A SIRT1-LSD1 Co-repressor Complex Regulates Notch Target Gene Expression and Development (original) (raw)
. Author manuscript; available in PMC: 2012 Jun 10.
Summary
Epigenetic regulation of gene expression by histone-modifying co-repressor complexes is central to normal animal development. The NAD+-dependent deacetylase and gene repressor SIRT1 removes histone H4K16 acetylation marks and facilitates heterochromatin formation. However, the mechanistic contribution of SIRT1 to epigenetic regulation at euchromatic loci and whether it acts in concert with other chromatin-modifying activities to control developmental gene expression programs remain unclear. We describe here a SIRT1 co-repressor complex containing the histone H3K4 demethylase LSD1/KDM1A and several other LSD1-associated proteins. SIRT1 and LSD1 interact directly and play conserved and concerted roles in H4K16 deacetylation and H3K4 demethylation to repress genes regulated by the Notch signaling pathway. Mutations in Drosophila SIRT1 and LSD1 orthologs result in similar developmental phenotypes and genetically interact with the Notch pathway in Drosophila. These findings offer new insights into conserved mechanisms of epigenetic gene repression and regulation of development by SIRT1 in metazoans.
Keywords: SIRT1, LSD1, co-repressor, Notch, chromatin modification
Introduction
Epigenetic regulation of transcriptional programs is critical to normal cellular function and developmental processes, and is frequently aberrant in disease (Chi et al.; Kouzarides, 2007; Nottke et al., 2009). Central to this is the targeted post-translational modification of histone residues by enzymes that render specific chromatin loci either activating or repressive to transcription. Recent studies have shown that deacetylation of the conserved histone residue H4K16 is required for higher order chromatin compaction, a process strongly associated with gene repression (Shogren-Knaak et al., 2006; Vaquero et al., 2007b). The major H4K16 deacetylase in interphase mammalian cells is SIRT1, a highly conserved transcriptional co-repressor (Vaquero et al., 2007b). SIRT1 is also an important regulator of cellular and organismal processes, including metabolism, cellular differentiation, development and stress responses such as DNA repair and inflammation, and is implicated in cancer as both a candidate tumor suppressor and oncogene, in a context-dependent manner (Vaquero, 2009). However, the molecular mechanisms underlying the ability of SIRT1 to regulate physiologic and disease processes remain poorly understood.
SIRT1 is a member of the class III NAD+-dependent histone deacetylases (HDACs) and is predominantly localized in the nucleus. Members of the other major class of nuclear HDACs involved in gene repression, Class I, are typically associated with multi-protein complexes that target their activities to particular chromatin loci or regulate their activity. For example, the metazoan Sin3, Mi-2/NuRD and CoREST complexes contain HDAC1/2 orthologs, while HDAC3 has been found in the NCoR/SMRT complex in mammals and flies. These large co-repressor complexes may also contain additional histone-modifying activities that facilitate coordinated modification of individual or distinct histone residues at target gene loci (Yang and Seto, 2008). In contrast with class I HDACs, SIRT1 was not generally found as a component of co-repressor complexes. While SIRT1 can associate with Polycomb Repressive Complex 2 (PRC2) (Furuyama et al., 2004; Kuzmichev et al., 2005), and interacts with the H3K9 methyltransferase SUV39H1 and the linker histone H1 to promote facultative heterochromatin formation (Vaquero et al., 2007a; Vaquero et al., 2004), little is known of SIRT1-interacting proteins that may direct its repressor activity to particular target genes. Moreover, it is currently unclear whether SIRT1 associates with additional chromatin-modifying activities to regulate gene expression at euchromatic loci in a concerted manner. Indeed, recent efforts to identify SIRT1-associated proteins using affinity-purification approaches from cells constitutively expressing SIRT1 did not yield co-factors that facilitate SIRT1 functions, but rather uncovered an inhibitor of SIRT1 catalytic activity, Deleted in Breast Cancer 1 (DBC1) (Kim et al., 2008; Zhao et al., 2008). This may be linked to the emerging role of SIRT1 as a candidate tumor suppressor (Deng, 2009), in which case constitutive expression of SIRT1 in cancer cells might select against productive interactions with functional partners and favor associations with inhibitors such as DBC1. We have developed an inducible SIRT1 expression system aimed at identify SIRT1-interacting proteins that might yield new insights into its roles in epigenetic regulation of gene expression and physiological functions. This approach has led to the discovery of a SIRT1-LSD1 epigenetic repressor complex that regulates genes governed by the Notch signaling pathway.
Results
Identification and characterization of a nuclear SIRT1-LSD1 complex
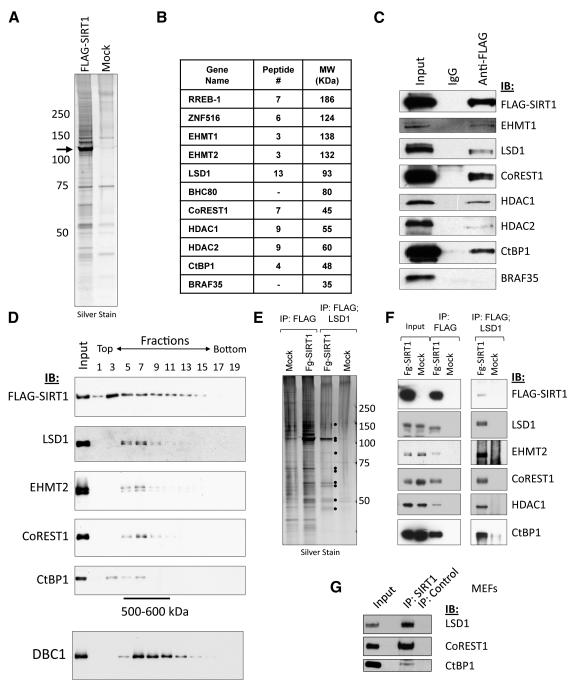
To increase the possibility of capturing interactions that may be lost upon constitutive overexpression of SIRT1 in cancer cells, we transiently expressed FLAG-tagged SIRT1 in a tetracycline-inducible manner in human embryonic kidney 293T cells (T-REx 293) and then isolated SIRT1 and associated polypeptides from nuclear extracts by anti-FLAG affinity chromatography. This led to the isolation of a number of FLAG-SIRT1-associated polypeptides that were not enriched in control purifications, as visualized by SDS-PAGE analysis and silver staining (Fig. 1A). Strikingly, identification of FLAG-SIRT1-interacting proteins by tandem mass spectrometry indicated an abundant association with components of the previously identified LSD1-CoREST1-CtBP1 co-repressor complex (Shi et al., 2004; Shi et al., 2003) (Fig. 1B). Indeed, immunoblotting of FLAG-SIRT1-immunopurified material confirmed the presence of many of these components, including the LSD1/KDM1A histone H3K4 demethylase, the HDAC1 and HDAC2 histone deacetylases, the EHMT1 and EHMT2 histone methyltransferases, as well as repressor adaptor proteins such as CoREST1 and CtBP1 (Fig. 1C). By contrast, we did not observe association with BHC80, a core component of the previously identified CtBP1 and BHC co-repressor complexes, which harbor overlapping subunits, nor with BRAF35, a component specific to the BHC complex (Hakimi et al., 2002; Shi et al., 2004; Shi et al., 2003) (Fig. 1B,C). Together with the fact that previous studies did not uncover SIRT1 as part of the LSD1 and CtBP1 complexes (Shi et al., 2004; Shi et al., 2003), these findings suggest that SIRT1 interacts with a variant form of the previously described LSD1-CoREST1-CtBP1 co-repressor complex.
Figure 1. Identification and characterization of the SIRT1-LSD1-CtBP1 complex.
(A) FLAG-tagged SIRT1 was transiently induced in a Tet-ON HEK293T stable cell line and nuclear extracts prepared. FLAG-SIRT1 and associated proteins were immunoaffinity-purified and eluted with FLAG peptide. Silver-stained SDS-PAGE gels showed that multiple polypeptides specifically associated with FLAG-SIRT1 as compared to control extracts from un-transfected cells.
(B) Tandem mass spectrometry (MS-MS) identified numerous interacting proteins, including multiple components of the LSD1/CtBP1/CoREST1 complexes.
(C) Immunoblotting was used to confirm the presence of core LSD1 complex components in FLAG-SIRT1 immunopurified material.
(D) Analysis of FLAG-SIRT1 purified material by glycerol gradient sedimentation revealed co-sedimentation of LSD1, EHMT2, CoREST1, and CtBP1 (fractions 5 – 9, corresponding to M.W. markers 500-600 kDa). By contrast, another FLAG-SIRT1-interacting protein, DBC1, peaked in fractions 7 – 13.
(E) FLAG-SIRT1 was purified as in (A), eluted under native conditions, followed by a second immunopurification using anti-LSD1 antibodies. Sequentially immunopurified proteins were eluted and analysed by SDS-PAGE/silver staining.
(F) Immunoblotting analysis of sequentially-purified material from (E).
(G) Immunoblot showing co-immunoprecipitation of endogenous LSD1, CoREST1 and CtBP1 with anti-SIRT1 in MEF nuclear extracts. Experiments in panels D – G were repeated three times with similar results.
In accord with the notion that LSD1 and other co-associated factors are indeed part of a SIRT1 co-repressor complex, glycerol gradient sedimentation of FLAG-SIRT1-associated polypeptides revealed that LSD1, EHMT2, CoREST1, and CtBP1 co-purify in a discrete peak with a predicted size of approximately 500-600 kDa (Fig. 1D). By contrast, DBC1, which also abundantly associated with FLAG-SIRT1, exhibited peak sedimentation in later fractions (Fig. 1D). This suggests that FLAG-SIRT1 associates with multiple, biochemically separable complexes. To further characterize the SIRT1-LSD1 complex, we employed sequential immunopurification of SIRT1-bound polypeptides using anti-LSD1 affinity-resin. These studies revealed co-purification of a discrete set of approximately 10 proteins as judged by silver staining (Fig. 1E). Immunoblotting confirmed the presence of LSD1, EHMT2, CoREST1, HDAC1, and CtBP1 in the sequentially purified material (Fig. 1F). Similar results were obtained when the second immunopurification step was performed using anti-CoREST1 resin (Fig. S1A,B). By contrast, sequential purification over anti-DBC1 affinity-resin did not co-purify LSD1 or CoREST1, and DBC1 was not present in the sequential FLAG/LSD1 or FLAG/CoREST1 purifications, in accord with the notion that DBC1 is part of a distinct SIRT1 complex (Fig. S1B and data not shown). Consistent with stable association of SIRT1 core complex components, conventional chromatography of nuclear extract from FLAG-SIRT1-expressing 293T cells over DEAE-Sepharose followed by anti-FLAG immunopurification also demonstrated co-purification of SIRT1 with LSD1 and CoREST1 (Fig. S1C,D). Importantly, immunopurification of endogenous SIRT1 from mouse embryonic fibroblasts (MEFs) revealed co-associated LSD1, CoREST1 and CtBP1 (Fig. 1G). These data together demonstrate that, in mammalian cells, SIRT1 forms a complex with multiple chromatin-directed activities, including histone-modifying enzymes LSD1, HDAC1/2 and EHMT1/2, and the repressor adaptor proteins CoREST1 and CtBP1.
We next asked whether SIRT1 was catalytically active in the context of the LSD1-CoREST1 complex, and whether SIRT1-associated LSD1 retained its histone H3K4me1/2 demethylase (HDM) activity in vitro. To address the first question, we stably transfected T-REx-293-FLAG-SIRT1 cells with a constitutive Myc-CoREST1 expression construct, added tetracycline to transiently induce FLAG-SIRT1 expression, and prepared nuclear extracts. These were then incubated with either anti-c-Myc antibody-coated resin, or control anti-HA resin, and bound Myc-CoREST1 complexes eluted from the washed resin using c-Myc peptide. Immuoblotting of Myc-CoREST1 purified material confirmed the presence of SIRT1 (Fig. S1E). Incubation of the Myc-CoREST1 eluate, but not the control IP material, with bulk histones extracted from butyrate-treated HeLa cells resulted in NAD+-dependent deacetylation of H4K16ac, a residue that is specifically targeted for deacetylation by SIRT1 in interphase cells (Fig. S1F). This indicates that SIRT1 associated with the Myc-CoREST1 complex is indeed catalytically active, at least on the H4K16ac substrate. To determine whether SIRT1-associated LSD1 is also catalytically active, we incubated FLAG-SIRT1 or control, mock-purified material (Fig. 1A) with histone H3 peptides containing monomethylated or dimethylated Lysine 4 (H3K4me1/2). Immunoblot analysis revealed that FLAG-SIRT1-associated LSD1 decreased the levels of both H3K4me1 and H3K4me2 marks, indicating that it is also catalytically active (Fig. S1G,H).
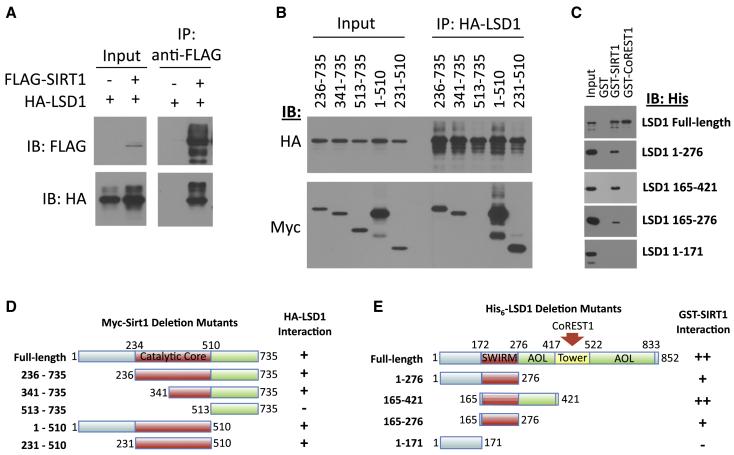
SIRT1 and LSD1 directly interact in vitro
The large number of LSD1 peptides identified in the mass spectrometry analysis relative to other complex components, as well as the strong association of endogenous SIRT1 with LSD1 from MEF nuclear extracts, suggested the possibility that LSD1 might be directly interacting with SIRT1. Indeed, we find that FLAG-SIRT1 strongly associates with HA-LSD1 in co-immunoprecipitation studies (Fig. 2A). Mapping of the SIRT1 sequences required for interaction with LSD1 revealed that the conserved catalytic core of SIRT1 is sufficient for binding LSD1 (Fig. 2B,D). Importantly, recombinant, purified GST-SIRT1 bound to recombinant, purified His6-LSD1, indicating a direct interaction of the two proteins (Fig. 2C). GST-CoREST1 also bound to His6-LSD1, serving as a positive control (Lee et al., 2005; Shi et al., 2004; Shi et al., 2005) (Fig. 2C). Although inclusion of additional C-terminal regions enhances the binding efficiency, LSD1 residues 165-276, corresponding to the LSD1 SWIRM domain, appear to represent a minimal SIRT1-binding domain (Fig. 2C,E). This suggests that in addition to its known function in binding histone H3 tails (Tochio et al., 2006), the highly conserved LSD1 SWIRM domain also plays a role in mediating direct interaction with a distinct histone modifying gene repressor, SIRT1. Furthermore, direct interaction of SIRT1 and LSD1 suggests that their distinct histone-modifying activities may function in a concerted manner to regulate transcription of certain target genes.
Figure 2. Direct interaction between SIRT1 and LSD1.
(A) Full-length FLAG-SIRT1 and HA-LSD1 co-immunopurify from transfected HEK293T cell nuclear extracts. Shown is an immunoblot analysis of FLAG-SIRT1 IP.
(B) The indicated Myc-mSirt1 deletion mutants (D) were examined for co-immunopurification with HA-LSD1 from co-transfected HEK293T cell nuclear extracts. SIRT1 catalytic core region alone (231 – 510) was sufficient, for LSD1 binding.
(C) Purified recombinant GST-SIRT1 interacted directly with purified recombinant His6-LSD1. GST-CoREST1 was used as a positive control and GST alone as a negative control. LSD1 amino acids 165-276, corresponding to the SWIRM domain, were sufficient for interaction with SIRT1.
(D) Schematic representation of the Myc-mSirt1 constructs tested for binding to LSD1 in (B). The minimal LSD1 interaction domain is highlighted in red.
(E) Schematic representation of LSD1 deletion mutants used to define the minimal SIRT1 binding domain (SWIRM, red). All binding assays were repeated at least three times with similar results.
A role for the SIRT1-LSD1 complex in Notch target gene repression
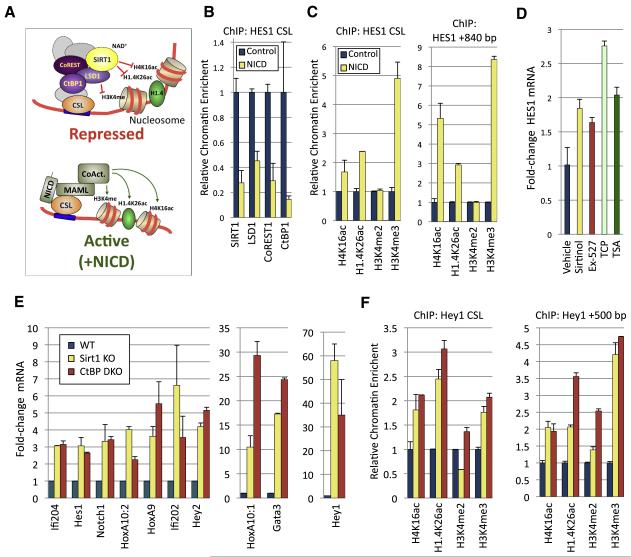
Next, we asked if the identification of SIRT1-binding partners might provide insights into its functions in euchromatic gene repression and biological regulation. Notably, the SIRT1-LSD1 complex contains two proteins, CoREST1 and CtBP1, which act as adaptors that recruit histone-modifying activities to DNA-binding repressors (Chinnadurai, 2007; Lakowski et al., 2006). One such repressor that utilizes CtBP to repress gene transcription and regulate developmental processes is CSL/RBP-Jḳ(Nagel et al., 2005; Oswald et al., 2005), which functions both in repression and activation of target genes in the Notch developmental signaling pathway (Kao et al., 1998; Oswald et al., 2005). Interestingly, LSD1 was also shown to directly bind the promoter of Notch target gene Hey1 in mouse pituitary cells and repress its transcription during embryonic development (Wang et al., 2007). Moreover, we recently found that the Drosophila LSD1 ortholog dLSD1 regulates fly development at least in part through control of Notch target genes (DiStefano et al., 2011). Accordingly, we have hypothesized that the SIRT1-LSD1-CtBP1 co-repressor complex described here might be recruited to Notch target genes to coordinately promote the establishment of a repressive pattern of histone marks and to repress their expression in the absence of Notch signaling. Supporting this hypothesis, chromatin immunoprecipitation (ChIP) analysis revealed that SIRT1, LSD1, CtBP1 and CoREST1 occupy the promoter-proximal CSL binding site at the archetypal Notch target gene HES1, but not the promoter of the DHFR control gene, in human IMR90 cells (Fig. S2A).
The Notch receptor is a transmembrane protein that is cleaved upon ligand binding to liberate the Notch intracellular domain (NICD), which then migrates to the nucleus and forms a complex with CSL, mastermind-like proteins, and co-activators such as CBP/p300 to promote activation of target genes (Fig. 3A) (Borggrefe and Oswald, 2009). It has been proposed that NICD induces dissociation of co-repressor proteins from CSL concomitant with the recruitment of co-activator complexes (Lai, 2002). We therefore asked if NICD binding to CSL might act as a molecular switch that causes displacement of the SIRT1-LSD1 complex from Notch target gene promoters (Fig. 3A). IMR90 cells were transduced with either control or NICD-expressing retroviruses. As expected, NICD expression strongly induced HES1 transcription (Fig. S2B). Strikingly, NICD expression also resulted in markedly diminished levels of SIRT1, LSD1, CoREST1 and CtBP1 at the HES1 promoter-proximal CSL site, as shown by ChIP analysis (Fig. 3B).
Figure 3. Notch target gene regulation by SIRT1-LSD1-CtBP1 complex.
(A) Schematic diagram indicating the proposed role of the SIRT1-LSD1-CtBP1 complex in Notch target gene repression in the absence of Notch Intracellular Domain (NICD), which forms a complex with CSL factors, MAML and co-activators to activate Notch target gene transcription.
(B) Diminished occupancy of SIRT1, LSD1, CoREST1, and CtBP1 at a promoter-proximal CSL site on the HES1 gene in IMR90 human fibroblasts overexpressing the Notch intracellular domain (NICD) or control, as shown by chromatin immunoprecipitation-qPCR.
(C) Expression of NICD resulted in increased levels of histone H4K16ac, H1.4K26ac, and H3K4me3 at both a promoter-proximal CSL site and a downstream region (+840 bp) of the HES1 gene. Samples were normalized to unmodified H3, or in the case of H1.4K26ac, unmodified H1.4.
(D) qRT-PCR analysis showing that HES1 expression was increased in IMR90 cells treated with SIRT1 inhibitors (Sirtinol, Ex-527), the LSD1 inhibitor tranylcypromine (TCP) or the HDAC1/2 inhibitor trichostatin A (TSA). Samples were normalized to 18S RNA and expressed as fold change relative to vehicle control.
(E) Notch target genes were de-repressed in Sirt1 knockout (KO) MEFs and Ctbp1/2 double-knockout (DKO) MEFs. Shown is qRT-PCR analysis of a panel of Notch target genes, normalized to 18S RNA.
(F) ChIP-qPCR analysis of the levels of modified histones H4K16ac, H1.4K26ac, H3K4me2 and H3K4me3 in WT, Sirt1 KO or Ctbp1/2 DKO MEFs at a promoter-proximal CSL site of the Hey1 promoter or 500 bp downstream. Samples were normalized to unmodified H3 or H1.4. In panels B-F, the experiments were repeated three times and mean values are shown; error bars, s.d.
SIRT1 has been shown to deacetylate histone residues H4K16 and H1K26, as well as increasing the deposition of repressive forms of H1 such as H1.4 onto chromatin loci, thereby promoting the assembly of higher-order repressive chromatin states (Vaquero, 2009). Accordingly, in response to NICD expression the levels of the SIRT1-targeted histone modifications H1.4K26ac and H4K16ac increased at the HES1 promoter-proximal CSL binding site and at a region 840 bp downstream, but not at the DHFR promoter (Fig. 3C and Fig. S2C). Additionally, we observed a strong increase in H3K4me3 at the promoter-proximal CSL site after NICD transduction, although levels of H3K4me2 were unaffected (Fig. 3C). In keeping with a role for CtBP1 in recruitment of the SIRT1-LSD1 complex to the HES1 CSL site, we found that shRNA depletion of CtBP1 in IMR90 cells led to de-repression of HES1 expression and decreased occupancy of the promoter-proximal CSL site by SIRT1, LSD1, CoREST1 and CtBP1 (Fig. S2D-F). Importantly, we also observed a concomitant increase in the levels of H4K16ac, H1.4K16ac and H3K4me3 at the CSL site in the CtBP1-knockdown cells (Fig. S2G). Taken together, these observations are consistent with a model where in the absence of Notch signaling the SIRT1-LSD1 core complex components are recruited in a CtBP1-dependent manner to the repressed HES1 promoter to coordinately deacetylate H4K16/H1.4K26 and demethylate H3K4. Upon NICD induction of HES1, their levels are diminished, facilitating H4K14/H1.4K26 acetylation and H4K16 methylation by histone-modifying coactivators to stimulate gene expression.
To further probe the requirement of the SIRT1-LSD1 complex for Notch target gene repression, we next investigated the effect of inhibition or loss-of-function of complex components on Notch target gene expression. Importantly, chemical inhibition of SIRT1 enzymatic activity using two validated compounds, Sirtinol, which was also shown to inhibit SIRT2, and the more specific EX-527, which has an IC50 for SIRT1 that is 200 and 497 times lower than for SIRT2 and SIRT3, respectively (Grozinger et al., 2001; Napper et al., 2005), led to significant, albeit modest, derepression of HES1 in IMR90 cells (Fig. 3D). Similarly, treatment of cells with the non-selective monoamine oxidase inhibitor tranylcypromine (TCP), which irreversibly inhibits the enzymatic activity of LSD1 (Lee et al., 2006a), as well as the class I/II HDAC inhibitor trichostatin A (TSA) (Hoshikawa et al., 1994), resulted in significant HES1 derepression (Fig. 3D). These data indicate that the catalytic activities of multiple chromatin-directed SIRT1 co-repressor complex components are involved in Notch target gene repression. However, we did not observe additive effects of LSD1 and SIRT1 inhibition on HES1 derepression (Fig. S2H,I). Interestingly, lentiviral-mediated RNAi-knockdown of SIRT1 or LSD1 did not lead to derepression of HES1 or several other Notch target genes, including HEY1, HEY2 and NOTCH1, in IMR90 cells (data not shown). A possible explanation for this observation is that chemical inhibition may achieve more robust inhibition of SIRT1, LSD1 and HDAC1/2 than RNAi knockdown in this cell line. In order to circumvent issues of RNAi efficiency, we next examined the expression of a panel of previously described Notch target genes in Sirt1 knockout (KO) mouse embryonic fibroblasts (MEFs). In accord with a critical role for SIRT1 in negative regulation of genes controlled by CSL/Notch, a broad panel of genes were found to be significantly de-repressed in MEFs ablated for Sirt1, including key downstream Notch effector genes such as Hes1, Hey1, and Hey2 (Fig. 3E). Similarly, Ctbp1/2 KO MEFs (Hildebrand and Soriano, 2002) also exhibited derepression of CSL/Notch-regulated genes (Fig. 3E). Collectively, these findings functionally implicate the SIRT1-LSD1 complex in repression of CSL/Notch target genes.
In keeping with an important role of histone deacetylation by SIRT1 in Notch target gene repression, ChIP studies revealed that the SIRT1-regulated H4K16ac and H1.4K26ac histone modifications were markedly increased at the key Notch target gene Hey1 in Sirt1 KO MEFs (Fig. 3F). Furthermore, we observed both increased H4K16ac and H1.4K26ac in Ctbp1/2 KO MEFs, consistent with the notion that Sirt1 represses Hey1 as part of a CtBP1-containing complex. To probe the possibility that CtBP1 is required for coordinated regulation of histone modifications by SIRT1 and LSD1, we examined if LSD1-regulated H3K4 methylation was also aberrant at Hey1 in the Ctbp1/2 KO MEFs. Indeed, we identified significantly elevated levels of H3K4 di- and trimethylation at the Hey1 gene in MEFs lacking Ctbp1/2 (Fig. 3F). Interestingly, H3K4me2/3 levels were also elevated at the Hey1 gene in Sirt1 KO MEFs, albeit to a lesser extent than in Ctbp1/2 KO MEFs (Fig. 3F). By contrast, the levels of H4K16ac, H1.4K26ac, and H3K4me2/3 were largely unaffected at the mouse Actb and Gapdh control promoters in the Sirt1 and Ctbp1/2 KO MEFs (Fig. S2J,K). These results in aggregate suggest that the SIRT1-LSD1-CtBP1 complex acts selectively as a co-repressor of CSL/Notch target genes in the absence of Notch signaling via the establishment of a pattern of histone modifications associated with repressive chromatin states.
Regulation of Notch target genes and development by dSir2 and dLsd1 in Drosophila melanogaster
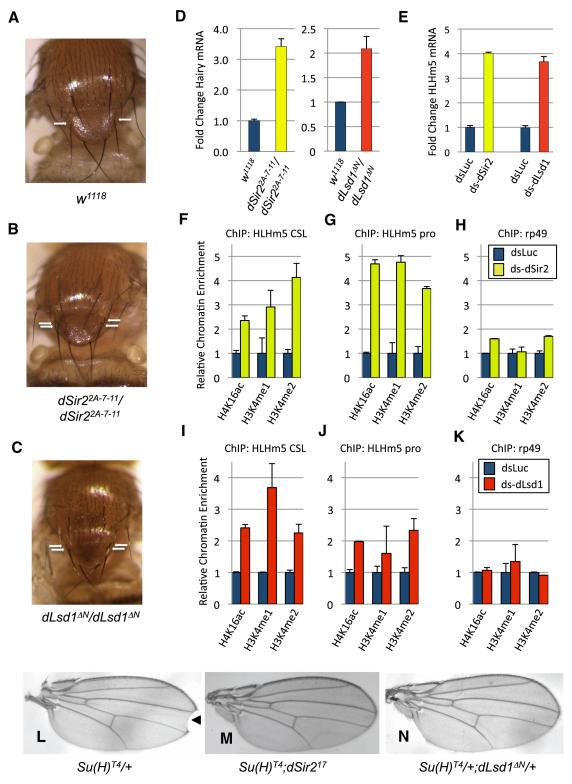
The identification of a broad role of the SIRT1-LSD1-CtBP1 complex in epigenetic regulation and transcriptional repression of Notch target genes in mammalian cells prompted us to ask if the SIRT1 complex also controls Notch-dependent developmental processes in vivo. The genetics of the Notch pathway has been most extensively studied in the fruit fly Drosophila melanogaster (Lai, 2004), and previous studies demonstrated that the Drosophila ortholog of CtBP1/2 (dCtBP) regulates Notch target genes and Notch-dependent developmental processes in flies (Castro et al., 2005; Morel et al., 2001; Nagel et al., 2005). Mutation of the dCtBP gene results in defects in sensory organ precursor cell fate and aberrant mechanosensory bristle development, the most severely affected of which are the anterior scutellar (ASC) bristles on the notum (Stern et al., 2009). Intriguingly, we report here that flies harboring null alleles of the Drosophila SIRT1 ortholog dSir2 (dSir22A-7-11) (Xie and Golic, 2004) or dLsd1 (dLsd1ΔN) (Di Stefano et al., 2007), also share this phenotype (Fig. 4C and Table S1A). In addition, both dSir2 and dLsd1 mutant flies display similar aberrations in posterior cross vein (PCV) patterning on the wing (Fig. S3A,B) (Di Stefano et al., 2011). The similarity of the effects of loss of dSir2 and dLsd1 function on ACS bristle and PCV development, the partial overlap with dCtBP phenotypes, and the fact that dLsd1 and dCtBP modulate Notch signaling in flies (Castro et al., 2005; Morel et al., 2001; Nagel et al., 2005; Di Stefano et al., 2011), suggests that they may together control Notch-dependent developmental processes in vivo as part of a conserved Drosophila dSir2-dLsd1-dCtBP complex. To probe this possibility further we examined the expression of the Notch target gene Hairy, which is aberrantly expressed in dCtBP mutant flies (Poortinga et al., 1998). Significantly, qRT-PCR analysis revealed that Hairy was de-repressed in both dSir22A-7-11 and dLsd1ΔN flies, consistent with our hypothesis (Fig. 4D).
Figure 4. Role of dSir2 and dLsd1 in Notch-regulated Drosophila development.
(A) and (B,) Homozygous-null dSir2 (dSir2 null) Drosophila melanogaster exhibit supernumerary anterior scutellar (ASC) bristles on the notum (B), as compared to wild-type flies (A).
(C) Similarly, flies homozygous for the dLsd1 loss-of-function dLsd1 _Δ_N allele also exhibit this phenotype. Shown are photographs of wild-type, dSir2 null and dLsd1 null fly nota, with the position of the ASC bristles indicated by arrows.
(D) The Notch target gene Hairy is upregulated in dSir22A-7-11 and dLsd1 _Δ_N flies, as determined by qRT-PCR analysis. Samples were normalized to control gene rp49.
(E) Double-strand RNA (ds)-mediated knockdown of either dSir2 or dLsd1 resulted in up-regulation of the Notch target gene HLHm5 in the D. melanogaster cell line S2, as determined by qRT-PCR analysis. dsRNA targeting luciferase (dsLuc) was used as a negative control. Samples were normalized to control gene rp49.
(F - H) ChIP analysis revealed that the levels of H4K16ac, H3K4me1 and H3K4me2 were increased at a CSL site and distal promoter region in the HLHm5 promoter in ds _dSir2_-treated S2 cells. As a control, the levels of these histone modifications were also analyzed at the control gene rp49. Samples were analyzed by qPCR and normalized to unmodified histone H3 ChIP signal.
(I - K) Similarly, the levels of H3K4me1, H3K4me2 and H4K16ac were increased at both genomic loci in the HLHm5 promoter, but not at the rp49 gene, in S2 cells treated with ds dLsd1. Samples were analyzed by qPCR and normalized to unmodified histone H3 ChIP signal.
(L) Flies heterozygous for the Suppressor of Hairless allele Su(H)T4 mutant allele exhibit wing margin defects at the wing tip (notches), indicated by arrowhead.
(M) When Su(H)T4 mutant flies were crossed onto the dSir217 mutant background, wing notching was substantially suppressed.
(N) Similar suppression was observed when Su(H)T4 mutant flies were crossed onto the dLsd1 _Δ_N mutant background. Shown are representative photographs of wings from flies of each genetic background. In panels (D - K), the experiments were repeated three times and mean values are shown; error bars, s.d.
Next, we investigated if dSir2 and dLsd1 repress Drosophila Notch target genes via regulation of histone modifications, as was observed for mammalian cells (Fig. 3). We found that RNAi-mediated depletion of dSir2 in Drosophila S2 cells induced derepression of several genes on the Enhancer of Split (E(Spl)) locus, which is regulated by CSL-dependent Notch signaling, including HLHm5 (Lecourtois and Schweisguth, 1995; Di Stefano et al., 2011) (Fig. 4E and Fig. S3C,D). Importantly, S2 cells do not express endogenous Notch receptor (Lake et al., 2009), suggesting that upregulation of CSL target genes upon dSir2 RNAi in these cells is due to loss of CSL repressor function and independent of its role as a Notch-dependent activator. Similar results were also observed with dLsd1 RNAi in S2 cells (Fig. 4E and Di Stefano et al., 2011). Individual roles for the histone-modifying activities of dSir2 and dLsd1 in mediating HLHm5 repression is indicated by observations that the levels of H4K16ac and H3K4me1/2 at the CSL binding site and promoter of the HLHm5 gene were elevated in response to dSir2 and dLsd1 RNAi, respectively but not at the unrelated gene rp49 (Fig. 4F-H). Interestingly, there also appears to be cross-talk between dSir2 and dLsd1 in regulating histone modifications, as RNAi knockdown of dSir2 increased the levels of both H3K4me1 and H3K4me2 at the HLHm5 CSL site and promoter (Fig. 4F-H), whereas knockdown of dLsd1 increased H4K16ac levels at both sites (Fig. 4I-K). This suggests a coordinated role of dSir2 and dLsd1 in regulating histone modifications at Notch target genes such as HLHm5; this concerted function may be conserved, as methylation of H3K4me was also increased at the Hey1 promoter in Sirt1 KO MEFs (Fig. 3F).
To further investigate the role of dSir2 and dLsd1 in regulation of Notch-dependent development in Drosophila, we asked whether dSir2 and dLsd1 mutant alleles genetically interact with a mutant allele of Suppressor of Hairless (Su(H)), the fly CSL DNA-binding factor mediating canonical Notch signaling (Fortini and Artavanis-Tsakonas, 1994). Strikingly, the wing notching phenotype characteristic of flies heterozygous for the Su(H) mutant allele Su(H)T4 (Fortini and Artavanis-Tsakonas, 1994), was substantially suppressed when crossed onto either dSir2 or dLsd1 mutant backgrounds (Fig. 4I-K and Table S1B,C), indicating genetic interaction. Collectively, these findings are consistent with a model where a dSir2-dLsd1-containing complex represses CSL/Notch target genes via coordinated regulation of H4K16 acetylation and H3K4 methylation, thereby modulating at least a subset of Notch-dependent developmental processes in Drosophila.
Discussion
The identification of a multi-functional SIRT1-LSD1 co-repressor complex has revealed an unanticipated coordination of SIRT1 with distinct histone-modifying activities in epigenetic regulation of Notch target gene expression. Previous studies have indicated that HDAC1 and HDAC2 also play a role in Notch target gene repression (Kao et al., 1998), however, these enzymes do not target the H4K16ac mark, a histone modification that is sufficient to inhibit chromatin compaction and is strongly linked to gene activation (Shogren-Knaak et al., 2006; Vaquero et al., 2007b), and which is increased at the Notch target gene HES1 upon NICD induction (Fig. 3C). Thus, the recruitment of SIRT1, the major interphase H4K16 deacetylase, appears to play a critical role in repression of Notch target genes. A key role for LSD1 in Notch target gene repression is also indicated, as its inhibition led to derepression of HES1 in IMR90 cells and mutation of dLsd1 derepressed Notch target genes such as Hairy and several genes in the Enhancer of Split (E(Spl)) locus in flies and in S2 cells (Fig. 4D,E) (Di Stefano et al., 2011). Furthermore, H3K4 dimethylation, which is mechanistically linked to gene activation and modified by LSD1 (Ruthenburg et al., 2007), was increased at the promoters of Hey1 in Ctbp1/2 KO MEFs and HLHm5 and other Enhancer of Split (E(Spl)) genes in S2 cells treated with dsRNA targeting dLsd1 (Fig. 3F, Fig. 4H) (Di Stefano et al., 2011). Both H4K16ac and H1.4K26ac were also increased at the Hey1 promoter in Sirt1 KO and Ctbp1/2 KO MEFs (Fig. 3F), and H4K16ac and H3K4me1/2 were elevated at the HLHm5 promoter in both ds-_dSir2_- and ds-_dLsd1_-treated S2 cells (Fig. 4F-K), consistent with concerted regulation of these genes by the histone-modifying activities of a SIRT1-LSD1-containing complex. The similarities of the ACS bristle and wing vein phenotypes of dSir2 and dLsd1 mutant flies, and their genetic suppression of Su(H)T4 wing notching phenotype further supports this model (Fig. 4A-C, L-N).
Previous studies have also identified co-repressors that may mediate epigenetic repression of Notch target genes in the absence of Notch signaling (Borggrefe and Oswald, 2009; Liefke et al., 2010; Moshkin et al., 2009). Notably, these include the H3K4 demethylase Lid/KDM5A (Liefke et al. 2010; Moshkin et al., 2009; Di Stefano et al., 2011). KDM5A removes di- and tri-, but not monomethyl marks from H3K4, whereas LSD1 removes H3K4 mono- and dimethyl marks (Shi et al., 2004), suggesting that the two enzymes may act in concert to convert H3K4me3 to the unmethylated state in a step-wise manner. This notion is consistent with our data showing elevated levels of both H3K4 di- and trimethylation at the Hey1 gene in MEFs lacking Ctbp1/2 (Fig. 3F). However, it is also possible that alternate corepressors are used to repress Notch target genes in distinct developmental stages, cell types or promoter contexts. In addition to regulating developmental processes in flies, Notch also plays a central role in mammalian development, and aberrant Notch signaling has been implicated in a number of cancers (Aster et al., 2008; Dotto, 2008). Thus, dysregulation of the mammalian SIRT1-LSD1 complex might also contribute to developmental abnormalities and Notch-dependent malignancies in humans. Furthermore, while the presence of CtBP1 in the complex led us to uncover a role in Notch target gene regulation, we speculate that the SIRT1-LSD1 complex may also facilitate epigenetic repression of other CtBP1 target genes via repressors such as the ZEB-family candidate tumor suppressors (Vandewalle et al., 2009). This possibility is particularly enticing, as previous studies have independently linked both LSD1 and SIRT1 to repression of the ZEB target gene E-cadherin/CDH1, itself a tumor suppressor (Lin et al., 2010; Pruitt et al., 2006; Shi et al., 2003). Likewise, the SIRT1-LSD1 complex may also regulate neuronal genes targeted by the repressor REST/NRSF, which uses the corepressor CoREST1 to repress specific subsets of target genes (Lakowski et al., 2006). This may have important implications for the emerging role of SIRT1 as a regulator of neuronal differentiation and function (Gao et al., 2010; Kim et al., 2007; Michan et al., 2010; Prozorovski et al., 2008). Significantly, a recent genomewide chromatin mapping study revealed widespread co-localization of dSir2, dCtBP and the HDAC1 ortholog dRpd3 at many euchromatic loci in fly cells (Filion et al. 2010). Along with our data showing physical association of the human orthologs of these proteins, this suggests conserved, widespread and concerted roles of SIRT1 orthologs, LSD1 and associated polypeptides in epigenetic regulation at euchromatic sites in metazoans.
Experimental Procedures
Cell culture
T-REx 293 cells (Invitrogen), HEK293T cells (ATCC), and mouse embryonic fibroblasts (MEFs) were maintained in DMEM supplemented with 5% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin/mL (all from Gibco). Wild-type, Sirt1 KO and Ctbp1/2 KO MEFs are described elsewhere (Cheng et al., 2003; Grooteclaes et al., 2003). IMR90 fibroblasts (ATCC) were maintained at low passage in MEM supplemented with 10% FBS, 1 mM sodium pyruvate, 100 U/mL penicillin and 100 μg/mL streptomycin/mL. Chemical inhibitor studies were performed by treating cells for 16h with 25 μM Sirtinol (Sigma) in ethanol vehicle, 1 - 10 μM EX-527 (Tocris) in DMSO, 25 - 200 μM tranylcypromine (TCP) (Sigma) in DMSO, or 1 μM trichostatin A (TSA) (Sigma) in DMSO. Paired vehicle controls were performed for each drug treatment. Drosophila S2 cells were maintained in Schneider’s medium (Sigma) supplemented with 5% FBS, 100 U/mL penicillin and 100 μg/mL streptomycin/mL. dsRNA knock-down was performed as previously described (Stevaux et al., 2002), using 50 μg dsRNA per 4 × 106 cells per well in 6-well plates. Cells treated with ds-dSir2 were harvested 3 days later, while cells treated with ds-dLsd1 were transfected again four days after the initial transfection, and samples harvested 3 days later.
Identification of the SIRT1-LSD1 complex by immunopurification
Tetracycline-inducible T-REx 293 cells (Invitrogen) were stably transfected with pcDNA4TO-FLAG plasmid containing full-length human SIRT1 cDNA. FLAG-SIRT1 expression was induced with tetracycline (0.2 μg/mL) for 40h and nuclear extracts prepared as previously described (Dignam et al., 1983). Mock-transfected cells were used as a negative control. Nuclear extracts were incubated with anti-FLAG-M2 agarose beads (Sigma) for 4h at 4°C and washed five times with Buffer A (20 mM Tris-HCl (pH 8.0), 10 μM ZnCl2, 10% glycerol, 0.1% NP-40, 1 mM DTT, 1 mM benzamidine, 0.25 mM PMSF, and 2 μg/ml aprotinin) containing 250 mM KCl, followed by two washes with 150 mM KCl Buffer A. FLAG-SIRT1 was eluted with 150 mM KCl Buffer A containing 0.2 mg/mL FLAG peptide (Sigma) and 200 mM Tris-HCl (pH 8.0), and associated polypeptides identified by SDS-PAGE and tandem mass spectrometry as previously described (Nakatani and Ogryzko, 2003).
Glycerol Gradient Sedimentation
FLAG-SIRT1 immunopurified material was analyzed as previously described (Naar et al., 2002), using 2 mL, 15 – 40% glycerol gradients in Buffer A containing 100 mM KCl, and centrifuged for 7 h at 55,000 r.p.m. in a TLS55 rotor (Beckmann). Fractions (100 μL) were collected from the top of the gradient, resolved by SDS-PAGE and analyzed by silver staining or immunoblotting.
Co-immunoprecipitation analysis
FLAG-SIRT1-immunopurified material eluted with FLAG peptide was incubated for 4 h at 4°C with Protein A/G beads (GE Healthcare) pre-coated with anti-LSD1, anti-CoREST1, anti-DBC1 or control IgG (all from Bethyl Laboratories). Beads were washed five times with Buffer A containing 200 mM KCl and 0.05% NP-40, and bound proteins were eluted in Buffer A containing 100 mM KCl and 0.3% Sarkosyl for 1h at 4°C, or by incubating at 65°C for 15 minutes in SDS-PAGE gel loading buffer. For endogenous Co-IP assays, nuclear extracts were prepared as previously described (Dignam et al., 1983), supplemented with 0.05% NP-40 and 10 μM ZnCl2 and pre-cleared with Protein A/G sepharose beads for 1h at 4°C. Pre-cleared nuclear extracts (1 mg) were then incubated with 5 μg of anti-SIRT1 (Cyclex), anti-LSD1 (Sigma) or control IgG overnight at 4°C. Immune complexes were collected by incubating with Protein A/G sepharose beads for 1h at 4°C and washed five times with Buffer A supplemented with 250 mM KCl followed by two washes with 100 mM KCl Buffer A. Bound proteins were eluted in SDS-PAGE gel loading buffer and analyzed by immunoblotting.
Protein interaction and domain mapping analysis
HEK293T cells were transiently co-transfected with pcDNA3-HA-LSD1 and either pcDNA3-FLAG-SIRT1 or pCS2-Myc-Sirt1 deletion mutants (gift of Vittorio Sartorelli). Nuclear extracts were prepared as previously described (Dignam et al., 1983), and immunoprecipitations performed using either FLAG-M2-agarose (Sigma) or Protein G sepharose (GE Healthcare) pre-coated with anti-HA antibody (Covance). Beads were washed five times in 150 mM KCl Buffer A, and bound proteins eluted in SDS-PAGE loading buffer and analyzed by immunoblotting. Recombinant full-length GST-SIRT1 and GST-CoREST1 were purified as previously described (Ouyang et al., 2009; Walker et al., 2010). LSD1 fragments were cloned into Gateway His6-tagging bacterial expression vectors (Invitrogen). Full-length His6-LSD1 and domain-mapping mutants were expressed in E. coli DE3-Rosetta bacteria (Novagen) and purified using HisPur Cobalt Resin (Pierce). Binding reactions containing 2 μg of GST or GST-fusion protein and 2 μg His6-LSD1 full-length or 1 μg of His6-LSD1 domain-mapping mutants were incubated at 4°C for 4 h in GST pull-down buffer (20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP-40, 10 mM ZnCl2, 1 mM DTT, 10% glycerol), centrifuged at 12000 r.c.f. for 15 min, and the supernatant incubated glutathione sepharose-4B (GE Healthcare) for 1 h. Beads were washed 5 times with GST pull-down buffer and bound proteins were eluted with SDS-PAGE loading buffer and analyzed by immunoblotting.
RT-PCR and Chromatin Immunoprecipitation
RNA was extracted using Trizol reagent (Invitrogen) and quantitative RT-PCR performed using the Lightcycler 480 (Roche). RT-PCR primer pairs are listed in Supplemental Information. Mammalian cell line ChIP was performed as previously described (Lee et al., 2006b), except that cross-linked nuclei were sonicated to 250 – 1000 bp fragments in buffer containing 1% SDS, 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1 mM PMSF and Complete protease inhibitors (Roche), and bound ChIP complexes washed according to the Upstate/Millipore protocol, as previously described (Mulligan et al., 2008). Drosophila S2 cell ChIP was performed as described previously, using micrococcal nuclease digestion to prepare chromatin fragments (Di Stefano et al., 2011). Antibodies and ChIP qPCR primer pairs used are listed in Supplemental Information.
dSir2 and dLsd1 genetic analyses
The w1118, dSir217, and dSir22A-7-11 fly stocks were obtained from the Bloomington stock collection, Dr. Stephen Helfand, and Dr. Kent Golic, respectively. The dLsd1ΔN allele was described previously (Di Stefano et al., 2007). Su(H)T4 allele was provided by the laboratory of Dr. Spyros Artavanis-Tsakonas. Flies were grown on standard Drosophila medium and maintained at 25°C. The effects on wing development were studied by crossing females heterozygous for the Su(H)T4 alleles with dLsd1 and dSir2 mutant males. The effect was quantified by counting the number of double heterozygous female flies that exhibit notched wings.
Supplementary Material
01
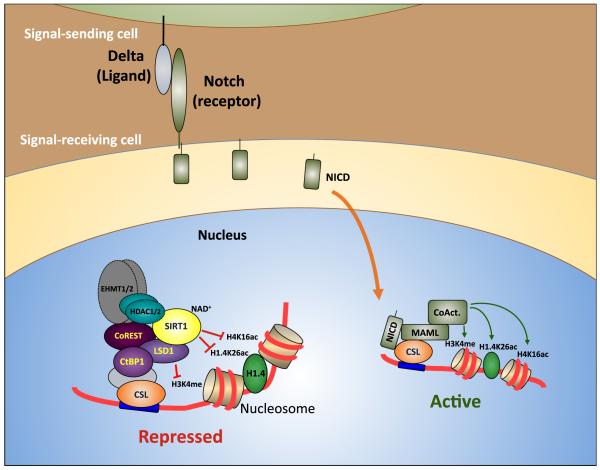
Figure 5. Model of regulation of Notch target genes by the SIRT1-LSD1 complex.
In the absence of Notch signaling, Notch target genes are maintained in a repressed state by the action of a co-repressor complex containing SIRT1, LSD1, CoREST1 and CtBP1 recruited via CSL DNA-binding factors. The enzymatic activity of SIRT1 removes H4K16ac and H1.4K26ac marks from chromatin, while LSD1 antagonizes H3K4 methylation. This helps to create a local chromatin environment repressive to transcription. Induction of Notch signaling by ligands such as Delta on adjacent cells leads to proteolytic cleavage of the Notch intracellular domain (NICD), which is then free to migrate to the nucleus and forms a transcriptional activator complex along with CSL, Mastermind-like (MAML) and co-activators including the histone lysine acetyltransferases p300/CBP. Concomitantly, the SIRT1-LSD1 complex becomes depleted at CSL binding sites. The combined effect of reduced levels of transcriptionally repressive histone-modifying enzymes, including SIRT1 and LSD1, and enrichment of activating histone-modifying enzymes such as p300/CBP contributes to the generation of a local chromatin environment conducive to Notch target gene activation.
Acknowledgments
We thank Amy Walker for critical reading of the manuscript and Luhan Yang for valuable assistance and discussion. We express our gratitude to the groups of Vittorio Sartorelli and Mark Leid for sharing with us SIRT1 deletion constructs, Stephen Helfand and Kent Golic for dSir2 mutant flies, Spyros Artavanis-Tsakonas for Notch and Su(H) mutant flies, the Bloomington Drosophila Stock Center for various fly strains, Jon C. Aster for providing the pMigRI-ICN1 plasmid, John R. Whetstine for kind assistance in performing LSD1 demethylase assays, and members of the MGH Cancer Center and Artavanis-Tsakonas’ lab for helpful discussions. These studies were supported by NIH grants R01GM071449 (A.M.N.), R01GM53203 (N.J.D.), and R01GM077689 (G.G.), R01GM093072-01 (R.M.), R01DK088190-01A1 (R.M.), the MGH-AstraZeneca Strategic Alliance (A.M.N.), the Sidney Kimmel Cancer Research Foundation (R.M.), a New Investigator Grant from the Massachusetts Life Sciences Center (R.M.) and an AFAR Research Grant (R.M.). P.M. was supported by a fellowship from the MGH ECOR Fund for Medical Discovery, L.D.S. was supported by a Leukemia and Lymphoma Society fellowship and D.T. was supported by the Brain Power for Israel Fund.
References
- Aster JC, Pear WS, Blacklow SC. Notch signaling in leukemia. Annu Rev Pathol. 2008;3:587–613. doi: 10.1146/annurev.pathmechdis.3.121806.154300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F. The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci. 2009;66:1631–1646. doi: 10.1007/s00018-009-8668-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro B, Barolo S, Bailey AM, Posakony JW. Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development. 2005;132:3333–3344. doi: 10.1242/dev.01920. [DOI] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnadurai G. Transcriptional regulation by C-terminal binding proteins. Int J Biochem Cell Biol. 2007;39:1593–1607. doi: 10.1016/j.biocel.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Deng CX. SIRT1, is it a tumor promoter or tumor suppressor? Int J Biol Sci. 2009;5:147–152. doi: 10.7150/ijbs.5.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L, Ji JY, Moon NS, Herr A, Dyson N. Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development. Curr Biol. 2007;17:808–812. doi: 10.1016/j.cub.2007.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano L, Walker JA, Burgio G, Corona DF, Mulligan P, Näär AM, Dyson NJ. Functional antagonism between histone H3K4 demethylases in vivo. Genes Dev. 2011;25:17–28. doi: 10.1101/gad.1983711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotto GP. Notch tumor suppressor function. Oncogene. 2008;27:5115–5123. doi: 10.1038/onc.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, van Steensel B. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini ME, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Banerjee R, Breen TR, Harte PJ. SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr Biol. 2004;14:1812–1821. doi: 10.1016/j.cub.2004.09.060. [DOI] [PubMed] [Google Scholar]
- Gao J, Wang WY, Mao YW, Graff J, Guan JS, Pan L, Mak G, Kim D, Su SC, Tsai LH. A novel pathway regulates memory and plasticity via SIRT1 and miR-134. Nature. 2010;466:1105–1109. doi: 10.1038/nature09271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grooteclaes M, Deveraux Q, Hildebrand J, Zhang Q, Goodman RH, Frisch SM. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc Natl Acad Sci U S A. 2003;100:4568–4573. doi: 10.1073/pnas.0830998100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinger CM, Chao ED, Blackwell HE, Moazed D, Schreiber SL. Identification of a class of small molecule inhibitors of the sirtuin family of NAD-dependent deacetylases by phenotypic screening. J Biol Chem. 2001;276:38837–38843. doi: 10.1074/jbc.M106779200. [DOI] [PubMed] [Google Scholar]
- Hakimi MA, Bochar DA, Chenoweth J, Lane WS, Mandel G, Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc Natl Acad Sci U S A. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand JD, Soriano P. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol Cell Biol. 2002;22:5296–5307. doi: 10.1128/MCB.22.15.5296-5307.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshikawa Y, Kwon HJ, Yoshida M, Horinouchi S, Beppu T. Trichostatin A induces morphological changes and gelsolin expression by inhibiting histone deacetylase in human carcinoma cell lines. Exp Cell Res. 1994;214:189–197. doi: 10.1006/excr.1994.1248. [DOI] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Nguyen MD, Dobbin MM, Fischer A, Sananbenesi F, Rodgers JT, Delalle I, Baur JA, Sui G, Armour SM, et al. SIRT1 deacetylase protects against neurodegeneration in models for Alzheimer’s disease and amyotrophic lateral sclerosis. EMBO J. 2007;26:3169–3179. doi: 10.1038/sj.emboj.7601758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002;3:840–845. doi: 10.1093/embo-reports/kvf170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Lake RJ, Grimm LM, Veraksa A, Banos A, Artavanis-Tsakonas S. In vivo analysis of the Notch receptor S1 cleavage. PLoS One. 2009;4:e6728. doi: 10.1371/journal.pone.0006728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B, Roelens I, Jacob S. CoREST-like complexes regulate chromatin modification and neuronal gene expression. J Mol Neurosci. 2006;29:227–239. doi: 10.1385/JMN:29:3:227. [DOI] [PubMed] [Google Scholar]
- Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006a;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006b;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefke R, Oswald F, Alvarado C, Ferres-Marco D, Mittler G, Rodriguez P, Dominguez M, Borggrefe T. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24:590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T, Ponn A, Hu X, Law BK, Lu J. Requirement of the histone demethylase LSD1 in Snai1-mediated transcriptional repression during epithelial-mesenchymal transition. Oncogene. 2010;29:4896–4904. doi: 10.1038/onc.2010.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michan S, Li Y, Chou MM, Parrella E, Ge H, Long JM, Allard JS, Lewis K, Miller M, Xu W, et al. SIRT1 is essential for normal cognitive function and synaptic plasticity. J Neurosci. 2010;30:9695–9707. doi: 10.1523/JNEUROSCI.0027-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel V, Lecourtois M, Massiani O, Maier D, Preiss A, Schweisguth F. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol. 2001;11:789–792. doi: 10.1016/s0960-9822(01)00224-x. [DOI] [PubMed] [Google Scholar]
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35:782–793. doi: 10.1016/j.molcel.2009.07.020. [DOI] [PubMed] [Google Scholar]
- Mulligan P, Westbrook TF, Ottinger M, Pavlova N, Chang B, Macia E, Shi YJ, Barretina J, Liu J, Howley PM, et al. CDYL bridges REST and histone methyltransferases for gene repression and suppression of cellular transformation. Mol Cell. 2008;32:718–726. doi: 10.1016/j.molcel.2008.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 2002;16:1339–1344. doi: 10.1101/gad.987602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel AC, Krejci A, Tenin G, Bravo-Patino A, Bray S, Maier D, Preiss A. Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol Cell Biol. 2005;25:10433–10441. doi: 10.1128/MCB.25.23.10433-10441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Napper AD, Hixon J, McDonagh T, Keavey K, Pons JF, Barker J, Yau WT, Amouzegh P, Flegg A, Hamelin E, et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang J, Shi Y, Valin A, Xuan Y, Gill G. Direct binding of CoREST1 to SUMO-2/3 contributes to gene-specific repression by the LSD1/CoREST1/HDAC complex. Mol Cell. 2009;34:145–154. doi: 10.1016/j.molcel.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, Siegert E, Bendix I, Brustle O, Nitsch R, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Shi Y, Sawada J, Sui G, Affar el B, Whetstine JR, Lan F, Ogawa H, Luke MP, Nakatani Y. Coordinated histone modifications mediated by a CtBP co-repressor complex. Nature. 2003;422:735–738. doi: 10.1038/nature01550. [DOI] [PubMed] [Google Scholar]
- Shi YJ, Matson C, Lan F, Iwase S, Baba T, Shi Y. Regulation of LSD1 histone demethylase activity by its associated factors. Mol Cell. 2005;19:857–864. doi: 10.1016/j.molcel.2005.08.027. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Stern MD, Aihara H, Roccaro GA, Cheung L, Zhang H, Negeri D, Nibu Y. CtBP is required for proper development of peripheral nervous system in Drosophila. Mech Dev. 2009;126:68–79. doi: 10.1016/j.mod.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Stevaux O, Dimova D, Frolov MV, Taylor-Harding B, Morris E, Dyson N. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. EMBO J. 2002;21:4927–4937. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tochio N, Umehara T, Koshiba S, Inoue M, Yabuki T, Aoki M, Seki E, Watanabe S, Tomo Y, Hanada M, et al. Solution structure of the SWIRM domain of human histone demethylase LSD1. Structure. 2006;14:457–468. doi: 10.1016/j.str.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Vandewalle C, Van Roy F, Berx G. The role of the ZEB family of transcription factors in development and disease. Cell Mol Life Sci. 2009;66:773–787. doi: 10.1007/s00018-008-8465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A. The conserved role of sirtuins in chromatin regulation. Int J Dev Biol. 2009;53:303–322. doi: 10.1387/ijdb.082675av. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007a;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007b;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, Boss O, Hirsch ML, Ribich S, Smith JJ, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Scully K, Zhu X, Cai L, Zhang J, Prefontaine GG, Krones A, Ohgi KA, Zhu P, Garcia-Bassets I, et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature. 2007;446:882–887. doi: 10.1038/nature05671. [DOI] [PubMed] [Google Scholar]
- Xie HB, Golic KG. Gene deletions by ends-in targeting in Drosophila melanogaster. Genetics. 2004;168:1477–1489. doi: 10.1534/genetics.104.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat Rev Mol Cell Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01