Cross-linking, ligation, and sequencing of hybrids reveals RNA–RNA interactions in yeast (original) (raw)
Abstract
Many protein–protein and protein–nucleic acid interactions have been experimentally characterized, whereas RNA–RNA interactions have generally only been predicted computationally. Here, we describe a high-throughput method to identify intramolecular and intermolecular RNA–RNA interactions experimentally by cross-linking, ligation, and sequencing of hybrids (CLASH). As validation, we identified 39 known target sites for box C/D modification-guide small nucleolar RNAs (snoRNAs) on the yeast pre-rRNA. Novel snoRNA-rRNA hybrids were recovered between snR4-5S and U14-25S. These are supported by native electrophoresis and consistent with previously unexplained data. The U3 snoRNA was found to be associated with sequences close to the 3′ side of the central pseudoknot in 18S rRNA, supporting a role in formation of this structure. Applying CLASH to the yeast U2 spliceosomal snRNA led to a revised predicted secondary structure, featuring alternative folding of the 3′ domain and long-range contacts between the 3′ and 5′ domains. CLASH should allow transcriptome-wide analyses of RNA–RNA interactions in many organisms.
Keywords: pre-rRNA, RNA structure, ribosome synthesis, UV cross-linking
The identification of RNA–RNA interactions is essential for detailed understanding of many biological processes. Almost all RNAs must be correctly folded to function, whereas base-pairing between different RNA molecules underlies many pathways of RNA metabolism, including pre-mRNA splicing, ribosome synthesis, and the regulation of mRNA stability by microRNAs (miRNAs), among many others. Even for RNAs for which the final structure is known (e.g., rRNA), the folding pathway in precursors is generally unclear. RNA-RNA interactions were previously analyzed by X-ray crystallography, NMR, psoralen cross-linking, and genetics, but all these methods are labor-intensive and typically require prior knowledge of the interacting partners. Because of these technical difficulties, RNA base-pairing is more commonly inferred from a combination of bioinformatic and evolutionary analyses. However, computational methods are applicable only to evolutionarily conserved interactions and provide little information about the physiological context of the interaction.
UV cross-linking methods have been developed to map protein interaction sites precisely on RNA molecules, including cross-linking and immunoprecipitation (CLIP) and cross-linking and analysis of cDNAs (CRAC) (1, 2). CRAC analyses have been performed on proteins (Nop1, Nop56, and Nop58) that are associated with all members of the box C/D class of small nucleolar RNAs (snoRNAs). Most box C/D snoRNAs base-pair with the rRNA to select sites of RNA 2′-O-methylation by the methyltransferase fibrillarin (Nop1). In contrast, the U3 snoRNA base-pairs to multiple sites on the pre-rRNA. These interactions probably facilitate correct folding of the pre-rRNA and are required for pre-rRNA processing (3, reviewed in refs. 4, 5). Pre-mRNA splicing requires five snRNAs that assemble the complex structure of the spliceosome, within which the U2 snRNA binds and activates an intronic sequence (the intron branch point) for cleavage of the 5′ splice site (6).
During the analysis of low-throughput sequence CRAC data obtained for the RNA helicase Prp43 (7), we identified a chimeric cDNA containing the methylation-guide region of the snR52 box C/D snoRNA fused to an rRNA region that included its cognate target site at A420 in the 18S rRNA (8). The CRAC procedure includes the ligation of oligonucleotide linkers to RNA fragments (2), and we hypothesized that base-paired RNA molecules could also be ligated together, generating chimeric RNAs (Fig. 1_A_). The two remaining ends of the fused RNAs would remain available for linker ligation, allowing cDNA generation, amplification, and recovery. The analysis of such chimeric cDNAs can identify sites of in vivo RNA–RNA interactions, and the approach should be widely applicable.
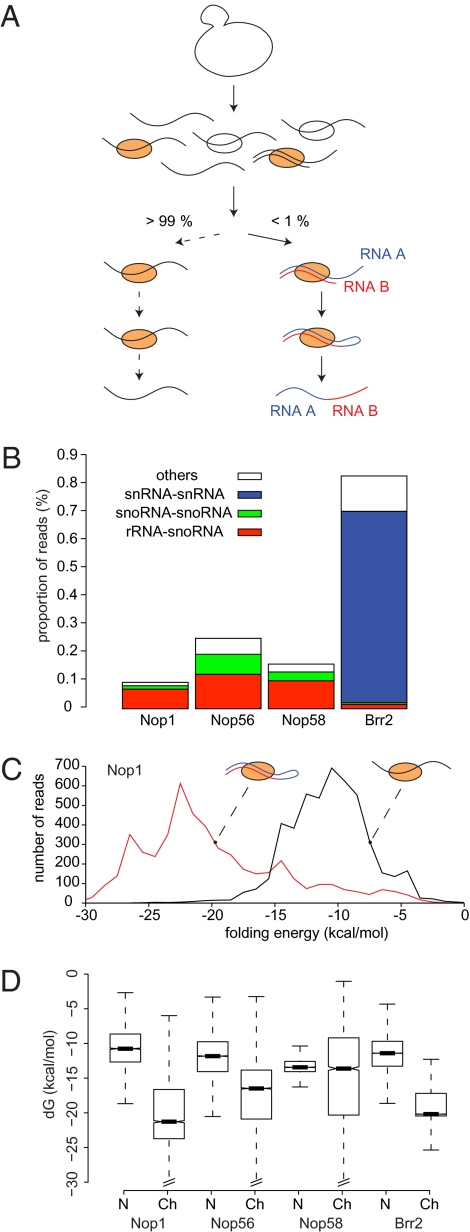
Fig. 1.
CLASH identifies RNA–RNA duplexes. (A) Schematic representation of the CLASH protocol. Following UV cross-linking, RNA–protein complexes were affinity-purified, RNA–RNA hybrids were ligated and sequenced, and chimeric reads were identified bioinformatically. (B) Classification of chimeric reads recovered with the snoRNP proteins Nop1, Nop56, and Nop58 and with the splicing factor Brr2. (C) Predicted minimum folding energies of chimeric reads (red trace) and nonchimeric reads (black trace) recovered with Nop1. (D) Distribution of minimum folding energies of nonchimeric and chimeric reads in all experiments. Dark bars, boxes, and whiskers represent the median, the first through third quartile ranges, and 1.5-fold the interquartile range, respectively. Ch, chimeric; N, nonchimeric; dG, mimimal free energy upon folding.
Results
Identification of Chimeric Sequences.
To identify chimeric reads, we analyzed high-throughput Illumina–Solexa sequence data derived from CRAC analyses of the box C/D snoRNA-associated proteins Nop1, Nop56, and Nop58 (2) as well as the spliceosome-associated helicase Brr2. Some 25 million 50-nt reads were analyzed using stringent quality filters (Materials and Methods). A total of 0.46% of all reads were composed of two distinct fragments that could be mapped separately, either to different RNA molecules or to distinct regions of the same molecule (Fig.1_B_). In most cases, the two mapped fragments were directly fused in the read. None of the chimeric reads could be fully aligned to a database of spliced yeast transcripts, indicating that the chimeras do not represent conventional splicing events.
Chimeric reads previously identified in high-throughput sequencing have been attributed to reverse transcriptase (RT) template switching (9, 10). However, the two regions of the chimeras did not show the short sequence duplications indicative of template switching (9). Notably, the patterns of chimeric reads were strongly dependent on the protein analyzed, confirming that they were not random events (Fig. 1_B_).
If chimeras result from the ligation of two base-paired RNAs, they should form stable stem structures (Fig. 1_A_). Consistent with this prediction, in silico folding analysis indicated that most chimeras form strong secondary structures, with mean folding energies between −14 and −20 kcal/mol. In contrast, the mean folding energies of nonchimeric reads ranged between −10 and −13 kcal/mol. In all four datasets, the predicted folding of chimeric RNAs was significantly stronger than the folding of nonchimeric RNAs of the same length (P < 10−15 for Nop1, Nop56, and Brr2 and P < 10−11 for Nop58, Wilcoxon rank sum test) (Fig. 1 C and D). Moreover, a negative correlation was observed between folding energy and number of chimeras (Spearman ρ = −0.40, P < 10−10), showing that stable chimeras tend to be recovered more frequently. This supports the hypothesis that most chimeric cDNAs originate from ligation events between stably base-paired RNA molecules.
Base-Pairing Between snoRNA and rRNA.
In yeast, all known methylation targets of the box C/D snoRNAs are in 18S and 25S rRNAs, and binding sites were previously defined for most but not all C/D snoRNAs (5, 8). Analysis of the Nop1, Nop56, and Nop58 datasets yielded 24,822 chimeric sequences, of which 56% were box C/D snoRNA-rRNA chimeras and 39% were snoRNA-snoRNA or rRNA-rRNA chimeras. Yeast has 47 box C/D snoRNAs (counting separately 39, 39b, U3a, and U3b) (11), and all these, except snR78, were found fused with rRNA in at least one experiment.
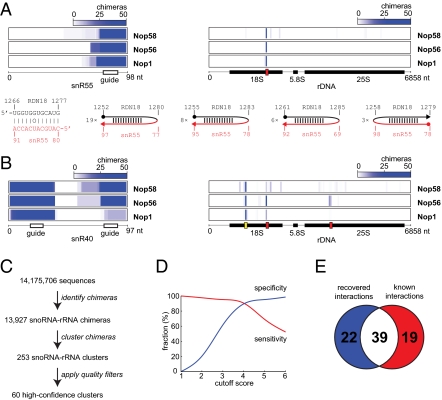
Most snoRNA-rRNA chimeras consisted of a snoRNA guide sequence fused to the corresponding known rRNA target sequence. These were separated by a stretch of four or more nucleotides derived from the flanking sequence of either the snoRNA or rRNA (Fig. 2_A_). The gap presumably reflects the need for a loop to form to permit ligation; any stem that is truncated precisely at its ends during RNase digestion will not be recovered as a chimera. The loops were up to 20-nt long and could be located on either side of the stem (Fig. 2_A_). Some snoRNAs have two guide sequences that base-pair with different positions on the rRNA, and for several snoRNAs, we recovered sets of chimeras corresponding to both interactions (shown for snR40; Fig. 2B, red boxes). In total, chimeras were found for 43 of the 58 known box C/D snoRNA-rRNA interactions in at least one experiment. Notably, some chimeras did not match known interactions, and we hypothesized that these represent previously unidentified sites of snoRNA-rRNA association (shown for snR40; Fig. 2_B_, yellow box).
Fig. 2.
Identification of box C/D snoRNA-rRNA interactions. (A) Analysis of snR55-rRNA interaction. (Upper) Density of snR55-rRNA chimeras along snR55 (Left) and along rRNA (Right). The red box represents the known snR55 target site in 18S rRNA. (Lower Left) Known base-pairing interaction between snR55 and rRNA. (Lower Right) Examples of chimeras supporting the snR55-rRNA interaction and numbers of times each chimera was found. Filled circles represent starts of reads, and arrowheads indicate ends of reads. (B) Density of snR40-rRNA chimeras along snR40 (Left) and along rRNA (Right). Red boxes indicate known snR40 target sites in 18S and 25S rRNA. The yellow box indicates the predicted novel site. (C) Numbers of reads, chimeric reads, chimeric read clusters, and high-confidence clusters in the combined Nop1, Nop56, and Nop58 datasets. (D) Scoring snoRNA-rRNA clusters. For each cutoff score N, the red line (“sensitivity,” or true-positive rate) represents the number of known targets with a score ≥N, divided by the total number of known targets, and the blue line (“specificity,” or true-negative rate) is the number of previously unknown targets with a score <N, divided by the total number of previously unknown targets. Only targets with corresponding clusters were used in the calculation. (E) Venn diagram showing the overlap between the set of previously identified interactions, and the set of called interactions at a cutoff score of 4.
To distinguish genuine snoRNA targets from background generated by nonbiologically relevant RNA ligation in vitro, we first clustered the chimeras to define the putative interactions (Fig. 2_C_). We then applied a scoring system that takes into account the number of replicate experiments and sequencing reads supporting each putative interaction; the predicted binding energy between the snoRNA and its target; and the location of the predicted binding site in the snoRNA (i.e., whether it was located within the guide region). The top-scoring category included 84% of all snoRNA-rRNA chimeric reads and represented 25 predicted snoRNA-rRNA interactions. Of these, 22 interactions (88%) were identified in previous studies, and 3 were novel (Fig. 2_D_ and Dataset S1). At the cutoff score of 4, we recovered 39 known snoRNA targets, corresponding to 67% of all known targets, or to 90% of the known targets found in our data (sensitivity; Fig. 2 D and E). At the same cutoff score, almost 90% of the previously unknown interactions we found were rejected (specificity; Fig. 2_D_).
Binding of snR190 to 25S rRNA (nucleotides 2,392–2,404) was previously predicted but could not be experimentally confirmed because it does not correspond to a modification site, and it was detected with a high score in the data (Dataset S1). The scoring gave strong support for putative novel interactions between the guide region of snR40 (nucleotides 19–30) and 18S rRNA (nucleotides 559–569) (Fig. 2_B_) and between the guide region of U14 (nucleotides 101–120) and 25S rRNA (nucleotides 2,647–2,666) (Fig. S1). Notably, previous data had indicated base-pairing between U14 and 27S pre-rRNA, the precursor to 25S rRNA, and several large subunit assembly factors have been identified in purified U14–small nucleolar ribonucleoprotein (snoRNP) complexes (12, 13), but the significance of this was unclear. However, inspection of published data (8) strongly indicates that methylation does not occur at the predicted sites (18S G562 for snR40 and 25S C2653 for U14).
Base-pairing was detected between the region flanking box D of snR75 (nucleotides 64–73) and the 25S rRNA (nucleotides 2,307–2,316). This is in close proximity to the known 25S methylation site at G2288, which is directed by the snR75 box D′ element. The novel base-pairing does not itself guide methylation, but it might stabilize the interaction between the box D′-associated guide and its target.
Yeast snR4 is an “orphan” snoRNA without known targets. None of the snR4 interactions were reproducibly found in all experiments (reducing the score). However, the Nop56 dataset included 146 snR4-5S rRNA chimeras, indicative of a perfect 11-nt stem formed between snR4 (nucleotides 155–165) and 5S (nucleotides 22–32) (Fig. S1), whereas the Nop1 dataset suggested a stem between snR4 (nucelotides 175–186) and 25S rRNA (nucleotides 1,866–1,878).
To validate the potential novel interactions, we analyzed the binding of U14 and snR4 to ribosomal RNA by native gel electrophoresis. Following deproteinization under conditions that retain RNA base-pairing, slow migrating bands were observed for U14 (Fig. S1). These were lost following heat denaturation at 95 °C before electrophoresis, consistent with the proposed interaction with the 27S pre-rRNA. snR4 migrated as three major bands, with mobilities consistent with the presence of free snR4 plus snR4-5S and snR4-27S complexes that were lost from heat-denatured samples (Fig. S1). Thus, native electrophoresis supported the novel interactions inferred from cross-linking, ligation, and sequencing of hybrids (CLASH) data analysis.
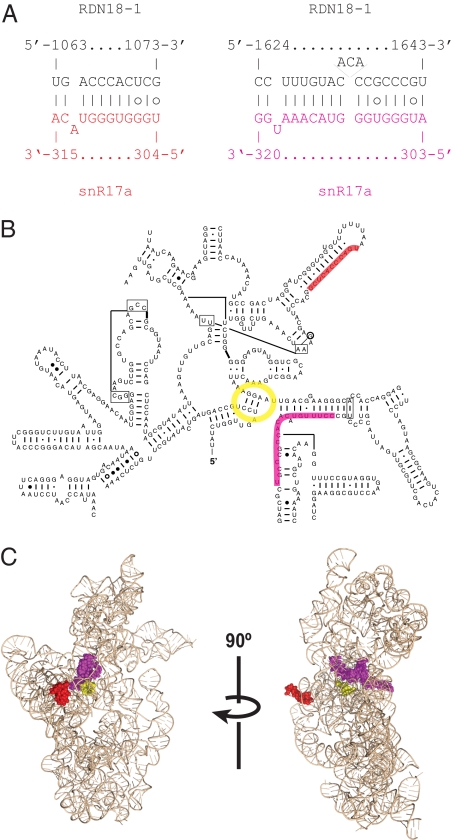
U3 is a box C/D snoRNA that does not direct pre-rRNA modification but is required for pre-rRNA processing on the pathway of 18S rRNA synthesis in all eukaryotes tested, including those in yeast and humans. A key structural feature of the small ribosomal subunit is the central pseudoknot, and yeast U3 is implicated in the mechanism and/or timing of pseudoknot formation (14–17). CLASH recovered hybrids between a U3 sequence close to box D at the 3′ end of the snoRNA (nucleotides 304–315) and two different regions in 18S rRNA (nucleotides 1,063–1,073 and 1,624–1,643) (Fig. 3_A_). Notably, this region of U3 is located at the same position as the modification guide sequences of other box C/D snoRNAs (reviewed in ref. 4). Both rRNA targets lie in close proximity to the central pseudoknot in the predicted 3D structure of the 40S ribosomal subunit (Fig. 3 B and C). The rRNA sequences are invariant, precluding the identification of compensatory base-changes that might have confirmed the interaction. However, homologous interactions are predicted in the distantly related fungi Neurospora crassa and Aspergillus nidulans but not in humans (Fig. S2). Shuffling the sequences of the predicted U3 guide region or its target (18S nucleotides 1,624–1,643) significantly decreases their predicted interaction strength (Fig. S3), suggesting that the U3 guide is evolutionarily adapted for binding to 18S rRNA.
Fig. 3.
Novel U3-rRNA interaction site close to the small subunit central pseudoknot. (A) Predicted U3-rRNA base-pairing. (B) Predicted interaction sites mapped onto the secondary structure of yeast 18S rRNA. (C) Predicted interaction sites mapped onto the Thermus thermophilus mature small ribosomal subunit structure. Central pseudoknot is shown in yellow, and interaction sites are shown in red and magenta.
In addition to snoRNA-rRNA chimeras, large numbers of snoRNA-snoRNA chimeras were identified. Ninety-three percent of such chimeras included two different fragments of the same snoRNA, suggestive of intramolecular interactions. Most reads corresponded to known contacts within U3 or to predicted contacts between boxes C and D in other snoRNAs. Folding energies for intramolecular snoRNA-snoRNA interactions ranged from −9 to −14 kcal⋅mol−1, lower than snoRNA-rRNA interactions and consistent with previous data showing that the C and D boxes of snoRNAs form short imperfect stems.
rRNA-rRNA chimeras were recovered in all datasets, including the untagged control, suggesting that they are not necessarily associated with the snoRNP proteins. We mapped these chimeras onto 3D structure models of ribosomal subunits and found that close to half of the sequences corresponded to known base-pairing interactions. Most reflected local stems, with a smaller number corresponding to interactions between more distant regions of individual ribosomal subunits (Figs. S4 and S5).
Mapping the Secondary Structure of the U2 snRNA.
The identification of intramolecular chimeras suggested that this approach could be used to identify features of RNA secondary structure. The yeast U2 spliceosomal snRNA is 1,177 nt in length, much larger than human U2 (187 nt) (18, 19), and offered a suitable target for these analyses.
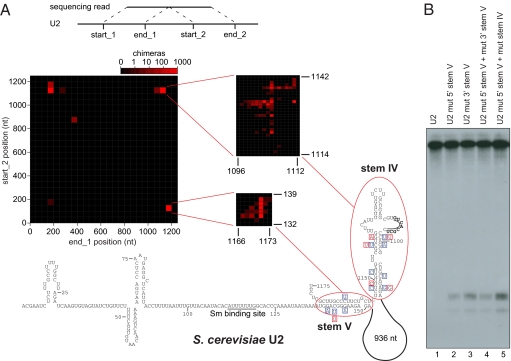
Brr2 is a spliceosome-associated DEIH-box helicase (20), and CLASH analyses yielded large numbers of chimeric reads, including 74,585 U2-U2 sequences. The distribution and frequency of chimeras identified within U2 are shown in Fig. 4_A_. We interpreted the three major clusters in this graph to represent two intramolecular stems within U2 (stems IV and V), with stem V recovered in both orientations. This interpretation is supported by the propensity of chimeric reads within U2 to form stable stems in silico. We applied the hybrid-ss-min folding algorithm (21) to yeast U2, using the predicted stems as structural constraints, and obtained the structure shown in Fig. 4_A_. The novel structure is substantially different from previously proposed structures for the 3′ domain of yeast U2 but significantly more similar to mammalian U2 (19, 22). In the new folding, the 3′ region is engaged in a stable stem structure, a feature found in many U2 orthologs, including humans and Trypanosoma spp., which is expected to help stabilize the RNA (23). An internal bulge in stem IV contains a stretch of nucleotides homologous to the loop sequence of the 3′ stem in human U2, which binds the hU2B′′ protein (24) (shown in bold in Fig. 4_A_). Stem V is formed by a long-range interaction that brings the 3′ and 5′ domains together, with the long ∼950 nt insertion sequence “looped-out” of the structure.
Fig. 4.
Structure analysis of yeast U2. (A) (Top) Line diagram indicating the positions of fragments found in chimeras. (Middle) Heat-map representation of intramolecular chimeras within U2. The x axis represents the position in U2 where the first fragment of the chimera was mapped, and the y axis shows the position of the second fragment. The red color intensity increases with the number of chimeric reads. The insets show the major peaks at higher resolution. The peaks in the lower right and upper left corners correspond to the same stem ligated at the opposite ends. (Bottom) Secondary structure of U2 inferred from the chimeras. The boxed nucleotides represent compensatory base-changes in S. mikatae (blue boxes) and S. kudriavzevii (red boxes). The conserved nucleotides in the internal bulge of stem IV are in bold and underlined. (B) Northern blot analysis of U2 carrying mutations in stem IV or V (details of mutations are provided in Fig. S6_B_).
To validate this structure, we analyzed its evolutionary conservation. U2 sequences from three yeast species could be aligned to the Saccharomyces cerevisiae U2, based on primary sequence alone. Applying the pfold secondary structure prediction algorithm to these alignments supported the conservation of these stems in all three species [shown for _Saccharomyces mikatae_ (blue boxes) and _Saccharomyces kudriavzevii_ (red boxes) in Fig. 4_A_]. None of the base substitutions are predicted to interfere with stem formation, whereas the occurrence of compensatory base-changes strongly supports the predicted secondary structure. In contrast, the previously proposed S. cerevisiae U2 structure (19) is not supported by evolutionary analysis (Fig. S6_A_).
For further validation, we analyzed the phenotypes of mutations predicted to disrupt the U2 stems. Single-point mutations did not have a measurable effect on yeast growth rates or U2 snRNA stability (Table S1). However, scrambling or deleting (Table S1) the 5′ branch of the predicted stem V resulted in the appearance of a truncated 5′ fragment ∼130 nt in size (Fig. 4_B_). Combining mutations in stems IV and V exacerbated this effect, and restoring the stems with compensatory mutations led to a weaker phenotype. The size of the truncated form of U2 is consistent with termination downstream of the Sm protein-binding site. To identify the 3′ ends of U2 with mutations in stems IV and V (mut 5′ stem V + mut stem IV), a linker (miRCat33) was ligated to the 3′ ends of all RNA molecules present in total RNA and used for cDNA synthesis. PCR amplification used a U2-specific primer, and cloned PCR products were individually sequenced (SI Materials and Methods). From 20 clones sequenced, 8 had an insert, each of which contained a truncated U2 fragment. The 3′ ends were located 9–18 nt downstream of the Sm protein-binding site (Fig. S7_A_), forming a ladder with single-nucleotide size differences. This strongly suggests that misfolded U2 is degraded by 3′ to 5′ exonuclease activity that is impeded by the Sm proteins. A fragment of U2 containing the 5′ ∼132 nt was previously detected when large segments within the nonessential region of U2 were deleted (19), but the basis of this effect was unclear. Inspection of the sequences deleted now indicates that they interfered with formation of the terminal stem V. The extent to which the truncated U2 fragment is generated likely coincides with the degree to which secondary structure in the U2 3′ domain is disturbed.
Deletion of the nonessential U2 region (nucleotides 123–1,068) leads to growth and splicing defects when combined with a U23G substitution (19, 25). The U23G mutation reduces base-pairing between U2 and U6 snRNAs in helix Ib, and it was therefore proposed that the nonessential region in U2 stabilizes U2/U6 helix Ib. To determine whether the observed synergistic phenotype was in fact attributable to disruption of stem IV and/or V, stem mutations were combined with the U23G mutation (Fig. S7_B_). Mild temperature sensitivity was seen on combination of the U23G mutation with either the deletion or mutation of the 5′ side of stem V. Strong temperature sensitive lethality was seen when stem IV was additionally destabilized by mutations in the 3′ side of the stem. Taken together, these results confirm that the stems IV and V form in vivo and act to stabilize U2.
Discussion
We have described and validated a method to map RNA–RNA binding directly in living cells by CLASH. The CLASH technique is conceptually related to the 3C chromosome mapping method (26): Both techniques use ligation to record the spatial arrangement of nucleic acids in living cells. CLASH provides a fast and reliable alternative to existing experimental and bioinformatic methods. It allows high-throughput identification of interacting partners and interaction sites in physiological conditions. Instead of detecting downstream consequences of interactions, such as methylation of snoRNA targets or down-regulation of micro-RNA targets, we directly recover RNA duplexes as chimeric sequences. This allows precise mapping of interactions for which the downstream consequences are unknown and/or difficult to measure. Because cross-linking is performed in living cells, the dynamic state of the RNA interactome can be probed as a function of physiological conditions.
Other high-throughput methods for mapping RNA secondary structures have recently been described. In selective 2′-hydroxyl acylation and primer extension (27, 28), RNA is modified with hydroxyl radicals and analyzed by primer extension and capillary electrophoresis. In parallel analysis of RNA structure (PARS) (29) and fragmentation sequencing (FragSeq) (30), RNA is partially digested with ribonuclease and analyzed by deep sequencing. Both methods reveal whether a certain position in RNA is engaged in a base-paired interaction but do not directly identify the base-pairing partner. In contrast, the CLASH approach reported here allows the locations of RNA stems present in vivo to be identified but probably with lower coverage. The data provided are therefore very complementary.
Applying CLASH to snoRNAs confirmed that they only significantly associate with rRNA in yeast. In addition to many known binding sites, we detected interactions between U14-25S, snR4-5S, and U3-18S, none of which appear to direct methylation. U14 depletion did not clearly inhibit 60S synthesis, but it was previously reported to interact with pre-60S particles (12, 13), supporting the hybrids recovered. No pre-rRNA binding site was previously identified for snR4, and no growth phenotype was detected following SNR4 deletion (31). Notably, homologous RNAs are present in several sequenced fungal genomes, indicating that snR4 is functional; however, the region of snR4 that base-pairs with 5S is not highly conserved.
The U3 snoRNA forms multiple interactions with the pre-rRNA, in the 5′ external transcribed spacer region and the 18S rRNA, which were shown by the requirement of compensatory mutations for pre-rRNA processing (14, 16). U3/pre-rRNA interactions were also implicated in formation of the central pseudoknot (16, 32), a key long-range interaction in the 18S rRNA. U3 binds to 18S rRNA on the 5′ side of the pseudoknot (16), whereas the novel U3-18S interactions lie close to the 3′ side, potentially facilitating pseudoknot formation. The U3 sequence involved occupies the same position as the modification guide in other box C/D snoRNAs, and deletion analyses showed that this region and the flanking C/D and C′/D′ boxes are essential (33). Substitution of the U3 guide sequence was tolerated, but this is also the case for other U3 sequences shown or predicted to base-pair to the pre-rRNA. Multiple U3/pre-rRNA interactions may render them individually dispensable. Hybrids between the 5′ domain of U3 and the pre-rRNA were not recovered, probably because Nop1, Nop56, and Nop58 do not bind this region (2). Repeating the experiment with other U3-bound proteins, Imp3, Imp4, or Mpp10 (34–36), might recover further U3/pre-rRNA interactions.
In previous secondary structure models for yeast U2 (19), the 5′ region closely resembles metazoan U2, whereas the 3′ domain appeared quite different, with a long 3′ single-stranded sequence. The revised model for yeast U2 shows greater similarity in overall fold, with the large additional domain clearly looped-out and a structured 3′ domain. It is possible that Brr2 is involved in establishment of the U2 structure, but we think it more likely interaction occurs during the splicing process. Extending these analyses to other U2-associated proteins may reveal conformational changes during the splicing cycle.
The CLASH method should be applicable to many different RNA–RNA interactions, including, for example, the identification of sRNA targets in bacteria and miRNA targets in eukaryotes. In the present approach, the affinity purification steps recover only RNA–RNA hybrids located close to the protein-binding site. This limits the interactions that can be identified in any specific experiment, but many important RNA–RNA interactions take place in the context of ribonucleoprotein complexes. Moreover, the UV cross-linking and protein purification steps could, in principle, be omitted to generate a transcriptome-wide list of RNA–RNA interactions for all abundant cellular RNAs.
Materials and Methods
Strains and Plasmids.
Strain and plasmids are described in SI Materials and Methods and Table S1.
Cross-Linking and Library Construction.
CRAC on Nop1, Nop56, and Nop58 was described previously (2). Briefly, intact cells expressing His6-tobacco etch virus protease (TEV)-cleavage site-Protein A (HTP)–tagged snoRNP proteins were UV-irradiated. HTP-tagged proteins were bound to IgG Sepharose beads and denatured in 6 M guanidine and bound to nickel resin following TEV protease cleavage. Under these conditions, noncovalent protein–protein interactions are normally disrupted; however, RNA duplexes appear to be stable. Protein–RNA complexes were immobilized on nickel beads, and linker ligation reactions were performed at either 16 °C or 25 °C, largely preserving RNA stem structures and allowing formation of chimeras. Cross-linking of Brr2-HTP was performed essentially as described (2), with minor modifications to allow actively growing cells to be irradiated in culture medium. Two high-speed spins were included to sediment polysomes before protein purification.
Bioinformatic Analyses.
To identify chimeras, we mapped the deep-sequencing reads to a database of nonprotein coding S. cerevisiae transcripts, using BLAST (SI Materials and Methods). We extracted those reads with two BLAST hits that were either directly adjacent in the read or with up to a 4-nt gap or overlap between hits. The majority (61%) of such hits were directly adjacent or overlapped by exactly 1 nt. This suggests that the chimeras did not result from RT template switching, which requires homology of several nucleotides between the start and landing molecules and would generate reads with larger overlaps. We discarded hits mapped in the antisense orientation, which represented less than 1% of chimeras. Statistical tests were performed using R (The R Project for Statistical Computing), minimum folding energies were calculated at 30 °C using mfold with default parameters (mfold web server: 1995–2010, Michael Zuker, Nick Markham, Rensselaer Polytechnic Institute), heat maps were drawn using Java TreeView (version 1.1.3, created by Alok J. Saldanha, 2004), 3D structures were rendered with MacPyMOL (Schrödinger, LLC), and all other graphs were done using gnuplot (www.gnuplot.info, version 4.2) and R. More details on the bioinformatics analyses can be found in SI Materials and Methods.
Clustering and Ranking of Chimeras.
To call RNA–RNA interactions, we first clustered the chimeras by iteratively merging the reads for which the mapped positions of both fragments overlapped by at least 1 nt. For each cluster, we then calculated a score by adding one point for each experiment that was represented in the cluster (Nop1, Nop56, and Nop58 experiments were clustered together), one point if there were at least four total reads in the cluster, and one point if the average minimum folding energy of reads in the cluster was below the threshold of −14 kcal/mol. In snoRNP experiments, one point was added for clusters that included a known snoRNA guide region (associated with the D or D′ box). Sequence, scoring, and folding information for the 253 snoRNA-rRNA clusters and folding of the 15 top snoRNA-snoRNA clusters are shown in Dataset S1. Raw sequence data are available on request from the authors.
Northern Blotting of U2 snRNA.
Isolation of total RNA from yeast and Northern blotting were carried out according to standard procedures. RNA was separated on denaturing 6% (wt/vol) polyacrylamide gels. 32P-labeled oligonucleotide U2 (5′-CTACACTTGATCTAAGCCAAAAG, complementary to nucleotides 15–37 of yeast U2 snRNA) (37) was used to detect U2 snRNA by autoradiography.
Native Gel Electrophoresis of snoRNA-rRNA Duplexes.
BY4741 yeast was grown in YPD (1% wt/vol yeast extract, 2% wt/vol peptone, 2% wt/vol dextrose) medium to OD600 of 0.8, washed, resuspended in lysis buffer [150 mM sodium acetate, 1 mM magnesium acetate, 40 mM Tris-base, 0.1% Triton X-100 (pH 7.5)], and lysed by vortexing with zirconia beads on ice. The lysate was treated with 0.2% SDS and protease K overnight at 18 °C; RNA was extracted at 18 °C, twice with phenol and once with chloroform, and was ethanol-precipitated. RNA was dissolved in resuspension buffer [80 mM Tris-base, 50 mM acetic acid, 10 mM sodium acetate, 0.5 mM magnesium acetate (pH 8.5)], and glycerol was added to a final concentration of 20% (vol/vol). For heat denaturation, EDTA was added to 10 mM and the sample was incubated at 95 °C for 5 min. Six micrograms of RNA was separated on a 2% (wt/vol) agarose gel in Tris-acetate buffer with 10 mM sodium acetate but without magnesium. Total RNA was visualized with ethidium bromide. RNA was transferred to a positively charged nylon membrane, which was sequentially probed with oligonucleotide probes.
Supplementary Material
Supporting Information
Acknowledgments
We thank Markus Bohnsack for communicating unpublished results, Al Kerr and Shaun Webb for bioinformatics support, and Olex Dybkov for helpful discussions. G.K. was supported by a European Molecular Biology Organization Long Term Fellowship and the Wellcome Trust, S.G. was supported by the European Union and the Wellcome Trust, D.H. was supported by the Darwin Trust of Edinburgh, J.D.B. was supported by the Royal Society, and D.T. was supported by the Wellcome Trust.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: All relevant chimeric sequences recovered are listed in Dataset S1.
References
- 1.Licatalosi DD, Darnell RB. RNA processing and its regulation: Global insights into biological networks. Nat Rev Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci USA. 2009;106:9613–9618. doi: 10.1073/pnas.0901997106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kiss-László Z, Henry Y, Bachellerie J-P, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell. 1996;85:1077–1088. doi: 10.1016/s0092-8674(00)81308-2. [DOI] [PubMed] [Google Scholar]
- 4.Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol. 1997;9:337–342. doi: 10.1016/s0955-0674(97)80005-1. [DOI] [PubMed] [Google Scholar]
- 5.Kiss T. Small nucleolar RNAs: An abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- 6.Parker R, Siliciano PG, Guthrie C. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987;49:229–239. doi: 10.1016/0092-8674(87)90564-2. [DOI] [PubMed] [Google Scholar]
- 7.Bohnsack MT, et al. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell. 2009;36:583–592. doi: 10.1016/j.molcel.2009.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowe TM, Eddy SR. A computational screen for methylation guide snoRNAs in yeast. Science. 1999;283:1168–1171. doi: 10.1126/science.283.5405.1168. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Zhao L, Jiang H, Wang W. Short homologous sequences are strongly associated with the generation of chimeric RNAs in eukaryotes. J Mol Evol. 2009;68:56–65. doi: 10.1007/s00239-008-9187-0. [DOI] [PubMed] [Google Scholar]
- 10.Houseley J, Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS ONE. 2010;5:e12271. doi: 10.1371/journal.pone.0012271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samarsky DA, Fournier MJ. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res. 1999;27:161–164. doi: 10.1093/nar/27.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zagorski J, Tollervey D, Fournier MJ. Characterization of an SNR gene locus in Saccharomyces cerevisiae that specifies both dispensible and essential small nuclear RNAs. Mol Cell Biol. 1988;8:3282–3290. doi: 10.1128/mcb.8.8.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piekna-Przybylska D, Liu B, Fournier MJ. The U1 snRNA hairpin II as a RNA affinity tag for selecting snoRNP complexes. Methods Enzymol. 2007;425:317–353. doi: 10.1016/S0076-6879(07)25014-1. [DOI] [PubMed] [Google Scholar]
- 14.Beltrame M, Tollervey D. Base pairing between U3 and the pre-ribosomal RNA is required for 18S rRNA synthesis. EMBO J. 1995;14:4350–4356. doi: 10.1002/j.1460-2075.1995.tb00109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hughes JMX. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J Mol Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- 16.Sharma K, Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol Cell Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Méreau A, et al. An in vivo and in vitro structure-function analysis of the Saccharomyces cerevisiae U3A snoRNP: Protein-RNA contacts and base-pair interaction with the pre-ribosomal RNA. J Mol Biol. 1997;273:552–571. doi: 10.1006/jmbi.1997.1320. [DOI] [PubMed] [Google Scholar]
- 18.Ares M., Jr U2 RNA from yeast is unexpectedly large and contains homology to vertebrate U4, U5, and U6 small nuclear RNAs. Cell. 1986;47:49–59. doi: 10.1016/0092-8674(86)90365-x. [DOI] [PubMed] [Google Scholar]
- 19.Shuster EO, Guthrie C. Two conserved domains of yeast U2 snRNA are separated by 945 nonessential nucleotides. Cell. 1988;55:41–48. doi: 10.1016/0092-8674(88)90007-4. [DOI] [PubMed] [Google Scholar]
- 20.Hahn D, Beggs JD. Brr2p RNA helicase with a split personality: Insights into structure and function. Biochem Soc Trans. 2010;38:1105–1109. doi: 10.1042/BST0381105. [DOI] [PubMed] [Google Scholar]
- 21.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33(Web Server issue):W577–W581. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Igel AH, Ares M., Jr Internal sequences that distinguish yeast from metazoan U2 snRNA are unnecessary for pre-mRNA splicing. Nature. 1988;334:450–453. doi: 10.1038/334450a0. [DOI] [PubMed] [Google Scholar]
- 23.Guthrie C, Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- 24.Scherly D, Boelens W, Dathan NA, van Venrooij WJ, Mattaj IW. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B′′ and their cognate RNAs. Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- 25.Hilliker AK, Staley JP. Multiple functions for the invariant AGC triad of U6 snRNA. RNA. 2004;10:921–928. doi: 10.1261/rna.7310704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 27.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J Am Chem Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 28.Shuster EO, Guthrie C. Human U2 snRNA can function in pre-mRNA splicing in yeast. Nature. 1990;345:270–273. doi: 10.1038/345270a0. [DOI] [PubMed] [Google Scholar]
- 29.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–107. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Underwood JG, et al. FragSeq: Transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7:995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parker R, Simmons T, Shuster EO, Siliciano PG, Guthrie C. Genetic analysis of small nuclear RNAs in Saccharomyces cerevisiae: Viable sextuple mutant. Mol Cell Biol. 1988;8:3150–3159. doi: 10.1128/mcb.8.8.3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hughes JMX, Konings DAM, Cesareni G. The yeast homologue of U3 snRNA. EMBO J. 1987;6:2145–2155. doi: 10.1002/j.1460-2075.1987.tb02482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Samarsky DA, Fournier MJ. Functional mapping of the U3 small nucleolar RNA from the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:3431–3444. doi: 10.1128/mcb.18.6.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baserga SJ, Agentis TM, Wormsley S, Dunbar DA, Lee S. Mpp10p, a new protein component of the U3 snoRNP required for processing of 18S rRNA precursors. Nucleic Acids Symp Ser. 1997;36:64–67. [PubMed] [Google Scholar]
- 35.Lee SJ, Baserga SJ. Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol Cell Biol. 1999;19:5441–5452. doi: 10.1128/mcb.19.8.5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wormsley S, Samarsky DA, Fournier MJ, Baserga SJ. An unexpected, conserved element of the U3 snoRNA is required for Mpp10p association. RNA. 2001;7:904–919. doi: 10.1017/s1355838201010238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boon KL, Norman CM, Grainger RJ, Newman AJ, Beggs JD. Prp8p dissection reveals domain structure and protein interaction sites. RNA. 2006;12:198–205. doi: 10.1261/rna.2281306. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information