Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats (original) (raw)
Abstract
The specific silencing of the gene of interest is the major objective of RNA interference technology; therefore, unique sequences but not abundant sequence repeats are targeted by silencing reagents. Here, we describe the targeting of expanded CAG repeats that occur in transcripts derived from the mutant allele of the gene implicated in Huntington’s disease (HD) in the presence of the normal allele and other human mRNAs containing CAG and CUG repeat tracts. We show that a high degree of silencing selectivity may be achieved between the repeated sequences. We demonstrate preferential suppression of the mutant huntingtin allele and concomitant activation of the normal huntingtin allele in cell lines derived from HD patients that were transfected with short RNA duplexes composed of CAG and CUG repeats containing mutations at specific positions. These effects may lead to promising therapeutic modalities for HD, a condition for which no therapy presently exists.
INTRODUCTION
Almost 20 human genetic diseases are caused by specific trinucleotide repeat expansions in different single genes (1). The best known of these diseases are fragile X syndrome (FXS), myotonic dystrophy type 1 (DM1), Huntington’s disease (HD) and a number of spinocerebellar ataxias (SCA). The mechanisms of pathogenesis differ between these diseases and include impaired transcription (FXS), transcript toxicity (DM1) and protein toxicity (HD and SCAs) (2,3). In several SCAs and HD, the expanded CAG repeats present in the open reading frame (ORFs) of the implicated genes (ATXN and HTT, respectively) are translated into polyglutamine tracts in the encoded, functionally unrelated proteins. This subgroup is commonly referred as the polyglutamine (polyQ) diseases. Pathogenesis in polyQ diseases is triggered by a single mutant allele of the particular gene, which acts in a dominant ‘gain-of-function’ fashion. Therefore, the allele-specific inhibition of mutant gene expression is considered a promising strategy for establishing causative therapies for these diseases. Mutant transcript silencing by RNA interference (RNAi) has been attempted in several studies. Single nucleotide polymorphisms (SNPs) linked to repeat expansions were targeted (4–6). The efficiency of this approach has increased with the advances in single nucleotide difference discrimination by RNAi reagents (7–9). SNP targeting, however, is limited by the low frequencies of suitable SNPs in the human population, and prospective treatments need to be tailored to individual patients. The recent identification of specific SNPs associated with CAG repeat expansion in HTT may increase the potential of this strategy (10). Non-allele-specific inhibition of HTT gene expression by RNAi has also been shown to offer some advantages (11–14).
The specific inhibition of mutant allele expression by targeting expanded CAG repeats could be developed into a more universal therapeutic approach that would potentially be applicable to all polyQ diseases. But how could it be possible to selectively silence a mutant allele containing ∼40–100 CAG repeats and discriminate it from both the normal allele and the numerous other human transcripts typically containing <20 CAG repeats? The RNAi approach was initially abandoned after discouraging results were obtained using repeat-targeting siRNAs. Both alleles of HTT and ATXN3 genes were shown to be silenced in cell culture in response to 21-nt siRNA duplexes composed of CUG/CAG repeats (5,15). Recently, a high degree of selectivity in mutant HTT allele inhibition has been achieved using repeat-targeting PNA and LNA antisense reagents (15). The allele-discriminating abilities of these reagents were considerably stronger than those of repeat-targeting siRNA.
In this study, we explored the potential of repeat-targeting RNA duplexes to discriminate between the mutant and the normal HTT transcript to achieve the desired allele selectivity. We also analyzed the gene selectivity of CAG/CUG duplexes, which is understood as discrimination between HTT and other mRNAs containing CAG and CUG repeat tracts. We observed some modest discrimination between mutant and normal HTT alleles by repeat-targeting siRNA. Then, we introduced mutations at specific positions of the repeat-targeting duplex what improved its gene and allele selectivity. Gene selectivity was further improved through CAG strand inactivation, which was achieved by reducing its length. We also provide first clues for the mechanism by which preferential mutant huntingtin inhibition and concomitant normal huntingtin activation may occur.
MATERIALS AND METHODS
Cell culture and transfection
Fibroblasts from HD patients (GM04281—17/68 CAG, GM09197—21/151 CAG, GM04208—21/44 CAG, GM01187—18/47 CAG) and SCA3 patients (GM06153—18/69 CAG), obtained from the Coriell Cell Repositories, were grown in minimal essential medium (Lonza) supplemented with 8% fetal bovine serum (Sigma), antibiotics (Sigma) and non-essential amino acids (Sigma). Cell transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Transfection efficiency was monitored using a BlockIT fluorescent siRNA (Invitrogen).
All RNA oligonucleotides were synthesized by Metabion. The sequences of the synthetic RNAs used in this study are presented in Figures.
RNA isolation and RT–PCR
Total RNA was isolated from fibroblast cells using TRI Reagent (BioShop) according to the manufacturer’s instructions. The RNA concentration was measured using a NanoDrop spectrophotometer. A total of 500 ng RNA was reverse transcribed using Superscript II (Invitrogen) and random hexamer primers (Promega). The quality of the reverse transcription (RT) reaction was assessed using amplification of the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) housekeeping gene. Primer sequences are listed in Supplementary Table S1. The reaction products were separated on 1.5% agarose gels in TBE buffer and stained with ethidium bromide.
Protein isolation and western blot
Fibroblast cells were lysed in buffer containing 60 mM Tris-base, 2% SDS, 10% sucrose and 2 mM PMSF. The protein concentration was estimated using a NanoDrop spectrophotometer. A total of 20 μg of protein was diluted in sample buffer containing 2-mercaptoethanol and boiled for 5 min. For ataxin-3 (∼40 kDa) and huntingtin (∼350 kDa) level analyses, different proteins were selected as controls of total protein levels: GAPDH (∼35 kDa) and plectin (∼500 kDa), respectively. Plectin is not a standard protein loading control, but in our experiments, we demonstrated that the level of plectin remained unchanged after cell transfection with RNAs. This was done by comparing the GAPDH and plectin levels from the same protein lysate. Therefore, this protein was used as a reliable gel loading control for huntingtin detection. For ataxin-3, RPL14, LRP8, EIF2AK3 and GAPDH detection the proteins were separated using Tris–HCl SDS–polyacrylamide gel electrophoresis (5% stacking/12% resolving gel) in Laemmli buffer. For huntingtin and plectin electrophoresis a Tris–acetate SDS–polyacrylamide gel was used (5% stacking/4% resolving gel) and run in XT Tricine buffer (Bio-Rad) with cooling to 15°C. After electrophoresis, the proteins were wet-transferred to a nitrocellulose membrane. The membranes were cut according to protein ladders for separate detection of respective proteins. All steps of immunodetection were performed on a SNAPid (Millipore) in buffer containing 0.25% non-fat milk in PBS/0.9% NaCl/0.1% Tween-20. For ataxin-3/GAPDH or RPL14, LRP8, EIF2AK3/GAPDH detection, the blots were probed with the primary antibodies anti-ataxin-3 (1:1000), anti-RPL14 (1:500), anti-LRP8 (1:500), anti-EIF2AK3 (1:500) and anti-GAPDH (1:5000) and then with a biotinylated secondary antibody (1:500); they were then incubated with a streptavidin–AP conjugate (1:2000, Millipore). The immunoreaction was detected using the BCIP/NBT substrate (Sigma). For huntingtin/plectin detection the blots were probed with the primary antibodies: anti-huntingtin (1:1000) and anti-plectin (1:1000) and then with HRP-conjugated secondary antibodies (1:500). The immunoreaction was detected using ECL Western Blotting Substrate (ThermoScientific). Examples of huntingtin alleles separation and examples of full blots for ataxin-3 and huntingtin level analysis are shown in Supplementary Figure S1. All antibodies are listed in Supplementary Table S2.
Cell viability assay
The effect of RNA reagents on cell mortality was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega) based on quantitation of adenosine-5′-triphosphate (ATP).
5′-RACE
Total RNA from transfected SCA3 or HD cells was ligated with 45 nt adaptor RNA using T4 RNA Ligase (Ambion) and then reverse transcribed using random hexamers (Promega) and SuperScript II (Invitrogen). First step amplifications were performed with Ad_ex and ATXN3_ex primer pair. Then, after 20× dilution of the product, nested PCR was performed with Ad_int and ATXN3_int primer pair. Primer sequences are listed in Supplementary Table S1. Prior to nested PCR the Ad_int primer was 5′-end-labeled with [γ-32P]ATP using T4 polinucleotide kinase (Epicentre). Final PCR products were diluted 20× in formamide buffer with dyes and analyzed on a 6 % polyacrylamide–7.5 M urea gel. The electrophoresis was run in a 1× TBE buffer. The products were detected by autoradiography, eluted from a gel, re-amplified and sequenced.
Statistical analysis
All experiments were repeated at least three times. The statistical significance of silencing or up-regulation was assessed with a one-sample _t-_test with the hypothetical value 1 assigned to untreated cells (0) or cells treated with control siRNA (C). Selected data were compared with each other with the use of an unpaired _t_-test. _P_-values (two-tailed) that were < 0.05 were considered significant.
RESULTS
Both strands of the d7-duplex silence transcripts having complementary sequences
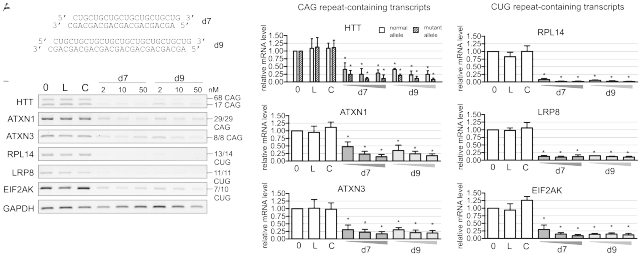
Duplex d7 is composed of a pure CUG and CAG repeat sequences and its 21 nt strands are predicted to form 1 nt 3′ overhangs (Figure 1A). When d7 is transfected into cells either the CUG or the CAG strand may activate RISC to become functional in RNAi. To demonstrate this, we analyzed the silencing activities of the CUG and CAG guide strands against selected transcripts containing at least seven CAG (HTT, ATXN1 and ATXN3) or CUG repeats (RPL14, LRP8 and EIF2AK3) in HD fibroblast cell line GM04281 (CAG 17/68 in HTT mRNA). Similar results were obtained for the 21-nt (d7) and 27-nt (d9) siRNAs: both strands of tested reagents were found to be active in silencing transcripts, even at the lowest siRNA concentrations used (Figure 1B and C). On the other hand, the activity of the shorter duplex formed by the 17-nt long CAG and CUG repeat strands was low (data not shown). We also analyzed the efficiency of HTT transcript silencing by d7 reagent in broader range of concentration (Supplementary Figure S2). We observed some preference for mutant transcript silencing at all concentrations tested, and the transcript level decreased considerably at concentrations as low as 0.25 nM.
Figure 1.
CUG repeat and CAG repeat strands are active in transcript silencing. (A) Sequences of d7 and d9 duplexes (B) RT-PCR analysis of cellular levels of CAG-containing (HTT, ATXN1 and ATXN3) and CUG-containing transcripts (RPL14, LRP8 and EIF2AK3) 24 h after transfection of HD fibroblasts (GM04281) with d7 (CAG/CUG-21 nt) or d9 (CAG/CUG-27 nt) at 2, 10 and 50 nM. (C) Bar graph shows cellular levels of analyzed transcripts for the experiment described in (B). Reference bars indicate transcript expression levels in non-transfected cells (0), cells treated with Lipofectamine 2000 only (L) and cells transfected with control siRNA (C). In all graphs, signal intensities were normalized to GAPDH mRNA and compared using a one-sample _t_-test. Error bars represent standard deviations. The _P_-value is indicated by an asterisk (* P<0.001).
Specific mutations in d7 increase its preference for mutant HTT allele silencing
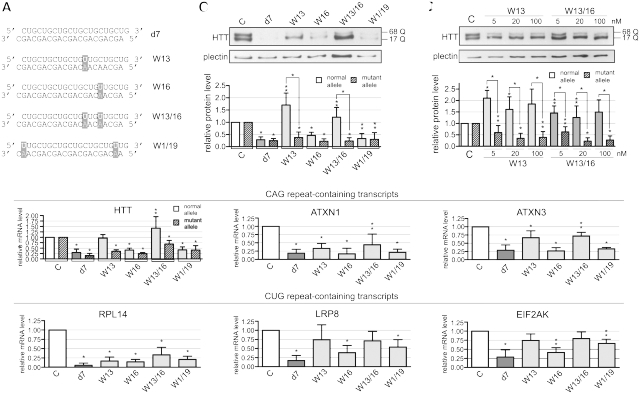
We attempted to increase both the gene selectivity and the allele selectivity of repeat-targeting duplex by selecting the mutations that decreased the strength of programmed RISC interactions with respective targets. To achieve this goal, we introduced either single or double C>U substitutions at positions 13 (W13), 16 (W16) and both 13 and 16 (W13/16) as well as at positions 1 and 19 (W1/19) of the d7 CUG strand (Figure 2A). Both siRNA strands were mutated to maintain their complementarities. This required G>A substitutions at positions 2, 5, 8 and 20 in the CAG strand. The mutant duplexes were transfected at 20 nM to HD fibroblasts (GM04281). We tested the activities of the duplexes on both HTT allelic variants and the same set of previously mentioned CAG repeat- and CUG repeat-containing transcripts. All these duplexes showed decreased activities as compared to d7, and the strongest decreases were shown for W13 and W13/16 by reverse transcriptase polymerase chain reaction (RT–PCR) analysis (Figure 2B). Protein analysis showed that W16 and W1/19 were similar to d7 with respect to their silencing efficiencies and allele selectivities (Figure 2C). In contrast, W13 and W13/16 showed strong preferences for mutant huntingtin inhibition. These reagents resulted in decreased mutant protein levels to ∼25% of the untreated control level (Figure 2C). Similar results were obtained for the testing of the set of duplexes in the other HD cell line GM09197 (CAG 21/151) (Supplementary Figure S3). Protein inhibition was also tested after the transfection of HD cells with different concentrations of W13 and W13/16. For both duplexes, 5 nM effectively inhibited mutant protein levels, and concentrations as high as 100 nM did not reduce normal protein levels (Figure 2D). Moreover, we observed reproducible up-regulation of normal huntingtin by W13 and W13/16 transfection (Figure 2C and D). A similar trend of W13/16 activity was observed for other tested HD cell lines, GM4208 (CAG 21/44) and GM01187 (CAG 18/47), representing the lengths of CAG repeat tracts which are more typical for HD patients (Supplementary Figure S4).
Figure 2.
Silencing activity of mutated CAG/CUG duplexes. (A) Sequences of modified duplexes with mutated positions indicated. (B) Results from the RT-PCR analysis of CAG-containing (HTT, ATXN1 and ATXN3) and CUG-containing transcript (RPL14, LRP8 and EIF2AK3) levels 24 h after transfection of HD fibroblasts (GM04281, 17/68 CAG in HTT) with 20 nM of modified duplexes or d7. (C) Western blot analysis of huntingtin levels for the experiment described in (B) After 72 h transfection. (D) Western blot analysis of huntingtin levels 72 h after transfection of HD fibroblasts with 5, 20 and 100 nM W13 or W13/16. Reference bar ‘C’ indicates HTT expression levels in cells transfected with control siRNA. In all graphs, signal intensities were normalized to GAPDH mRNA or plectin protein levels and compared using a one-sample _t_-test. Error bars represent standard deviations. The _P_-value is indicated by an asterisk (*P < 0.001; **0.001 < P < 0.05). More examples of raw data are presented in Supplementary Figure S10.
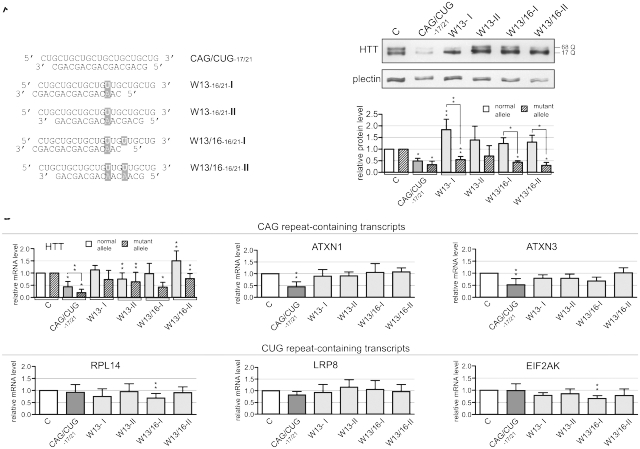
Elimination of CAG strand activity improves the gene selectivity of mutant duplexes
The mutations present in W13 and W13/16 strongly increased the allele selectivity and improved the gene selectivity of HTT silencing, but they did not sufficiently reduce the silencing of transcripts other than HTT. To retain the advantageous features of W13 and W13/16 and further improve their gene selectivity, we inactivated the CAG strand by shortening it to 17 or 16 nt. We tested the asymmetric siRNAs (Figure 3A) for their HTT protein inhibitory activity, and for their ability to silence the CUG repeat-containing transcripts. These duplexes inhibited the HTT protein with allele selectivities similar to that of their symmetric counterparts when used at 100 nM (Figure 3B). At 20 nM, their activities were considerably lower than those of the symmetric duplexes (data not shown). The asymmetric duplexes were considerably less effective in silencing the CUG repeat-containing transcripts, so an improvement in their gene selectivity (reduction of their off-target effect) was achieved (Figure 3C).
Figure 3.
Silencing activity of improved mutated CAG/CUG duplexes. (A) Sequences and preferential structures formed by d7-, W13- or W13/16-based duplexes with a shortened CAG strand. Mutated positions are indicated. (B) Western blot analysis of huntingtin level 72 h after transfection of HD fibroblasts (GM04281) with 100 nM of the reagents listed in (A). (C) Results from the RT–PCR analysis of CAG-containing (HD, ATXN1 and ATXN3) and CUG-containing transcript (RPL14, LRP8 and EIF2AK3) for the experiment described in (B) 24 h post-transfection. Reference bar ‘C’ indicates HTT expression levels in cells transfected with control siRNA. In all graphs, signal intensities were normalized to GAPDH mRNA or plectin protein levels and compared using a one-sample _t_-test. Error bars represent standard deviations. The _P_-value is indicated by an asterisk (*P < 0.001; **0.001 < P < 0.05). More examples of raw data are presented in Supplementary Figure S10.
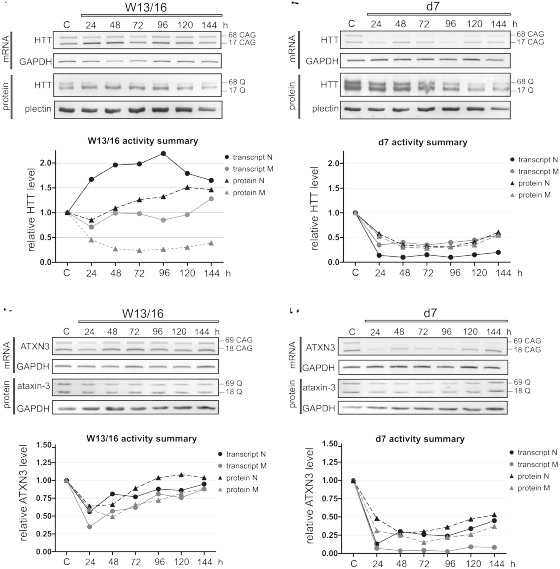
Different RNA duplexes silence the HTT gene through different mechanisms
The preferential inhibition of mutant HTT protein synthesis by W13/16, which was observed 72 h post-transfection (Figure 2), required confirmation using a more detailed analysis of the silencing process. The changes of both HTT protein and transcript levels were analyzed for 6 days after W13/16 transfection in 24-h intervals (Figure 4A) and compared to the effects brought about by d7 (Figure 4B). The mutant protein was effectively silenced starting from Day 2 after W13/16 transfection, but cellular levels of the mutant transcript remained unchanged (Figure 4A and Supplementary Figure S5A). At the same time, an ∼2-fold increase in normal transcript levels was observed, and resulted in smaller increase in normal HTT protein levels. In contrast, both mutant and normal transcript and protein levels decreased after d7 treatment (Figure 4B and Supplementary Figure S5B), confirming our observations made at the selected single time points. The same pair of reagents was then tested for any preferential silencing of mutant or normal ATXN3 transcripts or proteins in the cellular model of SCA3. The d7 transfection displayed similar silencing activities in SCA3 cells as compared to the HD cells (Figure 4D and Supplementary Figure S5D), but no allele discrimination comparable to that achieved in HD cells was induced by W13/16 (Figure 4C and Supplementary Figure S5C). Nevertheless, we demonstrated up-regulation of normal HTT transcript also in SCA3 cells (Supplementary Figure S6). W13/16 showed reduced silencing of proteins encoded by CUG repeat-containing transcripts: RPL14, LRP8 and EIF2AK3, as compared to d7 (Supplementary Figure S7). We then performed 5′-RACE analysis to assess whether transcript cleavage occurred in d7 and W13/16 treated cells due to RNAi mechanism. We used three repeat-targeting duplexes d7, d7-2 and d7-3, each having guide sequences that were shifted by 1 nt. Sequences of the corresponding 5′-RACE products and established cleavage sites showed that the AGO2-mediated cleavage mechanism contributed to the activity of all tested reagents (Supplementary Figure S8). This contribution, however, is very likely to be substantial in case of d7 activity and marginal in case of W13/16 activity. For W13/16-activity duplex structure was required, as the transfection of HD fibroblasts with W13/16 antisense or sense strand alone, caused neither downregulation nor up-regulation of mutant and normal huntingtin (Supplementary Figure S9).
Figure 4.
Time course analysis of d7 and W13/16 activity. RT–PCR and western blot analyses of HTT (A and B) and ATXN3 (C and D) transcript and protein levels after transfection of HD (GM04281) or SCA3 fibroblasts (GM06153) with 20 nM W13/16 (A, C) or d7 (B and D) at indicated time points. Reference point ‘C’ indicates HTT expression levels in cells transfected with control siRNA. In the graphs mean values from signal intensities are presented, for statistical analysis see Supplementary Figure S5.
Duplexes with better gene selectivity are less toxic to cells
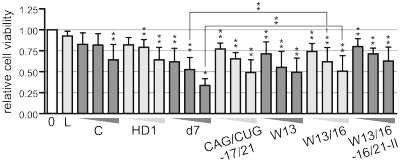
An important issue regarding the application of repeat-targeting reagents is their potential toxicity to cells, which may primarily be caused by their compromised gene selectivity. We analyzed the toxicity of the different types of reagents tested in this study, using a luminescent cell viability assay. As shown in Figure 5, the greatest decrease in viable cells was detected in response to d7. Transfection with this reagent at 20 nM resulted in ∼50% reduction in the cell number. Cell viability was positively correlated with increased gene selectivity of tested reagents, as W13/16 and its asymmetric version (W13/16-16/21-II) induced significantly lower mortality of cells.
Figure 5.
Toxicity of specific and repeat-targeting reagents. Results from cell viability assay 72 h after transfection of HD fibroblasts (GM04281) with control siRNA (C), specific siRNA (HD1) and repeat-targeting duplexes at 5, 20 and 100 nM. (0) reference bar, non-transfected cells, (L) cells treated with Lipofectamine 2000 only. Normalized signal intensities were compared using a one-sample _t_-test. Error bars represent standard deviations. The _P_-value is indicated by an asterisk (*P < 0.001; **0.001 < P < 0.05).
DISCUSSION
CAG and CUG repeats are often polymorphic in length and are among the most frequently occurring triplet repeats in the human transcriptome (16). Nearly 200 mRNAs contain CAG repeat tracts, and >100 mRNAs contain tracts of CUG repeats composed of at least six repeated units (17). This makes the repeat-targeting therapeutic strategy quite challenging. We showed that RISC, programmed by either strand of the repeat-targeting CAG/CUG siRNA (d7), was highly active in the silencing of several normal transcripts containing complementary sequences, what resulted in significant loss of cell viability.
In recent years, several rules have been proposed for the design of efficient allele-specific siRNAs, targeting single nucleotide polymorphism sites in RNAs. These rules showing the importance of specific nucleotides and nucleotide positions for allele discrimination (7,8) have provided some clues on how to increase the selectivity of the CAG repeat-targeting duplex. Our strategy was to introduce specific mutations at selected d7 positions to modulate RISC activity. The mutations at positions 13 and 16 of the antisense strand were selected as these positions were among the ones providing the strongest allele discrimination (8,9). By applying this strategy in the design of the W13 and W13/16 duplexes, we achieved improved gene selectivity and allele selectivity of mutant HTT allele silencing. The gene selectivity of the W13 and W13/16 duplexes was further increased by taking advantage of recent developments in asymmetric siRNA design (18,19). However, to achieve efficient silencing of mutant huntingtin, the asymmetric duplexes had to be used at a higher concentration than their symmetric counterparts. Increased gene selectivity was positively correlated with cell viability after transfection with reagents composed of CAG/CUG repeats. It may be speculated that the similar toxicity order of the reagents composed of repeats will be observed in neurons which are predominant sites of HD pathology.
Notably, d7, which possessed full complementarity to its HTT target, and the W13 and W13/16 duplexes, having either one or two G:U mismatches, silenced the expression of the mutant HTT allele very likely through different mechanisms, as shown by transcript and protein analyses from the same silencing reactions. The d7 siRNA acts according to the typical RNAi mechanism, in which mutant transcript level decrease is followed by a decrease in mutant huntingtin. On the contrary, the silencing of the mutant HTT allele by W13/16 seems to occur predominantly, through translational repression, what is suggested by the data presented in Figure 4. We cannot exclude, however, some contribution from mRNA cleavage mechanism (Supplementary Figure S5), and deadenylation and degradation mechanism (not addressed in this study), which was recently shown to be the major mechanism by which miRNAs regulate their targets (20).
As previously demonstrated in miRNA and siRNA studies, the increased number of target sites located in the 3′-UTR of exogenous transcripts facilitates gene silencing (21,22). Here, we show that this phenomenon occurs in overlapping target sites formed by CAG repeats located in the ORF of endogenous HTT transcript. The observed preferential silencing of the expanded HTT allele also indicates that the possession of a higher number of target sites (rCAG units) is more significant than the effect of a more stable secondary structure, which is likely to be formed by the expanded CAG repeats in cells as it is in vitro (23,24).
The observation that W13/16 duplex does not discriminate between the normal and mutant mRNAs of HTT and ATXN3 genes with similar allele selectivity is intriguing and the reason for this difference remains unknown. The HTT and ATXN3 mRNAs differ in their length, the localization of CAG repeat within ORF and in the structural details of their CAG repeat regions. These structural differences (23,25) are, however, unlikely to account for differences in the silencing selectivity of HTT and ATXN3 mRNAs as none of these structures inhibits mutant transcript targeting by RISC.
Little is known about the antisense transcriptomes of HTT and ATXN3 genes (26) which may be implicated in generating differences in allele selectivity of their silencing. This issue needs further exploration as silencing duplexes composed of repeats may target both sense and antisense transcripts spanning the repeat region and thus influence their normal regulatory functions.
Finally, it was shown earlier that miRNA, fully complementary to target sequence, can induce mRNA cleavage, and that siRNA forming mismatches with its target in the ORF sequence can induce translational inhibition (27,28). We demonstrated here that such mechanistic transition also occurs for repeat-targeting reagents, which substantially improves their gene and allele discriminating properties.
The most intriguing discovery of this study was the observation that the transcriptional up-regulation of normal huntingtin gene expression occurred together with translational mutant huntingtin inhibition. The transcriptional activation of gene expression using duplex RNAs have recently been demonstrated, and it has been shown that RNAi comes in many types and that its activity is not restricted to silencing effects (29–32). The upregulation of normal HTT level could also result from some unknown compensatory mechanism triggered by mutant HTT silencing. The transcriptional activation and concomitant translational repression of the two alleles of the same gene using repeat-targeting dsRNA is novel from a mechanistic viewpoint and should be considered a beneficial feature from the perspective of HD therapy using duplex RNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Higher Education (grant numbers: N N301 284837, N N301 569340); European Regional Development Fund within Innovative Economy Programme (POIG.01.03.01-30-098/08); A.F. was a scholarship holder within the project ‘Scholarship support for PhD students specializing in majors strategic for Wielkopolska’s development’, Sub-measure 8.2.2 Human Capital Operational Programme, co-financed by European Union under the European Social Funding for open access charge: European Regional Development Fund within Innovative Economy Programme (POIG.01.03.01-30-098/08).
Conflict of interest statement. None declared.
Supplementary Material
Supplementary Data
REFERENCES
- 1.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 2.Bauer PO, Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J. Neurochem. 2009;110:1737–1765. doi: 10.1111/j.1471-4159.2009.06302.x. [DOI] [PubMed] [Google Scholar]
- 3.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann. Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alves S, Nascimento-Ferreira I, Auregan G, Hassig R, Dufour N, Brouillet E, Pedroso de Lima MC, Hantraye P, Pereira de Almeida L, Deglon N. Allele-specific RNA silencing of mutant ataxin-3 mediates neuroprotection in a rat model of Machado-Joseph disease. PLoS ONE. 2008;3:e3341. doi: 10.1371/journal.pone.0003341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc. Natl Acad. Sci. USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pfister EL, Kennington L, Straubhaar J, Wagh S, Liu W, DiFiglia M, Landwehrmeyer B, Vonsattel JP, Zamore PD, Aronin N. Five siRNAs targeting three SNPs may provide therapy for three-quarters of Huntington's disease patients. Curr. Biol. 2009;19:774–778. doi: 10.1016/j.cub.2009.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang H, Qiao R, Zhao D, Zhang T, Li Y, Yi F, Lai F, Hong J, Ding X, Yang Z, et al. Profiling of mismatch discrimination in RNAi enabled rational design of allele-specific siRNAs. Nucleic Acids Res. 2009;37:7560–7569. doi: 10.1093/nar/gkp835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarz DS, Ding H, Kennington L, Moore JT, Schelter J, Burchard J, Linsley PS, Aronin N, Xu Z, Zamore PD. Designing siRNA that distinguish between genes that differ by a single nucleotide. PLoS Genet. 2006;2:e140. doi: 10.1371/journal.pgen.0020140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warby SC, Montpetit A, Hayden AR, Carroll JB, Butland SL, Visscher H, Collins JA, Semaka A, Hudson TJ, Hayden MR. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am. J. Hum. Genet. 2009;84:351–366. doi: 10.1016/j.ajhg.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M, Yoder J, Reeves P, Pandey RK, Rajeev KG, et al. Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc. Natl Acad. Sci. USA. 2007;104:17204–17209. doi: 10.1073/pnas.0708285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Machida Y, Okada T, Kurosawa M, Oyama F, Ozawa K, Nukina N. rAAV-mediated shRNA ameliorated neuropathology in Huntington disease model mouse. Biochem. Biophys. Res. Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- 13.Wang YL, Liu W, Wada E, Murata M, Wada K, Kanazawa I. Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neurosci. Res. 2005;53:241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 14.Boudreau RL, McBride JL, Martins I, Shen S, Xing Y, Carter BJ, Davidson BL. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in Huntington's disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu J, Matsui M, Gagnon KT, Schwartz JC, Gabillet S, Arar K, Wu J, Bezprozvanny I, Corey DR. Allele-specific silencing of mutant huntingtin and ataxin-3 genes by targeting expanded CAG repeats in mRNAs. Nat. Biotechnol. 2009;27:478–484. doi: 10.1038/nbt.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozanska M, Sobczak K, Jasinska A, Napierala M, Kaczynska D, Czerny A, Koziel M, Kozlowski P, Olejniczak M, Krzyzosiak WJ. CAG and CTG repeat polymorphism in exons of human genes shows distinct features at the expandable loci. Hum. Mutat. 2007;28:451–458. doi: 10.1002/humu.20466. [DOI] [PubMed] [Google Scholar]
- 17.Kozlowski P, de Mezer M, Krzyzosiak WJ. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang CI, Yoo JW, Hong SW, Lee SE, Kang HS, Sun X, Rogoff HA, Ban C, Kim S, Li CJ, et al. Asymmetric shorter-duplex siRNA structures trigger efficient gene silencing with reduced nonspecific effects. Mol. Ther. 2009;17:725–732. doi: 10.1038/mt.2008.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X, Rogoff HA, Li CJ. Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat. Biotechnol. 2008;26:1379–1382. doi: 10.1038/nbt.1512. [DOI] [PubMed] [Google Scholar]
- 20.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saetrom P, Heale BS, Snove O, Jr, Aagaard L, Alluin J, Rossi JJ. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Mezer M, Wojciechowska M, Napierala M, Sobczak K, Krzyzosiak WJ. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1323. doi: 10.1093/nar/gkq1323 [Epub ahead of print, 18 January 2011] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krol J, Fiszer A, Mykowska A, Sobczak K, de Mezer M, Krzyzosiak WJ. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol. Cell. 2007;25:575–586. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Michlewski G, Krzyzosiak WJ. Molecular architecture of CAG repeats in human disease related transcripts. J. Mol. Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Batra R, Charizanis K, Swanson MS. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum. Mol. Genet. 2010;19:R77–82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 28.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Place RF, Jia ZJ, Pookot D, Dahiya R, Li LC. Antitumor effect of dsRNA-induced p21(WAF1/CIP1) gene activation in human bladder cancer cells. Mol. Cancer Ther. 2008;7:698–703. doi: 10.1158/1535-7163.MCT-07-2312. [DOI] [PubMed] [Google Scholar]
- 30.Janowski BA, Younger ST, Hardy DB, Ram R, Huffman KE, Corey DR. Activating gene expression in mammalian cells with promoter-targeted duplex RNAs. Nat. Chem. Biol. 2007;3:166–173. doi: 10.1038/nchembio860. [DOI] [PubMed] [Google Scholar]
- 31.Li LC, Okino ST, Zhao H, Pookot D, Place RF, Urakami S, Enokida H, Dahiya R. Small dsRNAs induce transcriptional activation in human cells. Proc. Natl Acad. Sci. USA. 2006;103:17337–17342. doi: 10.1073/pnas.0607015103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morris KV, Santoso S, Turner AM, Pastori C, Hawkins PG. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data