S, Kaufmann I, Gentzel M, Foerstner KU, Gantzert AS, Gehring NH, Neu-Yilik G, Bork P, Keller W, Wilm M, Hentze MW, Kulozik AE (2007) Splicing factors stimulate polyadenylation via USEs at non-canonical 3′ end formation signals. EMBO J 26: 2658–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR (2003) A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532 [DOI] [PubMed] [Google Scholar]
- de la Mata M, Lafaille C, Kornblihtt AR (2010) First come, first served revisited: factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA 16: 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson MM, Tavares AA, Ohkura H, Deak P, Glover DM (2001) Metaphase arrest with centromere separation in polo mutants of Drosophila. J Cell Biol 153: 663–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, O’Farrell PH (1989) Genetic control of cell division patterns in the Drosophila embryo. Cell 57: 177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmonds M (2002) A history of poly A sequences: from formation to factors to function. Prog Nucleic Acid Res Mol Biol 71: 285–389 [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Veraldi KL, Milcarek C (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res 25: 2547–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley RL Jr, Thomas BJ, Zipursky SL, Brent R (1996) Isolation of Drosophila cyclin D, a protein expressed in the morphogenetic furrow before entry into S phase. Proc Natl Acad Sci USA 93: 3011–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawande B, Robida MD, Rahn A, Singh R (2006) Drosophila Sex-lethal protein mediates polyadenylation switching in the female germline. EMBO J 25: 1263–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh T, Soni K, Scaria V, Halimani M, Bhattacharjee C, Pillai B (2008) MicroRNA-mediated up-regulation of an alternatively polyadenylated variant of the mouse. Nucleic Acids Res 36: 6318–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin GM, Fleming ES, Oetjen J (1992) Activation of HIV-1 pre-mRNA 3′ processing in vitro requires both an upstream element and TAR. EMBO J 11: 4419–4428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour DS (2009) Promoter proximal pausing on genes in metazoans. Chromosoma 118: 1–10 [DOI] [PubMed] [Google Scholar]
- Glover DM (2005) Polo kinase and progression through M phase in Drosophila: a perspective from the spindle poles. Oncogene 24: 230–237 [DOI] [PubMed] [Google Scholar]
- Gompel N, Carroll SB (2003) Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature 424: 931–935 [DOI] [PubMed] [Google Scholar]
- Graber JH, Cantor CR, Mohr SC, Smith TF (1999) In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci USA 96: 14055–14060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf AL, Borsett LM, Jiamachello PF, Coulter DE (1979) Alpha-amanitin-resistant D. melanogaster with an altered RNA polymerase II. Cell 18: 613–622 [DOI] [PubMed] [Google Scholar]
- Greenleaf AL, Weeks JR, Voelker RA, Ohnishi S, Dickson B (1980) Genetic and biochemical characterization of mutants at an RNA polymerase II locus in D. melanogaster. Cell 21: 785–792 [DOI] [PubMed] [Google Scholar]
- Hall-Pogar T, Zhang H, Tian B, Lutz CS (2005) Alternative polyadenylation of cyclooxygenase-2. Nucleic Acids Res 33: 2565–2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Hirose S, Metcalfe T, Shirras AD (1993) Control of imaginal cell development by the escargot gene of Drosophila. Development 118: 105–115 [DOI] [PubMed] [Google Scholar]
- Herrmann S, Amorim I, Sunkel CE (1998) The POLO kinase is required at multiple stages during spermatogenesis in Drosophila melanogaster. Chromosoma 107: 440–451 [DOI] [PubMed] [Google Scholar]
- Hirose Y, Manley JL (2000) RNA polymerase II and the integration of nuclear events. Genes Dev 14: 1415–1429 [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B (2005) Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Carmichael GG (1996) Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol 16: 1534–1542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inohara N, Nunez G (1999) Genes with homology to DFF/CIDEs found in Drosophila melanogaster. Cell Death Differ 6: 823–824 [DOI] [PubMed] [Google Scholar]
- Jacobs HW, Knoblich JA, Lehner CF (1998) Drosophila Cyclin B3 is required for female fertility and is dispensable for mitosis like Cyclin B. Genes Dev 12: 3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson A, Peltz SW (1996) Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem 65: 693–739 [DOI] [PubMed] [Google Scholar]
- Ji Z, Tian B (2009) Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS ONE 4: e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Gunderson SI, Mattaj IW (1995) The influence of 5′ and 3′ end structures on pre-mRNA metabolism. J Cell Sci Suppl 19: 13–19 [DOI] [PubMed] [Google Scholar]
- Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE (1991) polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev 5: 2153–2165 [DOI] [PubMed] [Google Scholar]
- Lou H, Gagel RF, Berget SM (1996) An intron enhancer recognized by splicing factors activates polyadenylation. Genes Dev 10: 208–219 [DOI] [PubMed] [Google Scholar]
- Lutz CS (2008) Alternative polyadenylation: a twist on mRNA 3′ end formation. ACS Chem Biol 3: 609–617 [DOI] [PubMed] [Google Scholar]
- Lutz CS, Moreira A (2011) Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip Rev RNA 2: 22–31 [DOI] [PubMed] [Google Scholar]
- Madhavan MM, Madhavan K (1980) Morphogenesis of the epidermis of adult abdomen of Drosophila. J Embryol Exp Morphol 60: 1–31 [PubMed] [Google Scholar]
- Madhavan MM, Schneiderman HA (1977) Histological analysis of the dynamics of growth of imaginal discs and histoblast nests during the larval development of Drosophila melanogaster. Wilhelm Roux's Arch Dev Biol 186: 269–305 [DOI] [PubMed] [Google Scholar]
- Makarova O, Kamberov E, Margolis B (2000) Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques 29: 970–972 [DOI] [PubMed] [Google Scholar]
- Martins T, Maia AF, Steffensen S, Sunkel CE (2009) Sgt1, a co-chaperone of Hsp90 stabilizes Polo and is required for centrosome organization. EMBO J 28: 234–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP (2009) Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt C, Rasoloson D, Ko D, Seydoux G (2008) 3′ UTRs are the primary regulators of gene expression in the C. elegans germline. Curr Biol 18: 1476–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minvielle-Sebastia L, Keller W (1999) mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol 11: 352–357 [DOI] [PubMed] [Google Scholar]
- Mirouse V, Formstecher E, Couderc J-L (2006) Interaction between Polo and BicD proteins links oocyte determination and meiosis control in Drosophila. Development 133: 4005–4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Proudfoot NJ (2009) Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136: 688–700 [DOI] [PubMed] [Google Scholar]
- Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley JL, Proudfoot NJ (1998) The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev 12: 2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortin MA, Kim WJ, Huang J (1988) Antagonistic interactions between alleles of the RpII215 locus in Drosophila melanogaster. Genetics 119: 863–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Santos T, Sampaio P, Amorim I, Costa M, Sunkel CE (1999) In vivo localisation of the mitotic POLO kinase shows a highly dynamic association with the mitotic apparatus during early embryogenesis in Drosophila. Biol Cell 91: 585–596 [PubMed] [Google Scholar]
- Neilson JR, Sandberg R (2010) Heterogeneity in mammalian RNA 3′ end formation. Exp Cell Res 316: 1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnham CM, Hall-Pogar T, Liang S, Wu J, Tian B, Hu J, Lutz CS (2010) Alternative polyadenylation of MeCP2: Influence of cis-acting elements and trans-acting factors. RNA Biol 7: 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninov N, Chiarelli DA, Martín-Blanco E (2007) Extrinsic and intrinsic mechanisms directing epithelial cell sheet replacement during Drosophila metamorphosis. Development 134: 367–379 [DOI] [PubMed] [Google Scholar]
- Ninov N, Manjon C, Martin-Blanco E (2009) Dynamic control of cell cycle and growth coupling by ecdysone, EGFR, and PI3K signaling in Drosophila histoblasts. PLoS Biol 7: e1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson ML (2007) Mechanisms controlling production of membrane and secreted immunoglobulin during B cell development. Immunol Res 37: 33–46 [DOI] [PubMed] [Google Scholar]
- Peterson ML, Perry RP (1986) Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci USA 83: 8883–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Muckenthaler M, Hentze MW (1998) Poly(A)-tail-promoted translation in yeast: implications for translational control. RNA 4: 1321–1331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N (2004) New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol 16: 272–278 [DOI] [PubMed] [Google Scholar]
- Qu X, Qi Y, Qi B (2002) Generation of multiple mRNA transcripts from the novel human apoptosis-inducing gene hap by alternative polyadenylation utilization and the translational activation function of 3′ untranslated region. Arch Biochem Biophys 400: 233–244 [DOI] [PubMed] [Google Scholar]
- Rehwinkel J, Letunic I, Raes J, Bork P, Izaurralde E (2005) Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 11: 1530–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F (2006) Similarities and differences of polyadenylation signals in human and fly. BMC Genomics 7: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Manley JL (2009) Transcription termination by nuclear RNA polymerases. Genes Dev 23: 1247–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DB (ed) (1986) Drosophila: A Practical Approach. Oxford, UK: IRL Press at Oxford University Press [Google Scholar]
- Roseland CR, Schneiderman HA (1979) Regulation and metamorphosis of the abdominal histoblasts of Drosophila melanogaster. Wilhelm Roux's Arch Dev Biol 186: 235–265 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D (eds) (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sanchez-Herrero E, Vernos I, Marco R, Morata G (1985) Genetic organization of Drosophila bithorax complex. Nature 313: 108–113 [DOI] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB (2008) Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz BE, Larochelle S, Suter B, Lis JT (2003) Cdk7 is required for full activation of Drosophila heat shock genes and RNA polymerase II phosphorylation in vivo. Mol Cell Biol 23: 6876–6886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Di Giammartino DC, Taylor D, Sarkeshik A, Rice WJ, Yates JR, Frank J, Manley JL (2009) Molecular architecture of the human pre-mRNA 3′ processing complex. Mol Cell 33: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH (2009) Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res 69: 9422–9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, Crosby MA, Rasmussen MD, Roy S, Deoras AN, Ruby JG, Brennecke J, curators HF, Project BDG, Hodges E, Hinrichs AS, Caspi A, Paten B, Park S-W, Han MV et al. (2007) Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature 450: 219–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W, Ashburner M, Hawley RS (eds) (2000) Drosophila Protocols. Cold Spring Harbor, New York, USA: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sunkel CE, Glover DM (1988) polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci 89 (Part 1): 25–38 [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Seipelt RL, Peterson ML, Manley JL (1996) The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87: 941–952 [DOI] [PubMed] [Google Scholar]
- Venkataraman K, Brown KM, Gilmartin GM (2005) Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev 19: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E, Ruegsegger U (1999) 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev 23: 277–295 [DOI] [PubMed] [Google Scholar]
- Weaver TA, White RA (1995) headcase, an imaginal specific gene required for adult morphogenesis in Drosophila melanogaster. Development 121: 4149–4160 [DOI] [PubMed] [Google Scholar]
- Wickens M, Anderson P, Jackson RJ (1997) Life and death in the cytoplasm: messages from the 3′ end. Curr Opin Genet Dev 7: 220–232 [DOI] [PubMed] [Google Scholar]
- Wojcik E, Murphy AM, Fares H, Dang-Vu K, Tsubota SI (1994) Enhancer of rudimentaryp1, e(r)p1, a highly conserved enhancer of the rudimentary gene. Genetics 138: 1163–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Sha H, Gao Y, Zeng H, Zhu M, Gao X (2006) Alternative 3′ UTR polyadenylation of Bzw1 transcripts display differential translation efficiency and tissue-specific expression. Biochem Biophys Res Commun 345: 479–485 [DOI] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C (1999) Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Information
Review Process File