Reconstruction of Biochemical Networks in Microbial Organisms (original) (raw)
. Author manuscript; available in PMC: 2011 Jun 21.
Published in final edited form as: Nat Rev Microbiol. 2008 Dec 31;7(2):129–143. doi: 10.1038/nrmicro1949
Abstract
Systems analysis of metabolic and growth functions in microbial organisms is rapidly developing and maturing. Such studies are enabled by the reconstruction, at the genomic-scale, of the biochemical reaction networks that underlie cellular processes. The network reconstruction process is organism-specific and is based on an annotated genome sequence, high-throughput network-wide data sets, and bibliomic data on the detailed properties of individual network components. This review describes the details of the process that is currently implemented to achieve comprehensive network reconstructions and how they are curated and validated. The reconstruction process for genome-scale metabolic networks is well developed, while the process for the reconstruction of transcriptional regulation and for transcriptional / translational processes at the genome-scale is now developing. This review should accelerate the progress of the growing number of researchers that are carrying out reconstructions for particular target organisms.
Additional Glossary Terms (if desired): Automated Reconstruction, Manual Curation, Computational Validation, Constraint-based Methods (COBRA)
Introduction
Reconstructed networks of biochemical reactions are at the core of systems analysis of cellular processes. They form a common denominator for both experimental data analysis and computational studies in systems biology. The conceptual basis for the reconstruction process has been outlined2, and computational methods and tools used to characterize them have been reviewed3, 4. Furthermore, the number of available well-curated organism-specific network reconstructions is growing (see Supplementary Table 1) and the spectrum of their uses is broadening5.
This review describes the detailed workflows that form the basis of the reconstruction process and provide key procedural information needed for the growing number of researchers performing organism-specific reconstructions. We describe the procedures in which various experimental data types are integrated to reconstruct biochemical networks, the current status of network reconstructions, and how network reconstructions can be used in a prospective manner to discover new interactions and pathways. We will focus on the networks that underlie three key cellular processes: i) metabolism, ii) transcription/translation, and iii) transcriptional regulation. In addition, we will briefly discuss the impact of network content on modeling, integration of these types of networks, as well as the prospects of reconstructing other types of networks such as signaling and small RNA pathways.
Metabolic networks
Before annotated genomic sequences were available, primary literature and biochemical characterization of enzymes provided the major source of information for reconstructing metabolic networks in a select number of organisms. Accordingly, some of the earliest metabolic reconstructions that were subsequently used in modeling applications were for Clostridium acetobutylicum6, Bacillus subtilis7 and Escherichia coli8–11.
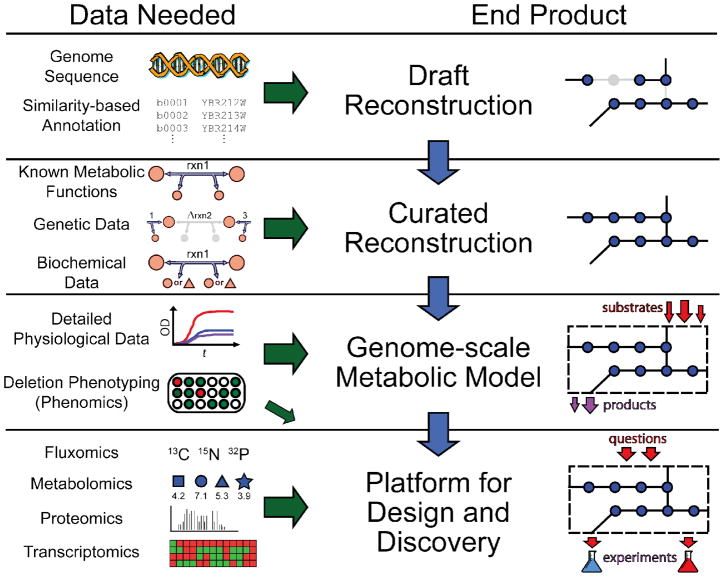
Today, with the ability to sequence and annotate whole genomes, we can generate metabolic network reconstructions at a genomic scale, even for organisms for which little direct biochemical information is available in the published literature. To implement the metabolic reconstruction process, we need to answer the following questions for each of the enzymes in a metabolic network: i) what substrates and products does an enzyme act on, ii) what are the stoichiometric coefficients for each metabolite participating in the reaction(s) catalyzed by an enzyme, iii) are the outlined reactions reversible, and iv) where does the reaction occur in the cell (e.g., cytoplasm, periplasm, etc.)? This data comes from a variety of sources. The establishment of a set of the chemical reactions that comprise a reaction network culminates in a database of proper chemical equations. Each reaction also has additional information associated, such as its cellular localization, thermodynamics, and genetic/genomic information. The genome-scale metabolic network reconstruction process is comprised of four fundamental steps (see Figure 1).
Figure 1. The phases and data utilized for generating a metabolic reconstruction.
Genome-scale metabolic reconstruction can be summarized in four major phases, each of the latter phases building off the previous. Also characteristic of the reconstruction process is the iterative refinement of reconstruction content that is driven by experimental data and occurs in the three latter phases. For each phase, specific data types are necessary and these range from high-throughput data types (e.g., phenomics, metabolimics, etc.), to detailed studies characterizing individual components (e.g., biochemical data for a particular reaction). For example, the genome annotation can provide a parts list of a cell, whereas genetic data can provide information about the contribution of each gene product towards a phenotype (e.g., when removed or mutated). The product generated from each reconstruction phase can be utilized and applied to examine a growing number of questions with the final product having the broadest applications.
Step 1: Automated genome-based reconstruction
The starting point for reconstructions is the annotated genome for a particular target organism and strain (Box 1). Genome annotations can be found in organism-specific databases, such as EcoCyc12 for E. coli and SGD13 or CYGD14 for Saccharomyces cerevisiae, or in databases with collections of genome annotations, such as EntrezGene15, Comprehensive Microbial Resource (CMR)16, Genome Reviews (through EBI)17 or the Integrated Microbial Genomes (IMG)18. The genome annotation provides unique identifiers for the reconstruction content and a list of the metabolic enzymes thought to be present in the target organism and can indicate how the gene products interact (as subunits, protein complexes or isozymes) to form active enzymes that catalyze metabolic reactions. The next step in the reconstruction process is to determine which biochemical reactions these enzymes carry out, and this can be determined manually or by using automated tools.
Box 1. Detailed process of reconstruction, validation and utilization of a metabolic reconstruction.
The process of metabolic reconstruction can be performed in a sequential fashion. The process is initiated by obtaining the genetic content (i.e., a parts list of the cell) from the genome annotation. Upon this scaffold, active enzymes are associated to the genetic content through utilizing information from databases and published literature. Further, the metabolic reactions that these enzymes catalyze are then delineated with the ultimate generation of a gene to protein to reaction association (GPR). Automated reconstruction tools are available to aid in this process and several databases posses the necessary information for each data type (see text for references).
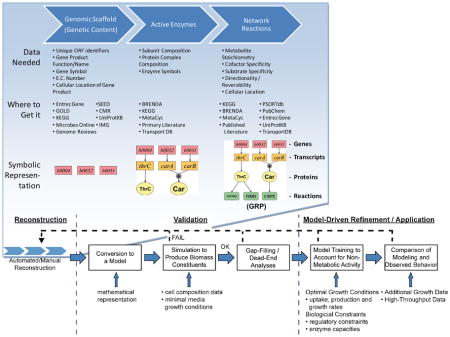
Following the initial reconstruction process, a reconstruction is converted to a model in a mathematical format that can be used for computation. Further in the validation phase, the ability of the organism to produce biomass constituents / grow is examined using a biomass objective function (Box 3). This analysis functionally tests the reconstruction for an experimentally observed phenomenon. A dead-end analysis should follow to examine reactions on a pathway basis for their physiological role and computational algorithms are available to aid in this process (see text).
For predictions of physiological behavior, a training data set is needed to examine non-metabolic energy needs and organism-specific components (e.g., the electron transport system). In this phase, additional known key network properties can be applied beyond the metabolic functions outlined in the reconstruction (e.g., key regulatory interactions under a given condition) to improve predictive capabilities. For prospective use, high- and low-throughput data can also be compared to modeling simulations to validate the content and make predictions or to find specific areas of disagreement between the functionality of the currently characterized content and experimental observations. BRENDA20, CMR16, Entrez Gene15, Genome Reviews17, GOLD109, IMG18, KEGG19, MetaCyc21, Microbes Online110, PSORTdb111, PubChem112, SEED113, Transport DB23, UniProtKB114
Metabolic databases such as KEGG19, BRENDA20, MetaCyc21, SEED22 and Transport DB23 contain collections of metabolic and transport reactions which have been shown to occur across a variety of organisms. Many of these databases link enzyme commission (EC) number(s) or transport commission (TC) numbers to individual or sets of reactions which have been observed biochemically in other organisms. However, substrate specificities and enzyme activities can vary between enzymes with the same EC/TC number, so the actual reactions that are catalyzed by the enzyme in the target organism may differ from that of the analogous enzyme in a reference organism. In addition, some information such as sub-cellular localization and reaction directionality might be missing but is needed for the metabolic reconstruction (see Supplementary Table 2).
Information from metabolic databases can be extracted manually, where each active enzyme and reaction given for an organism is examined, or automated tools can be used to piece together reactions from the metabolic databases. A number of such automated tools to facilitate the reconstruction process have appeared (Box 2).
Box 2. The state of automated reconstruction of metabolic networks.
To date, most metabolic reconstructions are generated based on a combination of genome annotation, database and literature mining, with heavy reliance on genome annotations for less studied organisms. Methods have also been developed to help automate this process, but the resulting reconstructions still require manual curation if the goal is to convert them to mathematical models32, 115.
A number of automated methods have appeared which facilitate the reconstruction process. Some are used to map genes in the genome to reactions forming a draft metabolic network (PathwayTools116, GEM System117, metaShark118, SEED22, and others32, 33, 119) and others are used to refine the networks by filling in missing reactions (SMILEY algorithm45, GapFind / GapFill120, PathoLogic121) or by evaluating reaction directionality122, 123. The later methods improve draft reconstructions built from gene to reaction mapping via databases, as they can correct incorrect or missing information from metabolic databases and/or genome annotations. Since automated methods rely heavily on metabolic and transport databases, along with genome annotations, errors will propagate into the reconstructed networks. A table of common issues encountered during automated network reconstruction is presented as a guide for use of such methods and should enable further advancement of such tools.
| Genome Annotations | ||
|---|---|---|
| Issue | Description | Methods |
| Annotations are not continuously updated with new information | As new genes are found, older genome annotations are not updated. The result is incorrectly annotated genes. For example, slr0788 in Synechocystis is annotated in most databases as a pre-B cell enhancing factor (a mammalian function assigned to a bacterial gene), but in SEED22, it is correctly annotated as nicotinamide phosphoribosyltransferase. | Automated annotation pipelines can be used to reanalyze older genome annotations124. |
| Incorrect annotations | Incorrect annotations can be either missing genes (due to sequencing or gene finding algorithm errors) or incorrect gene annotations. This can occur for a number of reasons; one example is illustrated above when new sequences are not used to update older genome annotations. This can also occur when weak homology is used as sole evidence for functional assignment. | Analysis of reconstructed networks can help identify some of these errors45, 96, 120, 121. |
| Missing functionalities | Around 30% of enzyme activities with EC numbers lack sequence data125.Therefore, not all reactions will have gene or protein sequences associated with them. For example, the 6-phosphogluconlactonase gene (pgl) in E. coli was discovered in 2005126. Prior to this, there was no pgl gene in the genome annotation even though the enzymatic activity was observed in cell extracts. | Automated tools have been developed to find missing reactions (e.g., SMILEY algorithm45, GapFind / GapFill120, PathoLogic121). |
| Transporter specificity | Annotations for transporters often lack sufficient detail to determine what substrate(s) they transport, even though the mechanism (e.g., proton symport or ATP hydrolysis) is known. | Methods for improving transporter functional annotations are needed. |
| Databases | ||
| Issue | Description | Methods |
| Gene-Protein-Reaction (GPR) associations | Relationships between genes, enzymes and reactions are not always clearly defined (e.g., subunits versus isozymes). | Can be automated based on comparisons of sequences and known GPRs32. |
| Reaction specificity | Reactions are often defined as acting on a general class of compounds. This can result in ambiguity for making connections in a network. Common general classes are electron carriers (e.g., quinones, NAD, vs. NADP) or alcohols (e.g., ethanol, methanol, vs. butanol). | Changes in databases are needed or automated tools could be developed. |
| Reaction imbalances | Reactions are not elementally balanced with respect to H, C, P, N, O or S. This means that substrates and products are missing from imbalanced reactions. For example, analysis of the KEGG database in 200497 found only 51% of the reactions were balanced with respect to C, P, N, O, H and S. | Automated procedures are available to check elemental reaction balancing97. |
| Reaction directionality | Reactions are generally defined as reversible. This can be a problem, for example, if cycles between reactions occur allowing the free conversion of ADP into ATP (i.e., free energy equivalents). | Automated procedures have been developed122, 123. |
| Compound protonation states | Reactions are generally written for the neutral from of molecules and do not account for the protonation state of compounds (e.g., carboxylic acid groups are deprotonated at pH 7). This affects the stoichiometric coefficients for proton across the network. | pKa prediction software is available, so automation is possible. |
| Coenzyme availability | Enzymes often need coenzymes (e.g., pyridoxal 5-phosphate, vitamin B12, and biotin). For enzymes to be functional, the cell has to be able to produce them or get them from the environment. BRENDA20 contains this type of information, and is available for download. | Automation is possible since data is available. |
| Organism specific pathways | The cell membrane(s) are often made up of macromolecules (e.g., phospholipids and peptidoglycans) that can vary across organisms and species. As a result, the biosynthesis pathways for these compounds are often unique. | This requires experimental data and is likely not subject to automation. |
Step 2: Curating the draft reconstruction
While the automated extraction of metabolic reactions from databases gives an initial set of candidate biochemical reactions encoded on a genome, they cannot establish certain organism-specific features such as substrate or cofactor specificity and sub-cellular localization. Such information requires domain-specific knowledge of the organism. Therefore, the draft network reconstruction needs to be manually curated, ideally with input from organism-specific experts. An automatically reconstructed metabolic network will be incomplete, and it will have gaps and may also contain mistakenly included reactions that may actually not occur in the target organism. Manual curation is thus necessary to add and correct information that the automatic procedures misses or misplaces in the initial network reconstruction. While the automated reconstruction step is rapid, the manual curation process is labor intensive and at times tedious.
Organism-specific databases, textbooks24–27, primary publications, review articles and experts familiar with the legacy data for an organism are the main sources of information for the manual curation step. These detailed sources contain information about properties such as reaction directionality and location that is not always found in more general databases. For example, protein localization studies28 can be used to assign metabolic reactions to sub-cellular compartments. Similarly, biochemical studies of enzymes from the target organism (or a closely related organism) can provide information on reversibility and substrate specificity specific to that organism. These sources of information provide more direct evidence for the inclusion of specific reactions in the metabolic reconstruction. The availability of such sources for a given organism is highly variable29. The goal of manual reconstruction is to fill in gaps or holes in the network by inference or through direct evidence in the available literature on the organism or its close relatives. Gap-filling is further discussed below and examples of gap-filling in metabolic networks are presented in Box 2 and Supplemental Table 2.
A high-quality network reconstruction is thus, based on a combination of automated genome-based procedures coupled with detailed and laborious literature-based manual curation. This process effectively creates a biochemically, genomically and genetically (BiGG) structured knowledge base that is both organism-specific and available to all researchers working with the target organism. All the reactions placed in a BiGG knowledge base form a genome-scale network reconstruction (GENRE). GENREs are formed in an iterative fashion (for example, E. coli30, 31) as the corresponding BiGG knowledge base grows for the target organism, based on new experimental data or new genome-annotation.
Step 3: Converting a genome-scale reconstruction to a computational model
Before a reconstruction can be used for computations of network and/or physiological capabilities, there is a subtle, but critical step where a reconstruction is converted to a mathematical representation32, 33 (Box 1). This conversion translates a GENRE into a mathematical format that becomes the basis for a genome-scale model (GEM). Subsequent computations serve as a way to interrogate data consistency and to compute which functions a reconstructed network can and cannot carry out.
Representation of a network in a mathematical format enables the deployment of a large range of computational tools to analyze network properties. These computational tools focus on the evaluation of network systemic properties and which functions a network can perform under the physico-chemical constraints placed on the cell. This step competes the so called constraint-based reconstruction and analysis (COBRA) framework2 for the target organism. Multiple computational platforms have been developed, which apply constraint-based methods to metabolic GEMs4, 34, 35. In addition to the stoichiometric representation, metabolic networks are commonly analyzed as graphs36 or using a pathway or subsystem-based approach37, but these essentially non-parametric approaches are not discussed further here.
With a mathematical representation and computational platform, the generation of a biomass objective function is necessary to compute a network’s ability to support growth (Box 3). Here, the macromolecular composition of the cell (and the building blocks which are used to generate them) is utilized to define a necessary functionality that the network must be able to execute. A useful consistency check performed on reconstructed networks is to use them to compute growth rates under a given condition. The set of experimental data necessary to perform such analysis includes, i) the composition of cellular biomass, ii) the composition of the minimal growth media necessary to support growth in vivo, and iii) a training data set including growth rate and substrate uptake rates. Phenotypic data (growth rates and uptake and secretion rates) can be obtained through growth experiments in minimal or complex media by monitoring media components. This data is typically available in published cell characterization studies, but may need to be generated for a specific organism-of-interest. Cellular biomass composition data is obtainable through experimental assays which determine overall cellular composition and further experimentation cataloging the breakdown of each macromolecule of the cell (this information has been cataloged extensively for E. coli38). With essentiality data (gene and/or cellular content), this equation can be refined31. Genome-scale gene essentiality data sets are appearing for model organisms (listed in39), and these data sets are often times available through specific projects or organism-specific databases, such as the SGD yeast online database40. Overall, the analysis and testing of a network’s ability to produce biomass components is often utilized to curate metabolic networks (see Supplementary Table 1).
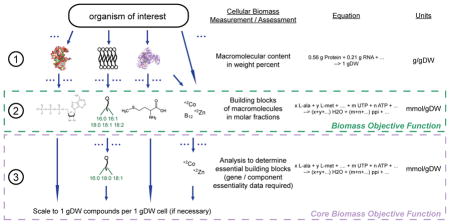
Box 3. Procedure to generate a biomass objective function.
An organism-specific biomass objective function (BOF) can be used to test the functionality of a network by examining the fundamental property of cellular growth / regeneration. The BOF, along with a known growth-supporting media condition and the reconstruction represented in a mathematical format is necessary for this test. Starting with the organism-of-interest, the macromolecular weight percent contribution of each component is determined (1). This data can be generated with readily available assay kits. Each macromolecule is then broken down into the cellular building blocks that comprise the macromolecule or those that are necessary to synthesize each it in terms of molar fractions (2). Often times, the building block will be physiologically present in the network (e.g., lipid molecules), but in some cases, the most appropriate metabolite that is in the network is used to generate the BOF (e.g., protein is broken down into individual amino acids and water; water being the net product of protein synthesis) . With the availability of gene and/or component essentiality data, a core BOF can be generated possessing different metabolites than the typical wild-type BOF. In formulating the core BOF, gene essentiality data is used along with pathway context to find the most basic essential macromolecule necessary for cell viability (3). Alternatively, published data determining minimally essential biomass components can also be incorporated into the generation of the core BOF. A core BOF can be used in simulations to more accurately examine essential components / aspects on the network. This process ultimately results in a BOF(s) that are in terms of mmol gDW−1 and be used to evaluate an organism-specific network.
Aside from simulations to produce biomass constituents, i) additional gap-filling analyses can be performed to add missed pathways or to remove any pathways that have been incorrectly included from the automated reconstruction process, and ii) additional cellular objective functions can be evaluated computationally to understand cellular behavior41, 42. The current state of gap-filling of metabolic networks has been recently reviewed43.
Once gap-filling analyses are complete, additional steps are necessary to account for strain-specific parameters and non-metabolic activities in modeling simulations. In this phase, growth data is necessary to understand and quantify these key physiological parameters. Two major factors to consider during this phase are the stoichiometry for translocation (or energy-coupling) reactions and maintenance parameters31, 44. Translocation reactions differ than other reactions in the network because the mass and energy balances around these ion pumping components are difficult to measure experimentally. Therefore characterizing reactions of this type is challenging, but can be accomplished given the proper experimental data (see Supplementary Table 2). After this phase is complete, a model can be applied to study the specific growth condition from which the training data was based and can be used to explore additional environmental conditions.
Step 4: Reconstruction uses and integration of high-throughput data
High-throughput data sets which evaluate a large number of interactions across different growth or genetic conditions can be utilized to refine and expand the metabolic content of a network. These types of comparisons and analyses have the potential to truly evaluate genome-scale omics data sets in an integrated manner by placing them in a functional and structured context. Several successful studies have been conducted for microbial species to uncover new metabolic knowledge using systematic data-driven discovery (Table 1). The necessary data types to support studies of discovery and expansion, as well as pilot studies for discovery have been recently reviewed43. Briefly, these studies fall into three categories, i) studies that have utilized a reconstruction to examine topological network properties, ii) those that have utilized a reconstruction in constraint-based modeling for quantitative or qualitative analyses, and iii) studies that are purely data driven.
Table 1.
Approaches for systematic data-driven discovery of new pathways or enzymes.
| Data type | Discovery type | References |
|---|---|---|
| Growth in diverse media conditions | New substrate utilization pathways | 45 |
| Deletion strain growth phenotyping | Alternative pathway discovery | 98, 103 |
| Synthetic lethal interactions | ||
| Systematic in vitro enzymatic assays | New metabolic reactions and pathways | 104 |
| Metabolomics | New metabolite utilization/production pathways | 105 |
| Proteomics | Candidate genes for filling network gaps | 106–108 |
| Transcriptomics | ||
| Genomic neighborhood |
One particular example of systematic data-driven discovery integrated a number of data types and GEM modeling to annotate unknown gene functions in E. coli45. In this analysis, an iterative process was utilized to, i) identify discrepancies between modeling predictions and high-throughput growth phenotyping data (Biolog data, http://www.biolog.com), ii) determine potential reactions which remedy disagreements (and the ORFs that might encode proteins to catalyze them) through a computational analysis, and iii) characterize targeted ORFs experimentally to confirm their function. To drive discovery, this approach analyzed a variety of data types (i.e., phenotyping, gene expression, and enzyme activity) to hypothesize and validate computational predictions. This one example demonstrates the promise of integrating modeling results and experimental, data and will likely become a key approach to expanding current metabolic knowledge along with aiding discovery of new components and interactions in cellular processes.
Transcription and translation processes
Reconstructions of transcriptional and translation (tr/tr) networks at genome-scale will follow a similar procedure as has been established for metabolism. Tr/tr network reconstructions can be generated using a genome annotation and the genome sequence as a scaffold. A tr/tr network reconstruction will contain sequence-specific synthesis reactions for every included gene and gene product participating in transcriptional and translational functions (Box 4D). Such tr/tr reconstructions do not contain transcriptional regulators and their functions (see the following section). Furthermore, the presented stoichiometric tr/tr reconstructions are different from kinetic, small-scale or sequence-independent formulations of transcriptional and/or translational networks46–50 whose are not discussed here. The scope of the tr/tr reconstruction is the synthesis of all proteins, tRNA and rRNA involved in the functions listed in Box 4D. This scope ranges from the metabolites that are consumed by the network to the functional proteins (e.g., ribosomes), mRNA, and tRNAs. This type of tr/tr network has recently been developed for E. coli51.
Box 4. Content and process to reconstruct transcriptional and translational (tr/tr) networks.
The reconstruction of a tr/tr network can be performed in an algorithmic manner, as illustrated in this flow chart (generated for E. coli). First, the network components responsible for every transcriptional or translational step need to be identified from different resources (e.g., primary and review literature, genome annotation, EcoCyc) (A). For each component, a function is then translated into a stoichiometric, mass- and charge-balanced reaction using mainly primary and review literature. The resulting set of reactions can additionally be separated into two groups, i) component-specific reactions (e.g., dimerization reaction of a protein); and ii) template reactions (e.g., a transcription initiation reaction). Template reactions are specified for each gene by considering the information listed in (B) to produce active gene products for the different subsystems (or pathways) listed in (D). The active form of some gene products may require post-translational modifications, protein folding, covalent binding of metallo-ions or co-enzymes (C). The resulting reaction list is subsequently converted into a mathematical format (e.g., a stoichiometric matrix) and tested for, i) functionality, ii) completeness, iii) correctness, and iv) predictive potential compared to known cellular phenotypes. Discrepancies are elucidated by repeating the procedure starting back at (A). Utilizing this process, the overall structure of the reconstructed network resembles that of a metabolic reconstruction (see text). The quality control and quality assurance (QC/QA) procedures help to guaranty self-consistency and correctness of the network through, i) mass and charge balancing of all possible network reactions, ii) analysis and filling of network gaps, and iii) functionally testing for the production of every network component and its intermediate form. Note that the different resources in (A) are color coded and correspond to their use in (B)-(D).
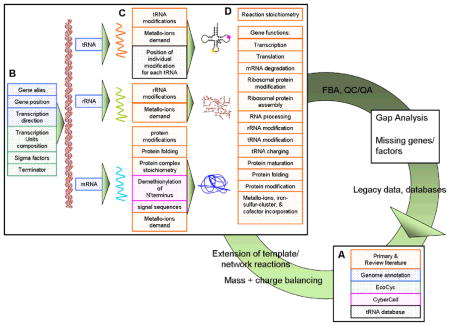
Step 1: Automated genome-based reconstruction
Information about the components of the tr/tr network can be directly extracted from the genome annotation. This step should provide details for, i) gene function, ii) gene type (e.g., protein coding, tRNA), iii) start and stop codons, iv) direction of transcription, and, v) transcription unit association (for prokaryotes). Some genome annotations and databases (e.g., Regulon DB48 and BioCyc52) provide information about the type of transcription terminator (e.g., rho-dependent, attenuation, etc.) and sigma factors for transcriptional initiation (e.g., sigma 70, sigma H, etc.). Tr/tr reactions can be formulated in an automated fashion using this information, the genome sequence and template reactions (see Box 4). These manually formulated template reactions make use of the fact that the tr/tr reactions are very similar for most genes or gene products. For example, in E. coli, the transcriptional initiation requires the binding of the holo-enzmye RNA polymerase (α2ββ’) to a sigma factor (e.g., σ70), followed by the binding of this complex to a promoter site of a gene that has a recognition site for this sigma factor. Other gene-specific features such as amounts of different amino acids for polypeptide or nucleotide triphosphates (NTPs) for mRNAs replace placeholders in the template reaction based on gene information. Subsequently, corresponding reactions can thus be formulated accurately and in a gene-specific manner, by utilizing information about sigma factors, amino acids and NTPs in conjunction with template reactions.
Step 2: Curation and formulation based on bibliomic data
Using primary literature articles, template reactions need to be manually formulated and curated. Manual curation is also required for protein complex stoichiometry information and the presence and stoichiometry of metallo-ions or involved co-enzymes (e.g., flavins), as most databases do not contain this information. Challenges unique to reconstruction of the tr/tr network include reaction mechanisms of certain modifications (e.g., tRNA modifications53) or certain pathways (e.g., iron-sulfur cluster formation) and these are not well established51. Reactions and pathways generated based on debated knowledge need to be tracked in the reconstruction, for example, by using notes or a confidence score, to facilitate their update as new information become available.
Step 3: Converting a genome-scale reconstruction to a computational model
The reactions list generated through step 1 and 2 can be readily converted into a mathematical format using bioinformatically-driven programming that extract the stoichiometric coefficients from each network reaction and transfers them into the matrix. The network boundaries in tr/tr networks typically border metabolism, i.e., metabolic components are imported or exported across these boundaries. The uptake constraints for these metabolites can be derived from experimental data (e.g., overall protein content) as a function of growth rate. For example, these parameters have been directly measured for E. coli cells with 40 minute doubling times54.
Step 4: Reconstruction uses and integration of high-throughput data
The reconstruction of tr/tr networks is a first step towards a new generation of cellular network models which will account quantitatively for mRNA and protein abundance. They have the potential to increase the scope of modeling and understanding cellular processes, such as the calculation of ribosome productions capabilities at different growth rates and of functional interactions of the network proteins by determining functional modules. Furthermore, such tr/tr networks will increase our understanding of the relationship between mRNA and protein abundance and will enable consideration of the cost of the cellular machinery synthesis in in silico modeling. The reconstruction of tr/tr networks should also enable quantitative integration of high-throughput data to both expand and refine the knowledge about tr/tr networks and its components. With this, there is a need to develop approaches to map relative or absolute molecule concentration data onto network reactions. While the integration of transcriptomic and proteomic data may be more straightforward, the integration procedure of ChIP-Chip data quantifying binding affinities of the RNA polymerase or other transcription factors needs to be established. Lastly, integration of the tr/tr network with other cellular processes should enable a mechanistically detailed and comprehensive description of organism’s capabilities.
Transcriptional regulatory networks (TRNs)
The basic structure of TRNs involves the interactions between transcription factors (TFs) and their target promoters that lead to activation or repression of transcription. This definition of network boundary leaves out upstream environmental and intracellular signals that regulate TF activity or any additional regulatory mechanisms that may influence gene expression levels (e.g., compacting DNA by various proteins that influence DNA structure so that it cannot be efficiently transcribed). Most of the experimental and computational activities to elucidate TRNs have so far focused on mapping the basic structure of the network and hence this review will concentrate on the network of TF-promoter interactions. ChIP-chip has also been applied to mapping genome-wide locations of proteins that are involved in the packaging of DNA55, 56 (e.g. histones and histone-like proteins), and it is expected that future reconstructions of TRNs could include global regulation of DNA accessibility and thus transcription in addition to local regulation at specific promoters by specific TFs.
Step 1. Automated reconstruction
In contrast to the case for metabolic networks, where experimental methods to measure system-wide levels of metabolites and fluxes are not yet fully developed, methods for large-scale measurement of TRN interactions and components are already well established. This capacity has enabled the development of top-down approaches for TRN reconstruction that integrate multiple high-throughput datasets in order to reconstruct TRNs. The types of experimental approaches that are used for high-throughput studies of TRNs are typically multiplexed versions of classical low throughput assays for gene expression, in vitro DNA binding and in vivo DNA binding.
The most direct way to experimentally map TRNs is to determine genome-wide in vivo binding sites of a TF using high-throughput versions of the chromatin immunoprecipitation (ChIP) assay. The most commonly used method is ChIP-chip that uses a microarray-based approach to detect genomic loci where a given TF binds under a given condition57. ChIP-chip data can now be generated in diverse microbial organisms and this type of data created for a large number of TFs has been used as the basis for comprehensive mapping of TRNs especially in yeast58. However, challenges remain in applying ChIP-chip (e.g., TF antibody availability). In order to fully map TRNs, ChIP-chip experiments have to be performed for the same TF under multiple conditions as the set of target genes may vary from one condition to another59. Analogously to the development of multiplexed ChIP assays, high-throughput in vitro DNA binding assays using both microarray60 and microfluidic platforms61 have been developed. In vitro methods have the benefit of allowing the determination of potential TF binding sites in a condition independent manner. These in vitro methods require using purified proteins and thus can be challenging to apply in practice; however, they have been shown to provide valuable complementary data to in vivo experiments62.
Array-based genome-wide gene expression profiling-based approaches are perhaps the most widely used methods to characterize TRN function. Of particular utility to TRN mapping are expression profiling studies of strains where specific TFs have been deleted63, 64 or overexpressed65. In addition, large compendia of gene expression data measured in response to different genetic and environmental perturbations can also be used to identify candidate regulatory interactions66, transcriptional modules as well as potential regulators for these modules67. However, gene expression profiling alone is not sufficient to differentiate between direct TF binding on a given promoter and indirect effects.
A major challenge remains in integrating all the available experimental data types as well as _cis_-regulatory motif information derived from sequence conservation in order to systematically reconstruct TRNs58, 68. ChIP-chip data alone is sufficient to reconstruct the connectivity of the TRN, but expression profiling data on TF deletion or overexpression or time course expression profiling studies are required to establish the mode of regulation (activation or repression). Furthermore, combinatorial interactions between TFs on promoters can only be mapped by performing expression profiling experiments in multiple TF deletion strains63 or by performing ChIP-chip experiments for one TF in strains where another TF is deleted69. While these approaches have shown impressive performance in reconstructing individual sub-networks, the ability of, for example, ChIP-chip to detect binding sites varies significantly depending on the TF and experimental condition utilized to perform the assay58, 70.
Fully automated TRN reconstruction would ideally require ChIP-chip experiments targeting all major TFs and gene expression profiling of TF deletion strains under a set of representative experimental conditions. If these types of data are available and are of sufficiently high quality, TRN reconstruction can be done in a largely automated fashion. Recent developments in massively parallel sequencing technologies promise to further improve our ability to automatically reconstruct TRNs by providing higher resolution, sensitivity and quality data on both gene expression71 and DNA binding72 than array-based methods. As an alternative to full mapping of TF-target interactions, a number of approaches have been developed to 1) identify condition-dependent co-regulated gene clusters or modules based on large gene expression data sets, and 2) assign regulators to these clusters based on a combination of ChIP-chip data, expression response to TF deletions, cis-regulatory motifs, and time dependent gene expression profiling data67, 73. These types of approaches may not allow mapping all individual regulatory interactions, but they reduce the complexity of the TRN reconstruction problem significantly.
Step 2. Reconstruction based on bibliomic data
TRNs can be reconstructed in a bottom-up fashion based on both genomic data and bibliomic data analogous to the reconstruction of metabolic networks. Genomic data can be used to identify potential TFs as well as potential TF target sites through comparative genomics of closely related species74. However, genomic information alone is insufficient to obtain predictions of TF function or targets75 and thus, significant amount of additional experimental information is required. The reconstruction of TRNs based on bibliomic data relies on individual studies on transcriptional regulation of single promoters that typically aim to dissect the role of different binding sites on the promoter using gene expression assays (e.g., northern blots, RT-PCR, or reporter gene approaches) in response to TF deletions or partial deletions of promoter regions, in vivo DNA-binding assays (e.g., chromatin immunoprecipitation), and in vitro DNA-binding assays. The challenges in utilizing literature data are that only a subset of all the promoters have been subjected to extensive characterization, in even well-characterized organisms such as E. coli, and the conditions, methods and strains utilized in different studies can be quite variable. For these reasons, bottom-up reconstructions are only expected to represent a partial picture of the full TRN and their role in most species would be primarily to provide validation data for more comprehensive top-down reconstruction approaches. There are a limited number of databases that currently store literature-derived information on transcriptional regulation – the most comprehensive one is RegulonDB for E. coli76.
Step 3. Converting a genome-scale reconstruction to a computational model
TRNs reconstructed using either automated or bibliomic methods are typically represented in two alternative ways: 1) as graphs where each TF node is connected to its target gene nodes by a directed edge or 2) as co-regulated gene modules with candidate TFs and environmental (e.g. carbon source) regulators associated with each module. However, in order to allow using the network for predicting expression responses to environmental or genetic perturbations, these network reconstructions have to be converted to computational models using one of the possible modeling frameworks. While stochastic and kinetic models provide a good starting point for small-scale regulatory network modeling, these approaches do not scale to larger and genome-scale networks. Large-scale regulatory network models have been built so far primarily using 1) Boolean network approaches, and 2) variety of probabilistic modeling frameworks including simplified additive kinetic modeling approaches using e.g. log-linear kinetics68, 77, 78.
The choice of the modeling framework is largely determined by the type of network reconstruction that is used as a starting point to build the model, the type of data that is available to parameterize the model, and by what types of predictions one wants to make. Boolean representations provide a good starting point for building qualitative models based on TRNs reconstructed primarily using bibliomic data63, 70. Boolean models have been so far built for E. coli and yeast, and these representations can also be further converted to a matrix formalism that allows more straightforward integration with metabolic network models79. Many different probabilistic modeling frameworks including probabilistic Boolean networks77, physical network models68 as well as more complex types of models78 have been applied to reconstruct large-scale TRNs. However, most of these approaches have primarily been used as tools for systematic data-based TRN reconstruction, and have not yet been used to build large-scale predictive models.
In contrast, recent studies have used additive kinetic modeling approaches to model genome-scale TRNs either in settings where the network structure is known based on e.g. ChIP-chip or bibliomic data80 or in conjunction with methods that identify co-regulated gene clusters73. Unlike Boolean models, these simplified kinetic models can be used to predict quantitative dynamic expression changes, but significant amount of time course gene expression data is usually needed to parameterize the models. Recently, a predictive additive kinetic model of the Halobacterium salinarum TRN GENRE was build using a combination of computational methods73. First, condition dependent regulatory modules were built using bi-clustering of a well-designed gene expression data compendium together with _cis_-regulatory motif information, and then quantitative effects of TFs and environmental factors on expression of these modules were identified based on dynamic gene expression data. The H. salinarum study also demonstrated that predictive TRN models can be built even for species with relatively poorly characterized TRNs given that sufficient quantities of relevant high-throughput data can be generated in a systematic manner.
Step 4. Applications of TRN models
Analyses performed using TRN models have resulted in identification of novel regulatory interactions as well as prediction of general patterns cellular behavior. For example, a previous effort combined a comprehensive literature-based reconstruction of the TRN controlling metabolism in E. coli with expression profiling of single and double TF deletions strains in order to improve the ability of an integrated regulatory/metabolic network model to predict phenotypes and expression changes63. Similarly, comparing the H. salinarum model73 predictions with experimental data resulted in identification of a number of novel regulators for key cellular processes in this archaeon.
Technologies for mapping of TRNs are maturing rapidly and promise to allow largely automated reconstruction of these types of networks in the near future. Major challenges still remain in modeling TRNs in a physico-chemically realistic fashion and in integrating TRNs with other cellular processes. The signaling pathways leading to activation of TFs are also much less well understood than the TRN itself and the experimental techniques for mapping these pathways are not as well developed as TRN mapping methods (see below).
Expected expansion in scope of networks reconstruction efforts
The metabolic, transcriptional regulation, translation and transcription processes together represent a sizable fraction of the genes in a microbial genome. However, there are other networks that are currently the subject of intensive study that will likely be the subject of future network reconstruction efforts. Such efforts are likely to develop four-step reconstruction processes, paralleling those described above.
Two-component signaling systems are an example of this type of network. Current models of TRNs in E. coli already include some of the known two-component signaling pathways that respond to metabolic stimuli63. While the components of two-component signaling pathways (histidine kinases and response regulators) can be identified relatively easily by sequence homology, the connectivity of these pathways is not completely known, even in E. coli. Progress has been made recently to systematically map the connectivity of two-component pathways in E. coli81 as well as in other bacteria82 using a variety of experimental methods. It is expected that in the future comprehensive reconstructions of two-components systems can be achieved by combining literature-based information with these types of high-throughput data83.
The second type of network that has attracted increasing attention in recent years is the translational regulatory network controlled by small non-coding RNAs (sRNA). The most common mechanism for sRNA action is repression of translation of specific mRNAs by binding to translation initiation regions, but other mechanisms including regulation of protein expression or activity also exist84. It has been estimated that typical bacterial genomes would carry up to 300 sRNA genes and that these sRNAs play a crucial role in controlling cellular functions including metabolism and virulence85. The process of finding sRNAs in bacterial genomes is reasonably well-established86, but finding mRNA targets for these sRNAs is still quite challenging. A number of experimental and computational techniques have been devised to determine the targets of sRNAs at the genome-wide scale (reviewed in84, 87) promising to accelerate the process of mapping comprehensive sRNA regulatory networks. Initial systems studies of the known sRNA regulatory network in E. coli have indicated that the most likely function of sRNA regulation is to act in concert with transcriptional regulation to provide mechanisms that allow tight condition-dependent regulation of the target protein levels88.
Integration of network reconstructions
Once two or more of the five different types of networks described above have been reconstructed for a target organism, they can be integrated to form computational GENREs and computational GEMs spanning a large number of cellular activities.
The integration of TRN and metabolic networks has received the most attention to date, given the fact that comprehensive reconstructions of these two network types exist63, 70, 89 (see Supplementary Table 1 for metabolic networks). TRNs regulate metabolism through modulating active enzyme concentrations and subsequently by controlling the maximum flux levels through reactions. The levels of metabolites, in turn, regulate gene expression and thus the two networks are an integrated process.
Integration of metabolism and transcription/translation processes is, in principle, quite straightforward. Transcription and translation requires energy and building blocks such as nucleotides and amino acids as inputs and hence these processes are constrained by the ability of the metabolic network to produce these precursors49. On the other hand, the transcription and translation processes can be seen to exert demands on the metabolic network function and thus limit other metabolic functions49. Furthermore, the transcription/translation network feeds back to the metabolic network by controlling the levels of the enzymes in the metabolic network.
Although, GENREs including three or more different networks have not appeared, they should be achievable. In principle, each network can be described by a stoichiometric matrix once the underlying reactions have been determined. Stoichiometric matrices for metabolism (M) have appeared. A stoichiometric matrix format for tr/tr reactions (E) which form the expression state of networks is achievable51. The TRN reconstruction in bacteria can be based on the operon structure of a genome and could be described by a corresponding stoichiometric matrix (O) as well, once the underlying chemical reactions have been defined. Given that stoichiometric matrices can be integrated in a onestep process, an ‘OME’ matrix describing the integrated network can be formulated. Currently however, TRNs are described by a set of logistical statements and although a matrix format has been developed for Boolean statements79 enabling network integration, we ultimately need to seek chemical representation of TRNs. Working towards this effort, a small-scale integration of the three networks has appeared89 foreshadowing what is to come at the genomic scale.
The conversion to a computational model
Integrated network reconstructions, that effectively are two-dimensional annotations1, can be used to build GEMs that represent the functions of integrated networks to make phenotypic predictions (Figure 2). This conversion has been described2, and it basically entails the mathematical description of the reactions that have been identified to take place in a network of interest; representing the conversion of a BiGG knowledge base into a GEM. The use of computational approaches to interrogate the properties of GEMs has been described3. Three uses of GEMs for experimentation have appeared5; i) use for the discovery of missing content in a reconstruction, ii) use of GEMs to understand integrated physiological process, and iii) use to prospectively design experiments and physiological processes. The first topic is germane to this review as it is aimed at systematically discovering the missing content of a reconstruction.
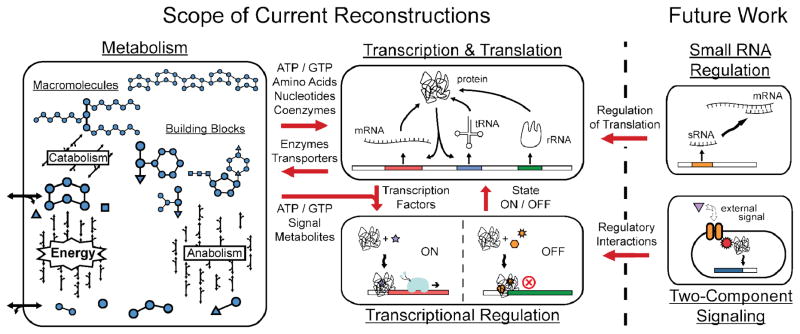
Figure 2. Network integration: the interface between different types of reconstructions.
The ultimate goal of network reconstruction is to fully represent every component of the cell and define the interactions between them. Reconstruction of metabolism, transcriptional regulation, and transcription and translation networks is currently possible (as outlined in the text), with the most emphasis, to date, on metabolic reconstruction. Incorporation of small RNAs and two-component signaling interactions are future areas of reconstruction where reconstruction technologies and development are needed. For integration of networks, the interplay between each of the processes needs to be defined to fully connect each of the major cellular functions.
The effects of missing network content
An important issue in the conversion of a network reconstruction into a predictive computational model is the coverage and accuracy of available data from which the network was reconstructed. Therefore, it is important to understand the impact and influence network components can have on computational results. Intended use examples of in silico models are used to help understand this issue.
Qualitative predictions obtained using GEMs (e.g., will an organism grow given an environmental or genetic perturbation, or does expression of gene increase or decrease) are likely to be less sensitive than quantitative predictions (e.g., what is the cellular growth rate or what level of gene expression is expected) to errors in the network content. This expectation is due to the fact that qualitative predictions are compared to binary outcomes (i.e. digital outcomes), rather than a range of numerical values (i.e., analog outcomes). If one is generating qualitative predictions regarding growth phenotypes, the effect of omitting an individual reaction from a network does not greatly affect your results. For example, removing approximately 87% of the 2077 reactions individually from an E. coli metabolic model (_i_AF126031) did not affect the qualitative growth predictions for a given environmental condition.
In depth studies have been performed to assess the influence of individual network components, input parameters, and the querying methods used to probe GEMs on computational predictions. The results from these studies can be used to gauge the influence of the content of reconstructions. These analyses include examining input/output values31, 44, 90, 91, BOF composition31, 41, 44, 92, 93, querying methods94, 95, and network components91, 96–101. For TRNs and tr/tr networks, the situation is comparatively less developed and is expected to be more difficult to assess than for metabolic networks; due to i) the highly non-linear structure of some components of the network, ii) the typically larger number of interactions per component, and iii) the larger amount of missing knowledge in these networks. Missing regulatory interactions between TFs and metabolic target genes in the network would thus be expected to have a relatively moderate effect on predictive abilities as the regulation is likely to be highly redundant.
These initial studies demonstrate the necessity to identify the scope and intention of GENRE applications a priori and further show how computational analysis can help to identify missing components and errors when computational results are compared to biological functions. The latter model-driven gap-filling approach is expected to continue to develop and lead to GEMs with improved predictive capabilities.
Conclusions
The reconstruction process relies on workflows that organize and integrate various data types and other relevant information about the network of interest. Over the past ten years, such workflows have been developed for genome-scale metabolic networks to the point where they represent BiGG knowledge bases and are in wide use. More recently, similar methods are being developed for other cellular processes such as transcriptional regulation, and for transcription and translation. The implementation of these workflows for a growing number of organisms should accelerate the systems analysis in a single organism, in communities of organisms, and through phyla. The workflows reviewed herein have been implemented and enabled a wide variety of analyses5. To facilitate wider use and the development of additional analysis procedures, improvements in the distribution of GENREs in needed. Two areas that will aid distribution and usage are the standardization of a reconstruction format (e.g., SBML102) and available reconstruction database where they can be accessed.
It is expected that the reconstruction process will continue to grow in scope, depth and accuracy, and it should continue to enable a broadening spectrum of basic and applied studies. The availability of high-quality comprehensive reconstructions will accelerate the implementation of the systems biology paradigm (i.e., biological components to networks to computational models to phenotypic studies) and will thus help realize the broad transformative potential of this paradigm in the life sciences. Network reconstructions are a key factor in building a mechanistic genotype-phenotype relationship. Quantitative genotype-phenotype relationships have been best established for bacterial metabolism5 to date and this review should aid new practitioners to build such relationships for their target organisms.
Box 5. Challenges in network reconstruction: The case of Plasmodium falciparum.
There is now a highly systematic process to build a metabolic network reconstruction and model for any given microbial organism starting with an annotated genome and ending with a predictive model of microbial physiology. For well characterized model organisms, this overall process has already moved to the step where models can aid in discovering new metabolic functionalities. However, there are many organism of practical interest for which only initial steps towards building comprehensive metabolic network reconstructions and models have been taken. These include pathogens such as P. falciparum or S. aureus as well as many microbes that are relevant to bioprocessing or bioenergy applications. In the following, we will outline some of the unique challenges that have to be addressed when metabolic network reconstructions for these organisms are built. We will use the malaria parasite P. falciparum as an example organism to discuss these challenges127.
The most fundamental type of challenge for reconstruction is that in which a genome encodes for proteins that have a low degree of sequence homology to any other organism (e.g., due to severe sequence biases, such as high A/T content, as is the case for P. falciparum). For these organisms automated homology-based function prediction tools will result in a highly incomplete initial reconstruction of metabolic networks with numerous gaps. While more sophisticated sequence analysis methods allow building more complete initial reconstructions for organisms such as the malaria pathogen, the initial networks still require significant manual curation just to define the comprehensive set of metabolic capabilities the organism possesses118. In the case of P. falciparum the identification of metabolic functions experimentally is further complicated by inefficient methods for genetic manipulation. However, these methods have significantly improved in recent years enabling systematic validation of putative metabolic functions as well as development of strain collections that can be used for general functional genomics studies128.
Our understanding of metabolic physiology of P. falciparum and many other organisms of practical importance is also limited by our inability to cultivate these organisms in defined conditions. In most challenging cases, the organisms cannot be readily cultivated outside the host organism at all. P. falciparum can be cultured in vitro in red blood cells, but the presence of two different cell types in the culture poses problems to understanding the physiology of the parasite. For example, the transport of nutrients from the media to the parasite is only partially understood129. Further complications arise from the fact that the typical in vitro cultivation conditions require using non-specific media components such as serum albumin. This makes it challenging to perform the types of auxotrophy experiments that are commonly used to establish metabolic functions in microbial organisms such as E. coli or yeast. Even if well defined in vitro cultivation conditions can be established, it is likely that the metabolic behavior in these conditions fails to capture relevant features of in vivo physiology. This has been shown to be the case for P. falciparum by comparing in vivo expression profiles derived from patient blood samples with expression profiles obtained from in vivo cultivation of the parasite130.
Despite these challenges, much progress has been made in understanding the metabolic physiology of pathogens such as P. falciparum. The development of metabolic network reconstructions and models for these challenging organisms enables systematic evaluation of current knowledge gaps as well as utilization of the types of model-based gap filling strategies that were previously discussed in this review. Progress in reconstructing other types of networks including TRNs for pathogens is more severely affected by the lack of facile genetic systems. For example, despite extensive profiling with gene and protein expression technologies, the mechanisms that regulate gene and protein expression in P. falciparum have remained elusive131.
Supplementary Material
Acknowledgments
The authors would like to thank A. Osterman and N. Jamshidi for their insights. Authors AMF, IT were supported by a grant from the by National Institutes of Health R01 GM057089 and author MJH by NIH grant R01 GM071808. BOP serves on the scientific advisory board of Genomatica Inc.
Suggested Glossary Terms
GENRE
Genome-scale network reconstruction. Applies to a particular organism, for example, GENRE of Escherichia coli. A GENRE (e.g., metabolic reconstruction) contains a list of all the chemical transformations that are known to take place in the particular network (usually the entire metabolic network in an organism). These transformations can be represented by a stoichiometric matrix. A reconstruction is effectively a two-dimensional genome annotation1
GEMs
Genome-scale models in silico of a particular organism, for example, GEMS of E. coli. GEMS are a mathematical format of a network reconstruction that can be computationally interrogated and subsequently used for experimental design
STOICHIOMETRIC MATRIX
A matrix containing the stoichiometric coefficients for the reactions that comprise a network. The rows of which represent the compounds, the columns of which represent the chemical transformations and the entries of which are the stoichiometric coefficients
BiGG KNOWLEDGE BASE
The collection of established biochemical, genetic, and genomic data (BiGG) represented by a network reconstruction
BIBLIOMIC DATA
Legacy data that is contained in peer reviewed scientific publications
References
- 1.Palsson BO. Two-dimensional annotation of genomes. Nat Biotechnol. 2004;22:1218–9. doi: 10.1038/nbt1004-1218. [DOI] [PubMed] [Google Scholar]
- 2.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nat Rev Genet. 2006;7:130–41. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 3.Price ND, Reed JL, Palsson BO. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nat Rev Microbiol. 2004;2:886–897. doi: 10.1038/nrmicro1023. [DOI] [PubMed] [Google Scholar]
- 4.Becker SA, et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox. Nat Protocols. 2007;2:727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 5.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotech. 2008;26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papoutsakis ET. Equations and calculations for fermentations of butyric acid bacteria. Biotechnology and Bioengineering. 1984;26:174–187. doi: 10.1002/bit.260260210. [DOI] [PubMed] [Google Scholar]
- 7.Papoutsakis E, Meyer C. Fermentation equations for propionic-acid bacteria and production of assorted oxychemicals from various sugars. Biotechnology and Bioengineering. 1985;27:67–80. doi: 10.1002/bit.260270109. [DOI] [PubMed] [Google Scholar]
- 8.Papoutsakis E, Meyer C. Equations and calculations of Product yields and preferred pathways for butanediol and mixed-acid fermentations. Biotechnology and Bioengineering. 1985;27:50–66. doi: 10.1002/bit.260270108. [DOI] [PubMed] [Google Scholar]
- 9.Majewski RA, Domach MM. Simple constrained optimization view of acetate overflow in E. coli. Biotechnology and Bioengineering. 1990;35:732–738. doi: 10.1002/bit.260350711. [DOI] [PubMed] [Google Scholar]
- 10.Varma A, Boesch BW, Palsson BO. Stoichiometric interpretation of Escherichia coli glucose catabolism under various oxygenation rates. Appl Environ Microbiol. 1993;59:2465–73. doi: 10.1128/aem.59.8.2465-2473.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varma A, Boesch BW, Palsson BO. Biochemical production capabilities of Escherichia coli. Biotechnology and Bioengineering. 1993;42:59–73. doi: 10.1002/bit.260420109. [DOI] [PubMed] [Google Scholar]
- 12.Karp PD, et al. Multidimensional annotation of the Escherichia coli K-12 genome. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christie KR, et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32(Database issue):D311–4. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guldener U, et al. CYGD: the Comprehensive Yeast Genome Database. Nucleic Acids Res. 2005;33:D364–8. doi: 10.1093/nar/gki053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peterson JD, Umayam LA, Dickinson T, Hickey EK, White O. The Comprehensive Microbial Resource. Nucleic Acids Res. 2001;29:123–5. doi: 10.1093/nar/29.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoesser G, Tuli MA, Lopez R, Sterk P. The EMBL Nucleotide Sequence Database. Nucleic Acids Res. 1999;27:18–24. doi: 10.1093/nar/27.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowitz VM, et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34:D344–8. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schomburg I, et al. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–3. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krieger CJ, et al. 2004:D438–442. [Google Scholar]
- 22.DeJongh M, et al. Toward the automated generation of genome-scale metabolic networks in the SEED. BMC Bioinformatics. 2007;8:139. doi: 10.1186/1471-2105-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–9. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neidhardt FC, editor. Escherichia coli and Salmonella: cellular and molecular biology. ASM Press; Washington, D.C: 1996. [Google Scholar]
- 25.Dickinson JR, Schweizer M. The metabolism and molecular physiology of Saccharomyces cerevisiae. Taylor & Francis Ltd; London ; Philadelphia: 2004. [Google Scholar]
- 26.Marre R. Legionella. ASM Press; Washington, D.C: 2002. [Google Scholar]
- 27.Mobley HLT, Mendz GL, Hazell SL. Helicobacter Pylori. ASM Press; Washington, D.C: 2001. [PubMed] [Google Scholar]
- 28.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 29.Janssen P, Goldovsky L, Kunin V, Darzentas N, Ouzounis CA. Genome coverage, literally speaking. The challenge of annotating 200 genomes with 4 million publications. EMBO Rep. 2005;6:397–9. doi: 10.1038/sj.embor.7400412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed JL, Palsson BO. Thirteen Years of Building Constraint-Based In Silico Models of Escherichia coli. J Bacteriol. 2003;185:2692–9. doi: 10.1128/JB.185.9.2692-2699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feist AM, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3 doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Notebaart RA, van Enckevort FH, Francke C, Siezen RJ, Teusink B. Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics. 2006;7:296. doi: 10.1186/1471-2105-7-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borodina I, Nielsen J. From genomes to in silico cells via metabolic networks. Curr Opin Biotechnol. 2005;16:350–5. doi: 10.1016/j.copbio.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Lee SY, et al. Systems-level analysis of genome-scale in silico metabolic models using MetaFluxNet. Biotechnol Bioproc Eng. 2005;10:425–431. [Google Scholar]
- 35.Klamt S, Saez-Rodriguez J, Gilles ED. Structural and functional analysis of cellular networks with CellNetAnalyzer. BMC Syst Biol. 2007;1:2. doi: 10.1186/1752-0509-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–13. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 37.Kwast KE, et al. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J Bacteriol. 2002;184:250–65. doi: 10.1128/JB.184.1.250-265.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neidhardt FC, Umbarger HE. In: Escherichia coli and Salmonella : cellular and molecular biology. Neidhardt FC, editor. ASM Press; Washington, D.C: 1996. pp. 13–16. [Google Scholar]
- 39.Joyce AR, et al. Experimental and Computational Assessment of Conditionally Essential Genes in Escherichia coli. J Bacteriol. 2006;188:8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cherry JM, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Research. 1998;26:73–9. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuetz R, Kuepfer L, Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol. 2007;3 doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knorr AL, Jain R, Srivastava R. Bayesian-based selection of metabolic objective functions. Bioinformatics. 2007;23:351–7. doi: 10.1093/bioinformatics/btl619. [DOI] [PubMed] [Google Scholar]
- 43.Breitling R, Vitkup D, Barrett MP. New surveyor tools for charting microbial metabolic maps. Nat Rev Microbiol. 2008;6:156–61. doi: 10.1038/nrmicro1797. [DOI] [PubMed] [Google Scholar]
- 44.Varma A, Palsson BO. Parametric sensitivity of stoichiometric flux balance models applied to wild-type Escherichia coli metabolism. Biotechnology and Bioengineering. 1995;45:69–79. doi: 10.1002/bit.260450110. [DOI] [PubMed] [Google Scholar]
- 45.Reed JL, et al. Systems approach to refining genome annotation. Proc Natl Acad Sci U S A. 2006;103:17480–4. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roussel MR, Zhu R. Stochastic kinetics description of a simple transcription model. Bull Math Biol. 2006;68:1681–713. doi: 10.1007/s11538-005-9048-6. [DOI] [PubMed] [Google Scholar]
- 47.Mehra A, Hatzimanikatis V. An algorithmic framework for genome-wide modeling and analysis of translation networks. Biophys J. 2006;90:1136–46. doi: 10.1529/biophysj.105.062521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weitzke EL, Ortoleva PJ. Simulating cellular dynamics through a coupled transcription, translation, metabolic model. Comput Biol Chem. 2003;27:469–80. doi: 10.1016/j.compbiolchem.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Allen TE, Palsson BO. Sequenced-Based Analysis of Metabolic Demands for Protein Synthesis in Prokaryotes. Journal of Theoretical Biology. 2003;220:1–18. doi: 10.1006/jtbi.2003.3087. [DOI] [PubMed] [Google Scholar]
- 50.Drew DA. A mathematical model for prokaryotic protein synthesis. Bull Math Biol. 2001;63:329–51. doi: 10.1006/bulm.2000.0225. [DOI] [PubMed] [Google Scholar]
- 51.Thiele I, Jamshidi N, Fleming RMT, Palsson BO. Genome-scale reconstruction of E. coli's transcriptional and translational machinery: A knowledge-base and its mathematical formulation. 2008 doi: 10.1371/journal.pcbi.1000312. Under Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karp PD, et al. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–9. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sprinzl M, Vassilenko KS. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–40. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neidhardt FC, Ingraham JL, Schaechter M. Physiology of the bacterial cell: a molecular approach. Sinauer Associates; Sunderland, Mass: 1990. [Google Scholar]
- 55.Cho BK, Knight EM, Barrett CL, Palsson BØ. Genome-wide Analysis of Fis Binding in Escherichia coli Indicates a Causative Role for A-/AT-tracts. Genome Res. 2008;18:900–910. doi: 10.1101/gr.070276.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–8. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 57.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–9. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 58.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Buck MJ, Lieb JD. A chromatin-mediated mechanism for specification of conditional transcription factor targets. Nat Genet. 2006;38:1446–51. doi: 10.1038/ng1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mukherjee S, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–9. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–7. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 62.Liu X, Lee CK, Granek JA, Clarke ND, Lieb JD. Whole-genome comparison of Leu3 binding in vitro and in vivo reveals the importance of nucleosome occupancy in target site selection. Genome Res. 2006;16:1517–28. doi: 10.1101/gr.5655606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–6. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 64.Hu Z, Killion PJ, Iyer VR. Genetic reconstruction of a functional transcriptional regulatory network. Nat Genet. 2007;39:683–7. doi: 10.1038/ng2012. [DOI] [PubMed] [Google Scholar]
- 65.Chua G, et al. Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci U S A. 2006;103:12045–50. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Faith JJ, et al. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segal E, et al. Module networks: identifying regulatory modules and their condition- specific regulators from gene expression data. Nat Genet. 2003;34:166–76. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 68.Workman CT, et al. A systems approach to mapping DNA damage response pathways. Science. 2006;312:1054–9. doi: 10.1126/science.1122088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeitlinger J, et al. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell. 2003;113:395–404. doi: 10.1016/s0092-8674(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 70.Herrgard MJ, Lee BS, Portnoy V, Palsson BO. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Research. 2006;16:627–635. doi: 10.1101/gr.4083206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim JB, et al. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–4. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 72.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–7. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 73.Bonneau R, et al. A predictive model for transcriptional control of physiology in a free living cell. Cell. 2007;131:1354–65. doi: 10.1016/j.cell.2007.10.053. [DOI] [PubMed] [Google Scholar]
- 74.Perez-Rueda E, Collado-Vides J. The repertoire of DNA-binding transcriptional regulators in Escherichia coli K-12. Nucleic Acids Res. 2000;28:1838–47. doi: 10.1093/nar/28.8.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Price MN, Dehal PS, Arkin AP. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput Biol. 2007;3:1739–50. doi: 10.1371/journal.pcbi.0030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salgado H, et al. RegulonDB (version 5.0): Escherichia coli K-12 transcriptional regulatory network, operon organization, and growth conditions. Nucleic Acids Res. 2006;34:D394–7. doi: 10.1093/nar/gkj156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shmulevich I, Dougherty ER, Kim S, Zhang W. Probabilistic Boolean Networks: a rule-based uncertainty model for gene regulatory networks. Bioinformatics. 2002;18:261–74. doi: 10.1093/bioinformatics/18.2.261. [DOI] [PubMed] [Google Scholar]
- 78.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–40. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 79.Gianchandani EP, Papin JA, Price ND, Joyce AR, Palsson BO. Matrix Formalism to Describe Functional States of Transcriptional Regulatory Systems. PLoS Comput Biol. 2006;2:e101. doi: 10.1371/journal.pcbi.0020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kao KC, et al. Transcriptome-based determination of multiple transcription regulator activities in Escherichia coli by using network component analysis. Proc Natl Acad Sci U S A. 2004;101:641–6. doi: 10.1073/pnas.0305287101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yamamoto K, et al. Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli. J Biol Chem. 2005;280:1448–56. doi: 10.1074/jbc.M410104200. [DOI] [PubMed] [Google Scholar]
- 82.Skerker JM, Prasol MS, Perchuk BS, Biondi EG, Laub MT. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 2005;3:e334. doi: 10.1371/journal.pbio.0030334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seshasayee AS, Bertone P, Fraser GM, Luscombe NM. Transcriptional regulatory networks in bacteria: from input signals to output responses. Curr Opin Microbiol. 2006;9:511–9. doi: 10.1016/j.mib.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 84.Vogel J, Wagner EG. Target identification of small noncoding RNAs in bacteria. Curr Opin Microbiol. 2007;10:262–70. doi: 10.1016/j.mib.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 85.Romby P, Vandenesch F, Wagner EG. The role of RNAs in the regulation of virulence-gene expression. Curr Opin Microbiol. 2006;9:229–36. doi: 10.1016/j.mib.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 86.Vogel J, Sharma CM. How to find small non-coding RNAs in bacteria. Biol Chem. 2005;386:1219–38. doi: 10.1515/BC.2005.140. [DOI] [PubMed] [Google Scholar]
- 87.Altuvia S. Identification of bacterial small non-coding RNAs: experimental approaches. Curr Opin Microbiol. 2007;10:257–61. doi: 10.1016/j.mib.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 88.Shimoni Y, et al. Regulation of gene expression by small non-coding RNAs: a quantitative view. Mol Syst Biol. 2007;3:138. doi: 10.1038/msb4100181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee JM, Gianchandani EP, Eddy JA, Papin JA. Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Comput Biol. 2008;4:e1000086. doi: 10.1371/journal.pcbi.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cakir T, et al. Flux balance analysis of a genome-scale yeast model constrained by exometabolomic data allows metabolic system identification of genetically different strains. Biotechnol Prog. 2007;23:320–6. doi: 10.1021/bp060272r. [DOI] [PubMed] [Google Scholar]
- 91.Vemuri GN, Eiteman MA, McEwen JE, Olsson L, Nielsen J. Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2007;104:2402–7. doi: 10.1073/pnas.0607469104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pramanik J, Keasling JD. Stoichiometric model of Escherichia coli metabolism: Incorporation of growth-rate dependent biomass composition and mechanistic energy requirements. Biotechnology and Bioengineering. 1997;56:398–421. doi: 10.1002/(SICI)1097-0290(19971120)56:4<398::AID-BIT6>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 93.Pramanik J, Keasling JD. Effect of Escherichia coli biomass composition on central metabolic fluxes predicted by a stoichiometric model. Biotechnology and Bioengineering. 1998;60:230–238. doi: 10.1002/(sici)1097-0290(19981020)60:2<230::aid-bit10>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 94.Segre D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A. 2002;99:15112–7. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shlomi T, Berkman O, Ruppin E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc Natl Acad Sci U S A. 2005;102:7695–700. doi: 10.1073/pnas.0406346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Feist AM, Scholten JCM, Palsson BO, Brockman FJ, Ideker T. Modeling methanogenesis with a genome-scale metabolic reconstruction of Methanosarcina barkeri. Mol Syst Biol. 2006;2:1–14. doi: 10.1038/msb4100046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pharkya P, Burgard AP, Maranas CD. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 2004;14:2367–76. doi: 10.1101/gr.2872004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Harrison R, Papp B, Pal C, Oliver SG, Delneri D. Plasticity of genetic interactions in metabolic networks of yeast. Proc Natl Acad Sci U S A. 2007;104:2307–12. doi: 10.1073/pnas.0607153104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Almaas E, Kovacs B, Vicsek T, Oltvai ZN, Barabasi AL. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427:839–843. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- 100.Burgard AP, Vaidyaraman S, Maranas CD. Minimal reaction sets for Escherichia coli metabolism under different growth requirements and uptake environments. Biotechnol Prog. 2001;17:791–7. doi: 10.1021/bp0100880. [DOI] [PubMed] [Google Scholar]
- 101.Mahadevan R, Schilling CH. The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng. 2003;5:264–76. doi: 10.1016/j.ymben.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 102.Hucka M, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 103.Shlomi T, et al. Systematic condition-dependent annotation of metabolic genes. Genome Res. 2007 doi: 10.1101/gr.6678707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Saito N, et al. Metabolomics approach for enzyme discovery. J Proteome Res. 2006;5:1979–87. doi: 10.1021/pr0600576. [DOI] [PubMed] [Google Scholar]
- 105.Chiang KP, Niessen S, Saghatelian A, Cravatt BF. An enzyme that regulates ether lipid signaling pathways in cancer annotated by multidimensional profiling. Chem Biol. 2006;13:1041–50. doi: 10.1016/j.chembiol.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 106.Fuhrer T, Chen L, Sauer U, Vitkup D. Computational prediction and experimental verification of the gene encoding the NAD+/NADP+-dependent succinate semialdehyde dehydrogenase in Escherichia coli. J Bacteriol. 2007;189:8073–8078. doi: 10.1128/JB.01027-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Popescu L, Yona G. Automation of gene assignments to metabolic pathways using high-throughput expression data. BMC Bioinformatics. 2005;6:217. doi: 10.1186/1471-2105-6-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rodionov DA, et al. Genomic identification and in vitro reconstitution of a complete biosynthetic pathway for the osmolyte di-myo-inositol-phosphate. Proc Natl Acad Sci U S A. 2007;104:4279–84. doi: 10.1073/pnas.0609279104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bernal A, Ear U, Kyrpides N. Genomes OnLine Database (GOLD): a monitor of genome projects world-wide. Nucleic Acids Res. 2001;29:126–7. doi: 10.1093/nar/29.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alm EJ, et al. The MicrobesOnline Web site for comparative genomics. Genome Res. 2005;15:1015–22. doi: 10.1101/gr.3844805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rey S, et al. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 2005;33:D164–8. doi: 10.1093/nar/gki027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wheeler DL, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Apweiler R, et al. UniProt: the Universal Protein knowledgebase. Nucleic Acids Res. 2004;32:D115–9. doi: 10.1093/nar/gkh131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Borodina I, Krabben P, Nielsen J. Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res. 2005;15:820–9. doi: 10.1101/gr.3364705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18 (Suppl 1):S225–32. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 117.Arakawa K, Yamada Y, Shinoda K, Nakayama Y, Tomita M. GEM System: automatic prototyping of cell-wide metabolic pathway models from genomes. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pinney JW, Shirley MW, McConkey GA, Westhead DR. metaSHARK: software for automated metabolic network prediction from DNA sequence and its application to the genomes of Plasmodium falciparum and Eimeria tenella. Nucleic Acids Res. 2005;33:1399–409. doi: 10.1093/nar/gki285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goesmann A, Haubrock M, Meyer F, Kalinowski J, Giegerich R. PathFinder: reconstruction and dynamic visualization of metabolic pathways. Bioinformatics. 2002;18:124–9. doi: 10.1093/bioinformatics/18.1.124. [DOI] [PubMed] [Google Scholar]
- 120.Satish Kumar V, Dasika MS, Maranas CD. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Green ML, Karp PD. A Bayesian method for identifying missing enzymes in predicted metabolic pathway databases. BMC Bioinformatics. 2004;5:76. doi: 10.1186/1471-2105-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Henry CS, Jankowski MD, Broadbelt LJ, Hatzimanikatis V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys J. 2006;90:1453–61. doi: 10.1529/biophysj.105.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kümmel A, Panke S, Heinemann M. Systematic assignment of thermodynamic constraints in metabolic network models. BMC Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Aziz RK, et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pouliot Y, Karp PD. A survey of orphan enzyme activities. BMC Bioinformatics. 2007;8:244. doi: 10.1186/1471-2105-8-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Thomason LC, Court DL, Datta AR, Khanna R, Rosner JL. Identification of the Escherichia coli K-12 ybhE gene as pgl, encoding 6-phosphogluconolactonase. J Bacteriol. 2004;186:8248–53. doi: 10.1128/JB.186.24.8248-8253.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pinney JW, et al. Metabolic reconstruction and analysis for parasite genomes. Trends Parasitol. 2007;23:548–54. doi: 10.1016/j.pt.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 128.Balu B, Adams JH. Advancements in transfection technologies for Plasmodium. Int J Parasitol. 2007;37:1–10. doi: 10.1016/j.ijpara.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 129.Kirk K, Saliba KJ. Targeting nutrient uptake mechanisms in Plasmodium. Curr Drug Targets. 2007;8:75–88. doi: 10.2174/138945007779315560. [DOI] [PubMed] [Google Scholar]
- 130.Daily JP, et al. Distinct physiological states of Plasmodium falciparum in malaria-infected patients. Nature. 2007 doi: 10.1038/nature06311. [DOI] [PubMed] [Google Scholar]
- 131.Deitsch K, et al. Mechanisms of gene regulation in Plasmodium. Am J Trop Med Hyg. 2007;77:201–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.