Sequence-specific error profile of Illumina sequencers (original) (raw)
Abstract
We identified the sequence-specific starting positions of consecutive miscalls in the mapping of reads obtained from the Illumina Genome Analyser (GA). Detailed analysis of the miscall pattern indicated that the underlying mechanism involves sequence-specific interference of the base elongation process during sequencing. The two major sequence patterns that trigger this sequence-specific error (SSE) are: (i) inverted repeats and (ii) GGC sequences. We speculate that these sequences favor dephasing by inhibiting single-base elongation, by: (i) folding single-stranded DNA and (ii) altering enzyme preference. This phenomenon is a major cause of sequence coverage variability and of the unfavorable bias observed for population-targeted methods such as RNA-seq and ChIP-seq. Moreover, SSE is a potential cause of false single-nucleotide polymorphism (SNP) calls and also significantly hinders de novo assembly. This article highlights the importance of recognizing SSE and its underlying mechanisms in the hope of enhancing the potential usefulness of the Illumina sequencers.
INTRODUCTION
The emergence of next-generation sequencing (NGS) technologies is yielding a revolutionary impact on biological research (1–3). Of the three current major platforms [Illumina/Solexa Genome Analyser (4), Life Technologies/ABI SOLiD System (5) and Roche/454 Genome Sequencer FLX (6)], the Illumina Genome Analyser (GA) is, at the moment, the most popular choice for the analysis of genomic information (7). The Illumina/Solexa sequencers are characterized by: (i) solid-phase amplification and (ii) a cyclic reversible termination (CRT) process, also termed sequencing-by-synthesis (SBS) technology (8). The sequencer can generate hundreds of millions of relatively short (30–100 bp) read sequences per run.
The application of data obtained from this NGS technology can be roughly categorized into the following three groups. First, genomic data can be used for resequencing. When an almost identical genome sequence is available as the master reference, relatively small mutations of the sample sequence such as single-nucleotide polymorphisms (SNPs) and short indels can be identified by comparison. This analysis is often carried out in order to differentiate species of the same family or members of the same species. Second, the data can be applied to seq-based techniques such as those designed to determine the transcriptome (RNA-seq) or protein–DNA interaction regions (ChIP-seq). The data also can be used for quantitative analysis based on the number of sequence segments (9,10). Third, the data are useful for the reconstruction of the original genome sequence after sample read assembly. This is done for species for which a reference genome has not been sequenced or for the metagenomic analysis of mixed bacterial cultures. NGS data are also used to reconstruct transcript sequences by assembling reads obtained by RNA-seq when the reference sequence is unavailable. Several software packages are available for assembly, including Velvet, SOAPdenovo and ABySS (2,11,12).
A computational procedure called mapping is often initially employed in the resequencing and seq-based methods to determine the position for each read within the reference. Mapping often accompanies gapped alignment relative to the regular sequence alignments performed by BLAST or ClustalW (13,14) or more rigorously by dynamic programming algorithms (15,16). Various software programs, including BWA, MAQ, BLAT, SHRiMP, SOAP2 and BFAST, have been designed for the mapping of short read sequences generated by NGS technologies (17–22).
In this article, we describe a newly identified error profile for Illumina sequencers. Several reports have described the Illumina sequencer data profiles in detail. An often-described property of these profiles is coverage variation, which partly results from the inherent bias of polymerase chain reaction (PCR) amplification during sample preparation (23). Stein et al. (24) suggested that this bias is mainly caused by the formation of secondary structures in single-stranded DNA (ssDNA). Harrismendy et al. (25) reported lower coverage of the short read platforms (Illumina and Life Technologies/ABI SOLiD) at AT-rich repetitive sequences.
Hoffman et al. (26) reported that the Illumina sequencers result in more substitution-type miscalls than indel-type miscalls, while the Roche/454 sequencers result in more indel-type miscalls than substitution-type miscalls. Kircher et al. (27) reported that miscalls are more frequent during the first and last cycles and proposed that Illumina-specific miscalls result from cycle-dependent variations of the cross-talk matrix, declining intensities, pre-phasing and phasing and T accumulation. According to Dohm et al. (28), miscalls are more frequently distributed in the GC-rich regions. The authors also claimed that the base-specific miscalls A to C and C to G are observed more often than the others, suggesting that this type of miscall is due to the inhibition of base elongation during SBS. Various researchers agree that the quality of the Illumina sequencer reads are significantly lower in the later cycles. Lagging-strand dephasing, caused by the incomplete extension of the template ensemble, has been suggested as one of the main reasons for this problem (7).
We determined that the error profile we observed was caused by the sequence-specific occurrence of lagging-strand dephasing. To our knowledge, there has been no report that describes lagging-strand dephasing triggered by specific sequence patterns. This error profile is observed in all the Illumina sequencing data that we examined, including the public data from the Short Read Archive (SRA), regardless of the source organisms or the sample preparation methods used. We believe this knowledge is important in order to fully exploit the potential of the Illumina sequencing technology, re-evaluate past experimental conditions and computational procedures, and aid the development of future sequencing tools. This article outlines the newly found sequence-specific error (SSE) profile, discusses the underlying mechanism of the error and proposes potential countermeasures.
MATERIALS AND METHODS
Sequencing
Sequencing of the Bacillus subtilis genome was performed using an Illumina GA II. The genomic DNA of B. subtilis was extracted with a DNeasy Blood and Tissue kit (Qiagen). Libraries of this genomic DNA were prepared according to the manufacturer’s protocol (Illumina) (8). Five micrograms of genomic DNA were fragmented to an average length of 200 bp using a Covaris S2 system (Covaris). The fragmented DNA was repaired using T4 polynucleotide kinase and Klenow fragment (New England Biolabs); the 3′-end of the end-repaired DNA was adenylated using Klenow fragment (New England Biolabs). Next, Index PE Adapters Oligo Mix (Illumina) was ligated to the fragments using Quick T4 DNA Ligase (New England Biolabs). The 5′-end adaptor extension and enrichment of the library were performed using 18 cycles of PCR with the primers InPE1.0, InPE2.0 and PCR index primer (Illumina). Cluster generations were performed on an Illumina cluster station using a Paired-End Cluster Generation Kit v4. Seventy-six cycles of multiplexed paired-end sequencing were carried out using an Illumina GA II system with an SBS 36-cycle Sequencing Kit v4, according to the manufacturer’s specifications. After the sequencing reactions were complete, the Illumina analysis pipeline (CASAVA 1.6.0) was used to process the raw sequencing data. The reference sequence of the mapping was B. subtilis str. 168 (NC_000964.3). The read data (DRX000504) were deposited in DRA (DDBJ Sequence Read Archive).
Data analysis
We created a new software program for mapping Illumina sequencer reads (MPSmap) and visualizing the mapping results (PSmap). Detailed description and evaluation of the software will appear elsewhere; here, we describe our method briefly. Initially, a simple index of _k_-mers was prepared for the reference sequence. Then all bases of the read were compared with that of the reference for each index match of the read. This comparison was performed for all the index matches, and the best-matched position for each read was identified. A limitation of the index approach is that some of the close-match positions may not be identified if any mismatches are present within the index. To minimize this problem, we repeated the index search while shifting the index position on read sequences. For instance, we repeated the index search three times to correctly locate the read positions while allowing two mismatches. Similarly, we repeated the index search (n + 1) times, where n is the number of mismatches per read allowed in the search. Each index hit is aligned on the reference in order to look for the best location, allowing up to the specified number of mismatches without a gap. The index approach is fast but does not guarantee sensitivity for reads shorter than k (n + 1), where, k is the index length. For the mapping of B. subtilis allowing 35 mismatches, we compared searches with index lengths of k = 2 and k = 10 in order to confirm that the difference in results is small (Supplementary Table S1). We also performed mapping with BWA and BFAST using Tablet (29) for visualization, in order to confirm that multiple mapping algorithms detect the SSE (Supplementary Data S1). The visualization program (PSmap) converts the mapping results to a PostScript file. The programs, executable on Linux (CentOS5.3) and MacOSX (ver. 10.6.6) systems, are available for download on our website (http://metalmine.naist.jp/maps/).
Public data
We analyzed several public data sets downloaded from the SRA database server at the National Center for Biotechnology Information (NCBI). The accession numbers of these samples and the corresponding reference sequences are ERX006616 (NC_02945.3; Mycobacterium bovis AF2122/97), SRX007714 (NC_010079, NC_012417.1 and NC_010063.1; Staphylococcus aureus USA300) and ERX002218 (NC_002929.2; Bordetella pertussis Tohama I).
RESULTS
Sequence mapping
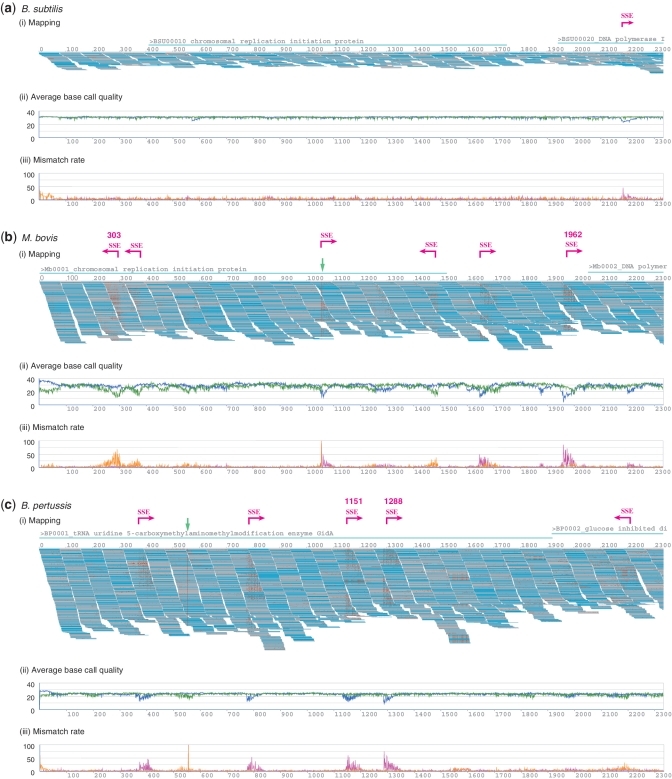
During the course of analysis for B. subtilis genome sequencing, we noticed a conspicuous cluster of errors localized in specific regions. To determine whether this phenomenon is common to all Illumina sequencing data, we also examined three short-read data sets (M. bovis, S. aureus and B. pertussis) from the SRA. Mapping parameters allowed 35 mismatches per read. The initial segments of the mapping for (i) B. subtilis, (ii) M. bovis and (iii) B. pertussis are shown in Figure 1(i). The first pages of the mapping results for each species are available (Supplementary Figure S5).
Figure 1.
(i) First segment of the mapping results obtained from Illumina sequencing runs for (a) B. subtilis, (b) M. bovis and (c) B. pertussis, generated using MPSmap and PSmap allowing 35 mismatches per read. Pale blue lines associated with the gene ID and name indicate gene areas. Magenta arrows with SSE signs indicate the positions of visually identified SSE. Green arrows indicate the positions of SNPs. SSE positions automatically detected are accompanied by numbers, which indicate the reference positions. For (b) and (c), mappings with the first 10 million reads are displayed. (ii) The average base call quality for all aligned bases at each reference position. The blue plot indicates forward reads, and the green plot, reverse reads. (iii) Ratio of the number of mismatches between reference and reads to the number of all mapped bases at each reference position. The magenta plot indicates forward reads, and the orange plot, reverse reads.
The mapping results obtained from our program (MPSmap) and some of the most popular open programs (BWA, BFAST and SOAP2) are shown in Table 1. Table 1 shows the number of reads mapped, using the default criteria for each program. For each sample, we also performed mapping of the first 35 bases of the read. For instance, 35/76 indicates that the first 35 bp was used out of a 76 bp read sequence. More detailed comparison of reads mapped by each program is available (Supplementary Table S1).
Table 1.
Number of reads mapped, and percentages of the total number of reads
| Species | Read length | BWA (%) | BFAST | SOAP2 | MPSmap mismatch 2 | MPSmap mismatch 35 | Total number of reads |
|---|---|---|---|---|---|---|---|
| Bacillus subtilis | 75 | 3 002 667 (96.3) | 3 049 778 (97.8) | 2 928 634 (94.0) | 2 921 625 (93.8) | 3 095 021 (99.3) | 3 115 816 |
| 35/75 | 3 073 992 (98.7) | 3 031 661 (97.2) | 3 074 149 (98.7) | 3 074 206 (98.7) | – | ||
| Mycobacterium bovis ERX006616 | 76 | 43 179 403 (82.0) | 51 451 693 (97.7) | 38 168 792 (72.5) | 38 527 210 (73.2) | 52 419 100 (99.6) | 52 654 994 |
| 35/76 | 51 279 692 (97.4) | 51 128 891 (97.1) | 51 235 406 (97.3) | 51 286 573 (97.4) | – | ||
| Staphylococcus aureus SRX007714 | 101 | 20 690 080 (67.6) | 27 940 969 (91.3) | 16 846 089 (55.1) | 16 965 251 (55.4) | 28 549 148 (93.3) | 30 597 352 |
| 35/101 | 25 496 252 (83.3) | 25 766 116 (84.2) | 25 284 229 (82.6) | 25 350 718 (82.9) | – | ||
| Bordetella pertussis ERX002218 | 76 | 9 735 459 (81.6) | 10 078 041 (84.5) | 8 958 762 (75.1) | 9 014 765 (75.5) | 11 575 310 (97.0) | 11 928 310 |
| 35/76 | 10 783 704 (90.4) | 9 902 510 (83.0) | 10 771 662 (90.3) | 10 786 830 (90.4) | – |
SSE in Illumina sequencing
Figure 1(i) shows the reference position (top line) and mapped reads (short horizontal bars below the top line). Mismatches are indicated by red dots. The reads were mapped to the reference sequence either in the originally sequenced direction (gray) or in the opposite direction as a reverse complement (cyan).
The most striking observation from the mapping results is that mismatches represented as red dots are not randomly distributed, but are obviously localized in specific regions, whose shape resembles a triangle. More importantly, we identified the starting positions of consecutive mismatches on the reference sequence (the side edges of the triangles). An interesting observation was that mismatches in these regions are concentrated in reads sequenced in the same direction. For instance, in Figure 2(a), all the mismatches are found in the reverse reads (cyan). We also found that positions dominated by mismatches often contain several matches. The position specificity, directionality and sporadic occurrence of mismatches implied an increase in miscall probability triggered at specific reference positions. As we will describe in more detail, these are the positions where sequence-specific interference of base elongation in the cyclic reversible termination induces a drastic lowering of base call quality, hence increasing the probability of miscalls. We refer to this error profile as the ‘sequence specific error (SSE)’ of the Illumina sequencer. In Figure 1, we have also plotted (ii) the average value of base call quality from each base calling software and (iii) the ratio of the number of base mismatches between reference and mapped reads to the number of all mapped bases, for each reference position. A strong correlation between the average base call quality and the mismatch rate is observed.
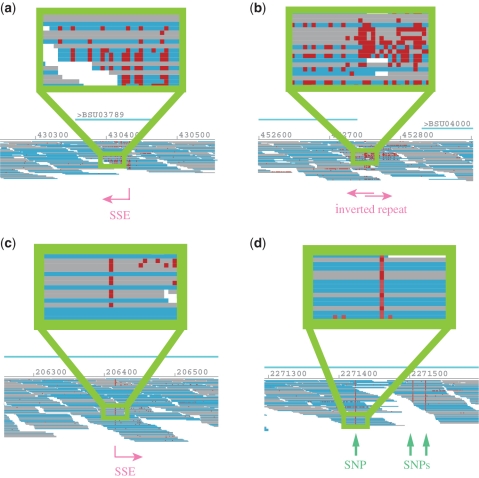
Figure 2.
Examples of SSE and SNP positions in mapping of B. subtilis. Each drawing displays areas with (a) an SSE position, (b) two overlapping SSE positions with inverted repeat, (c) an SSE resembling an SNP and (d) true SNPs.
Identification of SSE positions and sequence patterns
To identify the sequence common to all SSE positions, we first established a criterion to uniquely identify those positions. On the basis of the findings from Figure 1(iii), we identified the reference positions wherein: (i) mismatches occurred in >30% of the reads in the same direction, with (ii) four other such positions being present within 40 bases downstream and (iii) none within 40 bases upstream. This criterion is not completely rigorous, for it detects some non-SSE positions and fails to detect some SSE positions, especially when they are close in sequence. Nevertheless, it allows detection of most of the apparent SSE positions for further analysis. In Figure 1, visually identified SSE positions are marked with magenta arrows and an SSE sign, and SSE positions detected automatically by the aforementioned criterion are indicated by magenta numbers corresponding to the reference position.
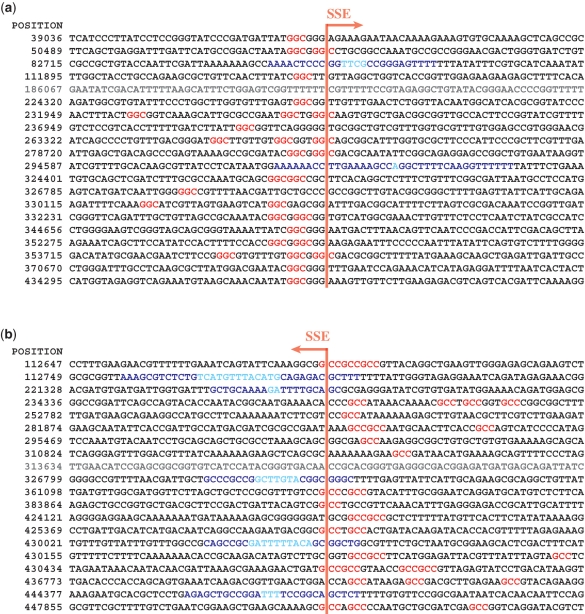
The number of SSE positions identified by this criterion is listed in Table 2. For instance, B. subtilis genome sequencing data has a total of 574 such positions (287 per sequencing direction). Full lists of SSE positions detected in all experiments are available (Supplementary Figure S6). Figure 3 shows the first 20 SSE positions in each direction with the neighboring sequences (±40 bp) for B. subtilis. In Figure 3, the SSE positions are aligned at the center of each sequence. We found GGC base triplets (colored red) within the upstream 10 bases of most of the SSE positions in the forward direction (a). Similarly, the reverse complement of GGC (GCC) was common within the upstream 10 bases of the SSE positions in the reverse direction (b). For some of the sequences, SSE positions were located in close vicinity to inverted repeats longer than 8 bp (colored cyan), which are likely to be associated with gene terminators. Table 3 summarizes the number of SSE positions with or without GGC and/or inverted repeats, showing that the majority of SSE positions were found to be associated with either GGC or inverted repeat sequences. GGC, as a base trimer, may occur once every 64 bases by chance. However, the SSE positions appeared to be less common. Therefore, we attempted to establish more restrictive rules for SSE-associated GGC sequences; we found weak evidence that C or T was more often found to precede GGC, and that G or T was more often found to follow GGC.
Table 2.
Number of SSE positions detected automatically
| Species | Forward | Backward | Total | Ref. length | SSE occurrence (one per bp) | GC contents (%) |
|---|---|---|---|---|---|---|
| Bacillus subtilis | 287 | 287 | 574 | 215 606 | 7344 | 43.5 |
| Mycobacterium bovis | 4374 | 4273 | 8647 | 4 345 492 | 502 | 65.4 |
| Staphylococcus aureus | 353 | 329 | 682 | 2 903 081 | 4256 | 32.7 |
| Bordetella pertussis | 2747 | 2675 | 5422 | 4 086 189 | 754 | 67.7 |
Figure 3.
First 20 SSE positions of B. subtilis automatically detected in the (a) forward and (b) backward directions. The numbers in the left column indicate the genome coordinate of each SSE position. For each row, the base next to the vertical red line is the SSE position.
Table 3.
Number of SSE positions associated with GGC and/or inverted repeat sequences longer than 8 bp
| Species | GGC | Inverted repeat | GGC and inverted repeat | None (%) | Total |
|---|---|---|---|---|---|
| Bacillus subtilis | 270 | 168 | 78 | 58 (10) | 574 |
| Mycobacterium bovis | 6001 | 409 | 1306 | 931 (11) | 8647 |
| Staphylococcus aureus | 488 | 9 | 35 | 150 (22) | 682 |
| Bordetella pertussis | 4141 | 62 | 603 | 531 (10) | 5422 |
The second-rightmost column of Table 2 shows the frequency of SSE positions in each data set. Smaller numbers indicate higher frequencies. For instance, SSE in B. subtilis mapping was detected once every 7344 bases. M. bovis and B. pertussis showed higher SSE frequencies than B. subtilis or S. aureus. Mycobacterium bovis and B. pertussis had higher GC content (65.4 and 67.7%, respectively) than B. subtilis or S. aureus. Presumably, higher GC contents results in a higher number of GGC sequences, which, in turn, may be responsible for higher SSE frequency. Notably, the genome of M. bovis contains genes of the PE-PGRS protein family, which have a high density of GGC repeats that significantly hinder sequencing with Illumina sequencers [Supplementary Figure S2 (c)].
SSE-induced mismatch patterns
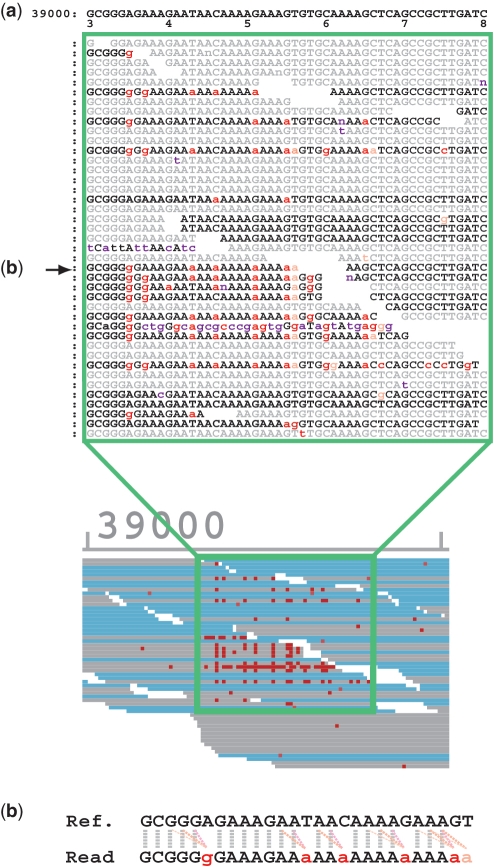
To identify the underlying mechanisms of SSE, we further dissected the mismatch profile of the SSE regions. Figure 4 shows one of the SSE positions of B. subtilis. The top line corresponds to the reference sequence starting at base 39 030, and the sample read sequences are aligned underneath. Forward reads are black and reverse reads are light gray. All mismatches are indicated in lowercase letters and colored. The majority of the mismatches in this region were A or G miscalls. However, this observation cannot be generalized. Interestingly, the mismatches tended to appear after a sequence of identical base calls in all SSE regions. In other words, the mismatched base was often similar to a preceding reference base. In Figure 4(b), we indicate the influence of preceding reference base calls to the mismatches observed in the read sequence. To clearly show this, mismatches are colored red if they match the corresponding preceding reference base, or orange if they match the second previous reference base; otherwise, they are colored magenta. In most cases, the mismatched bases matched the immediately preceding or the second preceding reference base. This observation was valid for all detected SSE positions. Table 4 shows the total number of bases in mapped reads, the total number of mismatches and the number of mismatches in all SSE regions. The SSE region was defined as the 40 bases downstream from each detected SSE position. Table 5 shows the percentage of mismatches in the SSE regions that match the reference base (1–5 preceding bases). If these were random errors, there is only a 25% chance that they would match an upstream base. However, the percentages shown in the first column of Table 5 (49–64%) are significantly higher. This result suggests that SSE-associated mismatches originate from contamination by lagged sequences (as discussed below).
Figure 4.
(a) Base-wise view of a part of the B. subtilis mapping result and (b) the alignment of the reference and the read in the middle row indicated by an arrow. The gray dotted lines show the match, whereas the pink dotted lines show the influence of previous base calls on mismatches.
Table 4.
Total number of base counts in reads mapped with MPSmap allowing 35 mismatches; total number of mismatches; mismatches as a percentage of total base calls; number of SSE mismatches; and SSE mismatches as a percentage of total mismatches
| Species | Total base calls | Total mismatches (%) | SSE mismatches (%) |
|---|---|---|---|
| Bacillus subtilis | 232 126 275 | 2 500 234 (1.1) | 215 088 (8.6) |
| Mycobacterium bovis | 3 983 850 916 | 140 028 534 (3.5) | 54 874 169 (39.2) |
| Staphylococcus aureus | 2 883 461 928 | 142 819 880 (4.9) | 8 526 781 (5.9) |
| Bordetella pertussis | 879 723 180 | 37 036 504 (4.2) | 8 427 651 (22.8) |
Table 5.
Percentage of mismatches in SSE regions that match the reference base positioned 1–5 bp before the mismatch position
| Species | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| Bacillus subtilis | 61.2 | 19.7 | 7.4 | 3.5 | 1.9 |
| Mycobacterium bovis | 61.6 | 22.3 | 7.7 | 3.4 | 1.7 |
| Staphylococcus aureus | 48.9 | 20.9 | 9.7 | 5.5 | 3.5 |
| Bordetella pertussis | 54.4 | 20.6 | 8.7 | 4.3 | 2.7 |
The base conversion ratio for all SSE mismatches is shown in Table 6. Values in Table 6 are normalized, so that the value of each cell is 1.00 in the absence of bias. The large influence of GC content is apparent. For instance, in species with higher GC content (M. bovis and B. pertussis), conversions between G and C (G–C) were significantly more frequent than conversions between A and T (A–T). Likewise, for species with low GC content (B. subtilis and S. aureus), A–T conversions are more frequent than G–C conversions. On the other hand, conversions from A/T (A or T) to G/C (G or C) were more frequent than conversions from G/C to A/T, regardless of the species or GC content. Among A/T to G/C and G/C to A/T conversions, T–G and A–C conversions were more frequent than T–C and A–G conversions, presumably reflecting the effects of optical signal crosstalk.
Table 6.
Base conversion ratio of all SSE mismatches
| ref. | read | |||
|---|---|---|---|---|
| A | T | G | C | |
| B. subtilis (44%) | ||||
| A | – | 1.05 | 1.04 | 1.14 |
| T | 1.02 | – | 1.21 | 1.06 |
| G | 0.92 | 0.93 | – | 0.87 |
| C | 0.92 | 0.93 | 0.87 | – |
| M. bovis (65%) | ||||
| A | – | 0.24 | 1.13 | 1.50 |
| T | 0.24 | – | 1.56 | 1.13 |
| G | 0.43 | 0.58 | – | 2.07 |
| C | 0.59 | 0.44 | 2.08 | – |
| S. aureus (33%) | ||||
| A | – | 1.55 | 0.98 | 1.44 |
| T | 1.58 | – | 1.51 | 1.04 |
| G | 0.66 | 0.84 | – | 0.44 |
| C | 0.84 | 0.66 | 0.43 | – |
| B. pertussis (68%) | ||||
| A | – | 0.35 | 0.98 | 1.20 |
| T | 0.35 | – | 1.29 | 1.00 |
| G | 0.67 | 0.73 | – | 2.01 |
| C | 0.74 | 0.66 | 2.00 | – |
DISCUSSION
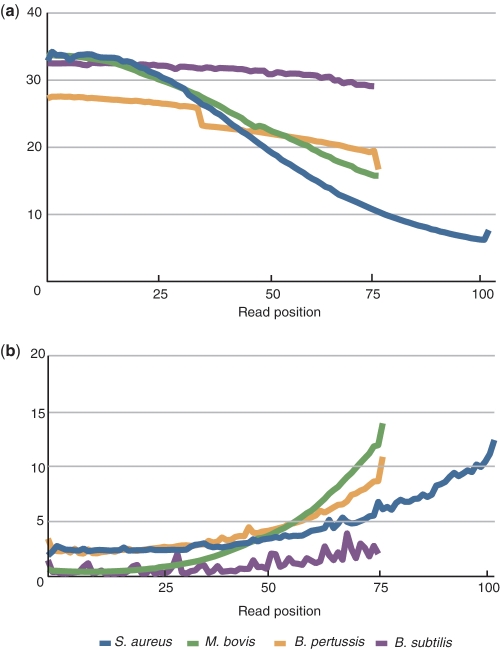
There are two types of mismatches in short-read mapping: sequencer-originated miscalls and actual differences between the sample and reference sequences. The latter consists of biological mutations and contamination by undesired sequences (for instance, adapters and primers). Since only the biological differences are relevant, it is desirable to exclude the other effects as much as possible. Contamination by undesired sequences can be excluded by computational filtering when the sequence is known. On the other hand, to deal with sequencer-originated miscalls, it is necessary to pinpoint the miscall mechanism and then improve the experimental procedure and/or base call algorithm. Some of the errors originate during the sample preparation steps using PCR, which partly explains the bias toward under-representation in GC-rich regions. On the other hand, other sequencer-originated miscalls in general have previously been considered to occur randomly. In the present report, we identified a mechanism that induces systematic miscalls in NGS data obtained using Illumina sequencers. In general, miscalls during sequencing occur when similar signal intensities for correct and false bases are observed. This situation is represented as the quality score for the base call. A correlation between the quality value and mismatch ratio is represented in Figures 1 and 5. Figure 5 also exhibits reduction of the quality score and increase of the mismatch ratio during later cycles. It has been suggested that the increase in the miscall ratio in later cycles is due to fading intensity, decreasing purity of nascent strands in a cluster, and accumulation of residual dyes (27). Independent of the generic regression of the base-call quality, there is a significant and systematic lowering of quality due to dephasing caused by specific sequence patterns. The average base-call quality and miscall ratio plotted in Figure 1 (ii) and (iii) indicate that particular sequence positions are associated with particularly low base-call quality and high mismatch rate in one direction. SSE-induced decreases in quality occur independent of the background variance of quality, but miscalls are the consequence of an overall reduction in quality.
Figure 5.
Plots of (a) average base call quality and (b) mismatch ratio along the sequencing cycle. Quality value of B. subtilis is based on the Illumina/Solexa standard protocol, while other data are PHREAD-type scores (30).
SSE is essentially cycle independent. However, it is more frequently observed for longer cycle sequencing, because the general lowering of quality in later cycles enhances miscall occurrence. One of the reasons why previous studies did not report SSE is probably that they only considered analysis of shorter reads. During the past few years, the standard read length of Illumina sequencers has increased from 30–50 to 70–100 bp. With the latest TruSeq reagents, it is now possible to run up to 150 cycles (31). We anticipate that SSE will become more evident as the reads become longer.
The mechanism of SSE
Alignment of the automatically collected SSE sequences (Figure 3) show that most SSE sites are associated with GGC or long inverted repeats. A long inverted repeat enhances the folding of DNA single strands, thereby inhibiting nucleotide elongation in both directions. On the other hand, GGC sequence-associated SSE occurs on reads in one direction only. Most of the miscalled bases in these regions match the preceding one or two reference bases. The similarity between the mismatch patterns associated with GGC and inverted repeat sequences suggests that they are based on the same mechanism. We do not have enough information on Illumina technology to further dissect the underlying mechanism of inhibition of the cyclic reversible termination process by GGC sequences. However, we suspect the preference of DNA polymerase is most likely to be responsible.
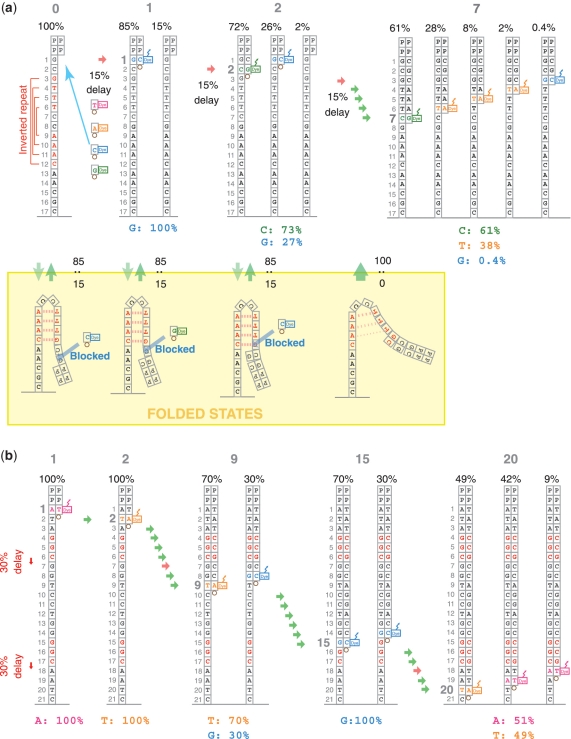
Figure 6 shows a schematic representation of the hypothetical models for the underlying SSE mechanisms. In model (a), inverted repeat-induced secondary structure formation causes a delay in nucleotide elongation. Single-stranded DNA can fold at the inverted repeat sequence (yellow box). In this model, a cluster of nascent single-stranded sequences is assumed to be in equilibrium between unfolded and folded states at a ratio of, for instance, 85:15. The folded structure blocks the addition of nucleotides for several of the steps prior to the inverted repeat, and causes contamination by lagged sequences. As the nascent strand grows, the stabilization by complementary hydrogen bonding is lost, and the CRT process returns to the normal rate (100% in this model). Some of the later cycles suffer more significantly from this contamination by lagged sequences. For instance, at the seventh cycle of this example, the probability of a false base call increases because the majority of the lagged sequences share the same base.
Figure 6.
Schematic representation of the (a) inverted repeat and (b) enzyme preference for the SSE hypothetical mechanistic models. The gray numbers at the top indicate the cycle number and the numbers below indicate the relative population of each single-stranded DNA during the cycle. The colored bases and numbers below the drawings show the relative intensity of signals during that cycle. For instance, the second cycle of model (a) emits signals for C and G with an intensity of 73 and 27%, respectively.
Model (b) of Figure 6 illustrates the hypothetical mechanism of dephasing caused by the preferential inhibition of an enzyme by a specific sequence of nucleic acids. Here, we assumed that single-nucleotide elongation is inhibited at the end of the GGC sequence by 30%. The sequence of model (b) includes two such positions; after the second GGC, the relative population of the correct sequence is reduced to less than half, and the rest consists of lagged sequences one or two steps behind.
If a cluster is dominated by one type of nascent strand after a certain cycle (one-step lagged, for instance), the read will simply appear to have a base deletion. Instead, based on our observations, we consider that a group of several lagged sequences is likely to be formed, so that the signal intensities generated by several types of bases become comparable. Consequently, the following base calls fluctuate under influence of the sequence context and several other factors.
In the standard base-calling procedure, dephasing is considered to be the consequence of incomplete CRT cycle or contamination of reagents or enzymes at each cycle (27). Therefore, dephasing is treated with just two (phasing and pre-phasing) parameters for all clusters in each tile. However, this regular dephasing differs from the SSE discussed here, which is a sequence-specific dephasing that only affects the template clusters incorporating specific parts of the genome sequence. Therefore, this sequence-specific dephasing should be addressed independently from the Illumina standard phasing and pre-phasing treatments.
So far, we have discussed some of the characteristics of SSE sequences. However, several questions remain unresolved. For instance, we found that some, but not all, GGC sequences or inverted repeats are associated with SSE. Furthermore, about 10% of the SSE positions are associated with neither GGC nor inverted repeat sequences (Table 3). This observation suggests the presence of other factors involved in SSE. A possible hypothesis is that the nascent single-stranded DNA forms secondary structures other than those formed by the gene terminator. The stability of the ssDNA folding structure is not only determined by the number of complementary base pairs, but also by the type of base pairs formed and the nature of the loop region. Moreover, the secondary structures of single-stranded nucleic acid sequences do not necessarily consist of a single stretch of complementary sequences. Some non-Watson–Crick base pairs may form, and the secondary structure may even include unpaired regions, yet these sequences may still provide large overall free energy stabilization. Some sequences may have more than one complementary counterpart, and a folded state that exists as an ensemble of multiple folded structures may be stabilized in terms of free energy. We are currently investigating potential secondary structures. This detailed analysis may provide clues to the unresolved sequence specificity of SSE.
Problems inherent to SSE
SSE hinders Illumina sequencing analysis. Despite this limitation, Illumina GA is one of the most powerful tools for nucleic acid sequence analysis. In the following sections, we focus on the three major associated problems (coverage variance, false SNP call and assembling gaps) and discuss possible solutions.
Coverage variance
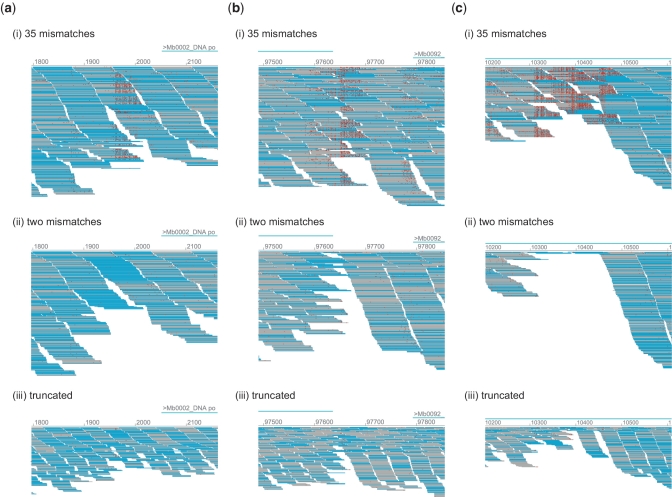
The most obvious problem associated with SSE is the coverage variance for the mapping. Figure 7 demonstrates some of the typical cases representing SSE-associated coverage loss. Each of the three sets of pictures (a–c) shows the mapping results of M. bovis obtained by MPSmap using three different conditions: (i) allowing 35 mismatches, (ii) allowing two mismatches and (iii) mapping of truncated sequences, using the first 35 bp allowing two mismatches. Figure 7a illustrates an area containing an SSE position. The comparison between (i) and (ii) of Figure 7a indicates that the number of reads mapped to this region is almost halved by the mapping allowing only two mismatches, which is the default setting of the most common mapping programs. Most of the remaining reads at this position in (ii) of Figure 7a share the same read direction (cyan). Because of the higher error rates observed in the later cycles, read truncation has become a common practice during Illumina read mapping (32). The mapping with read truncation (iii) levels out the decrease of coverage in this region, yet the overall depth is halved, since the amount of information is also decreased by the truncation. Figure 7b shows particular cases in which a couple of SSE positions in the opposite directions overlap with the terminator inverted repeat. In Figure 7c, several SSE in both directions overlap in a small area. In these cases, the number of reads obtained by regular mapping allowing only two mismatches, or by mapping of truncated reads, suffers from a significant decrease of coverage. The mismatch-tolerant mapping also suffers from the loss of coverage, yet more than half of the reads can still be mapped. Here, the mapped reads include a high number of mismatches, and sequence information around the region is significantly reduced. Nevertheless, we consider that these reads belong to the determined position, with a significant number of matches to the reference. Considering the balance of sensitivity and accuracy (Supplementary Data S1), mapping by BFAST would be one of the most appropriate choices for population-targeted sequencing.
Figure 7.
Comparison of coverage between (i) mapping allowing 35 mismatches, (ii) mapping allowing 2 mismatches and (iii) mapping of truncated reads using the first 35 bp, allowing 2 mismatches. Each drawing shows areas of the M. bovis genome including (a) an SSE position, (b) overlapping SSE positions in opposite directions associated with inverted repeats, and (c) multiple overlapping SSE positions. Mappings were carried out with MPSmap and PSmap for the first 10 million reads.
False SNP calls
SSE potentially causes false SNP calls. There are several cases where SSE-tailing mismatches are rather sparse and most mismatches appear in a few specific base positions (Figure 2c). In such cases, almost half of the reads mapped to the position appear to share the same mutation and are likely to be identified as a SNP. Especially in eukaryotic species, this can be erroneously identified as a SNP within a haplotype. Therefore, to make a SNP call, it is necessary to confirm that a significant number of specific mutations are found in reads in both directions. Moreover, it is essential to confirm the base call quality of suspicious SNP calls in order to avoid the identification of false SNPs induced by the SSE.
Gaps in assembled sequences
Assembling sequencer reads poses a challenging task especially in the presence of the long and persistent repeats often encountered in eukaryotic genomes. Even with bacterial sequencing, state-of-the-art assemblers still provide a mere collection of hundreds of contigs, and are unable to produce a single cyclic genome sequence without gaps (33,34). The presence of gapped regions caused by SSE (Figure 7c) provides another explanation for the difficulties of assembling the complete genome with the Illumina sequencer, even when the overall coverage is high. It may be possible to reproduce the genome structure by speculating the connectivity between contigs using paired-end information, if the gaps are short enough (35). There have also been several efforts to correct errors in reads prior to assembly (32). Nevertheless, at present, the only sensible way of filling the gaps of the Illumina sequencer reads appears to be combining the information from other experimental methods, including other NGS platforms that carry mutually complementary information (36,37). SSE not only causes gaps but also leads to the differentiation of partially overlapping read sequences (Figure 4). This situation leads to problems when considering the elongation of a contig from one direction, since it would appear as if the sequence were branching in multiple directions after the SSE site. Therefore, it is important to recognize the presence of these SSE-induced branches to avoid useless searches. Identification and prediction of SSE-causing sequences would greatly enhance the efficiency of Illumina/Solexa read assembly by performing SSE-specific phasing correction.
Base call improvement
Efforts have been made to improve the error rate of the Illumina sequencers. Besides the improvement in experimental procedures, several base call programs have been developed. These include Ibis, Alta-Cyclic and Rolexa (27,38,39). These programs consider a simple model for the regression of quality and improve base calls in terms of mismatch ratio. We examined the Ibis base calls for PhiX and found that it considerably improves randomly distributed miscalls. The systematic miscalls originating from SSE also appear to be improved, reflecting the decrease of background quality regression. It may be possible to further improve the reliability of base calls by explicitly incorporating the effect of the sequence specific, lagging-strand dephasing in the model.
Emerging NGS technologies produce increasing amounts of data. A majority of researchers believe that one should only adopt high-quality data, and should discard low-quality data that appears to contain a large number of errors. Here, we demonstrated that low-quality reads are not randomly distributed or unbiased, but are instead localized to specific mapping regions. Therefore, discarding low-quality data would result in loss of precious information in specific sequence regions. This issue needs to be properly addressed in order to obtain the maximum benefit from the Illumina sequencer. Fundamentally, it is necessary to revise the experimental procedures and redesign the base-calling algorithms. Meanwhile, we suggest that the re-evaluation and utilization of error-prone reads may provide an effective solution.
ACCESSION NUMBER
DRX000504.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant-in-aid for Scientific Research on Innovative Areas ‘Genome Science’ (221S0002) from Ministry of education, culture, sports, science and technology (MEXT), Japan; Center for Frontier Science and Technology at Nara Institute of Science and Technology. Funding for open access charge: Institute budget from Nara Institute of Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
Supplementary Data
ACKNOWLEDGEMENTS
K.N. thanks Dr Takeshi Kawabata from the Osaka University for stimulating discussions.
REFERENCES
- 1.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center’s improvements to the Illumina sequencing system. Nat. Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463:311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujimoto A, Nakagawa H, Hosono N, Nakano K, Abe T, Boroevich KA, Nagasaki M, Yamaguchi R, Shibuya T, Kubo M, et al. Whole-genome sequencing and comprehensive variant analysis of a Japanese individual using massively parallel sequencing. Nat. Genet. 2010;42:931–936. doi: 10.1038/ng.691. [DOI] [PubMed] [Google Scholar]
- 4.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 5.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y-J, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pandey V, Nutter RC, Prediger E. Applied Biosystems SOLiD system: ligation-based sequencing. In: Janitz M, editor. Next Generation Genome Sequencing: Toward Personalized Medicine. NJ: Wiley, Hoboken; 2008. pp. 29–41. [Google Scholar]
- 7.Metzker ML. Sequencing technologies—the next generation. Nat. Rev. Genet. 2010;11:31–46. doi: 10.1038/nrg2626. [DOI] [PubMed] [Google Scholar]
- 8.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pepke S, Wold B, Mortazavi A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods. 2009;6:S22–S32. doi: 10.1038/nmeth.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zerbino DR, Birney E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJM, Birol I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 15.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 16.Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent WJ. BLAT–The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rumble SM, Lacroute P, Dalca AV, Fiume M, Sidow A, Brudno M. SHRiMP: Accurate mapping of short color-space reads. PLoS Comput. Biol. 2009;5:e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li R, Yu C, Li Y, Lam T-W, Yiu S-M, Kristiansen K, Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 22.Homer N, Merriman B, Nelson SF. BFAST: An alignment tool for large scale genome resequencing. PLoS ONE. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozarewa I, Ning Z, Quail MA, Sanders MJ, Berriman M, Turner DJ. Amplification-free Illumina sequencing-library preparation facilitates improved mapping and assembly of (G+C)-biased genomes. Nat. Methods. 2009;6:291–295. doi: 10.1038/nmeth.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stein A, Takasuka TE, Collings CK. Are nucleosome positions in vivo primarily determined by histone-DNA sequence preferences? Nucleic Acids Res. 2010;38:709–719. doi: 10.1093/nar/gkp1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman S, Otto C, Kurtz S, Sharma CM, Khaitovich P, Vogel J, Stadler PF, Hackermuller J. Fast mapping of short sequences with mismatches, insertions and deletions using index structures. PLoS Comput. Biol. 2009;5:e1000502. doi: 10.1371/journal.pcbi.1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 2009;10:R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. Tablet–next generation sequence assmbly visualization. Bioinformatics. 2010;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cock PJA, Fields CJ, Goto N, Heuer ML, Rice PM. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38:1767–1771. doi: 10.1093/nar/gkp1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodrigue S, Materna AC, Timberlake SC, Blackburn MC, Malmstrom RR, Alm EJ, Chisholm SW. Unlocking short read sequencing for metagenomics. PLoS ONE. 2010;5:e11840. doi: 10.1371/journal.pone.0011840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaisson MJ, Brinza D, Pevzner PA. De novo fragment assembly with short mate-paired reads: Does the read length matter? Genome Res. 2009;19:336–346. doi: 10.1101/gr.079053.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Farrer RA, Kemen E, Jones JDG, Studholme DJ. De novo assembly of the Pseudomonas syringae pv.syringae B728a genome using Illumina/Solexa short sequence reads. FEMS Microbiol. Lett. 2009;291:103–111. doi: 10.1111/j.1574-6968.2008.01441.x. [DOI] [PubMed] [Google Scholar]
- 34.Tsai IJ, Otto TD, Berriman M. Improving draft assemblies by iterative mapping and assembly of short reads to eliminate gaps. Genome Biol. 2010;11:R41. doi: 10.1186/gb-2010-11-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 36.DiGuistini S, Liao NY, Platt D, Robertson G, Seidel M, Chan SK, Docking TR, Birol I, Holt RA, Hirst M, et al. De novo genome sequence assembly of a filamentous fungus using Sanger, 454 and Illumina sequence data. Genome Biol. 2009;10:R94. doi: 10.1186/gb-2009-10-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaisson MJ, Pevzner PA. Short read fragment assembly of bacterial genomes. Genome Res. 2008;18:324–330. doi: 10.1101/gr.7088808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erlich Y, Mitra PP, delaBastide M, McCombie WR, Hannon GJ. Alta-Cyclic: a self-optimizing base caller for next-generation sequencing. Nat. Methods. 2008;5:679–682. doi: 10.1038/nmeth.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rougemont J, Amzallag A, Iseli C, Farinelli L, Xenarios I, Naef F. Probabilistic base calling of Solexa sequencing data. BMC Bioinformatics. 2008;9:431–442. doi: 10.1186/1471-2105-9-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data