Deep-sequencing of human argonaute-associated small RNAs provides insight into miRNA sorting and reveals argonaute association with RNA fragments of diverse origin (original) (raw)
Abstract
While several studies have focused on the relationship between individual miRNA loci or classes of small RNA with human Argonaute (AGO) proteins, a comprehensive, global analysis of the RNA content associating with different AGO proteins has yet to be performed. We have compared the content of deep sequenced RNA extracted from immunoprecipitation experiments with the AGO1, AGO2 and AGO3 proteins. Consistent with previous observations, sequence tags derived from miRNA loci globally associate in approximately equivalent amounts with AGO1, AGO2 and AGO3. Exceptions include miR-182, miR-222 and miR-223*, which could be coupled to processes targeting the loci for interaction with specific AGO proteins. A closer inspection of the data, however, supports the presence of an unusual sorting mechanism wherein a subset of miRNA loci give rise to distinct isomirs which preferentially associate with distinct AGO proteins in a significantly differential manner. We also identify the complete set of short RNA derived from non-miRNA sources including tRNA, snRNA, snoRNA, vRNA and mRNA associating with the AGO proteins, many of which are predicted to play roles in post-transcriptional gene silencing. We also observe enrichment of tags mapping to promoter regions of genes, suggesting that a fraction of the recently-identified promoter-associated small RNAs in humans could function through interaction with AGO proteins. Finally, we observe antisense miRNA transcripts are frequently present in low copy numbers across a range of diverse miRNA loci and these transcripts appear to associate with AGO proteins.
Key words: AGO, argonaute, AGO1, AGO2, AGO3, tiRNA, anti-sense miRNA, miR-338, miR-182, snRNA
Introduction
In animals, an important layer of post-transcriptional regulation involves microRNAs (miRNAs), small RNA hairpin-like structures lacking perfect complementarity between a 5′ arm and 3′ arm. miRNAs are typically transcribed from the genome as primary (pri-miRNA) transcripts which are then processed into premiRNA hairpins of approximately 60–80 nucleotides in length via the action of the microprocessor complex containing the RNase III enzyme DROSHA and the RNA binding protein DGCR8.1,2 Pre-miRNAs are then exported to the cytoplasm3–5 and further processed into a duplex strand by DICER1 RNase III endonuclease-mediated removal of the hairpin region.6,7 Duplexes generated from DICER1 cleavage associate with Argonaute (AGO) proteins.8,9 One strand of the DICER1-generated duplex, known as the mature or guide strand, is selected for incorporation into the RNA-induced silencing complex (RISC) while the remaining star (miRNA*) or passenger strand is typically discarded but on occasion can also be incorporated into a functional RISC.10 Once bound, an miRNA that associates with a RISC complex contains a “seed” sequence of approximately seven nucleotides in length which binds complementary stretches of nucleotides in messenger RNA (mRNA) resulting in direct AGO-mediated endonuclease cleavage of the bound mRNA or a reduction in protein translation efficiency of the mRNA (reviewed in ref. 9 and 11). Collectively, this process is referred to as the miRNA-mediated gene silencing pathway.
Like many genes involved in post-transcriptional gene silencing pathways, genes encoding the AGO proteins have a complex evolutionary history punctuated by lineage-specific expansion and lineage-specific loss events.11–13 Consistent with this, the number and functional role of AGO-like proteins can vary greatly across animal species. One such functional difference concerns the sorting of miRNA into distinct AGO proteins. In Drosophila, several elegant studies have demonstrated that miRNA sorting into AGO proteins is coupled to strand selection and depends on specific structural and sequence criteria, with mature miRNA sequences tending to associate with AGO1 and miRNA* sequences tending to associate with AGO2.14–17 Similarly, in C. elegans, structural features intrinsic to miRNA lead to association with the ALG-1 and ALG-2 AGO-like proteins and not the RDE-1 AGO-like protein.18,19 In humans, previous studies have indicated such dramatic biases in miRNA association are not present across AGO proteins, at least in a handful of tested sequences20,21 or across miRNA profiles often generated for pairs of AGO proteins.22–24 Intriguingly, a recent study has suggested the possible presence of a mammalian-specific small-scale miRNA sorting system in which differentially-processed sequences from the same miRNA locus may preferentially associate with distinct AGO proteins.22
In parallel to studies analyzing the miRNA content across AGO proteins in different organisms, recent studies have discovered the association of small RNA fragments not derived from miRNA precursor transcripts with human AGO proteins. Fragments derived from small nucleolar RNA (snoRNA),23 vault RNA (vRNA),25 and tRNA26 undergo DICER1-mediated cleavage of the precursor RNA before association with AGO proteins and formation of functional RISCs. These non-miRNA-containing RISCs appear capable of regulating expression of target mRNAs analogous to miRNA-containing RISCs. Another recent study has characterized the association of an unprocessed pre-miRNA with human and mouse AGO2;27 the implication of this finding is that AGO proteins are capable of binding to RNAs of much longer length than mature miRNA sequences.
Despite the previous generation of AGO-associating miRNA profiles, the current literature in the field lacks a systematic approach addressing global miRNA sorting across distinct human AGO proteins and identifying the complete complement of non-miRNA short RNAs associating with AGO proteins. In this study we analyzed a recently generated dataset which captured short RNA profiles following immunoprecipitation (IP) of AGO1, AGO2 and AGO3 proteins alongside total RNA from the complete cell. Our findings confirm previous research indicating that with few exceptions, no bias across AGO proteins could be detected for the sorting of miRNA, either in the form of differential strand association or relative abundance of miRNA derived from the same locus in humans. At the same time, the findings presented here support the existence of a unique sorting system operating on a small scale that likely provides a means of expanding the set of mRNA substrates targeted by miRNA. Additionally, we identify novel classes of AGO-associating small RNA beyond vRNA, snoRNA and tRNA fragments, including sequence fragments derived from small nuclear RNA (snRNA), a single fragment derived from a predicted rRNA pseudogene and several unusual sequences of length 25–27 nucleotides derived from sense and anti-sense transcription of exonic regions of protein-coding genes. We also observed enrichment in the AGO-IP RNA for sequences mapping to the promoter regions of coding genes. Finally, we observed broad, low-level AGO-association of sequences mapping anti-sense to known miRNA loci.
Results
AGO-associated small RNA libraries.
We analyzed the short RNA dataset previously generated28 from complete THP-1 monocytic cells and from immunoprecipitated AGO1, AGO2 and AGO3 proteins isolated from THP-1 cells with antibodies from the Wako Company. Raw tags from the generated dataset were mapped to the hg18 human genome assembly using the procedures outlined in Materials and Methods.
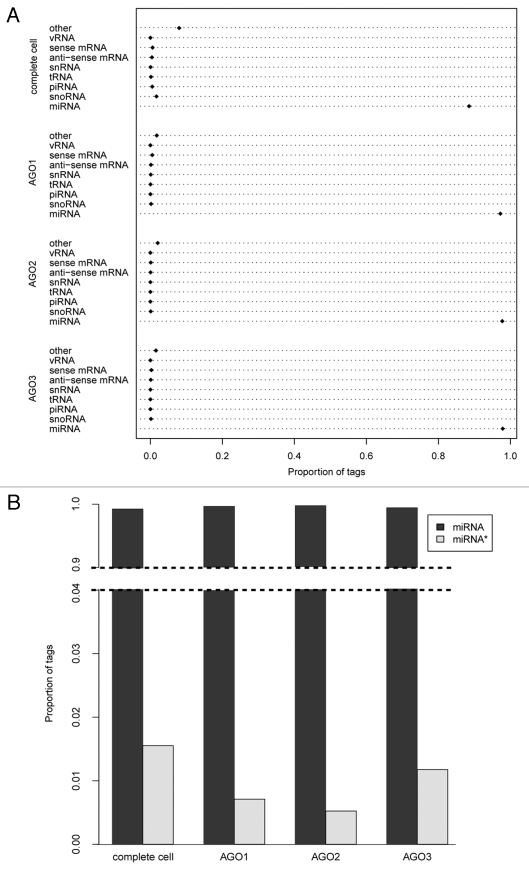
The percentage of tags mapping to distinct classes of RNA across the different libraries is depicted in Figure 1A and the total numbers of mapped tags are provided in Supplemental Table S1. Total mapped tag counts for the libraries ranged from 1.1 to 5.2 million tags, providing unprecedented sequencing depth for AGO-IP libraries.22–24,29 In each library, miRNA is the dominant class of RNA detected (Fig. 1A). The percentage of total miRNA increases in the three AGO protein-associated libraries relative to the library prepared from complete cell RNA, an indication that the IP experiments were successful. Additionally, the percent of miRNA observed across the three AGO proteins is constant, suggesting that no significant contaminations occurred during extraction in any single library.
Figure 1.
RNA associating with AGO proteins. (A) Overview of the classes of miRNA found in complete cell and associating with AGO proteins depicted as Cleveland dot plots. Classes of RNA are listed to the left, and proportion of total RNA derived from those classes is labeled along the x-axis. (B) Proportion of tags found across all libraries derived from the miRNA and miRNA* arm of the miRNA/miRNA* duplex.
Human miRNA loci typically lack strand selection or sorting bias when associating with distinct AGO proteins.
We first investigated whether differential strand partitioning occurred in miRNA/miRNA* strands similar to what is observed in Drosophila14–17 by determining their relative percentages in the AGO-associated libraries (Fig. 1B). In contrast to Drosophila,17 no difference is observed in uptake frequency across different AGO proteins. In each AGO-associating short RNA library, the total percentage of miRNA tags derived from the miRNA* arm of the miRNA/miRNA* duplex is lower than the total percentage observed in the complete cell (Fig. 1B). This modest accumulation of miRNA* sequences in the complete cell is consistent with the previous observation that miRNA* strand cleavage is not essential for miRNA AGO loading in mammalian cells.30 Interestingly, despite a lack of functional roles associated with human miRNA* strands in the current literature, approximately one percent of all sequences associating with AGO proteins descend from the miRNA* arm of the miRNA/miRNA* duplex. This may support findings of a recent study suggesting that miRNA* strands may play a greater role than previously assumed in affecting downstream gene expression in humans10 or it could represent the error rate for mis-association of miRNA/miRNA* duplex arms with AGO proteins.
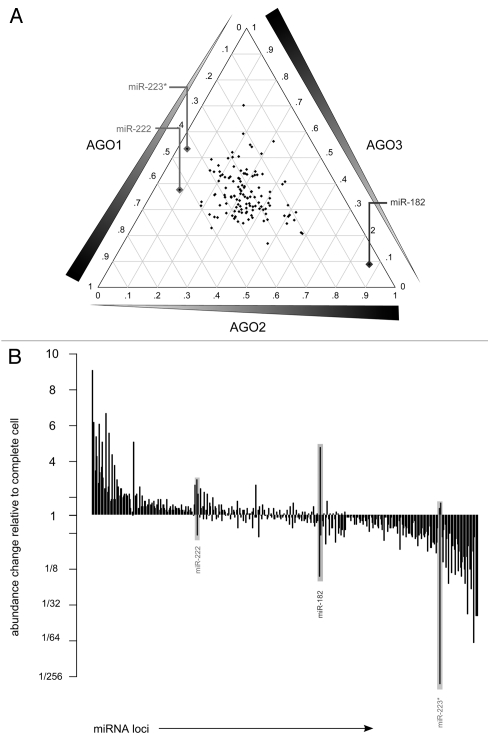
With no general differences in miRNA/miRNA* partitioning observed, the tag counts mapping to individual miRNA loci were compared against each other to ascertain the extent of total differential sorting across AGO proteins. The percentage of relative incorporation for each locus is depicted in a ternary plot in Figure 2A. Most miRNA loci are located at approximately equal (33%) abundance across the three AGO libraries on the plot, consistent with previous experimental observations20,21 and a recent large-scale sequencing experiment performed to complement miRNA PAR-CLIP sequencing24 (Fig. 2A and see Sup. Table S2 for tag counts). However, notable exceptions were observed. Two loci with at least moderate expression in THP-1, mir-222 and mir-223*, are significantly depleted (p < 2.2e−16) in the AGO2 library which contains less than 10% of all the sequences associating across the AGO libraries. The mir-182 locus, also moderately expressed in THP-1, is significantly enriched in the AGO2 library, containing over 85% of all tags found in the libraries (p < 2.2e−16) (Fig. 2A). These observations suggest that in THP-1 miR-222 and miR-223* are less likely to regulate downstream targets through mRNA transcript cleavage, as AGO2 is the sole Argonaute protein with demonstrated slicer activity.20,21 The relative abundance of miR-182 in the AGO2 library is most likely related to differential incorporation of specific miRNA isomir types, described in the section below.
Figure 2.
Sorting of individual miRNA loci across different AGO libraries. (A) Ternary plot of the relative proportion of counts mapping to miRNA loci across the three AGO libraries, with each diamond representing a single miRNA locus. Each side of the ternary plot measures the relative proportion for a single labeled library. Gridlines corresponding to a library are plotted at a 120° angle from the side of the ternary plot with the library name. Right triangles along each axis illustrate the direction of increasing proportion. Less than 10% of the total counts derived from the miR-182 locus are found in both the AGO1 and AGO3 libraries, this diamond is outlined and labeled with the name. miRNA loci with less than 10% of the total counts in a single library are outlined and labeled. (B) Barplot measuring expression value of miRNA loci associating with each AGO library relative to the expression of the same loci in the complete cell. Each locus has three distinct bars for the AGO1, AGO2 and AGO3 libraries. Loci are ordered by summing fold change across the three libraries, with the loci showing the most increase on the left-hand side. Complete list of fold changes are provided in Supplemental Table S2. Loci displaying differential sorting in (A) are outlined and labeled.
Despite a lack of differential sorting observed between the libraries, it is possible for simultaneous enrichment or depletion of a miRNA sequences to occur during the loading process. We calculated the relative abundance of individual miRNA loci in the complete cell against the abundance in individual AGO libraries (Fig. 2B). miRNA loci were typically found at roughly the same level of abundance in the complete cell as in the AGO-associating libraries (Fig. 2B). Instances of enrichment (fold change >2) or depletion (fold change <½) in one library almost always corresponded to respective enrichment or depletion in the other two libraries (Fig. 2B and see Sup. Table S3 for tag counts). Notable exceptions to this observation were consistent with the loci mentioned above (Fig. 2A).
The observation that miRNA sequences derived from the same locus can be simultaneously enriched or depleted across the three AGO proteins (Fig. 2B) could suggest systematic preferences govern the AGO loading process for all AGO proteins. Structural and sequence features of miRNA/miRNA* duplexes were investigated to identify any possible factors contributing to uniform enrichment/depletion. Paired probability values, previously used to establish structural determinants for the partitioning of miRNA/miRNA* duplexes in Drosophila AGO loading,17 were calculated at every nucleotide position in all miRNA/miRNA* duplexes whenever an miRNA sequence was enriched (fold change >2) across all three AGO libraries (Fig. 2A and Sup. Table S3) and this was compared to the same values for the set of miRNA/miRNA* duplexes depleted across all libraries (fold change <2). No significant differences were observed across the two sets of profiles in any AGO-associated library (Sup. Fig. S1). Nucleotide composition was also compared at each position in the enriched and depleted sets of miRNA loci and no differences were observed (data not shown). The lack of clear structural or sequence determinants for collective enrichment or depletion of miRNA loci in AGO-associated miRNA libraries may indicate that the regulation leading to enrichment/depletion of a given miRNA loci occurs upstream of the DICER1-mediated duplex formation step. Instances of upstream, miRNA-specific regulation have previously been reported, an example is seen in the blockage of let-7b pre-miRNA processing through the combined action of the LIN28A RNA-binding protein and the ZCCHC11 nucleotidyltransferase enzyme.31
Differential sorting of miRNA isomirs across AGO proteins.
Animal miRNA loci often give rise to multiple, distinct miRNA or miRNA* transcripts termed “isomirs” which can vary at the 5′ or 3′ ends of the miRNA sequence.32 At least some of the observed variation at the 3′ ends of miRNAs arises from post-transcriptional nucleotide addition,28,33 but instances of 5′ variation can be attributed to differential DICER1 endonuclease cutting34 and recent findings based on nucleotidyltransferase knockdown suggest that most observed 3′ miRNA variation which aligns to the genome sequence may often be the result of differential cutting.28 Often within an individual miRNA locus, a single isomir dominates the respective count at a given locus and is termed the “primary isomir”. On several occasions, however, the difference in expression between a primary isomir and a less-frequent variant is quite small.
Differential loading of distinct isomirs from the same miRNA locus into different AGO proteins has been previously reported for miR-142-5p.22 Differential cutting of this locus gives rise to two distinct isomirs, with one isomir containing a pair of nucleotides at the 5′ end not present in the other isomir. These isomirs are preferentially incorporated into different AGO proteins and target distinct mRNA substrates due to seed sequence differences.22 To investigate this on a global scale, we identified the primary isomir from each miRNA locus with the greatest abundance in each AGO-associated small RNA library. Loci with distinct primary isomirs across one or more libraries are reported in Table 1, along with the sequences for each isomir and the relative percentage of the observed primary isomirs in each library. Primary isomirs from 22 miRNA loci varied in at least one AGO protein, with the observed difference in relative abundance significantly differing across at least one pair of AGO libraries in 17 of these loci (Table 1). We observed the same 5′ isomir differences as previously reported for miR-142-5p and also observed 5′ differences at three additional loci: mir-140-3p, mir-16 and mir-29a (Table 1). Isomirs originating from these loci would also presumably target distinct mRNAs for gene silencing.
Table 1.
AGO-specific selective incorporation of distinct miRNA isomirs
| miRNA locus | Primary miRNA isomirs | AGO1%† | AGO2%† | AGO3%† | Significance††,‡ | Addition‡‡ |
|---|---|---|---|---|---|---|
| Differences in 5′ end | ||||||
| miR-140-3p | ACCACAGGGUAGAACCACGGAC | 42.75 | 61.24 | 63.54 | *^ | |
| UACCACAGGGUAGAACCACGGA | 57.25 | 38.76 | 36.46 | *^ | ||
| miR-142-5p | CAUAAAGUAGAAAGCACUACU | 21.15 | 81.09 | 31.55 | *^+ | |
| CCCAUAAAGUAGAAAGCACUAC | 78.85 | 18.91 | 68.45 | *+ | ||
| miR-16 | GCACGUAAAUAUUGGCG | 50.30 | 48.46 | 53.56 | + | |
| UAGCAGCACGUAAAUAUUGGCG | 49.70 | 51.54 | 46.44 | + | ||
| miR-29a | CUAGCACCAUCUGAAAUCGGUU | 28.99 | 20.38 | 59.94 | *^+ | |
| UAGCACCAUCUGAAAUCGGUU | 71.01 | 79.62 | 40.06 | ^+ | ||
| Differences in 3′ end | ||||||
| miR-10a | UACCCUGUAGAUCCGAAUUUGU | 36.84 | 53.15 | 37.32 | ||
| UACCCUGUAGAUCCGAAUUUGUG | 63.16 | 46.85 | 62.68 | |||
| miR-125a-5p | UCCCUGAGACCCUUUAACCUGU | 27.43 | 53.53 | 31.30 | *+ | |
| UCCCUGAGACCCUUUAACCUGUG | 72.57 | 46.47 | 68.70 | *+ | ||
| miR-15a | UAGCAGCACAUAAUGGUUUGUGG | 52.00 | 44.53 | 48.42 | * | |
| UAGCAGCACAUAAUGGUUUGUGGA | 48.00 | 55.47 | 51.58 | * | ||
| miR-182 | UUUGGCAAUGGUAGAACUCACACUGG | 100.00 | 27.20 | 100.00 | *+ | |
| UUUGGCAAUGGUAGAACUCACACUG | 0.00 | 72.80 | 0.00 | *+ | ||
| miR-221 | AGCUACAUUGUCUGCUGGGUUU | 55.80 | 41.08 | 59.07 | *+ | |
| AGCUACAUUGUCUGCUGGGUUUC | 44.20 | 58.92 | 40.93 | *+ | ||
| miR-222 | AGCUACAUCUGGCUACUGGG | 57.36 | 25.41 | 35.72 | *^+ | |
| AGCUACAUCUGGCUACUGGGUCUC | 42.64 | 74.59 | 64.28 | *^+ | ||
| miR-223 | UGUCAGUUUGUCAAAUACCCCA | 49.60 | 56.82 | 64.15 | *^+ | |
| UGUCAGUUUGUCAAAUACCCCAA | 50.40 | 43.18 | 35.85 | *^+ | ||
| miR-223* | CGUGUAUUUGACAAGCUGAGUUG | 44.44 | 18.18 | 39.70 | ||
| CGUGUAUUUGACAAGCUGAGUUGG | 40.28 | 19.32 | 44.24 | |||
| CGUGUAUUUGACAAGCUGAGUUGGACACU | 15.28 | 62.50 | 16.06 | *+ | ||
| miR-23a | AUCACAUUGCCAGGGAUUUCC | 36.58 | 66.77 | 41.11 | *+ | |
| AUCACAUUGCCAGGGAUUUCCAA | 63.42 | 33.23 | 58.89 | *+ | ||
| miR-23b | AUCACAUUGCCAGGGAUUACC | 36.64 | 55.79 | 33.44 | ||
| AUCACAUUGCCAGGGAUUACCAC | 63.36 | 44.21 | 66.56 | |||
| miR-24 | UGGCUCAGUUCAGCAGGAACAG | 41.25 | 51.71 | 47.76 | *^+ | |
| UGGCUCAGUUCAGCAGGAACAGU | 58.75 | 48.29 | 52.24 | *^+ | u | |
| miR-24-2* | GUGCCUACUGAGCUGAAACACAG | 55.26 | 41.05 | 45.67 | ||
| GUGCCUACUGAGCUGAAACACAGU | 44.74 | 58.95 | 54.33 | |||
| miR-30b | UGUAAACAUCCUACACUCAGC | 45.81 | 63.59 | 37.13 | *+ | |
| UGUAAACAUCCUACACUCAGCU | 54.19 | 36.41 | 62.87 | *+ | ||
| miR-374b | AUAUAAUACAACCUGCUAAGU | 36.68 | 52.40 | 35.81 | ||
| AUAUAAUACAACCUGCUAAGUG | 63.32 | 47.60 | 64.19 | |||
| miR-454 | UAGUGCAAUAUUGCUUAUAGGG | 60.44 | 20.80 | 28.30 | * | |
| UAGUGCAAUAUUGCUUAUAGGGUU | 39.56 | 79.20 | 71.70 | * | ||
| miR-484 | UCAGGCUCAGUCCCCUCCCGAU | 55.70 | 49.30 | 79.10 | ||
| UCAGGCUCAGUCCCCUCCCGAUU | 44.30 | 50.70 | 20.90 | + | u | |
| miR-577 | GUAGAUAAAAUAUUGGUACCUG | 60.00 | 34.46 | 58.39 | + | |
| GUAGAUAAAAUAUUGGUACCUGU | 40.00 | 65.54 | 41.61 | u | ||
| miR-660 | UACCCAUUGCAUAUCGGAGUUG | 54.31 | 37.37 | 40.22 | ||
| UACCCAUUGCAUAUCGGAGUUGU | 45.69 | 62.63 | 59.78 |
Interestingly, the remaining variations occur at the 3′ end. Given the frequency of 3′ variation in differential AGO-loading and the significant differences in incorporation tendencies for these distinct isomirs, it is possible that 3′ variation may also affect regulation in some as-yet undetermined manner. This could be particularly true in variations with large length differences such as miR-222 in which the isomir more likely to associate with AGO2 or AGO3 contains four additional nucleotides at the 3′ end relative to the isomir more commonly associating with AGO1 (Table 1). It is important to note that the 3′ end variations listed in Table 1 almost exclusively match the genome sequence encoded at the respective miRNA locus and predominantly involve nucleotide combinations that have not been linked to post-processing nucleotidyltransferase addition, suggesting that variations associated with differential isomir sorting are largely driven by DICER1-mediated cutting and not nucleotidyltransferase activity.28
One specific pair of interesting isomir variants is observed at the mir-182 locus (Table 1). mir-182 is one of three miRNA loci showing strong enrichment/depletion across AGO libraries (Fig. 2). Enrichment in the AGO2 library corresponds to enrichment of a shorter primary isomir lacking the additional 3′ guanine nucleotide of the longer primary isomir, which is the only isomir present in the AGO1 and AGO3 libraries (Table 1). It is likely for this locus that production of the shorter primary isomir is tightly linked with AGO2 uptake, resulting in both differential incorporation of this specific isomir and enrichment of the miRNA locus in the AGO2-associated library.
Expansion of the complement of RNA sequences associating with AGO proteins.
Several classes of small RNAs derived from diverse non-coding RNA progenitors have been recently reported in references 23, 25, 26, 35–38; some of these classes, including those derived from vRNA, snoRNA and tRNA, have also been shown to associate with human AGO proteins.23,25,26 We extracted all sequences from the AGO-associated deep-sequenced libraries which failed to map to known miRNA loci and checked for the presence of the same sequence in the complete cell library. The genomic location of all sequences from the AGO-associated libraries which were found to contain similar or greater abundance (Materials and Methods) as their corresponding sequences in the complete cell were inspected for concordance with known non-coding RNA sequences (Materials and Methods). In addition to sequences mapping to snoRNA, vRNA and tRNA, we identified tags mapping to snRNA and an rRNA pseudogene (Table 2). Several unusual sequences were also retrieved which mapped to exonic regions of mRNA. A small but significant number of sequences derived from the promoter regions of genes were also observed. Sequences mapping to these classes of RNA are discussed in detail below.
Table 2.
Association of unexpected RNA fragments in AGO libraries.
Sequences mapping to mRNA. Several tags, typically of length 21–23, were observed mapping to the intronic and exonic regions of mRNA in both the sense and anti-sense direction (Sup. Table S4). Some of these sequences have previously been identified in screens for novel miRNAs, suggesting that many of these tags are potential novel, lowly-expressed miRNAs. In addition, several tags of similar length which mapped to regions lacking annotation are included in Supplemental Table S4 and could represent novel miRNA loci.
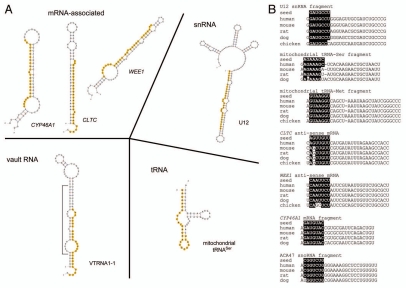
Three mRNA-mapping sequences were of particular interest due to their unusual length. Two of these sequences are 27 nucleotides in length and mapped anti-sense to the exonic regions of the CLTC and WEE1 genes while the other sequence, length 25 nucleotides, mapped in the sense direction to an exon of the CYP46A1 gene (Table 2). Despite a length longer than canonical miRNAs, these fragments are predicted to form hairpin-like structures resembling pre-miRNA (Fig. 3A), suggesting they could be processed by the DICER1 enzyme. Additionally, the predicted “seed sequence” for these fragments (bases 2–8 from the 5′ end) are well-conserved across vertebrate species (Fig. 3B) and many potential targets for such a seed sequence are recognized (see Sup. Table S5 for target information for all Table 2 entries). Taken together with recent experimental evidence that the mammalian AGO2 protein is capable of binding fragments larger than the standard miRNA length of 21–23 nucleotides,27 the preceding observations suggest that despite an unusual length, these sequences are performing miRNA-like gene-silencing functions and may be processed in a step analogous to DICER1 processing of pre-miRNAs. Notably, however, we failed to uncover a single sequence in any library mapping to the opposite arm in any predicted hairpin-like structure (Fig. 3A). Given the high number of total counts observed, particularly for sequences derived from CLTC and CYP46A1 (Table 2), at least a small number of star sequences would likely be recovered if biogenesis involved DICER1-mediated cleavage. Additionally, it is unclear how DICER1 could mediate cleavage of fragments of this longer length,39,40 although DICER1 appears capable of cutting longer fragments from tRNA precursors (see below).26 Expression of the _CYP46A1_-derived fragment was verified via qRT-PCR in RNA derived from the complete cell and also from AGO-IP experiments (Sup. Fig. S2, Materials and Methods).
Figure 3.
AGO association with sequences derived from various classes of non-coding RNA. (A) Two-dimensional structural diagrams of sequences identified in AGO libraries forming hairpin-like structures that conceivably are processed via DICE R1-mediated cleavage. Class of RNA is labeled at top of diagram. Name of specific RNA gene is given below diagram. Sequences identified in AGO libraries are highlighted by orange circles. In the diagram for VTRNA1-1, the sequence of the identified putative star sequence is enclosed in a bracket. (B) Conservation of putative seed sequences across vertebrate species for selected sequences listed in Table 2 (for complete list of alignments, see Sup. Fig. S3). Conserved regions in the seed sequence are colored in white and shaded in black.
Sequences mapping to snRNA. Three sequences mapping to U12, U1 and U2 snRNA and an additional sequence mapping to a predicted U2-like snRNA pseudogene were identified. These sequences range in length with sizes of 23, 27 and 30 nucleotides (Table 2) and at least one sequence (mapping to the U12 snRNA) was previously recovered through deep-sequencing with a different sequencing platform and library preparation technique.41 Unlike the sequences above which were recovered in all three libraries, sequences derived from snRNA were enriched in the AGO1 and AGO3 libraries. Of these four, only the sequences derived from the U12 snRNA locus appear to lie in a hairpin-like structure that would be a likely candidate for DICER1-mediated cleavage (Fig. 3A). However, similar to the above-mentioned sequences, we did not detect any potential candidate star sequences for the snRNA-derived sequences in any library again suggesting a processing mechanism independent of DICER1. Interestingly, three of the four sequences are located at the 3′ end of the precursor snRNA gene, possibly indicating a uniform cleavage mechanism (Table 3). With the exception of the U12-derived sequence, the predicted seed regions are poorly conserved across vertebrate species (Fig. 3B and see Sup. Fig. S3 for all putative seed alignments). Nevertheless, several candidate 3′UTR target sites for the sequences are predicted to occur in humans (Sup. Table S5), suggesting a role for at least some of the snRNA-derived sequences in gene-silencing.
Table 3.
Start and stop positions of AGO-associating sequences relative to genome position of non-coding RNA precursor.
| Precursor type | Precursor name | Chromosome | Strand | Tag start | Tag stop | Precursor stop | Precursor stop | Offset | 5′/3′ |
|---|---|---|---|---|---|---|---|---|---|
| snRNA | U12 | chr22 | + | 41341316 | 41341343 | 41341195 | 41341343 | 0 | 3′ |
| U1 | chr1 | − | 16865866 | 16865893 | 16865866 | 16866030 | 0 | 3′ | |
| U2 | chr10 | − | 103114594 | 103114617 | 103114594 | 103114782 | 0 | 3′ | |
| U2-like | chr6 | − | 89830006 | 89830036 | 89829940 | 89830128 | −8 | 5′ | |
| vault RNA | VTRNA1-1 | chr5 | + | 140071115 | 140071141 | 140071045 | 140071142 | −1 | 3′ |
| snoRNA | ACA45 | chr15 | + | 81221814 | 81221836 | 81221750 | 81221877 | −41 | 3′ |
| ACA25 | chr11 | − | 93103369 | 93103391 | 93103328 | 93103460 | −41 | 3′ | |
| U17a | chr1 | + | 28706464 | 28706489 | 28706464 | 28706670 | 0 | 5′ | |
| U17b | chr1 | + | 28707657 | 28707681 | 28707657 | 28707861 | 0 | 5′ | |
| HBI-100 | chr1 | − | 174204158 | 174204180 | 174204155 | 174204299 | −3 | 3′ | |
| ACA47 | chr17 | + | 72596984 | 72597009 | 72596984 | 72597170 | 0 | 5′ | |
| ACA17 | chr9 | − | 138741088 | 138741111 | 138741021 | 138741152 | −41 | 5′ | |
| tRNA | mitochondrial tRNA-Ser | chrM | + | 12207 | 12233 | 12207 | 12266 | 0 | 5′ |
| mitochondrial tRNA-Cys | chrM | − | 5797 | 5827 | 5762 | 5827 | 0 | 5′ | |
| tRNA-His-GTG | chr6 | + | 27233892 | 27233918 | 27233884 | 27233956 | −8 | 5′ | |
| mitochondrial tRNA-Met | chrM | + | 4403 | 4433 | 4403 | 4470 | 0 | 5′ | |
| tRNA-Glu-CTC | chr1 | − | 159698475 | 159698504 | 159698433 | 159698504 | 0 | 5′ | |
| tRNA-Leu-CAA | chr1 | + | 247134714 | 247134736 | 247134676 | 247134782 | −38 | 5′ | |
| rRNA | 28S-like | chr1 | − | 91625396 | 91625415 | 91625370 | 91625736 | 326 | 3′ |
Sequences mapping to vRNA. Recently, small RNA fragments derived from vRNA (svRNAs) were found to associate with AGO2 and regulate expression of the CYP3A4 gene.25 We were unable to detect the same sequences in our libraries, perhaps unsurprising given that svRNAs display tissue-specific expression patterns.25 However, we did detect marginal enrichment of a novel, low-expressed svRNA in the AGO3 library (Table 2). While the sequence does not match the previously reported svRNA sequences, it closely resembles the svRNAa* sequence but appears to have been cleaved from the vRNA precursor a single nucleotide upstream (Table 2 and Fig. 3A). In this case, we did detect a single “star” sequence in the AGO3-associated RNA library, consistent with previously observed DICER1-mediated vRNA cleavage (Fig. 3A).25 These observations have two important implications for svRNAs: (1) the choice of which “arm” is loaded after DICER1 processing of vault RNA may vary across cell types and (2) the DICER1 cleavage point may also vary across cell types. Variation at the 5′ end is particularly noteworthy, as this would provide the cell with a flexible means of targeting a range of substrates depending on variable processing, similar to what we characterize in Table 1 and what has been previously reported in reference 22.
Sequences mapping to tRNA. Several recent studies have identified different classes of tRNA-derived short RNA fragments produced through distinct biogenesis pathways which possibly occupy distinct functional niches in the cell.26,35–38 We detected the presence of several sequences mapping to tRNA precursors that were primarily enriched in the AGO1 library (Table 2) and verified the expression of one of these sequences in RNA derived from AGO-IP using qRT-PCR (Sup. Fig. S2). These sequences most closely match the so-called tRNA-derived RNA fragments which map to the 5′ end of tRNAs (tRF-5 series).37 Interestingly, only the sequence derived from tRNAGlu-CTC matches a previously identified tRF-5 and the most abundant sequence identified in these libraries is considerably longer, 26 vs. 21 nucleotides (Table 2).37 In fact, the most abundant 5′ end-derived tRNA fragments have longer lengths than the previously reported length range (Table 3).37 Interestingly, we also observed a sequence mapping to the 5′ end of mitochondrial tRNAMet enriched in the AGO1 library which could be related to previous research describing the mitochondrial export and subsequent association of the tRNAMet precursor with AGO1 and AGO2 proteins.42 A number of important inferences can be drawn from these observations. First, the cleaved length of tRF-5 sequences may be controlled in a cell-specific manner and could even be processed through distinct mechanisms. Additionally, selection of precursor tRNA for processing and consequent tRF-5 expression may also be controlled in a cell-specific manner. Finally, as this is the first report of AGO protein association, it is reasonable to predict that tRF-5 sequences are involved in gene-silencing.
The mechanism for processing tRF-5-like sequences has yet to be elucidated. At least one of the observed tRNA sequence fragments in the data analyzed here, derived from the mitochondrial tRNASer gene, contains a severely abbreviated D-loop making it a possible candidate for canonical DICER1 processing (Fig. 3A). Interestingly, we observed that the unusual three-dimensional structural properties of tRNA result in the tRF-5 AGO-associating fragments having approximately equivalent lengths with processed miRNA sequence lengths when mapped on the tRNA structure (Sup. Fig. S4). Superimposition of a solved tRNAGlu structure on the solved structure of the DICER1 enzyme from Giardia40 suggests that the tRNA cleavage point identified in the analyzed sequencing data aligns with the DICER1 active site. In this scenario, the 3′ end of the tRNA could be positioned to interact with the PAZ domain, consistent with proposed miRNA processing (Sup. Fig. S5).40 Thus, the intrinsic structural properties of tRNA could facilitate the processing of longer tRF-5-like sequences through the pre-miRNA cleavage machinery.
RNA fragments derived from the 3′ end of tRNA, the so-called “tRF-3” sequences,37 also referred to as DICER1-dependent type I tsRNAs, were first identified by Babiarz and colleagues36 and later investigated in detail by Haussecker and colleagues,26 who persuasively show these sequences associate with Argonaute proteins and participate in gene silencing.26 Genome mapping of these sequences is complicated by 3′ CCA modifications which introduce several mismatches (see Sup. Discussion). However, manual searching of known tRF-3 sequences in the raw, unprocessed data identified the presence of this class of tRNA fragments and also confirmed their association with AGO proteins (data not shown).26
We were unable to detect the presence of the shorter, RNaseZ-derived and AGO-associating tRF-1 series, also referred to as type II tsRNA sequences, although we did detect these in the complete cell library26 (data not shown) suggesting that AGO-association with these fragments may occur at a level too low to be detected by deep-sequencing following IP. Additionally, we were unable to detect the recently reported class of DICER1-dependent RNA fragments of approximately 18–19 nucleotides in length, consistent with a lack of observed association between these fragments and AGO proteins.37 It is also important to note that the size fraction of the prepared short RNA libraries precludes detection of short RNA fragments with a length of greater than 30 nucleotides. Previously, a class of tRNA fragments centered on a length of 37 nucleotides was observed in humans.38 Given the observations above, it is possible these fragments also associate with AGO proteins. Similarly observed fragment classes derived from both snoRNA and snRNA with a length greater than 30 nucleotides could also associate with AGO proteins.38
Sequences mapping to promoter regions of coding genes. At least three possibly overlapping classes of promoter-derived non-coding RNA sequences with varying lengths of between 18 and 200 nucleotides have been described in animals: the transcription initiation RNAs (tiRNAs),43 transcription start site associated RNAs (TSSa-RNAs),44 and promoter-associated small RNAs (PASRs).45,46 At the same time, AGO proteins have been implicated in the regulation of transcript expression in the nucleus47 via interaction between exogenously introduced, promoter-targeting siRNA duplexes (sometimes referred to as antigene RNAs or agRNAs) with endogenously-transcribed non-coding RNA transcripts which overlap promoter regions in sense or antisense directions.48–52 The size of these overlapping non-coding RNA transcripts is typically at least several hundred nucleotides in length. Combining these two observations, we reasoned that some promoter-associated small RNAs could function as endogenous agRNAs and the AGO-IP libraries were searched for tags mapping to promoter regions (Materials and Methods).
Sequence tag counts for recovered sequences are given in Supplemental Table S6 and the mapping locations and nucleotide composition of all promoter-mapping sequences recovered from the AGO-IP libraries are provided in Supplemental Table S7. The percent of promoter-mapping tags observed in the complete cell RNA library is comparable to percentages observed in other previously-studied deep-sequenced libraries.43 Surprisingly, a combined 1,044 tags were also observed in the AGO-IP libraries, with individual libraries containing approximately 10× fewer total tags than the complete cell library with the AGO1-IP and AGO3-IP libraries containing higher total percentages of these tags relative to the AGO2-IP library (Sup. Table S6). Comparison to tag counts obtained from randomized trials suggests promoter regions are significantly enriched for tags (p < 2.2e−16 for all AGO-IP libraries) (see Sup. Fig. S6, Materials and Methods).
Characteristics of the promoter-derived sequences were investigated in an effort to assess which class of promoter-derived non-coding RNA they most closely resemble. As PASRs are thought to be primarily modified at their 5′ end through capping and the deep-sequencing library construction protocol is not compatible with 5′ capped transcripts, it seems unlikely that these tags are related to PASRs.53 The distribution of calculated distances from the transcriptional start site (TSS) is similar to both tiRNAs and TSSa-RNAs (Sup. Fig. S7).43,44 The length distributions of both tiRNAs and TSSa-RNAs derived from deep-sequencing data are reported to be centered around 18–20 nucleotides. Promoter-mapping sequences from the complete RNA dataset here appear slightly larger; shifted to around 20–21 nucleotides although a clear peak is visible at 18–19 nucleotides (Sup. Fig. S7). Interestingly, the peak of the length distribution of the AGO-associating promoter-derived RNAs is markedly shifted to 21–24 nucleotides in length, with the 18–19 nucleotide sequences absent (Sup. Fig. S7). As the tiRNA data was initially discovered in the THP-1 cell line, we directly compared identified tiRNA sequences43 with the AGO-IP libraries. Several tags were exactly the same and others overlapped substantially with identified tiRNAs (Sup. Table S7).
tiRNAs identified in Drosophila deep-sequenced short RNA datasets were previously compared to short RNAs sequenced after immunoprecipitation and extraction of Drosophila AGO-associating short RNAs and no overlap between the two populations was reported.43 The findings presented here, however, suggest at least a fraction of tiRNA-like transcripts with lengths resembling that of mature miRNA sequences may associate with human AGO proteins, perhaps indicative of a novel, endogenous agRNA-like function for a subset of tiRNA-like transcripts as well as pointing to another potential functional difference between Drosophila and human AGO proteins.
Sequences mapping to snoRNA and other locations. The incorporation of sequences derived from snoRNA into AGO proteins has previously been investigated in detail in referene 23, and may be an ancient feature of eukaryotic genomes.54 While we identified several sequences associating with AGO proteins, these sequences were not enriched (fold change >2) in the AGO libraries relative to the complete cell but instead were present in roughly the same abundance levels (Table 2 and Fig. 1A). The observation that the ACA45 miRNA-like snoRNA sequence is present in roughly equivalent amounts in the complete cell and in AGO-IP libraries and yet has clearly been shown to be an active regulator of gene expression through association with AGO proteins strongly supports an active functional role for other sequences we identify here as being considerably more enriched in the AGO libraries (Table 2).
An additional lowly-expressed sequence present in the AGO1 library maps to a predicted 28S-like rRNA pseudogene. This, along with the sequence mapping to the U2-like snRNA pseudogene, raises the intriguing possibility that at least some non-coding RNA pseudogenes are under selective pressure to be retained in the genome and transcribed due to a functional role as a precursor to an RNA fragment utilized by the post-transcriptional gene-silencing machinery, a process likely distinct from the derivation of siRNAs from pseudogenes of coding RNA.55
Widespread, non-sporadic association of anti-sense miRNA transcripts with AGO proteins.
The strand opposite of a genomic locus encoding a miRNA locus could necessarily be capable of forming a hairpin-like structure. A series of studies described anti-sense miRNA (asmiRNA) transcription at the Drosophila mir-iab-4 locus and demonstrated that the anti-sense transcripts are involved in repression of gene expression.56–58 Two of the studies also found evidence in the form of ESTs or cDNA transcripts, computational predictions or tags extracted from deep-sequenced data sets that some miRNA loci in mouse and humans might also produce anti-sense tags.56,58 We discovered widespread but lowly-expressed association of anti-sense miRNA sequences with AGO proteins in our pull-down libraries in addition to sequences found in the complete cell RNA library (Table 4).
Table 4.
asmiRNA raw tag counts across libraries
| miRNA locus | Sequence† | complete THP1 | AGO1 | AGO2 | AGO3 | FANTOM4 THP141 | H52059 | A54959 | MCF759 | U2OS59 |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-24 | primary sense isomir | 36561 | 9249 | 39194 | 17669 | 21604 | 24189 | 26911 | -- | 22886 |
| GUUCCUGCUGAACUGAGCCAGU | 1 | 1 | 1 | 4 | 1 | 1 | 1 | -- | 5 | |
| GUUCCUGCUGAACUGAGCCAG | 0 | 1 | 2 | 0 | 1 | 0 | 0 | -- | 0 | |
| UUCCUGCUGAACUGAGCCAGU | 2 | 0 | 1 | 0 | 0 | 0 | 0 | -- | 2 | |
| UGUUCCUGCUGAACUGAGCCAG | 0 | 0 | 1 | 1 | 1 | 0 | 0 | -- | 1 | |
| miR-24-1* | primary sense isomir | 5 | 2 | 5 | 13 | 6 | 13 | -- | 14 | 24 |
| UGCCUACUGAGCUGAUAUCAGU | 2 | 1 | 0 | 0 | 0 | 0 | -- | 0 | 0 | |
| UGAUAUCAGCUCAGUAGGCACCG | 0 | 0 | 1 | 0 | 1 | 0 | -- | 0 | 2 | |
| GAUAUCAGCUCAGUAGGCACCG | 0 | 0 | 0 | 1 | 0 | 0 | -- | 1 | 3 | |
| UGAUAUCAGCUCAGUAGGCA | 0 | 0 | 0 | 0 | 0 | 3 | -- | 0 | 0 | |
| GAUAUCAGCUCAGUAGGCACC | 0 | 0 | 0 | 0 | 0 | 0 | -- | 2 | 0 | |
| AUAUCAGCUCAGUAGGCACCG | 0 | 0 | 0 | 0 | 0 | 0 | -- | 0 | 1 | |
| miR-126 | primary sense isomir | 3050 | -- | 7327 | -- | -- | -- | -- | -- | 354 |
| CAUUAUUACUCACGGUACGAGU | 1 | -- | 1 | -- | -- | -- | -- | -- | 1 | |
| CGCAUUAUUACUCACGGUACGA | 0 | -- | 0 | -- | -- | -- | -- | -- | 1 | |
| miR-126* | primary sense isomir | -- | -- | 1838 | -- | -- | 85 | -- | -- | -- |
| CGCGUACCAAAAGUAAUAAUGU | -- | -- | 1 | -- | -- | 0 | -- | -- | -- | |
| CGCGUACCAAAAGUAAUAAUGUC | -- | -- | 0 | -- | -- | 2 | -- | -- | -- | |
| miR-181a | primary sense isomir | -- | -- | -- | -- | -- | 632 | -- | -- | -- |
| ACUCACCGACAGCGUUGAAUGUUC | -- | -- | -- | -- | -- | 1 | -- | -- | -- | |
| miR-181a-2* | primary sense isomir | -- | -- | -- | -- | -- | -- | 11 | 1 | 16 |
| UACAGUCAACGGUCAGUGGUUU | -- | -- | -- | -- | -- | -- | 4 | 0 | 1 | |
| UACAGUCAACGGUCAGUGGUU | -- | -- | -- | -- | -- | -- | 0 | 1 | 0 | |
| miR-183* | primary sense isomir | -- | -- | -- | -- | -- | -- | -- | -- | 30 |
| UUAUGGCCCUUCGGUAAUUCACUG | -- | -- | -- | -- | -- | -- | -- | -- | 1 | |
| UUAUGGCCCUUCGGUAAUUCACU | -- | -- | -- | -- | -- | -- | -- | -- | 1 | |
| miR-191* | primary sense isomir | 20 | -- | 50 | 36 | -- | -- | -- | -- | -- |
| GGACGAAAUCCAAGCGCAGCUG | 1 | -- | 1 | 1 | -- | -- | -- | -- | -- | |
| GGACGAAAUCCAAGCGCAGCU | 0 | -- | 1 | 0 | -- | -- | -- | -- | -- | |
| miR-199a/199b | primary sense isomir | -- | -- | -- | -- | 63 | 154 | -- | -- | -- |
| UAACCAAUGUGCAGACUACUGU | -- | -- | -- | -- | 1 | 6 | -- | -- | -- | |
| miR-210-3p | primary sense isomir | -- | -- | -- | -- | 6068 | -- | -- | -- | -- |
| UCAGCCGCUGUCACACGCAC | -- | -- | -- | -- | 1 | -- | -- | -- | -- | |
| miR-219-1-3p | primary sense isomir | -- | -- | 5 | -- | -- | -- | -- | -- | -- |
| ACGUCCAGACUCAACUCUCGG | -- | -- | 1 | -- | -- | -- | -- | -- | -- | |
| miR-219-5p | primary sense isomir | -- | -- | 11 | -- | -- | -- | -- | 5 | 53 |
| UCGAGAAUUGCGUUUGGACA | -- | -- | 1 | -- | -- | -- | -- | 0 | 0 | |
| UCGAGAAUUGCGUUUGGACAAU | -- | -- | 0 | -- | -- | -- | -- | 0 | 1 | |
| AGAAUUGCGUUUGGACAAUCAG | -- | -- | 0 | -- | -- | -- | -- | 1 | 0 | |
| AGAAUUGCGUUUGGACAAUCAGU | -- | -- | 0 | -- | -- | -- | -- | 1 | 1 | |
| miR-26a | primary sense isomir | -- | 498 | -- | -- | -- | -- | -- | -- | -- |
| CCUAUCCUGGAUUACUUGAACG | -- | 1 | -- | -- | -- | -- | -- | -- | -- | |
| miR-30a | primary sense isomir | -- | -- | -- | -- | -- | -- | -- | -- | 474 |
| UCCAGUCGAGGAUGUUUACAGU | -- | -- | -- | -- | -- | -- | -- | -- | 2 | |
| miR-33b | primary sense isomir | 45 | -- | -- | -- | 1621 | -- | 159 | -- | 210 |
| AAUGCAACAGCAAUGCACCGC | 1 | -- | -- | -- | 0 | -- | 1 | -- | 2 | |
| CAAUGCAACAGCAAUGCACCGC | 0 | -- | -- | -- | 1 | -- | 0 | -- | 2 | |
| AAUGCAACAGCAAUGCACCGCG | 0 | -- | -- | -- | 0 | -- | 0 | -- | 2 | |
| miR-33b* | primary sense isomir | -- | -- | -- | -- | -- | -- | -- | -- | 54 |
| GCUGCACUGCCGAGGCACUGC | -- | -- | -- | -- | -- | -- | -- | -- | 1 | |
| miR-34b | primary sense isomir | 5 | 6 | -- | -- | -- | -- | -- | -- | -- |
| UGGCAGUGGAGUUAGUGAUUGU | 1 | 0 | -- | -- | -- | -- | -- | -- | -- | |
| UGGCAGUGGAGUUAGUGAUUGUAA | 0 | 1 | -- | -- | -- | -- | -- | -- | -- | |
| miR-338-5p | primary sense isomir | 51 | 45 | 51 | 45 | 5 | 8 | -- | 1 | -- |
| UCAGCACCAGGAUAUUGUUGGA | 18 | 16 | 44 | 8 | 2 | 9 | -- | 3 | -- | |
| UCAGCACCAGGAUAUUGUUGGAG | 7 | 3 | 3 | 1 | 3 | 1 | -- | 1 | -- | |
| UCAGCACCAGGAUAUUGUUGG | 2 | 1 | 1 | 1 | 2 | 0 | -- | 4 | -- | |
| UCAGCACCAGGAUAUUGUU | 0 | 0 | 0 | 0 | 0 | 0 | -- | 2 | -- | |
| CAGCACCAGGAUAUUGUUGGAGA | 0 | 0 | 0 | 0 | 1 | 0 | -- | 0 | -- | |
| CAGCACCAGGAUAUUGUUGGAG | 0 | 0 | 0 | 0 | 1 | 0 | -- | 0 | -- | |
| CAGCACCAGGAUAUUGUUGGA | 0 | 0 | 0 | 0 | 1 | 0 | -- | 1 | -- | |
| miR-338-3p | primary sense isomir | 193 | 118 | 291 | 85 | 191 | 25 | 7 | 6 | 12 |
| UCAACAAAAUCACUGAUGCUGG | 21 | 13 | 47 | 11 | 5 | 6 | 0 | 16 | 6 | |
| UCAACAAAAUCACUGAUGCUGGA | 29 | 9 | 46 | 22 | 4 | 32 | 0 | 27 | 10 | |
| UCAACAAAAUCACUGAUGCUGGAG | 11 | 3 | 57 | 25 | 5 | 15 | 25 | 10 | 5 | |
| UCAACAAAAUCACUGAUGCUGGAGU | 3 | 0 | 18 | 6 | 4 | 25 | 0 | 25 | 5 | |
| UCAACAAAAUCACUGAUGCUG | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 3 | 0 | |
| UCAACAAAAUCACUGAUGCU | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| AACAAAAUCACUGAUGCUGGA | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | |
| AACAAAAUCACUGAUGCUGG | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | |
| CAACAAAAUCACUGAUGCUGGA | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | |
| CAAAAUCACUGAUGCUGGAGU | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| UUCAACAAAAUCACUGAUGCUGGA | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | |
| miR-499-5p | primary sense isomir | -- | -- | -- | 3 | -- | -- | -- | -- | 44 |
| AAACAUCACUGCAAGUCUUAAC | -- | -- | -- | 1 | -- | -- | -- | -- | 0 | |
| AACAUCACUGCAAGUCUUAACA | -- | -- | -- | 0 | -- | -- | -- | -- | 2 | |
| miR-93 | primary sense isomir | -- | -- | -- | -- | -- | -- | -- | -- | 9435 |
| CUGCACGAACAGCACUUUGGA | -- | -- | -- | -- | -- | -- | -- | -- | 1 | |
| miR-96-3p | primary sense isomir | -- | -- | -- | -- | -- | -- | -- | 3 | 3 |
| AUUGGCACUGCACAUGAUUGCU | -- | -- | -- | -- | -- | -- | -- | 1 | 1 | |
| AUUGGCACUGCACAUGAUUGC | -- | -- | -- | -- | -- | -- | -- | 0 | 1 |
The counts for these sequences are low enough at most loci to dismiss as possible artifacts. However, these tags consistently appear at the same loci across the four libraries and are often found at both the 3′ and 5′ arms of the same miRNA locus (Table 4). We also searched publicly-available short RNA libraries with comparable sequencing depth constructed in different human cell types with different library preparation techniques and produced on different sequencing platforms.41,59,60 Again we observed anti-sense miRNA tags; many mapping to the same miRNA loci suggesting that low-level anti-sense miRNA transcription is widespread and occurs in a non-sporadic fashion (Table 4).
We extended this systematic analysis to different tissues from different organisms and discovered similar low-level expression of anti-sense tags mapping to miRNA loci across mouse, chicken, Drosophila, C. elegans and Arabidopsis (Sup. Table S8). Notably, anti-sense transcription in chicken embryo was extremely high at several loci (Sup. Table S8). While miRNA homology across more distantly-related species can be difficult to ascertain, we did observe overlap between loci affected in mouse and human (mir-24, mir-126, mir-219, mir-34b and mir-338) and in human, mouse and chicken (mir-24, mir-126 and mir-219) (Table 4, Sup. Tables S8). Interestingly, anti-sense miRNA tags in C. elegans mapped almost exclusively to the miRNA* arm of the miRNA loci. This was in sharp contrast to distribution patterns of other species which were found evenly across the two arms of the miRNA/miRNA* duplex and were often found at both arms of an miRNA locus. This could suggest a distinct mechanism for generation of asmiRNA sequences in C. elegans. In humans, we observed that anti-sense transcripts tend to originate from miRNA loci found in the introns or anti-sense to the introns of mRNA transcripts. However, this trend was not conserved across other species in which the genomic location of anti-sense generally followed the expected distribution based on the location of all known miRNA loci (Sup. Fig. S8).
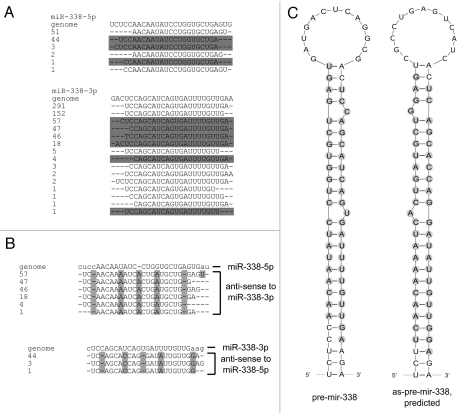
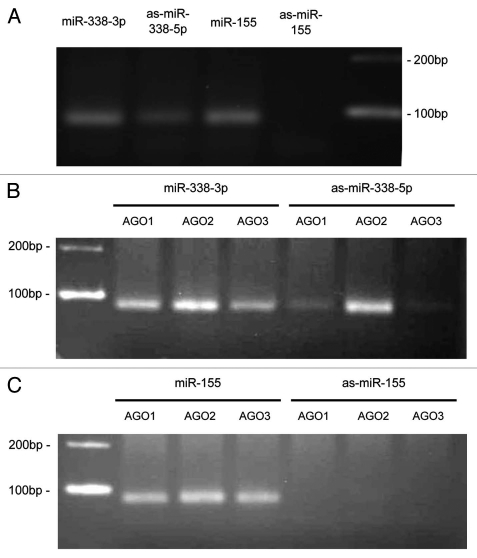
We investigated the human mir-338 locus in more detail due to its higher expression and because it has previously been singled out as a likely candidate for anti-sense transcription58 based on high levels of tags recovered from a deep-sequenced short RNA library.32 An overview of the tags mapping exactly sense and anti-sense to mir-338 are provided in Figure 4. We confirmed the existence of the relatively-highly expressed anti-sense miR-338-5p mature miRNA sequences and also verified expression in the HeLa cell line (Fig. 5A and Sup. Fig. 2) (see Materials and Methods). We also confirmed association of this asmiRNA sequence with the AGO1, AGO2 and AGO3 proteins (Fig. 5B and Sup. Fig. 2), suggesting that asmiRNA sequences are actively involved in gene repression in humans.
Figure 4.
Sequences from the AGO2-associating short RNA library mapping anti-sense to the mir-338 locus. (A) Alignments of tags mapping to both the miR-338-5p and miR-338-3p mature miRNA loci. For mapping procedures, see Materials and Methods. Tags that map to the opposite (anti-sense) strand are shown as their complement sequence, and are shaded in gray. (B) Mapping the perfectly matching anti-sense tags to the opposite strand of the miRNA locus. In order for these tags to be derived from the opposite strand, several distinct post-transcriptional editing, insertion and deletion events would be required. (C) Mapping the tags to predicted anti-sense pre-miRNA-like secondary structures reveals 3′ overhangs, indicative of DICE R1-mediated processing of the mir-338 locus.
Figure 5.
as-miR-338-5p is present in the complete cell and associates with AGO proteins. (A) Detection of as-miR-338-5p in the complete cell. (B) Detection of as-miR-338-5p in the IP extract of AGO1, AGO2 and AGO3. (C) Same as (B), but showing the absence of the negative control asmiR-155 in IP extracts.
Combining the above observations suggests that despite low expression levels, anti-sense miRNA transcription is more widespread than currently acknowledged and the products of this transcription are active through association with AGO proteins. Based on previous experiments56–58 it appears likely that these asmiRNA transcripts could silence gene transcription through direct incorporation into RISCs. However, an endogenous antagomir functionality at the RISC cannot be ruled out, wherein asmiRNA interacts with the AGO-bound complementary sense miRNA, “silencing” the function of the miRNA sense transcript. miR-338 has been linked to several cell-specific regulatory roles including progression of hepatocellular carcinoma,61 basolateral polarity in epithelial cells62 and regulation of axonal respiration in neural cells.63,64 Investigating the effects of the _asmir-338_-derived asmiRNA sequences in these processes could distinguish between possible modes of function for asmiRNAs.
Discussion
Human Argonaute proteins have been shown to be functionally redundant in miRNA-mediated gene silencing. Knockout of the AGO1-4 proteins and subsequent re-introduction of single AGO protein rescued miRNA-mediated silencing activity,65 which would imply that partitioning biases in miRNA strands or loading biases for miRNA across different AGO proteins does not exist on a broad scale similar to what is observed in Drosophila or C. elegans.14–16,18,19 Previous studies have indicated for a handful of miRNA that association with human AGO proteins occurs in an unbiased manner.20,21 A recent, detailed biochemical study confirmed a lack of bias across AGO proteins during RISC formation with synthetic miRNA.66 Here, through analysis of deep sequencing of short RNA from complete THP-1 cell and short RNA associating with the AGO1-3 proteins, we show that with a few notable exceptions, miRNA globally associate in largely equivalent amounts with AGO1, AGO2 and AGO3. The exceptions to this observation, which include miR-182, miR-222 and miR-223*, could be coupled to processes targeting these miRNA loci for interaction with specific AGO proteins. Recent reports have indicated that perfectly complementary duplexes display association biases with different AGO proteins.26,65 We were unable to confirm this observation, primarily because perfectlymatching miRNA duplexes have low expression levels in THP-1.
A distinct form of miRNA sorting has previously been suggested based on the observation that differentially-processed isomirs derived from the same miRNA locus were incorporated into distinct AGO-like proteins and the corresponding RISCs formed from these isomirs targeted separate mRNA substrates for gene silencing.22 Global characterization of differences in primary isomir uptake reveals that a subset of miRNA loci give rise to distinct isomirs which preferentially associate with distinct AGO proteins in a significantly differential manner (Table 1). Despite this, only on rare occasions does one isomir associate with one AGO protein at the complete expense of another; instead, differential incorporation of distinct isomirs favors one at varying percentages over another (Table 1).
Tremendous variation in isomirs derived from the same miRNA locus, both in the number of distinct isomirs and their respective frequencies have previously been observed in mammals.32 While the mechanism of expansion of diversity at a specific miRNA locus appears to relate to the symmetricity of the pre-miRNA precursor,34 the functional advantages derived from this diversity remain largely unknown. Combining the observation of AGO functional redundancy65 with the evidence for gradated preferential incorporation of miRNA isomirs into distinct AGO proteins suggests a possible explanation. AGO functional redundancy establishes a framework wherein mild to moderate, but not absolute, frequency alterations in AGO-bound miRNA isomir content is not likely to be deleterious. The production of a novel isomir which confers a selective advantage, presumably through addition of a novel mRNA target, can then be fixed in the cell and even associate significantly more with one AGO protein relative to another to increase the targeting effectiveness of that isomir. The novel isomir is kept from becoming the only isomir from a locus to associate with an AGO protein because this would presumably disrupt AGO functional redundancy, a selective disadvantage that would prevent further exploration of mRNA target space through differential isomir generation. As such intra-locus sorting has not been observed in other organisms, this may represent a uniquely mammalian post-transcriptional regulatory mechanism.
It is becoming clear that small transcripts derived from larger non-coding RNA genes distinct from miRNA play a role in RISC-mediated post-transcriptional gene regulation.23,25,26 We searched for sequences in AGO-associated short RNA libraries not descending from miRNA loci and discovered sequences derived from snRNA precursors and unusually long sequences derived from sense or anti-sense exonic mRNA regions which appear to be processed for incorporation into the RISC. Additionally, we show for the first time association of the tRF-5 series of tRNA-derived fragments with AGO proteins and confirmed association with AGO for other classes of transcripts derived from tRNA, vRNA and snoRNA. The association of these diverse fragments with AGO proteins strongly suggests or otherwise confirms a role for these fragments in gene silencing; however, it is also possible that they are involved in other functional roles. AGO1 has previously been implicated in directing transcriptional gene silencing through heterochromatin formation.67 As many of the non-miRNA-derived sequences identified in this study are specifically enriched in their association with AGO1 (Table 2), there is the possibility of a tantalizing link between these sequences and silencing of genes through chromatin remodeling. Alternatively, the association of a small number of promoter-derived small RNA transcripts with AGO proteins described here (Sup. Tables S6 and S7, Sup. Figs. S6 and S7) provides another possible mechanism for AGO-mediated gene silencing. Transfected exogenous siRNAs/agRNAs which mimic promoter regions participate in transcriptional regulation after formation of RNA-protein complexes with AGO proteins; these complexes are then recruited to interact with long non-coding transcripts which overlap with promoter regions, resulting in either gene silencing or activation through reasons not yet entirely clear.50,51 Certain miRNAs may similarly silence transcriptional activity by seed sequence recognition of promoter regions.68 The promoter-derived, AGO-associating transcripts identified here could represent a kind of endogenous human siRNA/agRNA which interact with non-coding transcripts at promoter regions to affect gene expression. As previously reasoned,49 while the tag count for individual sequences is quite low, analogous to the low abundance of many transcription factors in the cell, a small number of these endogenous promoter-derived agRNA tags could still substantially impact gene expression at the point of transcription.
Two recent studies demonstrated the remarkable presence of an unusually long miRNA associating with AGO2 generated by the intrinsic nuclease activity of the AGO protein and not through canonical DICER1 processing.69,70 As this locus, mir-451, is expressed in extremely low levels in THP-1, we did not observe association of the longer transcript form in any of the short RNA libraries. We assiduously searched our data for other possible instances of AGO-generated long miRNA transcripts matching the characteristics reported for miR-451 but failed to recover any likely candidates. While the evolutionary implications of this finding are potentially enormous through suggestion that gene silencing could occur with just an AGO-like protein and a hairpin, its presence may be rather limited in mammals indicating that this unique form of miR-451 processing is either a vestige of an ancestral mechanism (as DICER1 has been coupled to miRNA production at least prior to the emergence of the crown group eukaryotes71) or miR-451 itself contains specific structural or sequence features lending it to such processing. Several of the sequences identified in this study as being derived from non-miRNA precursors, including those sequences mapping sense or anti-sense to mRNA, exceed typical miRNA size with lengths of 26–30 nucleotides, raising the possibility that in some cases they may be processed and associate with AGO proteins through a mechanism distinct from the canonical DROSHA/DICER1 pathway. Intrinsic AGO-processing activity is one possible option for generation of these sequences. While such processing would explain the lack of recovered star sequences for these fragments, we do not observe the same distribution of nucleotidyltransferase-modified 3′ ends seen in the miR-451 processing.69,70
We also demonstrate that anti-sense miRNA transcription is more widespread than previously reported. While asmiRNAs were observed in extremely low concentrations, their distribution is decidedly non-sporadic and similar levels are seen across phylogenetically diverse animals and also Arabidopsis (Table 3, see Sup. Table S8). Association of asmiRNAs with AGO proteins suggests these transcripts are functionally active in gene repression, consistent with the previously demonstrated silencing activity of the Drosophila as-miR-iab-4 sequence. It is also possible that some asmiRNA, particularly those expressed at an extremely low level, instead of forming active RISCs, act as endogenous antagomirs following RISC formation to modulate functionality of the sense miRNA transcript. It is additionally possible that mechanisms of asmiRNA transcript biogenesis vary across species given the distinct pattern of antisense transcription observed in C. elegans.
Materials and Methods
Short RNA libraries.
The short RNA libraries for each AGO library and THP-1 whole cell are publicly-available at the DDBJ Sequence Read Archive (DRA) (trace.ddbj.nig.ac.jp/dra/index_e.shtml) with the accession number DRA000205.28 These libraries were sequenced using the Genome Analyzer GA-II (Illumina) with a maximum read length of 30 nucleotides, precluding discovery of longer fragments. Specific binding of the Anti-AGO antibodies to their respective Argonaute proteins was demonstrated previously in reference 28. AGO4 was not included in the original dataset due to lack of availability when the research was planned and data collected. While AGO4 IP-seq experiments are unlikely to have much impact on the observed lack of a widespread miRNA-AGO sorting mechanism, the data derived from such experiments could help complete the known complement of RNAs associating with human AGO proteins and also clarify isomir-specific sorting biases.
Analysis of short RNA libraries.
Linker sequences attached during short RNA library construction were computationally filtered following tag extraction from sequencing runs. Artifacts, often taking the form of linker-formed dimers, were removed with the TagDust program.72 Resulting sequences were aligned to the genome using the Nexalign program (genome.gsc.riken.jp/osc/english/software).60 Genome alignments underwent additional refinement by correcting for cross-mapping of sequences mapping to multiple loci, which weights the set of all potential mapping sites for a tag based on frequency and positional error likelihood.60 Tags mapping to mirBase-defined miRNA loci73 (version 12.0) were included in strand partitioning, AGO-associated enrichment and differential isomir association analyses. The remaining tags were collected and used for identification of non-miRNA tags associating with AGO proteins. Several of the analyses described below incorporated scripts from the bedtools software package.74
Comparisons of miRNA loci across individual libraries were performed after tallying counts of all tags mapping exactly to an miRNA locus allowing for up to two mismatches at the 3′ end of the sequence to account for the reported action of animal miRNA nucleotidyltransferases.28,31,33 Raw tag counts were normalized to tag per million (tpm) counts prior to comparisons. To reduce noise, all pairwise comparisons across short RNA libraries required 100 tpm in at least one condition. Comparisons across multiple libraries required 100 tpm in at least one library. Significant differences in tpm counts across the three AGO-IP libraries were determined using a chi-square goodness-of-fit test; differences were considered significant at p < 0.05 with a Bonferroni post hoc correction.
Pairing probability profiles for comparison of secondary structural characteristics were constructed essentially as described in reference 17, utilizing the RNAcofold program from the ViennaRNA-1.8.4 package (www.tbi.univie.ac.at/RNA/) with profiles based on the primary isomir associating with a given AGO protein and the corresponding miRNA* sequence with the highest counts in the same library. Comparisons were performed between the set of miRNA loci enriched in all AGO proteins versus the set depleted in all AGO proteins across a range of fold change cut-off values. Sequence nucleotide composition bias was calculated using a weighted scheme accounting for differences in isomir frequency while guarding against overrepresentation of any single loci. Briefly, sequence compositions of individual isomirs were weighted by their percent frequency in a locus. The combination of the weighted isomirs from a given locus was then weighted equally when comparing with all other loci.
Significance in differential isomir association with AGO proteins was determined using Fisher's exact test; differences were considered significant at p < 0.05 with a Bonferroni post hoc correction. Raw read counts for a given isomir associating with an AGO protein and the total raw read counts in the same library were compared against raw read counts for the same isomir in another AGO library. Calculations were performed for all variant primary isomirs in a pairwise manner across all three AGO libraries (Table 1). For analyses in this research, unique isomirs of a miRNA locus were taken to be any tag mapping exactly to the genome sequence regardless of 5′ or 3′ end variation while allowing for up to two mismatches at the 3′ terminus.
When searching for non-miRNA sequences associating with AGO proteins, we initially required the fold change increase relative to the normalized tag count in the complete cell to be greater than two in order to reduce detection of false positives. Upon realizing that all snoRNA-derived fragments, which have previously been experimentally demonstrated to associate with AGO proteins,23 fell well below this cut-off we relaxed the restriction to include any tags with a fold change greater than ½ to accommodate tags present in roughly equivalent proportions in the complete cell and in association with AGO proteins (see Table 2). Tags in these searches were required to map exactly to the genome, although we made exceptions when circumstances warranted (for a discussion of inclusion of the sequences mapping to CYP46A1, see Sup. Discussion/Methods). If a cluster of closely overlapping sequences was present, sequence composition and counts are reported for only the most abundant sequence. Potential mRNA targets for sequences identified in Table 2 were predicted using TargetScan Custom 5.1.75 Cross-species alignments of the N-terminal regions of Table 2 sequences were constructed using the axtChain program76 which utilizes genome alignments generated with BLASTZ.77 RNA secondary structure predictions were constructed using the Mfold web server78,79 and, where applicable, manually adjusted to be consistent with RNA molecule-specific, experimentally-determined structural features.
Anti-sense miRNA tags were subjected to additional rounds of filtering to guard against incorrect assignment. asmiRNA tags were required to match exactly to the genome and in only one location, except for cases when an asmiRNA tag maps to an miRNA loci that has undergone a duplication event in the human genome (for example, miR-24). Also, potential asmiRNA tags mapping to miRNA predicted to form perfectly complementary duplexes (for example, miR-625) were discarded.
Searches for potential promoter-derived tags were restricted by filtering tags which mapped uniquely and exactly to the genome in promoter regions, defined as +500 nucleotides upstream and −100 nucleotides downstream of Refseq-defined TSSs. Significance of tag enrichment in promoter regions was determined by randomly selecting equivalently-sized regions of the genome (prefiltered to avoid overlap with known classes of RNA) over 1,000 trials and summing the total tag counts for these regions in each trial to obtain a distribution which was compared to the observed counts for promoter regions using student's t test (Sup. Fig. S6). Tags derived from all three libraries were pooled to calculate length distributions and average start distance from defined TSSs (Sup. Fig. S7).
Reverse transcription PCR (RT-PCR).
Either complete cellular RNA or RNAs from IP with AGO proteins was isolated as previously described in reference 28 and subjected to RT-PCR. The reverse transcription reaction was performed with the miScript Reverse Transcription Kit (Qiagen) according to the manufacturer's instructions using 2 µg of complete cellular RNA or 100 ng of each AGO-IP RNA in 20 µl reactions, and PCR was immediately performed with 2 µl of the RT reaction using the miRNA-specific primer and miScript Universal primer (32 cycles: 94°C, 15 s; 60°C, 30 s; 70°C, 30 s). The following primers were used as the miRNA-specific primer: miR338-3p-F (5′-TCC AGC ATC AGT GAT TTT GTT-3′), as-miR338-5p-F (5′-TCA ACA AAA TCA CTG ATG CTG G-3′), miR155-F (5′-CCC TAT CAC GAT TAG CAT TAA-3′), as-miR155-F (5′-TTA ATG CTA ATC GTG ATA GGG-3′). Please see Supplemental Discussion/Methods section for description of qRT-PCR methods.
Acknowledgements
We thank Takahiro Nishibu, Ryo Ukekawa, Taku Funakoshi and Tsutomu Kurokawa of Wako Pure Chemical Industries Ltd., for supplying the human AGO1 and AGO3 IP kits for an earlier study which generated the data used in this analysis. This work was supported by a grant of the Genome Network Project from the Ministry of Educations, Culture, Sports, Science and Technology (MEXT) of Japan to Y.H., a research grant for the RIKEN Omics Science Center from MEXT to Y.H., and a grant for the RIKEN Frontier Research System, Functional RNA research program to Y.H.
Abbreviations
AGO
argonaute
RISC
RNA-induced silencing complex
vRNA
vault RNA
svRNA
small RNA fragments derived from vRNA
asmiRNA
anti-sense miRNA
DRA
DDBJ sequence read archive
Supplementary Material
Supplementary Material
References
- 1.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 2.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okada C, Yamashita E, Lee SJ, Shibata S, Katahira J, Nakagawa A, et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 4.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 7.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 8.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 10.Jazdzewski K, Liyanarachchi S, Swierniak M, Pachucki J, Ringel MD, Jarzab B, et al. Polymorphic mature microRNAs from passenger strand of pre-miR-146a contribute to thyroid cancer. Proc Natl Acad Sci USA. 2009;106:1502–1505. doi: 10.1073/pnas.0812591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hock J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006;50:81–99. doi: 10.1007/s00294-006-0078-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Czech B, Zhou R, Erlich Y, Brennecke J, Binari R, Villalta C, et al. Hierarchical rules for Argonaute loading in Drosophila. Mol Cell. 2009;36:445–456. doi: 10.1016/j.molcel.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamura K, Liu N, Lai EC. Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell. 2009;36:431–444. doi: 10.1016/j.molcel.2009.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawamata T, Seitz H, Tomari Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat Struct Mol Biol. 2009;16:953–960. doi: 10.1038/nsmb.1630. [DOI] [PubMed] [Google Scholar]
- 17.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of Drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jannot G, Boisvert ME, Banville IH, Simard MJ. Two molecular features contribute to the Argonaute specificity for the microRNA and RNAi pathways in C. elegans. RNA. 2008;14:829–835. doi: 10.1261/rna.901908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steiner FA, Hoogstrate SW, Okihara KL, Thijssen KL, Ketting RF, Plasterk RH, et al. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat Struct Mol Biol. 2007;14:927–933. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- 20.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 22.Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, et al. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci USA. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, et al. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 24.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol. 2009;11:1268–1271. doi: 10.1038/ncb1972. [DOI] [PubMed] [Google Scholar]
- 26.Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA. 2010;16:673–695. doi: 10.1261/rna.2000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tan GS, Garchow BG, Liu X, Yeung J, Morris JPt, Cuellar TL, et al. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burroughs AM, Ando Y, de Hoon MJ, Tomaru Y, Nishibu T, Ukekawa R, et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 31.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh T, Sakaguchi Y, Miyauchi K, Suzuki T, Kashiwabara S, Baba T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009;23:433–438. doi: 10.1101/gad.1761509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Starega-Roslan J, Krol J, Koscianska E, Kozlowski P, Szlachcic WJ, Sobczak K, et al. Structural basis of microRNA length variety. Nucleic Acids Res. 2011;39:257–268. doi: 10.1093/nar/gkq727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs) Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, et al. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA. 2009;15:2147–2160. doi: 10.1261/rna.1738409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawaji H, Nakamura M, Takahashi Y, Sandelin A, Katayama S, Fukuda S, et al. Hidden layers of human small RNAs. BMC Genomics. 2008;9:157. doi: 10.1186/1471-2164-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 40.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, et al. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 41.Suzuki H, Forrest AR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nat Genet. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maniataki E, Mourelatos Z. Human mitochondrial tRNAMet is exported to the cytoplasm and associates with the Argonaute 2 protein. RNA. 2005;11:849–852. doi: 10.1261/rna.2210805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 45.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 46.Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat Struct Mol Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 48.Napoli S, Pastori C, Magistri M, Carbone GM, Catapano CV. Promoter-specific transcriptional interference and c-myc gene silencing by siRNAs in human cells. EMBO J. 2009;28:1708–1719. doi: 10.1038/emboj.2009.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736–7748. doi: 10.1093/nar/gkq648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwartz JC, Younger ST, Nguyen NB, Hardy DB, Monia BP, Corey DR, et al. Antisense transcripts are targets for activating small RNAs. Nat Struct Mol Biol. 2008;15:842–848. doi: 10.1038/nsmb.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hawkins PG, Santoso S, Adams C, Anest V, Morris KV. Promoter targeted small RNAs induce long-term transcriptional gene silencing in human cells. Nucleic Acids Res. 2009;37:2984–2995. doi: 10.1093/nar/gkp127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle. 2009;8:2332–2338. doi: 10.4161/cc.8.15.9154. [DOI] [PubMed] [Google Scholar]
- 54.Saraiya AA, Wang CC. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stark A, Bushati N, Jan CH, Kheradpour P, Hodges E, Brennecke J, et al. A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev. 2008;22:8–13. doi: 10.1101/gad.1613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bender W. MicroRNAs in the Drosophila bithorax complex. Genes Dev. 2008;22:14–19. doi: 10.1101/gad.1614208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyler DM, Okamura K, Chung WJ, Hagen JW, Berezikov E, Hannon GJ, et al. Functionally distinct regulatory RNAs generated by bidirectional transcription and processing of microRNA loci. Genes Dev. 2008;22:26–36. doi: 10.1101/gad.1615208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mayr C, Bartel DP. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.De Hoon MJ, Taft RJ, Hashimoto T, Kanamori-Katayama M, Kawaji H, Kawano M, et al. Cross-mapping and the identification of editing sites in mature microRNAs in high-throughput sequencing libraries. Genome Res. 2010;20:257–264. doi: 10.1101/gr.095273.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang XH, Wang Q, Chen JS, Fu XH, Chen XL, Chen LZ, et al. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol Res. 2009;39:786–794. doi: 10.1111/j.1872-034X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- 62.Tsuchiya S, Oku M, Imanaka Y, Kunimoto R, Okuno Y, Terasawa K, et al. MicroRNA-338-3p and microRNA-451 contribute to the formation of basolateral polarity in epithelial cells. Nucleic Acids Res. 2009;37:3821–3827. doi: 10.1093/nar/gkp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaplan BB, Gioio AE, Hillefors M, Aschrafi A. Axonal protein synthesis and the regulation of local mitochondrial function. Results Probl Cell Differ. 2009;48:225–242. doi: 10.1007/400_2009_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S, Liu Q, et al. ATP-dependent human RISC assembly pathways. Nat Struct Mol Biol. 17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793–797. doi: 10.1038/nsmb1142. [DOI] [PubMed] [Google Scholar]
- 68.Kim DH, Saetrom P, Snove O, Jr, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230–16235. doi: 10.1073/pnas.0808830105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cifuentes D, Xue H, Taylor DW, Patnode H, Mishima Y, Cheloufi S, et al. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muljo SA, Kanellopoulou C, Aravind L. MicroRNA targeting in mammalian genomes: genes and mechanisms. Wiley Interdiscip Rev Syst Biol Med. 2010;2:148–161. doi: 10.1002/wsbm.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lassmann T, Hayashizaki Y, Daub CO. TagDust—a program to eliminate artifacts from next generation sequencing data. Bioinformatics. 2009;25:2839–2840. doi: 10.1093/bioinformatics/btp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 76.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion and rearrangement in the mouse and human genomes. Proc Natl Acad Sci USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, et al. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material