Optogenetic Manipulation of Neural Circuitry In Vivo (original) (raw)
. Author manuscript; available in PMC: 2012 Jun 1.
Published in final edited form as: Curr Opin Neurobiol. 2011 Mar 21;21(3):433–439. doi: 10.1016/j.conb.2011.02.010
Abstract
Recent advances in optogenetics have permitted investigations of specific cell types in the nervous system with unprecedented precision and control. This review will discuss the use of optogenetic techniques in the study of mammalian neural circuitry in vivo, as well as practical and theoretical considerations in their application.
Introduction
For hundreds of years, neuroscientists have gleaned information about the function of specific brain regions by observing what happens when they are injured. More recently, advances in imaging and electrophysiology have allowed scientists to characterize the activity of living brains while animals and humans are exposed to stimuli or perform tasks. These approaches have convincingly determined what different brain regions are doing, but have revealed little about how they function. For example, the striatum has been linked to voluntary movement for over a century [1], and since the 1960s it has been identified as a major site of dysfunction in Parkinson’s disease [2–4]. However, these facts reveal little about how the striatum controls movement or how striatal dysfunction contributes to Parkinson’s disease. In order to understand how our brains function, new technologies are needed to probe the function of specific cell types and circuits in behaving animals. Historically, the application of pharmaceuticals or electrical stimulation has been used for testing the function of specific cell types and circuits, but these techniques are often not very specific. The recent development of optogenetic methods in neuroscience has allowed researchers to sensitize distinct cell types to light, enabling the non-invasive activation, inhibition, and modulation of specific neuronal populations in living animals with millisecond precision. Techniques like these will lead to a richer understanding of how specific brain regions function, and how their dysfunction might be corrected.
Optogenetic tools
Optogenetic tools for stimulating neuronal activity
Although a number of methods for optically stimulating specific cell types have been developed over the past two decades [5–10], they have generally relied on expression of multiple proteins or application of exogenous cofactors, limiting their utility in vivo. With the discovery that expression of a single protein—channelrhodopsin-2 (ChR2)—can mediate light-sensitive cation currents [11] that enable robust and temporally-precise control of neural activity in vitro [12,13] and in vivo [14–18], the use of optogenetic approaches in neuroscience has exploded. Since its initial discovery, many channelrhodopsin variants have been discovered or engineered to confer additional properties on the cells in which they are expressed [19–22]. At the time of this writing, at least 12 channelrhodopsin variants have been reported, each with a unique set of properties [23].
Optogenetic tools for inhibiting neuronal activity
Whereas activating specific neural subtypes can reveal whether neurons are sufficient for driving a behavior, inhibiting specific neural types can determine whether they are necessary for that behavior. Methods for achieving this include physical or toxigenic methods to kill targeted cell types [24–26], expression of modified potassium or chloride channels that are sensitive to light or exogenous compounds [27–29], and expression of engineered or non-mammalian G-protein-coupled receptors [13,30–33]. Despite the utility of these techniques, newer optogenetic manipulations that are rapidly reversible and do not require a cofactor can be more useful in vivo, especially in applications that benefit from fast kinetics. In 2007, a chloride pump from the archaeon Natronomonas pharaonis (NpHR) was engineered to express in mammalian cells. When illuminated with yellow (~590nm) light, this pump inhibited spiking in neurons [34,35]. More recently, proton pumps have also been used to inhibit neurons [36]. Expressing combinations of these proteins may allow for rapid inhibitory control over multiple neural populations using different frequencies of light. In addition, the activation maxima of certain inhibitory proteins is spectrally separated from ChR2, enabling bidirectional control of individual neurons using yellow and blue light [34].
Optogenetic tools for modulating neural activity
In addition to modulating neural activity with light-activated ion channels or pumps, it is possible to control intracellular signaling pathways with light [37]. Adenyl cyclase is a ubiquitous signaling molecule in many tissues of the body. While most forms of adenyl cyclase are not light sensitive, a light-activated adenyl cyclase was discovered in the protist Euglena gracilis [38]. This light-activated cyclase was subsequently expressed in neurons, where it rapidly modulated cAMP levels with illumination [39,40]. Another approach for controlling intracellular signaling involves engineering G-protein coupled receptors to respond to light by integrating their intracellular domains with the light-sensing domain of rhodopsin. This approach has been successfully applied with Gq coupled adrenergic α1a, Gi coupled β2 receptors, and Gi/o coupled serotonin 5-HT1A receptors [41,42]. Finally, expressing naturally occurring opsins such as melanopsin (a Gq coupled receptor) can also confer optical control over G-protein signaling [43].
Optimizing Optical Control
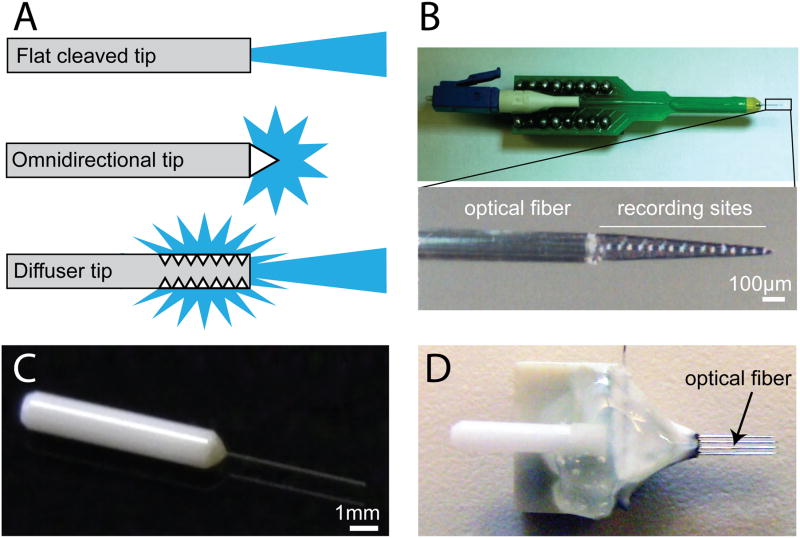
Although optogenetics has numerous advantages over other techniques, a number of practical and conceptual issues arise when these techniques are applied to mammals in vivo. These include relatively simple issues such as effectively illuminating large volumes of tissue, as well as more complex issues such as interpreting experimental results. Gross estimations have revealed that tissue can be safely and effectively illuminated up to about 1.5mm from the tip of an optical fiber [18]. Most mouse brain regions are small enough to illuminate with one fiber, but experiments in larger rodents or primates may require a larger area of illumination. One way to achieve this is to implant multiple optical fibers, but this is not ideal as it increases tissue damage. This can be mitigated to some extent by etching fiber tips to reduce their size [44]. An elegant alternative is to use a fiber with a machined tip that either distributes light omnidirectionally or diffuses light along the length of the fiber (Figure 1A). These machined fibers can be coupled to higher laser powers without increasing tissue damage, as they release light over a broader surface area than a flat-cleaved fiber. Alternatively, illumination can be achieved with head mounted light emitting diodes (LEDs) [45,46]. LEDs have several advantages over laser-coupled optical fibers, as they are much cheaper than lasers, more durable than optical fibers, and can be powered by small head mounted batteries that do not require the animal to be tethered throughout the experiment.
Figure 1. Advances in optrode design.
A. Schematic of a flat cleaved fiber (top), as well as two machined fiber tips (bottom) that release light in different configurations (illustration adapted from Polymicro Technologies, Phoenix, AZ). B. Photograph of a silicon-probe-based optrode that can be used for anesthetized recording. C. Photograph of a short fiber and ferrule that can be attached to a microwire recording array. D. Photograph of a microwire array with an integrated ferrule and optical fiber for multiunit recording and optogenetic identification of cell types in awake behaving animals.
A complementary problem to light delivery is achieving abundant and specific expression of optogenetic proteins. Cell-type specific expression has been accomplished in mammalian tissue with transgenic mice, viral infection strategies, and in utero electroporation. In one group of transgenic lines, ChR2 was expressed in multiple brain regions under control of the Thy1 promoter [17,47]. In another line, ChR2 was expressed in the spinal cord and hind brain under control of the VGlut2 promoter, and used to drive locomotion [48]. New transgenic mice are under development, including mice that contain optogenetic proteins such as ChR2 in floxed configurations that can be crossed into cre driver lines to drive cell type specific expression [49], an advantage that had previously been limited to viral expression strategies [50–55]. Transgenic mice are advantageous for expressing optogenetic proteins throughout a large brain structure with highly-reproducible expression patterns, although achieving sufficient expression levels for stimulation in vivo remains a challenge.
Although transgenic mice offer a convenient and reliable expression system, viral-based methods are more versatile than mouse lines for many applications. Viruses are easier to generate than transgenic mice, can be engineered to control expression level and cell-type specificity, and can drive expression in species that are not as genetically tractable as mice, such as rats or primates. Furthermore, viral expression is restricted to injected sites, making it possible to inject a virus in one structure and illuminate in another to investigate projections between two structures [16,55–57]. Multiple viruses can also be injected into the same brain structure to achieve multimodal control of distinct neuronal populations [19,34]. An alternative to viral-based methods is in utero electroporation, which can be used to introduce optogenetic proteins into specific cell types or layers, typically in the neocortex [45,58–60].
Optical stimulation in vivo
Many laboratories have used ChR2 to probe the function of specific cell types and circuits in mammalian brains. Among others, these include exploring the minimal number of neurons required to represent a perception [45], demonstrating that phasic firing of dopamine neurons can mediate conditioned learning [54], that parvalbumin-expressing cells in cortex contribute to gamma rhythms [51,52], that hypocretin expressing cells in the lateral hypothalamus contribute to sleep to wake transitions [61], and that striatal output pathways control motor output [53]. Despite strong behavioral results, none of these studies conclusively showed that the targeted cell types were, in fact, efficiently excited by illumination in the awake animal. Nearly all in vivo validation of ChR2 stimulation has been performed in anesthetized animals, which does not accurately reflect the awake state. For example, neurons in awake animals are typically more depolarized and receive more synaptic drive. Under these conditions, neurons can enter a depolarization block when activated too strongly, which could reduce spiking of ChR2-expressing cells. Additionally, network effects such as lateral inhibition could result in a net inhibition of a targeted cell type, despite activation of ChR2. To investigate these issues, it is necessary to combine optogenetics with awake electrophysiology and record from target neurons while they are illuminated. Briefly, this approach involves implanting single or multiple ‘optrodes’ (combined electrode and optical fiber) into the brain of an animal that expresses an excitable optogenetic protein such as ChR2. By examining recordings from this optrode it is possible to conclusively determine what happens to the target cell type during illumination.
Construction of optrodes
The first optrodes were made by simply gluing an optical fiber to a single tungsten microelectrode for use in anesthetized recordings [62]. We, and others, have found that a slightly evolved design in which an optical fiber was glued on a 16-site silicon probe is ideal for anesthetized recording, as it allows 16 channels of recording in a spatially defined manner with minimal tissue damage (Figure 1B) [53]. A similar silicon-probe based approach was recently used in chronic awake recordings [44]. For awake recordings, we have had success with a simple design utilizing a zirconia ferrule and a short piece of optical fiber (Figure 1C). This ferrule assembly can be attached to a traditional microwire array, resulting in a compact optrode array for chronic awake recording and stimulation (Figure 1D). It is also possible to place the electrodes in tetrode configurations or mount optrodes in microdrives to record from different depths in the same animal. While optrodes based on silicon probes allow for the highest density recordings with the smallest damage to the brain, optrodes based on microwires are cheaper to build and can record neurons for months after implantation.
Identifying neuronal subtypes during awake recordings
Optrode recordings can be used for more than validating ChR2 activation. Awake electrophysiology has historically been limited by a lack of reliable methods for identifying specific cell types in extracellular recordings. By combining optogenetics with awake recordings, specific cell types can be identified by testing their responsiveness to light. Once identified, the activity of these subtypes can be tracked to determine their behavioral correlates [44,63,64].
Several issues need to be addressed when identifying cell types based on ChR2 expression. The first concerns stimulation parameters for identifying ChR2-expressing neurons. It is possible to use brief (<10msec), high powered (>10mW) laser pulses to drive single action potentials in neurons [63,64]. However, illuminations like this can cause large populations of ChR2-expressing neurons to fire at once, which can increase synchronous multi-unit activity on a recording electrode, making spike sorting difficult [44]. This multi-unit activity can also infiltrate the recording of a well isolated unit and spuriously lead a researcher to conclude that the isolated unit expresses ChR2. Finally, high powered laser pulses can cause photoelectric artifacts on recording electrodes [44,63,65]. In the striatum, we have found that longer (1s), low power (~0.1 to 3.0mW) laser pulses are sufficient to drive spiking in ChR2 expressing neurons, while avoiding both large increases in multi-unit activity and photoelectric effects. Useful stimulation parameters are dependent on the recorded brain structure, and different stimulation parameters have been used to mitigate these issues in the hippocampus [44].
After determining useful stimulation parameters, identifying ChR2-expressing neurons can still be difficult due to the high interconnectivity among neurons in most brain regions. For example, a non-ChR2 expressing neuron could appear to be light-responsive if it receives excitatory input from a neighboring ChR2-expressing neuron. In brain regions that are interconnected by excitatory synapses exhibiting short-term depression of glutamate release, this issue can be addressed with trains of moderate frequency laser pulses. Due to the rapid kinetics of ChR2, neurons that express ChR2 reliably respond to each light pulse in a train (at rates <40Hz). In contrast, synaptically activated neurons respond unreliably after the first few pulses, presumably as the readily releasable pool of glutamate is depleted [64].
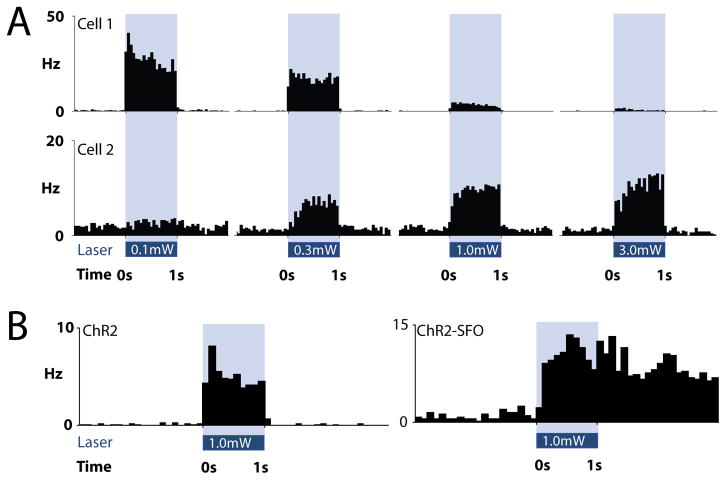
This specific confound is less likely in inhibitory structures. For example, the striatum does not contain any excitatory neurons, so increases in spiking that follow the laser pulse cannot be due to local excitatory drive. However, inhibitory structures like the striatum often exhibit lateral inhibition which can overpower light-activated currents in ChR2-expressing neurons. In these cases, a lower laser power can reveal ChR2-mediated responses. For example, some striatal neurons respond most strongly to low power laser illumination, because higher laser powers either depolarize neurons too much (inactivating sodium channels and driving neurons into a depolarization block) or drive more lateral inhibition from other ChR2-expressing cells (Figure 2A). Other striatal neurons respond better to higher laser powers (Figure 2A), likely because they express lower levels of ChR2 or do not receive adequate illumination at lower laser power. In such cases, it can be useful to illuminate with a series of pulses of incrementing laser powers to determine which neurons express ChR2. As a final note, while it is possible to identify ChR2 expressing neurons with these techniques, it is difficult to conclude anything about the cell type of neurons that do not respond to the light. Non-responsive neurons may not express high enough levels of ChR2 to drive spiking, may not be effectively illuminated, or could be synaptically inhibited.
Figure 2. Optogenetic identification of specific cell types in vivo.
A. The responses of two simultaneously-recorded striatal medium spiny neurons to four incrementing laser powers in an awake mouse. Cell 1 is best excited by low power (0.1mW at fiber tip), while cell 2 is best excited by high power (3.0mW at fiber tip) laser light. B. Examples of neuronal responses to ChR2, and ChR2-SFO. Note that the neuron expressing ChR2-SFO continues firing after the light is extinguished, giving it a unique kinetic signature.
Theoretically, it should also be possible to identify and track more than one cell type simultaneously. This would involve targeting multiple optogenetic proteins to different cell types, and using the unique characteristics of these proteins to identify each cell type. For example, spectrally-distinct channelrhodopsin variants could be targeted to two cell populations, which can be selectively stimulated with different wavelengths of light [19]. The unique kinetics of different optogenetic proteins can also be used to identify different cell types. We have expressed ChR2 in one neuron type in the striatum and a step-function variant of ChR2 (ChR2-SFO) in another. Both of these proteins are rapidly activated by blue light, but the ChR2 expressing neurons stop firing when the light turns off, while ChR2-SFO expressing neurons continue firing after the light is shut off (Figure 2B). Using combinations of spectrally and kinetically distinct optogenetic proteins under control of cell-type specific promoters, it may be possible to identify more than two cell types in the same recording.
Future of optogenetics
Optogenetics has evolved extremely rapidly in the years since light was first used to modulate the activity of neurons. Future years are expected to bring additional optogenetic tools, as well as new applications for these tools. Some of these tools may include proteins with new functionalities such as more restricted activation spectra, activation by additional frequencies such as infrared that better penetrate through tissue, and better targeting to subcellular domains or organelles. It is also likely that newer optrode designs will allow for higher density recordings and efficient illumination of all recording sites with less damage to the brain. Finally, in addition to tagging neurons based on genetic expression patterns, it may eventually be possible to control expression of optogenetic proteins using activity-sensitive promoters for immediate early genes such as _c_-fos or arc. Using this strategy, neurons that are activated during a certain behavior would express ChR2 and could be re-activated at a later time, potentially re-activating that behavior. These types of experiments may allow researchers to tease apart different functional ensembles of neurons (cell assemblies) that drive a particular behavior. For example, it may be possible to identify a functional ensemble early in a behavioral learning paradigm, and track the activity of that ensemble as the animal learns and improves the behavior. The growing application of optogenetic techniques promises to increase our understanding of how different brain regions function, and how to intervene when they malfunction.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wilson S. An experimental research into the anatomy and physiology of the corpus striatum. Brain. 1914;36:427–492. [Google Scholar]
- 2.Ehringer H, Hornykiewicz O. Distribution of noradrenaline and dopamine (3-hydroxytyramine) in the human brain and their behavior in diseases of the extrapyramidal system. Parkinsonism Relat Disord. 1998;4:53–57. doi: 10.1016/s1353-8020(98)00012-1. [DOI] [PubMed] [Google Scholar]
- 3.Ehringer H, Hornykiewicz O. Verteilung Von Noradrenalin Und Dopamin (3-Hydroxytyramin) Im Gehirn Des Menschen Und Ihr Verhalten Bei Erkrankungen Des Extrapyramidalen Systems. Klin Wochenschr. 1960;38:1236–1239. doi: 10.1007/BF01485901. [DOI] [PubMed] [Google Scholar]
- 4.Carlsson A. Evidence for a Role of Dopamine in Extrapyramidal Functions. Acta Neuroveg (Wien) 1964;26:484–493. doi: 10.1007/BF01252144. [DOI] [PubMed] [Google Scholar]
- 5.Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Callaway EM, Katz LC. Photostimulation using caged glutamate reveals functional circuitry in living brain slices. Proc Natl Acad Sci U S A. 1993;90:7661–7665. doi: 10.1073/pnas.90.16.7661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szobota S, Gorostiza P, Del Bene F, Wyart C, Fortin DL, Kolstad KD, Tulyathan O, Volgraf M, Numano R, Aaron HL, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–545. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 9.Zemelman BV, Lee GA, Ng M, Miesenbock G. Selective photostimulation of genetically chARGed neurons. Neuron. 2002;33:15–22. doi: 10.1016/s0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- 10.Zemelman BV, Nesnas N, Lee GA, Miesenbock G. Photochemical gating of heterologous ion channels: remote control over genetically designated populations of neurons. Proc Natl Acad Sci U S A. 2003;100:1352–1357. doi: 10.1073/pnas.242738899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, Bamberg E. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci U S A. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 13.Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, Chiel H, Hegemann P, Landmesser LT, Herlitze S. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci U S A. 2005;102:17816–17821. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–2284. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 15.Schroll C, Riemensperger T, Bucher D, Ehmer J, Voller T, Erbguth K, Gerber B, Hendel T, Nagel G, Buchner E, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–1747. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- 16.Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–668. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- 17*.Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, Augustine GJ, Ehlers MD, Feng G. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–218. doi: 10.1016/j.neuron.2007.03.005. The authors describe transgenic mouse lines that express ChR2 under control of the Thy1 promoter. Due to randomness in gene insertion, several lines were generated that each exhibit different expression patterns. Across these lines, ChR2 expression was detected in hippocampus, cortex, cerebellum, thalamus, midbrain, brainstem and retina. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18**.Aravanis AM, Wang LP, Zhang F, Meltzer LA, Mogri MZ, Schneider MB, Deisseroth K. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:S143–156. doi: 10.1088/1741-2560/4/3/S02. This paper describes the construction and use of a simple fiber optic system for in vivo illumination in rodents. This system has subsequently served as a model for optogenetic behavioral experiments in multiple laboratories. [DOI] [PubMed] [Google Scholar]
- 19.Zhang F, Prigge M, Beyriere F, Tsunoda SP, Mattis J, Yizhar O, Hegemann P, Deisseroth K. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat Neurosci. 2008;11:631–633. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–1814. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–392. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- 22.Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–234. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- 23.Lin JY. A User’s Guide to Channelrhodopsin Variants: Features, Limitations and Future Developments. Exp Physiol. 2010 doi: 10.1113/expphysiol.2009.051961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Kane CJ, Moffat KG. Selective cell ablation and genetic surgery. Curr Opin Genet Dev. 1992;2:602–607. doi: 10.1016/s0959-437x(05)80179-0. [DOI] [PubMed] [Google Scholar]
- 25.Bulina ME, Chudakov DM, Britanova OV, Yanushevich YG, Staroverov DB, Chepurnykh TV, Merzlyak EM, Shkrob MA, Lukyanov S, Lukyanov KA. A genetically encoded photosensitizer. Nat Biotechnol. 2006;24:95–99. doi: 10.1038/nbt1175. [DOI] [PubMed] [Google Scholar]
- 26.Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- 27.Chambers JJ, Banghart MR, Trauner D, Kramer RH. Light-induced depolarization of neurons using a modified Shaker K(+) channel and a molecular photoswitch. J Neurophysiol. 2006;96:2792–2796. doi: 10.1152/jn.00318.2006. [DOI] [PubMed] [Google Scholar]
- 28.Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci. 2004;7:1381–1386. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Slimko EM, McKinney S, Anderson DJ, Davidson N, Lester HA. Selective electrical silencing of mammalian neurons in vitro by the use of invertebrate ligand-gated chloride channels. J Neurosci. 2002;22:7373–7379. doi: 10.1523/JNEUROSCI.22-17-07373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alexander GM, Rogan SC, Abbas AI, Armbruster BN, Pei Y, Allen JA, Nonneman RJ, Hartmann J, Moy SS, Nicolelis MA, et al. Remote control of neuronal activity in transgenic mice expressing evolved G protein-coupled receptors. Neuron. 2009;63:27–39. doi: 10.1016/j.neuron.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coward P, Wada HG, Falk MS, Chan SD, Meng F, Akil H, Conklin BR. Controlling signaling with a specifically designed Gi-coupled receptor. Proc Natl Acad Sci U S A. 1998;95:352–357. doi: 10.1073/pnas.95.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Armbruster BN, Li X, Pausch MH, Herlitze S, Roth BL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci U S A. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lechner HA, Lein ES, Callaway EM. A genetic method for selective and quickly reversible silencing of Mammalian neurons. J Neurosci. 2002;22:5287–5290. doi: 10.1523/JNEUROSCI.22-13-05287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. The authors demonstrated that the light activated Cl− pump from the archeal Natronomonas pharaonis can rapidly and reversibly inhibit mammalian neurons. Although other methods can inhibit neurons in a cell-type specific manner, this method has gained popularity as it does not require any exogenous cofactors, and is therefore ideal for use in vivo. [DOI] [PubMed] [Google Scholar]
- 35.Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–139. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chow BY, Han X, Dobry AS, Qian X, Chuong AS, Li M, Henninger MA, Belfort GM, Lin Y, Monahan PE, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masseck OA, Rubelowski JM, Spoida K, Herlitze S. Light- and Drug-activated G-Protein coupled Receptors to Control intracellular Signaling. Exp Physiol. 2010 doi: 10.1113/expphysiol.2010.055517. [DOI] [PubMed] [Google Scholar]
- 38.Iseki M, Matsunaga S, Murakami A, Ohno K, Shiga K, Yoshida K, Sugai M, Takahashi T, Hori T, Watanabe M. A blue-light-activated adenylyl cyclase mediates photoavoidance in Euglena gracilis. Nature. 2002;415:1047–1051. doi: 10.1038/4151047a. [DOI] [PubMed] [Google Scholar]
- 39.Schroder-Lang S, Schwarzel M, Seifert R, Strunker T, Kateriya S, Looser J, Watanabe M, Kaupp UB, Hegemann P, Nagel G. Fast manipulation of cellular cAMP level by light in vivo. Nat Methods. 2007;4:39–42. doi: 10.1038/nmeth975. [DOI] [PubMed] [Google Scholar]
- 40.Nagahama T, Suzuki T, Yoshikawa S, Iseki M. Functional transplant of photoactivated adenylyl cyclase (PAC) into Aplysia sensory neurons. Neurosci Res. 2007;59:81–88. doi: 10.1016/j.neures.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 41.Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–1029. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- 42.Oh E, Maejima T, Liu C, Deneris E, Herlitze S. Substitution of 5-HT1A receptor signaling by a light-activated G protein-coupled receptor. J Biol Chem. 2010;285:30825–30836. doi: 10.1074/jbc.M110.147298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melyan Z, Tarttelin EE, Bellingham J, Lucas RJ, Hankins MW. Addition of human melanopsin renders mammalian cells photoresponsive. Nature. 2005;433:741–745. doi: 10.1038/nature03344. [DOI] [PubMed] [Google Scholar]
- 44.Royer S, Zemelman BV, Barbic M, Losonczy A, Buzsaki G, Magee JC. Multi-array silicon probes with integrated optical fibers: light-assisted perturbation and recording of local neural circuits in the behaving animal. Eur J Neurosci. 2010;31:2279–2291. doi: 10.1111/j.1460-9568.2010.07250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huber D, Petreanu L, Ghitani N, Ranade S, Hromadka T, Mainen Z, Svoboda K. Sparse optical microstimulation in barrel cortex drives learned behaviour in freely moving mice. Nature. 2008;451:61–64. doi: 10.1038/nature06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernstein JG, Han X, Henninger MA, Ko EY, Qian X, Franzesi GT, McConnell JP, Stern P, Desimone R, Boyden ES. Prosthetic systems for therapeutic optical activation and silencing of genetically-targeted neurons. Proc Soc Photo Opt Instrum Eng. 2008;6854:68540H. doi: 10.1117/12.768798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Peca J, Matsuzaki M, Matsuzaki K, Noguchi J, Qiu L, Wang D, Zhang F, Boyden E, Deisseroth K, et al. High-speed mapping of synaptic connectivity using photostimulation in Channelrhodopsin-2 transgenic mice. Proc Natl Acad Sci U S A. 2007;104:8143–8148. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hagglund M, Borgius L, Dougherty KJ, Kiehn O. Activation of groups of excitatory neurons in the mammalian spinal cord or hindbrain evokes locomotion. Nat Neurosci. 2010;13:246–252. doi: 10.1038/nn.2482. [DOI] [PubMed] [Google Scholar]
- 49*.Katzel D, Zemelman BV, Buetfering C, Wolfel M, Miesenbock G. The columnar and laminar organization of inhibitory connections to neocortical excitatory cells. Nat Neurosci. 2010 doi: 10.1038/nn.2687. The authors investigate the organization of interneuron connections onto pyramidal cells in multiple cortical regions. They also describe a novel transgenic expression strategy, crossing mice that express ChR2 in a floxed configuration with a cre driver line. This enables the benefits of transgenic expression of ChR2 while maintaining the versatility of cre driver lines for targeting specific cell types. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cardin JA, Carlen M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, Moore CI. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–667. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kravitz AV, Freeze BS, Parker PR, Kay K, Thwin MT, Deisseroth K, Kreitzer AC. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–626. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai HC, Zhang F, Adamantidis A, Stuber GD, Bonci A, de Lecea L, Deisseroth K. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–1084. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stuber GD, Hnasko TS, Britt JP, Edwards RH, Bonci A. Dopaminergic terminals in the nucleus accumbens but not the dorsal striatum corelease glutamate. J Neurosci. 2010;30:8229–8233. doi: 10.1523/JNEUROSCI.1754-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Lee JH, Durand R, Gradinaru V, Zhang F, Goshen I, Kim DS, Fenno LE, Ramakrishnan C, Deisseroth K. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465:788–792. doi: 10.1038/nature09108. The authors integrated optogenetic stimulation with fMRI imaging to investigate the mechanism underlying the BOLD response. In addition, the authors showed that this approach can reveal long range projections by optically stimulating motor cortex and observing changes in BOLD response in thalamus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cruikshank SJ, Urabe H, Nurmikko AV, Connors BW. Pathway-specific feedforward circuits between thalamus and neocortex revealed by selective optical stimulation of axons. Neuron. 2010;65:230–245. doi: 10.1016/j.neuron.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saito T, Nakatsuji N. Efficient gene transfer into the embryonic mouse brain using in vivo electroporation. Dev Biol. 2001;240:237–246. doi: 10.1006/dbio.2001.0439. [DOI] [PubMed] [Google Scholar]
- 59.Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–872. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- 60.Borrell V, Yoshimura Y, Callaway EM. Targeted gene delivery to telencephalic inhibitory neurons by directional in utero electroporation. J Neurosci Methods. 2005;143:151–158. doi: 10.1016/j.jneumeth.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 61.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–424. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, Schneider MB, Deisseroth K. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J Neurosci. 2007;27:14231–14238. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63*.Cardin JA, Carlen M, Meletis K, Knoblich U, Zhang F, Deisseroth K, Tsai LH, Moore CI. Targeted optogenetic stimulation and recording of neurons in vivo using cell-type-specific expression of Channelrhodopsin-2. Nat Protoc. 2010;5:247–254. doi: 10.1038/nprot.2009.228. The authors discuss the use of ChR2 for identifying specific cell types in in vivo electrophysiological recordings, as well as problems that can arise with this approach. Notably, they describe how to reduce optically-induced artifacts which may occur on recording electrodes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lima SQ, Hromadka T, Znamenskiy P, Zador AM. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PLoS One. 2009;4:e6099. doi: 10.1371/journal.pone.0006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Han X, Qian X, Bernstein JG, Zhou HH, Franzesi GT, Stern P, Bronson RT, Graybiel AM, Desimone R, Boyden ES. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–198. doi: 10.1016/j.neuron.2009.03.011. The authors describe the first use of optogenetics in a nonhuman primate brain. Recordings of ChR2-driven spiking were made in the frontal cortex of awake rhesus macaques. Notably, these recordings took place over many months, indicating that ChR2 expression was not damaging to macaque cortical neurons over this time span. [DOI] [PMC free article] [PubMed] [Google Scholar]