The transcription factor PU.1 is required for the development of interleukin 9-producing T cells and allergic inflammation (original) (raw)
. Author manuscript; available in PMC: 2011 Jul 14.
Published in final edited form as: Nat Immunol. 2010 May 2;11(6):527–534. doi: 10.1038/ni.1867
Abstract
CD4+ T helper cells acquire effector phenotypes that promote specialized inflammatory responses. We show that the ETS family transcription factor, PU.1 was required for the development of an interleukin 9 (IL-9)-secreting subset of TH cells. Decreasing PU.1 expression either by conditional deletion in murine T cells or siRNA in human T cells impaired IL-9 production, while ectopic PU.1 expression promoted IL-9 production. Mice with PU.1-deficient T cells developed normal TH2 responses in vivo, but exhibited attenuated allergic pulmonary inflammation corresponding to decreased Il9 and chemokine expression in peripheral T cells and in lungs as compared to wild-type mice. Together, these data suggest a critical role for PU.1 in generating the TH9 phenotype and in the development of allergic inflammation.
Introduction
T helper (TH) cells promote the development of specific types of inflammation following differentiation into various effector subsets. The cytokine microenvironment present when a naive TH cell is activated determines the developmental pathway. Interleukin 4 (IL-4 [http://www.signaling-gateway.org/molecule/query?afcsid=A001262]) promotes the differentiation of TH2 cells and allergic inflammation. IL-12 promotes TH1 differentiation, while inflammatory cytokines, including transforming growth factor-β (TGF-β1), IL-6, IL-1 and IL-23, stimulate the development of TH17 cells, both cell types contribute to autoimmune inflammation1–3. TH differentiation and the establishment of effector phenotypes are governed by a network of transcription factors that regulate the expression of cytokines and other effector genes. For example, TH2 development requires the IL-4–induced activation of STAT6, followed by the subsequent induction of GATA-3, a TH2 lineage-promoting factor. The ability of TH2 cells to respond to subsequent T cell receptor (TCR) stimulation with a rapid induction of TH2-specific cytokines further requires NFAT family members, Jun family members, c-maf, and IRF4, likely among other factors, to transactivate cytokine loci4–6.
We have described that the transcription factor PU.1 [http://www.signaling-gateway.org/molecule/query?afcsid=A001950] also contributes to the TH2 phenotype by promoting heterogeneity of TH2 cytokine secretion7,8. PU.1 was specifically expressed in subpopulations of TH2 cells that had low IL-4 expression. Ectopic expression of PU.1 reduced production of most TH2 cytokines, while shRNA that decreased PU.1 expression, or PU.1-deficiency, increased overall cytokine production specifically in populations of cells that secreted two or more cytokines7,8. PU.1 functions, at least in part, by limiting the ability of GATA-3 and IRF4 to interact with cytokine loci targets7–9.
IL-9 [http://www.signaling-gateway.org/molecule/query?afcsid=A001559] has long been thought to be a TH2 cytokine as it promotes allergic inflammation and is associated with various TH2 responses10. Transgenic expression of Il9 results in allergic inflammation11–13. IL-9 promotes mucus production from lung epithelial cells and pulmonary mastocytosis14–17. While allergic inflammation can develop in _Il9_−/− mice using a standard sensitization-challenge model, blockade of IL-9 in a similar model results in decreased allergic inflammation18,19. IL-9 expression is increased in lungs of asthmatic patients20,21, and blocking antibodies are being developed as a therapy for atopic disease22. IL-9 may also contribute to extracellular parasite immunity23,24. Most recently, IL-9 has been shown to promote the development of TH17 cells, while increasing Treg activity25,26, suggesting that IL-9 biology may be more complex.
In addition to established TH subsets, reports have described an IL-9-secreting population of T cells27. TH2 cells and Treg cells can produce IL-9 (ref. 28), and culture with TGF-β1 may increase IL-9 production from multiple cell types26. A TH subset putatively called TH9 cells are derived in culture with a combination of TGF-β1 and IL-4 (refs. 24,29). These cells are related to TH2 cells in that they require STAT6 and GATA-3 for development, but have reduced expression of TH2 cytokines24,29. Transcription factors that specifically control the IL-9–secreting T cell phenotype have not been identified.
In this report, we identified PU.1 as a regulator of IL-9 production in T cells. PU.1-deficient T cells had diminished IL-9 production and ectopic expression of PU.1 increased IL-9 production from TH2 or TH9 cultures. Diminishing PU.1 expression in human IL-9–secreting T cell cultures also decreased IL-9 production. In mice that lack PU.1 only in T cells, allergic airway inflammation was attenuated and this observation correlated with decreased IL-9 and PU.1-induced chemokine production in the lung. These results suggest that PU.1 is a critical regulator of the IL-9–secreting T cell phenotype and is important for the development of allergic inflammation.
RESULTS
PU.1 promotes the development of IL-9–secreting T cells
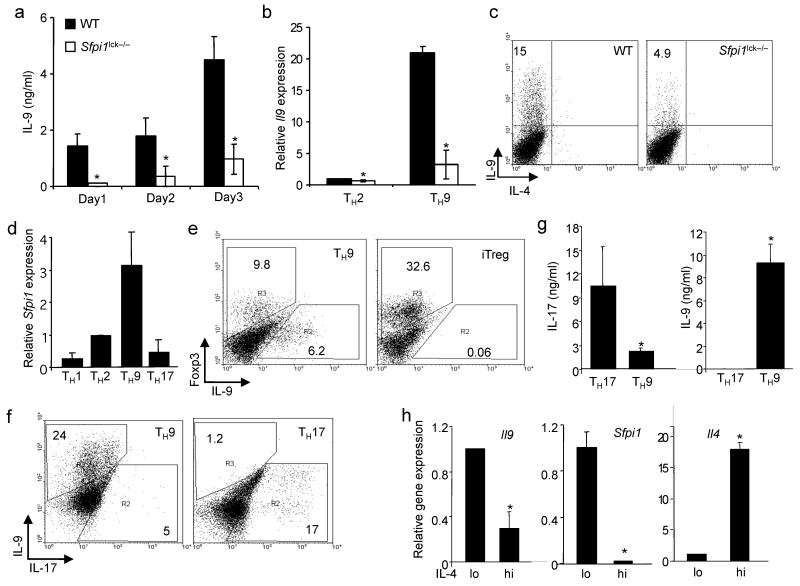
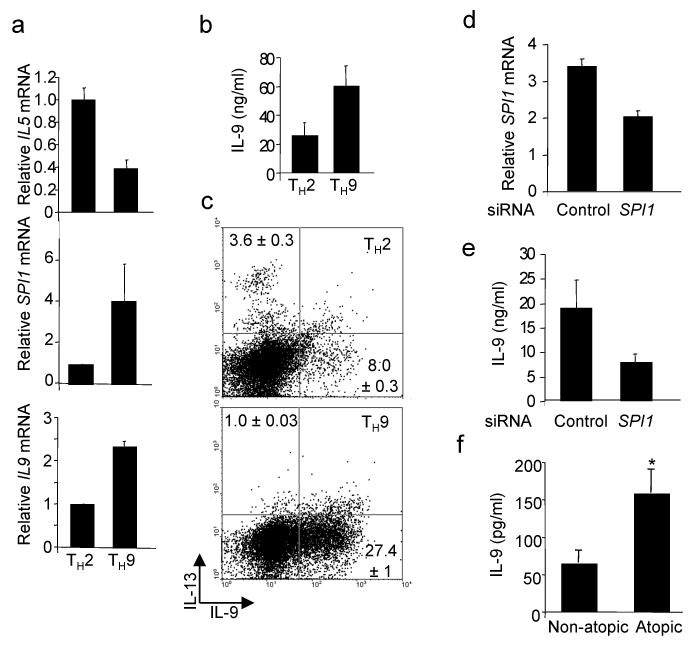
Initial descriptions of the IL-9–secreting TH cells (hereafter called TH9 for simplicity) did not identify a transcription factor that distinguished these cells as a distinct lineage24,29. We have shown that PU.1 limits TH2 cytokine production in T cells7,8, suggesting that it could be involved in the TH9 phenotype. To test this hypothesis, we differentiated naive T cells from wild-type and PU.1-deficient (_Sfpi1_lck−/−) mice under TH9 conditions (TGF-β1 plus IL-4). We observed that IL-9 secretion, mRNA expression and IL-9 intracellular staining were decreased in TH9 cultures that lacked PU.1 expression (Fig. 1a–c).
Figure 1.
PU.1 is required for optimal IL-9 production in murine T cells. Wild-type and _Sfpi1_lck−/− naïve CD4+ T cells were cultured under TH9 conditions. (a) After five days of culture, cells were stimulated with anti-CD3 and supernatants after 1, 2 or 3 days of stimulation were tested for IL-9 using ELISA. (b) RNA was isolated from wild-type and _Sfpi1_lck−/− TH2 and TH9 cultures after stimulation with anti-CD3. Il9 mRNA was assessed using qPCR. (c) Wild-type and _Sfpi1_lck−/– TH9 cultures were stimulated with PMA-ionomycin for 5 h before intracellular staining for IL-9. Results in (a-c) are the average of 3 mice and representative of more than four experiments. (d) Naïve CD4+ T cells cultured under TH1, TH2, TH9 or TH17 conditions were analyzed for Sfpi1 expression using qPCR. Results are an average of 3-5 experiments. (e) Naïve CD4+ T cells were cultured under TH9 or iTreg conditions for five days and stimulated with anti-CD3 for 6 h before intracellular staining for IL-9 and Foxp3. (f,g) Naïve CD4+ T cells were cultured under TH9 or TH17 conditions for five days and stimulated with (f) PMA-ionomycin for 6 h before intracellular staining for IL-9 and IL-17 or (g) anti-CD3 for 24 h before supernatants were collected for analysis using ELISA. Results in (e-g) are representative of at least three experiments. (h) TH2 cultures were separated into IL-4lo and IL-4hi populations using magnetic selection. RNA was isolated from separated cells to examine expression of Il4, Il9 and Sfpi1. Results are an average of three experiments. *, P < 0.05 using two-tailed student t test.
PU.1 expression was low in TH1 cells, and in TH2 populations was expressed in IL-4lo but not IL-4hi cells8. To compare the expression of PU.1 in additional TH subsets, we generated TH1, TH2, TH17 and TH9 cells and analyzed RNA for expression of PU.1. We observed that TH1 and TH17 had the lowest expression of Sfpi1 mRNA, TH2 cells had 3–5-fold more abundant expression, and TH9 cells expressed approximately 3-fold more Sfpi1 than TH2 cells (Fig. 1d). Inducible Treg cultures expressed even more Sfpi1 mRNA than in TH9 cultures (Supplementary Fig. 1a), suggesting that Sfpi1 might be a TGF-β-induced gene. The expression of Sfpi1 was not STAT6-dependent as determined by comparing wild-type and _Stat6_−/− TH9 cultures, (data not shown). As both iTreg and TH17 cultures have been suggested to produce IL-9 (refs. 25,26,28), we also directly compared cells cultured under these conditions to TH9 cultures. By intracellular staining and ELISA we observed low or undetectable amounts of IL-9 produced by iTreg cultures (Fig. 1e and data not shown). In comparing TH9 and TH17 cultures, we observed little IL-9 production in TH17 cultures assays by intracellular staining or ELISA, though there was a low amount of IL-17 produced from TH9 cultures (Fig. 1f,g). As IL-9 was observed in TH2 cultures, we separated TH2 populations into IL-4hi and IL-4lo populations8, and observed that Il9 expression segregated with Sfpi1 expression in the IL-4lo cells and segregated away from Il4 expression (Fig. 1h).
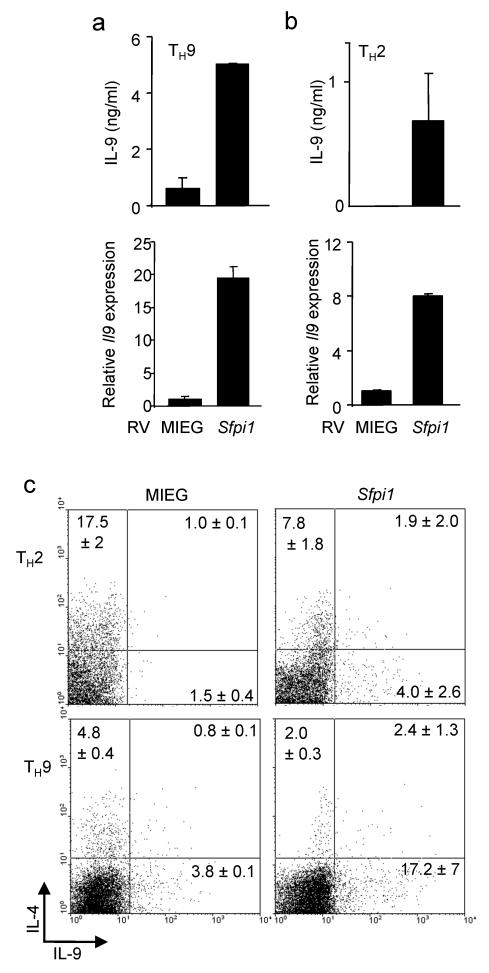
To define if PU.1 was a determining factor in promoting IL-9 secretion, we ectopically expressed PU.1 using a bicistronic retrovirus in TH2, TH9 and iTreg cultures and examined cytokine secretion and gene expression in sorted EGFP+ cells. Increased PU.1 expression in TH2 or TH9 cultures further induced IL-9 production, assessed by both ELISA and intracellular cytokine staining, and measurement of Il9 mRNA in these cultures (Fig. 2a-c). Ectopic PU.1 expression in TH2 cells, as previously described, reduced the expression of TH2 cytokines (Fig. 2c). While ectopic PU.1 expression could decrease Foxp3 expression in iTreg cultures (Supplementary Fig. 1b), it did not induce Il9 expression in these cells (data not shown). Inducible Treg generation was normal in PU.1-deficient cells, and the ability of IL-4 to repress Foxp3 was not significantly impaired in the absence of PU.1 expression (Supplementary Fig. 1c,d). Moreover, there were no significant increases in CD4+Foxp3+ cells in a variety of _Sfpi1_lck−/− lymphoid organs, compared to wild-type mice (Supplementary Fig. 1e,f), suggesting that PU.1-deficiency does not affect Foxp3 expression. These results suggest that PU.1 promotes the IL-9–secreting T cell phenotype, but that additional IL-4-induced signals are required to generate TH9 cells.
Figure 2.
PU.1 promotes IL-9 production. TH9 (a,c) or TH2 (b,c) cultures were transduced with control (MIEG) or PU.1-expressing retroviruses. (a,b) EGFP-positive cells were sorted and stimulated to determine secretion of IL-9 (top) and Il9 expression (bottom). (c) IL-4 and IL-9 production was determined by intracellular staining in the EGFP+ populations of control and PU.1-transduced cells. Percentages in each quadrant are the average of results from 2-3 mice. Data are representative of 2-3 experiments.
Distinct populations secrete IL-10 and IL-9
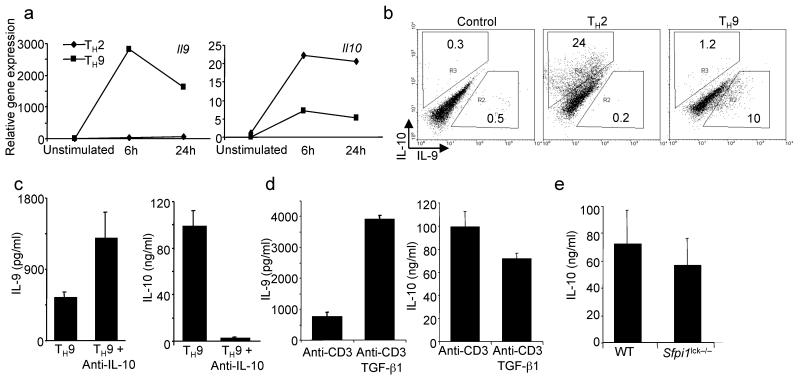
Previous reports on IL-9–secreting T cells suggested that they also express IL-10 (refs. 24,29). In contrast, our previous report demonstrated that IL-9 and IL-10 expression in TH2 cells were reciprocally regulated9. To further examine this issue, we differentiated TH2 and TH9 cells from cultures of naïve CD4+ T cells using IL-4 or a combination of IL-4 and TGF-β1, respectively. While IL-10 was detected in TH9 cells, production was much lower than from TH2 cultures (Fig. 3a,b). Moreover, additional treatments of TH9 cultures had reciprocal effects on IL-9 and IL-10 production. We observed an increase of IL-9 production from TH9 cultures when they were differentiated in the presence of anti-IL-10 concomitant with a decrease in IL-10 production (Fig. 3c). Addition of IL-9 to TH2 cultures did not affect IL-10 production (data not shown). Addition of TGF-β1 to differentiated TH9 cultures, during anti-CD3 restimulation, increased IL-9 production, but decreased IL-10 production (Fig. 3d). While PU.1-deficiency resulted in less IL-9 production (Fig. 1), it did not significantly affect IL-10 production from TH9 cultures (Fig. 3e). Thus, while TH9 cultures may produce IL-10, IL-9 and IL-10 are not coordinately regulated.
Figure 3.
IL-9 and IL-10 are not coordinately regulated in TH9 cells. (a) Naïve CD4+ T cells were cultured under TH2 or TH9 conditions for five days before cells were left unstimulated or stimulated with plate-bound anti-CD3 for 6 or 24 hours and RNA was isolated for qPCR analysis of Il9 and Il10. (b) TH2 and TH9 cultures derived as in a were stimulated for 5 hours before intracellular cytokine analysis of IL-10 and IL-9. (c) Naïve CD4+ T cells were cultured under TH9 conditions in the presence or absence of anti-IL-10. After five days in culture cells were restimulated with anti-CD3 and supernatants were analyzed for cytokines using ELISA. (d) Naïve CD4+ T cells were cultured under TH9 conditions. After five days in culture, cells were restimulated with anti-CD3 in the presence or absence of TGF-β1 and supernatants were analyzed for cytokines using ELISA. (e) Wild-type and _Sfpi1_lck−/− naïve CD4+ T cells were cultured under TH9 conditions for five days, and stimulated with anti-CD3 for 24 hours before supernatants were tested for IL-10 using ELISA. Results are representative of at least three experiments.
PU.1 is required for chromatin modifications of Il9
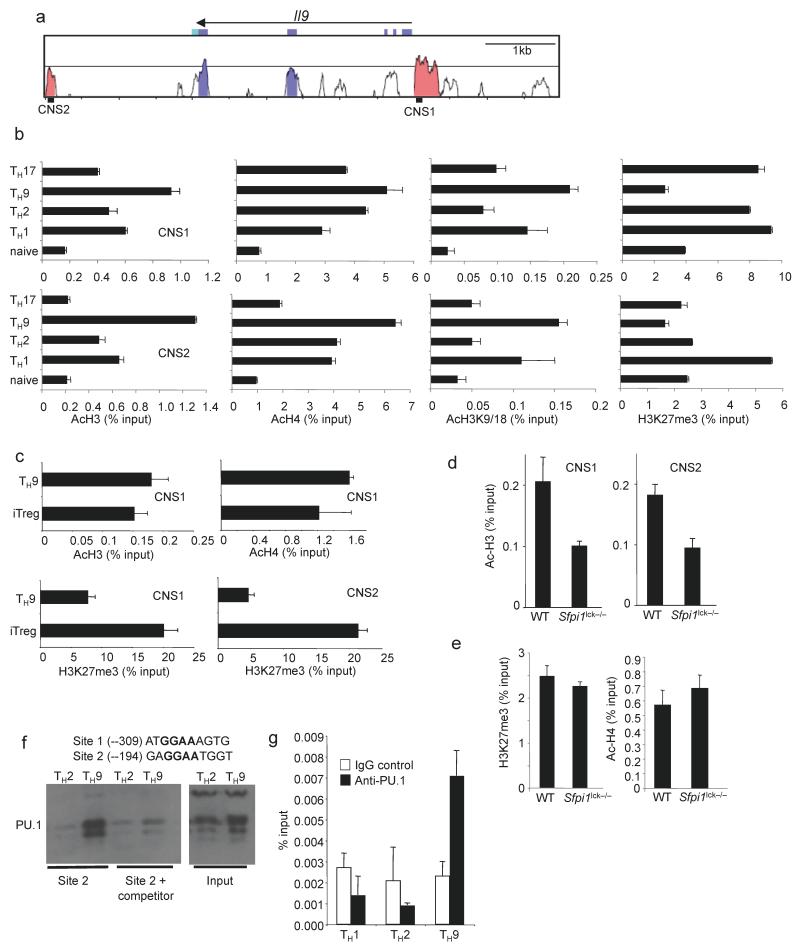
As T helper specification is associated with specific chromatin structure, we examined chromatin modifications at the Il9 locus in naive T cells, and naive T cells cultured for five days under TH1, TH2, TH9 or TH17 conditions using chromatin immunoprecipitation (ChIP) assays. Total acetylation of both histone H3 and H4, and acetylation of H3K9 and H3K18 at two Il9 CNS (Fig. 4a) were highest in TH9 cultures compared to all other subsets (Fig. 4b), though all subsets demonstrated an increase in acetylation compared to naïve cells. TH9 cells also had the lowest amount of trimethylated H3K27 (H3K27me3), a modification associated with repressed genes (Fig. 4b). Cultures of iTreg cells displayed histone acetylation comparable to TH9 cells (Fig. 4c), in agreement with data on the Il9 locus derived from genome-wide studies30. However, the Il9 locus had increased amounts of H3K27me3 in iTreg cultures compared to TH9 cultures (Fig. 4c). Thus, active Il9 expression was associated with a combination of high H3 acetylation and low H3K27 methylation. To test if PU.1 was required for the establishment of these chromatin modifications, we differentiated wild-type and PU.1-deficient naive T cells under TH9 conditions and analyzed chromatin modifications. We observed H3 acetylation, the modification that showed the greatest difference between TH9 and other TH cultures, was decreased in the absence of PU.1 at both CNS sites (Fig. 4d) and that this effect was specific for TH9 cultures since PU.1-deficiency did not affect H3 acetylation of Il9 in TH2 or TH17 cells (Supplementary Fig. 2). Moreover, the presence of the Cre transgene did not decrease Il9 H3 acetylation (Supplementary Fig. 2). In contrast, H4 acetylation or H3K27 methylation were not affected by the absence of PU.1 (Fig. 4e). These results suggest that PU.1 is required for programming the Il9 locus but plays a restricted role in the histone modifications that are affected.
Figure 4.
Histone modifications at the Il9 locus. (a) VISTA plot of conserved non-coding sequences adjacent to the Il9 locus. (b,c) Naïve CD4+ T cells were isolated and analyzed directly or differentiated under TH1, TH2, TH9, TH17 or iTreg culture conditions for five days. Cells were analyzed for histone modifications using chromatin immunoprecipitation for the indicated histone modifications and primers for Il9 CNS. (d,e) Wild-type and _Sfpi1_lck−/− naïve CD4+ T cells were cultured under TH9 conditions and used for ChIP analysis as described in b performed for AcH3 at CNS1 and CNS2 (d), or for AcH4 and me3H3K27 at CNS1 (e). (f) Top, consensus PU.1 binding sites in the Il9 CNS1. Bottom, DAPA analysis of PU.1 binding from TH2 or TH9 extracts to site 2. For competition, extracts were incubated with a 5-fold excess of unlabelled PU.1 consensus double-stranded oligonucleotide. Expression of PU.1 in each cell extract is shown as input. Results are representative of three experiments. (g) Naïve CD4+ T cells were differentiated under TH1, TH2, or TH9 culture conditions for five days. Cells were analyzed for PU.1 binding by ChIP with primers for _Il9_CNS1. Results of ChIP experiments are shown as average ± SD of percent input with control IgG subtracted (a-e) or with anti-PU.1 and IgG control ChIP values shown separately (g) and are representative of 2-4 experiments.
To determine if PU.1 was acting directly on the Il9 gene we examined the CNS1 sequence for consensus PU.1 binding sites (5′-GGAA-3′ core sequence). We found two potential sites within sequences that were conserved among human, mouse and dog Il9 promoters (Fig. 4f). Using DNA affinity precipitation assay (DAPA) with cell extracts from TH2 or TH9 cells, followed by immunoblot, we observed PU.1 binding to both sites, with stronger binding to site 2 and binding was decreased by incubation with a non-biotinylated competitor oligonucleotide (Fig. 4f and data not shown). More PU.1 binding was observed using TH9 extracts, parallel to increased PU.1 protein expression in TH9 extracts (Fig. 4f). We then tested if PU.1 bound to the Il9 promoter using ChIP assays. Following differentiation of TH1, TH2 and TH9 cells, PU.1 ChIP assays detected PU.1 binding to the Il9 promoter in TH9 cells, with no detectable binding in TH1 or TH2 cells (Fig. 4g). Thus, PU.1 binds to sites in the Il9 promoter specifically in TH9 cells.
PU.1 is required for IL-9 production in human T cells
The ability of conditions that promote the development of murine TH9 cells to promote a human TH9 phenotype has not been tested. To assess the derivation of human TH9 cells, we cultured naive CD4+ T cells purified from peripheral blood under TH2 or TH9 conditions. As with murine cells, the addition of TGF-β1 to TH2 cultures decreased TH2 cytokine production and increased production of IL-9 (Fig. 5a–c). Human TH9 cultures also expressed more SPI1 than TH2 cultures (Fig. 5a). We then determined if PU.1 was required for IL-9 expression in human TH9 cells by transfecting differentiated human TH9 cultures with control or _SPI1_-specific siRNA before restimulation. Inhibiting SPI1 expression resulted in a decrease in IL-9 production (Fig. 5d,e). These results suggest that the ability of PU.1 to promote the development of IL-9–secreting T cells is conserved in human and mouse cells.
Figure 5.
PU.1 promotes IL-9 production in human T cells. (a) Naïve human CD4+ T cells were cultured under TH2 or TH9 conditions and after five days in culture restimulated to analyze gene expression by qPCR. (b,c) TH2 and TH9 cultures were derived as in a and b supernatants from cells stimulated for 24 hours were tested for IL-9 production using ELISA or (c) cells were stimulated with anti-CD3 for 6 hours for analysis of IL-9 and IL-13 by intracellular cytokine staining. (d,e) Th9 cultures derived as in a were transfected with control or _SPI1_-specific siRNA. Twenty-four hours after transfection cells were analyzed for SPI1 expression by qPCR (d) and IL-9 production using ELISA (e). Results in (a-e) are representative data from 3-5 experiments with different donors. (f) PBMCs from infants (age 18-30 months) classified as atopic (n= 49) or non-atopic (n= 33) based on positive allergen-specific serum IgE, were stimulated with anti-CD3 for 48 hours and supernatants were tested for production of IL-9 using multiplex analysis. *, p<0.04 using an unpaired student t test.
TH9 cells and allergic inflammation
The role of IL-9 in allergic inflammation is still unclear. To begin to explore this area further we examined IL-9 production from anti-CD3–stimulated peripheral blood mononuclear cells (PBMCs) from atopic (positive for at least one allergen-specific IgE of ten examined) and non-atopic (no allergen-specific IgE) children with ages ranging from 18-30 months. This cohort has been described in more detail elsewhere31. Importantly, while these children show signs of developing atopy, they have not had episodes of wheezing and are not considered to be asthmatic. IL-9 production was significantly higher from atopic patients than from non-atopic patients (Fig. 5f).
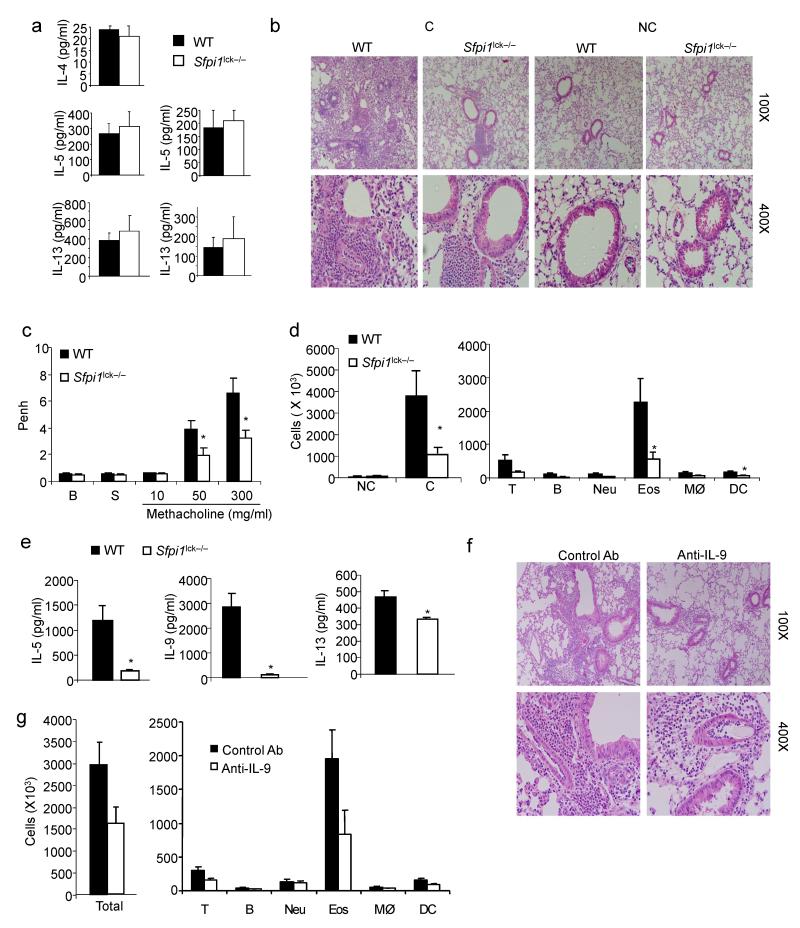
To further examine a role for PU.1-dependent T cell function in allergic inflammation, wild-type and _Sfpi1_lck−/− mice were subjected to a standard protocol for the development of ovalbumin (Ova)-induced allergic airway inflammation. Mice were sensitized with Ova in alum and challenged intranasally with Ova in PBS. We first tested the generation of TH responses in the periphery by stimulating splenic cells with Ova or anti-CD3. Consistent with our previous in vitro analyses7,8 there was no impairment of TH2 cytokine responses in _Sfpi1_lck−/− mice, compared to wild-type mice (Fig. 6a). Production of TH17 cytokines was also unimpaired by the absence of PU.1 (Supplementary Fig. 3). The generation of antigen-specific IgE and IgG1 was not significantly different between wild-type and _Sfpi1_lck−/− mice (data not shown).
Figure 6.
PU.1 expression in T cells is required for the development of allergic inflammation. Wild-type and _Sfpi1_lck−/− mice were sensitized and challenged intranasally with ovalbumin. Forty-eight hours after the last intranasal challenge mice were sacrificed for analysis. (a) Splenocytes were stimulated with anti-CD3 (left) or Ova (right) for 72 h and supernatants were tested for the amounts of TH2 cytokines by ELISA. (b) Lungs of challenged (C) or non-challenged (NC) mice were embedded in paraffin and analyzed by H/E staining. Magnification is indicated in the panel. (c) Wild-type and _Sfpi1_lck−/− mice were analyzed for airway hyperreactivity using whole body plethysmography 24 h after the last intranasal challenge. Airway function was tested at baseline (B), with saline inhalation (S) and following inhalation of the indicated doses of methacholine. (d) Numbers of cells recovered by bronchoalveolar lavage in challenged or non-challenged mice (left). Numbers of specific cell types in BAL from challenged mice, as determined by flow cytometry, are indicated (right). (e) Amounts of IL-9 and TH2 cytokines present in BAL fluid were determined using ELISA. (f, g) Wild type mice were sensitized and challenged intranasally with ovalbumin daily for five days. Mice were injected intravenously with control Ig or anti-IL-9, 30 minutes before the first, third and fifth intranasal challenge. Forty-eight hours after the last intranasal challenge, lungs were processed for histological analysis (f) as in b, and BAL cells were harvested and analyzed (g) as in d. *, p<0.05 using two-tailed student t test. All results are the average ± SEM of 5-7 mice and are representative of three experiments.
In contrast to the normal development of peripheral responses, inflammation in the lung was decreased in mice with PU.1-deficient T cells, compared to wild-type mice (Fig. 6b). _Sfpi1_lck−/− mice also demonstrated significantly decreased airway hyperresponsiveness in response to methacholine challenge than wild-type mice (Fig. 6c). Analysis of bronchoalveolar lavage cellular composition demonstrated increased infiltration in wild-type and _Sfpi1_lck−/− mice compared to non-challenged mice, but total cell numbers, and the cell numbers of specific cell populations, including eosinophils, were decreased in _Sfpi1_lck−/− mice compared to wild-type mice (Fig. 6d). To confirm that Lck-Cre-mediated deletion of the Sfpi1 allele was not affecting PU.1-dependent development of dendritic cells important for the development of inflammation, we examined CD11b+ and CD103+ subpopulations of CD11c+ MHC class II+ cells in bronchoalveolar lavage (BAL) and lung cell suspensions of non-sensitized and non-challenged mice and observed no significant differences between wild-type and _Sfpi1_lck−/− mice (data not shown). There were decreased amounts of IL-9 and TH2 cytokines present in BAL fluid (Fig. 6e). In contrast, amounts of interferon-γ and IL-17 present in the BAL were not significantly affected (data not shown).
To demonstrate that decreases in IL-9 could result in a similar phenotype, we used the Ova-induced allergic airway inflammation model for wild-type mice treated with control or anti-IL-9 administered intravenously at the first, third and fifth challenges. We observed decreased inflammation and cell numbers of allergic inflammatory cells in mice treated with anti-IL-9, compared to control Ab-treated mice (Fig. 6f,g). This result confirms a role for IL-9 in this model established using a similar protocol in BALB/c mice18.
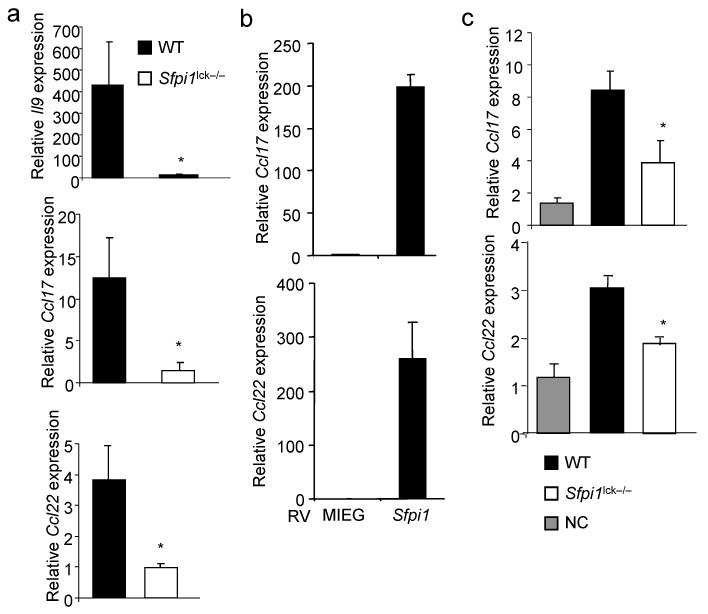
To determine if Il9 expression was lower in T cells from sensitized _Sfpi1_lck−/− mice than in wild-type mice, we used qPCR to assess mRNA abundance in antigen-stimulated splenocyte cultures. We observed diminished Il9 mRNA in _Sfpi1_lck−/− cultures (Fig. 7a). It was possible that PU.1 was regulating other genes associated with a TH9 phenotype. We have previously shown that PU.1 expression segregated with expression of Ccl22 in IL-4lo cells, and that ectopic PU.1 expression increased CCL22 production8. Consistent with those observations, we observed that chemokines associated with allergic inflammation, Ccl17 and Ccl22, were less abundantly produced from antigen-stimulated _Sfpi1_lck−/− cultures as compared to wild-type cultures (Fig. 7a). Conversely, ectopic expression of PU.1 induced the expression of both chemokines in TH9 cultures (Fig. 7b). To determine if the deficiency of chemokine production from T cells was reflected in the allergic inflammation model, we analyzed whole lung homogenates for Ccl17 and Ccl22 mRNA. While mRNA for both chemokines was present in tissue from sensitized and challenged mice, both were decreased in _Sfpi1_lck−/− lung tissue as compared to wild-type (Fig. 7c). These results suggest that PU.1 in T cells regulates allergic inflammation by inducing the expression of IL-9 and pro-allergic chemokines.
Figure 7.
PU.1 is required for IL-9 and chemokine expression in allergen sensitized mice. (a) RNA isolated from Ova-stimulated splenocytes as described in (Fig. 6a) was analyzed for expression of Il9 and chemokines associated with allergic inflammation. (b) TH9 cultures were transduced with control (MIEG) or PU.1 expressing retroviruses. EGFP-positive cells were sorted and stimulated to determine expression of Ccl17 and Ccl22. (c) Total lung RNA was isolated from sensitized and challenged WT and _Sfpi1_lck−/− mice, or WT non-challenged (NC) mice, and analyzed for Ccl17 and Ccl22 mRNA by qPCR. *, p<0.05 using two-tailed student t test.
DISCUSSION
The defined phenotypes of effector T helper subsets, once classified simply as TH1 and TH2, have been expanded to include TH17, TFH and inducible Treg cells. Most recently, an IL-9–secreting T cell, putatively termed TH9, was identified as arising from a cytokine combination of TGF-β1 and IL-4 (refs. 24,29). Distinguishing these subsets has become more difficult as the plasticity between the phenotypes has become apparent32. However, each of the lineages express transcription factors that are lineage-determining factors or that promote particular aspects of the lineage genetic program. The TH9 lineage, if it is indeed a separate lineage, is related to TH2 by the developmental requirement for IL-4 and the downstream transcription factors STAT6 and GATA3 (refs. 24,29). In this report, we have identified PU.1 as a transcription factor that induces the TH9 phenotype by promoting the expression of IL-9 and pro-allergic chemokines.
In our previous analyses of PU.1 in TH2 cells we noted that expression was segregated to a subpopulation of TH2 cells that expressed low amounts of IL-4 and other TH2 cytokines8. This observation corresponded with the ability of ectopically expressed PU.1 to inhibit the production of TH2 cytokines, and of decreasing PU.1 expression, either by siRNA or gene deletion, to increase TH2 cytokine production7,8. This function was at least partly by interfering with factors that promote the TH2 phenotype including GATA3 and IRF4 (ref. 8,9). The data presented in this report suggests that the IL-4lo TH2 subset that we previously characterized might contain some TH9 cells. Given the apparent plasticity of TH2 cells to acquire a TH9 phenotype24, these two cell types might exist in a continuum where increasing PU.1 expression would lead to decreasing TH2 cytokines and increasing characteristics of the TH9 phenotype, with intermediate phenotypes being possible.
Our data indicate that PU.1 can bind the Il9 promoter directly in TH9 cells, and that conserved PU.1 binding sites exist within CNS1. While ectopic expression of PU.1 increased characteristics of TH9 cells and converted TH2 cells to a TH9 phenotype, it had more modest effects on the induction of Il9 in iTreg, TH1 or TH17 cultures (data not shown). These results might suggest that while PU.1 promotes the TH9 phenotype, it may be most active in cells where additional factors are present to mediate these effects. This scenario is not uncommon in the TH differentiation paradigm. As described above, multiple factors contribute to the TH2 phenotype, and multiple factors including T-bet, STAT4, Hlx and Runx3 contribute to the TH1 genetic program33–35. One possible context requirement might be decreased expression of IRF4, which we have demonstrated represses Il9 in TH2 cultures and is further required for the development of TH17 cells9,36. Preliminary observations suggest that Irf4 expression is less in TH9 cells than in TH2 cells (data not shown). The chromatin modifications present at the Il9 locus in each of these cell types may also reflect the ability of PU.1 to induce Il9 mRNA. The activity of the Il9 locus correlated with the combination of increased histone acetylation and reduced H3K27 methylation. However, in iTreg cultures that had a comparable extent of histone acetylation but abundant H3K27 methylation at the Il9 locus, Il9 induction by PU.1 was not detected. The ability of PU.1 to induce IL-9 in TH2 but not iTreg cultures suggests that additional IL-4-induced factors exist that are required for acquisition of the IL-9-secreting phenotype. While GATA-3 is required for TH9 development29, it seems an unlikely candidate since it is expressed only at low amounts in TH9 cultures24,29 and since we have shown PU.1 antagonizes GATA-3 activity in TH cells7,8. IRF4 also seems unlikely as it represses Il9 expression in TH2 cells, and PU.1 antagonizes its function9. It will be interesting to determine if other transcription factors, in the presence of PU.1, promote Il9 expression.
The relatedness between TH2 and TH9 phenotypes, and the requirement for STAT6 and GATA3 in the development of both subsets24,29, raises some interesting questions about phenotypes that have been described in mice deficient in either STAT6 or GATA3, specifically whether phenotypes in allergic inflammation and parasite immunity are a result of defects in the development of one or both subsets. A previous report demonstrated that mice with defective TGF-β1 signaling had decreased parasite clearance that correlated with decreased IL-9 (ref. 24). Similarly, in this report we show defects in the development of allergic inflammation in mice that have decreased IL-9-secreting T cells, but have normal development of TH2 immunity. These findings suggest that TH9 cells might be required for what has classically been termed TH2 immunity and there is ample data supporting a role for IL-9 in allergic disease. Transgenic models have demonstrated that IL-9 can promote12,13 and blockade of IL-9 inhibits18 the development of allergic inflammation. Moreover, increased IL-9 is found in lungs of patients with asthma20,21, and T cells from patients with atopic disease secreted more IL-9 than non-atopic patient T cells. Importantly, this observation does not diminish the role of TH2 cytokines in the development of allergic inflammation, rather suggesting that these two TH subsets work in concert to initiate allergic inflammation.
IL-9 can promote the development of TH17 cells and was reported to also be produced by TH17 cells25,26. Thus the decreased allergic inflammation in mice with PU.1-deficient T cells might be interpreted to result from the decreased generation of TH17, which are required for the development of inflammation in this model37. However, we did not see decreases in the ability of PU.1-deficient T cells to acquire the TH17 phenotype in vitro and ectopic expression of PU.1 did not induce IL-17 production (data not shown). Moreover, IL-17 production from mice that are sensitized and challenged was higher in Ova-stimulated splenocyte cultures from _Sfpi1_lck−/− mice than from wild-type mice. This finding suggests that PU.1 is not required for TH17 development. TGF-β1 appears to be an important factor in promoting IL-9 production from cells cultured with various cytokines26,27. It is possible that while TGF-β1 promotes IL-9 production, the addition of IL-4 alters the phenotype such that TH17 cytokine production is decreased and resulting cells are predominantly producers of IL-9 alone. In our experiments, TH9 cultures produce IL-9, which was barely detectable in TH17 cultures, and TH9 cultures produced only low amounts of IL-17 compared to TH17 cells. Thus, while there may be overlap in the cytokine signatures of these subsets, highly polarized cells might still have distinct functional properties.
The precise contribution of PU.1-expressing T cells to allergic inflammation remains undefined, and is likely a combination of altered expression of IL-9 and chemokines. Our data confirm that blocking IL-9 with antibodies decreases allergic inflammation in the OVA-induced model. As the previous report used BALB/c mice18, and our study utilized C57BL/6 mice, our data demonstrate that the effects of blocking IL-9 are not strain-specific. The chemokines CCL17, CCL22 and their receptor CCR4, are each important in the development of allergic inflammation38–40. One possibility is that PU.1-dependent chemokine production from T cells is responsible for the diminished allergic inflammation in _Sfpi1_lck−/− mice. This possibility represents an addition to the current paradigm wherein T cells in the lung secrete cytokines, such as IL-13, that promote chemokine production from resident lung cells including epithelial cells, smooth muscle cells and fibroblasts41. Our results suggest that T cell chemokine production also contributes to the overall extent of inflammation. PU.1-dependent IL-9 production, or the expression of other PU.1-dependent genes, may also contribute to inflammation in this model, and the phenotype we observe may indeed be caused by several different factors. As the PU.1-dependent genetic program in T cells becomes better defined, the role of these factors can be assessed more directly.
The recent description of an IL-9-secreting T cell population added further complexity to effector TH phenotypes. One factor that prevented this population from initially being termed a lineage was the absence of an identified transcription factor that conferred lineage-specific expression. Our work has identified PU.1 as a factor that promotes the TH9 phenotype by both repressing TH2 cytokine production and increasing IL-9 production. PU.1 is likely not the only factor that promotes this lineage, but will be an important component of continuing investigations into TH heterogeneity.
METHODS
Methods and any associated references are available in the online version of the paper.
Supplementary Material
1
02
ACKNOWLEDGEMENTS
Research was supported by Public Health Service Grants RO1 AI57459 and U19 AI070448 (MHK), RO1 CA118118 (MJR), and RO1 HL080071 (RST). GLS was supported by T32 AI060519.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare no competing financial interests.
References
- 1.Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–348. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- 2.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 3.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 4.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 5.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 7.Chang HC, et al. PU.1 regulates TCR expression by modulating GATA-3 activity. J Immunol. 2009;183:4887–4894. doi: 10.4049/jimmunol.0900363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang HC, et al. PU.1 expression delineates heterogeneity in primary Th2 cells. Immunity. 2005;22:693–703. doi: 10.1016/j.immuni.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 9.Ahyi AN, Chang HC, Dent AL, Nutt SL, Kaplan MH. IFN Regulatory Factor 4 Regulates the Expression of a Subset of Th2 Cytokines. J Immunol. 2009;183:1598–1606. doi: 10.4049/jimmunol.0803302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hauber HP, Bergeron C, Hamid Q. IL-9 in allergic inflammation. Int Arch Allergy Immunol. 2004;134:79–87. doi: 10.1159/000078384. [DOI] [PubMed] [Google Scholar]
- 11.Forbes EE, et al. IL-9- and mast cell-mediated intestinal permeability predisposes to oral antigen hypersensitivity. J Exp Med. 2008;205:897–913. doi: 10.1084/jem.20071046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Temann UA, Geba GP, Rankin JA, Flavell RA. Expression of interleukin 9 in the lungs of transgenic mice causes airway inflammation, mast cell hyperplasia, and bronchial hyperresponsiveness. J Exp Med. 1998;188:1307–1320. doi: 10.1084/jem.188.7.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Temann UA, Ray P, Flavell RA. Pulmonary overexpression of IL-9 induces Th2 cytokine expression, leading to immune pathology. J Clin Invest. 2002;109:29–39. doi: 10.1172/JCI13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steenwinckel V, et al. IL-9 promotes IL-13-dependent paneth cell hyperplasia and up-regulation of innate immunity mediators in intestinal mucosa. J Immunol. 2009;182:4737–4743. doi: 10.4049/jimmunol.0801941. [DOI] [PubMed] [Google Scholar]
- 15.Steenwinckel V, et al. IL-13 mediates in vivo IL-9 activities on lung epithelial cells but not on hematopoietic cells. J Immunol. 2007;178:3244–3251. doi: 10.4049/jimmunol.178.5.3244. [DOI] [PubMed] [Google Scholar]
- 16.Temann UA, Laouar Y, Eynon EE, Homer R, Flavell RA. IL9 leads to airway inflammation by inducing IL13 expression in airway epithelial cells. Int Immunol. 2007;19:1–10. doi: 10.1093/intimm/dxl117. [DOI] [PubMed] [Google Scholar]
- 17.Townsend JM, et al. IL-9-deficient mice establish fundamental roles for IL-9 in pulmonary mastocytosis and goblet cell hyperplasia but not T cell development. Immunity. 2000;13:573–583. doi: 10.1016/s1074-7613(00)00056-x. [DOI] [PubMed] [Google Scholar]
- 18.Cheng G, et al. Anti-interleukin-9 antibody treatment inhibits airway inflammation and hyperreactivity in mouse asthma model. Am J Respir Crit Care Med. 2002;166:409–416. doi: 10.1164/rccm.2105079. [DOI] [PubMed] [Google Scholar]
- 19.McMillan SJ, Bishop B, Townsend MJ, McKenzie AN, Lloyd CM. The absence of interleukin 9 does not affect the development of allergen-induced pulmonary inflammation nor airway hyperreactivity. J Exp Med. 2002;195:51–57. doi: 10.1084/jem.20011732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erpenbeck VJ, et al. Segmental allergen challenge in patients with atopic asthma leads to increased IL-9 expression in bronchoalveolar lavage fluid lymphocytes. J Allergy Clin Immunol. 2003;111:1319–1327. doi: 10.1067/mai.2003.1485. [DOI] [PubMed] [Google Scholar]
- 21.Shimbara A, et al. IL-9 and its receptor in allergic and nonallergic lung disease: increased expression in asthma. J Allergy Clin Immunol. 2000;105:108–115. doi: 10.1016/s0091-6749(00)90185-4. [DOI] [PubMed] [Google Scholar]
- 22.White B, Leon F, White W, Robbie G. Two first-in-human, open-label, phase I dose-escalation safety trials of MEDI-528, a monoclonal antibody against interleukin-9, in healthy adult volunteers. Clin Ther. 2009;31:728–740. doi: 10.1016/j.clinthera.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 23.Faulkner H, Renauld JC, Van Snick J, Grencis RK. Interleukin-9 enhances resistance to the intestinal nematode Trichuris muris. Infect Immun. 1998;66:3832–3840. doi: 10.1128/iai.66.8.3832-3840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veldhoen M, et al. Transforming growth factor-beta ‘reprograms’ the differentiation of T helper 2 cells and promotes an interleukin 9-producing subset. Nat Immunol. 2008;9:1341–1346. doi: 10.1038/ni.1659. [DOI] [PubMed] [Google Scholar]
- 25.Elyaman W, et al. IL-9 induces differentiation of TH17 cells and enhances function of FoxP3+ natural regulatory T cells. Proc Natl Acad Sci U S A. 2009;106:12885–12890. doi: 10.1073/pnas.0812530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nowak EC, et al. IL-9 as a mediator of Th17-driven inflammatory disease. J Exp Med. 2009;206:1653–1660. doi: 10.1084/jem.20090246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmitt E, et al. IL-9 production of naive CD4+ T cells depends on IL-2, is synergistically enhanced by a combination of TGF-beta and IL-4, and is inhibited by IFN-gamma. J Immunol. 1994;153:3989–3996. [PubMed] [Google Scholar]
- 28.Lu LF, et al. Mast cells are essential intermediaries in regulatory T-cell tolerance. Nature. 2006;442:997–1002. doi: 10.1038/nature05010. [DOI] [PubMed] [Google Scholar]
- 29.Dardalhon V, et al. IL-4 inhibits TGF-beta-induced Foxp3+ T cells and, together with TGF-beta, generates IL-9+ IL-10+ Foxp3(−) effector T cells. Nat Immunol. 2008;9:1347–1355. doi: 10.1038/ni.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei G, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tepper RS, et al. Expired nitric oxide and airway reactivity in infants at risk for asthma. J Allergy Clin Immunol. 2008;122:760–765. doi: 10.1016/j.jaci.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 33.Djuretic IM, et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 34.Mullen AC, et al. Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat Immunol. 2002;3:652–658. doi: 10.1038/ni807. [DOI] [PubMed] [Google Scholar]
- 35.Thieu VT, et al. Signal transducer and activator of transcription 4 is required for the transcription factor T-bet to promote T helper 1 cell-fate determination. Immunity. 2008;29:679–690. doi: 10.1016/j.immuni.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brustle A, et al. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–966. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- 37.Schnyder-Candrian S, et al. Interleukin-17 is a negative regulator of established allergic asthma. J Exp Med. 2006;203:2715–2725. doi: 10.1084/jem.20061401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalo JA, et al. Mouse monocyte-derived chemokine is involved in airway hyperreactivity and lung inflammation. J Immunol. 1999;163:403–411. [PubMed] [Google Scholar]
- 39.Kawasaki S, et al. Intervention of thymus and activation-regulated chemokine attenuates the development of allergic airway inflammation and hyperresponsiveness in mice. J Immunol. 2001;166:2055–2062. doi: 10.4049/jimmunol.166.3.2055. [DOI] [PubMed] [Google Scholar]
- 40.Mikhak Z, et al. Contribution of CCR4 and CCR8 to antigen-specific T(H)2 cell trafficking in allergic pulmonary inflammation. J Allergy Clin Immunol. 2009;123:67–73. e63. doi: 10.1016/j.jaci.2008.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004;202:175–190. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 42.Dakic A, et al. PU.1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stritesky GL, Yeh N, Kaplan MH. IL-23 mediates stability but not commitment to the Th17 lineage. J. Immunol. 2008;181:5948–5955. doi: 10.4049/jimmunol.181.9.5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Rijt LS, et al. A rapid flow cytometric method for determining the cellular composition of bronchoalveolar lavage fluid cells in mouse models of asthma. J Immunol Methods. 2004;288:111–121. doi: 10.1016/j.jim.2004.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
02